Abstract
Unrepaired DNA double-strand breaks (DSBs) typically result in G2 arrest. Cell cycle progression can resume following repair of the DSBs or through adaptation to the checkpoint, even if the damage remains unrepaired. We developed a screen for factors in the yeast Saccharomyces cerevisiae that affect checkpoint control and/or viability in response to a single, unrepairable DSB that is induced by HO endonuclease in a dispensable yeast artificial chromosome containing human DNA. SIR2, -3, or -4 mutants exhibit a prolonged, RAD9-dependent G2 arrest in response to the unrepairable DSB followed by a slow adaptation to the persistent break, leading to division and rearrest in the next G2. There are a small number of additional cycles before permanent arrest as microcolonies. Thus, SIR genes, which repress silent mating type gene expression, are required for the adaptation and the prevention of indirect lethality resulting from an unrepairable DSB in nonessential DNA. Rapid adaptation to the G2 checkpoint and high viability were restored in sir− strains containing additional deletions of the silent mating type loci HML and HMR, suggesting that genes under mating type control can reduce the toleration of a single DSB. However, coexpression of MATa1 and MATα2 in Sir+ haploid cells did not lead to lethality from the HO-induced DSB, suggesting that toleration of an unrepaired DSB requires more than one Sir+ function.
In the yeast Saccharomyces cerevisiae, chromosomal double-strand breaks (DSBs) are predominantly repaired by a recombinational repair pathway involving interaction between the broken molecule and a homologous chromosome or sister chromatid. This repair pathway is largely dependent on the RAD52 epistasis group gene products (56, 61). To a lesser extent and in the absence of a homologous template, yeast can repair plasmid or chromosomal DSBs using a nonhomologous, end-joining (NHEJ) repair pathway that depends on YKU70, YKU80, and the MRE11-RAD50-XRS2 complex as well as DNL4 and LIF1 (summarized in references 40, 41, 76, 79, and 80).
The products of the SIR genes have also been proposed to function in DSB repair and NHEJ, but their role is unclear. The Sir2, Sir3, and Sir4 proteins form a complex that has a direct role in producing transcriptionally repressed chromatin structures at both telomeres and the silent mating type loci HML and HMR. Recent studies using immunolocalization and immunoprecipitation have suggested that DSB damage leads to the dissociation of Sir and Ku proteins from telomeres and relocation in complexes at the DSB site (48, 49, 50). Furthermore, the enhanced sensitivity to γ-ray or methyl methanesulfonate (MMS) damage in the absence of Sir4 supports a direct role for this protein in the toleration of DSBs (48, 80, 81).
Although some studies using plasmid end-joining assays have reported a direct effect of the Sir4 gene product on NHEJ repair (29, 79, 80), other studies have shown little or no direct effect of the Sir gene products on NHEJ (5, 39). The latter studies suggest that, in some strains, NHEJ repair is inhibited to similar extents in sir− a1/α2 haploids and diploids, in which the transcriptional regulators a1 and α2 are expressed within both MATa and MATα (5, 73). In sir− cells, expression of these regulators from within the silent mating loci HML and HMR determines the diploid mating type and confers an a1/α2 haploid phenotype. Furthermore, enhanced resistance to the killing effects of ionizing radiation is observed in MATa/MATα heterozygous diploids compared to the killing effects in either MATa/MATa or MATα/MATα homozygous diploids (24, 43). These results indicate that a gene(s) under MAT regulation influences both NHEJ and recombinational repair pathways in yeast.
Both haploid Rad+ cells in the G1 phase of the cell cycle as well as diploid rad52 cells are sensitive to ionizing radiation, and it appears that approximately one unrepaired DSB is sufficient to kill these cells (62). It has been assumed that death results from the direct effects of an unrepaired DSB, namely, the loss of essential genetic material. However, in a previous study (25), one or a small number of unrepaired DSBs induced by ionizing radiation caused killing in polyploid rad52 strains, suggesting that lethality need not be due simply to the direct effects of an unrepaired DSB.
Unrepaired lesions, particularly DSBs, can have a severe impact on cells, and this has led to studies examining their effects on cell cycle progression and viability. The underlying mechanisms appear to be relevant to many disease processes (20, 83). DNA damage, including unrepaired DSBs, can lead to a transient arrest of cells in the G1, S (2, 70), and G2 (22, 84, 85, 86) phases of the cell cycle. This arrest involves many genes that act in multiple pathways in the budding yeast Saccharomyces cerevisiae (2, 26, 69, 86). The RAD9, RAD17, RAD24, MEC3, and SFP1 genes are required for arrest in the G2 phase of the cell cycle (22, 26, 44, 85, 86, 89). Specific kinases, including MEC1 (rad3+ of Schizosaccharomyces pombe) and MEC2 (SAD1 or RAD53), are required for arrest in S and G2 phases (30, 75, 86). RAD9 and RAD24 genes are also required for arrest in the G1 phase of the cell cycle (70). The RAD9, RAD17, RAD24, and DUN2 genes are also required for arrest in the S phase of the cell cycle following DNA damage (54, 58). These checkpoint control genes appear to function in a complex network to assess the integrity of chromosomal DNA and delay cell progression after damage, thereby increasing the opportunity for DNA repair.
While a DSB produced in asynchronously growing cells leads to G2 arrest, cells can eventually proceed past the G2 checkpoint and undergo mitosis even if the damage is not repaired. This toleration mechanism has been termed adaptation (20, 38, 58, 78, 83) and has been observed in cells that experienced a single unrepairable DSB in a dispensable plasmid, chromosome, or artificial chromosome derived from lambda DNA or human DNA (60, 65, 67). Both CDC5 and CKB2 have been shown to be important for adaptation in a rad52 mutant (78). A single enzymatically induced DSB in a dispensable disomic chromosome that remained unrepaired, due to the absence of the RAD52 recombinational repair pathway, caused permanent G2 arrest in strains defective in either of these genes (78). These results must be interpreted with regard to other effects that RAD52 mutations might have on DNA metabolism (6), including cell cycle arrest and subsequent growth inhibition. Recently, it was shown that the long G2 arrest induced in yku70 mutants by a DSB in an essential chromosome can be suppressed by rad50 or mre11 deletions or by a mutation in the single-strand binding protein RPA (38).
We have taken an alternative approach to identifying genes that influence cell cycle progression in response to unrepaired DSBs. Previously, we had shown that, in Rad+ cells, the presence of a single DSB, which is not subject to recombinational repair, could lead to cell death (7, 8, 9). Induction of a single unrepairable DSB by HO endonuclease at a YZ target site in a dispensable plasmid or yeast artificial chromosome (YAC) caused prolonged but transient G2 arrest. Subsequently, many of the cells slowly adapted to the persistent damage, resumed cycling, and formed microcolonies (<50 cells) which did not progress further. Since the DNA containing the DSB was dispensable (7, 8, 9), this process was called indirect lethality because the cells died by a mechanism(s) that does not involve direct loss of essential chromosomal material. While a persistent DSB in dispensable DNA always results in G2 arrest, in some strain backgrounds such as LS20 (60, 65, 67) there was no reduced viability from the unrepaired DSB. This finding indicates that there may be genetic differences in the types of adaptation to DSB damage. The CDC5 and CKB2 adaptation genes were identified in a repair-deficient rad52 version of the LS20 strain (78).
We initiated a screen utilizing our DSB break system in a repair-proficient LS20 strain to identify adaptation mutants that exhibit extended G2 delay and indirect lethality. We found that the SIR genes play an important and indirect role in the adaptation response to persistent DSB damage. Sir+ cells adapted quickly to a persistent DSB and rapidly formed viable microcolonies after G2 arrest. The single unrepaired DSB led to a more prolonged G2 arrest in cells that are deficient for SIR2, SIR3, or SIR4. In contrast to the adaptation mutants described previously (cdc5 and ckb2 mutants), these sirΔ Rad+ mutant cells eventually adapted to the damage and slowly divided. They produced mainly microcolonies that did not proceed further, resulting in clonal death. Toleration of the persistent YAC DSB does not appear to be due to a direct effect of SIR gene products on the NHEJ repair functions at the DSB site since the responses were not affected by RAD50. Checkpoint adaptation and viability were restored in sir2Δ, sir3Δ, and sir4Δ strains with the silent mating type loci HML and HMR deleted, but viability was not reduced in Sir+ cells that express the a1 and α2 transcriptional regulators, which confer an a1/α2 haploid phenotype. Therefore, there are at least two functions of the SIR genes that determine the biological effects of a persistent, HO-induced DSB. One role is indirect in that SIR is required for silencing HMR and HML, which can regulate a gene(s) that is involved in checkpoint adaptation and survival following induction of a persistent DSB. However, this effect can occur only in the absence of SIR4, suggesting that SIR can have an additional role, possible through direct interactions at the break site not involving end joining.
MATERIALS AND METHODS
Strains, plasmids, and YACs.
The haploid S. cerevisiae strain LS20 (Table 1) (65) was transformed (66) with the selectable (URA3) low-copy-number plasmid (YZ-CEN) that contains a 45-bp HO endonuclease target sequence (YZ) flanked by nonhomologous DNA sequences (7). In strain LS20 the HO endonuclease was fused to the GAL promoter and integrated at the ade3 locus (65, 67). Strain LS20 was also spheroplasted and transformed with the u8 and u17 YACs, which contain human DNA (8, 9), by methods previously described (36). The u8 YAC is a 365-kb derivative of YAC12 with the YZ site (and URA3 marker) positioned 70 kb from the telomere within the human DNA. The u17 YAC is a derivative of YAC12 with a 230-kb terminal deletion and with the YZ site (and URA3 marker) positioned 5 kb from the telomere within the human DNA. These YACs are hereinafter collectively termed YZ YACs. Strain LS20 containing the VS8 lambda DNA-based YAC has also been described previously (67). In strains CBY and NR85 (Table 1) the GAL::HO fusion was on the centromere plasmid pGALHOT, as previously described (7, 8, 9). Strain NR85 was transformed with the VS8 YAC by methods previously described (36). Strains CBY and NR85 are both Sir+. A series of isogenic strains were constructed using strain LS20 containing the u8 YAC (see below). These strains and their relevant genotypes have been listed in Table 1.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Reference(s) or Source |
|---|---|---|
| LS20 | GAL lys5 ura3,52 leu2 his3 trp1 matΔ | 61 |
| NR85 | GAL ho HMLα leu2 mat::LEU2 hmr-Δ3 mal2 ura3-52 thr4 trp1 | 7 |
| CBY | GAL MATa ura3-52 trp1Δ1 lys2-801 his3Δ200 ade2-101 RAD52 | 8, 9 |
| gal MATα ura3-52 trp1Δ63 lys2-Δ202 his3Δ200 ade2-1 (oc) rad52::LEU2 | ||
| BY4741 | GAL MATa his3Δ1 leu2Δ met15Δ ura3Δ | K. Lewis |
| BY4742 | GAL MATα his3Δ1 leu2Δ lys2Δ ura3Δ | K. Lewis |
| CBY09 | GAL lys5 ura3 leu2 his3 trp1 matΔ hml::G418rhmr::TRP1a | This study |
| CBY10 | GAL lys5 ura3 leu2 his3 trp1 matΔ hml::G418rhmr::TRP1 sir2Δ::LEU2 | This study |
| CBY11 | GAL lys5 ura3 leu2 his3 trp1 matΔ hml::G418rhmr::TRP1 sir3Δ::LEU2 | This study |
| CBY12 | GAL lys5 ura3 leu2 his3 trp1 matΔ hml::G418rhmr::TRP1 sir4Δ::LEU2 | This study |
| CBY13 | GAL lys5 ura3 leu2 his3 trp1 matΔ hml::G418rhmr::TRP1 rad50Δ::LEU2 | This study |
| CBY14 | GAL lys5 ura3 leu2 his3 trp1 matΔ hml::G418rhmr::TRP1 sir4Δ::LEU2 rad50Δ::HygBr | This study |
| CBY15 | GAL lys5 ura3 leu2 his3 trp1 matΔ hml::G418rhmr::TRP1 rad9Δ::HygBr | This study |
| CBY16 | GAL lys5 ura3 leu2 his3 trp1 matΔ hml::G418rhmr::TRP1 rad9Δ::HygBrsir4Δ::LEU2 | This study |
| CBY17 | GAL lys5 ura3 leu2 his3 trp1 matΔ chr.III::[hmlΔ hmrΔ]b | This study |
| CBY18 | GAL lys5 ura3 leu2 his3 trp1 matΔ chr.III::[hmlΔ hmrΔ] sir2Δ::LEU2 | This study |
| CBY19 | GAL lys5 ura3 leu2 his3 trp1 matΔ chr.III::[hmlΔ hmrΔ] sir3Δ::LEU2 | This study |
| CBY20 | GAL lys5 ura3 leu2 his3 trp1 matΔ chr.III::[hmlΔ hmrΔ] sir4Δ::LEU2 | This study |
| CBY21 | GAL lys5 ura3 leu2 his3 trp1 matΔ chr.III::[hmlΔ hmrΔ] rad50Δ::HygBr | This study |
| CBY22 | GAL lys5 ura3 leu2 his3 trp1 matΔ chr.III::[hmlΔ hmrΔ] sir4Δ::LEU2 rad50Δ::HygBr | This study |
Precise deletions of the HO-cut sites were made within HML and HMR such that nonsilenced strains (sir−) retained the ability to express the regulatory proteins a1 and α2 and confer a nonmating, a1/α2 haploid phenotype. These strains have been designated HM+ in the text.
