Abstract
Cells generate and sense mechanical forces that trigger biochemical signals to elicit cellular responses that control cell fate changes. Mechanical forces also physically distort neighboring cells and the surrounding connective tissue, which propagate mechanochemical signals over long distances to guide tissue patterning, organogenesis, and adult tissue homeostasis. As the largest and stiffest organelle, the nucleus is particularly sensitive to mechanical force and deformation. Nuclear responses to mechanical force include adaptations in chromatin architecture and transcriptional activity that trigger changes in cell state. These force-driven changes also influence the mechanical properties of chromatin and nuclei themselves to prevent aberrant alterations in nuclear shape and help maintain genome integrity. This review will discuss principles of nuclear mechanotransduction and chromatin mechanics and their role in DNA damage and cell fate regulation.
MECHANICAL FORCES IN CELLS AND TISSUES
Tissue and cell dynamics and movement generate physical forces, including compression or stretching, which are transferred to the nucleus and chromatin, resulting in their deformation. Substantial deformation occurs not only in mechanically active tissues, such as the muscle, where cells and nuclei are directly exposed to constant contractile forces, but also in tissues such as the lung, skin, and vasculature whose surface area changes during respiration, body motion, and blood flow (Fig. 1). In addition to these high-amplitude changes that affect the tissue as a whole, most cells experience deformation at smaller scales. For instance, cell migration, especially within confined spaces of the connective tissue, causes substantial cell deformation (Friedl et al. 2011). Cell motility, death, division, and extrusion also inflict dynamic changes in cell shape, which can simultaneously trigger deformation of neighboring cells (Chen et al. 2018; Wickström and Niessen 2018). Importantly, local compression and stretching events have profound effects on cell state. Cell compression can trigger cell differentiation and extrusion, whereas stretching can prevent differentiation and promote cell division (Ruprecht et al. 2015; Le et al. 2016; Gudipaty et al. 2017; Miroshnikova et al. 2018; Lam et al. 2020). Thus, mechanical forces relay information on tissue dynamics across length scales and provide efficient means to communicate tissue needs to individual cells.
Figure 1.
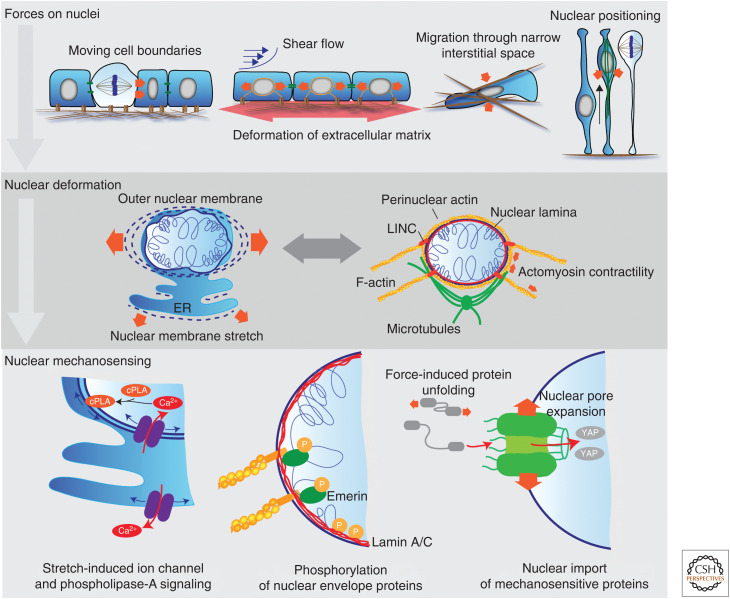
The nucleus as a mechanosensor. The nucleus is subject to deformation forces (red block arrows) when cells are compressed, stretched, or actively contracting during various tissue processes (upper panel). Nuclear deformation causes stretching of the nuclear envelope and the associated endoplasmic reticulum (ER). The amount of force transmitted, and the degree of deformation is modulated by the nuclear-cytoplasmic linkage and the stiffness of the nuclear lamina and chromatin. Perinuclear actin and microtubule networks exert contractile and compressive forces on the nucleus to counteract deformation and regulate nuclear shape (middle panel). Nuclear deformation and stretching of nuclear membranes activate stretch-induced ion channels causing elevated intracellular Ca2+ levels and incorporation of phospholipase A2 (cPLA) into the nuclear membrane (bottom left). Mechanical force transmitted by the linker of nucleoskeleton and cytoskeleton (LINC) complex induces phosphorylation of emerin and lamin A/C through unknown mechanisms (bottom middle). Nuclear deformation can lead to expansion of the nuclear pore to facilitate nuclear import. Import of mechanically unstable and mechanosensitive proteins such as YAP are induced by force (bottom right).
PRINCIPLES OF MECHANOTRANSDUCTION
The dynamic deformation of cells is transferred to various intracellular, nuclear, and nucleus-associated macromolecular complexes to trigger mechanochemical signaling cascades that regulate cell morphology, metabolism, and gene expression. These processes are collectively termed mechanotransduction. Nuclear mechanotransduction refers to specific events that are triggered through structures associated with or located within the nucleus and that specifically impact nuclear components, most importantly chromatin itself. Upon particularly large-scale deformations, such as immune cell infiltration and cancer cell invasion through the narrow space of the interstitium, mechanical stresses have the potential to physically damage the nucleus and the genetic material (Denais et al. 2016; Raab et al. 2016; Irianto et al. 2017). Recent studies have also identified mechanisms by which cells are able to counteract mechanical stress to prevent damage by altering nuclear mechanical properties (Stephens et al. 2019; Nava et al. 2020). This review will address both of these aspects of nuclear mechanotransduction.
Until recently, mechanosensing has been thought to occur mainly at the plasma membrane through transmembrane receptor complexes, such as integrin-based cell–matrix adhesions and cadherin-based cell–cell adhesions (Iskratsch et al. 2014). These adhesive complexes are connected with the contractile actomyosin cytoskeleton, through which cells can both exert forces on their surroundings, and sense mechanical properties or dynamic deformation of neighboring cells or the connective tissue substrate (Charras and Yap 2018; Gauthier and Roca-Cusachs 2018). Mechanical forces applied on these multiprotein adhesion complexes can be converted into biochemical signals, for example through their mechanical unfolding and subsequent activation of signaling molecules (Hu et al. 2017). Stretch-induced ion channels, such as the Piezo channels, can also be activated at the plasma membrane to trigger Ca2+-dependent intracellular mechanosignaling (Murthy et al. 2017). Further downstream, remodeling of the actomyosin cytoskeleton that occurs in response to force also controls the activity of a number of signaling molecules and transcriptional regulators, of which the MRTF/SRF and YAP signaling pathways are best understood. For more detailed insights on these mechanochemical signaling pathways, which are potent regulators of cell behavior, we refer to other recent reviews (Totaro et al. 2018; Sidorenko and Vartiainen 2019).
Mechanical forces and deformation of the plasma membrane are directly transmitted to the nucleus through the contractile actomyosin cytoskeleton, which connects adhesion complexes to the nucleus and thus can cause nuclear deformation. This connection occurs through specialized receptors collectively termed the linker of nucleoskeleton and cytoskeleton (LINC) complex (Starr and Fridolfsson 2010; Luxton and Starr 2014). The LINC complex is composed of cytoplasmic components of the nesprin family members. These proteins link the actomyosin and microtubule cytoskeletons to the nuclear membrane. On the inner side of the nuclear membrane, nesprins bind SUN domain proteins, which in turn provide a direct mechanochemical link to chromatin through their association with components of the nuclear lamina and various inner nuclear membrane proteins, such as emerin, torsinA, lamina-associated polypeptide 1, and spectrin-repeat-containing proteins (Rothballer and Kutay 2013; Hao and Starr 2019).
