Abstract
Aconitum piepunense belonging to the family Ranunculaceae is an endangered herb species in southwestern China. In this study, the complete chloroplast genome of A. piepunense was sequenced, and the results revealed a typical quadripartite structure with a length of 155,836 bp, including a large single-copy region (LSC, 86,433 bp), a small single-copy region (SSC, 16,945 bp), and a pair of inverted repeat (IR) regions (IRa and IRb, 26,229 bp, respectively). A total of 130 genes were identified in the A. piepunense chloroplast genome, containing 85 protein-coding genes, 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. Phylogenetic analysis based on the maximum likelihood method indicated that A. piepunense formed a monophyletic group, which was sister to A. contortum and A. vilmorinianum.
Keywords: Aconitum piepunense, complete chloroplast genome, phylogenetic analysis
The genus Aconitum includes approximately 400 species that belong to the Ranunculaceae family (Park et al. 2017). Even though Aconitum possesses substantial toxicity, it is one of the most significant medicinal plants globally (Li and Kadota 2001). Modern pharmacological studies have shown that Aconitum has various pharmacological effects, especially analgesic and anti-inflammatory activities (Zhou et al. 2015). Aconitum piepunense is a critical medicinal plant, mainly distributed in tropical areas with an altitude of around 3000 m. At least sixteen active compounds of this species were discovered in the previous studies (Cai, Chen, Liu, et al. 2006; Cai, Chen, Liu 2006; Cai et al. 2010). Due to the apparent pharmacological activities, wild resources of A. piepunense are on the verge of extinction. Given this situation, to unravel the complete chloroplast genome of A. piepunense would pave the way for its molecular authentication and resource conservation.
Fresh leaves of A. piepunense were collected from the Alpine Botanical Garden, Zhongdian County, Yunnan Province, China (99°63′29″E, 27°91′62″N) and dried with silica gel. The plant specimen was deposited in the herbarium of Dali University, under the voucher number ZD201909. Total genomic DNA was extracted using a plant DNA kit (OMEGA, Shanghai, China). Approximately 1 ng of DNA was used to construct the library with a DNA sample preparation kit (Illumina Inc., San Diego, CA, USA). The library was sequenced on an Illumina NovaSeq platform at 2 × 150 bp using the Trusrq SBS Kit (Illumina Inc., San Diego, CA, USA). The obtained raw data were filtered using Trimmomatic v0.39 to ensure the accuracy of the following assembly results (Bolger et al. 2014). The chloroplast genome of A. piepunense was assembled using Novoplasty 4.0 (Dierckxsens et al. 2017), and the A. contortum chloroplast genome (NC038098) was used as the reference. To explore the phylogenetic relationship of A. piepunense, 26 complete chloroplast genomes were downloaded from Genbank (https://www.ncbi.nlm.nih.gov/). All chloroplast sequences were aligned using MAFFT v7 (Katoh and Standley 2013). The maximum likelihood (ML) method was adopted to construct a phylogenetic tree using MEGA X (Kumar et al. 2018) based on 1,000 replicates and the best-fit nucleotide model TVM + F + R4. The complete chloroplast genome sequence of A. piepunense was submitted to GenBank under the accession number MZ169393.
The chloroplast genome of A. piepunense was 155,836 bp in length and showed a typical quadripartite structure, including a large single-copy region (LSC, 86,433 bp), a small single-copy region (SSC, 16,945 bp), and a pair of inverted repeat (IR) regions (IRa and IRb, 26,229 bp, respectively). In addition, the coding regions (78,342 bp) accounted for 50.27%, while the intergenic regions (77,494 bp) accounted for 49.73%. The total GC content of this chloroplast genome was 37.8%. A total of 130 genes were annotated, containing 85 protein-coding genes (PCGs), 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes.
