Abstract
Ty1 retrotransposons in the yeast Saccharomyces cerevisiae are maintained in a genetically competent but transpositionally dormant state. When located in the ribosomal DNA (rDNA) locus, Ty1 elements are transcriptionally silenced by the specialized heterochromatin that inhibits rDNA repeat recombination. In addition, transposition of all Ty1 elements is repressed at multiple posttranscriptional levels. Here, we demonstrate that Sgs1, a RecQ helicase required for genome stability, inhibits the mobility of Ty1 elements by a posttranslational mechanism. Using an assay for the mobility of Ty1 cDNA via integration or homologous recombination, we found that the mobility of both euchromatic and rDNA-Ty1 elements was increased 32- to 79-fold in sgs1Δ mutants. Increased Ty1 mobility was not due to derepression of silent rDNA-Ty1 elements, since deletion of SGS1 reduced the mitotic stability of rDNA-Ty1 elements but did not stimulate their transcription. Furthermore, deletion of SGS1 did not significantly increase the levels of total Ty1 RNA, protein, or cDNA and did not alter the level or specificity of Ty1 integration. Instead, Ty1 cDNA molecules recombined at a high frequency in sgs1Δ mutants, resulting in transposition of heterogeneous Ty1 multimers. Formation of Ty1 multimers required the homologous recombination protein Rad52 but did not involve recombination between Ty1 cDNA and genomic Ty1 elements. Therefore, Ty1 multimers that transpose at a high frequency in sgs1Δ mutants are formed by intermolecular recombination between extrachromosomal Ty1 cDNA molecules before or during integration. Our data provide the first evidence that the host cell promotes retrotransposition of monomeric Ty1 elements by repressing cDNA recombination.
DNA helicases catalyze the unwinding of duplex DNA into individual DNA strands (42). A plethora of DNA helicases within cells is involved in DNA replication, repair, recombination, and transcription. Members of the RecQ family of DNA helicases are involved in the maintenance of genome stability in all organisms characterized, from bacteria to humans (7). Mutations in the SGS1 gene, which encodes the only RecQ homologue in Saccharomyces cerevisiae, result in elevated levels of mitotic homologous and illegitimate recombination, increased rates of chromosomal nondisjunction, and accelerated aging (21, 56, 61, 62, 65). Similarly, mutations in human genes encoding the RecQ homologues RecQL4 (35, 49), WRN (67), and BLM (18) give rise to rare hereditary disorders that are characterized by genome instability and a pronounced predisposition to cancer. Notably, expression of either WRN or BLM in yeast complements the hyperrecombination phenotypes of an sgs1 mutant (65). These findings suggest that the mechanisms by which Sgs1 preserves genetic stability in yeast will serve as a paradigm for the role of RecQ homologues in human disease.
The SGS1 gene was originally isolated in a screen for genetic interaction with DNA topoisomerase III (21). Both topoisomerase III and Sgs1 repress recombination of DNA repeats, and inactivation of either Sgs1 or topoisomerase III results in S-phase-specific defects. Therefore, it was proposed that the combined activity of Sgs1 and topoisomerase III is required to initiate DNA repair events during replication (7). In support of this model, Sgs1 was shown to colocalize with the cell cycle checkpoint kinase, Rad53, in S-phase-specific foci and to participate in activation of the intra-S checkpoint (19). In the absence of Sgs1, DNA lesions that arise during DNA replication may be diverted into a homologous recombination pathway. This view is supported by recent reports that inactivation of Sgs1 and Srs2, another DNA helicase with a partially redundant role in genome maintenance, results in a severe growth defect involving a high incidence of mitotic arrest (22, 40, 46). The growth defect is suppressed by inactivation of homologous recombination proteins, including Rad51, Rad52, Rad55, and Rad57, suggesting that mitotic arrest in sgs1 srs2 mutants results from DNA lesions that arise from abortive recombination events (22, 46). Taken together, these findings suggest that DNA lesions that block the replication fork act as triggers for homologous recombination in sgs1Δ mutants.
Sgs1 is critical for the stability of repeated DNA sequences such as those in the RDN1 locus. RDN1 contains 100 to 200 tandem repeats of a 9.1-kb region encoding the RNA polymerase I (Pol I)-transcribed 35S rRNA gene and the Pol III-transcribed 5S rRNA gene. Both recombination between ribosomal DNA (rDNA) repeats and Pol II-mediated transcription are inhibited in the RDN1 locus (6, 25, 57, 58). Recombination in the rDNA can give rise to extrachromosomal rDNA circles (ERCs), which accumulate in aging cells and may trigger senescence (55). Many trans-acting factors that repress recombination in rDNA have been identified. Where analyzed, genes that inhibit rDNA recombination, including SIR2, TOP1, UBC2, and ZDS2, have been found to also be required for transcriptional silencing in rDNA (6, 8, 20, 25, 52, 57). Furthermore, both SIR2 and ZDS2 forestall aging (31, 52). These and other findings have led to models in which a specific type of heterochromatin mediates the repression of recombination and transcription in the rDNA, and the integrity of this heterochromatin is a primary determinant of longevity (26, 29). In sgs1 mutants, mitotic recombination in the rDNA is increased sevenfold (21) and longevity is reduced by 60% (55, 56). Paradoxically, ERCs accumulate to the same level in the presence and absence of Sgs1 (28, 46). The role of Sgs1 in rDNA silencing has not been investigated previously, despite its potential to provide insight into the mechanism by which Sgs1 represses rDNA recombination and aging.
In this study, we determined the effect of Sgs1 on the expression and transposition of Ty1 elements in rDNA and euchromatic DNA. Ty1 elements constitute one of five families of retrovirus-like transposons (Ty1 to Ty5) in yeast. Approximately 30 Ty1 elements reside at dispersed sites in the haploid genome, and the majority are transpositionally competent (12, 34). Ty1 elements consist of a central coding domain flanked by long terminal repeats (LTRs). The coding domain contains two overlapping open reading frames: TYA1, which encodes a capsid protein, and TYB1, which encodes protease, integrase (IN), and reverse transcriptase (RT). Ty1 elements are transcribed by Pol II to form a terminally redundant transcript. Ty1 RNA is translated into two proteins, TyA1 and the TyA1-TyB1 fusion protein, which assemble into virus-like particles (VLPs) that encapsulate Ty1 RNA during assembly. Maturation of VLPs occurs by protease-mediated processing of Ty1 precursor proteins, and it is required for synthesis of a full-length linear cDNA by RT, using Ty1 RNA as a template. Subsequently, the Ty1 cDNA and IN protein are transported to the nucleus, presumably as components of a preintegration complex (33, 48). IN mediates transposition of Ty1 cDNA at a nonhomologous target in the genome. Alternatively, Ty1 cDNA can recombine with genomic Ty1 elements, which occurs by IN-independent and Rad52-dependent mechanisms (53). The introduction of Ty1 cDNA into the genome by transposition or recombination is referred to as Ty1 cDNA-mediated mobility.
Several host mechanisms limit the potentially mutagenic effects of Ty1 transposition. First, Ty1 elements are preferentially targeted to genomic regions that do not encode proteins, including the upstream regions of Pol III-transcribed genes (16, 30). Second, Ty1 elements that are located in the rDNA array are subject to transcriptional silencing (6). Third, transposition is inhibited by host factors that block posttranscriptional steps in Ty1 replication. For example, Fus3, a mitogen-activated protein kinase, destabilizes VLP-associated proteins via its negative regulation of the invasive growth pathway (10). In addition, Ssl2 and Rad3, two helicase components of transcription factor TFIIH, promote degradation of Ty1 cDNA, thereby repressing transposition 100-fold or more (39). Finally, members of the Rad52 epistasis group, including Rad50, Rad51, Rad52, Rad54, and Rad57, reduce Ty1 cDNA levels and transposition by inhibiting cDNA synthesis or stability (50).
In this work, we demonstrate that Sgs1 inhibits the mobility of Ty1 elements by a novel mechanism. Initially, we proposed that Sgs1 might specifically repress transcription (and therefore transposition) of rDNA-Ty1 elements, since Sgs1 is required for the repression of rDNA recombination. To our surprise, we found that Sgs1 is not involved in the transcriptional silencing of Ty1 elements in the rDNA, although it is required for their mitotic stability. Furthermore, Sgs1 is a global inhibitor of Ty1 mobility that acts primarily at a step following cDNA synthesis. In the absence of Sgs1, extrachromosomal Ty1 cDNA molecules recombine at a high frequency, forming multimeric cDNA arrays that are subsequently integrated into the genome. We demonstrate that transposition of multimeric cDNA is the major cause of increased Ty1 mobility in sgs1Δ mutants.
MATERIALS AND METHODS
Yeast strains, media, and genetic procedures.
Standard yeast culture medium was prepared as described previously (51). The yeast strains used in this study are listed in Table 1. The mapping of Ty1his3AI-236r and Ty1his3AI-816r to the rDNA and Ty1his3AI-242 to a locus outside of rDNA on chromosome XII was described previously (6). Strain JC1109 is a haploid spore derived from a cross between strain JC1078 [MATa ura3(-52 or -167) trp1(-289 or :hisG) his3Δ200 ade2Δ:hisG Ty1ade2AI-515] and strain JC816 (6). The leu2:hisG allele was introduced into strains JC236, JC242, and JC1109 by two-step transplacement using plasmid pNK58, as described by Alani et al. (1). Subsequently, sgs1ΔLEU2 derivatives were constructed by one-step transplacement using a DNA fragment of plasmid pPWΔSGS1 (62). The sgs1ΔLEU2 strain JC3161 was constructed by the same method of transformation in strain YH8 (64). The sgs1ΔLEU2 disruption alleles were verified by Southern analysis. Strains containing the rad52:hisG allele were constructed by two-step transplacement using plasmid pBDG542 (12). To construct strains containing a HIS3-marked Ty1 element, the his3AI marker was replaced in strains JC236, JC242, and JC1109 and isogenic leu2:hisG sgs1ΔLEU2 derivatives by integrative transformation of a 0.8-kb ClaI fragment of plasmid pGEM-HIS3 containing the HIS3 allele (6).
TABLE 1.
