Abstract
The neuraminidase (NA) of influenza A and B viruses plays a distinct role in viral replication and has a highly conserved catalytic site. Numerous sialic (neuraminic) acid analogs that competitively bind to the NA active site and potently inhibit enzyme activity have been synthesized and tested. Four NA inhibitors are now licensed in various parts of the world (zanamivir, oseltamivir, peramivir, and laninamivir) to treat influenza A and B infections. NA changes, naturally occurring or acquired under selective pressure, have been shown to reduce drug binding, thereby affecting the effectiveness of NA inhibitors. Drug resistance and other drawbacks have prompted the search for the next-generation NA-targeting therapeutics. One of the promising approaches is the identification of monoclonal antibodies (mAbs) targeting the conserved NA epitopes. Anti-NA mAbs demonstrate Fab-based antiviral activity supplemented with Fc-mediated immune effector functions. Antiviral Fc-conjugates offer another cutting-edge strategy that is based on a multimodal mechanism of action. These novel antiviral agents are composed of a small-molecule NA inhibitor and an Fc-region that simultaneously engages the immune system. The significant advancements made in recent years further support the value of NA as an attractive target for the antiviral development.
NEURAMINIDASE AS A TARGET
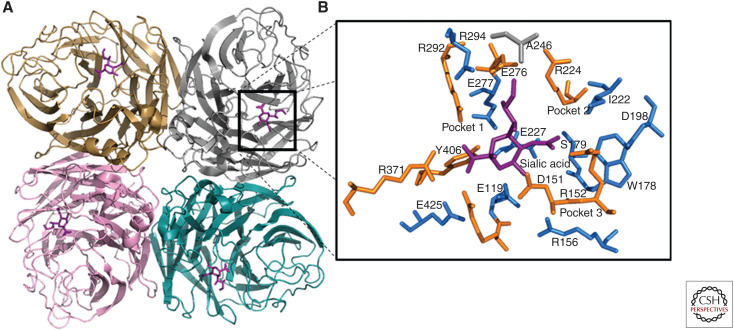
Neuraminidase (NA) is an appealing target for the development of therapeutics against influenza A and B viruses (Air and Laver 1989). This viral enzyme is expressed on the surface of type A and B virus particles and infected cells, which makes it readily accessible to antiviral agents and antibodies. NA, a glycoside hydrolase (EC 3.2.1.18), cleaves the terminal sialic (N-acetylneuraminic) acids from glycoproteins and glycolipids. NA is a type II integral membrane glycoprotein and is formed by four identical monomers. Each monomer consists of a globular head that carries the enzyme active site (Fig. 1A), a stalk of variable length, a membrane-anchoring hydrophobic transmembrane domain, and a cytoplasmic tail (Varghese et al. 1983; Colman et al. 1987; Air and Laver 1989).
Figure 1.
Crystal structure of neuraminidase (NA) catalytic head with sialic acid. The structure was visualized in PyMOL using information from Protein Data Bank code 4GZX and 4GZQ (A/Tanzania/205/2010 N2 NA). (A) A ribbon representation of an NA tetramer, in which each monomer is individually colored and sialic acid is shown in purple. (B) Black-boxed region is magnified and shows the catalytic (brown sticks) and the framework (blue sticks) residues in the three major binding pockets.
The NA active site has been directly identified by soaking neuraminic acid substrate into NA (N2 subtype) crystals (Colman et al. 1983). It is a shallow pocket formed by highly conserved amino acid residues, some of which bind directly to the substrate and participate in catalysis, whereas others provide the framework (Colman et al. 1993). A remarkable feature of the NA active site is the large number of charged residues. The residues R118, D151, R152, R224, E276, R292, R371, and Y406 are defined as catalytic (or functional), whereas E119, R156, W178, S179, D198, I222, E227, N294, and E425 belong to the framework (N2 amino acid numbering is used throughout the text) (Fig. 1B; Colman et al. 1993). Of note, aspartic acid (D) is present at residue 198 in all type A NAs, except subtypes N7 and N9, which have asparagine (N) at this position. Based on the crystal structure, the NA active site and adjacent residues can be divided into three major binding pockets, which could be explored when designing inhibitors. Pocket 1 is formed by E276, E277, R292, and N294 residues and interacts with the glycerol moiety of sialic acid. Pocket 2 is formed by A246, I222, and R224 residues and is not utilized by sialic acid for binding. Pocket 3 is surrounded by E119, D151, R152, W178, S179, I222, and E227 residues and participates in sialic acid binding (Fig. 1B; Kim et al. 1997). The role of the individual residues has been investigated to elucidate the underlying enzymatic mechanism (Lentz et al. 1987; Ghate and Air 1998; Watson et al. 2003; Chan et al. 2012). Notably, during catalysis, amino acid residues of the active site bind more tightly to the transition state of the substrate (oxocarbenium ion) than to its ground state form (neuraminic acid). This feature has been exploited extensively in designing the structure-based NA inhibitors representing chemical analogs of the substrate's transition state.
INHIBITORS OF NA ENZYME ACTIVITY
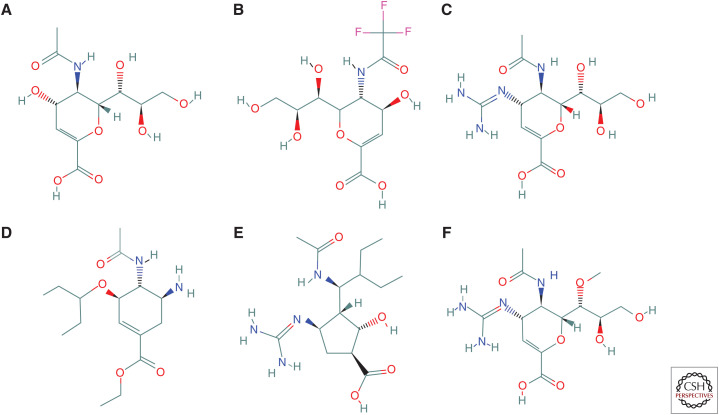
NA inhibitors compete with neuraminic acid (substrate) by reversibly binding to the NA active site. The earliest transition state analog of the substrate, 2,3-dehydro-2-deoxy-N-acetylneuraminic acid (DANA) (Fig. 2A), binds rapidly to NA and is a weak inhibitor of NA enzyme activity. Modifications that conferred a time-dependent or slow binding can improve potency of this inhibitor (Smith et al. 2001). Its trifluoroacetyl derivative (FANA) (Fig. 2B) was shown to be a potent inhibitor in vitro; however, it failed to protect animals against influenza challenge (Palese et al. 1974; Schulman and Palese 1975; Palese and Compans 1976).
Figure 2.
Chemical structure of neuraminidase (NA) inhibitors. Structures were drawn using the PubChem compound database. (A) DANA (2-deoxy-2,3-dehydro-N-acetylneuraminic acid), (B) FANA (2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid), (C) zanamivir (4-guanidino-Neu5Ac2en), (D) oseltamivir carboxylate (GS-4071), (E) peramivir, and (F) laninamivir (R-125489).
Years later, the first generation of potent influenza NA inhibitors was rationally designed using information gained from extensive NA sequence analysis, detailed enzyme crystal structures, and computational chemistry tools (Colman and Ward 1985; Burmeister et al. 1992; von Itzstein et al. 1993; Kim et al. 1997; Kobasa et al. 1999; Moscona 2005).
LICENSED NA INHIBITORS
Four NA inhibitors are currently licensed in various parts of the world for the treatment and prevention of influenza A and B infections: zanamivir, oseltamivir, peramivir, and laninamivir. The first two NA inhibitors, zanamivir and oseltamivir, were approved in 1999. Peramivir and laninamivir were approved in 2014 and 2010, respectively; the latter is available only in Japan. The four NA inhibitors share a certain degree of similarity in chemical structure, but differ by the route of administration, treatment regimen, and other attributes.
Zanamivir (4-guanidino-Neu5Ac2en) (Fig. 2C) was designed using a minimalistic approach (Colman 1999). It is an analog of DANA, where a hydroxyl group linked to C-4 was replaced with a positively charged guanidine group. This modification resulted in a significant increase in binding affinity. It has been demonstrated that guanidinium formed a hydrogen bond to E119 (Taylor et al. 1998); however, some researchers suggested that it formed an ionic contact with this framework residue through a stacking interaction (Varghese et al. 1983, 1995). Zanamivir has low bioavailability (2%) and therefore requires topical administration. Zanamivir is approved as an inhaled drug that needs to be taken twice a day for 5 d. Over the years, an intravenous formulation of zanamivir has been developed and provided for compassionate use. In 2019, it received a marketing authorization in the European Union to be used under exceptional circumstances (e.g., resistance to other anti-influenza medications). The transdermal delivery system of zanamivir in the treatment of influenza using microneedle patch has also been investigated (NIH-R43AI129122).
Oseltamivir carboxylate (GS-4071) (Fig. 2D) was designed with the aim to simplify synthesis and improve bioavailability (Lew et al. 1999). This compound utilizes a carbocyclic scaffold instead of the pyran of zanamivir, where the 3-pentyl ether side chain replaced the hydrophilic glycerol side chain and the amino group replaced the guanidine group (von Itzstein and Thomson 2009). Unlike zanamivir, oseltamivir relies on strong hydrophobic interactions rather than polar interactions. The oseltamivir–NA complex crystal structure indicates that one branch of the 3-pentyl side chain of oseltamivir contacts with A246, I222, and R224, resulting in a significant increase in binding affinity. In contrast, the other branch of the 3-pentyl side chain of oseltamivir binds to the highly polar pocket 1. Such binding becomes possible because the E276 side chain moves away from the middle of NA pocket 1 and adopts an alternative confirmation. This conformational change effectively makes pocket 1 larger and less polar to accommodate the hydrophobic moiety of oseltamivir. Oseltamivir is a potent inhibitor of NA activity; however, its bioavailability was lower than desired. Therefore, oseltamivir phosphate (GS-4104) was developed as a prodrug of oseltamivir carboxylate. Oseltamivir phosphate is prescribed as an oral medication, to be taken twice a day for five days. It is converted to the active metabolite (GS-4071) by host hepatic esterases (Kim et al. 1997; He et al. 1999).
Peramivir (Fig. 2E) is a cyclopentane (five-membered ring) derivative, making it structurally the most different among the other approved NA inhibitors. However, it shares similar functional groups—namely, a guanidino moiety of zanamivir and a hydrophobic side chain of oseltamivir (Babu et al. 2000). Its performance as an oral medication was not very effective, and this facilitated the development of an intravenous formulation. Because of a prolonged half-life, peramivir was approved as a single-dose treatment. It may provide a substitute for patients unable to take medication via the oral or inhalation routes.
Laninamivir (R-125489) (Fig. 2F) was derived by methylation of the C-7 hydroxyl group of zanamivir. It is administered as an octanoyl ester prodrug, laninamivir octanoate (CS-8958). After absorption by the respiratory tract epithelial cells, laninamivir octanoate is rapidly hydrolyzed to an active form (R-125489) (Kiso et al. 2010). Laninamivir acts as a long-lasting NA inhibitor and showed therapeutic efficacy after a single nasally inhaled dose (Yamashita et al. 2009; Kubo et al. 2010a,b; Sugaya and Ohashi 2010). Different mechanisms leading to the retention of the drug in target organs have been proposed, but not yet fully elucidated (Koyama et al. 2009; Yamashita et al. 2009; Ikematsu and Kawai 2011).
OTHER NA INHIBITORS
The licensure of the first-generation NA inhibitors has inspired a search for compounds with improved properties (Laborda et al. 2016). Among those, multimeric derivatives and diversely decorated analogs of zanamivir and oseltamivir have been reported (Macdonald et al. 2004, 2005; Watson et al. 2004).
Dimeric derivatives of zanamivir conjugates with various linking groups have been shown to be more potent than zanamivir both in vitro and in vivo (Macdonald et al. 2005). Trimeric and tetrameric compounds of zanamivir were significantly more active than the monomeric ones and, more importantly, showed long-lasting protective activity in a mouse model (Watson et al. 2004). Zanamivir derivatives, such as zanamivir-amino acid prodrugs and zanamivir phosphate congeners, have also been patented (Han and Amidon 2000; Shie et al. 2011; Dahan et al. 2014). Among other modifications, the additions of the biodegradable polymer and poly-L-glutamine were also explored (Watson et al. 2004; Weight et al. 2011; Lee et al. 2012).