Chromosome III (chr.III) was circularized using a one-step targeted deletion of HML and HMR using G418r as the selectable marker (see Materials and Methods). These strains did not retain a1 or α2 and were unable to confer a nonmating haploid phenotype following the loss of silencing. They have been designated hm− in the text.
A plasmid, pCB115, that contained both of the transcriptional regulators MATa1 and MATα2 under the control of their native promoters was created. They were cloned into the centromeric, LEU2-based plasmid pRS315. PCR amplification of MATa1 from plasmid p54-45 (containing the HMRa allele) and MATα2 from a second plasmid, LCIII (containing the MATα allele), utilized the following primer sets designed with PstI (MATa1) and BamHI (MATα2) restriction sites at the termini: a1Pst-L, 5′-AAAATTTTACT GCAGCGGAAA GCTGAAACTA AAAGAAAAAC CCGACTATGC-3′; a1Pst-R, 5′-CATTTCTTTC TGCAGTACAG TAATGGTAGT AGTGAGTTGA GATGTTGTTT GC-3′; alpha2Bam-L, 5′-CTTCGAAGTG GATCCATTGT GAATGTCGTC TATTAAGTTA TTATATATGG TTGATAC-3′; and alpha2Bam-R, 5′-CATATTTGTG GATCCGGCTC ATTCTTTCTT CTTTGCCAGA GGCTCACCGC TCAAGAGGTC CGC-3′. Following PCR amplification, the products were digested with the appropriate enzyme and sequentially inserted in opposite orientations at the BamHI and PstI sites within pRS315. Transcriptional activities of the cloned inserts were determined by the introduction of plasmids pCB115 and pRS315 into the haploid strains BY4741, BY4742, and CBY09. The presence of pCB115 rendered the haploid strains nonmaters, whereas strains containing pRS315 retained their respective haploid mating types.
Survival.
To determine the effects of a HO-induced DSB, cells were grown to log phase in synthetic complete (SC)-glucose (GLU) medium (88) with selection for the individual YACs or plasmids. Selection for the pGALHOT TRP1 plasmid in strains CBY and NR85 was maintained throughout the experiment. (The differences in a strain's ability to express indirect lethality [CBY and NR85 versus LS20] was not due to selection for the pGALHOT plasmid in CBY and NR85, since LS20 containing pGALHOT and YZ-CEN exhibited high levels of survival on galactose (GAL) when selection for pGALHOT was maintained [data not shown].) Growth in GLU medium repressed HO expression, and the YZ vectors remained intact. Cells were then washed in sugarless SC medium and plated to SC-GLU and SC-GAL plates. Survival was determined by a strain's colony-forming ability on GAL compared to that on GLU plates after 72 h of incubation at 30°C. No selection was placed on the YZ YACs (8, 9) or the YZ-CEN (7) plasmid so that they would be dispensable following HO induction of a DSB at the YZ target site. HO-induced cutting at the YZ site was an efficient process since most of the surviving colonies (>95%) did not retain the YAC or plasmid URA3 marker, as determined by replica plating (data not shown).
To determine radiation-induced killing, cells cured of the u8 YAC were grown to stationary phase in liquid yeast extract-peptone-dextrose (YEPD) medium. G1 (unbudded) cells were visually identified using a microscope, and their overall percentage was determined. The cells were washed and irradiated in water using a 137Cs irradiator (1.25 krad/min). They were then plated to YEPD plates, and survival relative to that of the unirradiated control was determined. The dose-modifying factor corresponds to the increase in dose required to achieve a comparable level of survival (in the exponential survival range). This is an indicator of the relative amount of increased damage that can be tolerated.
To determine MMS-induced killing, cells that had been cured of the u8 YAC were grown to logarithmic phase in SC-GLU medium and MMS (Boehringer) was added to the growing cultures at a concentration of either 0.01 or 0.03%. Cells were incubated at 30°C with vigorous shaking. At various times, cells were washed in water and plated to YEPD to determine survival.
Commitment to death and kinetics of marker loss following induction of a DSB.
To determine the times at which the URA3 marker was lost and cells were committed to death, cells were grown and washed as described above. In the pullback experiments, cells were resuspended in SC medium-uracil (URA)-sugar and aliquots of between 200 and 300 μl were delivered to plastic petri dishes. Cells were imbedded in agar by adding 15 ml of liquid SC medium plus 2% GAL with or without URA agar medium (45°C). After various periods of incubation at 30°C, we added a 15-ml overlay containing 2% agar and SC plus 4% GLU with or without URA. The diffusion of GLU into the bottom layer rapidly repressed the expression of HO endonuclease.
Library screening.
The LS20 strain containing the selectable (URA3) YZ-CEN plasmid was transformed with a high-copy-number, selectable (LEU2) yeast genomic library (ATCC 37323 [53]), and 3,100 transformants were examined. Since this library contained genomic fragments ranging from 5 to 20 kb in length, this screen corresponded to coverage of the yeast genome one to two times. Library transformants were grown overnight in 96-chambered multiwell dishes in 200 μl of SC-GLU medium lacking URA and leucine. Approximately 2 μl from each well was placed with a 48-pin pronged device on either SC-GLU or SC-GAL plates lacking leucine to maintain selection for the library plasmids. Candidate factors were identified by their ability to cause slow growth (and possibly lethality) on GAL following induction of a DSB in the YZ-CEN plasmid.
Precise deletion of the YZ sites within HML and HMR.
The YZ sites within the HMR and HML loci of LS20 strains were deleted using a two-step procedure. HMR was first deleted using a PCR-mediated disruption procedure. The primers HMR-Z (5′-GAAAGATAAA CAACCTCCGC CACGACCACA CTCTATAAGG CCAAATGTAC AAACACATCT TCCCAAATATC CAGAGCAGAT TGTACTGAGA GTGCACC-3′) and HMR-Y (5′-GAGTTTGGGT ATGTAATATG AGAATCAAAC TTAAATATAT CCTATACTAA CAATTTGTAG TTCATAAATA CGCATCTGTG CGGTATTTCA CACCGC-3′) were obtained from Bioserve Biotechnologies and contain sequences that closely flank the YZ site within the Y and Z domains of HMR as well as sequences (italics) homologous to plasmid pRS314 (71) that flank the TRP1 marker contained within that plasmid. Following PCR amplification of TRP1 within pRS314, the LS20 or LS20 sir4Δ strains containing u8 were transformed with the PCR amplification product. Deletion of the YZ junction within HMR was confirmed by Southern blotting (data not shown). The YZ junction within HML was then disrupted. The primers HML-Z3G (5′-CTTCCCAATA TCCGTCACCA CGTACTTCAG CATAATTATT CGTCAACCAC TCTACAAAAC CAAAACCAGGG CGTACGCTGCCAGGTCGAC-3′) and HML-Y3G (5′-CACAGTTTGG CTCCGGTGTA AAACAAAATG TCTTGTCTTC TCTGCTCGCT GAAGAATGGC ACGCGGACAA ATCGATGAAT TCGAGCTCG-3′) were also obtained from Bioserve Biotechnologies and contain sequences flanking the YZ junction within the Y and Z domains of HML as well as sequences (italics) that flank the G418 marker within plasmid pFA6a (82). Following PCR amplification of the G418-resistant (G418r) marker in plasmid pFA6a, strains containing the u8 YAC and the deleted YZ site within HMR were transformed with the PCR product. Transformants were selected on plates containing G418 as described previously (82), and disruption of HML was confirmed by Southern blot analysis (data not shown).
One-step deletion of HML and HMR through targeted PCR-mediated circularization of chromosome III.
The LS20 strain containing the u8 YAC was completely deleted for the HML and HMR loci using a one-step, PCR-mediated targeted deletion that resulted in circularization of chromosome III. To do this, primers derived from sequences obtained using the Saccharomyces Genome Database (Stanford Genomic Resources) were obtained from Bioserve Biotechnologies. Since the HM loci and sequences distal to them are dispensable for growth, one primer (Omega3 [5′-CCTCGCACTA TCGCTGTTAT ACATGATGTC CCCAAAGCGT GTACAAATAA TTTTGTAGTA TTGTATCGGT AATATCATACA CAGAGCAGAT TGTACTGAGA GTGCACC-3′]) contained yeast sequences (nonitalicized) between HMR and the next open reading frame (ORF) (omega 3) proximal to the centromere. Another primer (CHA1 [5′-GTAAGCATCA ACATATCCAA AACGTTGACA TATTTCTAGG CCGGCAATGC ACAGAATTTG TATAAAGGGG GACATGCTGCAG CGCATCTGTG CGGTATTTCA CACCGC-3′]) contained yeast sequences (nonitalicized) between HML and the next ORF (CHA1) proximal to the centromere. Both primers also contained sequences (italics) that flanked the G418r marker in plasmid pFA6a. Following PCR amplification of the G418r marker from plasmid pFA6a, the LS20 strain was transformed with the PCR product and G418-resistant colonies were selected as described above. Circularization of chromosome III and retention of the u8 YAC were confirmed by transverse alternating field electrophoresis (TAFE) gel analysis as previously described (8, 9). Since circular chromosomes or YACs are unable to enter TAFE gels, the absence of chromosome III from the normal migration position of ∼360 kb confirmed circularization in the G418-resistant colonies.