Lamins are intermediate filament proteins that form the nuclear lamina, and have been directly implicated in establishing the mechanical properties and stability of the nucleus. Together with B-type lamins, the A-type lamins, lamin A and C, form a filamentous network underlying the inner nuclear membrane. The nuclear lamina, and specifically lamin A, are critical for determining nuclear shape, stiffness, and deformability (Liu et al. 2000). Lamin A/C levels positively correlate with nuclear stiffness, whereas reduced lamin A levels result in softer, more deformable, and fragile nuclei (Lammerding et al. 2004). Lamins also interact directly and indirectly, through chromatin-binding proteins, with specific genomic regions to generate lamina-associated domains (LADs) (Guelen et al. 2008), which are rich in silenced heterochromatin and are involved in regulating chromatin organization and gene expression (Briand and Collas 2020). Reflecting their central function in nuclear mechanics and chromatin organization, mutations in the LINC complex and lamins are linked to developmental disorders such as Emery–Dreifuss muscular dystrophy, dilated cardiomyopathy, and Hutchinson–Gilford progeria syndrome. On the cellular level, these diseases manifest with nuclear shape abnormalities and fragility, gene regulation defects, and DNA damage (Davidson and Lammerding 2014). Intriguingly, despite ubiquitous expression of the mutant proteins, the diseases specifically affect mechanically active or mechanically loaded tissues such as skeletal muscle, heart, and skin (Davidson and Lammerding 2014; Miroshnikova et al. 2019). DNA damage has also been shown to occur as a response to mechanically induced nuclear rupture, at least in some muscular dystrophies (Earle et al. 2020), highlighting the role of nuclear deformation and nuclear mechanics in tissue development and function, and the important but incompletely understood relationship between chromatin and mechanical stress.
THE NUCLEUS AS A MECHANOSENSOR
Through its unique elastic properties and compressibility, the nucleus functions as a mechanosensor by detecting dynamic changes in cell volume and by acting as a mechanical shock absorber (Dahl et al. 2004, 2005; Lomakin et al. 2020). While the plasma membrane, cytoplasm, and the associated cytoskeleton are all highly deformable, the nucleus is up to tenfold stiffer than the rest of the cell, making its deformation a potentially rate-limiting process and thus ideal for mechanosensing (Harada et al. 2014; Davidson et al. 2015; Renkawitz et al. 2019).
Sensing Nuclear Deformation
Migrating cells use the nucleus as a “ruler” to measure both the dimensions of the surrounding microenvironment and acute changes in their own volume due to extrinsic forces, such as physical confinement (Fig. 1; Renkawitz et al. 2019; Lomakin et al. 2020). For example, migrating leukocytes apply cytoskeletal forces to insert their nuclei into multiple adjacent connective tissue pores to measure pore sizes, after which the largest pore will be selected as the path for migration (Renkawitz et al. 2019). The proposed ruler function is beautifully simple: the nucleus can deform until the membrane reservoirs of the outer nuclear membrane are fully unfolded. Any further deformation will stretch and tense the outer nuclear membrane and most likely its continuous endoplasmic reticulum (ER), activating stretch-induced calcium channels and subsequent downstream effects such as actomyosin contractility (Lomakin et al. 2020). A similar nuclear membrane deformation pathway operates in epithelial monolayers in response to substrate stretch (Nava et al. 2020). Here, stretch-activated calcium signaling, activated by nuclear membrane stretching via Piezo-1 ion channel, reduces the chromatin occupancy of trimethylation of lysine 9 on histone 3 (H3K9me3) genome wide resulting in nucleus softening, which protects it from mechanical damage. Nuclear deformation in response to cell spreading or osmotic swelling of the nucleus has also been shown to trigger perinuclear calcium release, which results in elevated nuclear calcium levels that change gene expression (Itano et al. 2003). Osmotic swelling of the nucleus also triggers activation of phospholipase A2 by promoting its hydrophobic membrane insertion, which together with raised calcium signals enhance inflammatory signaling and cell contractility (Fig. 1; Enyedi and Niethammer 2017; Lomakin et al. 2020).
Regulation of Nuclear Transport and Signaling by Mechanical Forces
Nuclear pore complexes, which are large protein complexes that traverse the nuclear envelope and regulate nuclear transport of RNA and proteins, are also sensitive to mechanical force (Donnaloja et al. 2019; Infante et al. 2019). These structures have been shown to dilate and assume a more open conformation in response to mechanical force, which enhances the import of transcriptional regulators such as YAP (Elosegui-Artola et al. 2017). Nuclear transport can further be modulated by mechanical unfolding of proteins to enhance their translocation rate into the nucleus (Infante et al. 2019), leading to changes in gene expression. Finally, phosphorylation of components of the nucleus, such as lamin A/C or emerin, is also altered by mechanical forces acting on nuclei, through molecular mechanisms that remain unclear. Lamin A/C phosphorylation is triggered by low cytoskeletal tension on soft adhesive substrates, resulting in increased lamin A/C mobility and turnover (Swift et al. 2013; Buxboim et al. 2014; Kochin et al. 2014). Emerin phosphorylation, on the other hand, is triggered by high tension, resulting in strengthening interactions of the lamina with the cytoskeleton and in mechanosensitive signaling thorough YAP (Fig. 1; Guilluy et al. 2014).
Cytoskeletal Modulation of Nuclear Mechanosensing
Direct deformation of perinuclear membranes and/or the nuclear pore by extrinsic forces can trigger downstream signaling, and thus nuclear mechanosensing may occur independent of the cytoskeleton and other mechanosensitive signaling pathways. However, the lamina and the LINC complex are likely involved in modulating “nuclear mechanosensitivity” also in cases where signaling is triggered through nuclear deformation. The LINC complex is critical for force transmission from the cytoskeleton to the nucleus and can control the amplitude of nuclear deformation in response to cell contractility (Fig. 1; Lombardi et al. 2011). Further, as lamin A is central to determining nuclear elasticity, its levels influence the degree of force-induced nuclear deformation, and help adjust nuclear membrane tension and membrane reservoir (Enyedi and Niethammer 2017; Lomakin et al. 2020; Nava et al. 2020). Importantly, cell-type-specific effects of nuclear mechanotransduction can be modulated through the lamina, especially considering marked differences in lamin A levels between cell types and differentiation states, by defining the nuclear deformation threshold that triggers downstream signaling (Nava et al. 2020).
The actin cytoskeleton, in particular perinuclear actin, is also involved in nuclear mechanosensing. Perinuclear actin is arranged in a cell-type- and cell-state-specific manner and can consist of (1) an actin cap formed by dorsal actin stress fibers that are attached to adhesions in both ends and to the nuclear envelope through the LINC complex, or (2) a perinuclear actin ring that associates with perinuclear membranes and the ER. Both structures are dynamic, and form rapidly in response to force application through Ca2+ or small GTPase signaling (Woroniuk et al. 2018; Wang et al. 2019), thus most likely directly responding to the stretching of nuclear membranes. The central functions of perinuclear actin are to stabilize nuclear shape and volume in the presence of mechanical stress, and to regulate mechanosensitive gene expression through controlling nuclear and cytoplasmic pools of free G-actin and MAL/SRF and YAP pathways (Fig. 1; Le et al. 2016; Wales et al. 2016; Kim et al. 2017; Shiu et al. 2018; Nava et al. 2020). Importantly, both G- and F-forms of actin also exist in the nucleus and are critical mediators of chromatin architecture, as will be discussed in the following section.
Direct Chromatin Deformation by Force
Mechanical forces transmitted via the cytoskeleton to the nucleus may directly stretch chromatin and activate gene expression. Application of ∼20 kPa shear stress at the cell surface using a magnetic bead leads to chromatin displacement and rapid transcriptional activation of a set of mechanosensitive genes (Tajik et al. 2016; Sun et al. 2020). While direct force-mediated regulation of chromatin is an exciting concept, the mechanism of specificity that render certain genes sensitive to force without activating others remains a key open question.