A series of oseltamivir derivatives carrying a phosphonate and alkyl substituents at carboxylic acid and 4-amido and/or 5-amino groups, respectively, were reported but did not show better efficacies than oseltamivir (Rungrotmongkol et al. 2009; Hussain Basha and Prasad 2012; Venkatramani et al. 2012). Oseltamivir derivatives bearing a pyridyl group were explored and some of the compounds exhibited more potent inhibitory activity against A(H5N1) NA than oseltamivir (Wang et al. 2019). A panel of other oseltamivir derivatives with modifications at carboxyl-modified or C1 displayed robust anti-influenza potencies in vitro against A(H5N1) or A(H5N6) and both viruses, respectively (Ju et al. 2018; Wang et al. 2018). The latter is potentially propitious because of the improved membrane permeability and oral drug absorption (Wang et al. 2018). Many other oseltamivir derivatives designed through structure–activity relationship studies were also investigated (Gong and Xu 2008; Schade et al. 2014; Li et al. 2017; Zhang et al. 2018; Ye et al. 2019).
Among NA inhibitors with a different scaffold, cyclopentane amide and pyrrolidine derivatives named A-192558 and A-315675, respectively, were synthesized and tested (Wang et al. 2001; Kati et al. 2002). Both compounds effectively inhibited NA activity of influenza A and B viruses (Abed et al. 2008). A-315675 exhibited comparable potencies against a panel of A(H1N1) and A(H3N2) influenza viruses and was found to be significantly more potent than oseltamivir against type B isolates (Kati et al. 2002). Prodrug of A-315675 (A-322278) was used for oral administration and showed promising anti-influenza activity in animal models (Mishin et al. 2005a). An aromatic compound with an oxopyridine scaffold was discovered to inhibit NA activity of A(H1N1) influenza virus in the micromolar range (Karpf and Trussardi 2001).
Instead of focusing on compounds that bind reversibly, a research team applied a different approach to develop NA inhibitors that bind covalently to NA (Kim et al. 2013). In this study, 2,3-difluorosialic acid (DFSA) and its derivatives were used as a substrate exhibiting a slow turnover. These compounds function through the formation of a stabilized covalent intermediate with NA, which can permanently inactivate this viral enzyme. DFSA inhibitors demonstrated comparable efficacies to that of zanamivir and showed broad-spectrum activity against drug-resistant strains. They provide valuable insight into the design of next-generation covalent mechanism-based influenza therapeutics (Kim et al. 2013). Notably, to prove the possibility of covalent inhibition of NA, the role of conserved Y406 residue in the NA active site was highlighted (Vavricka et al. 2013). They demonstrated that Y406 acts as a catalytic nucleophile and plays an integral role in the catalytic mechanism. The drug-resistant mutations conferred by substitution of Y406 were enormously unfavorable for the virus (Lentz et al. 1987; Ghate and Air 1998; Vavricka et al. 2013). Therefore, targeting conserved Y406 with covalent inhibitors may be a promising strategy for NA inhibitor development (Vavricka et al. 2013).
The advancements made in the analysis of crystal structures of NA from various subtypes also stimulated the search for improved antiviral compounds. With the exception of recently identified N10 and N11 in bats (Tong et al. 2012, 2013; Mehle 2014), NAs of type A viruses are divided into two major groups: group 1 (N1, N4, N5, and N8) and group 2 (N2, N3, N6, N7, and N9) (Sui et al. 2009; Nabel and Fauci 2010). The NAs of group 1 possess a flexible loop (known as the 150-loop) and a cavity (the 150-cavity formed by amino acids 147–152 of the 150-loop), which is adjacent to the NA active site. After binding with inhibitors, NAs of group 1 can render the conformational change from the “open” to the “closed” form, whereas group 2 NAs assume a “closed” form at all time (Russell et al. 2006). Different oseltamivir derivatives targeting the 150-cavity (e.g., modifications at C-5 NH2 position, addition of N/O/C fragment/linker, and lipophilic side chains at C-5) were synthesized. These derivatives had significantly improved potency (Wang et al. 2010; Xie et al. 2014; Lin et al. 2018; Zhang et al. 2018). As these inhibitors target the 150-cavity and can specifically inhibit only the group 1 NAs, including the A(H1N1)pdm09, they may provide a prospective way to design selective NA inhibitors.
NA CHANGES THAT AFFECT INHIBITOR BINDING
All four approved NA inhibitors potently reduce influenza NA activity and display low IC50 values (subnanomolar–nanomolar range) in NA inhibition assays (Gubareva et al. 2001b; Ikematsu et al. 2015; Lackenby et al. 2018). Overall, peramivir shows the lowest IC50 values across influenza types and subtypes and thus is the most potent NA inhibitor in vitro (Okomo-Adhiambo et al. 2010; Lackenby et al. 2018). Despite the high conservation of amino acid residues of the NA active site, the potency of NA inhibitors can be influenced by a variance in NA sequence and structure. For example, all three NA inhibitors that are approved in the United States (zanamivir, oseltamivir, and peramivir) more potently inhibit the NA activity of type A viruses compared to type B. Moreover, certain NA changes, naturally occurring or acquired during virus replication in the presence of NA inhibitor, can reduce the potency of a NA inhibitor. Notably, these NA changes interrupt the slow binding or faster dissociation of NA inhibitors (McKimm-Breschkin and Barrett 2015).
The common change associated with zanamivir resistance in N2-subtype viruses is E119G substitution, although A and D substitutions were also reported (Gubareva et al. 1997; Zurcher et al. 2006; L'Huillier et al. 2015; Tamura et al. 2015). E119 is critical for interactions with the C4-guanidino of zanamivir and the C4-amino group of oseltamivir. E119G confers high-level resistance only to zanamivir, whereas E119D affects interaction with all NA inhibitors (McKimm-Breschkin et al. 1996; Colacino et al. 1997; Tamura et al. 2015). A recent study implicated the 150-loop in oseltamivir resistance caused by the E119D substitution (Hanpaibool et al. 2019). Noteworthy, a different substitution at the same residue, E119V, confers oseltamivir resistance in viruses carrying the N2 subtype; however, it does not affect binding of zanamivir (Mishin et al. 2005a; Pizzorno et al. 2011; Eshaghi et al. 2014; Gaymard et al. 2016a).
For oseltamivir, the most common changes conferring resistance are H274Y (H275Y in N1 amino acid numbering) in the N1 subtype and R292K in the N2 subtype (Whitley et al. 2013; Roosenhoff et al. 2019).
In the N1 background, H274Y reduces binding of oseltamivir to NA by several 100-fold. Tyrosine at 274 prevents the conformational change needed to accommodate the bulky pentyl ether side chain of oseltamivir, whereas zanamivir and sialic acid can still bind to the mutant NA. It was proposed that tyrosine at 274 stabilizes the E276, so that there is a high-energy penalty for the movement of E276 and this results in weak oseltamivir binding and high resistance. Although most of the published studies on H274Y focused only on the drug-bound system, the direct effects of this substitution on NA itself prior to drug binding remain unclear. The structural changes in the NA binding site caused by H274Y substitution were examined (Yusuf et al. 2016). They observed that the H274Y substitution disturbed the NA binding site because of the repulsive effect of Y274 in the 250-loop (a loop formed in the region of residue 250 in NA). The result may be an adverse effect in the flexibility and positioning of this loop and its contribution in the hydrogen bonding network involved in binding of NA inhibitors (Yusuf et al. 2016; Wan et al. 2018). H274Y was shown to cause a slower association and faster dissociation rate of oseltamivir, whereas it does not affect the kinetics for binding of zanamivir (Barrett et al. 2011). Of note, the H274Y substitution has also been found to affect the binding of peramivir (Mishin et al. 2005a).
Interestingly, H274Y oseltamivir-resistant A(H1N1) viruses first broadly circulated in Europe (prior to the 2009 pandemic) (Lackenby et al. 2008), where oseltamivir was not commonly used, have subsequently became prevalent worldwide. The NA of these resistant A(H1N1) viruses has shown a slightly higher activity and affinity for the substrate than that of previously circulating sensitive viruses (Rameix-Welti et al. 2008). Furthermore, in addition to the H274Y substitution, some possibly permissive changes in the NA and hemagglutinin (HA) proteins were also implicated (Bloom et al. 2010; Ginting et al. 2012). Thus, these two causes may have permitted oseltamivir-resistant A(H1N1) viruses to acquire the capacity for efficient human-to-human transmission. These oseltamivir-resistant viruses exhibited cross-resistance to peramivir but remained sensitive to zanamivir and laninamivir (McKimm-Breschkin 2000; Baum et al. 2003; Baz et al. 2007; Collins et al. 2009; Hurt et al. 2009; Takashita et al. 2013). Notably, the transmission of oseltamivir-resistant H274Y A(H1N1)pdm09 strains to date has been limited. The clusters of oseltamivir-resistant A(H1N1)pdm09 viruses in Australia (2011) (Hurt et al. 2012b) and in Japan (2013–2014) (Takashita et al. 2015a) were reported. Other studies also confirmed that transmission most commonly occurred in closed and near-contact settings (Le et al. 2010; Wolfe et al. 2010; Chen et al. 2011; Graitcer et al. 2011; Storms et al. 2012).
In the N2 background, R292K confers oseltamivir resistance by introducing an obstacle to the structural transition through which E276 engages R224 to create the novel hydrophobic binding pocket. It was suggested that a hydrogen bond between E276 and K292 is responsible for this failure (Varghese et al. 1998). R292K has a greater effect on binding of oseltamivir and peramivir compared to other NA inhibitors (Varghese et al. 1998; Gubareva et al. 2017). This substitution also affects the NA enzyme activity (Sleeman et al. 2013; Yen et al. 2013). This is not surprising as R292 is a catalytic residue and is a part of the highly conserved arginine triad (R118, R292, R371) shared by all sialidases (Colman et al. 1983; Roggentin et al. 1989; Chavas et al. 2005). The R292K substitution confers the same profound effect among all group 2 NAs, including N6 and N9, as per the random mutagenesis and reverse genetic approaches (Song et al. 2015; Gaymard et al. 2016a).
Over the years, numerous NA changes have been implicated in the decreased potency of one or more NA inhibitors, at least in vitro (Thorlund et al. 2011; Samson et al. 2013; WHO 2018). Among those, N294S (N295S in N1 amino acid numbering) confers resistance to oseltamivir by virtue of hydrogen bonding to E276 and restraining the necessary conformational change associated with oseltamivir binding (Collins et al. 2008). The D198E (D197E in B amino acid numbering) in type B affected the interaction of R152 with the N-acetyl group, thus reducing binding of oseltamivir and peramivir (Oakley et al. 2010). A combination of NA changes has been shown to enhance drug resistance (Nguyen et al. 2010). For example, a dual H274Y and I222K/R change (H275Y and I223K/R in N1 amino acid numbering) has been reported to greatly reduce the drug binding (Singh and Soliman 2015; Hsu et al. 2017). Noteworthy, the long-timescale dynamics simulations demonstrated that the mechanism of oseltamivir resistance of dual I222R/H274Y mutations was due to the loss of key hydrogen bonds between the inhibitor and residues in the 150-loop (Woods et al. 2012).
Furthermore, changes affecting the 250-loop have been reported to reduce the binding of zanamivir, laninamivir, and oseltamivir. One such NA substitution, S247P, was detected in the influenza A(H3N2) variant virus (also known as “A(H3N2)v” subtype, according to World Health Organization [WHO] nomenclature) (Sleeman et al. 2014). It was speculated that having proline at position 247 increases rigidity of the 250-loop by imposing strict torsion angles on this NA portion. This disturbance may in turn alter the hydrogen-bonding network involved in drug binding (Sleeman et al. 2014). Notably, not only substitutions, but also deletions, can affect drug binding. It was shown that a 4-amino acid deletion, such as 245–248 or 247–250 in the N2 subtype, adversely affect binding of oseltamivir and zanamivir (Abed et al. 2009; Tamura et al. 2015). Moreover, substitutions at remote residues that are neither conserved nor having direct interaction with the substrate can also confer resistance. For example, the Y155H substitution conferred reduced susceptibility to both oseltamivir and zanamivir in NA inhibition assay (McKimm-Breschkin et al. 2013).
Notably, influenza A viruses of animal origin can cross the interspecies barrier causing sporadic infections and large outbreaks in humans or even trigger pandemics (Vincent et al. 2014; Nelson and Vincent 2015; Subbarao 2019). A recent example is the emergence of avian A(H7N9) viruses in China since 2013 (Gao et al. 2013). Avian influenza A viruses of the H5N1, H5N6, H6N1, and H10N8 subtypes are also known to cause human infections (WHO 2014a, 2016b, 2019). Therefore, analysis of the identified substitutions has been expanded to NAs of different subtypes. These studies were conducted using recombinant NA proteins and reverse genetically engineered viruses (Pizzorno et al. 2011, 2013; Baek et al. 2015; Gubareva et al. 2017; Choi et al. 2018; Kwon et al. 2018; Tewawong et al. 2018). It was observed that NA proteins from the same group share certain similarities in their drug resistance profiles. For example, R292K caused a drastic reduction in oseltamivir potency among all group 2 NAs, including N9 (R289K in H7N9 amino acid numbering) and N6 (Song et al. 2015; Gaymard et al. 2016a). There were also apparent differences in drug resistance profiles within the group. Notably, H274Y in N2 did not affect oseltamivir binding, whereas in N9, it did reduce the drug potency by 100-fold in NA inhibition assay (Wang et al. 2002; Gubareva et al. 2017).