Yeast strains
| Name | Relevant genotype | Reference or source |
|---|---|---|
| GRF167 | MATα his3Δ200 ura3-167 | 5 |
| DG789 | GRF167 spt3-101 | 14 |
| JC236 | GRF167 Ty1his3AI-236r | 14 |
| JC280 | GRF167 Ty1his3AI-236r ubc2Δ:hisG | 12 |
| JC1427 | GRF167 Ty1his3AI-236r leu2:hisG | This work |
| JC2359 | GRF167 Ty1his3AI-236r leu2:hisG sgs1ΔLEU2 | This work |
| MBY1487 | GRF167 Ty1HIS3-236r leu2:hisG | This work |
| MBY1049 | GRF167 Ty1HIS3-236r leu2:hisG sgs1ΔLEU2 | This work |
| JC242 | GRF167 Ty1his3AI-242 | 14 |
| JC544 | GRF167 Ty1his3AI-242 ubc2Δ:hisG | 12 |
| JC1430 | GRF167 Ty1his3AI-242 leu2:hisG | 9 |
| JC2360 | GRF167 Ty1his3AI-242 leu2:hisG sgs1ΔLEU2 | This work |
| JC2698 | GRF167 Ty1his3AI-242 leu2:hisG sgs1ΔLEU2 rad52:URA3 | This work |
| JC2712 | GRF167 Ty1his3AI-242 leu2:hisG rad52:URA3 | 9 |
| JC2148 | GRF167 Ty1his3AI-242 leu2:hisG tec1:URA3 | 10 |
| JC959 | GRF167 Ty1HIS3-242 | 6 |
| MBY1050 | GRF167 Ty1HIS3-242 leu2:hisG sgs1ΔLEU2 | This work |
| JC1109 | MATα his3Δ200 ura3(−52 or −167) trp1(−289 or :hisG) ade2Δ:hisG Ty1ade2AI-515 Ty1his3AI-816r | This work |
| JC2102 | MATα his3Δ200 ura3(−52 or −167) trp1(−289 or :hisG) ade2Δ:hisG Ty1ade2AI-515 Ty1his3AI-816r leu2:hisG | This work |
| JC2378 | MATα his3Δ200 trp1-289 ade2Δ:hisG ura3− Ty1ade2AI-515 Ty1his3AI-816r leu2:hisG sgs1 ΔLEU2 | This work |
| MBY1008 | MATα his3Δ200 trp1-289 ade2Δ:hisG ura3− Ty1ade2AI-515 Ty1HIS3-816r | This work |
| MBY1047 | MATα his3Δ200 trp1-289 ade2Δ:hisG ura3− Ty1ade2AI-515 Ty1HIS3-816r leu2:hisG sgs1 ΔLEU2 | This work |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Research Genetics |
| BY4742-10775 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sgs1ΔkanMX4 | Research Genetics |
| JS210-1 | MATα his3Δ200 ura3-167 trp1Δ63 leu2Δ1:mURA3-LEU2 | 57 |
| JS215-10 | MATα his3Δ200 ura3-167 trp1Δ63 RDN1:mURA3-LEU2 | 57 |
| YH8 | MATα his3Δ200 ura3-167 leu2Δ1 trp1Δ1 | 64 |
| JC3161 | MATα his3Δ200 ura3-167 leu2Δ1 trp1Δ1 sgs1ΔLEU2 | This work |
Plasmid construction.
Plasmid pJC573, a URA3-based integrating vector containing ≈1.2 kb of yeast genomic DNA sequences from the BIK1-HIS4 intergenic region (chromosome III, nucleotides 68462 to 69636) adjacent to a Ty1his3AI[Δ1] element, was constructed in two steps. First, an 8.9-kb XhoI-EagI fragment of plasmid pOY1 (38) including the BIK1-HIS4 intergenic region adjacent to a Ty1-H3/912 element marked with his3AI was subcloned into pRS406 (54). Second, a 0.9-kb ClaI fragment containing his3AI was deleted from this plasmid and replaced with a 0.9-kb ClaI fragment containing the modified retrotranscript indicator gene (RIG), his3AI[Δ1]. The his3AI[Δ1] RIG contains the same 104-bp artificial intron (AI) that is present in his3AI, but the intron has been relocated to position +440 in the HIS3 open reading frame. Consequently, the AI in his3AI[Δ1] is included within sequences that are deleted in the his3Δ1 allele in yeast strain BY4742 and derivatives. Use of the his3AI[Δ1] RIG in strains carrying the his3Δ1 allele prevents formation of His+ prototrophs by ectopic DNA-mediated recombination. The his3AI[Δ1] RIG was generously provided by D. Garfinkel (Frederick Cancer Research and Development Center, National Cancer Institute), and its construction will be described elsewhere.
Plasmid pGEM-URA3-HIS3 contains a URA3-HIS3 cassette in the vector pSP70 (Promega). It was constructed by subcloning a 1.3-kb BamHI-BglII fragment containing the URA3 allele into the unique BamHI site of plasmid pGEM-HIS3 (24). Plasmid pGEM-TYB1 contains the 934-bp HindIII-BglII fragment of Ty1-H3 (4) cloned into plasmid vector pSP70 (Promega).
Northern blot analysis.
Total RNA was isolated from yeast strains JC236, JC280, JC2359, JC242, JC544, and JC2360 as described previously (6). Northern blot analysis was performed as described previously (59). 32P-labeled RNA probes were used to detect Ty1his3AI, total Ty1, and PYK1 transcripts (13). Quantification was performed on a Storm 8600 phosphorimager using ImageQuant software.
Mitotic stability of Ty1HIS3 elements.
The mitotic stability of Ty1HIS3 elements was assayed as described previously (6). Briefly, single His+ colonies were inoculated into 10 ml of yeast extract-peptone-dextrose (YPD) medium and grown overnight at 30°C. Cultures were diluted 1:10,000 in fresh YPD and grown to saturation. A dilution of the ninth serial culture was plated on YPD medium and replicated to synthetic complete (SC) medium lacking histidine to determine the fraction of His− auxotrophs.
Phenotypic assay for expression of mURA3-LEU2.
SGS1 and congenic sgs1Δ strains containing the mURA3-LEU2 marker in rDNA or at the leu2Δ1 locus were constructed using standard genetic techniques (51). Strain BY4742-10775 (sgs1ΔkanMX4) was crossed to strain JS210-1 (mURA3-LEU2 at leu2Δ1) and strain JS215-10 (mURA3-LEU2 in rDNA) (57) to obtain SGS1 and sgs1ΔkanMX4 spores from the same tetrad. Individual spores were used to seed 10-ml cultures of YPD medium, which were grown to saturation at 30°C. Tenfold serial dilutions of each culture were made in sterile distilled water, and 5 μl of each dilution was spotted onto SC, SC-Ura, and 5-fluoro-orotic acid (5-FOA) agar.
Ty1 cDNA-mediated mobility assay.
The rate of His+ prototroph formation in strains containing a Ty1his3AI element was determined by the maximum-likelihood method (37). Strains JC236, JC2359, JC1109, JC2378, JC242, JC2360, JC2712, and JC2698 were grown to saturation in YPD medium at 30°C. For each strain, 9 or 11 tubes containing 2 ml of YPD medium were inoculated with ≈500 cells and grown to saturation at 20 or 23°C. The cell count in three cultures was determined by determining the titers on YPD plates. All cultures were subsequently plated to SC-His medium and grown at 30°C to quantify His+ prototrophs. SGS1 and sgs1ΔLEU2 congenic pairs were tested in the same experiment.
Protein analysis.
Western blot analyses were performed on whole-cell yeast extracts prepared from exponential-phase cultures as described previously (2), except that 50 μg of protein was analyzed per lane on a sodium dodecyl sulfate–10% polyacrylamide gel. Proteins were transferred to a polyvinylidene difluoride membrane and then incubated overnight with TyA1 antiserum (9) diluted 1:5,000. Horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (Amersham) diluted 1:2,500 was used as a secondary antibody. Subsequently, the membrane was incubated with goat antisera raised against N- and C-terminal peptides of Fus3 (Santa Cruz Biotechnology, Inc.), which were combined and used at a 1:2,000 dilution for 1 to 2 h. HRP-conjugated anti-goat antibody (Santa Cruz Biotechnology, Inc.) diluted 1:5,000 was used as a secondary antibody.
Subcellular fractions enriched for Ty1 VLPs were obtained by fractionating 105 A260 units of cell lysate from each strain on 20 to 75% sucrose step gradients, as described previously (9). Approximately 50 μg of protein from each VLP-enriched fraction was analyzed on a sodium dodecyl sulfate–8% polyacrylamide gel and then transferred to a polyvinylidene difluoride membrane. The membrane was incubated sequentially with TYA1 antiserum diluted 1:5,000, B2 antiserum (66) diluted 1:5,000, and B8 antiserum (23) diluted 1:5,000. HRP-conjugated anti-rabbit antibody diluted 1:2,500 was used as a secondary antibody.
Immune complexes were detected by incubating membranes with 1.25 mM Luminol (Sigma) in 0.1 M Tris-HCl (pH 8.5)–0.68 mM para-coumaric acid–0.09% hydrogen peroxide for 1 min, followed by autoradiography. Quantitation of specific bands detected by autoradiography was performed by scanning densitometry using a Howtek Scanmaster3+ and Scanalytics software.
Quantitation of Ty1 cDNA.
Two independent colonies of each yeast strain were grown on YPD medium at 20°C and then inoculated into a 10-ml culture of YPD medium and grown to saturation at 20°C. Genomic DNA was prepared from each culture as described previously (9). DNA samples were digested with PvuII and subjected to electrophoresis on a 1% Seakem-GTG agarose (FMC) gel, followed by transfer of the DNA to a Hybond N+ membrane (Amersham). A 32P-labeled riboprobe containing sense-strand TYB1 sequences was synthesized using plasmid pGEM-TYB1. Bands were quantified by phosphorimage analysis using a Molecular Dynamics Phosphorimager and ImageQuant software.
Ty1 integration into the CAN1 locus.
Strains JC242 and JC2360 were grown overnight in YPD medium at 30°C. Twenty 2-ml cultures of YPD were inoculated with ≈500 cells and grown at 20°C to saturation. The titers of four cultures were determined by plating dilutions on YPD medium, and the number of canavanine-resistant (Canr) cells in each culture was determined by plating on SC-Arg+Can medium. The rate of resistance to canavanine was calculated by the maximum-likelihood method (37). To determine the fraction of can1 mutations caused by insertion of a Ty1 element, one Canr colony was obtained from 40 independent cultures. Genomic DNA was prepared from each strain, and ≈250 ng of DNA was used as a template in PCR analysis with AX016 (GAAAATTTCGAGGAAGACGATAAGG) and AX019 (CAAATGCTTCTACTCCGTCTGC) to amplify a 2,265-bp fragment of the CAN1 gene (9). DNA samples that yielded no CAN1-specific PCR product were subjected to two additional PCR amplifications with the CAN1-specific primer AX016 or AX019 and a Ty1 LTR-specific oligomer, AX015 (GCCTTTATCAACAATGGAATCCC), to amplify a Ty1:can1 junction fragment, if present. The rate of Ty1 transposition into CAN1 in strain JC2360 was calculated by multiplying the rate of resistance to canavanine by the fraction of Canr strains that contained a Ty1 element within CAN1.
