ABSTRACT
In order to assess the risk of SARS-CoV-2 infection, transmission and reservoir development in swine, we combined results of an experimental and two observational studies. First, intranasal and intratracheal challenge of eight pigs did not result in infection, based on clinical signs and PCR on swab and lung tissue samples. Two serum samples returned a low positive result in virus neutralization, in line with findings in other infection experiments in pigs. Next, a retrospective observational study was performed in the Netherlands in the spring of 2020. Serum samples (N =417) obtained at slaughter from 17 farms located in a region with a high human case incidence in the first wave of the pandemic. Samples were tested with protein micro array, plaque reduction neutralization test and receptor-binding-domain ELISA. None of the serum samples was positive in all three assays, although six samples from one farm returned a low positive result in PRNT (titers 40–80). Therefore we conclude that serological evidence for large scale transmission was not observed. Finally, an outbreak of respiratory disease in pigs on one farm, coinciding with recent exposure to SARS-CoV-2 infected animal caretakers, was investigated. Tonsil swabs and paired serum samples were tested. No evidence for infection with SARS-CoV-2 was found. In conclusion, Although in both the experimental and the observational study few samples returned low antibody titer results in PRNT infection with SARS-CoV-2 was not confirmed. It was concluded that sporadic infections in the field cannot be excluded, but large-scale SARS-CoV-2 transmission among pigs is unlikely.
KEYWORDS: Swine, Coronavirus, SARS-CoV-2, antibody, One Health
Knowledge on host species susceptibility and transmission of emerging pathogens is essential to assess the risk for reservoir development. SARS-CoV-2 can infect multiple animal species, which has become clear from both experimental [1,2] and observational studies [3]. Previous experimental infection studies in pigs (Appendix 1-1) reported no evidence for SARS-CoV-2 replication [1,2,4–7], with the exception of one study that reported the isolation of SARS-CoV-2 from a single lymph node 13 days post-infection (DPI) [4]. Neutralizing antibodies have been occasionally detected in pigs [4–6], possibly indicating infection. The validity of experimental transmission studies for transmission under field conditions can be limited. Therefore, additional observational studies need to be considered [4,8]. In this letter, we report the results of an experimental challenge study combined with a repeated cross-sectional serosurvey of pigs in the Netherlands in 2020 and an outbreak investigation of pigs with respiratory signs linked to two human SARS-CoV-2 cases. The aim was to investigate susceptibility, replication, and transmissibility of SARS-CoV-2 in pigs.
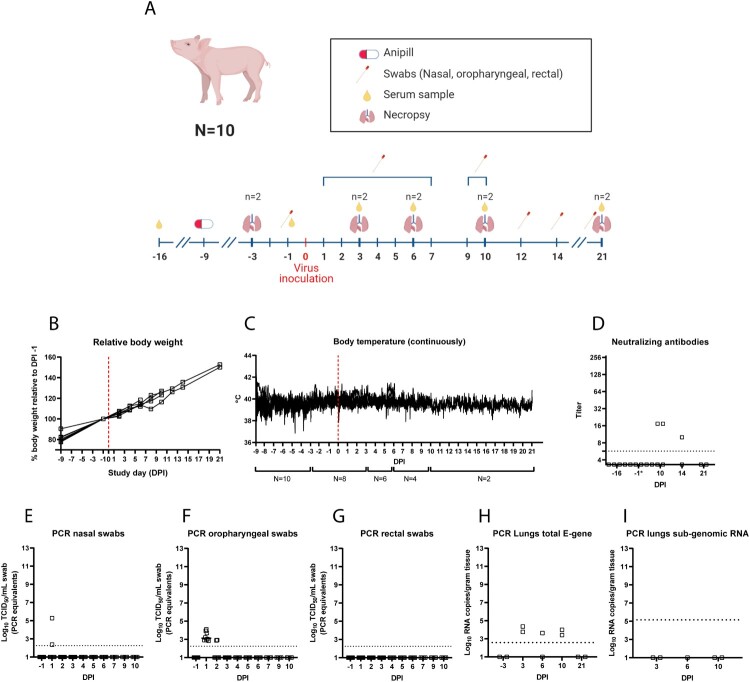