Genomic deletions of SIR2, SIR3, and SIR4
The SIR2, SIR3, and SIR4 loci were deleted in two LS20-derived strains containing the u8 YAC. These two LS20 strains lacked the genomic HO-cut sites either by sequential deletion of the YZ sites within HML and HMR or through a single-step-PCR-mediated complete deletion of HML and HMR that resulted in the circularization of chromosome III (see above). SIR2, SIR3, and SIR4 were deleted using the disruption plasmids pJH103.1, pDP91 (derived from pJH107.1), and pDM610.23, respectively (28). Plasmids pJH103.1 and pDP91were digested with the restriction endonucleases HindIII and EcoRI, respectively. Plasmid pDM610.23 was digested with the restriction endonucleases PvuII and PspAI. Restriction endonuclease-digested disruption plasmids were reintroduced into the appropriate host strains by transformation (66), and colonies were selected on leucine-deficient medium. Deletion of SIR2, SIR3, and SIR4 resulted in a nonmating phenotype in the LS20 strain that still retained the a1 and α2 transcription regulators within the HM loci (those deleted sequentially at each YZ junction). Deletions in this strain were confirmed by mating to the appropriate MATa and MATα tester strains as well as by Southern blot analysis (data not shown). Deletions of SIR2, SIR3, and SIR4 loci in the LS20 strain containing circular chromosome III were confirmed by Southern blot analysis alone.
Genomic deletions of RAD9 and RAD50.
The RAD9 locus was deleted in a Sir+ strain using a one-step-PCR-mediated procedure (see above). Subsequently, SIR4 deletions were made in the rad9Δ strain using plasmid pDM610.23 as described above. The RAD50 locus was also deleted from Sir+ and sir4Δ strains using a one-step-PCR-mediated procedure. The following primers contain yeast sequences that flank the RAD9 and RAD50 loci, as well as sequences (italicized) that flank the hygromycin resistance gene within plasmid pAG32 (19): RAD9-LG (5′-CGTGGATATT TGCAACGATG AGCAATGTGA AGTGACCAAGATA GAGAAACGCC ATGTGACTGT CGCCCGTACA TT-3′), RAD9-RG (5′-CCAATCTTGA ACATTAACCA CTCCTGGCGT GTGGGAGGAT GTTCTTAGAC T GACAAGTTCT TGAAAACAAG AATC-3′), gr50c (5′-GCATGAGCGC TATCTATAAA TTATCTATTC AGGGCATACG GTCTT CGTACGCTG CAGGTCGAC-3′), and gr50d (5′-CGCAGTCTTA TAGGAGAGCT CCGTTTCTTC CAGGACATCA TTATA ATCGATGAAT TCGAGCTCG-3′).
Following PCR-mediated amplification of the hygromycin resistance gene, the appropriate strains were transformed with ∼1 to ∼2 μg of the amplified DNA as described above and transformants were selected on hygromycin-YEPD plates (19). Hygromycin-resistant colonies that had retained the u8 YAC were screened for radiation sensitivity by growing putative isolates in SC-GLU-URA overnight at 30°C in multiwell dishes and replica plating them with a 48-pin pronged device on SC-GLU-URA and YEPD plates. These cells were immediately UV irradiated (60 J/m2; for putative rad9 deletions only) or γ irradiated (40 krad). Irradiated isolates that contained RAD9 or RAD50 deletions demonstrated greatly reduced colony-forming ability after 48 h of growth compared to that of Rad+ controls.
RESULTS
Multiple copies of the silent mating locus HMR enhance indirect lethality resulting from a DSB.
To identify genetic factors that influence the cellular response to a DSB, we screened a high-copy-number yeast genomic library for factors that would reduce the growth of an LS20 strain following induction of a single, HO-induced, unrepairable DSB at a YZ junction in a centromere-based plasmid (YZ-CEN). We chose this strain background because previously it had been shown that the in vivo induction of a nonrepairable DSB at a YZ site in a dispensable lambda DNA-based YAC (VS8) or chromosome did not lead to lethality (65, 67). However, a persistent DSB in YZ-CEN or a YZ YAC could lead to prolonged cell cycle delay at G2 and indirect lethality in other strain backgrounds (CBY and NR85 [7, 8, 9]). We first established that the same persistent HO-induced DSB in the YZ-CEN plasmid or the YZ YACs did not cause lethality in the LS20 strain (Table 2). Contrary to results with the CBY and NR85 strains, the unrepairable break did not reduce the viability of LS20, indicating that the strain background may markedly affect the cellular response to an unrepaired DSB.
TABLE 2.
Indirect lethality resulting from a single persistent DSB in cells of various strain backgrounds
| Strain | YZ-containing vectora | Survivalb |
|---|---|---|
| LS20c | u8 YAC (human) | 0.95 ± 0.17 (21) |
| u17 YAC (human) | 0.89 ± 0.22 (9) | |
| VS8 YAC (lambda) | 0.98 ± 0.20 (4) | |
| YZ-CEN (plasmid) | 1.02 ± 0.22 (4) | |
| CBYd | u8 | 0.19 ± 0.07 (8) |
| u17 | 0.18 ± 0.09 (7) | |
| NR85e | VS8 | 0.12 ± 0.09 (14) |
| YZ-CEN | 0.15 ± 0.09 (12) |
The vectors are efficiently cut in vivo by the induced HO endonuclease at the YZ site in the vector; almost all surviving colonies lose the vector (8, 65, 67).
Cells were plated to GAL-containing SC medium (to induce the HO endonuclease) and GLU-containing SC medium, which does not select for the YAC or the plasmid. Survival corresponds to relative colony-forming ability on GAL medium versus that on GLU medium. Error values are ±1 standard deviation about the mean. Numbers in parentheses are numbers of experiments.
A persistent DSB induced in various dispensable vectors in the LS20 strain does not affect survival, whereas the DSB decreased survival when it was present in the same vectors in the NR85 and CBY strains (7, 8, 9).
Results obtained with the CBY strain containing the u8 or u17 YAC have been previously described (8, 9).
Data for the NR85 strain containing the YZ-CEN plasmid have been previously described (7), and the results with the VS8 vector are from this study.
Among approximately 3,100 clones, one plasmid (p54-45) resulted in slow growth and reduced viability following HO endonuclease induction (Table 3). This plasmid, which contained a 6-kb insert, was isolated and retransformed back into the LS20 strain containing either the original YZ-CEN plasmid or YZ YACs (see below). As with the original clone, slow growth and indirect lethality occurred upon induction of the HO endonuclease.
TABLE 3.
DSB-enhanced lethality in strain LS20 mediated by high-copy-number plasmids that can deplete proteins required for silencing
| High-copy-number plasmid | YZ-containing vector | Survivala |
|---|---|---|
| None | YZ-CEN | 1.02 ± 0.22 (4) |
| u8 YAC | 0.95 ± 0.22 (21) | |
| VS8 YAC | 0.98 ± 0.21 (4) | |
| p54-45b | YZ-CEN | 0.33 ± 0.04 (4) |
| u8 | 0.10 ± 0.08 (6) | |
| None | 0.60 ± 0.10 (5) | |
| HMRc | u8 | 0.37 ± 0.09 (6) |
| None | 0.97 ± 0.11 (6) | |
| SIR4 truncationd | VS8 | 0.42 ± 0.13 (6) |
| u8 | 0.27 ± 0.21 (12) | |
| None | 0.95 ± 0.17 (8) |
Relative colony-forming ability on GAL-versus GLU-containing SC medium that selects for the high-copy-number plasmid but not the YZ-containing vector; over 95% of the surviving colonies lack the vector. Error values are ±1 standard deviation about the mean. Numbers of experiments are indicated in parentheses.
p54-45 is described in the text. The genomic insert contains a complete copy of the silent mating type locus HMR and a sequence that extends towards the centromere to include a number of uncharacterized ORFs.
HMR is a high-copy-number YEP13-based plasmid containing a cloned HMR (34) and the selectable marker LEU2.
The SIR4 truncation plasmids used in this study, pKR61 and pKR62, are derivatives of pKR60 (64); these plasmids constitutively overexpress a truncated C-terminal fragment of the Sir4 protein but differ in the selectable markers used (TRP1 LEU2 and URA3, respectively). pKR61 was used in conjunction with VS8, whereas in the other two experiments u8 and (with no vector), the strain contained pKR62.
Sequencing revealed that the fragment in p54-45 originated from chromosome III and included a complete copy of the nontranscribed (silent) mating type locus HMR (data not shown). Retransformation of LS20 containing the u8 or u17 YZ YAC with p54-45 also resulted in indirect lethality following HO induction (Table 3), demonstrating that the original clone was not a genomic mutation picked up during the screening process. The 6-kb genomic DNA fragment within p54-45 also contained approximately six small uncharacterized ORFs extending rightward from HMR (proximal to the centromere). The HMR segment was examined first since it might be a target for HO endonuclease. Transformation of LS20 containing a YZ YAC with a different high-copy-number plasmid containing only the HMR region (34) also resulted in indirect lethality following induction of a DSB (Table 3). The high-copy-number plasmid containing HMR and the DSB appeared to be required for lethality, since the viability remained high when either the YZ YAC or the HMR plasmid was absent (Table 3). The high-copy-number vector lacking HMR did not affect survival.
Depletion of Sir silencing complexes enhances indirect lethality.
Plasmids containing transcriptionally inactive (silenced) mating type loci have been shown to require SIR silencing for plasmid replication and segregation (33), suggesting that high-copy-number plasmid expression may titrate SIR protein complexes away from other genomic sites. The results obtained with the high-copy-number HMR plasmid may therefore be due to (i) induction of additional DSBs at the YZ junction within HM loci in the plasmid or in the genome, (ii) a direct role of silencing proteins at the site of the DSB, or (iii) expression of MAT-controlled genes if the silent HML or HMR locus is activated by titration of Sir complexes.
The somewhat reduced survival of a strain containing p54-45 but lacking a YZ-containing vector (Table 3) might suggest cutting of a YZ site within the HMR region of the plasmid, which would result in loss of this plasmid and a corresponding lack of growth under selection for the plasmid marker. However, induction of HO did not result in significantly enhanced p54-45 loss (Table 3). If cutting occurs, it must be infrequent or efficiently repaired using undamaged homologues.
It is possible that multiple copies of HMR titrate Sir proteins, which are normally required to maintain silencing at telomeres and the HM loci. To test this, LS20 containing a human DNA YAC with a poorly repairable DSB site (u8 YZ YAC [8]) was transformed with a high-copy-number plasmid expressing a C-terminally truncated form of the Sir4 protein (47, 64). Since this plasmid does not contain a YZ site, it is not subject to HO cutting. The truncated Sir4 protein forms an inactive complex with Sir2 and Sir3 proteins, thereby depleting cells of the proteins required to maintain silencing at the HML and HMR chromosomal loci as well as at telomeres (47, 64). Isolates containing these plasmids clearly showed indirect lethality following the induction of a DSB by the HO endonuclease (Table 3). There was no decrease in survival if the YAC was absent, demonstrating that the effect of truncated Sir4 depended on the induction of a DSB in the YAC and was not due to nonspecific cleavage within the yeast genome. These results suggest that the lethality observed in the cells containing p54-45 or HMR must be due to titration of Sir complexes.
A persistent DSB causes indirect lethality in sir2Δ, sir3Δ, and sir4Δ mutants.