HA AND NA FUNCTIONAL BALANCE
Although the approved antiviral agents potently reduce NA enzyme activity, numerous studies indicate that their effect on virus replication can be less consistent. Influenza NA is not vital for virus entry, replication, assembly, or budding (Liu et al. 1995). Furthermore, some NA-lacking viruses can replicate in cell culture to appreciable titers. Such viruses were selected in cell culture, generated by reverse genetics, and even detected in clinical specimens (Gubareva et al. 2002; Mishin et al. 2005c; Moules et al. 2010). Taken together, these findings seem to challenge the appropriateness of NA as the target for drug design. On the other hand, a large body of evidence highlights the importance of NA during virus replication in vivo. Indeed, reduced virus replication in the respiratory tract of animals and humans treated with NA inhibitors was observed (von Itzstein et al. 1993; Hayden et al. 2000; Nishikawa et al. 2012). Moreover, the NA-lacking virus displayed severe attenuation in a ferret model (Mishin et al. 2005b).
To better understand the antiviral effect of NA inhibitors, it is essential to consider the roles played by both NA and HA (Wagner et al. 2002; Du et al. 2019; McAuley et al. 2019). Both surface glycoproteins bind to neuraminic acid–containing receptors that are used by influenza A and B viruses to enter cells. It was postulated that HA enables virus attachment, whereas NA facilitates release of nascent virus particles by destroying receptors (Gaymard et al. 2016b). However, the HA–NA interplay is a more complex matter. To optimize virus replication, the opposing HA and NA activities must be balanced at all stages. NA's role in preventing the virus entrapment by mucin-rich secretions and in virus entry was recognized in several studies (Matrosovich et al. 2004; Cohen et al. 2013; Zanin et al. 2016). By using R18-labeled viruses and a fetuin-coated surface, it was discovered that influenza viruses can move via the HA-receptor exchange mechanism (Sakai et al. 2017). Moreover, NA contributes to the initiation of crawling and gliding and elongation of gliding. Together HA and NA act as novel motile machinery, and this movement accelerates virus entry into cells. Blocking this viral movement resulted in the suppression of virus endocytosis by cells (Sakai et al. 2017). The contribution of NA to virus-receptor binding was revealed using the biolayer interferometric analysis (Guo et al. 2018). They also observed that NA activity strongly stimulates virus rolling on surfaces covered with receptors. It was concluded that after attaching to the mucus layer in vivo, virus particles reside in a receptor-bound state while rolling through the mucus layer and over epithelial cells driven by the HA–NA-receptor balance. The receptor gradient is likely to serve as the driving force for virus rolling. Dynamic and motile interactions between HA and NA are crucial for penetration of the mucus layer and subsequent infection of cells (Guo et al. 2018). Further insights into how HA and NA balance works to meet both needs—to penetrate mucus and to attach to cells—were also provided (Vahey and Fletcher 2019). They demonstrated that the asymmetrical distribution of receptor-binding and receptor-destroying activities on the surface of the viral particle facilitates its persistent (directional) motion while maintaining stable attachment (Vahey and Fletcher 2019).
The HA affinity has a profound effect on NA enzyme activity and specificity toward sialic acid–containing receptors and receptor analogs (Lai et al. 2019). The results of this study have direct implications for numerous laboratory methods used for virus characterization, including the assessment of NA inhibitor susceptibility (Lai et al. 2019). The influence of HA–receptor specificity may explain why NA inhibition results (small substrate-based assays) better correlate with the outcomes of studies in ferrets than with the traditional cell culture–based assays. Although traditional cell cultures (e.g., Madin–Darby canine kidney [MDCK]) predominantly express receptors linked to the adjacent carbohydrate moiety via α-(2,3) linkage, the HA of human viruses prefers receptors linked with α-(2,6) linkage. The ferret upper respiratory tract epithelium predominantly expresses α-(2,6) receptors, which makes ferrets a suitable model for assessing the NA inhibitor sensitivity of human viruses (Gubareva et al. 1998; Yen et al. 2005). Conversely, the HA of avian and egg-adapted human viruses preferably binds to the receptors with α-(2,3) linkage, and this manifests in an apparent higher sensitivity of avian viruses versus human viruses in traditional cell culture–based assays. Viruses exhibiting a drug-resistant phenotype in humans and ferrets can display drug sensitivity in cell culture and vice versa (Blick et al. 1998; Gubareva et al. 1998, 2001a; Abed et al. 2002a). Moreover, the role of HA glycosylation in virus susceptibility to NA inhibitors was evidenced and thus provides a further insight into the functional balance of HA and NA (Mishin et al. 2005c). Based on many observations made over the years since the approval of the first NA inhibitor, cell culture–based assays are not recommended for the assessment of virus susceptibility to NA inhibitors.
For the purpose of virological surveillance, sensitivity to NA inhibitors is assessed using biochemical NA inhibition assays that utilize small synthetic substrates (i.e., 4-methylumbelliferyl N-acetyl-α-D-neuraminic acid [MUNANA] and NA-Star) (Potier et al. 1979; Buxton et al. 2000; Takashita et al. 2015b). However, there are also NA assays that utilize a large substrate (e.g., fetuin) in enzyme-linked lectin assay (ELLA) (Lambré et al. 1990; Gao et al. 2016). Despite the clear advantages, there are also limitations to this approach. Namely, these assays cannot accurately predict the drug's effect on virus replication, and it has been challenging to establish laboratory correlates of clinically relevant NA inhibitor resistance.
Another manifestation of the HA and NA interplay is the emergence of NA mutants during virus culturing. The HA of human A(H3N2) viruses has evolved in such a way that it displays a low affinity for receptors expressed on the surface of MDCK cells (Asaoka et al. 2006; Yang et al. 2015). As a result, culturing of these viruses often leads to the emergence of NA mutants with substitution at D151 in the NA active site (Lin et al. 2010; Tamura et al. 2013). The substitution at D151 allows switching from the receptor-destroying to the receptor-binding activity (Xue et al. 2018). A virus isolate may contain two NA subpopulations: One (D151) acts as an enzyme, whereas the other (N151) acts as the receptor-binding protein. This cell culture artifact interferes with drug susceptibility monitoring (Mishin et al. 2014). To delay the selection of NA mutants, it is recommended to use cell cultures that were genetically modified to overexpress α-(2,6)-containing receptors, such as MDCK SIAT1 (Matrosovich et al. 2003) and AX-4 cells (Hatakeyama et al. 2005). Moreover, the recently developed hCK cell line may further delay the selection of NA mutants as it additionally expresses extremely low levels of α-(2,3) sialoglycans (Takada et al. 2019). Interestingly, the wild-type NA genotype can be restored by passage of mutated viruses in human primary respiratory cell cultures (Brown et al. 2019).
ANTI-NA ANTIBODIES
Since the 1980s (Webster et al. 1988), there has been growing experimental evidence supporting the role of antibodies raised against NA as an important correlate of protection against influenza (Memoli et al. 2016; Krammer et al. 2018). Because of a lower rate of evolution compared to HA and its highly conserved active site, NA is viewed as an attractive target for the development of therapeutic monoclonal antibodies (mAbs) (Abed et al. 2002b; Doyle et al. 2013c; Clark et al. 2017; Tewawong et al. 2017). Unlike antibodies targeting the HA, anti-NA antibodies do not seem to directly neutralize the infectivity of the virus, although they can impede virus spread. They may also help to clear virus by engaging the Fc region, thereby mediating antibody-dependent cellular cytotoxicity (ADCC), antigen-dependent cellular phagocytosis (ADCP), and complement-dependent cytotoxicity (CDC) (Hashimoto et al. 1983; Co et al. 2012, 2014; He et al. 2016; Wohlbold et al. 2017). Anti-NA antibodies can also act like NA inhibitors by sterically interfering with NA's access to sialic acid moieties on receptors. In recent years, there have been numerous reports describing the generation and characterization of mAbs that broadly and potently inhibit NA activity.
Human infections with A(H1N1)pdm09 and the highly pathogenic avian influenza (HPAI) A(H5N1) viruses have stimulated research toward the antigenic properties of N1 NA. The murine mAb 2B9, generated against a plant-derived recombinant N1 NA, was shown to broadly inhibit NA activity in the MUNANA-based assay (Shoji et al. 2011). Repeated treatment with mAb 2B9 significantly decreased morbidity and mortality of mice and ferrets infected with HPAI A(H5N1) viruses. Additionally, a panel of murine mAbs was generated to map strain-specific and conserved N1 antigenic domains (Wan et al. 2013). Given prophylactically, the cross-reactive mAb fully protected mice against a lethal challenge with A(H1N1) and A(H1N1)pdm09 viruses and provided partial protection against A(H5N1) virus. In another study, a unique 30-amino acid epitope that spans the lateral side of a NA dimer was identified with the use of CD6 mAb. Because this lateral CD6 epitope is conserved among circulating A(H1N1)pdm09 viruses, it may be used as an effective therapeutic target against this subtype (Wan et al. 2015). Furthermore, two mouse mAbs, N1-7D3 and N1-C4, were identified to bind to NAs of multiple N1 subtype viruses (Job et al. 2018). MAb N1-7D3, which recognizes a linear epitope, cross-reacted with NAs of N1 subtype but failed to inhibit NA activity and protect mice against lethal challenge. Conversely, N1-C4 mAb, which recognizes a conformational epitope, displayed NA inhibitory activity by ELLA. However, this was not seen in the MUNANA-based assay, indicating that this mAb did not block access of a small substrate to the enzyme active site. mAb N1-C4 showed partial protective potential without cross protection in mice.
A research team developed two mAbs (IgG2a isotype) against A(H1N1)pdm09 virus; 3C02 was strain-specific, whereas 3C05 cross-reacted with multiple N1 strains (DiLillo et al. 2016). The former was 100-fold more potent in vitro (DiLillo et al. 2016). Using a mouse model, they demonstrated that 3C05 relied on Fc-FcγR interactions to mediate protection, whereas 3C02 did not (DiLillo et al. 2016). Furthermore, the authors emphasized that for broadly acting mAbs with in vitro antiviral activity (e.g., prevention of egress), the FcγR requirement for protection can be circumvented by increasing the dose of mAb.
The emergence of avian A(H7N9) viruses in China represents a significant public health concern and has been a focus of intense research since 2013 (WHO 2017a, 2017b). A murine mAb 3c10-3 directed against A(H7N9) NA showed prophylactic and therapeutic efficacy in mice; it inhibited N9 NA activity in MUNANA-based assay by binding across the enzyme active site. Notably, affinity analysis revealed that mAb 3c10-3 binds equally well to wild-type NA and NA carrying the R292K substitution that confers cross resistance to NA inhibitors. This mAb also restricted the cell-to-cell spread of the virus. As mAb 3c10-3 belongs to IgG1/κ isotype, the question was posed whether a version with the IgG2a Fc would be more protective (Wilson et al. 2016, 2017).
Influenza A viruses of N8 subtype also present a concern because of evidence suggesting their circulation in humans in the nineteenth century (Worobey et al. 2014), recent zoonotic infections in China by avian A(H10N8) viruses (Chen et al. 2014; WHO 2014b), and the spread of HPAI A(H5N8) viruses in birds (WHO 2016a). A panel of murine N8-reactive mAbs that displayed broad binding, encompassing both North American and Eurasian N8 lineages, was generated and characterized (Wohlbold et al. 2016). The 2E11 mAb showed strong antiviral activity in vitro and protection in mice.
Studies also included the NA of influenza B viruses, in which broadly reactive epitopes were elucidated. A panel of mAbs (1F2, 1F4, 3G1, 4B2, and 4F11) that can bind to NAs of viruses from both lineages was generated (Wohlbold et al. 2017). The conserved epitopes targeted by these mAbs are located on the NA head and are distant from the active site. These mAbs demonstrated broad binding to NAs spanning more than 70 years of type B evolution. They broadly inhibited NA activity in ELLA, albeit with relatively high IC50 values. When administered prophylactically at high dose (5 mg/kg), mAbs protected mice against lethal challenge with viruses from both the lineages (Wohlbold et al. 2017).