Ty1HIS3 cDNA-genomic Ty1his3AI element recombination assay.
Plasmid pJC573 was linearized at the PacI site in BIK1-HIS4 intergenic DNA and used to transform strains BY4742, BY4742-10775, YH8, and JC3161. Ura+ transformants were grown as large patches on SC-Ura medium at 30°C and then replicated to two YPD plates, one of which was grown at 20°C and the other of which was grown at 30°C. Following growth for 3 days, patches of cells were replicated to SC-Ura-His medium to select for His+ Ura+ colonies that sustained a Ty1HIS3 insertion and retained the Ty1his3AI[Δ1]-URA3 cassette. No His+ Ura+ prototrophs were detected following growth on YPD at 30°C, indicating that His+ Ura+ colonies that arose at 20°C were independent. His+ Ura+ colonies were transferred as small patches to YPD medium. Following growth at 30°C for 2 to 3 days, patches of cells were replicated to 5-FOA–His medium and grown for 3 days at 30°C. The fraction of His+ colonies that failed to generate His+ Ura− derivatives was determined.
PCR-based detection of multimeric Ty1HIS3 transposition events.
Independent His+ colonies that sustained an insertion of Ty1HIS3 by retrotransposition were obtained as follows. Yeast strain JC236 and isogenic sgs1Δ, rad52, and sgs1Δ rad52 derivatives were single-colony purified and then spread onto YPD plates and grown at 30°C. After 2 days, the cells were replicated to SC-His to check that no preexisting His+ colonies were present. Cells were also replicated to a fresh YPD plate and grown at 20°C for three days to induce transposition. The cells were subsequently replicated to SC-His medium and grown at 30°C. His+ papillae were single-colony purified, and cells in a medium-sized colony were lysed using Lyse-N-Go PCR Reagent (Pierce Chemical Co.), according to the manufacturer's specifications. Lysates were used in PCR amplification reactions with oligomers HISOUT-2 (GTACTAGAGGAGGCCAAGAG) and TYA1OUT-2 (TCTCTGGAACAGCTGATGAAG). Oligomers TEL1-F (CGGATTTCTGACGATATGGAC) and TEL1-R (ACCAACGTACTGAAGGTATCC), which amplify a 475-bp fragment of the TEL1 locus, were included in each PCR amplification to ensure that the genomic DNA was PCR competent. PCR mixtures were incubated at 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s, followed by 29 cycles in which the annealing temperature (63°C) was successively lowered by 0.3°C. A portion of each reaction mixture was analyzed on a 2% agarose gel.
Sequence analysis of 3′ Ty1HIS3:genomic DNA junctions.
Plasmid pGTy1-H3his3AI (14) was introduced into sgs1Δ strain JC3161. Ura+ transformants were grown on SC-Ura–2% galactose medium at 20°C for 4 days to induce expression of the pGTy1-H3his3AI element and subsequently plated on SC-His–2% glucose to isolate colonies containing Ty1HIS3 transpositions. Ura− segregants were identified following single-colony purification of His+ isolates on YPD plates. Independent His+ Ura− colonies containing a single >3.0-kb PvuII fragment that hybridized to a 32P-labeled HIS3 riboprobe in Southern blot analysis were identified. The 3′ Ty1HIS3:genomic DNA junction was cloned by integration and eviction of plasmid pGEM-URA3-HIS3 DNA linearized with NheI. The Ty1HIS3:genomic DNA junction fragments were evicted from pGEM-URA3-HIS3 transformants by digestion of genomic DNA that was prepared with AatII and ligation at <0.1 μg of DNA/μl. Ligation reactions were used to transform Escherichia coli to ampicillin resistance. DNA sequencing was performed with the HIS3-specific primer AX009 (CTTTATCAACAATGGAATCCC), and sequencing data were analyzed by BLASTN homology search with the Saccharomyces Genome Database (http://genome-www2.stanford.edu/cgi-bin/SGD/).
RESULTS
Mitotic stability and transcriptional silencing of Ty1 elements in rDNA.
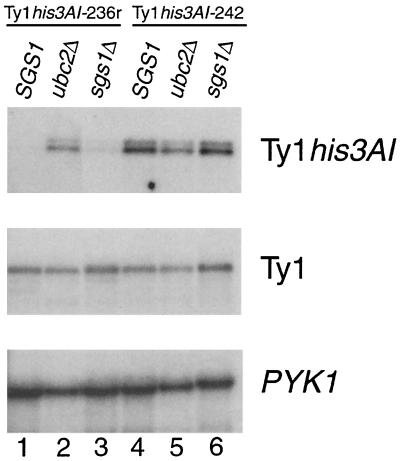
We demonstrated previously that Ty1 elements located in the rDNA are subject to transcriptional silencing (6). To monitor the effect of deleting SGS1 on transcriptional silencing of Ty1 elements in the rDNA, we quantified RNA from individual Ty1 elements marked with the his3AI gene and from genomic Ty1 elements collectively in wild-type, sgs1Δ, and ubc2Δ cells (Fig. 1). RNA of the rDNA element Ty1his3AI-236r was found at very low abundance in the SGS1 strain JC236 because of rDNA silencing, and it was increased only twofold in an isogenic sgs1Δ strain (Fig. 1, lanes 1 and 3). In contrast, deletion of UBC2, which is required for rDNA-specific transcriptional silencing (6), increased Ty1his3AI-236r RNA 13.3-fold (lanes 1 and 2). RNA of the euchromatic element Ty1his3AI-242 was not significantly altered by deletion of SGS1 (lanes 4 and 6) or UBC2 in strain JC242 (lanes 4 and 5). Deletion of SGS1 caused a minor increase in total Ty1 RNA levels in both strains (lanes 1, 3, 4, and 6), as did deletion of UBC2 (lanes 1, 2, 4, and 5). In summary, the data show that SGS1 is not involved in transcriptional silencing of rDNA Ty1 elements or in the regulation of total Ty1 RNA levels.
FIG. 1.
SGS1 is not required for transcriptional silencing of Ty1his3AI-236r in rDNA. Total RNA samples from strains JC236 and JC242 and sgs1Δ and ubc2Δ derivatives were analyzed by Northern blotting. The blot was hybridized to a 32P-labeled sense-strand HIS3 riboprobe to detect Ty1his3AI RNA (top), an antisense Ty1 riboprobe to detect total Ty1 RNA (middle), and an antisense PYK1 riboprobe as a loading control (bottom). The ratios of Ty1his3AI-236 to PYK1 RNA, normalized to the SGS1 strain (lane 1), were 13.3 (ubc2Δ; lane 2) and 2.0 (sgs1Δ; lane 3); the Ty1his3AI-242/PYK1 RNA ratios, normalized to SGS1 (lane 4), were 1.1 (ubc2Δ; lane 5) and 1.5 (sgs1Δ; lane 6); the Ty1/PYK1 RNA ratios, normalized to SGS1 (lane 1), were 1.5 (ubc2Δ; lane 2) and 1.7 (sgs1Δ; lane 3); and the Ty1/PYK1 RNA ratios, normalized to SGS1 (lane 4), were 1.5 (ubc2Δ; lane 5) and 1.7 (sgs1Δ; lane 6).
Because the mitotic stability of marker genes in rDNA is dependent on Sgs1 (21, 61), we expected that recombination of Ty1 elements in rDNA would also be repressed by Sgs1. To confirm this, the mitotic stability of Ty1 insertions was analyzed by replacing the his3AI marker in different Ty1 elements with a HIS3 allele. We then determined the frequency of HIS3 marker loss in isogenic SGS1 and sgs1Δ strains (Table 2). The rate of loss per generation of the rDNA element Ty1HIS3-236r was increased 2.9-fold, and that of the rDNA element Ty1HIS3-816r was increased 7.0-fold, in sgs1Δ mutants. In contrast, the rate of loss of Ty1HIS3-242, located outside the rDNA, was decreased fivefold in an sgs1Δ derivative. These data demonstrate that unlike previously analyzed regulators of rDNA silencing and recombination, Sgs1 represses recombination without affecting transcriptional silencing at the rDNA. Therefore, silencing of Pol II transcription and repression of rDNA repeat recombination are separable functions.
TABLE 2.
Mitotic stability of Ty1HIS3 elements
| Strain | Ty1HIS3 element | Relevant genotype | No. of His− auxotrophs/no. of cells analyzed (fraction)a | HIS3 loss/generation | Loss relative to SGS1 strain |
|---|---|---|---|---|---|
| MBY1487 | -236r | SGS1 | 105/1,124 (0.093) | 7.8 × 10−4 | |
| MBY1049 | -236r | sgs1Δ | 297/1,093 (0.272) | 2.3 × 10−3 | 2.9 |
| MBY1008 | -816r | SGS1 | 19/380 (0.050) | 4.2 × 10−4 | |
| MBY1047 | -816r | sgs1Δ | 367/1,053 (0.348) | 2.9 × 10−3 | 7.0 |
| JC959 | -242 | SGS1 | 137/3,079 (0.044)b | 3.7 × 10−4 | |
| MBY1050 | -242 | sgs1Δ | 10/972 (0.010) | 8.6 × 10−5 | 0.2 |
Determined after 120 generations of nonselective growth in YPD liquid medium. Fractions are averages from two or three independent cultures.
Includes data from reference 6.
Silencing of the mURA3 marker in rDNA.
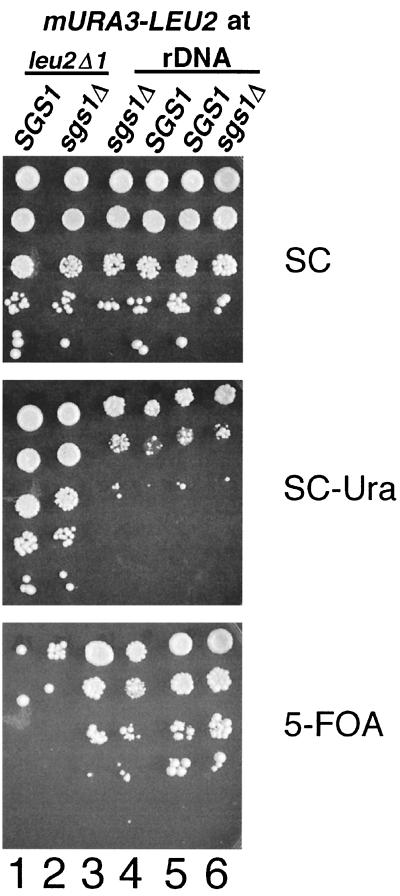
To corroborate the finding that SGS1 does not regulate transcriptional silencing of Ty1 elements in rDNA, we analyzed the effect of the sgs1Δ mutation on silencing of the mURA3 gene in rDNA using quantitative growth assays (Fig. 2). An mURA3-LEU2 cassette was integrated in rDNA or at the leu2Δ1 locus, and serial dilutions of cells from SGS1 and sgs1Δ spores containing the mURA3-LEU2 cassettes were plated on different media. In SGS1 strains, expression of mURA3 in rDNA was reduced relative to the same marker at leu2Δ1, resulting in relatively weaker growth on SC-Ura and stronger growth on 5-FOA medium. This result reflects the fact that mURA3 is subject to transcriptional silencing in the rDNA (57). Deletion of SGS1 did not relieve silencing of mURA3 in the rDNA (compare growth on SC, SC-Ura, and 5-FOA plates in columns 3 and 4 or 5 and 6 of Fig. 2). As expected, deletion of SGS1 also did not affect expression of mURA3 at leu2Δ1 (compare growth on SC, SC-Ura, and 5-FOA plates in columns 1 and 2). In summary, our data demonstrate that Sgs1 is not required for transcriptional silencing of the mURA3-LEU2 cassette or Ty1his3AI elements in rDNA.