FORCE-MEDIATED REGULATION OF TRANSCRIPTION AND CELL STATE
The nucleus lies in the direct path of cellular force sensing and transduction. Recent data show that mechanical forces transmitted to the nucleus regulate chromatin architecture and transcription to guide cell fate changes. Dynamic changes in cell and nuclear geometry are powerful regulators of cell states. This connection is highly relevant, as changes in shape/geometry occur downstream of most mechanical forces and are core to many developmental and homeostatic processes that involve fate transitions. Perhaps the clearest illustration of this phenomenon can be observed during early embryonic development where mechanical forces are critical for the establishment of the anterior–posterior axis, as well as for the sorting of the germ layers (Vining and Mooney 2017).
Simplest experimental demonstrations of the direct effects of cell geometry on chromatin and cell state come from in vitro experiments using adhesive micropatterns that can be used to force cultured cells to assume specific shapes. Among the first studies, restricting adhesive area of mammary epithelial cells to induce “rounded” shapes, which are reminiscent of the cellular shape observed in 3D versus flat 2D culture conditions, resulted in global histone deacetylation, chromatin condensation, and reduction in gene expression (Le Beyec et al. 2007). Similarly, culturing mesenchymal stem cells on anisotropic, elongated micropatterns led to increased histone deacetylase activity and decreased histone acetylation (Li et al. 2011), whereas increasing cell spread area of fibroblasts triggered increased histone acetylation and modifies gene expression patterns related to actin cytoskeleton (Jain et al. 2013). Further, restriction of adhesive area, or even a change from an isotropic circular geometry to an anisotropic elongated geometry in epidermal stem cells induced transcription of epidermal differentiation genes (Connelly et al. 2010; Miroshnikova et al. 2018).
The mechanisms by which cell geometry changes drive epigenetic and transcriptional adaptation are beginning to unravel. For instance, stretch-induced ion channels are triggered by changes in cell and nuclear shape, implicating the role of intracellular Ca2+ signaling in heterochromatin regulation to occur downstream of mechanical deformation (Stephens et al. 2017; Nava et al. 2020). Calcium signaling can trigger chromatin and gene expression changes through various mechanisms, where the source and the duration of the calcium signals are of critical importance. Imaging-based studies have shown that artificially elevating nuclear calcium levels by ionophore treatment induces chromatin hypercompaction (Phengchat et al. 2016). However, genome-wide chromatin accessibility studies find changes in only relatively few sites in response to artificial calcium influx, compared to the effects of biochemical signaling events that trigger elevation of intracellular Ca2+ (Brignall et al. 2017), indicating a more complex involvement of Ca2+ in chromatin regulation. While the precise mechanisms are yet to be elucidated, the nucleus is thought to have autonomy in regulating its own calcium levels (Leite et al. 2003; Bootman et al. 2009; Rodrigues et al. 2009). Going forward, it will thus be important to define the specific Ca2+ oscillation patterns within the cytoplasmic and nuclear compartments that are directly triggered by nuclear deformation, and to dissect the precise intranuclear effects of Ca2+, considering the myriad mechanisms by which Ca2+ signaling impacts the core transcriptional machinery (Vilborg et al. 2016).
The actin cytoskeleton also plays an important role in communicating changes in cell geometry to the nucleus. Actin dynamically senses changes in geometry through regulation of the balance between filamentous F-actin and free, nuclear-import compatible G-actin, which communicates mechanoregulation between the cytoplasm and the nucleus. In particular, nuclear actin has important functions in chromatin organization and cellular transcriptional activity (Grosse and Vartiainen 2013). Nuclear actin is required for nuclear reprogramming of oocytes via its ability to transcriptionally reactivate the otherwise silenced Oct4 pluripotency gene (Miyamoto et al. 2011). Downstream of mechanical cues, nuclear actin regulates the mammary epithelial cell fate as these cells fluctuate between periods of quiescence and active growth in response to environmental and hormonal cues (Spencer et al. 2011). Specifically, signals from the basement membrane, which are absent in regions of active growth in the developing mammary end buds, decrease levels of nuclear G-actin, which destabilizes RNA polymerases II/III, in turn leading to reduced transcription and thereby triggering cell quiescence (Spencer et al. 2011). Further increased perinuclear F-actin polymerization, and the resulting decreased nuclear G-actin, attenuates RNA polymerase II–driven transcription in stretched epidermal stem cells, resulting in increased deposition of H3K27me3 on lineage commitment genes, thereby preventing differentiation (Le et al. 2016). Collectively, mechanical regulation of nuclear actin is implicated in determining stem cell quiescence and commitment to differentiation through its effects on global transcription levels. In general, low nuclear actin levels correlate with reduced transcription and promote quiescence/stemness, whereas high levels increase transcription and promote activated/differentiated states. Interestingly, the formation of perinuclear actin is Ca2+ dependent, providing an intriguing link between Ca2+ signaling induced by nuclear deformation and mechanotransduction pathways dependent on actin dynamics (Le et al. 2016; Wales et al. 2016; Nava et al. 2020).
Actin is not the only relevant force-responsive and force-transducing cytoskeletal element that plays a role in transcription. Compressive forces from the microtubules have also been recently shown to deform the nucleus, inducing lobulated nuclear shapes and local loss of H3K9me3-marked heterochromatin from within nuclear envelope invaginations. These local heterochromatin changes drive specific gene expression changes in human hematopoietic stem cells during their early differentiation (Biedzinski et al. 2020), although the precise mechanisms are unclear.
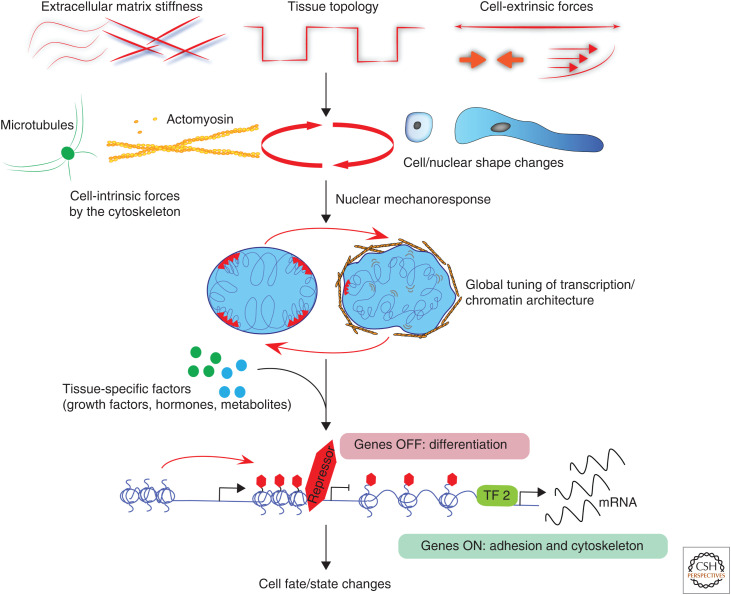
Thus, cellular cytoskeleton both efficiently relays cell-extrinsic mechanical forces into the nucleus through its dynamic assembly cycles, as well as applies direct mechanical forces to the nucleus to reorganize chromatin and regulate gene expression. In addition to these mechanisms, there is a potentially highly relevant, but currently understudied link between mechanotransduction, metabolism, and epigenetic regulation of gene expression. For instance, shear stress in endothelial cells activates AMP-activated protein kinase (AMPK), the master regulator of energy homeostasis (Bays et al. 2017), whereas matrix rigidity can control lipid metabolism and glycolysis (Romani et al. 2019; Park et al. 2020). Given the direct link between metabolites feeding into epigenetic reactions of methylation and acetylation (Su et al. 2016), it seems plausible that mechanical regulation of metabolism is involved in facilitating epigenetic changes caused by cell and nuclear deformation, although the precise mechanisms require further investigation. While the mechanisms by which cell and nuclear shape changes regulate chromatin and transcription are being unraveled, the question of specificity still remains. How do force-induced global changes in transcription or histone modifications lead to specific state transitions or cell-type-specific responses? We postulate that the specificity could arise from cross talk between mechanical and biochemical pathways, where mechanics, through its effect on transcription or chromatin state, plays a role in thresholding, amplifying, or attenuating the specific signals that are propagated by growth factors, hormones, and their downstream transcription factors (Fig. 2).