Influenza A and B NAs share a unique universally conserved sequence spanning amino acids 222 to 230, ILRTQESEC (Doyle et al. 2013a,b,c). Notably, it encompasses three amino acid residues (222, 224, and 227) of the NA active site. A rabbit mAb, HCA-2, was raised by immunization with this short peptide. In ELLA, this mAb caused a significant decrease in the enzyme activity of the N1–N9 subtypes and influenza B viruses from both lineages. Although the inhibitory effect was broad, it was not very potent (IC50 > 10 µg/mL). MAb HCA-2 provided heterosubtypic protection in mice lethally challenged with A(H1N1) and A(H3N2) viruses; however, high doses were required (Doyle et al. 2013a,b).
Recently, a panel of human NA (N1 and N2)-reactive antibodies was isolated from NA-reactive B cells of infected patients (Chen et al. 2018). Among anti-N2 mAbs, several displayed broad reactivity in both ELLA and NA-Star assay, spanning five decades of A(H3N2) antigenic drift. They also identified anti-N1 mAbs that were active in ELLA and NA-Star assays against distant strains of N1 subtype. There was an overlap between the mAb binding footprints and the site occupied by oseltamivir. These mAbs protected mice from lethal challenge, even against distant strains within the NA subtype (Chen et al. 2018).
A study in which broadly reactive NA epitopes for type A and B viruses were identified represents an important milestone (Stadlbauer et al. 2019). Human mAbs derived from plasmablasts of an individual infected with a seasonal A(H3N2) virus were characterized. One of them, mAb 1G01, showed the broadest binding activity toward NAs of type A (N1–N9 subtypes) and type B viruses. In ELLA, this mAb broadly inhibited NA activity of type A viruses (except for one N5 subtype virus) and type B/Victoria lineage viruses. In NA-Star assay, mAb 1G01 displayed a high potency in inhibiting NA activity of N2 viruses (IC50 ∼ 1 nM), indicating that this antibody directly targets the enzyme active site. Remarkably, mAb 1G01 fully protected animals against lethal challenge with different influenza A and B viruses (Stadlbauer et al. 2019).
It has been challenging to identify anti-NA mAbs displaying broad inhibitory activity, especially when their activity was measured using small substrates. This is not surprising considering that the NA active site presents a surface smaller than the footprint of a typical antibody-binding site (Colman et al. 1987). An innovative approach that utilizes novel molecules known as antiviral Fc-conjugates (AVCs) may overcome this challenge. The lead conjugate, CB-012, combines an antiviral molecule with the Fc-fragment of human IgG1. CB-012 was designed to target the virus and simultaneously engage the immune system (www.cidara.com/cloudbreak). In cell culture, this conjugate was 98- and 20-fold more potent than oseltamivir against influenza A and B viruses, respectively (Locke et al. 2019). Its efficacy was assessed in a lethal mouse model using oseltamivir-sensitive and -resistant influenza A(H1N1) and A(H3N2) viruses. A single intravenous dose administered 4 h prior to infection provided full protection in mice (Levin et al. 2019). Moreover, it was shown that the conjugate engages the immune system to accelerate elimination of the pathogen (Döhrmann et al. 2019). These findings validate the utility of multimodal mechanism of action—direct antiviral activity and immune-mediated clearance—for the prevention and treatment of influenza infections.
CONCLUDING REMARKS
The studies conducted around the world have contributed substantially to improve knowledge of NA structure, function, and interplay with HA (Colman et al. 1993). For two decades, NA inhibitors, the sialic acid analogs, have been the mainstay of antiviral therapy against influenza. Four NA inhibitors, oseltamivir, zanamivir, peramivir, and laninamivir, are currently licensed in different parts of the world to control influenza A and B infections (Ison 2017). Influenza viruses that display resistance to NA inhibitors typically contain amino acid substitutions in the NA active site. Because of the difference in NA inhibitors’ chemistry and subtle variance in NA structures, resistance can be NA inhibitor–specific and NA type/subtype–specific. Changes outside of the NA active site have also been shown to affect drug binding (Hurt et al. 2012a; Li et al. 2015). Resistance to NA inhibitors, especially to oseltamivir and peramivir, is a public health concern. Over the years, extensive studies have been conducted with the goal to identify NA inhibitors with improved characteristics (multimeric derivatives and diversely decorated analogs, covalent inhibitors, selective NA inhibitors, etc.).
A new and vibrant area of research is the identification of mAbs that can be used as broadly acting anti-influenza therapeutics. Anti-NA mAbs can prevent virus transmission and protect against lethal virus challenge in animal models (Doyle et al. 2013b,c; Wan et al. 2013, 2015; Liu et al. 2015; Jiang et al. 2016; Memoli et al. 2016; Wohlbold et al. 2016, 2017; Chen et al. 2018). The mAbs that display potent NA inhibitory activity in a small substrate-based NI assay seem to be most promising. Another innovative approach utilizes an AVC, a molecule that directly inhibits viral replication while simultaneously engaging the immune system. Although reports describing the outcomes of influenza AVC studies are not yet published, the data presented on AVC high potency and low clearance raise hopes of protection after a single dose, regardless of a patient's immune status.
Efforts made to improve the understanding of NA-based immunity and protection may pave the way for the development of new influenza therapeutics of greater cross protection and efficacy, while reducing the threat of drug resistance.
ACKNOWLEDGMENT
This article has been made freely available online courtesy of TAUNS Laboratories.
Footnotes
Editors: Gabriele Neumann and Yoshihiro Kawaoka
Additional Perspectives on Influenza: The Cutting Edge available at www.perspectivesinmedicine.org
REFERENCES
- Abed Y, Bourgault AM, Fenton RJ, Morley PJ, Gower D, Owens IJ, Tisdale M, Boivin G. 2002a. Characterization of 2 influenza A(H3N2) clinical isolates with reduced susceptibility to neuraminidase inhibitors due to mutations in the hemagglutinin gene. J Infect Dis 186: 1074–1080. 10.1086/344237 [DOI] [PubMed] [Google Scholar]
- Abed Y, Hardy I, Li Y, Boivin G. 2002b. Divergent evolution of hemagglutinin and neuraminidase genes in recent influenza A:H3N2 viruses isolated in Canada. J Med Virol 67: 589–595. 10.1002/jmv.10143 [DOI] [PubMed] [Google Scholar]
- Abed Y, Nehmé B, Baz M, Boivin G. 2008. Activity of the neuraminidase inhibitor A-315675 against oseltamivir-resistant influenza neuraminidases of N1 and N2 subtypes. Antiviral Res 77: 163–166. 10.1016/j.antiviral.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Abed Y, Baz M, Boivin G. 2009. A novel neuraminidase deletion mutation conferring resistance to oseltamivir in clinical influenza A/H3N2 virus. J Infect Dis 199: 180–183. 10.1086/595736 [DOI] [PubMed] [Google Scholar]
- Air GM, Laver WG. 1989. The neuraminidase of influenza virus. Proteins 6: 341–356. 10.1002/prot.340060402 [DOI] [PubMed] [Google Scholar]
- Asaoka N, Tanaka Y, Sakai T, Fujii Y, Ohuchi R, Ohuchi M. 2006. Low growth ability of recent influenza clinical isolates in MDCK cells is due to their low receptor binding affinities. Microbes Infect 8: 511–519. 10.1016/j.micinf.2005.08.006 [DOI] [PubMed] [Google Scholar]
- Babu YS, Chand P, Bantia S, Kotian P, Dehghani A, El-Kattan Y, Lin TH, Hutchison TL, Elliott AJ, Parker CD, et al. 2000. BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J Med Chem 43: 3482–3486. 10.1021/jm0002679 [DOI] [PubMed] [Google Scholar]
- Baek YH, Song MS, Lee EY, Kim YI, Kim EH, Park SJ, Park KJ, Kwon HI, Pascua PN, Lim GJ, et al. 2015. Profiling and characterization of influenza virus N1 strains potentially resistant to multiple neuraminidase inhibitors. J Virol 89: 287–299. 10.1128/JVI.02485-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett S, Mohr PG, Schmidt PM, McKimm-Breschkin JL. 2011. Real time enzyme inhibition assays provide insights into differences in binding of neuraminidase inhibitors to wild type and mutant influenza viruses. PLoS One 6: e23627. 10.1371/journal.pone.0023627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum EZ, Wagaman PC, Ly L, Turchi I, Le J, Bucher D, Bush K. 2003. A point mutation in influenza B neuraminidase confers resistance to peramivir and loss of slow binding. Antiviral Res 59: 13–22. 10.1016/S0166-3542(03)00011-1 [DOI] [PubMed] [Google Scholar]
- Baz M, Abed Y, Boivin G. 2007. Characterization of drug-resistant recombinant influenza A/H1N1 viruses selected in vitro with peramivir and zanamivir. Antiviral Res 74: 159–162. 10.1016/j.antiviral.2006.10.012 [DOI] [PubMed] [Google Scholar]
- Blick TJ, Sahasrabudhe A, McDonald M, Owens IJ, Morley PJ, Fenton RJ, McKimm-Breschkin JL. 1998. The interaction of neuraminidase and hemagglutinin mutations in influenza virus in resistance to 4-guanidino-Neu5Ac2en. Virology 246: 95–103. 10.1006/viro.1998.9194 [DOI] [PubMed] [Google Scholar]
- Bloom JD, Gong LI, Baltimore D. 2010. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328: 1272–1275. 10.1126/science.1187816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JC, Barclay WS, Galiano M, Harvey R. 2019. Passage of influenza A/H3N2 viruses in human airway cells removes artefactual variants associated with neuraminidase-mediated binding. J Gen Virol 10.1099/jgv.0.001348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister WP, Ruigrok RW, Cusack S. 1992. The 2.2 A resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J 11: 49–56. 10.1002/j.1460-2075.1992.tb05026.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton RC, Edwards B, Juo RR, Voyta JC, Tisdale M, Bethell RC. 2000. Development of a sensitive chemiluminescent neuraminidase assay for the determination of influenza virus susceptibility to zanamivir. Anal Biochem 280: 291–300. 10.1006/abio.2000.4517 [DOI] [PubMed] [Google Scholar]
- Chan J, Watson JN, Lu A, Cerda VC, Borgford TJ, Bennet AJ. 2012. Bacterial and viral sialidases: contribution of the conserved active site glutamate to catalysis. Biochemistry 51: 433–441. 10.1021/bi201019n [DOI] [PubMed] [Google Scholar]
- Chavas LM, Tringali C, Fusi P, Venerando B, Tettamanti G, Kato R, Monti E, Wakatsuki S. 2005. Crystal structure of the human cytosolic sialidase Neu2. Evidence for the dynamic nature of substrate recognition. J Biol Chem 280: 469–475. 10.1074/jbc.M411506200 [DOI] [PubMed] [Google Scholar]
- Chen LF, Dailey NJ, Rao AK, Fleischauer AT, Greenwald I, Deyde VM, Moore ZS, Anderson DJ, Duffy J, Gubareva LV, et al. 2011. Cluster of oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infections on a hospital ward among immunocompromised patients—North Carolina, 2009. J Infect Dis 203: 838–846. 10.1093/infdis/jiq124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, et al. 2014. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383: 714–721. 10.1016/S0140-6736(14)60111-2 [DOI] [PubMed] [Google Scholar]
- Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, et al. 2018. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173: 417–429.e10. 10.1016/j.cell.2018.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WS, Jeong JH, Kwon JJ, Ahn SJ, Lloren KKS, Kwon HI, Chae HB, Hwang J, Kim MH, Kim CJ, et al. 2018. Screening for neuraminidase inhibitor resistance markers among avian influenza viruses of the N4, N5, N6, and N8 neuraminidase subtypes. J Virol 92: e01580-17. 10.1128/JVI.01580-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AM, DeDiego ML, Anderson CS, Wang J, Yang H, Nogales A, Martinez-Sobrido L, Zand MS, Sangster MY, Topham DJ. 2017. Antigenicity of the 2015–2016 seasonal H1N1 human influenza virus HA and NA proteins. PLoS One 12: e0188267. 10.1371/journal.pone.0188267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co MD, Cruz J, Takeda A, Ennis FA, Terajima M. 2012. Comparison of complement dependent lytic, hemagglutination inhibition and microneutralization antibody responses in influenza vaccinated individuals. Hum Vaccin Immunother 8: 1218–1222. 10.4161/hv.21025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co MD, Terajima M, Thomas SJ, Jarman RG, Rungrojcharoenkit K, Fernandez S, Yoon IK, Buddhari D, Cruz J, Ennis FA. 2014. Relationship of preexisting influenza hemagglutination inhibition, complement-dependent lytic, and antibody-dependent cellular cytotoxicity antibodies to the development of clinical illness in a prospective study of A(H1N1)pdm09 Influenza in children. Viral Immunol 27: 375–382. 10.1089/vim.2014.0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Zhang XQ, Senaati HP, Chen HW, Varki NM, Schooley RT, Gagneux P. 2013. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J 10: 321. 10.1186/1743-422X-10-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colacino JM, Chirgadze NY, Garman E, Murti KG, Loncharich RJ, Baxter AJ, Staschke KA, Laver WG. 1997. A single sequence change destabilizes the influenza virus neuraminidase tetramer. Virology 236: 66–75. 10.1006/viro.1997.8709 [DOI] [PubMed] [Google Scholar]
- Collins PJ, Haire LF, Lin YP, Liu J, Russell RJ, Walker PA, Skehel JJ, Martin SR, Hay AJ, Gamblin SJ. 2008. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 453: 1258–1261. 10.1038/nature06956 [DOI] [PubMed] [Google Scholar]
- Collins PJ, Haire LF, Lin YP, Liu J, Russell RJ, Walker PA, Martin SR, Daniels RS, Gregory V, Skehel JJ, et al. 2009. Structural basis for oseltamivir resistance of influenza viruses. Vaccine 27: 6317–6323. 10.1016/j.vaccine.2009.07.017 [DOI] [PubMed] [Google Scholar]
- Colman PM. 1999. A novel approach to antiviral therapy for influenza. J Antimicrob Chemother 44: 17–22. 10.1093/jac/44.suppl_2.17 [DOI] [PubMed] [Google Scholar]
- Colman PM, Ward CW. 1985. Structure and diversity of influenza virus neuraminidase. Curr Top Microbiol Immunol 114: 177–255. [DOI] [PubMed] [Google Scholar]
- Colman PM, Varghese JN, Laver WG. 1983. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303: 41–44. 10.1038/303041a0 [DOI] [PubMed] [Google Scholar]
- Colman PM, Laver WG, Varghese JN, Baker AT, Tulloch PA, Air GM, Webster RG. 1987. Three-dimensional structure of a complex of antibody with influenza virus neuraminidase. Nature 326: 358–363. 10.1038/326358a0 [DOI] [PubMed] [Google Scholar]
- Colman PM, Hoyne PA, Lawrence MC. 1993. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J Virol 67: 2972–2980. 10.1128/JVI.67.6.2972-2980.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan A, Zimmermann EM, Ben-Shabat S. 2014. Modern prodrug design for targeted oral drug delivery. Molecules 19: 16489–16505. 10.3390/molecules191016489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo DJ, Palese P, Wilson PC, Ravetch JV. 2016. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest 126: 605–610. 10.1172/JCI84428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhrmann S, Donatelli J, Amundson K, Borchardt A, Cole J, Levin J, Tari L. 2019. Fc-mediated effector function contributes to potency of novel cloudbreak antiviral Fc-conjugate (AVC) CB-012. Presented at Options X, August 28–September 1, 2019, Singapore. Abstract 11094. [Google Scholar]
- Doyle TM, Hashem AM, Li C, Van Domselaar G, Larocque L, Wang J, Smith D, Cyr T, Farnsworth A, He R, et al. 2013a. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antiviral Res 100: 567–574. 10.1016/j.antiviral.2013.09.018 [DOI] [PubMed] [Google Scholar]
- Doyle TM, Jaentschke B, Van Domselaar G, Hashem AM, Farnsworth A, Forbes NE, Li C, Wang J, He R, Brown EG, et al. 2013b. The universal epitope of influenza A viral neuraminidase fundamentally contributes to enzyme activity and viral replication. J Biol Chem 288: 18283–18289. 10.1074/jbc.M113.468884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle TM, Li C, Bucher DJ, Hashem AM, Van Domselaar G, Wang J, Farnsworth A, She YM, Cyr T, He R, et al. 2013c. A monoclonal antibody targeting a highly conserved epitope in influenza B neuraminidase provides protection against drug resistant strains. Biochem Biophys Res Commun 441: 226–229. 10.1016/j.bbrc.2013.10.041 [DOI] [PubMed] [Google Scholar]
- Du R, Cui Q, Rong L. 2019. Competitive cooperation of hemagglutinin and neuraminidase during influenza A virus entry. Viruses 11: E458. 10.3390/v11050458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi A, Shalhoub S, Rosenfeld P, Li A, Higgins RR, Stogios PJ, Savchenko A, Bastien N, Li Y, Rotstein C, et al. 2014. Multiple influenza A (H3N2) mutations conferring resistance to neuraminidase inhibitors in a bone marrow transplant recipient. Antimicrob Agents Chemother 58: 7188–7197. 10.1128/AAC.03667-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368: 1888–1897. 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- Gao J, Couzens L, Eichelberger MC. 2016. Measuring influenza neuraminidase inhibition antibody titers by enzyme-linked lectin assay. J Vis Exp 10.3791/54573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard A, Charles-Dufant A, Sabatier M, Cortay JC, Frobert E, Picard C, Casalegno JS, Rosa-Calatrava M, Ferraris O, Valette M, et al. 2016a. Impact on antiviral resistance of E119V, I222L and R292K substitutions in influenza A viruses bearing a group 2 neuraminidase (N2, N3, N6, N7 and N9). J Antimicrob Chemother 71: 3036–3045. 10.1093/jac/dkw275 [DOI] [PubMed] [Google Scholar]
- Gaymard A, Le Briand N, Frobert E, Lina B, Escuret V. 2016b. Functional balance between neuraminidase and haemagglutinin in influenza viruses. Clin Microbiol Infect 22: 975–983. 10.1016/j.cmi.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Ghate AA, Air GM. 1998. Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design. Eur J Biochem 258: 320–331. 10.1046/j.1432-1327.1998.2580320.x [DOI] [PubMed] [Google Scholar]
- Ginting TE, Shinya K, Kyan Y, Makino A, Matsumoto N, Kaneda S, Kawaoka Y. 2012. Amino acid changes in hemagglutinin contribute to the replication of oseltamivir-resistant H1N1 influenza viruses. J Virol 86: 121–127. 10.1128/JVI.06085-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Xu W. 2008. Different synthetic strategies of oseltamivir phosphate: a potent influenza neuraminidase inhibitor. Curr Med Chem 15: 3145–3159. 10.2174/092986708786848497 [DOI] [PubMed] [Google Scholar]
- Graitcer SB, Gubareva L, Kamimoto L, Doshi S, Vandermeer M, Louie J, Waters C, Moore Z, Sleeman K, Okomo-Adhiambo M, et al. 2011. Characteristics of patients with oseltamivir-resistant pandemic (H1N1) 2009, United States. Emerg Infect Dis 17: 255–257. 10.3201/eid1702.101724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva LV, Robinson MJ, Bethell RC, Webster RG. 1997. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J Virol 71: 3385–3390. 10.1128/JVI.71.5.3385-3390.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva LV, Matrosovich MN, Brenner MK, Bethell RC, Webster RG. 1998. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J Infect Dis 178: 1257–1262. 10.1086/314440 [DOI] [PubMed] [Google Scholar]
- Gubareva LV, Kaiser L, Matrosovich MN, Soo-Hoo Y, Hayden FG. 2001a. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J Infect Dis 183: 523–531. 10.1086/318537 [DOI] [PubMed] [Google Scholar]
- Gubareva LV, Webster RG, Hayden FG. 2001b. Comparison of the activities of zanamivir, oseltamivir, and RWJ-270201 against clinical isolates of influenza virus and neuraminidase inhibitor-resistant variants. Antimicrob Agents Chemother 45: 3403–3408. 10.1128/AAC.45.12.3403-3408.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva LV, Nedyalkova MS, Novikov DV, Murti KG, Hoffmann E, Hayden FG. 2002. A release-competent influenza A virus mutant lacking the coding capacity for the neuraminidase active site. J Gen Virol 83: 2683–2692. 10.1099/0022-1317-83-11-2683 [DOI] [PubMed] [Google Scholar]
- Gubareva LV, Sleeman K, Guo Z, Yang H, Hodges E, Davis CT, Baranovich T, Stevens J. 2017. Drug susceptibility evaluation of an influenza A(H7N9) virus by analyzing recombinant neuraminidase proteins. J Infect Dis 216: S566–S574. 10.1093/infdis/jiw625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Rabouw H, Slomp A, Dai M, van der Vegt F, van Lent JWM, McBride R, Paulson JC, de Groot RJ, van Kuppeveld FJM, et al. 2018. Kinetic analysis of the influenza A virus HA/NA balance reveals contribution of NA to virus-receptor binding and NA-dependent rolling on receptor-containing surfaces. PLoS Pathog 14: e1007233. 10.1371/journal.ppat.1007233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HK, Amidon GL. 2000. Targeted prodrug design to optimize drug delivery. AAPS PharmSci 2: E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanpaibool C, Leelawiwat M, Takahashi K, Rungrotmongkol T. 2019. Source of oseltamivir resistance due to single E119D and double E119D/H274Y mutations in pdm09H1N1 influenza neuraminidase. J Comput Aided Mol Des 34: 27–37. 10.1007/s10822-019-00251-7 [DOI] [PubMed] [Google Scholar]
- Hashimoto G, Wright PF, Karzon DT. 1983. Antibody-dependent cell-mediated cytotoxicity against influenza virus-infected cells. J Infect Dis 148: 785–794. 10.1093/infdis/148.5.785 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Sakai-Tagawa Y, Kiso M, Goto H, Kawakami C, Mitamura K, Sugaya N, Suzuki Y, Kawaoka Y. 2005. Enhanced expression of an α2,6-linked sialic acid on MDCK cells improves isolation of human influenza viruses and evaluation of their sensitivity to a neuraminidase inhibitor. J Clin Microbiol 43: 4139–4146. 10.1128/JCM.43.8.4139-4146.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden FG, Gubareva LV, Monto AS, Klein TC, Elliot MJ, Hammond JM, Sharp SJ, Ossi MJ, Zanamivir Family Study Group. 2000. Inhaled zanamivir for the prevention of influenza in families. N Engl J Med 343: 1282–1289. 10.1056/NEJM200011023431801 [DOI] [PubMed] [Google Scholar]
- He G, Massarella J, Ward P. 1999. Clinical pharmacokinetics of the prodrug oseltamivir and its active metabolite Ro 64-0802. Clin Pharmacokinet 37: 471–484. 10.2165/00003088-199937060-00003 [DOI] [PubMed] [Google Scholar]
- He W, Tan GS, Mullarkey CE, Lee AJ, Lam MM, Krammer F, Henry C, Wilson PC, Ashkar AA, Palese P, et al. 2016. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc Natl Acad Sci 113: 11931–11936. 10.1073/pnas.1609316113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KC, Hung HC, HuangFu WC, Sung TY, Eight Lin T, Fang MY, Chen IJ, Pathak N, Hsu JT, Yang JM. 2017. Identification of neuraminidase inhibitors against dual H274Y/I222R mutant strains. Sci Rep 7: 12336. 10.1038/s41598-017-12101-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt AC, Holien JK, Parker M, Kelso A, Barr IG. 2009. Zanamivir-resistant influenza viruses with a novel neuraminidase mutation. J Virol 83: 10366–10373. 10.1128/JVI.01200-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt AC, Chotpitayasunondh T, Cox NJ, Daniels R, Fry AM, Gubareva LV, Hayden FG, Hui DS, Hungnes O, Lackenby A, et al. 2012a. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis 12: 240–248. 10.1016/S1473-3099(11)70318-8 [DOI] [PubMed] [Google Scholar]
- Hurt AC, Hardie K, Wilson NJ, Deng YM, Osbourn M, Leang SK, Lee RT, Iannello P, Gehrig N, Shaw R, et al. 2012b. Characteristics of a widespread community cluster of H275Y oseltamivir-resistant A(H1N1)pdm09 influenza in Australia. J Infect Dis 206: 148–157. 10.1093/infdis/jis337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain Basha S, Prasad RN. 2012. In-silico screening of Pleconaril and its novel substituted derivatives with neuraminidase of H1N1 influenza strain. BMC Res Notes 5: 105. 10.1186/1756-0500-5-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu H, Kawai N. 2011. Laninamivir octanoate: a new long-acting neuraminidase inhibitor for the treatment of influenza. Expert Rev Anti Infect Ther 9: 851–857. 10.1586/eri.11.112 [DOI] [PubMed] [Google Scholar]
- Ikematsu H, Kawai N, Iwaki N, Kashiwagi S. 2015. In vitro neuraminidase inhibitory activity of four neuraminidase inhibitors against clinical isolates of influenza virus in the Japanese 2012–2013 season. J Infect Chemother 21: 39–42. 10.1016/j.jiac.2014.08.030 [DOI] [PubMed] [Google Scholar]