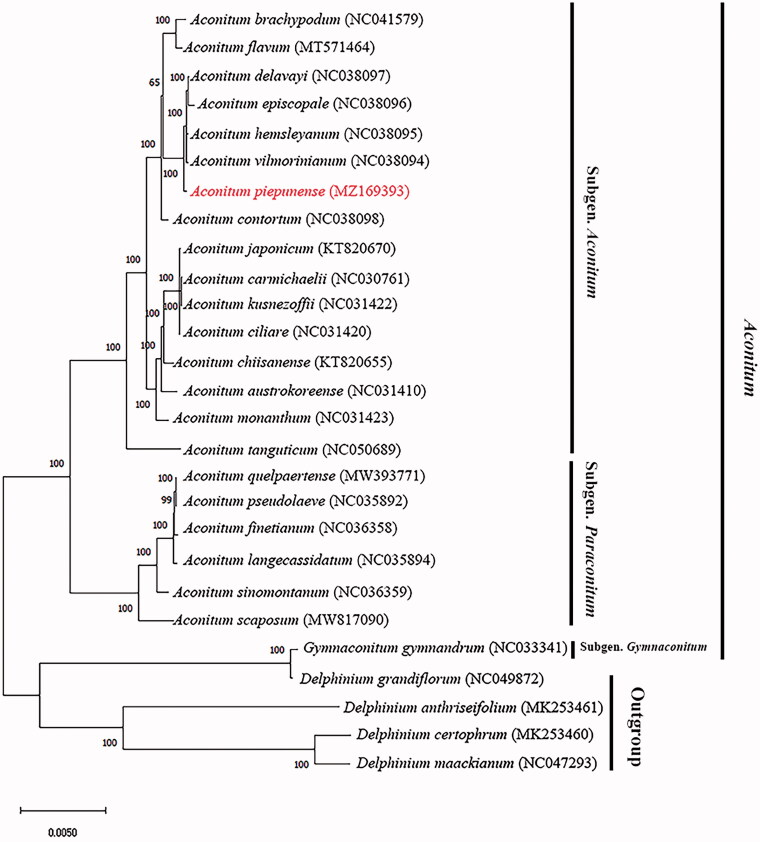
The ML tree showed that A. piepunense formed a monophyletic group that was sister to A. contortum and A. vilmorinianum; all the three species belonged to the subgenus Aconitum. It is worth mentioning that Gymnaconitum gymnandrum clustered into a clade with Delphinium grandiflorum, and sistered to the other three Delphinium species (Figure 1). The ML tree revealed a deep-level phylogenetic relationship of A. piepunense. These results contribute to the phylogeny and molecular authentication of this traditional medicinal plant.
Figure 1.
Phylogenetic relationships of the 27 species inferred from maximum likelihood (ML). The bootstrap value shown on each node was obtained based on 1000 replicates.
Funding Statement
This work was supported by the Major Science and Technology Project in Dali Prefecture [D2019NA03].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession MZ169393. The associated BioProject, SRA, and Bio-Sample ID are PRJNA748565, SRR15206984, and SAMN20338565, respectively.
References
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Chen DL, Liu SY, Wang FP.. 2006. New C19-diterpenoid alkaloids from Aconitum piepunense. Chem Pharm Bull. 54(6):779–781. [DOI] [PubMed] [Google Scholar]
- Cai L, Chen D, Peng F.. 2006. Two new C18-diterpenoid alkaloids from Aconitum piepunense. Nat Prod Commun. 1(3):191–194. [Google Scholar]
- Cai L, Song L, Chen QH, Liu XY, Wang FP.. 2010. Piepunine, a novel bis‐diterpenoid alkaloid from the roots of Aconitum piepunense. HCA. 93(11):2251–5255. [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LQ, Kadota Y.. 2001. Aconitum. Beijing: Science Press. [Google Scholar]
- Park I, Kim W, Yang S, Yeo S, Li H, Moon BC.. 2017. The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLOS One. 12(9):e184257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Tang L, Zhou X, Wang T, Kou Z, Wang Z.. 2015. A review on phytochemistry and pharmacological activities of the processed lateral root of Aconitum carmichaelii Debeaux. J Ethnopharmacol. 160:173–193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession MZ169393. The associated BioProject, SRA, and Bio-Sample ID are PRJNA748565, SRR15206984, and SAMN20338565, respectively.