Figure 2.
Force-mediated regulation of transcription and cell state. Cells sense a spectrum of cell-extrinsic forces and biophysical cues from their microenvironment, such as tissue stiffness, topology, shear stress, compression, and stretch. These cues induce cell and organelle shape changes, triggering cellular mechanosensing and mechanosignaling, facilitated by cell-intrinsic force generation and force distribution through the microtubule and actomyosin cytoskeletons. Nuclear mechanosignaling involves changes in global chromatin architecture and transcriptional activity. We propose that specificity in nuclear mechanoresponses is achieved via collaborative, bidirectional cross talk with tissue-specific factors to facilitate specific transcriptional changes to alter cell fate/state. As examples, extrinsic stretch on epidermal stem cells reduces transcription of differentiation genes but promotes transcription of cytoskeleton and adhesion genes.
MECHANICAL PROPERTIES OF CHROMATIN AND THEIR IMPLICATIONS IN GENOME ORGANIZATION AND FUNCTION
To understand how chromatin is impacted by mechanical forces, it is also important to consider their influence on core chromatin processes, such as transcription factor binding, enhancer engagement and DNA damage repair, all of which are tightly linked to 3D genome structure and influenced by the physical properties of chromatin. Chromatin is a disordered, variably compacted polymer chain that can interact with itself on multiple scales (compartments, topologically associating domains, loops) as well as with the lamina or nucleolar periphery (Pombo and Dillon 2015; Szabo et al. 2019). Initial chromatin rheological studies used particle nanotracking of injected beads, whereas recent work mainly use fluorescently labeled histones or specifically tagged genomic loci, such as telomeres, coupled to mechanical manipulation by micropipette aspiration or atomic force microscopy to map chromatin movement in intact cells (Tseng et al. 2004; Lammerding 2011; Spagnol and Dahl 2016; Stephens et al. 2017, 2018; Hobson et al. 2020). Collectively, these studies have revealed several intriguing features of chromatin. First, instead of being a purely viscous “melt” of liquid-like polymers, the nuclear interior is actually significantly stiffer than the cytoplasm, where chromatin displays elastic and solid-like behaviors. While isolated chromosomes respond elastically in response to force (Cui and Bustamante 2000; Marko 2008; Strickfaden et al. 2020), micropipette aspiration-based measurements from intact, locally deformed nuclei show that chromatin can also flow and locally condense as would be expected of a spongy polymer from which solvent is locally squeezed out (Pajerowski et al. 2007). It is further evident that chromatin motion is spatially correlated over length scales ranging up to several micrometers (Tseng et al. 2004; Zidovska et al. 2013). Thus, based on current data, chromatin can be considered viscoelastic (i.e., displaying mechanical properties of both an elastic solid and a viscous liquid).
When mechanically deformed, both nuclei and chromatin show an elastic deformation response (i.e., they have the ability to return to their original shape after the applied force is removed) (Maeshima et al. 2019). Interestingly, the mechanical resistance of chromatin governs elastic deformations of the nucleus under small (<3 μm) extensions, while the mechanical properties of lamins govern the elastic deformation under larger extensions (Stephens et al. 2018). Thus, it seems that the nucleus consists of at least two spring-like mechanical elements, the chromatin and the nuclear lamina, which operate on distinct deformation scales. This interesting phenomenon might be related to the fact that the nuclear lamina is wrinkled, and requires a large deformation to be fully stretched, together with the inherent mechanical properties of intermediate filaments, which are easy to bend but hard to stretch (Li et al. 2015; Turgay et al. 2017; Lomakin et al. 2020).
Importantly, mechanical properties of chromatin and thus nuclei are dictated by the chromatin compaction state. Nuclei with abundant heterochromatin are rigid, whereas the nuclei with decondensed chromatin are soft (Stephens et al. 2018; Maeshima et al. 2019; Nava et al. 2020). Further, less compacted chromatin is more mobile and deformable (Booth-Gauthier et al. 2012; Spagnol and Dahl 2016; Whitefield et al. 2018; Ghosh et al. 2019; Nava et al. 2020). The association of chromatin to the nuclear lamina also modulates nuclear mechanical properties, and untethering of chromatin from the nuclear lamina increases chromatin flow, enhances nuclear deformability and alters nuclear shape (Schreiner et al. 2015; Stephens et al. 2019; Hobson et al. 2020; Nava et al. 2020).
Collectively, these mechanical properties of chromatin have profound functional implications. The spring-like behavior of chromatin points to chromatin mechanics as a central stabilizer of chromatin architecture and nuclear shape in response to extrinsic mechanical forces that deform the nucleus. The specific mechanical properties of H3K9me3-containing and lamina-associated heterochromatin, and their significant contribution to bulk nuclear stiffness, indicate that chromatin and nuclear mechanical properties can be dynamically regulated to match the force environment of a given cell/tissue. Conversely, the fact that local deformation can be transmitted over micrometers implies that a sustained localized force acting on the nucleus and/or chromatin could, in principle, specifically modulate chromatin interactions and thus gene activity. Both of these aspects will be discussed in the following sections.
MECHANICAL FORCE AND DNA DAMAGE
As discussed above, nuclei undergo substantial deformation during physiological processes such as during migration of leukocytes or cancer cells through small pores of connective tissue. Importantly, nuclear deformation is also associated with DNA damage. The first observations of deformation-induced damage come from the field of cancer cell migration, where migration of tumor cells in confined environments triggers pressurization, which in turn induces formation of local inflations of the nuclear envelope as “blebs.” Blebs can eventually burst to cause leakage of nuclear factors and protrusion of chromatin into the cytoplasm (Denais et al. 2016; Raab et al. 2016; Deviri et al. 2017). The exposure of genomic DNA to cytoplasmic components such as nucleases leads to DNA damage, which is particularly enriched at these herniated sites. Consequently, depletion of cytoplasmic exonuclease TREX1 is sufficient to abolish DNA damage in disrupted nuclei in human breast cancer cells (Fig. 3; Nader et al. 2020).
Figure 3.