- Ison MG. 2017. Antiviral treatments. Clin Chest Med 38: 139–153. 10.1016/j.ccm.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Fantoni G, Couzens L, Gao J, Plant E, Ye Z, Eichelberger MC, Wan H. 2016. Comparative efficacy of monoclonal antibodies that bind to different epitopes of the 2009 pandemic H1N1 influenza virus neuraminidase. J Virol 90: 117–128. 10.1128/JVI.01756-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job ER, Schotsaert M, Ibanez LI, Smet A, Ysenbaert T, Roose K, Dai M, de Haan CAM, Kleanthous H, Vogel TU, et al. 2018. Antibodies directed toward neuraminidase N1 control disease in a mouse model of influenza. J Virol 92: e01584-17. 10.1128/JVI.01584-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju H, Zhang J, Sun Z, Huang Z, Qi W, Huang B, Zhan P, Liu X. 2018. Discovery of C-1 modified oseltamivir derivatives as potent influenza neuraminidase inhibitors. Eur J Med Chem 146: 220–231. 10.1016/j.ejmech.2018.01.050 [DOI] [PubMed] [Google Scholar]
- Karpf M, Trussardi R. 2001. New, azide-free transformation of epoxides into 1,2-diamino compounds: synthesis of the anti-influenza neuraminidase inhibitor oseltamivir phosphate (Tamiflu). J Org Chem 66: 2044–2051. 10.1021/jo005702l [DOI] [PubMed] [Google Scholar]
- Kati WM, Montgomery D, Carrick R, Gubareva L, Maring C, McDaniel K, Steffy K, Molla A, Hayden F, Kempf D, et al. 2002. In vitro characterization of A-315675, a highly potent inhibitor of A and B strain influenza virus neuraminidases and influenza virus replication. Antimicrob Agents Chemother 46: 1014–1021. 10.1128/AAC.46.4.1014-1021.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CU, Lew W, Williams MA, Liu H, Zhang L, Swaminathan S, Bischofberger N, Chen MS, Mendel DB, Tai CY, et al. 1997. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J Am Chem Soc 119: 681–690. 10.1021/ja963036t [DOI] [PubMed] [Google Scholar]
- Kim JH, Resende R, Wennekes T, Chen HM, Bance N, Buchini S, Watts AG, Pilling P, Streltsov VA, Petric M, et al. 2013. Mechanism-based covalent neuraminidase inhibitors with broad-spectrum influenza antiviral activity. Science 340: 71–75. 10.1126/science.1232552 [DOI] [PubMed] [Google Scholar]
- Kiso M, Kubo S, Ozawa M, Le QM, Nidom CA, Yamashita M, Kawaoka Y. 2010. Efficacy of the new neuraminidase inhibitor CS-8958 against H5N1 influenza viruses. PLoS Pathog 6: e1000786. 10.1371/journal.ppat.1000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobasa D, Kodihalli S, Luo M, Castrucci MR, Donatelli I, Suzuki Y, Suzuki T, Kawaoka Y. 1999. Amino acid residues contributing to the substrate specificity of the influenza A virus neuraminidase. J Virol 73: 6743–6751. 10.1128/JVI.73.8.6743-6751.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama K, Takahashi M, Oitate M, Nakai N, Takakusa H, Miura S, Okazaki O. 2009. CS-8958, a prodrug of the novel neuraminidase inhibitor R-125489, demonstrates a favorable long-retention profile in the mouse respiratory tract. Antimicrob Agents Chemother 53: 4845–4851. 10.1128/AAC.00731-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, Wilson PC, Compans RW, Skountzou I, Monto AS. 2018. NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? MBio 9: e02332-17. 10.1128/mBio.02332-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo S, Kakuta M, Yamashita M. 2010a. [In vitro and in vivo effects of a long-acting anti-influenza agent CS-8958 (laninamivir octanoate, Inavir) against pandemic (H1N1) 2009 influenza viruses]. Jpn J Antibiot 63: 337–346. [PubMed] [Google Scholar]
- Kubo S, Tomozawa T, Kakuta M, Tokumitsu A, Yamashita M. 2010b. Laninamivir prodrug CS-8958, a long-acting neuraminidase inhibitor, shows superior anti-influenza virus activity after a single administration. Antimicrob Agents Chemother 54: 1256–1264. 10.1128/AAC.01311-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JJ, Choi WS, Jeong JH, Kim EH, Lee OJ, Yoon SW, Hwang J, Webby RJ, Govorkova EA, Choi YK, et al. 2018. An I436N substitution confers resistance of influenza A(H1N1)pdm09 viruses to multiple neuraminidase inhibitors without affecting viral fitness. J Gen Virol 99: 292–302. 10.1099/jgv.0.001029 [DOI] [PubMed] [Google Scholar]
- Laborda P, Wang SY, Voglmeir J. 2016. Influenza neuraminidase inhibitors: synthetic approaches, derivatives and biological activity. Molecules 21: 1513. 10.3390/molecules21111513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackenby A, Hungnes O, Dudman SG, Meijer A, Paget WJ, Hay AJ, Zambon MC. 2008. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Surveill 13: 3–4. 10.2807/ese.13.05.08026-en [DOI] [PubMed] [Google Scholar]
- Lackenby A, Besselaar TG, Daniels RS, Fry A, Gregory V, Gubareva LV, Huang W, Hurt AC, Leang SK, Lee RTC, et al. 2018. Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors and status of novel antivirals, 2016–2017. Antiviral Res 157: 38–46. 10.1016/j.antiviral.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JCC, Karunarathna H, Wong HH, Peiris JSM, Nicholls JM. 2019. Neuraminidase activity and specificity of influenza A virus are influenced by haemagglutinin-receptor binding. Emerg Microbes Infect 8: 327–338. 10.1080/22221751.2019.1581034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambré CR, Terzidis H, Greffard A, Webster RG. 1990. Measurement of anti-influenza neuraminidase antibody using a peroxidase-linked lectin and microtitre plates coated with natural substrates. J Immunol Methods 135: 49–57. 10.1016/0022-1759(90)90255-T [DOI] [PubMed] [Google Scholar]
- Le QM, Wertheim HF, Tran ND, van Doorn HR, Nguyen TH, Horby P, Vietnam HNIT. 2010. A community cluster of oseltamivir-resistant cases of 2009 H1N1 influenza. N Engl J Med 362: 86–87. [DOI] [PubMed] [Google Scholar]
- Lee CM, Weight AK, Haldar J, Wang L, Klibanov AM, Chen J. 2012. Polymer-attached zanamivir inhibits synergistically both early and late stages of influenza virus infection. Proc Natl Acad Sci 109: 20385–20390. 10.1073/pnas.1219155109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz MR, Webster RG, Air GM. 1987. Site-directed mutation of the active site of influenza neuraminidase and implications for the catalytic mechanism. Biochemistry 26: 5351–5358. 10.1021/bi00391a020 [DOI] [PubMed] [Google Scholar]
- Levin J, Borchardt A, Lam T, Jiang W, Chen Z, Do Q, Brady T, Noncovich A, Fortier J, Akers-Rodriguez S, et al. 2019. Efficacy of CB-012, a novel antiviral Fc-conjugate, in lethal mouse models of oseltamivir-sensitive and -resistant influenza A H1N1 and H3N2 isolates. Presented at Options X, August 28–September 1, 2019, Singapore. Abstract 10979. [Google Scholar]
- Lew W, Escarpe PA, Mendel DB, Sweeny DJ, Kim CU. 1999. Stereospecific synthesis of a GS 4104 metabolite: determination of absolute stereochemistry and influenza neuraminidase inhibitory activity. Bioorg Med Chem Lett 9: 2811–2814. 10.1016/S0960-894X(99)00479-5 [DOI] [PubMed] [Google Scholar]
- L'Huillier AG, Abed Y, Petty TJ, Cordey S, Thomas Y, Bouhy X, Schibler M, Simon A, Chalandon Y, van Delden C, et al. 2015. E119D neuraminidase mutation conferring pan-resistance to neuraminidase inhibitors in an A(H1N1)pdm09 isolate from a stem-cell transplant recipient. J Infect Dis 212: 1726–1734. 10.1093/infdis/jiv288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TC, Chan MC, Lee N. 2015. Clinical implications of antiviral resistance in influenza. Viruses 7: 4929–4944. 10.3390/v7092850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Meng Y, Xu S, Shen W, Meng Z, Wang Z, Ding G, Huang W, Xiao W, Xu J. 2017. Discovery of acylguanidine oseltamivir carboxylate derivatives as potent neuraminidase inhibitors. Bioorg Med Chem 25: 2772–2781. 10.1016/j.bmc.2017.03.052 [DOI] [PubMed] [Google Scholar]
- Lin YP, Gregory V, Collins P, Kloess J, Wharton S, Cattle N, Lackenby A, Daniels R, Hay A. 2010. Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: a role in virus attachment? J Virol 84: 6769–6781. 10.1128/JVI.00458-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Qin-Hua C, Peng L, Chun-Lei L, Guang-De Y. 2018. The hydrophobic side chain of oseltamivir influences type A subtype selectivity of neuraminidase inhibitors. Chem Biol Drug Des 91: 105–115. 10.1111/cbdd.13060 [DOI] [PubMed] [Google Scholar]
- Liu C, Eichelberger MC, Compans RW, Air GM. 1995. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol 69: 1099–1106. 10.1128/JVI.69.2.1099-1106.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WC, Lin CY, Tsou YT, Jan JT, Wu SC. 2015. Cross-reactive neuraminidase-inhibiting antibodies elicited by immunization with recombinant neuraminidase proteins of H5N1 and pandemic H1N1 influenza A viruses. J Virol 89: 7224–7234. 10.1128/JVI.00585-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JB, Borchardt A, Lam T, Jiang W, Chen Z, Levin J, Martin-Sancho L, De Jesus P, Chanda S, Tari L. 2019. Novel antiviral Fc-conjugate CB-012 demonstrates potent activity in cytopathic effect (CPE) and viral growth inhibition assays against influenza A and B strains. Presented at Options X, August 28–September 1, 2019, Singapore. Abstract 10974. [Google Scholar]
- Macdonald SJ, Watson KG, Cameron R, Chalmers DK, Demaine DA, Fenton RJ, Gower D, Hamblin JN, Hamilton S, Hart GJ, et al. 2004. Potent and long-acting dimeric inhibitors of influenza virus neuraminidase are effective at a once-weekly dosing regimen. Antimicrob Agents Chemother 48: 4542–4549. 10.1128/AAC.48.12.4542-4549.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald SJ, Cameron R, Demaine DA, Fenton RJ, Foster G, Gower D, Hamblin JN, Hamilton S, Hart GJ, Hill AP, et al. 2005. Dimeric zanamivir conjugates with various linking groups are potent, long-lasting inhibitors of influenza neuraminidase including H5N1 avian influenza. J Med Chem 48: 2964–2971. 10.1021/jm040891b [DOI] [PubMed] [Google Scholar]
- Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk HD. 2003. Overexpression of the α-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol 77: 8418–8425. 10.1128/JVI.77.15.8418-8425.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol 78: 12665–12667. 10.1128/JVI.78.22.12665-12667.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JL, Gilbertson BP, Trifkovic S, Brown LE, McKimm-Breschkin JL. 2019. Influenza virus neuraminidase structure and functions. Front Microbiol 10: 39. 10.3389/fmicb.2019.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKimm-Breschkin JL. 2000. Resistance of influenza viruses to neuraminidase inhibitors—a review. Antiviral Res 47: 1–17. 10.1016/S0166-3542(00)00103-0 [DOI] [PubMed] [Google Scholar]
- McKimm-Breschkin JL, Barrett S. 2015. Neuraminidase mutations conferring resistance to laninamivir lead to faster drug binding and dissociation. Antiviral Res 114: 62–66. 10.1016/j.antiviral.2014.12.004 [DOI] [PubMed] [Google Scholar]
- McKimm-Breschkin JL, McDonald M, Blick TJ, Colman PM. 1996. Mutation in the influenza virus neuraminidase gene resulting in decreased sensitivity to the neuraminidase inhibitor 4-guanidino-Neu5Ac2en leads to instability of the enzyme. Virology 225: 240–242. 10.1006/viro.1996.0595 [DOI] [PubMed] [Google Scholar]
- McKimm-Breschkin JL, Williams J, Barrett S, Jachno K, McDonald M, Mohr PG, Saito T, Tashiro M. 2013. Reduced susceptibility to all neuraminidase inhibitors of influenza H1N1 viruses with haemagglutinin mutations and mutations in non-conserved residues of the neuraminidase. J Antimicrob Chemother 68: 2210–2221. 10.1093/jac/dkt205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A. 2014. Unusual influenza A viruses in bats. Viruses 6: 3438–3449. 10.3390/v6093438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, et al. 2016. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. MBio 7: e00417-16. 10.1128/mBio.00417-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishin VP, Hayden FG, Gubareva LV. 2005a. Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob Agents Chemother 49: 4515–4520. 10.1128/AAC.49.11.4515-4520.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishin VP, Nedyalkova MS, Hayden FG, Gubareva LV. 2005b. Protection afforded by intranasal immunization with the neuraminidase-lacking mutant of influenza A virus in a ferret model. Vaccine 23: 2922–2927. 10.1016/j.vaccine.2004.11.058 [DOI] [PubMed] [Google Scholar]