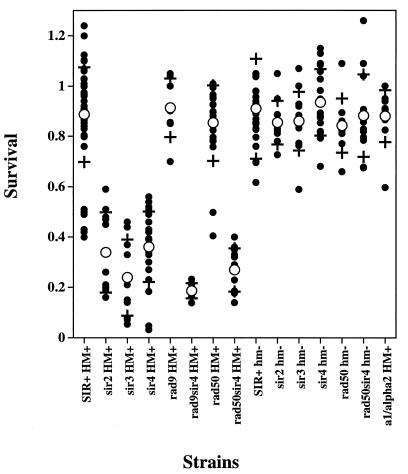
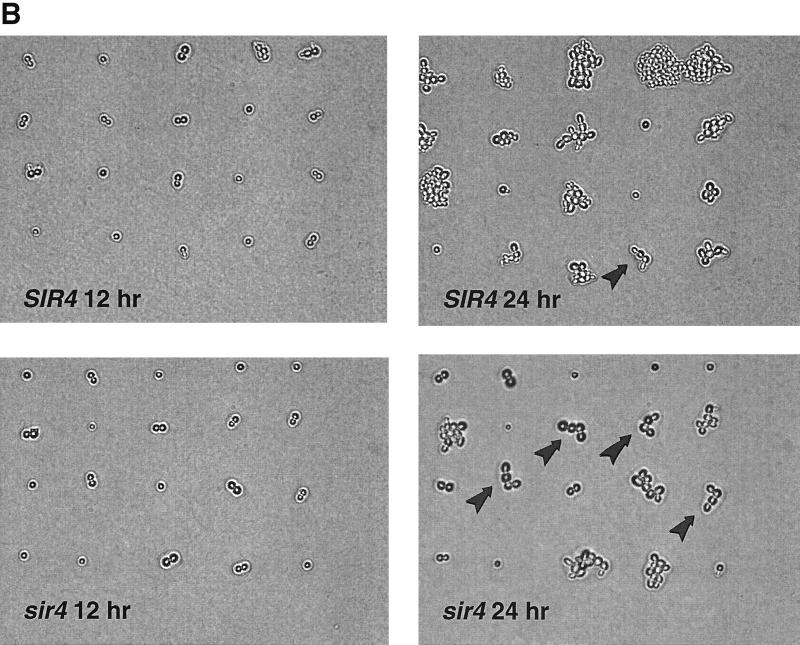
Since the SIR4 gene product appeared to play an important role in the cellular response to a DSB, we deleted each of the three genes required for telomere and mating type silencing (SIR2, SIR3, and SIR4). The Sir proteins silence the HML and HMR loci so that transcription of the mating type regulatory genes are repressed and the access of HO protein to the YZ target site is greatly inhibited (23, 37). Deletion of any of these genes did not affect growth rates of the LS20 cells (data not shown) but did eliminate silencing (data not shown). We also deleted the HO endonuclease target sites within the chromosomal silent mating loci HMR and HML at positions closely flanking the YZ junction. Therefore, the disruptions of the SIR genes could be examined for their effects on transcriptional silencing at HML and HMR versus their potential for inducing a DSB at these sites. The inability to make functional silencing complexes changed LS20 from an “a-like faker” phenotype (MAT had already been deleted [65]) into a nonmating, a1/α2 haploid phenotype (HM+ in the sir2Δ, sir3Δ, and sir4Δ strains [data not shown]). As shown in Fig. 1, indirect lethality was observed only in sir2Δ , sir3Δ, and sir4Δ strains containing the YZ YAC. Sir+ strains were unaffected by the DSB. These results demonstrate that the Sir protein complex plays a key role in the events proceeding from the appearance of an unrepairable DSB to indirect lethality.
FIG. 1.
Effects of a single persistent double-strand break within a dispensable YAC on survival. LS20 (with the MAT locus deleted) was transformed with the human artificial chromosome u8, which contains a YZ-URA3 sequence embedded in a cluster of ALU repeats 70 kb from the telomere. Either the HO target sites within the HML and HMR loci were precisely deleted (HM+) or HML and HMR were completely deleted (hm− by circularizing chromosome III in LS20 strains containing the u8 YAC. SIR2, SIR3, SIR4, RAD50, and both SIR4 and RAD50 were subsequently deleted from HM+ and hm− strains. RAD9 was deleted from a Sir+ HM+ strain, and SIR4 was subsequently deleted. A Sir+ HM+ strain expressing the a1 and α2 transcriptional regulators from pCB115 is denoted a1/α2 HM+. Cells were grown to logarithmic phase (1 × 107 to 3 × 107 cells/ml) in liquid SC-GLU medium (88) lacking URA to maintain selection for the YAC. In the case of the strain containing plasmid pCB115, the medium also lacked leucine to maintain plasmid selection. Cells were washed in sugarless medium, and appropriate dilutions were plated to SC-GLU and SC-GAL plates (with URA to remove selection for the YAC). Survival was determined by relative colony-forming abilities on GAL versus GLU plates after 4 days of incubation at 30°C. Each small filled circle represents the survival data from one experiment. Large open circles and crosses denote the mean survival ± 1 standard deviation for each strain.
SIR silencing of mating type provides toleration of a persistent DSB.
The toleration of a DSB in Sir+ strains might be due to the role that Sir proteins have on chromatin structure in the vicinity of the DSB site or alternatively from the repression of mating type regulatory genes within HML and HMR loci by Sir complexes. To address this issue we deleted HML and HMR (these strains are denoted hm−) using a one-step-PCR-mediated gene disruption that resulted in a circular chromosome III [see Materials and Methods]). Following circularization of chromosome III, the SIR2, SIR3, and SIR4 genes were individually deleted. As expected, the circularization of chromosome III had no effect on the growth of LS20 since naturally occurring circular derivatives have been previously shown to be stable during replication and segregation (72). As shown in Fig. 1, indirect lethality was not observed in any of these strains following induction of the persistent DSB. These results demonstrate that HML and HMR are required for indirect lethality to occur when Sir2, Sir3, and Sir4 products are absent. In the absence of silencing, HMR and HML normally produce the a1-α2 repressor complex that confers an a1/α2 haploid (nonmating) phenotype upon the LS20 strain. It therefore appears that a gene(s) under mating type regulation is responsible for the observed DSB responses.
To determine if changes in mating type regulation alone were sufficient for reduced viability following a DSB, we cloned the a1 and α2 transcriptional regulators into a plasmid (pCB115) and transformed a Sir+ strain carrying the u8 YAC. This plasmid confers a nonmating phenotype in the Sir+ strain. Following induction of a persistent DSB in the Sir+ strain containing this plasmid, no lethality was seen (Fig. 1). This indicates that it is the combined absence of SIR and expression of a1/α2 that are required for reduced viability following a persistent DSB.
Commitment to death in sir4Δ mutants occurs later than an HO-induced DSB in a dispensable YAC.
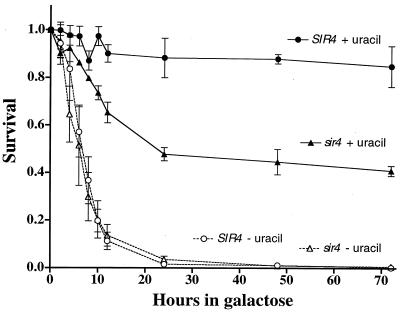
Since a DSB can initiate lethality in the Sir− mutants, we wanted to establish if the lethality is reversible and, if so, when the lethal effects of the break are irreversible. We used the previously described pullback approach (8) in which logarithmically growing cells are embedded in GAL-URA medium to induce the HO endonuclease and at various times the plates are overlaid with GLU-URA to shut off HO. Since the HO cut site in the YAC is closely linked to the URA3 marker and the break is not repaired, it is possible to genetically determine when the break occurs. Similarly, cells are plated within GAL+URA medium and overlaid with GLU+URA medium to determine when cells cannot be rescued and become committed to death.
As shown in Fig. 2, nearly 50% of the cells lost the URA3 marker of the YAC within 4 to 5 h after HO induction; by 12 h, only 10% of the cells retained the marker, with the sir4Δ strain being slightly slower. These kinetics were similar to those previously found for the same YAC in strain CBY (8). Physical measurements of the appearance of a break in the YAC using our previously described methods (8) established that the loss of the genetic marker corresponded to the time at which the single break was found in the YAC (data not shown). As expected, there was a time-dependent decrease in survival for the sir4Δ strain. The time required for the loss of colony-forming ability was threefold shorter for cells embedded in GAL plates lacking URA than in GAL plates with URA, suggesting that the processes leading to cell death on complete medium was substantially longer than the loss of the marker due to the DSB. Only 50% of the cells experienced a commitment to death, whereas nearly all the cells lost the marker. Possible reasons for the survival of the remaining cells are discussed below.
FIG. 2.
Commitment to death in the sir4Δ HM+ strain following induction of a persistent DSB within the u8 YAC. A pullback procedure (8) was used where sir4Δ (triangles) or Sir+ (circles) HM+ strains containing the u8 YAC were plated within GAL medium with URA (filled symbols) or without URA (open symbols) to induce expression of the HO endonuclease. At the indicated times, the plates were overlaid with either GLU medium with URA or GLU medium without URA to repress expression of the HO endonuclease. Decreased survival on plates with GAL medium lacking URA requires HO-induced cutting of the YAC and concomitant loss of the URA3 marker from DNA degradation. Each data point is the average of results from two to three experiments. Results with the sir4Δ rad50Δ strain and the sir4Δ strain were comparable (data not shown).
Adaptation to DSB-induced G2 arrest is decreased in Sir mutants.
To address the role of mating type on the ability of a DSB to trigger cell cycle arrest, we monitored microscopically wild-type, sir2Δ , sir3Δ, and sir4Δ cells that either contained HML and HMR (with HO sites deleted but a1 and α2 sites intact) or completely lacked HML and HMR (the strains contained circular chromosome III). Since the HML and HMR loci are derepressed in a sir− background, the HML HMR cells expressed the a1 and α2 repressors and therefore appeared as an a1/α2 (nonmating) haploid, while the hmlΔ hmrΔ strain retained an a-like faker haploid mating type phenotype.
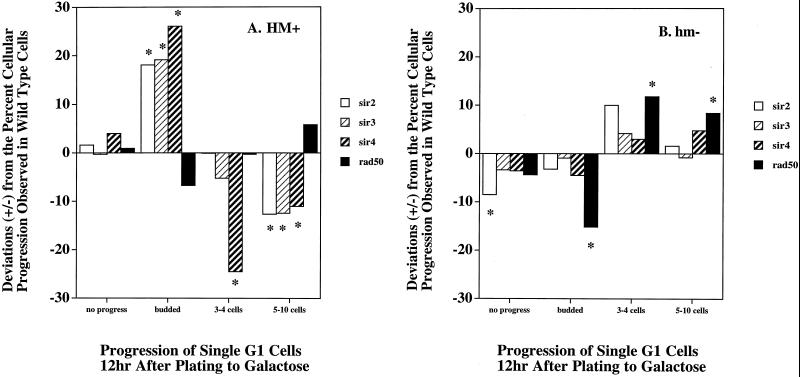
Shown in Fig. 3 are the results for cells that were initially plated to GAL in the G1 (unbudded) stage of the cell cycle and observed microscopically 12 h later. (Similar results were found with cells that were budded at the time of plating except that cell cycle arrest occurred in the subsequent G2 phase at the four-cell stage [data not shown].) Single cells could remain as plated (no progress) or proceed to G2 (large-budded cells). Cells could also adapt to the DSB, progress past the G2 checkpoint, and form a small microcolony, which we categorized as containing 3 to 4 or 5 to 10 cells. Cells in the three- to four-cell stage may have rearrested in the subsequent G2. The mean percentages for all of these classes were determined for the mutant strains and compared to those obtained with the isogenic wild-type cells. Deviations from results with the wild-type strain are depicted in Fig. 3 as bars extending in the positive direction (i.e., there was a greater percentage of mutant cells with the particular characteristic) or the negative direction. Values close to 0 indicate the same frequencies for both the wild-type and mutant strains.