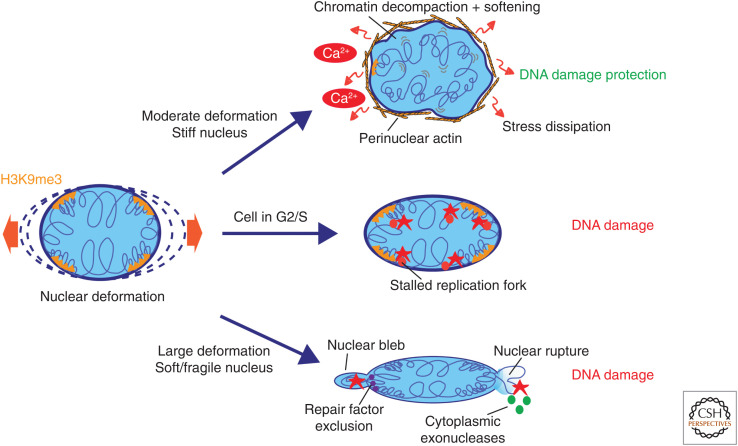
Mechanisms of nuclear deformation–induced DNA damage and damage protection. The impact of nuclear deformation on genome integrity depends on the mechanical properties of the nuclear lamina and on the cell cycle state of the cell. Cells with stiff nuclei respond to deformation by reducing lamina-associated H3K9me3 heterochromatin in a Ca2+-dependent manner, which increases chromatin mobility to facilitate dissipation of the mechanical energy to prevent DNA damage. Simultaneous formation of a perinuclear actin ring prevents further nuclear shape and volume changes. Nuclear deformation in cells in G2/S phase cause replication fork stalling and subsequent DNA damage. Particularly large deformations in cells with soft/fragile nuclei can cause blebbing and bleb rupture. Resulting exposure of herniating chromatin to cytoplasmic exonucleases triggers DNA damage. Exclusion of repair factors from blebs may hamper damage repair.
Depletion of lamins increases the likelihood of nuclear envelope ruptures, consistent with their importance in stabilizing the nuclear envelope (Schreiber and Kennedy 2013). On the other hand, increasing heterochromatin to increase nuclear stiffness can decrease blebbing, further underlining the important role of chromatin as a mechanical regulator of the nucleus (Stephens et al. 2019). Cells are able to rapidly repair the ruptured nuclear envelope using the ESCRT pathway (Denais et al. 2016; Raab et al. 2016), which is also used for sealing plasma membrane ruptures and the postmitotic nuclear envelope (Jimenez et al. 2014; Olmos et al. 2015; Vietri et al. 2015). However, in cases where deformation occurs repeatedly and the mechanical DNA damage becomes chronic, the long-term consequences include induction of senescence in nontransformed cells and induced invasive behavior in cancer cells (Nader et al. 2020). Cells that have repeatedly migrated through narrow pores have also been shown to display cell cycle arrest, chromosome copy number alteration, and loss of heterozygosity (Irianto et al. 2017; Pfeifer et al. 2018), although the direct evidence that this is caused by the physical damage, and not selection for cells particularly capable of migration through narrow pores, is missing.
DNA damage can also occur in deformed nuclei in the absence of nuclear envelope rupture (Denais et al. 2016; Nava et al. 2020; Shah et al. 2021), and even in the case of rupture, DNA damage can occasionally be observed distant from the damage site (Irianto et al. 2017). Two main mechanisms by which this rupture-independent damage occurs have been proposed: One posits that since chromatin is elastic and thus behaves like a solid, nuclear constriction may function to expel the liquid component that contains soluble repair factors. Thus, the mechanism of damage would be the limitation of repair factors (Irianto et al. 2017). This type of factor exclusion most likely operates only upon extreme deformation. Another proposed mechanism, which could also explain observed damage in moderately deformed nuclei, is replicative stress-induced DNA damage. Both confined migration and experimental compression of the nucleus have been shown to increase replication fork stalling, triggering DNA damage at these sites (Fig. 3; Shah et al. 2021). Going forward, it will be important to dissect the mechanisms by which deformation induces replication fork stalling and to understand whether certain genomic regions are more susceptible than others to force-induced DNA damage.
Interestingly, despite in vitro studies showing that nuclear deformation can also induce DNA damage in nontransformed cells, tissues that undergo large-scale deformation during normal physiology, such as muscle, heart, lung, and skin, do not display frequent DNA damage. This indicates that mechanisms exist to counteract nuclear deformation as well as deformation-induced DNA damage. Recent studies show that cells are able to dissipate mechanical stress both on the tissue scale and at the chromatin scale to prevent nuclear deformation and damage. Skin epithelial cell monolayers exposed to uniaxial stretch display moderate (1–3 µm scale) nuclear deformation in the direction of stretch. Despite this deformation, no DNA damage is induced. Instead, cells respond by decreasing levels of H3K9me3, in particular in the proximity of the nuclear lamina. This reduction in H3K9me3 has no immediate transcriptional consequences but renders the nucleus and chromatin more elastic, allowing dissipation of mechanical energy (Nava et al. 2020). Only if this chromatin remodeling is prevented, DNA becomes damaged, indicating that decreased H3K9me3 and chromatin softening is a mechanism to protect the genome from damage (Fig. 3). Interestingly, when the stretch persists over hours, the entire cell monolayer or even an intact skin tissue explant, aligns according to the direction of stretch, redistributing mechanical energy within the tissue to prevent nuclear deformation. This rearrangement allows chromatin to resume its steady-state architecture for more sustainable long-term mechanoprotection (Nava et al. 2020). The precise mechanism by which DNA is damaged in this scenario is unclear, but it is likely related to torsional stress and replication fork stalling. This notion is supported by studies on long-term (>6 h) biaxial stretch, where cells cannot reorient themselves due to the absence of a single direction of strain. This results not only in reduced H3K9me3 and repositioning of chromatin away from the lamina but also in global transcriptional repression (Le et al. 2016), which could collectively reduce torsional stress on chromatin (Nelson 1999).
CONCLUDING REMARKS
Mechanical forces reorganize chromatin and impact nuclear architecture and mechanics by engaging signaling cascades capable of modifying the levels and distribution of charged entities such as Ca2+ ions or ATP, and by modifying chromatin accessibility, compaction, and association with nuclear landmarks such as the lamina. With major advances in our understanding of the mechanisms by which mechanical forces mediate transcriptional regulation, it becomes increasingly important to study how forces organize chromatin and how specificity in chromatin rearrangements is achieved. Further technological developments will be needed, such as integrated systems capable of exerting calibrated forces on specific genomic loci or nuclear structures, while measuring both chromatin state/mechanics and transcriptional output in real time with high spatiotemporal resolution.
Major advances have been made in understanding of mechanotransduction into the cell nucleus to directly act on gene regulation and chromatin functions. A large body of in vitro experiments in multiple systems have demonstrated that dynamic, bidirectional cross talk between cell and nuclear mechanics and chromatin state modulate stem cell differentiation. The next step will be to construct genetic or otherwise manipulatable animal and organ models to challenge and refine these mechanisms in complex, multicellular tissues. Besides development and homeostasis, nucleomechanical regulation of cell states is likely relevant more broadly during processes such as aging, which leads to modified tissue mechanics, as well as many diseases, such as cancers, where abnormal nuclear mechanics has already been shown to be beneficial for aggression within the primary site and during invasion across a spectrum of tissues (Levental et al. 2009; Denais and Lammerding 2014; Bell and Lammerding 2016; Miroshnikova et al. 2016; Emon et al. 2018; Pfeifer et al. 2019). A better understanding of the dynamic relationship between cell-extrinsic forces and their effect on nuclear shape, mechanics, chromatin organization, and cell state during disease onset and evolution, is potentially of high clinical relevance in diagnosis and even treatment. To that effect, a number of high throughout imaging and screening efforts have begun to unravel the mechanisms regulating nuclear shape and mechanics in homeostasis and disease progression (Kume et al. 2017; Hwang et al. 2019; Tamashunas et al. 2020), but their clinical relevance remains to be fully explored.
ACKNOWLEDGMENTS
We apologize to all investigators whose work could not be cited due to space constraints, and thank Clementine Villeneuve and Matthias Rübsam for feedback on the manuscript. Work on nuclear mechanotransduction in the Wickström laboratory is supported by the Helsinki Institute of Life Science, Wihuri Research Institute, Academy of Finland, Juselius Foundation, and European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (Grant agreement 770877 - STEMpop). Y.A.M. is the recipient of the EMBO Long-Term fellowship ALTF 728-2017 and Human Frontier Science Program fellowship LT000861/2018.