FIG. 2.
Quantitative growth assay measuring the effect of sgs1Δ on silencing of the mURA3 gene in rDNA. Tenfold serial dilutions of saturated cultures of strains containing the mURA3-LEU2 cassette at leu2Δ1 (columns 1 and 2) or in rDNA (columns 3 to 6) were spotted onto SC, SC-Ura, and 5-FOA plates. The plates were incubated for 3 days at 30°C before being photographed. The strain pairs in columns 1 (SGS1) and 2 (sgs1Δ), columns 3 (sgs1Δ) and 4 (SGS1), and columns 5 (SGS1) and 6 (sgs1Δ) are progeny from the same tetrad.
Global increase in the cDNA-mediated mobility of Ty1his3AI elements in sgs1Δ mutants.
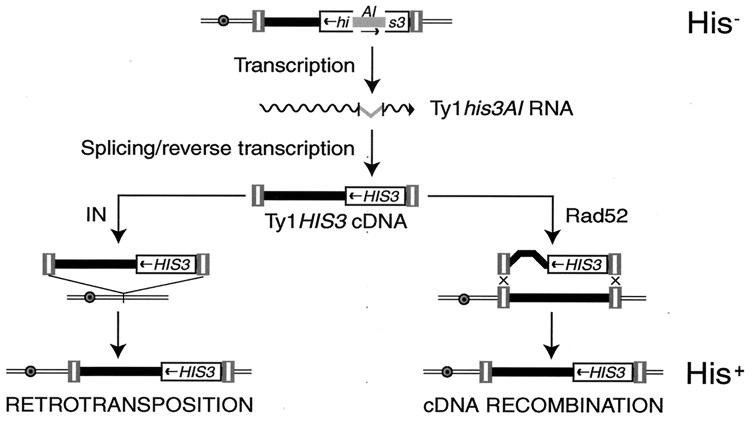
To determine if Sgs1 is involved in maintaining transpositional dormancy, we measured the effect of deleting SGS1 on the mobility of individual Ty1 elements marked with his3AI. The his3AI RIG allows the cDNA-mediated mobility of Ty1 elements to be detected in a quantitative phenotypic assay for His+ prototroph formation (Fig. 3). Because the formation of His+ prototrophs is absolutely dependent on splicing of the intron from the Ty1his3AI transcript (14), this assay detects only cDNA-mediated transposition or recombination events. The rate of His+ prototroph formation in a strain containing the rDNA element Ty1his3AI-236r was increased 79-fold as a result of deleting SGS1 (Table 3). Similarly, deletion of SGS1 resulted in a 43-fold increase in the cDNA-mediated mobility of a second element in the rDNA, Ty1his3AI-816r, and a 32-fold increase in the mobility of the euchromatic element, Ty1his3AI-242. These data demonstrate that Sgs1 is a global repressor of Ty1 mobility. Furthermore, the data are consistent with the finding that Sgs1 does not specifically affect expression of Ty1 elements in the rDNA. The increased Ty1 mobility in the absence of significantly elevated Ty1 RNA levels in sgs1Δ mutants (Fig. 1) suggests that Sgs1 inhibits a posttranscriptional step in Ty1 retrotransposition.
FIG. 3.
A phenotypic assay for Ty1 cDNA-mediated mobility events, which utilizes a genomic Ty1 element (tripartite rectangles flanking a black rectangle) marked with the his3AI RIG. his3AI consists of an AI interrupting the HIS3 coding sequence (broken rectangle). The AI is in the antisense orientation relative to HIS3 transcription; consequently, his3AI is nonfunctional and cells containing it, which also harbor a deletion of the chromosomal HIS3 locus, are phenotypically His−. The his3AI gene is placed within the Ty1 element in the opposing transcriptional orientation. Therefore, the AI is in the sense orientation in the Ty1his3AI RNA (wavy line) and can be removed by splicing. When the spliced transcript is used as a template for reverse transcription, a linear double-stranded Ty1 cDNA containing a functional copy of the HIS3 gene is formed. Insertion of Ty1HIS3 cDNA into the genome by IN-mediated transposition (left) or Rad52-mediated recombination with preexisting genomic Ty1 elements (right) results in formation of a His+ prototroph.
TABLE 3.
cDNA-mediated mobility of genomic Ty1his3AI elements
| Strain | Relevant genotype | His+ prototroph formation/cell/generation (mean ± SE)
|
Ratio of rates (sgs1Δ/SGS1) | |
|---|---|---|---|---|
| SGS1 | sgs1Δ | |||
| JC236 | Ty1his3AI-236r | (2.4 ± 1.6) × 10−8 | (1.9 ± 0.8) × 10−6 | 79 |
| JC1109 | Ty1his3AI-816r | (1.2 ± 0.88) × 10−8 | (5.1 ± 2.16) × 10−7 | 43 |
| JC242 | Ty1his3AI-242 | (2.4 ± 1.0) × 10−7 | (7.7 ± 3.0) × 10−6 | 32 |
| JC2712 | Ty1his3AI-242 rad52 | 5.9 × 10−6 | 1.0 × 10−5 | 1.7 |
Sgs1 affects a posttranslational step in retrotransposition.
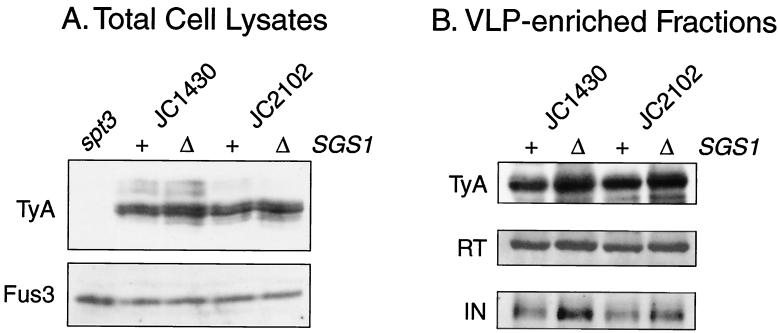
To determine if the sgs1Δ mutation affects the level of Ty1 proteins, total cell proteins isolated from two different SGS1 strains and isogenic sgs1Δ mutants were analyzed on a Western blot. The blot was probed with antiserum against TyA1 protein and subsequently with antiserum against Fus3, which served as a loading control (Fig. 4A). The levels of TyA1 protein relative to Fus3 in the sgs1Δ mutants were 1.0- and 2.0-fold that of TyA1 in the isogenic SGS1 strains. As a control, we showed that an spt3 mutant had undetectable levels of TyA1 protein. TyB1 proteins are present in very low levels in whole-cell extracts and are therefore difficult to detect. For that reason, we obtained subcellular fractions enriched for Ty1 VLPs from the same SGS1 and sgs1Δ strain pairs. Equal amounts of protein from each VLP-enriched fraction were analyzed with antisera directed against TyA1, IN, and RT (Fig. 4B). Only very minor increases in the levels of TyA1 (1.6- and 1.3-fold), p60-RT (1.5- and 1.6-fold), and p90-IN (2.3- and 2.0-fold) were observed in the sgs1Δ mutants. These data suggest that Sgs1 does not directly regulate the synthesis or stability of Ty1 proteins.
FIG. 4.
Ty1 protein levels in sgs1Δ mutants. Western blot analyses of Ty1 proteins in total cell lysates (A) and VLP-enriched subcellular fractions (B) from SGS1 strain JC1430, the isogenic sgs1Δ derivative JC2360, strain JC2102, and the isogenic sgs1Δ derivative JC2378 are shown. The spt3 strain DG789 was used as a negative control. (A) The Western blot of total cell lysates was probed sequentially with antisera specific for TyA1 and Fus3. (B) The Western blot of VLP fractions was probed sequentially with antisera against TyA1, RT, and IN.
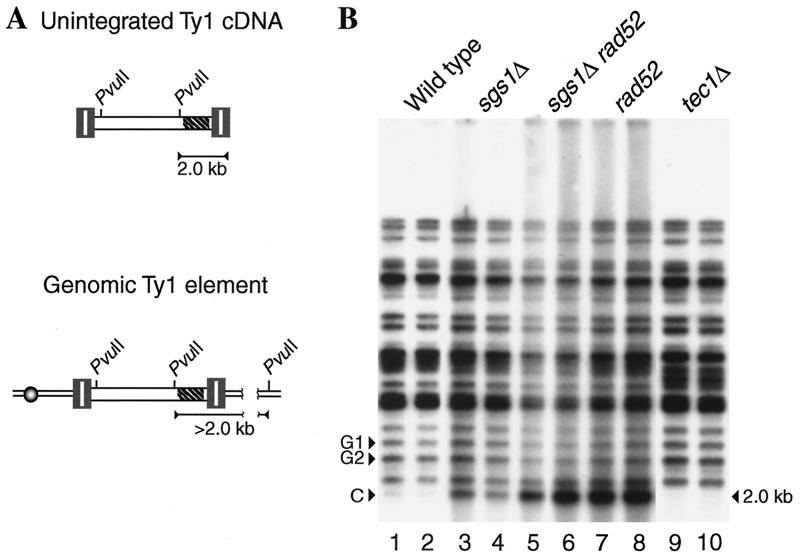
To determine if Sgs1 inhibits Ty1 transposition by promoting the degradation of Ty1 cDNA, we quantified unintegrated linear Ty1 cDNA in an sgs1Δ strain relative to that in an isogenic SGS1 strain by Southern analysis. A TYB1 probe hybridized to total cellular DNA digested with PvuII detected a 2.0-kb fragment of Ty1 linear extrachromosomal cDNA as well as multiple >2.0-kb PvuII fragments consisting of the junction between the 3′ end of a genomic Ty1 element and flanking DNA (Fig. 5A). Ty1 cDNA levels were determined by quantifying the intensity of the 2.0-kb Ty1 cDNA band (band C in Fig. 5B) relative to the intensities of two Ty1:genomic DNA junction bands (bands G1 and G2 in Fig. 5B) in two DNA samples from each strain. Deletion of SGS1 increased the level of Ty1 cDNA 2.6-fold. As a control, we showed that Ty1 cDNA was decreased 4.0-fold in a tec1 mutant, which has significantly reduced levels of Ty1 RNA (36). The simplest explanation for the 2.6-fold increase in cDNA is that it results from the minor increase in Ty1 proteins in the sgs1Δ mutant. Therefore, our results suggest that Sgs1 does not significantly inhibit the synthesis or stability of Ty1 cDNA.