- Mishin VP, Novikov D, Hayden FG, Gubareva LV. 2005c. Effect of hemagglutinin glycosylation on influenza virus susceptibility to neuraminidase inhibitors. J Virol 79: 12416–12424. 10.1128/JVI.79.19.12416-12424.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishin VP, Sleeman K, Levine M, Carney PJ, Stevens J, Gubareva LV. 2014. The effect of the MDCK cell selected neuraminidase D151G mutation on the drug susceptibility assessment of influenza A(H3N2) viruses. Antiviral Res 101: 93–96. 10.1016/j.antiviral.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscona A. 2005. Neuraminidase inhibitors for influenza. N Engl J Med 353: 1363–1373. 10.1056/NEJMra050740 [DOI] [PubMed] [Google Scholar]
- Moules V, Ferraris O, Terrier O, Giudice E, Yver M, Rolland JP, Bouscambert-Duchamp M, Bergeron C, Ottmann M, Fournier E, et al. 2010. In vitro characterization of naturally occurring influenza H3NA− viruses lacking the NA gene segment: toward a new mechanism of viral resistance? Virology 404: 215–224. 10.1016/j.virol.2010.04.030 [DOI] [PubMed] [Google Scholar]
- Nabel GJ, Fauci AS. 2010. Induction of unnatural immunity: prospects for a broadly protective universal influenza vaccine. Nat Med 16: 1389–1391. 10.1038/nm1210-1389 [DOI] [PubMed] [Google Scholar]
- Nelson MI, Vincent AL. 2015. Reverse zoonosis of influenza to swine: new perspectives on the human–animal interface. Trends Microbiol 23: 142–153. 10.1016/j.tim.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Fry AM, Loveless PA, Klimov AI, Gubareva LV. 2010. Recovery of a multidrug-resistant strain of pandemic influenza A 2009 (H1N1) virus carrying a dual H275Y/I223R mutation from a child after prolonged treatment with oseltamivir. Clin Infect Dis 51: 983–984. 10.1086/656439 [DOI] [PubMed] [Google Scholar]
- Nishikawa T, Shimizu K, Tanaka T, Kuroda K, Takayama T, Yamamoto T, Hanada N, Hamada Y. 2012. Bacterial neuraminidase rescues influenza virus replication from inhibition by a neuraminidase inhibitor. PLoS One 7: e45371. 10.1371/journal.pone.0045371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley AJ, Barrett S, Peat TS, Newman J, Streltsov VA, Waddington L, Saito T, Tashiro M, McKimm-Breschkin JL. 2010. Structural and functional basis of resistance to neuraminidase inhibitors of influenza B viruses. J Med Chem 53: 6421–6431. 10.1021/jm100621s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okomo-Adhiambo M, Sleeman K, Ballenger K, Nguyen HT, Mishin VP, Sheu TG, Smagala J, Li Y, Klimov AI, Gubareva LV. 2010. Neuraminidase inhibitor susceptibility testing in human influenza viruses: a laboratory surveillance perspective. Viruses 2: 2269–2289. 10.3390/v2102269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P, Compans RW. 1976. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. J Gen Virol 33: 159–163. 10.1099/0022-1317-33-1-159 [DOI] [PubMed] [Google Scholar]
- Palese P, Schulman JL, Bodo G, Meindl P. 1974. Inhibition of influenza and parainfluenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA). Virology 59: 490–498. 10.1016/0042-6822(74)90458-9 [DOI] [PubMed] [Google Scholar]
- Pizzorno A, Bouhy X, Abed Y, Boivin G. 2011. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J Infect Dis 203: 25–31. 10.1093/infdis/jiq010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno A, Abed Y, Rhéaume C, Bouhy X, Boivin G. 2013. Evaluation of recombinant 2009 pandemic influenza A (H1N1) viruses harboring zanamivir resistance mutations in mice and ferrets. Antimicrob Agents Chemother 57: 1784–1789. 10.1128/AAC.02269-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potier M, Mameli L, Bélisle M, Dallaire L, Melançon SB. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-D-N-acetylneuraminate) substrate. Anal Biochem 94: 287–296. 10.1016/0003-2697(79)90362-2 [DOI] [PubMed] [Google Scholar]
- Rameix-Welti MA, Enouf V, Cuvelier F, Jeannin P, van der Werf S. 2008. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog 4: e1000103. 10.1371/journal.ppat.1000103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggentin P, Rothe B, Kaper JB, Galen J, Lawrisuk L, Vimr ER, Schauer R. 1989. Conserved sequences in bacterial and viral sialidases. Glycoconj J 6: 349–353. 10.1007/BF01047853 [DOI] [PubMed] [Google Scholar]
- Roosenhoff R, Reed V, Kenwright A, Schutten M, Boucher CA, Monto A, Clinch B, Kumar D, Whitley R, Nguyen-Van-Tam JS, et al. 2019. Viral kinetics and resistance development in children treated with neuraminidase inhibitors: the Influenza Resistance Information Study (IRIS). Clin Infect Dis ciz939. 10.1093/cid/ciz939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungrotmongkol T, Frecer V, De-Eknamkul W, Hannongbua S, Miertus S. 2009. Design of oseltamivir analogs inhibiting neuraminidase of avian influenza virus H5N1. Antiviral Res 82: 51–58. 10.1016/j.antiviral.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443: 45–49. 10.1038/nature05114 [DOI] [PubMed] [Google Scholar]
- Sakai T, Nishimura SI, Naito T, Saito M. 2017. Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery. Sci Rep 7: 45043. 10.1038/srep45043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M, Pizzorno A, Abed Y, Boivin G. 2013. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res 98: 174–185. 10.1016/j.antiviral.2013.03.014 [DOI] [PubMed] [Google Scholar]
- Schade D, Kotthaus J, Riebling L, Kotthaus J, Müller-Fielitz H, Raasch W, Koch O, Seidel N, Schmidtke M, Clement B. 2014. Development of novel potent orally bioavailable oseltamivir derivatives active against resistant influenza A. J Med Chem 57: 759–769. 10.1021/jm401492x [DOI] [PubMed] [Google Scholar]
- Schulman JL, Palese P. 1975. Susceptibility of different strains of influenza A virus to the inhibitory effects of 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA). Virology 63: 98–104. 10.1016/0042-6822(75)90374-8 [DOI] [PubMed] [Google Scholar]
- Shie JJ, Fang JM, Lai PT, Wen WH, Wang SY, Cheng YS, Tsai KC, Yang AS, Wong CH. 2011. A practical synthesis of zanamivir phosphonate congeners with potent anti-influenza activity. J Am Chem Soc 133: 17959–17965. 10.1021/ja207892q [DOI] [PubMed] [Google Scholar]
- Shoji Y, Chichester JA, Palmer GA, Farrance CE, Stevens R, Stewart M, Goldschmidt L, Deyde V, Gubareva L, Klimov A, et al. 2011. An influenza N1 neuraminidase-specific monoclonal antibody with broad neuraminidase inhibition activity against H5N1 HPAI viruses. Hum Vaccin 7: 199–204. 10.4161/hv.7.0.14595 [DOI] [PubMed] [Google Scholar]
- Singh A, Soliman ME. 2015. Understanding the cross-resistance of oseltamivir to H1N1 and H5N1 influenza A neuraminidase mutations using multidimensional computational analyses. Drug Des Devel Ther 9: 4137–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman K, Guo Z, Barnes J, Shaw M, Stevens J, Gubareva LV. 2013. R292K substitution and drug susceptibility of influenza A(H7N9) viruses. Emerg Infect Dis 19: 1521–1524. 10.3201/eid1909.130724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman K, Mishin VP, Guo Z, Garten RJ, Balish A, Fry AM, Villanueva J, Stevens J, Gubareva LV. 2014. Antiviral susceptibility of variant influenza A(H3N2)v viruses isolated in the United States from 2011 to 2013. Antimicrob Agents Chemother 58: 2045–2051. 10.1128/AAC.02556-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BJ, Colman PM, Von Itzstein M, Danylec B, Varghese JN. 2001. Analysis of inhibitor binding in influenza virus neuraminidase. Protein Sci 10: 689–696. 10.1110/ps.41801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Marathe BM, Kumar G, Wong SS, Rubrum A, Zanin M, Choi YK, Webster RG, Govorkova EA, Webby RJ. 2015. Unique determinants of neuraminidase inhibitor resistance among N3, N7, and N9 avian influenza viruses. J Virol 89: 10891–10900. 10.1128/JVI.01514-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer D, Zhu X, McMahon M, Turner JS, Wohlbold TJ, Schmitz AJ, Strohmeier S, Yu W, Nachbagauer R, Mudd PA, et al. 2019. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science 366: 499–504. 10.1126/science.aay0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storms AD, Gubareva LV, Su S, Wheeling JT, Okomo-Adhiambo M, Pan CY, Reisdorf E, St. George K, Myers R, Wotton JT, et al. 2012. Oseltamivir-resistant pandemic (H1N1) 2009 virus infections, United States, 2010–11. Emerg Infect Dis 18: 308–311. 10.3201/eid1802.111466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K. 2019. The critical interspecies transmission barrier at the animal–human interface. Trop Med Infect Dis 4: 72. 10.3390/tropicalmed4020072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaya N, Ohashi Y. 2010. Long-acting neuraminidase inhibitor laninamivir octanoate (CS-8958) versus oseltamivir as treatment for children with influenza virus infection. Antimicrob Agents Chemother 54: 2575–2582. 10.1128/AAC.01755-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16: 265–273. 10.1038/nsmb.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K, Kawakami C, Fan S, Chiba S, Zhong G, Gu C, Shimizu K, Takasaki S, Sakai-Tagawa Y, Lopes TJS, et al. 2019. A humanized MDCK cell line for the efficient isolation and propagation of human influenza viruses. Nat Microbiol 4: 1268–1273. 10.1038/s41564-019-0433-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E, Fujisaki S, Kishida N, Xu H, Imai M, Tashiro M, Odagiri T, The Influenza Virus Surveillance Group of Japan. 2013. Characterization of neuraminidase inhibitor-resistant influenza A(H1N1)pdm09 viruses isolated in four seasons during pandemic and post-pandemic periods in Japan. Influenza Other Respir Viruses 7: 1390–1399. 10.1111/irv.12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E, Kiso M, Fujisaki S, Yokoyama M, Nakamura K, Shirakura M, Sato H, Odagiri T, Kawaoka Y, Tashiro M. 2015a. Characterization of a large cluster of influenza A(H1N1)pdm09 viruses cross-resistant to oseltamivir and peramivir during the 2013–2014 influenza season in Japan. Antimicrob Agents Chemother 59: 2607–2617. 10.1128/AAC.04836-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E, Meijer A, Lackenby A, Gubareva L, Rebelo-de-Andrade H, Besselaar T, Fry A, Gregory V, Leang SK, Huang W, et al. 2015b. Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2013–2014. Antiviral Res 117: 27–38. 10.1016/j.antiviral.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura D, Nguyen HT, Sleeman K, Levine M, Mishin VP, Yang H, Guo Z, Okomo-Adhiambo M, Xu X, Stevens J, et al. 2013. Cell culture-selected substitutions in influenza A(H3N2) neuraminidase affect drug susceptibility assessment. Antimicrob Agents Chemother 57: 6141–6146. 10.1128/AAC.01364-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura D, DeBiasi RL, Okomo-Adhiambo M, Mishin VP, Campbell AP, Loechelt B, Wiedermann BL, Fry AM, Gubareva LV. 2015. Emergence of multidrug-resistant influenza A(H1N1)pdm09 virus variants in an immunocompromised child treated with oseltamivir and zanamivir. J Infect Dis 212: 1209–1213. 10.1093/infdis/jiv245 [DOI] [PubMed] [Google Scholar]
- Taylor NR, Cleasby A, Singh O, Skarzynski T, Wonacott AJ, Smith PW, Sollis SL, Howes PD, Cherry PC, Bethell R, et al. 1998. Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B. J Med Chem 41: 798–807. 10.1021/jm9703754 [DOI] [PubMed] [Google Scholar]