Footnotes
Editors: Ana Pombo, Martin W. Hetzer, and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Bays JL, Campbell HK, Heidema C, Sebbagh M, Demali KA. 2017. Linking E-cadherin mechanotransduction to cell metabolism through force-mediated activation of AMPK. Nat Cell Biol 19: 724–731. 10.1038/ncb3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell ES, Lammerding J. 2016. Causes and consequences of nuclear envelope alterations in tumour progression. Eur J Cell Biol 95: 449–464. 10.1016/j.ejcb.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedzinski S, Agsu C, Vianay B, Delord M, Blanchoin L, Larghero J, Faivre LThéry M, Brunet S. 2020. Microtubules control nuclear shape and gene expression during stages of hematopoietic differentiation. EMBO J 39: e103957. 10.15252/embj.2019103957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth-Gauthier EA, Alcoser TA, Yang G, Dahl KN. 2012. Force-induced changes in subnuclear movement and rheology. Biophys J 103: 2423–2431. 10.1016/j.bpj.2012.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman MD, Fearnley C, Smyrnias I, MacDonald F, Roderick HL. 2009. An update on nuclear calcium signalling. J Cell Sci 122: 2337–2350. 10.1242/jcs.028100 [DOI] [PubMed] [Google Scholar]
- Briand N, Collas P. 2020. Lamina-associated domains: peripheral matters and internal affairs. Genome Biol 21: 85. 10.1186/s13059-020-02003-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignall R, Cauchy P, Bevington SL, Gorman B, Pisco AO, Bagnall J, Boddington C, Rowe W, England H, Rich K, et al. 2017. Integration of kinase and calcium signaling at the level of chromatin underlies inducible gene activation in T cells. J Immunol 199: 2652–2667. 10.4049/jimmunol.1602033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A, Swift J, Irianto J, Spinler KR, Dingal PCDP, Athirasala A, Kao YRC, Cho S, Harada T, Shin JW, et al. 2014. Matrix elasticity regulates lamin-A,C phosphorylation and turnover with feedback to actomyosin. Curr Biol 24: 1909–1917. 10.1016/j.cub.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G, Yap AS. 2018. Tensile forces and mechanotransduction at cell–cell junctions. Curr Biol 28: R445–R457. 10.1016/j.cub.2018.02.003 [DOI] [PubMed] [Google Scholar]
- Chen T, Saw TB, Mège RM, Ladoux B. 2018. Mechanical forces in cell monolayers. J Cell Sci 131: jcs218156. 10.1242/jcs.218156 [DOI] [PubMed] [Google Scholar]
- Connelly JT, Gautrot JE, Trappmann B, Tan DWM, Donati G, Huck WTS, Watt FM. 2010. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat Cell Biol 12: 711–718. 10.1038/ncb2074 [DOI] [PubMed] [Google Scholar]
- Cui Y, Bustamante C. 2000. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc Natl Acad Sci 97: 127–132. 10.1073/pnas.97.1.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Kahn SM, Wilson KL, Discher DE. 2004. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci 117: 4779–4786. 10.1242/jcs.01357 [DOI] [PubMed] [Google Scholar]
- Dahl KN, Engler AJ, Pajerowski JD, Discher DE. 2005. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys J 89: 2855–2864. 10.1529/biophysj.105.062554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson PM, Lammerding J. 2014. Broken nuclei—lamins, nuclear mechanics, and disease. Trends Cell Biol 24: 247–256. 10.1016/j.tcb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson PM, Sliz J, Isermann P, Denais C, Lammerding J. 2015. Design of a microfluidic device to quantify dynamic intra-nuclear deformation during cell migration through confining environments. Integr Biol (Camb) 7: 1534–1546. 10.1039/C5IB00200A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais C, Lammerding J. 2014. Nuclear mechanics in cancer. Adv Exp Med Biol 773: 435–470. 10.1007/978-1-4899-8032-8_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P. McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. 2016. Nuclear envelope rupture and repair during cancer cell migration. Science 352: 353–358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deviri D, Discher DE, Safran SA. 2017. Rupture dynamics and chromatin herniation in deformed nuclei. Biophys J 113: 1060–1071. 10.1016/j.bpj.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnaloja F, Jacchetti E, Soncini M, Raimondi MT. 2019. Mechanosensing at the nuclear envelope by nuclear pore complex stretch activation and its effect in physiology and pathology. Front Physiol 10: 896. 10.3389/fphys.2019.00896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle AJ, Kirby TJ, Fedorchak GR, Isermann P, Patel J, Iruvanti S, Moore SA, Bonne G, Wallrath LL, Lammerding J. 2020. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat Mater 19: 464–473. 10.1038/s41563-019-0563-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux AL, et al. 2017. Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171: 1397–1410.e14. 10.1016/j.cell.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Emon B, Bauer J, Jain Y, Jung B, Saif T. 2018. Biophysics of tumor microenvironment and cancer metastasis—a mini review. Comput Struct Biotechnol J 16: 279–287. 10.1016/j.csbj.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi B, Niethammer P. 2017. Nuclear membrane stretch and its role in mechanotransduction. Nucleus 8: 156–161. 10.1080/19491034.2016.1263411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Wolf K, Lammerding J. 2011. Nuclear mechanics during cell migration. Curr Opin Cell Biol 23: 55–64. 10.1016/j.ceb.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier NC, Roca-Cusachs P. 2018. Mechanosensing at integrin-mediated cell–matrix adhesions: from molecular to integrated mechanisms. Curr Opin Cell Biol 50: 20–26. 10.1016/j.ceb.2017.12.014 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Seelbinder B, Henderson JT, Watts RD, Scott AK, Veress AI, Neu CP. 2019. Deformation microscopy for dynamic intracellular and intranuclear mapping of mechanics with high spatiotemporal resolution. Cell Rep 27: 1607–1620.e4. 10.1016/j.celrep.2019.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse R, Vartiainen MK. 2013. To be or not to be assembled: progressing into nuclear actin filaments. Nat Rev Mol Cell Biol 14: 693–697. 10.1038/nrm3681 [DOI] [PubMed] [Google Scholar]
- Gudipaty SA, Lindblom J, Loftus PD, Redd MJ, Edes K, Davey CF, Krishnegowda V, Rosenblatt J. 2017. Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature 543: 118–121. 10.1038/nature21407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, De Klein A, Wessels L, De Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K. 2014. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol 16: 376–381. 10.1038/ncb2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Starr DA. 2019. SUN/KASH interactions facilitate force transmission across the nuclear envelope. Nucleus 10: 73–80. 10.1080/19491034.2019.1595313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PCDP, Ivanovska IL, Discher DE. 2014. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol 204: 669–682. 10.1083/jcb.201308029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson CM, Kern M, O'Brien ET, Stephens AD, Falvo MR, Superfine R. 2020. Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Mol Biol Cell 31: 1788–1801. 10.1091/mbc.E20-01-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Margadant FM, Yao M, Sheetz MP. 2017. Molecular stretching modulates mechanosensing pathways. Protein Sci 26: 1337–1351. 10.1002/pro.3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Williams JF, Kneissig M, Lioudyno M, Rivera I, Helguera P, Busciglio J, Storchova Z, King MC, Torres EM. 2019. Suppressing aneuploidy-associated phenotypes improves the fitness of trisomy 21 cells. Cell Rep 29: 2473–2488.e5. 10.1016/j.celrep.2019.10.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante E, Stannard A, Board SJ, Rico-Lastres P, Rostkova E, Beedle AEM, Lezamiz A, Wang YJ, Gulaidi Breen S, Panagaki F, et al. 2019. The mechanical stability of proteins regulates their translocation rate into the cell nucleus. Nat Phys 15: 973–981. 10.1038/s41567-019-0551-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Xia Y, Pfeifer CR, Athirasala A, Ji J, Alvey C, Tewari M, Bennett RR, Harding SM, Liu AJ, et al. 2017. DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr Biol 27: 210–223. 10.1016/j.cub.2016.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T, Wolfenson H, Sheetz MP. 2014. Appreciating force and shape—the rise of mechanotransduction in cell biology. Nat Rev Mol Cell Biol 15: 825–833. 10.1038/nrm3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itano N, Okamoto Si, Zhang D, Lipton SA, Ruoslahti E. 2003. Cell spreading controls endoplasmic and nuclear calcium: a physical gene regulation pathway from the cell surface to the nucleus. Proc Natl Acad Sci 100: 5181–5186. 10.1073/pnas.0531397100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Iyer KV, Kumar A, Shivashankar GV. 2013. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci 110: 11349–11354. 10.1073/pnas.1300801110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F. 2014. ESCRT machinery is required for plasma membrane repair. Science 343: 1247136. 10.1126/science.1247136 [DOI] [PubMed] [Google Scholar]