First, eight pigs were challenged with 106,2 TCID50 SARS-CoV-2 (SARS-CoV-2/human/NL/Lelystad/2020 (wild-type D614G), Genbank accession number MZ144583 [9]) both intranasally and intratracheally (Appendix 1-2, Figure 1A). Animals were sacrificed on DPI3 (n = 2), DPI6 (n = 2), DPI10 (n = 2) and DPI21 (n = 2). Clinical signs were not observed during the 21-day study period. Low levels of viral RNA were found in nasal swabs on DPI1 (n = 2; 2.4 and 5.3 Log10 TCID50/ml PCR equivalents), and in oropharyngeal swabs on DPI1 (n = 8; range 2.8–4.1 Log10 TCID50/ml PCR equivalents) and DPI2 (n = 2; 2.9 Log10 TCID50/ml PCR equivalents). Rectal swabs all tested negative. Low amount of RNA was found in lung samples (n = 2 on DPI3, n = 1 on DPI6 and n = 2 on DPI10), but no evidence for virus replication was found given the negative subgenomic RNA PCR results [10] (Figure 1I). Neutralizing antibodies were detected in two out of four pigs on DPI10 (Figure 1D). One of the two animals still had antibodies on DPI14, while the other was euthanized on DPI10.
Figure 1.
Study design and results for the SARS-CoV-2 pig challenge study. Challenge and sampling timeline (A), effect of SARS-CoV-2 challenge on pig body weight (B), and pig body temperature (C). Neutralizing antibodies (D) and (subgenomic) PCR results (E, F, G, H, I).
Second, a retrospective observational study was conducted in pigs reared in a region in the Netherlands both with a high incidence of human SARS-CoV-2 cases during the early phase of the epidemic (cumulative infection rate 318 cases/100,000 inhabitants on March 31st, 2020) (Appendix 1-4) as well as a high incidence of SARS-CoV-2 affected mink farms [3]. Twenty-one pig farms were selected (of which 17 participated), based on region and availability to collect blood samples during exsanguination at slaughter. Based on the assumption that potential transmission in a group of pigs would result in a final size of >50% infected pigs (expected value when R0 = 2 [11]), we aimed to obtain six blood samples of slaughter pigs at two timepoints per farm. Totally, 417 serum samples were collected from pigs of 17 farms between March and July 2020 (Appendix 1-4). A testing algorithm was designed and validated (Appendix 1-3), using a protein microarray (PMA) [12] with the SARS-CoV-2 S ectodomain antigen for screening, followed by a plaque reduction neutralization test (PRNT) and an receptor-binding domain (RBD)-ELISA to confirm positive results ([13], Appendices 1-3 and 1-4). A total of 29 sera, that showed reactivity in the PMA, were included in PRNT. In six out of 29 samples, derived from one farm at one timepoint, low titers were found in the PRNT (titer 40-80). None of these 29 samples tested above cut-off in the RBD-ELISA. Other serum samples of pigs from the same farm tested negative (n = 35) in PMA and PRNT, including serum samples obtained 2 months prior (n = 12) and 3 months after (n = 18) the timepoint with PRNT positive serum samples.
Finally, in February 2021 in another region in the Netherlands, two pig caretakers were confirmed SARS-CoV-2 positive. They had close contact with the pigs in the days before onset of symptoms, which coincided with an episode of nonspecific respiratory clinical signs in weaned pigs, rearing gilts and sows. Clinical signs were characterized by cough and hyperthermia. Tonsil swabs and paired serum samples (27 paired and 3 single sera) were collected from 30 pigs (18 exhibited respiratory signs), to detect a minimal prevalence of ∼10% with 95% confidence. All tonsil swabs tested negative for SARS-CoV-2 by E gene PCR [3,14]. In 5 out of 57 sera SARS-CoV-2 spike-binding binding antibodies were detected with PMA. These sera were negative in the PRNT and RBD-ELISA.