- Tewawong N, Vichiwattana P, Korkong S, Klinfueng S, Suntronwong N, Thongmee T, Theamboonlers A, Vongpunsawad S, Poovorawan Y. 2017. Evolution of the neuraminidase gene of seasonal influenza A and B viruses in Thailand between 2010 and 2015. PLoS One 12: e0175655. 10.1371/journal.pone.0175655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewawong N, Marathe BM, Poovorawan Y, Vongpunsawad S, Webby RJ, Govorkova EA. 2018. Neuraminidase inhibitor susceptibility and neuraminidase enzyme kinetics of human influenza A and B viruses circulating in Thailand in 2010–2015. PLoS One 13: e0190877. 10.1371/journal.pone.0190877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorlund K, Awad T, Boivin G, Thabane L. 2011. Systematic review of influenza resistance to the neuraminidase inhibitors. BMC Infect Dis 11: 134. 10.1186/1471-2334-11-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, et al. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci 109: 4269–4274. 10.1073/pnas.1116200109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9: e1003657. 10.1371/journal.ppat.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahey MD, Fletcher DA. 2019. Influenza A virus surface proteins are organized to help penetrate host mucus. Elife 8: e43764. 10.7554/eLife.43764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese JN, Laver WG, Colman PM. 1983. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 303: 35–40. 10.1038/303035a0 [DOI] [PubMed] [Google Scholar]
- Varghese JN, Epa VC, Colman PM. 1995. Three-dimensional structure of the complex of 4-guanidino-Neu5Ac2en and influenza virus neuraminidase. Protein Sci 4: 1081–1087. 10.1002/pro.5560040606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese JN, Smith PW, Sollis SL, Blick TJ, Sahasrabudhe A, McKimm-Breschkin JL, Colman PM. 1998. Drug design against a shifting target: a structural basis for resistance to inhibitors in a variant of influenza virus neuraminidase. Structure 6: 735–746. 10.1016/S0969-2126(98)00075-6 [DOI] [PubMed] [Google Scholar]
- Vavricka CJ, Liu Y, Kiyota H, Sriwilaijaroen N, Qi J, Tanaka K, Wu Y, Li Q, Li Y, Yan J, et al. 2013. Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors. Nat Commun 4: 1491. 10.1038/ncomms2487 [DOI] [PubMed] [Google Scholar]
- Venkatramani L, Johnson ES, Kolavi G, Air GM, Brouillette WJ, Mooers BH. 2012. Crystal structure of a new benzoic acid inhibitor of influenza neuraminidase bound with a new tilt induced by overpacking subsite C6. BMC Struct Biol 12: 7. 10.1186/1472-6807-12-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, Donis R, Culhane M, Hamilton K, Lewis N, et al. 2014. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health 61: 4–17. 10.1111/zph.12049 [DOI] [PubMed] [Google Scholar]
- von Itzstein M, Thomson R. 2009. Anti-influenza drugs: the development of sialidase inhibitors. Handb Exp Pharmacol 189: 111–154. [DOI] [PubMed] [Google Scholar]
- von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, Van Phan T, Smythe ML, White HF, Oliver SW, et al. 1993. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363: 418–423. 10.1038/363418a0 [DOI] [PubMed] [Google Scholar]
- Wagner R, Matrosovich M, Klenk HD. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12: 159–166. 10.1002/rmv.352 [DOI] [PubMed] [Google Scholar]
- Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, et al. 2013. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol 87: 9290–9300. 10.1128/JVI.01203-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Yang H, Shore DA, Garten RJ, Couzens L, Gao J, Jiang L, Carney PJ, Villanueva J, Stevens J, et al. 2015. Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nat Commun 6: 6114. 10.1038/ncomms7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Qi L, Gao J, Couzens LK, Jiang L, Gao Y, Sheng ZM, Fong S, Hahn M, Khurana S, et al. 2018. Comparison of the efficacy of N9 neuraminidase-specific monoclonal antibodies against influenza A(H7N9) virus infection. J Virol 92: e01588-17. 10.1128/JVI.01588-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GT, Chen Y, Wang S, Gentles R, Sowin T, Kati W, Muchmore S, Giranda V, Stewart K, Sham H, et al. 2001. Design, synthesis, and structural analysis of influenza neuraminidase inhibitors containing pyrrolidine cores. J Med Chem 44: 1192–1201. 10.1021/jm000468c [DOI] [PubMed] [Google Scholar]
- Wang MZ, Tai CY, Mendel DB. 2002. Mechanism by which mutations at his274 alter sensitivity of influenza A virus N1 neuraminidase to oseltamivir carboxylate and zanamivir. Antimicrob Agents Chemother 46: 3809–3816. 10.1128/AAC.46.12.3809-3816.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SQ, Cheng XC, Dong WL, Wang RL, Chou KC. 2010. Three new powerful oseltamivir derivatives for inhibiting the neuraminidase of influenza virus. Biochem Biophys Res Commun 401: 188–191. 10.1016/j.bbrc.2010.09.020 [DOI] [PubMed] [Google Scholar]
- Wang B, Wang K, Meng P, Hu Y, Yang F, Liu K, Lei Z, Chen B, Tian Y. 2018. Design, synthesis, and evaluation of carboxyl-modified oseltamivir derivatives with improved lipophilicity as neuraminidase inhibitors. Bioorg Med Chem Lett 28: 3477–3482. 10.1016/j.bmcl.2018.09.014 [DOI] [PubMed] [Google Scholar]
- Wang K, Lei Z, Zhao L, Chen B, Yang F, Liu K, Zhu H, Zhao H, Cao R, Zhang K, et al. 2019. Design, synthesis and biological evaluation of oseltamivir derivatives containing pyridyl group as potent inhibitors of neuraminidase for influenza A. Eur J Med Chem 185: 111841. [DOI] [PubMed] [Google Scholar]
- Watson JN, Dookhun V, Borgford TJ, Bennet AJ. 2003. Mutagenesis of the conserved active-site tyrosine changes a retaining sialidase into an inverting sialidase. Biochemistry 42: 12682–12690. 10.1021/bi035396g [DOI] [PubMed] [Google Scholar]
- Watson KG, Cameron R, Fenton RJ, Gower D, Hamilton S, Jin B, Krippner GY, Luttick A, McConnell D, MacDonald SJ, et al. 2004. Highly potent and long-acting trimeric and tetrameric inhibitors of influenza virus neuraminidase. Bioorg Med Chem Lett 14: 1589–1592. 10.1016/j.bmcl.2003.09.102 [DOI] [PubMed] [Google Scholar]
- Webster RG, Reay PA, Laver WG. 1988. Protection against lethal influenza with neuraminidase. Virology 164: 230–237. 10.1016/0042-6822(88)90640-X [DOI] [PubMed] [Google Scholar]
- Weight AK, Haldar J, de Cienfuegos L A, Gubareva LV, Tumpey TM, Chen J, Klibanov AM. 2011. Attaching zanamivir to a polymer markedly enhances its activity against drug-resistant strains of influenza A virus. J Pharm Sci 100: 831–835. 10.1002/jps.22338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley RJ, Boucher CA, Lina B, Nguyen-Van-Tam JS, Osterhaus A, Schutten M, Monto AS. 2013. Global assessment of resistance to neuraminidase inhibitors, 2008–2011: The Influenza Resistance Information Study (IRIS). Clin Infect Dis 56: 1197–1205. 10.1093/cid/cis1220 [DOI] [PubMed] [Google Scholar]
- WHO. 2014a. Weekly epidemiological record.
- WHO. 2014b. World Health Organization. 2014. Avian influenza A (H10N8). World Health Organization, Geneva, Switzerland. [Google Scholar]
- WHO. 2016a. Assessment of risk associated with influenza A(H5N8) virus. November 2016.
- WHO. 2016b. Weekly epidemiological record.
- WHO. 2017a. World Health Organization. Human infection with avian influenza A(H7N9) virus—China. Disease outbreak news 2017.
- WHO. 2017b. World Health Organization. Human infection with avian influenza A(H7N9) virus—China. Disease outbreak news 2017. Feb 20.
- WHO. 2018. A summary of neuraminidase amino acid substitutions associated with reduced inhibition by NAIs.
- WHO. 2019. Influenza at the human–animal interface.
- Wilson JR, Guo Z, Reber A, Kamal RP, Music N, Gansebom S, Bai Y, Levine M, Carney P, Tzeng WP, et al. 2016. An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody with prophylactic and therapeutic activity in vivo. Antiviral Res 135: 48–55. 10.1016/j.antiviral.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JR, Belser JA, DaSilva J, Guo Z, Sun X, Gansebom S, Bai Y, Stark TJ, Chang J, Carney P, et al. 2017. An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody protects mice from morbidity without interfering with the development of protective immunity to subsequent homologous challenge. Virology 511: 214–221. 10.1016/j.virol.2017.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbold TJ, Chromikova V, Tan GS, Meade P, Amanat F, Comella P, Hirsh A, Krammer F. 2016. Hemagglutinin stalk- and neuraminidase-specific monoclonal antibodies protect against lethal H10N8 influenza virus infection in mice. J Virol 90: 851–861. 10.1128/JVI.02275-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, Amanat F, Tan J, tenOever BR, Tan GS, et al. 2017. Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes. Nat Microbiol 2: 1415–1424. 10.1038/s41564-017-0011-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe C, Greenwald I, Chen L. 2010. Pandemic (H1N1) 2009 and oseltamivir resistance in hematology/oncology patients. Emerg Infect Dis 16: 1809–1811. 10.3201/eid1611.101053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CJ, Malaisree M, Pattarapongdilok N, Sompornpisut P, Hannongbua S, Mulholland AJ. 2012. Long time scale GPU dynamics reveal the mechanism of drug resistance of the dual mutant I223R/H275Y neuraminidase from H1N1-2009 influenza virus. Biochemistry 51: 4364–4375. 10.1021/bi300561n [DOI] [PubMed] [Google Scholar]
- Worobey M, Han GZ, Rambaut A. 2014. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci 111: 8107–8112. 10.1073/pnas.1324197111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Xu D, Huang B, Ma X, Qi W, Shi F, Liu X, Zhang Y, Xu W. 2014. Discovery of N-substituted oseltamivir derivatives as potent and selective inhibitors of H5N1 influenza neuraminidase. J Med Chem 57: 8445–8458. 10.1021/jm500892k [DOI] [PubMed] [Google Scholar]
- Xue KS, Greninger AL, Perez-Osorio A, Bloom JD. 2018. Cooperating H3N2 influenza virus variants are not detectable in primary clinical samples. mSphere 3: e00552-17. 10.1128/mSphereDirect.00552-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Tomozawa T, Kakuta M, Tokumitsu A, Nasu H, Kubo S. 2009. CS-8958, a prodrug of the new neuraminidase inhibitor R-125489, shows long-acting anti-influenza virus activity. Antimicrob Agents Chemother 53: 186–192. 10.1128/AAC.00333-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Carney PJ, Chang JC, Guo Z, Villanueva JM, Stevens J. 2015. Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins. Virology 477: 18–31. 10.1016/j.virol.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Yang X, Xu M, Chan PK, Ma C. 2019. Novel N-substituted oseltamivir derivatives as potent influenza neuraminidase inhibitors: design, synthesis, biological evaluation, ADME prediction and molecular docking studies. Eur J Med Chem 182: 111635. 10.1016/j.ejmech.2019.111635 [DOI] [PubMed] [Google Scholar]
- Yen HL, Herlocher LM, Hoffmann E, Matrosovich MN, Monto AS, Webster RG, Govorkova EA. 2005. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob Agents Chemother 49: 4075–4084. 10.1128/AAC.49.10.4075-4084.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HL, McKimm-Breschkin JL, Choy KT, Wong DD, Cheung PP, Zhou J, Ng IH, Zhu H, Webby RJ, Guan Y, et al. 2013. Resistance to neuraminidase inhibitors conferred by an R292K mutation in a human influenza virus H7N9 isolate can be masked by a mixed R/K viral population. MBio 4: e00396-13. 10.1128/mBio.00396-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf M, Mohamed N, Mohamad S, Janezic D, Damodaran KV, Wahab HA. 2016. H274Y's effect on oseltamivir resistance: what happens before the drug enters the binding site. J Chem Inf Model 56: 82–100. 10.1021/acs.jcim.5b00331 [DOI] [PubMed] [Google Scholar]
- Zanin M, Baviskar P, Webster R, Webby R. 2016. The interaction between respiratory pathogens and mucus. Cell Host Microbe 19: 159–168. 10.1016/j.chom.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Murugan NA, Tian Y, Bertagnin C, Fang Z, Kang D, Kong X, Jia H, Sun Z, Jia R, et al. 2018. Structure-based optimization of N-substituted oseltamivir derivatives as potent anti-influenza A virus agents with significantly improved potency against oseltamivir-resistant N1-H274Y variant. J Med Chem 61: 9976–9999. 10.1021/acs.jmedchem.8b01065 [DOI] [PubMed] [Google Scholar]
- Zurcher T, Yates PJ, Daly J, Sahasrabudhe A, Walters M, Dash L, Tisdale M, McKimm-Breschkin JL. 2006. Mutations conferring zanamivir resistance in human influenza virus N2 neuraminidases compromise virus fitness and are not stably maintained in vitro. J Antimicrob Chemother 58: 723–732. 10.1093/jac/dkl321 [DOI] [PubMed] [Google Scholar]