FIG. 3.
Effects of a single persistent double-strand break within a dispensable YAC on cell cycle progression of single (G1) cells. Either the HO target sites in sir2Δ , sir3Δ , sir4Δ, and rad50Δ LS20-derived strains containing the u8 YAC were precisely deleted (HM+) (A) or HML and HMR were completely deleted (hm−) (B). The HM+ cells retained the a1/α2 mating type gene regulators and expressed an a1/α2 haploid (nonmating) phenotype in sir− strains. Cells were grown and washed as described in the legend to Fig. 1 and then spread onto SC-GAL plates using a multipin pronging device. For each mutant strain, two isolates were examined microscopically using a Singer MSM dissecting microscope and compared to the corresponding wild-type parental strain (8, 9). Individual cells (>400 for each bar in the graph) were examined immediately after being plated to the SC-GAL medium and 12 h later. At the time of plating, all isolates had similar distributions of cells: ∼50% were G1 (single cells), ∼25% were S (small budded), and ∼25% were G2 (large budded). Presented are the results for the cells that were in G1 (unbudded cells) (A and B) at the time of plating. G1 cells either remained as initially plated (no progress), progressed to the budded stage, progressed to the 3- to 4-cell stage, or formed microcolonies (5 to 10 cells). The mean percentage of cells for the wild-type parental strain was subtracted from the mean percentage obtained for each mutant strain in each category. Mutant strains that contained a higher percentage of cells in a given category had positive values, whereas mutant strains that had fewer cells in a given category had negative values. The deviations from the wild-type value (baseline = 0) for these mean percentages are indicated by bars. An ∗ over a bar indicates a deviation from the wild-type value that is significant (±1 standard deviation). Cells that were in G2 at the time of plating either remained as initially plated (budded), progressed to the 3- to 4-cell stage, or formed microcolonies (5 to 10 cells and >10 cells; data not shown). For the sir2Δ, sir3Δ, and sir4Δ cells able to form an a1/α2 haploid phenotype (HM+) (A), a persistent DSB in the u8 YAC leads to the accumulation of cells at G2 (i.e., as budded cells [22, 84]). No G2 delay was observed in sir2Δ, sir3Δ, and sir4Δ cells lacking the u8 YAC or those without the HML and HMR loci (hm−) (B). Similarly, sir2Δ, sir3Δ, and sir4Δ cells able to form an a1/α2 haploid phenotype that were in S or G2 (data not shown) at the time of plating, also accumulated at the subsequent G2 stage (i.e., as budded cells [22, 84]).
In all strains that had an a1/α2 haploid phenotype (sir2Δ, sir3Δ, and sir4Δ cells in the HM+ background), there was an excess of cells (>18 to 26%) showing G2 arrest compared to the percentage of wild-type cells showing G2 arrest (Fig. 4A). There was a corresponding decrease in the number of cells that adapted to the G2 arrest checkpoint, with 13 to 27% fewer cells forming microcolonies. The increased percentage of large-budded cells seen in these sir− a1/α2 haploid strains also correlated with reduced viability resulting from the persistent YAC DSB in these strains (Fig. 1). In sir− mutants that did not have the a1/α2 haploid phenotype (sir2Δ, sir3Δ, and sir4Δ cells from which HML and HMR were also deleted), the mutant and wild-type cells were similar with the exceptions that fewer mutant cells were arrested at 12 h and that 5 to 10% more proceeded to the multiple-cell stage, suggesting a more rapid ability to adapt to the break. Thus, the extended G2 arrest associated with the loss of SIR function requires derepression of the HML and HMR loci in sir mutants. This finding suggests that a gene(s) under mating type control may alter cell cycle adaptation. In Sir+ strains of LS20, rapid cell cycle adaptation to a persistent DSB can occur since the HML and HMR loci are under SIR-mediated repression.
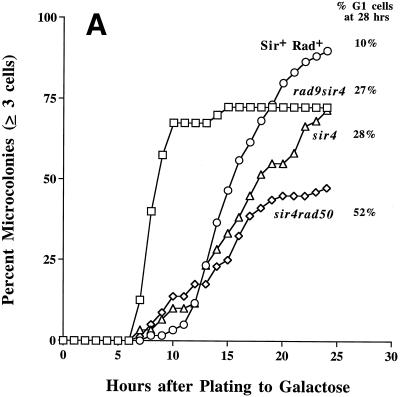
FIG. 4.
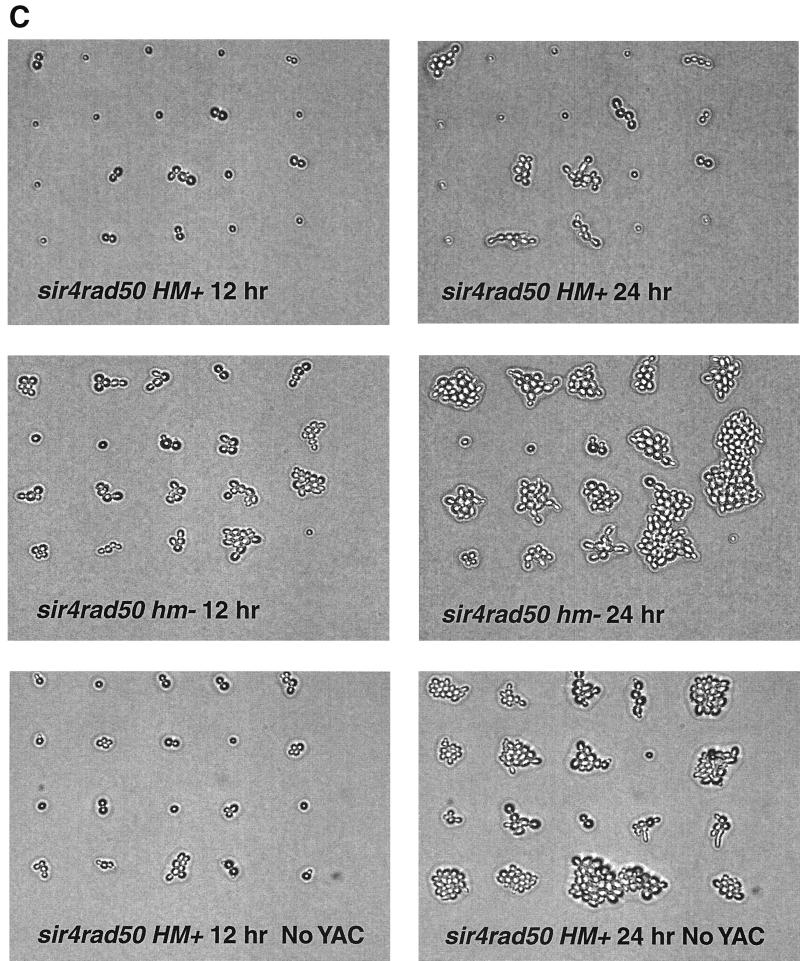
Delayed cell cycle progression of sir4Δ cells following induction of a persistent DSB in a dispensable YAC. Cells were grown as described in the legend to Fig. 1. (A) Single (G1) cells of SIR4 (circles), sir4Δ (triangles), rad9Δ sir4Δ (squares), and sir4Δ rad50Δ (diamonds) strains were micromanipulated into a grid pattern on an SC-GAL plate and photographed at hourly intervals. Cells with RAD9 deleted show no G2 delay. Cells were scored as percentages of cells that had progressed past the G2/M checkpoint and formed a microcolony containing three or more cells. Zero hour is the time of initial plating. (B) Representative photomicrographs at 12 h (left panels) and 24 h (right panels) for the SIR4 (upper panels) and sir4Δ (lower panels) HM+ cells are depicted. Cell cycle progression is delayed in the sir4Δ strain as the result of a more prolonged arrest in the initial and second (arrows) G2 phases. This is followed by repeated cycles of G2 arrest, adaptation, and rearrest in G2 (see the text). (C) Representative photomicrographs at 12 h (left panels) and 24 h (right panels) for the sir4Δ rad50Δ HM+ (upper panels), sir4Δ rad50Δ hm− (middle panels), and sir4Δ rad50Δ HM+ cells without a persistent DSB (no YAC in lower panels) are depicted. Cell cycle arrest in the sir4Δ rad50Δ strain is specific for a YAC DSB and requires HML and HMR.
Prolonged G2 arrest in sir4Δ a1/α2 haploid cells is dependent on RAD9.
To further characterize the response of various mutants to a DSB in terms of time of onset of cell division, time in G2, ability to adapt or rearrest after the first division, and dependence on the RAD9 checkpoint gene, individual cells were monitored microscopically at hourly intervals for 36 h. Cells of each strain were micromanipulated onto a GAL plate in a grid pattern within one microscopic field of view. These were photographed at hourly intervals, and the mean times for the onset of cell division, as well as the time spent as budded cells or as four-cell microcolonies, were calculated. Depicted in Fig. 4A are the time courses for the formation of microcolonies (at least three cells) of SIR4, sir4Δ, rad9Δ sir4Δ, and sir4Δ rad50Δ HM+ cells after single (G1) cells were plated to GAL. The times of onset of initial bud formation in all the strains examined were similar (∼6 h) (data not shown). Overall, the SIR4 cells were delayed in G2 for less time and had formed a higher percentage of large microcolonies (more than four cells; 70%) than the sir4Δ cells (20%) after 24 h (Fig. 4B). Half (50%) of the sir4Δ cells were still in the large-budded stage or had rearrested as a chain of four cells after 24 h of growth on GAL. These arrested sir4Δ cells could go on to form microcolonies of ∼50 to ∼100 nonviable cells, which did not progress further. Unlike wild-type cells, the individual sir− cells appeared to exhibit irregular growth patterns within the microcolonies, which would explain their distorted shapes (Fig. 4B). These microcolonies account for the increased lethality seen in this strain compared to the lethality of SIR4 cells (the relative plating efficiencies on GLU versus GAL in this experiment were 0.46 and 1.1 for the sir4Δ and SIR4 strains, respectively).
On average, the sir4Δ cells (Fig. 4AB) were arrested for 8.4 ± 0.5 (mean ± 1 standard error) h as large-budded (G2) cells and rearrested as four-cell chains for 4.0 ± 0.4 h. The average times of G2 arrest for SIR4 cells were 6.4 ± 0.4 and 2.9 ± 0.4 h in the four-cell stage. The increased time that cells spent in G2 arrest is reflected by the decreased percentage of cells that form microcolonies in the sir4Δ cells (Fig. 4A). Therefore, the sir4Δ cells were arrested significantly longer in G2 at both the first division (2.0 h longer) and at the second division (1.1 h longer) following induction of the DSB. The average time of initial bud emergence was the same for both strains. Strains without the u8 YAC did not arrest in G2 and formed viable microcolonies 24 h following plating to GAL (Fig. 4C). The sir4Δ or sir4Δ rad50Δ cells that were incapable of forming an a1/α2 haploid phenotype (hm−) arrested in a manner similar to that of the SIR4 HM+ strain following plating to GAL (Fig. 4C). As noted above, these cells did not die in response to the DSB (Fig. 1).
