FIG 6.
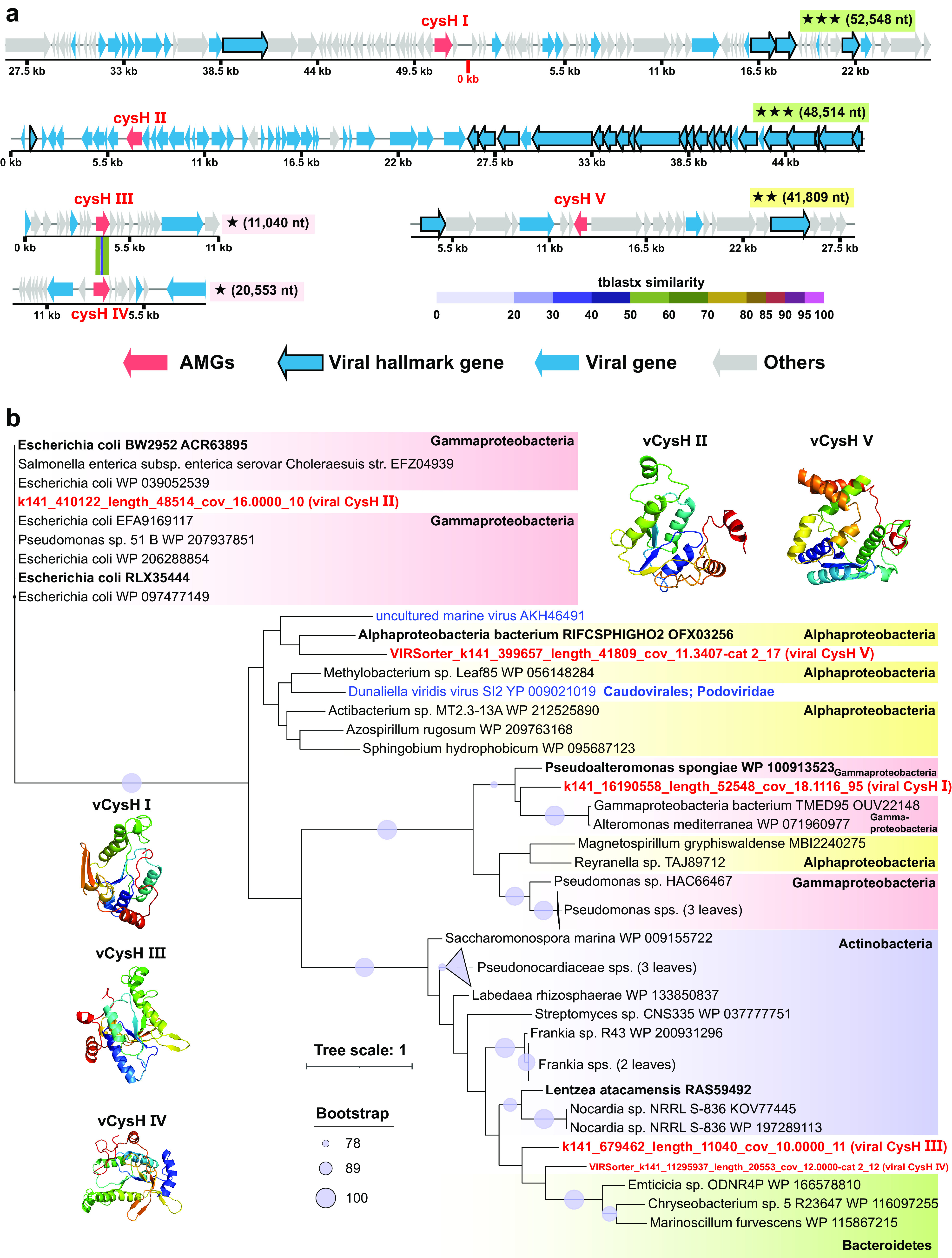
Genomic context, predicted protein structure, and phylogeny of the viral cysH gene. (a) Genomic maps of cysH-containing viral scaffolds. The genome quality (green rectangles with three stars for high quality, yellow rectangles with two stars for medium quality, and pink rectangles with one star for low quality or not determined) and length are shown near the maps. Viral cysH genes are in red, virus-like genes are in blue (viral hallmark genes are framed), and non-virus-like or uncharacterized genes are in gray. Between two similar cysH genes (III and IV), tBLASTx alignment is represented by colored lines, and their percent identity is represented by the color scale. Detailed annotations can be found in Table S10 at https://doi.org/10.6084/m9.figshare.c.5703367.v6. (b) Maximum likelihood tree (from an amino acid alignment) of the viral cysH gene, including 5 MTS viral cysH sequences (in red) and 39 reference sequences (two viral sequences are in blue), and tertiary structures of these viral CysH proteins. The most closely related reference sequences are highlighted in boldface type. The proportional circles represent internal nodes and bootstraps, and the clades where the average branch length distance to leaves is <0.4 are collapsed.