In conclusion, some pig sera of one farm had low neutralizing antibody titers in a virus neutralization assay, known to be very specific for human sera [13]. Low antibody titers were also seen in experimentally infected pigs (Appendix 1-2 and [5]). In the field, we neither found serological evidence for large-scale transmission among pigs from farms in a high-risk region, nor for human–pig transmission on a farm with a known outbreak among animal caretakers. Furthermore, no experimental evidence for viral replication in pigs was found, which is in line with the literature [1,2,4–7]. Therefore, we conclude that sporadic infections in the field cannot be excluded, but large-scale SARS-CoV-2 transmission among pigs in the field is unlikely. Neutralizing antibodies seen after experimental infection may be induced by the inoculum and cross-neutralization by porcine coronavirus infections needs to be investigated in more detail in future studies.
Serological surveys can be very useful to investigate previous exposure to SARS-CoV-2 in potentially susceptible animal species, as recommended by the Food and Agriculture Organization (FAO) (https://doi.org/10.4060/ca9959en). However, interpretation may sometimes be challenging, and it is essential that serological assays are appropriately validated, taking into account possible cross-reactivity due to the circulation of other known and unknown coronaviruses in many animal species. Research on SARS-CoV-2 variants and susceptibility of swine is not yet available, whereas with the emergence of new SARS-CoV-2 variants, also host range may change, as was seen in susceptibility of mice for some novel variants of concern [15]. Therefore, it is important to regularly monitor possible animal reservoirs. A One Health approach and vigilance of researchers, veterinarians and farmers are needed to detect and recognize future host species jumps.
Supplementary Material
Acknowledgements
The work described in this manuscript was funded by the Ministry of Agriculture, Nature and Food Safety of the Netherlands and by the Coalition for Epidemic Preparedness Innovations (CEPI). Positive control pig sera were obtained from the CBIG Consortium (constituted by IRTA-CReSA, BSC, & IrsiCaixa, ES) and supported by Grifols (ES). Hans Nauwynck (U-Ghent, BE) is acknowledged for providing PHEV serum.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Schlottau K, Rissmann M, Graaf A, et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. 2020 Sep;1(5):e218–e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi J, Wen Z, Zhong G, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020 May;368(6494):1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oreshkova N, Molenaar RJ, Vreman S, et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020 Jun;25(23):pii=2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickering BS, Smith G, Pinette MM, et al. Susceptibility of domestic swine to experimental infection with Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 2021 Jan;27(1):104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veragara-Alert J, Rodon J, Carrillo J, et al. Pigs are not susceptible to SARS-CoV-2 infection but are a model for viral immunogenicity studies. Transbound Emerg Dis. 2020;Oct:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meekins DA, Morozov I, Trujillo JD, et al. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckley A, Falkenberg S, Martins M, et al. Intravenous, intratracheal, and intranasal inoculation of swine with SARS-CoV-2. Viruses. 2021 Jul 30;13(8):1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerino P, Buonerba C, Brambilla G, et al. No detection of SARS-CoV-2 in animals exposed to infected keepers: results of a COVID-19 surveillance program. Future ScienceOA. 2021;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerhards NM, Cornelissen J, van Keulen LJM, et al. Predictive value of precision-cut lung slices for the susceptibility of three animal species for SARS-CoV-2 and validation in a refined Hamster model. Pathogens. 2021;10(7):824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. [DOI] [PubMed] [Google Scholar]
- 11.Kermack WO, McKendrick AG.. A contribution to the mathematical theory of epidemics. Proc R Soc Lond A. 1927;115(772):700–721. [Google Scholar]
- 12.Westerhuis BM, de Bruin E, Chandler FD, et al. Homologous and heterologous antibodies to coronavirus 229E, NL63, OC43, HKU1, SARS, MERS and SARS-CoV-2 antigens in an age stratified cross-sectional serosurvey in a large tertiary hospital in the Netherlands. medRxiv. 2020. doi: 10.1101/2020.08.21.20177857 [DOI] [Google Scholar]
- 13.Okba NMA, Müller MA, Li W, et al. Severe Acute Respiratory Syndrome Coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020 Jul;26(7):1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 Jan;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montagutelli X, Prot M, Levillayer L, et al. The B1.351 and P.1 variants extend SARS-CoV-2 host range to mice. bioRxiv. 2021:2021.03.18.436013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.