In order to determine if the prolonged G2 arrest in sir4Δ cells requires the RAD9 checkpoint gene, RAD9 was deleted in Sir+ and sir4Δ HM+ strains and individual G1 cells were microscopically examined for cell cycle progression following induction of the DSB in the dispensable YAC as described above (Fig. 4A). G2 arrest was greatly reduced among the population of single (G1) cells plated to GAL and observed at 12 or 24 h following the induction of the DSB in either Sir+ or sir4Δ strains in which RAD9 was deleted (Fig. 4A and data not shown). For the rad9Δ sir4Δ cells, the onset of microcolony formation was much more rapid (by >5 h) than for the SIR4 cells. The appearance of microcolonies took longer with either the sir4Δ or sir4Δ rad50Δ cells (Fig. 4). The cell cycle response of the rad9Δ Sir+ strain was comparable to that of the rad9Δ sir4Δ strain (data not shown); however, reduced viability was observed in the rad9Δ sir4Δ but not in the rad9Δ cells following the HO induction of the DSB (Fig. 1). Since indirect lethality occurs in the absence of Sir4, the mechanism of cell death is therefore independent of repeated cycles of G2 arrest and adaptation.
The rad9Δ sir4Δ cells were much smaller and the microcolonies had progressed through more divisions than the Rad+ sir4Δ cells (data not shown). This supports the conclusion that the prolonged DSB-induced G2 arrest is RAD9 dependent in the sir4Δ HM+ strain and that cells have the ability to rearrest at the four-cell stage following the first cell division. Furthermore, it also appears that the multiple rounds of G2 arrest, adaptation, and rearrest in the sir4Δ cells require the Rad9 protein. This finding suggests that the signal responsible for RAD9-dependent arrest becomes attenuated but that it can be reestablished in the next cell cycle. This suggestion implies that broken YAC fragments may persist in mother and daughter cells after division or that a diffusible nuclear factor that transmits the G2 arrest signal is still active in the undamaged daughter nuclei. Fragments of a broken YAC have been previously shown to persist for several rounds of cell division in this strain using another YAC (65).
The RAD50-dependent end-joining repair pathway is not required for toleration of a persistent DSB.
In some strain backgrounds the SIR gene products appear to play a direct role in NHEJ repair of DSBs. In addition to the Sir proteins, Yku70 or Yku80 and the Rad50-Mre11-Xrs2 complex are all required for NHEJ repair. Furthermore, deletion of RAD50 suppressed permanent arrest in a yku70Δ mutant by enhancing adaptation to an HO-induced, chromosomal DSB (38). We therefore deleted RAD50 to determine if the observed effects of SIR on cell cycle adaptation and toleration of a DSB were mediated through this pathway.
Deletion of RAD50 in Sir+ strains did not affect their toleration of the persistent DSB (Fig. 1). As expected, NHEJ repair of linearized plasmids transformed into the rad50Δ mutant was reduced (data not shown). Checkpoint adaptation responses to the YAC DSB in both the HM+ and hm− backgrounds were similar to or slightly enhanced compared to those in the wild type (Fig. 3). The percentage of rad50Δ cells that were in a large-budded (G2) stage of the cell cycle 12 h after being plated to GAL was decreased by 7% in HM+ cells (Fig. 3A) and by 15% in hm− strain backgrounds compared to the percentage in RAD50 cells (Fig. 3B). Conversely, the percentage of G1 cells that had progressed to the microcolony stage was increased by 6% in HM+ cells and by 18% in hm− strain backgrounds compared to the percentage in RAD50 cells. Since survival was not decreased in the rad50Δ strains following the DSB, this suggests that the Rad50-dependent NHEJ pathway is not required for toleration of a persistent DSB in a dispensable YAC.
We also constructed a sir4Δ rad50Δ double mutant to determine if deletion of RAD50 could suppress the prolonged G2 arrest and lethality observed in the sir4Δ HM+ strain in a manner similar to that observed for a yku70Δ strain (36). As shown in Fig. 1, the mean survival for the sir4Δ rad50Δ HM+ strain was 27% versus 36% for the sir4Δ HM+ strain. The cell cycle progression of single, G1 sir4Δ rad50Δ HM+ cells following induction of the DSB on GAL was delayed to an extent similar to (or slightly longer than) that seen for the sir4Δ HM+ strain (Fig. 4A). This was reflected in a low rate of survival (0.26) when plating efficiencies on GAL and GLU media were compared. The arrest observed in the sir4Δ rad50Δ HM+ strain was DSB dependent, since isogenic sir4Δ rad50Δ HM+ strains without the dispensable YZ-containing YAC did not arrest (Fig. 4C) and displayed a high (1.05) relative plating efficiency of colony formation on GAL versus GLU. A persistent DSB in a sir4Δ rad50Δ strain unable to form the a1/α2 phenotype (hm−) did not lead to prolonged G2 arrest (Fig. 4C) or reduced survival (Fig. 1). Interestingly, a large fraction of cells (52%) did not progress past the unbudded (G1) stage in the arrest seen with the sir4Δ rad50Δ HM+ strains (Fig. 4A and C), suggesting that RAD50 may be required for the G1-to-S transition following the formation of a persistent DSB in sir4Δ HM+ strains. This G1 arrest was not observed in either sir4Δ HM+ (Fig. 4B) or rad50Δ HM+ (data not shown) single-deletion mutants. Taken together, these results suggest that a gene(s) under mating type control regulates adaptation to the G2 checkpoint arrest arising from a persistent DSB in a pathway other than the NHEJ repair pathway.
Absence of Sir4 does not increase sensitivity to γ-ray or MMS-induced damage.
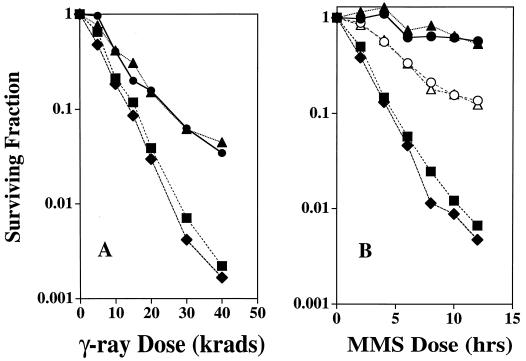
Recently it was proposed that the SIR4 gene might play a direct role in the repair of radiation-induced breaks (11, 81) or MMS-induced damage (48) via NHEJ. The sensitivity of rad52 cells, which are unable to undergo homologous recombinational repair of DSBs, was increased by a sir4 mutation (81). However, little effect could be discerned for the sir4 mutation in haploid Rad+ cells, which is surprising since haploid G1 cells are radiation sensitive due to a lack of opportunities for recombinational repair (12). We, therefore, examined the radiation sensitivity of stationary-phase cells (∼95% of cells in G1) (Fig. 5A) and the MMS sensitivity of logarithmically growing cells (Fig. 5B) with various combinations of SIR, sir4Δ, and RAD50 alleles.
FIG. 5.
Survival following γ-irradiation (A) or MMS treatment (B) of haploid Sir+ (circles), sir4Δ (triangles), rad50Δ (squares), and sir4Δ rad50Δ (diamonds) cells. (A) For γ-irradiation, the LS20-derived strains were grown to stationary phase in YEPD. Based on cell appearance, ∼95% of the cells were in G1 (i.e., unbudded) at the time of irradiation. Survival was determined by counting relative numbers of CFU after 3 days of growth at 30°C on YEPD plates. (B) For MMS treatment, survival was determined following treatment of logarithmically growing Sir+ (circles), sir4Δ (triangles), rad50Δ (squares), and sir4Δ rad50Δ (diamonds) cells. Cells in SC-GLU medium were treated with 0.01% (filled symbols) or 0.03% (open symbols) MMS and reincubated with shaking at 30°C. At the indicated times, cells were diluted in water and plated onto YEPD plates. Survival was determined by counting relative numbers of colonies after 4 days of growth at 30°C. Each data point is the mean of results of two to three replica experiments.
As seen in Fig. 5A, the survival curves were single-component curves and the presence or absence of the SIR4 gene had no impact on the ability of Rad+ cells to tolerate radiation-induced DSBs. While the rad50Δ strains exhibited an approximately 50% increase in sensitivity compared to that of wild-type strains at equal survival levels (i.e., a dose-modifying factor of 2), there was no apparent effect of the sir4 mutation. Furthermore, no differences were observed for sir4Δ cells irradiated in the logarithmic phase of growth, indicating that SIR4 does not play a role in the recombinational repair of G2 cells (data not shown). Also, there was no difference in MMS sensitivity for Sir+ versus sir4Δ mutants in either a Rad+ or rad50Δ background (Fig. 5B). We also examined Sir+ and sir4Δ strains for their plasmid end-joining capability using transformation of a restriction endonuclease-digested centromeric plasmid, pAG36 (19; data not shown). No decrease in end-joining capability was found in the LS20 strain background following deletion of SIR4. These results indicate that, in the LS20 strain background, there is no apparent role for SIR4 in either NHEJ or the recombinational repair of radiation- or MMS-induced breaks.
DISCUSSION
In addition to the elaborate systems involved in the repair of DNA damage, there are many genes that influence cell progression in response to chromosomal lesions. Checkpoint genes such as RAD9 serve to arrest cell cycle progression at G2 in the presence of persistent damage. Presumably, this allows additional time for repair (84). However, under conditions where a DSB remains unrepaired in an LS20 strain, either because of a defect in the DSB recombinational repair pathway (74) or lack of a homologue (65, 67), the arrest is only temporary. The reentry of cells into the cell cycle in the presence of unrepaired damage has been termed adaptation (20, 58, 83), and factors that are responsible for the adaptation response to an unrepaired chromosomal DSB in a rad52 mutant have been identified (78). We sought to identify additional factors that play a role in adaptation and lethality using a system where a persistent DSB is produced in a dispensable YAC.
An indirect role for SIR2, SIR3, and SIR4 in cell cycle adaptation and toleration of a DSB.
We found that SIR genes play an indirect role in adaptation. The approach that we took was motivated by our observations that an unrepairable DSB in some repair-proficient strain backgrounds (Rad+) leads to extended G2 arrest and indirect lethality but that there is only a modest delay and no lethality in other strains (references 65 and 67 and this report). An unrepairable DSB in the repair-proficient LS20 strain normally leads to several hours of delay in G2 (∼6 h) (Fig. 4A) and is well tolerated since it does not affect survival.
A high percentage of cells exhibit an extended G2 checkpoint delay and reduced survival in response to an unrepairable DSB when SIR2, SIR3, or SIR4 is deleted or when proteins of the Sir4 complex are depleted. Using the fluorescent DNA-specific stain DAPI (4′,6-diamidino-2-phenylindole), the cell nucleus was observed to be undivided at, or within, the bud neck (data not shown). The G2 delay was similar to that observed with other strains (7, 8, 9). Rather than stopping cells at the G2 checkpoint, the absence of Sir4 extended the G2 checkpoint to 8 h. Cells adapted to the damage and then rearrested in the subsequent G2. Subsequent limited and irregular cell division resulted in microcolonies with distorted shapes (compared to the smooth, rounded colonies of the wild type) that did not progress further. The reason for the reduced survival of these microcolonies remains to be determined; however, this response is also similar to our results with a persistent DSB in other strain backgrounds (7, 8, 9). The delayed adaptation and low rate of survival is an indirect effect determined by mating type control, since sir− strains lacking HML and HMR tolerate the break as well as Sir+ strains do.