- Kim KD, Bae S, Capece T, Nedelkovska H, De Rubio RG, Smrcka AV, Jun CD, Jung W, Park B, Kim TI, et al. 2017. Targeted calcium influx boosts cytotoxic T lymphocyte function in the tumour microenvironment. Nat Commun 8: 1–10. 10.1038/ncomms15365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochin V, Shimi T, Torvaldson E, Adam SA, Goldman A, Pack CG, Melo-Cardenas J, Imanishi SY, Goldman RD, Eriksson JE. 2014. Interphase phosphorylation of lamin A. J Cell Sci 127: 2683–2696. 10.1242/jcs.141820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Cantwell H, Neumann FR, Jones AW, Snijders AP, Nurse P. 2017. A systematic genomic screen implicates nucleocytoplasmic transport and membrane growth in nuclear size control. PLoS Genet 13: e1006767. 10.1371/journal.pgen.1006767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MSY, Lisica A, Ramkumar N, Hunter G, Mao Y, Charras G, Baum B. 2020. Isotropic myosin-generated tissue tension is required for the dynamic orientation of the mitotic spindle. Mol Biol Cell 31: 1370–1379. 10.1091/mbc.E19-09-0545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J. 2011. Mechanics of the nucleus. Compr Physiol 1: 783–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. 2004. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest 113: 370–378. 10.1172/JCI200419670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le HQ, Ghatak S, Yeung CYC, Tellkamp F, Günschmann C, Dieterich C, Yeroslaviz A, Habermann B, Pombo A, Niessen CM, et al. 2016. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol 18: 864–875. 10.1038/ncb3387 [DOI] [PubMed] [Google Scholar]
- Le Beyec J, Xu R, Lee SY, Nelson CM, Rizki A, Alcaraz J, Bissell MJ. 2007. Cell shape regulates global histone acetylation in human mammary epithelial cells. Exp Cell Res 313: 3066–3075. 10.1016/j.yexcr.2007.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite MF, Thrower EC, Echevarria W, Koulen P, Hirata K, Bennett AM, Ehrlich BE, Nathanson MH. 2003. Nuclear and cytosolic calcium are regulated independently. Proc Natl Acad Sci 100: 2975–2980. 10.1073/pnas.0536590100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, et al. 2009. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139: 891–906. 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chu JS, Kurpinski K, Li X, Bautista DM, Yang L, Paul Sung KL, Li S. 2011. Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys J 100: 1902–1909. 10.1016/j.bpj.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lovett D, Zhang Q, Neelam S, Kuchibhotla RA, Zhu R, Gundersen GG, Lele TP, Dickinson RB. 2015. Moving cell boundaries drive nuclear shaping during cell spreading. Biophys J 109: 670–686. 10.1016/j.bpj.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ben-Shahar TR, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. 2000. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell 11: 3937–3947. 10.1091/mbc.11.11.3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin AJ, Cattin CJ, Cuvelier D, Alraies Z, Molina M, Nader GPF, Srivastava N, Sáez PJ, Garcia-Arcos JM, Zhitnyak IY, et al. 2020. The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science 370: eaba2894. 10.1126/science.aba2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. 2011. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem 286: 26743–26753. 10.1074/jbc.M111.233700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GG, Starr DA. 2014. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol 28: 69–75. 10.1016/j.ceb.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Ide S, Babokhov M. 2019. Dynamic chromatin organization without the 30-nm fiber. Curr Opin Cell Biol 58: 95–104. 10.1016/j.ceb.2019.02.003 [DOI] [PubMed] [Google Scholar]
- Marko JF. 2008. Micromechanical studies of mitotic chromosomes. Chromosom Res 16: 469–497. 10.1007/s10577-008-1233-7 [DOI] [PubMed] [Google Scholar]
- Miroshnikova YA, Mouw JK, Barnes JM, Pickup MW, Lakins JN, Kim Y, Lobo K, Persson AI, Reis GF, McKnight TR, et al. 2016. Tissue mechanics promote IDH1-dependent HIF1α–tenascin C feedback to regulate glioblastoma aggression. Nat Cell Biol 18: 1336–1345. 10.1038/ncb3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnikova YA, Le HQ, Schneider D, Thalheim T, Rübsam M, Bremicker N, Polleux J, Kamprad N, Tarantola M, Wang I, et al. 2018. Adhesion forces and cortical tension couple cell proliferation and differentiation to drive epidermal stratification. Nat Cell Biol 20: 69–80. 10.1038/s41556-017-0005-z [DOI] [PubMed] [Google Scholar]
- Miroshnikova YA, Hammesfahr T, Wickström SA. 2019. Cell biology and mechanopathology of laminopathic cardiomyopathies. J. Cell Biol 218: 393–394. 10.1083/jcb.201805079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Pasque V, Jullien J, Gurdon JB. 2011. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev 25: 946–958. 10.1101/gad.615211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy SE, Dubin AE, Patapoutian A. 2017. Piezos thrive under pressure: mechanically activated ion channels in health and disease. Nat Rev Mol Cell Biol 18: 771–783. 10.1038/nrm.2017.92 [DOI] [PubMed] [Google Scholar]
- Nader GP, Agüera-Gonzalez S, Routet F, Gratia M, Maurin M, Cancila V, Cadart C, Gentili M, Yamada A, Lodillinsky C, et al. 2020. Compromised nuclear envelope integrity drives tumor cell invasion. bioRxiv 10.1101/2020.05.22.110122 [DOI] [PubMed] [Google Scholar]
- Nava MM, Miroshnikova YA, Biggs LC, Whitefield DB, Metge F, Boucas J, Vihinen H, Jokitalo E, Li X, García Arcos JM, et al. 2020. Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell 181: 800–817.e22. 10.1016/j.cell.2020.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. 1999. Transport of torsional stress in DNA. Proc Natl Acad Sci 96: 14342–14347. 10.1073/pnas.96.25.14342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. 2015. ESCRT-III controls nuclear envelope reformation. Nature 522: 236–239. 10.1038/nature14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. 2007. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci 104: 15619–15624. 10.1073/pnas.0702576104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Burckhardt CJ, Lazcano R, Solis LM, Isogai T, Li L, Chen CS, Gao B, Minna JD, Bachoo R, et al. 2020. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature 578: 621–626. 10.1038/s41586-020-1998-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer CR, Xia Y, Zhu K, Liu D, Irianto J, Morales García VM, Santiago Millán LM, Niese B, Harding S, Deviri D, et al. 2018. Constricted migration increases DNA damage and independently represses cell cycle. Mol Biol Cell 29: 1948–1962. 10.1091/mbc.E18-02-0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer CR, Irianto J, Discher DE. 2019. Nuclear mechanics and cancer cell migration. Adv Exp Med Biol 1146: 117–130. 10.1007/978-3-030-17593-1_8 [DOI] [PubMed] [Google Scholar]