FIG. 5.
Levels of unintegrated Ty1 cDNA in sgs1Δ, rad52, and sgs1Δ rad52 mutants. (A) The structures of unintegrated Ty1 cDNA and a genomic Ty1 element are shown. The hatched area within the Ty1 elements (tripartite rectangles bordering an open box) indicates the location of the TYB1 probe. The locations of relevant PvuII sites are indicated. The TYB1 riboprobe detects a 2.0-kb fragment from the PvuII site in Ty1 to the 3′ end of the unintegrated Ty1 cDNA and >2.0-kb PvuII fragments from genomic Ty1 elements. (B) Southern blot analysis of PvuII-digested genomic DNAs from two independent colonies of each strain grown at 20°C. The strains analyzed are wild-type strain JC1430 (lanes 1 and 2), an isogenic sgs1Δ derivative (JC2360; lanes 3 and 4), an sgs1Δ rad52 derivative (JC2698; lanes 5 and 6), a rad52 derivative (JC2712; lanes 7 and 8), and a tec1 derivative (JC2148; lanes 9 and 10). Two Ty1:genomic DNA junction fragments (G1 and G2) detected by the TYB1 riboprobe were used as controls in the quantification of the Ty1 cDNA band (C) by phosphorimage analysis.
Can the minor increase in Ty1 cDNA levels in the sgs1Δ mutant explain the 32- to 79-fold increases in transposition? We addressed this question by comparing the increase in Ty1 cDNA resulting from deletion of SGS1 to that resulting from loss of function of another characterized regulator of Ty1 transposition. Rad52 is an inhibitor of Ty1 transposition that reduces the level of Ty1 cDNA (50). Deletion of RAD52 in a strain harboring Ty1his3AI-242 resulted in a 25-fold increase in the rate of His+ prototroph formation, which was similar in magnitude to the 32-fold increase resulting from deletion of SGS1 in the same strain (Table 3). However, the level of Ty1 cDNA was increased 9.6-fold by deletion of RAD52, which was almost four times higher than the 2.6-fold increase resulting from deletion of SGS1 (Fig. 5B). Similar to a rad52 mutant, an isogenic sgs1Δ rad52 strain displayed an 11.3-fold increase in Ty1 cDNA, suggesting that deletion of SGS1 resulted in little or no increase in Ty1 cDNA in the absence of Rad52. In summary, even though the repression of Ty1 mobility by Rad52 was similar in magnitude to the repression caused by Sgs1, Rad52 mediated a significant reduction in Ty1 cDNA levels, whereas Sgs1 mediated a relatively minor reduction. The data suggest that Sgs1 inhibits the mobility of Ty1 elements at a step following cDNA accumulation. Therefore, Sgs1 may regulate the insertion of Ty1 cDNA into the genome.
Integration of Ty1 in sgs1Δ mutants.
The hypothesis that Sgs1 represses integration of Ty1 cDNA was explored using two different assays. First, a PCR-based assay was employed to detect de novo Ty1 integration events upstream of Pol III transcription units (38; M. Bryk and M. J. Curcio, unpublished results). Unselected integration events upstream of two types of targets, the 16 glycyl-tRNA genes or the 100 to 200 5S rRNA genes, were detected in DNA samples prepared from cells grown at 20°C, the permissive temperature for transposition. No significant change in the pattern or intensity of integration events was observed at either target set in an sgs1Δ mutant compared to an isogenic SGS1 strain (data not shown). The same conclusion was drawn from quantitation of spontaneous Ty1 insertions into the selectable target gene, CAN1, in SGS1 and isogenic sgs1Δ strains. Loss-of-function mutations in CAN1 cause resistance to canavanine. Deletion of SGS1 resulted in a 2.4-fold increase in the rate of resistance to canavanine; however, the fraction of canavanine-resistant mutants with a Ty1 insertion in CAN1 was not significantly increased in the sgs1Δ strain (1/40) relative to the isogenic SGS1 strain (0/40). The rate of Ty1 transposition into CAN1 in the sgs1Δ strain JC2360 (1.15 × 10−8) was equivalent to that previously measured in a wild-type strain and was significantly lower than that in a fus3Δ mutant, despite the fact that the mobility of Ty1his3AI elements is elevated to similar levels in sgs1Δ and fus3Δ mutants (9). In summary, these data indicate that deletion of SGS1 does not significantly increase the frequency of Ty1 integration at preferred or selected target sites.
Recombination between Ty1 cDNA and genomic Ty1 elements.
In the absence of an increase in Ty1 integration, enhanced recombination of Ty1 cDNA might explain how Ty1 cDNA-mediated mobility events are increased in sgs1Δ mutants. Rad52 is required for homologous recombination of cDNA (15, 53). Therefore, we determined whether Rad52 is required for the increase in Ty1HIS3 cDNA-mediated mobility events in an sgs1Δ mutant. Deletion of SGS1 in the rad52 strain JC2712, which contains Ty1his3AI-242, increased the rate of His+ formation 1.7-fold (Table 3). This is a minor increase compared to the 32-fold increase resulting from deletion of SGS1 in the isogenic RAD52 strain JC242. Hence, the data suggest that Rad52 is required for most of the stimulation of Ty1HIS3 cDNA-mediated mobility in sgs1Δ mutants. Therefore, Sgs1 may repress recombination of Ty1 cDNA with genomic Ty1 elements or recombination between Ty1 cDNA molecules prior to integration.
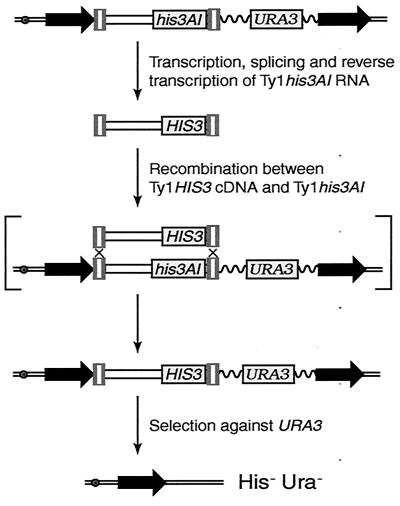
To differentiate between these models, we developed an assay to measure recombination between Ty1 cDNA and a genomic Ty1 element (Fig. 6). A cassette containing Ty1his3AI and URA3 between 1.2-kb direct repeats of BIK1-HIS4 intergenic DNA was introduced into yeast by integration of plasmid pJC573. The cassette was used to determine how frequently Ty1HIS3 cDNA recombines with the genomic Ty1his3AI element in SGS1 and sgs1Δ strains. Recombination of Ty1HIS3 cDNA with the Ty1his3AI element would give rise to a His+ cell that contained Ty1HIS3 adjacent to URA3 and flanked by the 1.2-kb direct repeats. Therefore, selection for loss of URA3 by recombination between the flanking direct repeats would result in concomitant loss of the functional HIS3 gene.
FIG. 6.
Assay for recombination between Ty1HIS3 cDNA and a genomic Ty1his3AI element. Yeast strains containing direct repeats of a 1.2-kb region of yeast DNA (horizontal black arrows) flanking a Ty1his3AI element and the URA3 gene were constructed as described in Materials and Methods. Splicing of AI from Ty1his3AI RNA and subsequent reverse transcription result in formation of Ty1HIS3 cDNA. The Ty1HIS3 cDNA can transpose into nonhomologous target sites (not illustrated) or recombine with genomic Ty1 elements, including the Ty1his3AI element that is linked to URA3. One possible mechanism of recombination in which Ty1HIS3 cDNA initiates gene conversion of the Ty1his3AI element is illustrated in brackets. Gene conversion would result in replacement of Ty1his3AI with a Ty1HIS3 element adjacent to URA3. Subsequent selection for the loss of URA3 by recombination between the flanking direct repeats would result in concomitant loss of the Ty1HIS3 element. Therefore, cosegregation of Ura− and His− phenotypes would be observed. Wavy line, plasmid vector sequences; double line with open circle, chromosome III.
Strains containing the Ty1his3AI-URA3 cassette were grown at 20°C on rich medium, and then independent His+ Ura+ colonies that sustained a Ty1HIS3 cDNA-mediated mobility event were selected. To determine if the His+ Ura+ strains could give rise to His+ Ura− derivatives that retain a genomic Ty1HIS3 element but have lost the Ty1his3AI-URA3 cassette, each isolate was transferred to 5-FOA–His medium. Inability of strains to grow on 5-FOA–His medium indicated that HIS3 was cosegregating with URA3, which was the phenotype expected if Ty1HIS3 cDNA had recombined with the Ty1his3AI element next to URA3. Out of 305 His+ Ura+ colonies screened from the SGS1 strain BY4742, 3 (1%) were His− Ura− cosegregants. Similarly, 3 out of 286 (1%) independent His+ Ura+ colonies of the isogenic sgs1Δ strain were His− Ura− cosegregants. When the Ty1his3AI-URA3 cassette was integrated in a second strain background, 4 out of 101 (4%) His+ Ura+ colonies of the SGS1 strain YH8 were His− Ura− cosegregants, whereas 5 out of 103 (5%) His+ Ura+ colonies of the isogenic sgs1Δ strain were His− Ura− cosegregants. In summary, these findings demonstrate that deletion of SGS1 does not stimulate recombination between Ty1 cDNA and genomic Ty1 elements.
Formation of multimeric Ty1 arrays during transposition in sgs1Δ mutants
Our results suggested that Sgs1 inhibits a Ty1 cDNA-mediated mobility process that is Rad52 dependent but does not involve recombination of Ty1 cDNA with genomic Ty1 elements. Therefore, we hypothesized that Ty1 cDNA molecules undergo intermolecular recombination in sgs1Δ mutants, forming tandem Ty1 arrays that transpose into the genome. This process would increase the number of Ty1 cDNA molecules per transposition event in sgs1Δ mutants.