The abilities of the sir4Δ mutants to slowly adapt to the G2 checkpoint, progress into mitosis, and then rearrest differs from those of a rad52 derivative of LS20 with mutations in both cdc5 and ckb2 (78), and their ability to rearrest differs from that of a ku70Δ strain (38) where a single DSB in chromosomal DNA leads to permanently arrested large-budded cells. Since rad52 mutants are known to grow more poorly than Rad+ cells and they are sensitive to alterations in DNA metabolism, particularly replication (discussed in reference 6), adaptation mutants might have a different effect in terms of preventing division.
There are several explanations for the defective adaptation response and the indirect lethality in the sir− mutants. The effects may be due to suppression of the RAD9-dependent signaling mechanism that transduces information about a DSB to the cell cycle apparatus as well as another RAD9-independent pathway that results in reduced viability. Possibly, there are stable diffusible factors that modify the transduction signal leading to RAD9-dependent cell cycle delay at G2 (22, 84). Following initial arrest, cells resume cycling, the nucleus divides, and the diffusible factor segregates to the undamaged daughter nuclei, causing rearrest in the next G2 phase. It seems more likely that, upon prolonged arrest in G2, the strength of the arrest signal decays and the cell cycle resumes. Since fragments of a YAC are likely to persist, these fragments segregate to the daughter nucleus (61) and the signal would be reestablished in the following division, thereby blocking cellular progression at the next G2.
SIR4 does not play a direct role in the repair of γ-ray- or MMS-induced damage.
The SIR gene products are part of a multiprotein complex that determines chromatin structure and enables transcriptional silencing of genes at both silent mating type loci and telomeres. In addition to silencing, Sir4 also has a key role in other aspects of DNA metabolism. Sir4 is required for mitotic chromosomal stability and efficient segregation of unstable plasmids (3, 57, 33). Recently, it was shown that Sir4 physically interacts with Hdf1 (now designated Yku70), a yeast homologue of Ku70 required for end-joining repair of DSBs (81). Furthermore, using plasmid end-joining repair assays, SIR2, SIR3, and SIR4 have all been shown to be essential for Ku-dependent DSB DNA repair (11).
In the experiments of Tsukamoto et al. (81), sir4 mutants were deficient in illegitimate recombination within plasmids and the protein was proposed to be part of a complex responsible for Ku-dependent recombination. However, it was not possible to assess the impact of an unrepaired break on cell viability. Transformation with a broken or dicentric plasmid led to loss of transformants, but this may have been due either to a lack of repair or to death resulting from the presence of a broken molecule in the sir4 strain. The sir4 mutation, as well as a Ku mutation, increased the sensitivity of a rad52 haploid mutant to ionizing radiation. There was an approximate dose-modifying factor of 3 between the survival response of Sir+ rad52 and sir4 rad52 strains. This sensitivity was proposed to be due to a lack of end-joining repair. However, there was an unusually high resistance of rad52 mutants. In the present experiments, we found that G1 cells of a Sir+ Rad+ strain were not more resistant to γ-irradiation than a sir4 Rad+ strain. Since G1 haploid cells do not exhibit recombinational DSB repair because there is no homologue present (12, 61), it appears that there is little, if any, SIR4-mediated nonhomologous end-joining repair of DSBs in Rad+ cells. In light of the unusually high resistance of rad52 mutants in that study (81), it is hard to draw general conclusions about sir4 effects.
It has also been reported that sir− mutants are hypersensitive when stationary-phase cells are replica plated to media containing the DNA-damaging agents bleomycin and MMS (48). Contrary to these results, we found that a sir− mutation did not increase the sensitivity of logarithmically growing LS20 cells to transient exposure to MMS or increase the sensitivity of a rad50Δ strain. Thus, under our conditions and in the LS20 background, Sir4 appears to play no role in the repair of MMS damage regardless of its NHEJ repair capabilities. This is consistent with the absence of a direct role in the repair of γ-ray-induced DSBs.
Deletion of RAD50 does not enhance adaptation to a DSB in sir4Δ strains.
A number of proteins, including Hdf1, Mre11, Rad1, Rad5, Rad9, Rad17, Rad24, Rad50, Xrs2, and Mec3, have been reported to bind to or interacting with the exposed ends of a DSB in yeast (1, 16, 44, 51, 81). Recently, Yku70 or Yku80, Mre11, and Rad50 were shown to function as a complex in both Ku-dependent NHEJ repair and telomere length maintenance (10, 11). In addition, both Rad50 and Mre11 are required for the initiation of spontaneous breaks in meiosis resulting in a reduction in meiotic recombination (52). Furthermore, Mre11, Rad50, and Xrs2 have been shown to interact as a complex. While null mutations are proficient in mating type switching and spontaneous mitotic recombination, they are sensitive to the killing effects of ionizing radiation (18, 27, 45, 46, 55) or restriction enzyme-induced DSBs (40). Deletion of RAD50, MRE11, or XRS2 resulted in decreased 5′-to-3′ degradation at HO-induced DSBs and also greatly reduced the level of NHEJ repair of this lesion (51, 74). Cells with YKU70 deleted expressed a permanent G2 arrest following induction of a single HO-induced chromosomal break, and this was attributed to enhanced 5′-3′ degradation at the break site (38). Deletion of either RAD50 or MRE11 in the yku70Δ strain enabled the cells to adapt to the DSB and resume cell cycle progression due to reduced degradation at the break site (38). In our system, deletion of RAD50 did not rescue logarithmically growing sir4Δ HM+ cells from repeated cell cycle arrests. Thus, it is unlikely that, in the LS20 strain background, the prolonged G2 arrest and lethality in sir4Δ strains is due to greatly enhanced 5′-3′ recision at the DSB site.
In comparison to previous reports of a direct effect of Sir4 on end-joining repair of MMS- or γ-ray-induced DSBs (11, 81), our data cannot exclude a direct DNA repair role for Sir4 in dealing with a persistent HO-induced break (see below). However, it should be noted that strain background may be a factor that influences the relative roles of Sir proteins in the repair of DSBs. Since the Ku class of proteins has been shown to bind directly to broken DNA ends and at telomeres (48), Sir4 may be part of the complex that binds directly to DSB ends. Possibly, the complex inhibits 5′-to-3′ nucleolytic processing that occurs at HO-cleaved YZ junctions (38, 74, 87). In some strains, in the absence of Sir4 protein, single-strand recision of a YZ break may be more extensive, resulting in longer single-stranded DNA tracts at the DSB site.
Mating type control of the repair of DSBs.
It has been suggested that the Ku, Sir2, Sir3, and Sir4 proteins are required for both DSB repair and silencing at telomeres (11). Furthermore, other studies have described a redistribution of Sir proteins from telomere regions to DSB damage sites that is dependent on MEC1 and RAD9 (48, 49, 50). However, recent studies suggest that the effects of SIR4 deletions may be mostly indirect, resulting from derepression of the silent mating loci and formation of an a1/α2 haploid phenotype (5, 39). These studies suggest that another gene(s) under mating type control may be involved in DSB repair. Therefore, a haploid-specific gene(s) may be turned off; alternatively, a diploid-specific gene(s) may be turned on.
Surprisingly, the expression of the a1/α2 regulators alone was insufficient to reduce viability following the HO induction of a persistent DSB. Reduced viability was observed only if SIR4 was also deleted. Possibly another function such as SIR-dependent silencing at a telomere (or at some other location) of an unidentified gene can prevent a persistent DSB from becoming lethal. Alternatively, SIR4 may have a direct role that is independent of NHEJ at the break site. Members of the SIR complex may bind to a DSB site to protect the DNA ends, as suggested from results with induced EcoRI (48, 49, 50). It is interesting that the level of recombinational repair of ionizing-radiation-induced DSBs is elevated in cells expressing a MATa/MATα phenotype, which is independent of ploidy (15, 24, 43). Possibly, in sir− haploid cells, the expression of MATa and MATα results in an unprotected DSB region being preferentially channeled into a recombinational repair pathway even when there is no available homologue. This in turn might lead to lethality through an, as yet, undefined process. The results of this study may help to reconcile the many disparate reports of either indirect (5, 39) or direct (29, 79, 81) involvement of SIR functions in NHEJ repair of DSBs. Depending on the experimental system employed and/or the yeast strain background used, either direct or indirect effects may predominate in the repair event. The dual nature of SIR involvement as revealed in this report suggests that under some conditions, both direct and indirect effects may be involved in DSB-induced processes.
Implications of SIR4-dependent toleration of DNA damage.
A response similar to that in sir− cells was found in the diploid CBY strain in that it also shows poor adaptation and prolonged G2 arrest following the appearance of an unrepaired DSB (8). The Sir4 silencing system may be part of an adaptive response that enables haploid cells to tolerate unrepaired DNA damage. Only in the absence of both SIR4 and the ability to express a1 and α2 is the lesion rendered lethal by virtue of delayed adaptation resulting in cell death. The redundant nature of this damage toleration system ensures that the adverse genetic consequences of an unrepaired break will be avoided.
This mechanism of damage toleration is analogous to p53-independent, mitotic cell death following ionizing radiation in mammalian cells (63). This process (where cells die after a varied number of cell cycles) appears to be the principal mode of death in solid tumors following irradiation (63). The similarities between indirect lethality in yeast and the senescence-like, p53-independent mitotic cell death seen in human cells following exposure to DNA-damaging agents (7, 8, 13, 63) lead us to suggest that there may be a Sir4 type of interpretation of damage in higher organisms that may also play a critical role in the radiation-induced killing response in human cancer cells.
Other silencing components in addition to Sir4 may also affect the cellular response to a single persistent DSB. The Sir4 protein is an essential component in a complex that includes Sir2, Sir3, Rap1, ABF1, and the highly conserved N termini of histones H3 and H4, all of which are required for transcriptional inactivation (silencing) at telomeres and the silent mating loci HML and HMR (4, 14, 31, 59, 68, 77). Silencing also blocks access of the HO endonuclease to the YZ cutting sites contained within the HM loci (6). Both transcriptional inactivation and blockage of the HO enzyme is thought to be mediated by alterations in chromatin structure which are dependent on the direct participation of the silencing-factor complex. Yeast can switch from a silenced to a nonsilenced state at regions near telomeres by a poorly understood process that is under epigenetic control (4, 59). It will be interesting to determine if epigenetic controls and/or subtle alterations of silencing factors are involved in the previously described differences in the adaptive response to a DSB observed for various strains (7, 8, 9, 65, 67).
A decreased life span has also been demonstrated in yeast sir4Δ mutants (32). It is interesting to speculate that this increased aging may be due to lack of adaptation and indirect lethality resulting from spontaneously occurring lesions. Considerable evidence from yeast and bacteria suggest that DSBs can occur during DNA replication (17, 35), especially at novel DNA structures (42; K. S. Lobachev and M. A. Resnick, unpublished). Furthermore, specific mutations in SIR4 extended, rather than decreased, the life span of yeast, and it was proposed that this occurred through a gene(s) that can affect aging (32). Such genes might also be expected to affect checkpoint adaptation and indirect lethality. The relatedness between yeast and mammalian cell repair and signaling systems provides opportunities for understanding the control of aging (32) and malignancy (21) in vertebrates. An examination of the roles of the gene products involved in adaptation and indirect lethality may therefore provide insight into strategies to limit uncontrolled malignant cell proliferation and modify life spans.