- Phengchat R, Takata H, Morii K, Inada N, Murakoshi H, Uchiyama S, Fukui K. 2016. Calcium ions function as a booster of chromosome condensation. Sci Rep 6: 1–10. 10.1038/srep38281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Dillon N. 2015. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 16: 245–257. 10.1038/nrm3965 [DOI] [PubMed] [Google Scholar]
- Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Duménil AM, Manel N, et al. 2016. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352: 359–362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- Renkawitz J, Kopf A, Stopp J, de Vries I, Driscoll MK, Merrin J, Hauschild R, Welf ES, Danuser G, Fiolka R, et al. 2019. Nuclear positioning facilitates amoeboid migration along the path of least resistance. Nature 568: 546–550. 10.1038/s41586-019-1087-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues MA, Gomes DA, Nathanson MH, Leite MF. 2009. Nuclear calcium signaling: a cell within a cell. Braz J Med Biol Res 42: 17–20. 10.1590/S0100-879X2008005000050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani P, Brian I, Santinon G, Pocaterra A, Audano M, Pedretti S, Mathieu S, Forcato M, Bicciato S, Manneville JB, et al. 2019. Extracellular matrix mechanical cues regulate lipid metabolism through Lipin-1 and SREBP. Nat Cell Biol 21: 338–347. 10.1038/s41556-018-0270-5 [DOI] [PubMed] [Google Scholar]
- Rothballer A, Kutay U. 2013. The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma 122: 415–429. 10.1007/s00412-013-0417-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht V, Wieser S, Callan-Jones A, Smutny M, Morita H, Sako K, Barone V, Ritsch-Marte M, Sixt M, Voituriez R, et al. 2015. Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell 160: 673–685. 10.1016/j.cell.2015.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber KH, Kennedy BK. 2013. When lamins go bad: nuclear structure and disease. Cell 152: 1365–1375. 10.1016/j.cell.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SGJ, King MC. 2015. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun 6: 7159. 10.1038/ncomms8159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Hobson CM, Cheng S, Colville MJ, Paszek MJ, Superfine R, Lammerding J. 2021. Nuclear deformation causes DNA damage by increasing replication stress. Curr Biol 31: 753–765.e6. 10.1016/j.cub.2020.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu JY, Aires L, Lin Z, Vogel V. 2018. Nanopillar force measurements reveal actin-cap-mediated YAP mechanotransduction. Nat Cell Biol 20: 262–271. 10.1038/s41556-017-0030-y [DOI] [PubMed] [Google Scholar]
- Sidorenko E, Vartiainen MK. 2019. Nucleoskeletal regulation of transcription: actin on MRTF. Exp Biol Med 244: 1372–1381. 10.1177/1535370219854669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnol ST, Dahl KN. 2016. Spatially resolved quantification of chromatin condensation through differential local rheology in cell nuclei fluorescence lifetime imaging. PLoS ONE 11: e0146244. 10.1371/journal.pone.0146244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer VA, Costes S, Inman JL, Xu R, Chen J, Hendzel MJ, Bissell MJ. 2011. Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J Cell Sci 124: 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN. 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 26: 421–444. 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF. 2017. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell 28: 1984–1996. 10.1091/mbc.e16-09-0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Banigan EJ, Almassalha LM, Backman Vadim, Adam SA, Goldman RD, Marko JF. 2018. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol Biol Cell 29: 220–233. 10.1091/mbc.E17-06-0410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Kandula V, Chen H, Almassalha LM, Herman C, Backman V, O'Halloran T, Adam SA, Goldman RD, et al. 2019. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol Biol Cell 30: 2320–2330. 10.1091/mbc.E19-05-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden H, Tolsma TO, Sharma A, Underhill DA, Hansen JC, Hendzel MJ. 2020. Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell 183: 1772–1784.e13. 10.1016/j.cell.2020.11.027 [DOI] [PubMed] [Google Scholar]
- Su X, Wellen KE, Rabinowitz JD. 2016. Metabolic control of methylation and acetylation. Curr Opin Chem Biol 30: 52–60. 10.1016/j.cbpa.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chen J, Mohagheghian E, Wang N. 2020. Force-induced gene up-regulation does not follow the weak power law but depends on H3K9 demethylation. Sci Adv 6: eaay9095. 10.1126/sciadv.aay9095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. 2013. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341: 1240104. 10.1126/science.1240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Bantignies F, Cavalli G. 2019. Principles of genome folding into topologically associating domains. Sci Adv 5: eaaw1668. 10.1126/sciadv.aaw1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N. 2016. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater 15: 1287–1296. 10.1038/nmat4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashunas AC, Tocco VJ, Matthews J, Zhang Q, Atanasova KR, Paschall L, Pathak S, Ratnayake R, Stephens AD, Luesch H, et al. 2020. High-throughput gene screen reveals modulators of nuclear shape. Mol Biol Cell 31: 1392–1402. 10.1091/mbc.E19-09-0520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A, Panciera T, Piccolo S. 2018. YAP/TAZ upstream signals and downstream responses. Nat Cell Biol 20: 888–899. 10.1038/s41556-018-0142-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y, Lee JSH, Kole TP, Jiang I, Wirtz D. 2004. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J Cell Sci 117: 2159–2167. 10.1242/jcs.01073 [DOI] [PubMed] [Google Scholar]
- Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, Medalia O. 2017. The molecular architecture of lamins in somatic cells. Nature 543: 261–264. 10.1038/nature21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H. 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522: 231–235. 10.1038/nature14408 [DOI] [PubMed] [Google Scholar]
- Vilborg A, Passarelli M, Steitz J. 2016. Calcium signaling and transcription: elongation, DoGs, and eRNAs. Recept Clin Investig 3: e1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining KH, Mooney DJ. 2017. Mechanical forces direct stem cell behaviour in development and regeneration. Nat Rev Mol Cell Biol 18: 728–742. 10.1038/nrm.2017.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales P, Schuberth CE, Aufschnaiter R, Fels J, García-Aguilar I, Janning A, Dlugos CP, Schäfer-Herte M, Klingner C, Wälte M, et al. 2016. Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. eLife 5: e19850. 10.7554/eLife.19850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sherrard A, Zhao B, Melak M, Trautwein J, Kleinschnitz EM, Tsopoulidis N, Fackler OT, Schwan C, Grosse R. 2019. GPCR-induced calcium transients trigger nuclear actin assembly for chromatin dynamics. Nat Commun 10: 5271. 10.1038/s41467-019-13322-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitefield DB, Spagnol ST, Armiger TJ, Lan L, Dahl KN. 2018. Quantifying site-specific chromatin mechanics and DNA damage response. Sci Rep 8: 18084. 10.1038/s41598-018-36343-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström SA, Niessen CM. 2018. Cell adhesion and mechanics as drivers of tissue organization and differentiation: local cues for large scale organization. Curr Opin Cell Biol 54: 89–97. 10.1016/j.ceb.2018.05.003 [DOI] [PubMed] [Google Scholar]
- Woroniuk A, Porter A, White G, Newman DT, Diamantopoulou Z, Waring T, Rooney C, Strathdee D, Marston DJ, Hahn KM, et al. 2018. STEF/TIAM2-mediated Rac1 activity at the nuclear envelope regulates the perinuclear actin cap. Nat Commun 9: 2124. 10.1038/s41467-018-04404-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidovska A, Weitz DA, Mitchison TJ. 2013. Micron-scale coherence in interphase chromatin dynamics. Proc Natl Acad Sci 110: 15555–15560. 10.1073/pnas.1220313110 [DOI] [PMC free article] [PubMed] [Google Scholar]