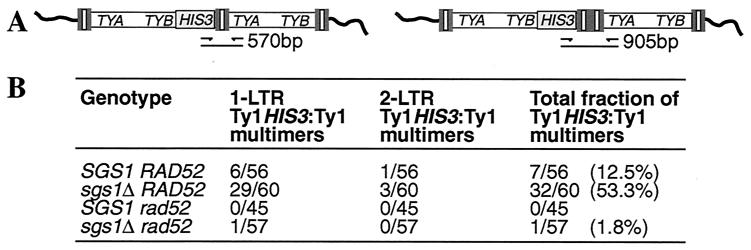
To test this hypothesis, we isolated spontaneous His+ prototrophs from strains containing the chromosomal Ty1his3AI-236r element and determined if the transposed Ty1HIS3 insertion was present in a Ty1 tandem array. His+ prototrophs generated from a genomic Ty1his3AI element typically contain only one HIS3-marked Ty1 at a dispersed genomic location (14). DNA derived from independent His+ strains was subjected to PCR analysis using a HIS3-specific primer and a TYA1-specific primer. These primers allowed detection of Ty1 arrays containing the HIS3 marker in the upstream Ty1 element (Fig. 7A). In the SGS1 strain, 12.5% of independent His+ isolates contained a Ty1HIS3:Ty1 multimer (Fig. 7B). The fraction of Ty1HIS3:Ty1 arrays was increased more than fourfold in an isogenic sgs1Δ derivative, to 53.3% of His+ isolates. The majority of PCR products derived from Ty1HIS3:Ty1 multimers were of a size consistent with the presence of one LTR at the junction between Ty1 elements. These multimers can be explained as arising by homologous recombination between LTRs of different Ty1 cDNA molecules. However, one His+ isolate from the SGS1 strain and three from the isogenic sgs1Δ strain gave rise to a PCR product indicative of two LTRs at the junction between Ty1 elements (Fig. 7). Two-LTR Ty1 arrays probably arose by end joining of LTR sequences.
FIG. 7.
Deletion of SGS1 increases genomic Ty1HIS3:Ty1 multimers. (A) Tandem arrays of Ty1 elements, consisting of LTRs (tripartite rectangles) bordering a central domain that carries TYA1 and TYB1 (open box), within chromosomal DNA (curved line). The left schematic represents a dimeric Ty1 array with a single shared LTR between each coding domain and the HIS3 marker adjacent to the upstream TYB1 domain (one-LTR Ty1HIS3:Ty1 multimer). The right schematic represents a Ty1 dimer with two joined LTRs between coding domains (two-LTR Ty1HIS3:Ty1 multimer). PCR primers that hybridize to the HIS3 marker gene and to the TYA1 domain amplify a 570-bp DNA fragment of a one-LTR Ty1HIS3:Ty1 element and a 905-bp DNA fragment of a two-LTR Ty1HIS3:Ty1 element. (B) Independent His+ revertants of the SGS1 RAD52 strain JC236 and isogenic mutant derivatives were analyzed by PCR to determine the fraction of His+ revertants that contained a one-LTR Ty1HIS3:Ty1 multimer, a two-LTR Ty1HIS3:Ty1 multimer, or either.
Rad52 is required for increased mobility of Ty1 elements in sgs1Δ mutants (Table 3). If transposition of Ty1HIS3:Ty1 multimers is responsible for the increased mobility, then we would expect multimer formation in an sgs1Δ mutant to be Rad52 dependent as well. In fact, formation of Ty1HIS3:Ty1 multimers was reduced 30-fold in the sgs1Δ strain when RAD52 was disrupted. Moreover, formation of Ty1 multimers was abolished in an SGS1 rad52 strain (Fig. 7B). Taken together, our results demonstrate that Ty1 cDNA forms multimers by a Rad52-dependent process that is stimulated in sgs1Δ mutants.
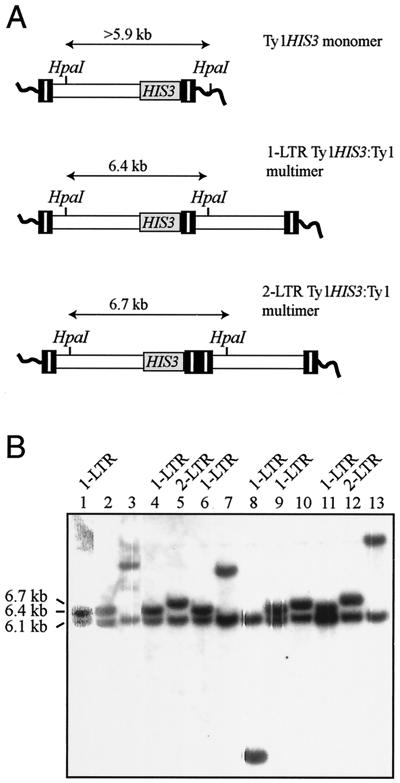
To extend our understanding of the fate of Ty1 cDNA in an sgs1Δ mutant, 13 of the His+ prototrophs that were analyzed by PCR were also subjected to Southern analysis. Genomic DNA was digested with HpaI, which cleaves once in Ty1 DNA. Hybridization to a HIS3 probe detected a 6.1-kb band derived from Ty1his3AI-236r and a second, variably sized band derived from the transposed Ty1HIS3 element in all 13 DNA samples (Fig. 8). The presence of two HIS3 bands in each DNA sample indicated that none of the 13 genomic Ty1HIS3 elements had replaced the Ty1his3AI-236r element. This finding was consistent with data presented above demonstrating that deletion of SGS1 does not stimulate gene conversion of genomic Ty1 elements. DNA from each of the five His+ prototrophs that contained a monomeric Ty1HIS3 element yielded a 6.5- to >12-kb band representing the 3′ junction of Ty1HIS3 with genomic DNA at different target sites (Fig. 8B, lanes 1, 2, 7, 10, and 13). DNAs from seven of the eight remaining strains yielded the predicted HpaI fragment for one-LTR Ty1HIS3:Ty1 multimers (6.4 kb) and two-LTR Ty1HIS3:Ty1 multimers (6.7 kb). The eighth strain had a 3.3-kb HpaI band, indicative of a deletion or rearrangement within the multimeric array (Fig. 8B, lane 8). The presence of only one Ty1HIS3:genomic DNA junction band in addition to the Ty1his3AI-236r band in all eight strains demonstrates that Ty1 multimers are heterogeneous arrays containing a Ty1HIS3 element and one or more unmarked Ty1 elements. The same conclusion was drawn from analysis of six isolates of the isogenic SGS1 strain containing Ty1HIS3:Ty1 multimers (data not shown). These data confirm the validity of the PCR assay for detecting Ty1HIS3 multimers. Furthermore, they demonstrate that Ty1HIS3:Ty1 multimers form by intermolecular recombination between a Ty1HIS3 cDNA molecule and unmarked Ty1 cDNA molecules prior to or during integration into the genome.
FIG. 8.
Southern blot analysis of sgs1Δ strains that sustained a Ty1HIS3 transposition event. (A) Three schematics showing the size of the HIS3-containing HpaI fragment from a monomeric Ty1HIS3 element within chromosomal DNA or a one-LTR or two-LTR Ty1HIS3:Ty1 multimer. (B) Results of hybridization of a 32P-labeled HIS3 riboprobe to HpaI-digested genomic DNA from 13 sgs1Δ Ty1his3AI-236 strains harboring a Ty1HIS3 element. DNA samples containing a one-LTR or two-LTR Ty1HIS3:Ty1 multimer, as demonstrated by PCR (Fig. 7), are labeled accordingly. The sizes of the HpaI bands derived from Ty1his3AI-236 (6.1 kb) and from one-LTR (6.4 kb) and two-LTR (6.7 kb) Ty1HIS3:Ty1 multimers are indicated. The sizes of the bands derived from monomeric Ty1HIS3 elements, which are expected to be >5.9 kb, are not indicated.
Preferred target sites for Ty1 transposition in sgs1Δ mutants.
We have observed a stimulation of Ty1 mobility in sgs1Δ mutants that is not associated with elevated Ty1 integration into preferred or selected target sites. However, it is possible that transposition into a novel target, which would not be detected by assays for Ty1 integration at glycyl-tRNA, 5S rRNA, or CAN1 genes, occurs at a high frequency in sgs1Δ mutants. To test this hypothesis, unselected genomic DNA targets of Ty1 transposition were identified in sgs1Δ mutants. Independent His+ colonies that harbored Ty1HIS3 transpositions were obtained following galactose induction of a plasmid-borne PGAL1Ty1-H3his3AI fusion element in the sgs1Δ strain JC3161. Plasmid segregants were analyzed by Southern blotting with a HIS3 probe to identify strains containing one Ty1HIS3 element in genomic DNA. Of 56 PvuII fragments derived from Ty1HIS3 elements that were detected, 21 (37.5%) were 3.0 kb, which is the length expected for Ty1HIS3 elements within multimeric Ty1 arrays. Five strains were identified that contained one >3.0-kb PvuII fragment, indicating the presence of a single 3′ Ty1HIS3:genomic DNA junction fragment. The junction fragment was cloned from each of the five strains by plasmid integration and eviction, and the site of Ty1HIS3 integration into genomic DNA was determined by sequencing and comparison to the Saccharomyces Genome Database. Four of the five Ty1HIS3 elements analyzed were located upstream of Pol III-transcribed genes (Table 4), which are preferred target sites for Ty1 transposition in SGS1 strains. The fifth Ty1HIS3 element analyzed was adjacent to the beginning of the TYA2 open reading frame of a Ty2 element. Ty2 elements are highly related to Ty1 elements and have nearly identical LTRs (34). Thus, this Ty1HIS3 element is a component of a heterogeneous Ty multimer containing an upstream Ty1HIS3 element and a downstream Ty2 element separated by a single LTR. In summary, this analysis failed to reveal a novel target site preference for Ty1 transposition in sgs1Δ mutants. As predicted by the effect of Sgs1 on spontaneous transposition, expression of the inducible PGAL1Ty1-H3his3AI element in an sgs1Δ mutant resulted in a high frequency of Ty1 multimer transposition. We conclude that transposition of Ty1 cDNA multimers into typically preferred transposition targets is primarily responsible for the increase in Ty1 cDNA-mediated mobility in sgs1Δ mutants.
TABLE 4.
Locations of Ty1HIS3 elements in an sgs1Δ strain
| Ty1HIS3 insertion | Chromosome | Nucleotide coordinate 3′ of Ty1 insertiona | Neighboring Pol III-transcribed gene (encoded RNA) | Distance (bp) upstream of Pol III-transcribed gene |
|---|---|---|---|---|
| -3 | V | 99793 | tM(CAU)E (tRNAMet) | 341 |
| -22 | XII | rDNA | RDN5 (5S rRNA) | 125 |
| -56 | VIII | 475922 | tV(CAC)H (tRNAVal) | 148 |
| -59 | XII | 1052246 | tI(AUU)L2 (tRNAIle) | 106 |
Chromosomal coordinates are those provided in the Saccharomyces Genome Database (http://genome-www2.stanford.edu/cgi-bin/SGD/).