ACKNOWLEDGMENTS
We thank Virginia Zakian, Jeffrey Strathern, Kurt Runge, Kevin Lewis, and Dan Gietz for strains and plasmids and Hiep Tran for sequencing plasmid p54-45. We thank Kerry Bloom, Jim Mason, Dmitry Gordenin, Jake Kirchner, Kevin Lewis, Beverly Errede, and Doug Thrower for reviewing the manuscript and helpful discussions. C.B.B. also thanks D. Downie and C. Whatley for their technical expertise, without which this paper could not have been completed.
Partial support was provided by an interagency agreement grant (DE-A105-94ER61940) from the Department of Energy.
REFERENCES
- 1.Ahne F, Jha B, Eckardt-Schupp F. The RAD5 gene product is involved in the avoidance of non-homologous end-joining of DNA double strand breaks in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1997;4:743–759. doi: 10.1093/nar/25.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2416–2428. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 3.Ansari A, Gartenberg M R. The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol Cell Biol. 1997;17:7061–7068. doi: 10.1128/mcb.17.12.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 5.Astrom S U, Okamura S M, Rhine J. Yeast cell-type regulation of DNA repair. Nature. 1999;397:310. doi: 10.1038/16833. [DOI] [PubMed] [Google Scholar]
- 6.Bennett C, Perkins E, Resnick M A. Chromosomal metabolism and organization in yeast: genetic and molecular approaches. In: Streips U, Yasbin R, editors. Modern microbial genetics. New York, N.Y: Wiley-Liss; 1991. pp. 389–430. [Google Scholar]
- 7.Bennett C B, Lewis A L, Baldwin K K, Resnick M A. Lethality induced by a single site-specific double strand break in a dispensable yeast plasmid. Proc Natl Acad Sci USA. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett C B, Westmoreland T J, Snipe J R, Resnick M A. A double-strand break within a yeast artificial chromosome (YAC) containing human DNA can result in YAC loss, deletion, or cell lethality. Mol Cell Biol. 1996;16:4414–4425. doi: 10.1128/mcb.16.8.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett C B, Snipe J R, Resnick M A. A persistent double-strand break destabilizes human DNA in yeast and can lead to G2 arrest and lethality. Cancer Res. 1997;57:1970–1980. [PubMed] [Google Scholar]
- 10.Boulton S J, Jackson S P. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulton S J, Jackson S P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunborg G, Resnick M A, Williamson D H. Cell-cycle-specific repair of DNA double strand breaks in Saccharomyces cerevisiae. Radiat Res. 1980;82:547–558. [PubMed] [Google Scholar]
- 13.Chang B D, Broude E V, Dokmanovic M, Zhu H, Ruth A, Xuan Y, Kandel E S, Lausch E, Christov K, Roninson I B. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761–3767. [PubMed] [Google Scholar]
- 14.Diffley J F, Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci USA. 1988;85:2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fasullo M, Dave P. Mating type regulates the radiation-associated stimulation of reciprocal translocation events in Saccharomyces cerevisiae. Mol Gen Genet. 1994;243:63–70. doi: 10.1007/BF00283877. [DOI] [PubMed] [Google Scholar]
- 16.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 17.Flores-Rozas H, Kolodner R D. Links between replication, recombination and genome instability in eukaryotes. Trends Biochem Sci. 2000;25:196–200. doi: 10.1016/s0968-0004(00)01568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Game J C, Mortimer R K. A genetic study of x-ray sensitive mutants in yeast. Mutat Res. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein A L, McCusker J H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 20.Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 21.Hartwell L, Szankasi P, Roberts C J, Murrary A W, Friend S H. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 22.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 23.Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heude M, Fabre F. a/alpha-control of DNA repair in the yeast Saccharomyces cerevisiae: genetic and physiological aspects. Genetics. 1993;133:489–498. doi: 10.1093/genetics/133.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho K S, Mortimer R K. Induction of dominant lethality by x-rays in a radiosensitive strain of yeast. Mutat Res. 1973;20:45–51. doi: 10.1016/0027-5107(73)90096-1. [DOI] [PubMed] [Google Scholar]
- 26.Humphery T, Enoch T. Keeping mitosis in check. Curr Biol. 1995;5:376–379. doi: 10.1016/s0960-9822(95)00077-7. [DOI] [PubMed] [Google Scholar]
- 27.Ivanov E L, Sugawara N, White C I, Fabre F, Haber J E. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivy J M, Klar A J, Hicks J B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson S P. Genomic stability silencing and DNA repair connect. Nature. 1997;388:829–830. doi: 10.1038/42136. [DOI] [PubMed] [Google Scholar]
- 30.Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kayne P S, Kim U J, Han M, Mullen J R, Yoshizaki F, Grunstein M. Extremely conserved histone H4N terminus is dispensable for growth but essential for repressing silent mating type loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy B K, Austriaco N R, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 33.Kimmerly W, Buchman A, Kornberg R, Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988;7:2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klar A J, Strathern J N, Broach J R, Hicks J B. Regulation of transcription in expressed and unexpressed mating type cassettes of yeast. Nature. 1981;289:239–244. doi: 10.1038/289239a0. [DOI] [PubMed] [Google Scholar]
- 35.Kowalczykowski S C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 36.Larionov V, Kouprina N, Graves J, Chen X-N, Korenberg J R, Resnick M A. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci USA. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laurenson P, Rhine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S E, Moore J K, Holmes A, Umeza K, Kolodner R D, Haber J E. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 39.Lee S E, Paques F, Sylvan J, Haber J E. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol. 1999;9:767–770. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- 40.Lewis L K, Kirchner J M, Resnick M A. Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA. Mol Cell Biol. 1998;18:1891–1902. doi: 10.1128/mcb.18.4.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis L K, Resnick M A. Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat Res. 2000;451:71–89. doi: 10.1016/s0027-5107(00)00041-5. [DOI] [PubMed] [Google Scholar]
- 42.Lobachev K S, Stenger J E, Kozyreva O G, Jurka J, Gordenin D A, Resnick M A. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lovett S T, Mortimer R K. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics. 1987;116:547–553. doi: 10.1093/genetics/116.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lydall D, Weinert T. Yeast checkpoint genes in DNA processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 45.Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- 46.Malone R E, Ward T, Lin S, Waring J. The RAD50 gene, a member of the double strand break repair epistasis group, is not required for spontaneous mitotic recombination in yeast. Curr Genet. 1990;18:111–116. doi: 10.1007/BF00312598. [DOI] [PubMed] [Google Scholar]
- 47.Marshall M, Mahoney D, Rose A, Hicks J B, Broach J R. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4441–4452. doi: 10.1128/mcb.7.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin S G, Laroche T, Suka N, Grunstein M, Gasser S M. Relocalization of telomeric Ku and Sir proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 49.McAinsh A D, Scott-Drew S, Murray J A, Jackson S P. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr Biol. 1999;9:963–966. doi: 10.1016/s0960-9822(99)80424-2. [DOI] [PubMed] [Google Scholar]
- 50.Mills K D, Sinclair D A, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 51.Moore J K, Haber J E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nairz K, Klein F. mre11S—a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nasmyth K A, Reed S I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci USA. 1980;77:2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navas T A, Zheng Z, Elledge S J. DNA polymerase ɛ links the DNA replication machinery to the S-phase checkpoint. Cell. 1995;74:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 55.Ogawa H, Johzuka K, Nakagawa T, Leem S H, Hagihara A H. Functions of the yeast meiotic recombination genes, MRE11 and MRE2. Adv Biophys. 1995;31:67–76. doi: 10.1016/0065-227x(95)99383-z. [DOI] [PubMed] [Google Scholar]
- 56.Orr-Weaver T L, Szostak J W. Yeast recombination: the association between double strand-gap repair and crossing-over. Proc Natl Acad Sci USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser S M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 58.Paulovich A G, Toczyski D P, Hartwell L. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 59.Pilus L, Rhine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- 60.Raghuraman M K, Brewer B J, Fangman W L. Activation of a yeast replication origin near a double-stranded DNA break. Genes Dev. 1994;8:554–562. doi: 10.1101/gad.8.5.554. [DOI] [PubMed] [Google Scholar]
- 61.Resnick M A. The repair of double-strand breaks in DNA: a model involving recombination. J Theor Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 62.Resnick M A, Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 63.Ross G M. Induction of cell death by radiotherapy. Endocr Relat Cancer. 1999;6:41–44. doi: 10.1677/erc.0.0060041. [DOI] [PubMed] [Google Scholar]
- 64.Runge K W, Zakian V A. TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3094–3105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandell L L, Zakian V A. Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 66.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as carriers. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 67.Schultz V P, Zakian V A. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1995;76:1–20. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 68.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 69.Siede W. Cell cycle arrest in response to DNA damage: lessons from yeast. Mutat Res. 1995;337:73–84. doi: 10.1016/0921-8777(95)00023-d. [DOI] [PubMed] [Google Scholar]
- 70.Siede W, Friedberg A S, Dianova I, Friedberg E C. Characterization of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA-damaging agents. Genetics. 1994;138:271–281. doi: 10.1093/genetics/138.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strathern J N, Newlon C S, Herskowitz I, Hicks J B. Isolation of a circular derivative of yeast chromosome III: implications for the mechanism of mating type interconversion. Cell. 1979;18:309–319. doi: 10.1016/0092-8674(79)90050-3. [DOI] [PubMed] [Google Scholar]
- 73.Strathern J, Hicks J, Herskowitz I. Control of cell type in yeast by the mating type locus. The alpha-1 alpha-2 hypothesis. J Mol Biol. 1981;147:357–372. doi: 10.1016/0022-2836(81)90488-5. [DOI] [PubMed] [Google Scholar]
- 74.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1 dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 76.Teo S H, Jackson S P. Lifp targets the DNA ligase Lig4p to sites of DNA double strand breaks. Curr Biol. 2000;10:165–168. doi: 10.1016/s0960-9822(00)00317-1. [DOI] [PubMed] [Google Scholar]
- 77.Thompson J S, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 78.Toczyski D P, Galgoczy D J, Hartwell L H. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 79.Tsukamoto Y, Kato J, Ikeda H. Effects of mutations of RAD50, RAD51, RAD52, and related genes on illegitimate recombination in Saccharomyces cerevisiae. Genetics. 1996;142:383–391. doi: 10.1093/genetics/142.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsukamoto Y, Kato J, Ikeda H. Hdf1, a yeast Ku-protein homologue, is involved in illegitimate recombination, but not homologous recombination. Nucleic Acids Res. 1996;24:2067–2072. doi: 10.1093/nar/24.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 82.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 83.Weinert T, Lydall D. Cell cycle checkpoints, genetic instability and cancer. Semin Cancer Biol. 1993;4:129–140. [PubMed] [Google Scholar]
- 84.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 85.Weinert T A, Hartwell L H. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol Cell Biol. 1990;10:6554–6564. doi: 10.1128/mcb.10.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–655. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 87.White C I, Haber J E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Whittaker S G, Rockmill B M, Blechl A E, Malone D H, Resnick M A, Fogel S. The detection of mitotic and meiotic aneuploidy in yeast using a gene dosage selection system. Mol Gen Genet. 1988;215:10–18. doi: 10.1007/BF00331296. [DOI] [PubMed] [Google Scholar]
- 89.Xu Z, Norris D. The SFP1 gene product of Sacharromyces cerevisiae regulates G2/M transitions during the mitotic cell cycle and DNA-damage response. Genetics. 1998;150:1419–1428. doi: 10.1093/genetics/150.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]