DISCUSSION
Although host-mediated regulation of Ty1 transposition has been evoked for some time, the identity and role of the host regulators have only recently begun to be elucidated. The findings presented here demonstrate that Sgs1 is involved in maintaining transpositional dormancy by a novel mechanism. Sgs1 does not regulate transcriptional silencing of Ty1 elements located in the rDNA, even though it represses rDNA recombination. This finding indicates that Sgs1 inhibits rDNA recombination by a mechanism that is independent of maintaining the rDNA heterochromatin. Moreover, Sgs1 does not have a major effect of the levels of Ty1 RNA, proteins, cDNA, or integration. However, Sgs1 was shown to inhibit recombination of extrachromosomal Ty1 cDNA molecules during transposition. Our data suggest that repression of cDNA recombination by Sgs1 limits the number of individual Ty1 cDNA molecules that are involved in each transposition event.
Sgs1 represses mitotic recombination without affecting gene silencing in the rDNA.
Our findings demonstrate that the mechanism by which Sgs1 represses recombination in the rDNA is independent from the mechanism of maintaining rDNA silencing. The three- to sevenfold increase in the loss of rDNA-Ty1HIS3 elements in sgs1Δ mutants (Table 2) is comparable to the sevenfold increase in loss of marker genes in the rDNA that was observed previously (21). Variation in the rate of loss may be due to the different locations of individual Ty1HIS3 elements within the rDNA gene array. In contrast to the requirement for Sgs1 in the mitotic stability of rDNA-Ty1 elements, Sgs1 was not required for and may inhibit the stability of euchromatic Ty1 elements.
Despite the role of Sgs1 in repressing rDNA recombination, deletion of SGS1 did not derepress transcription of Ty1 elements (Fig. 1) or the mURA3 marker gene (Fig. 2) in rDNA. Transcriptional silencing in rDNA is dependent on a unique type of heterochromatin (6, 57). Several modulators of chromatin structure have been shown to promote rDNA silencing and increase longevity (6, 20, 52, 57, 58). For example, the histone deacetylase Sir2 is a dosage-dependent regulator of rDNA silencing and longevity that also functions at sites of DNA damage (20, 31, 44, 47, 60). Arguably, loss of rDNA silencing promotes aging by allowing an increase in rDNA recombination and ERC formation (26, 29). However, our findings demonstrate that aging and rDNA recombination can be stimulated without perturbing rDNA silencing. Therefore, the role of Sgs1 in repressing rDNA recombination may be related to its function in initiating the repair of DNA lesions during replication (7). One model previously proposed to explain the stimulation of rDNA recombination in sgs1 mutants is that an accumulation of DNA damage outside the rDNA results in sequestration of Sir2 away from the nucleolus. Accordingly, rDNA silencing is perturbed and rDNA recombination is stimulated (26). Because this model predicts that rDNA silencing would be disrupted in sgs1 mutants, it is not consistent with our observations. An alternative model is that elevated rDNA recombination is a consequence of DNA lesions within the rDNA in sgs1 mutants. When Sgs1 cannot initiate repair of these lesions during DNA replication, recombination between rDNA repeats may provide an alternative mechanism to purge the lesions. A recent report suggests that much of the rDNA recombination in sgs1Δ mutants occurs via the single-strand annealing pathway, which could lead to enhanced rDNA recombination without stimulating ERC formation (46). Hence, this model provides an explanation for the stimulation of rDNA recombination in the absence of an effect on ERC formation and rDNA silencing in sgs1Δ mutants. It does not readily provide an explanation for the shortened life span of sgs1 mutants, which probably involves a high incidence of mitotic arrest (46).
Rad52 is required for increased Ty1 cDNA mobility in sgs1Δ mutants.
In strains containing Ty1his3AI elements at different locations, deletion of SGS1 caused a marked increase in the cDNA-mediated mobility of Ty1 elements (Table 3) accompanied by a twofold or lower increase in Ty1 RNA (Fig. 1). Hence, Sgs1 may be a weak inhibitor of the transcription or stability of Ty1 RNA. Ty1 protein levels (Fig. 4) and cDNA levels (Fig. 5) are also slightly elevated in sgs1Δ mutants, probably as a direct result of the small increase in Ty1 RNA, although this cannot be determined with certainty. Several findings indicate that modestly elevated Ty1 cDNA levels are not the major reason for the increase in Ty1 mobility in sgs1Δ mutants. First, no significant stimulation of Ty1 integration upstream of glycyl-tRNA genes or 5S rRNA genes or into the CAN1 locus was observed in sgs1 mutants. These findings do not support a simple model in which elevated levels of Ty1 cDNA result in an increase in the number of integration events in sgs1Δ mutants. Second, the level of Ty1 cDNA in an sgs1Δ mutant was about one-quarter of that in a rad52 mutant (Fig. 5), even though both mutations stimulated the mobility of Ty1his3AI-242 to an equivalent level (Table 3). This finding suggests that there is a second mechanism of increasing the cDNA-mediated mobility of Ty1 elements in sgs1Δ mutants aside from the small increase in Ty1 cDNA. Third, deletion of SGS1 in a rad52 strain did not cause a significant increase in the mobility of Ty1his3AI-242 or the level of Ty1 cDNA, indicating that Sgs1 inhibits a Rad52-dependent mechanism of Ty1 mobility.
Sgs1 inhibits transposition of multimeric Ty1 cDNA.
We envisaged two Rad52-dependent pathways for entry of Ty1 cDNA into the genome that might be stimulated in the absence of Sgs1. First, recombination of Ty1 cDNA with genomic Ty1 elements might be enhanced, resulting in introduction of Ty1HIS3 cDNA into the genome at an elevated rate. Notably, genomic multimers of Ty5 elements are commonly formed by “ends-out” recombination between Ty5 cDNA and the upstream LTR of a genomic Ty5 element (32). However, this possibility was excluded for Ty1 multimers, since recombination between Ty1HIS3 cDNA and a genomic Ty1his3AI element was not affected by deletion of SGS1. Second, intermolecular recombination between extrachromosomal Ty1 cDNA molecules including Ty1HIS3 cDNA might be stimulated in sgs1 mutants, leading to transposition of tandem Ty cDNA arrays. In fact, the fraction of His+ strains that contain Ty1HIS3 as a component of a multimeric array was fourfold higher in sgs1Δ mutants (Fig. 7). The PCR assay we used detected multimers minimally containing a Ty1HIS3:Ty1 array. However, the total number of Ty1 elements within each multimer could be higher, especially in sgs1Δ strains. Hence, elevated Ty1 mobility in sgs1Δ mutants could result from increases both in the fraction of multimeric transposition events and in the average number of Ty1 cDNA molecules per multimer. Formation of Ty1 multimers required Rad52 in both SGS1 and sgs1Δ strains. Since the increase in Ty1 mobility in sgs1Δ mutants is also dependent on Rad52, our data suggest that transposition of Ty1 multimers is the primary cause of the elevated Ty1 mobility in sgs1Δ mutants.
Independent Ty1 multimers in eight sgs1Δ strains and six SGS1 strains all contained one Ty1HIS3 element and one or more unmarked Ty1 elements. Therefore, Ty1 multimer formation involves two or more different cDNA molecules. This conclusion is also supported by the isolation of a Ty1HIS3:Ty2 multimer in an sgs1Δ mutant. The evidence for heterogeneous Ty arrays eliminates models in which multimeric cDNA is formed by rolling-circle replication of a monomeric Ty1HIS3 cDNA. Moreover, it supports a model in which intermolecular recombination between different Ty1 cDNA molecules results in the formation of linear cDNA arrays. Crossover recombination between the 3′ and 5′ LTRs of different Ty1 cDNA molecules would result in tandem cDNA arrays separated by one LTR. More rarely, end joining of cDNA molecules would result in tandem arrays separated by two LTRs. Since the Ty1HIS3 cDNA derived from a single genomic Ty1his3AI element is a small fraction of the total Ty1 cDNA from approximately 30 genomic Ty1 elements, multimers selected for the presence of Ty1HIS3 are likely to contain an unmarked Ty1 element. Ty1HIS3:Ty1, Ty1:Ty1HIS3, or larger heterogeneous multimers could form. Integration of linear multimeric cDNA at preferred target sites would then be carried out by Ty1 IN. This model explains how Ty1HIS3 mobility is elevated in sgs1Δ mutants in the absence of a significant increase in the frequency of Ty1 integration or a change in integration target specificity. Transposition occurs at far less than one event per cell in normal yeast strains. Because HIS3-marked cDNA is a small fraction of the total Ty1 cDNA, it undergoes transposition only a small percentage of the time. Concerted insertion of multiple cDNA molecules per transposition event in sgs1Δ mutants would increase the probability of a HIS3-marked Ty1 cDNA entering the genome.
The fact that Ty1 multimeric arrays are products of Ty1 mobility in both SGS1 and sgs1Δ cells suggests that a normal mechanism of multimer formation is enhanced in the absence of Sgs1. Previously, tandem arrays of Ty1 elements that transposed into the HMLα locus and upstream of a promoterless his3Δ4 allele were isolated (63). In the latter case, Ty multimers represented 5.4% of the transposition events and consisted of two or three Ty elements separated by one LTR. These findings support the notion that extrachromosomal multimeric Ty1 cDNA can be integrated into de novo target sites in normal yeast cells.
Another prediction of our model is that extrachromosomal multimeric Ty1 cDNA, the proposed intermediate in Ty1 multimer transposition, is present in sgs1Δ mutants. However, we have failed to detect extrachromosomal multimeric Ty1 cDNA in repeated Southern hybridization experiments (data not shown), which may indicate that this intermediate is short-lived. In contrast, unintegrated monomeric Ty1 cDNA is reproducibly detected in both sgs1Δ and SGS1 strains (Fig. 5). Perhaps most of the monomeric Ty1 cDNA is located in the cytoplasm, whereas multimeric cDNA may be present only in the nucleus, as it requires recombination or end joining for its formation. In the absence of physical evidence for the extrachromosomal multimeric Ty1 cDNA, we cannot rule out other models to explain Ty1 multimer formation. For example, integration of a monomeric Ty1 cDNA could result in formation of a favored recombination substrate in sgs1Δ mutants (63). The single-stranded gaps created when IN joins the cDNA to a staggered cut in target DNA may not be efficiently repaired in sgs1Δ mutants. Unrepaired integration lesions could then promote recombination between Ty1 cDNA and the LTRs of the newly integrated Ty1 elements, resulting in Ty1 multimer formation. This model of integration-coupled recombination still involves the use of multiple cDNA molecules in a single integration event and does not involve recombination of cDNA with preexisting genomic Ty1 elements. However, it cannot easily explain the formation of two-LTR Ty1 multimers, which occurs at a low frequency in both sgs1Δ and SGS1 strains (Fig. 7).
Direct or indirect interaction of Sgs1 with Ty1 cDNA?
Our finding that extrachromosomal Ty1 cDNA recombines at a high frequency in sgs1Δ mutants raises the possibility that Sgs1 interacts directly with Ty1 cDNA. Assuming the interaction is direct, we propose the following model to explain how Sgs1 represses recombination between Ty1 cDNA molecules. Although a fraction of Ty1 cDNA in normal cells enters the genome by integration or recombination, most of it is degraded (39). Hence, some of the nuclear cDNA may not be protected by Ty1 IN and instead may be recognized as a recombination substrate. By analogy to a chromosomal double-strand break, free cDNA ends are likely to be potent initiators of recombination. Since 5′-end resection is the initial step in all pathways of double-strand break repair (27), the cDNA ends would be resected by a 5′-to-3′ exonuclease activity, leaving 3′ single-stranded DNA tails. The single-stranded 3′ ends would be able to invade homologous Ty1 cDNA molecules and initiate recombination, thereby forming structures in which the DNA strands from different molecules are annealed. Sgs1 may bind these branched structures and unwind the annealed strands to prevent homologous recombination. In fact, purified Sgs1 has been shown to bind preferentially to forked DNA substrates (3). Following unwinding by Sgs1, partially degraded Ty1 cDNA molecules may lack the terminal nucleotides required for recognition by Ty1 IN and be completely degraded. In the absence of Sgs1, homologous recombination between the LTR sequences of extrachromosomal Ty1 cDNAs results in the formation of multimeric Ty1 arrays with double-stranded DNA ends restored by gap filling. Hence, these multimers would be suitable substrates for Ty IN to carry out integration.
Because Ty1 cDNA is extrachromosomal and does not contain autonomously replicating sequences, it is not expected to undergo DNA replication. Therefore, interaction of Sgs1 with Ty1 cDNA would constitute a novel role for Sgs1 outside of its characterized role as a component of the pathway that monitors DNA replication fork progression (19). The identification of factors that inhibit Ty1 mobility along with Sgs1 would help to determine how Sgs1 represses Ty1 cDNA recombination. One interesting candidate is the yeast Ku70 protein, a component of the Ku heterodimer that binds to double-strand breaks and is required for nonhomologous end joining (43). Ku70 has been shown to bind to Ty1 cDNA and to be required for the high levels of transposition that result from induction of a PGAL1Ty1 element (17). Notably, the human Ku heterodimer associates with the human RecQ homologue WRN and specifically stimulates its exonuclease activity (11, 41). Perhaps Sgs1 and Ku associate and bind the ends of Ty1 cDNA to prevent recombination and/or promote integration.
Alternatively, the effect of deleting SGS1 on Ty1 cDNA recombination may be indirect. An elevated level of chromosomal lesions that cannot be repaired in sgs1Δ mutants might signal the induction of recombination proteins that enhance intermolecular cDNA recombination. In fact, stochastic cell cycle arrest characteristic of sgs1Δ mutants is mediated through the RAD9-dependent DNA damage checkpoint pathway (46). Perhaps specific recombination functions are induced through this pathway in the absence of Sgs1.
Implications for the host cell and Ty1 retrotransposons.
The results presented here implicate a host mechanism in the repression of Ty1 cDNA recombination and the transposition of monomeric Ty1 elements. This is the first evidence that the host cell actively represses Ty1 cDNA recombination, and it suggests that transposition of Ty1 multimers may be disadvantageous to the host. Previous results have indicated the importance of the Ty1 IN protein in sequestering cDNA from homologous recombination pathways (53). When IN-mediated integration is blocked, Ty1 cDNA forms complex multimeric arrays by homologous recombination. Notably, strains containing genomic insertions of complex Ty1 arrays at independent locations had a growth defect, suggesting that large multimers can perturb some feature of chromosome organization. In addition, it has been demonstrated that multimeric Ty1 insertions can disrupt the silent chromatin domain of the HMLα locus, whereas single Ty1 or solo-LTR derivatives of the multimers restore silencing (45).
Transposition of monomeric cDNA is likely to be important for the maintenance of retrotransposons as well. Although our data indicate that multimeric Ty1 insertions arise spontaneously, they are not a stable component of the S. cerevisiae genome (34). Hence, they may be reduced to monomeric Ty1 elements or LTR derivatives by recombination at a relatively high frequency. Therefore, transposition of Ty1 multimeric arrays is not likely to be an efficient mechanism for Ty1 to spread throughout the genome.
ACKNOWLEDGMENTS
We thank Ed Louis, Jeff Smith, and David Garfinkel for supplying plasmids and strains used in this study, the Wadsworth Center Molecular Genetics Core Facility for oligonucleotide synthesis and DNA sequencing, and Abram Gabriel, John Mueller, and Keith Derbyshire for critical reading of the manuscript. Moreover, we are grateful to Fred Winston and Winston lab members for support and encouragement.
M. Bryk is the recipient of a Special Fellowship from the Leukemia & Lymphoma Society. M. Bryk's research in Fred Winston's lab was funded by grant GM32967 from the National Institutes of Health. This work was funded by National Institutes of Health grant GM52072 to M. J. Curcio.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkin A L, Altamura N, Leeds P, Culbertson M R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett R J, Sharp J A, Wang J C. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J Biol Chem. 1998;273:9644–9650. doi: 10.1074/jbc.273.16.9644. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Eichinger D, Castrillon D, Fink G R. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol Cell Biol. 1988;8:1432–1442. doi: 10.1128/mcb.8.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke J D, Garfinkel D J, Styles C A, Fink G R. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 6.Bryk M, Banerjee M, Murphy M, Knudsen K E, Garfinkel D J, Curcio M J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 7.Chakraverty R K, Hickson I D. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 8.Christman M F, Dietrich F S, Fink G R. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55:413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- 9.Conte D, Barber E, Banerjee M, Garfinkel D J, Curcio M J. Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol Cell Biol. 1998;18:2502–2513. doi: 10.1128/mcb.18.5.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conte D, Curcio M J. Fus3 controls Ty1 transpositional dormancy through the invasive growth MAPK pathway. Mol Microbiol. 2000;35:415–427. doi: 10.1046/j.1365-2958.2000.01710.x. [DOI] [PubMed] [Google Scholar]
- 11.Cooper M P, Machwe A, Orren D K, Brosh R M, Ramsden D, Bohr V A. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- 12.Curcio M J, Garfinkel D J. Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics. 1994;136:1245–1259. doi: 10.1093/genetics/136.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curcio M J, Garfinkel D J. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol Cell Biol. 1992;12:2813–2825. doi: 10.1128/mcb.12.6.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curcio M J, Garfinkel D J. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derr L K, Strathern J N. A role for reverse transcripts in gene conversion. Nature. 1993;361:170–173. doi: 10.1038/361170a0. [DOI] [PubMed] [Google Scholar]
- 16.Devine S E, Boeke J D. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 17.Downs J A, Jackson S P. Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Mol Cell Biol. 1999;19:6260–6268. doi: 10.1128/mcb.19.9.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis N A, Groden J, Ye T Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 19.Frei C, Gasser S M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 20.Fritze C E, Verschueren K, Strich R, Esposito R E. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 23.Garfinkel D J, Hedge A M, Youngren S D, Copeland T D. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991;65:4573–4581. doi: 10.1128/jvi.65.9.4573-4581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garfinkel D J, Mastrangelo M F, Sanders N J, Shafer B K, Strathern J N. Transposon tagging using Ty elements in yeast. Genetics. 1988;120:95–108. doi: 10.1093/genetics/120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottlieb S, Esposito R E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 26.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 27.Haber J E. Partners and pathways: repairing a double-strand break. Trends Genet. 2000;16:259–264. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 28.Heo S-J, Tatebayashi K, Ohsugi I, Shimamoto A, Furuichi Y, Ikeda H. Bloom's syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells. 1999;4:619–625. doi: 10.1046/j.1365-2443.1999.00288.x. [DOI] [PubMed] [Google Scholar]
- 29.Jazwinski S M. Metabolic control and ageing. Trends Genet. 2000;16:506–511. doi: 10.1016/s0168-9525(00)02119-3. [DOI] [PubMed] [Google Scholar]
- 30.Ji H, Moore D P, Blomberg M A, Braiterman L T, Voytas D F, Natsoulis G, Boeke J D. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 31.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ke N, Voytas D F. cDNA of the yeast retrotransposon Ty5 preferentially recombines with substrates in silent chromatin. Mol Cell Biol. 1999;19:484–494. doi: 10.1128/mcb.19.1.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenna M A, Brachmann C B, Devine S E, Boeke J D. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol. 1998;18:1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J M, Vanguri S, Boeke J D, Gabriel A, Voytas D F. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 35.Kitao S, Lindor N M, Shiratori M, Furuichi Y, Shimamoto A. Rothmund-thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics. 1999;61:268–276. doi: 10.1006/geno.1999.5959. [DOI] [PubMed] [Google Scholar]
- 36.Laloux I, Dubois E, Dewerchin M, Jacobs E. TEC1, a gene involved in the activation of Ty1 and Ty1-mediated gene expression in Saccharomyces cerevisiae: cloning and molecular analysis. Mol Cell Biol. 1990;10:3541–3550. doi: 10.1128/mcb.10.7.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 38.Lee B-S, Lichtenstein C P, Faiola B, Rinckel L A, Wysock W, Curcio M J, Garfinkel D J. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics. 1998;148:1743–1761. doi: 10.1093/genetics/148.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee B S, Bi L, Garfinkel D J, Bailis A M. Nucleotide excision repair/TFIIH helicases RAD3 and SSL2 inhibit short-sequence recombination and Ty1 retrotransposition by similar mechanisms. Mol Cell Biol. 2000;20:2436–2445. doi: 10.1128/mcb.20.7.2436-2445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S-K, Johnson R E, Yu S-L, Prakash L, Prakash S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- 41.Li B, Comai L. Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J Biol Chem. 2000;275:28349–28352. doi: 10.1074/jbc.C000289200. [DOI] [PubMed] [Google Scholar]
- 42.Lohman T M, Bjornson K P. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 43.Lustig A J. The Kudos of non-homologous end-joining. Nat Genet. 1999;23:130–131. doi: 10.1038/13755. [DOI] [PubMed] [Google Scholar]
- 44.Martin S G, Laroche T, Suka N, Grunstein M, Gasser S M. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 45.Mastrangelo M F, Weinstock K G, Shafer B K, Hedge A M, Garfinkel D J, Strathern J N. Disruption of a silencer domain by a retrotransposon. Genetics. 1992;131:519–529. doi: 10.1093/genetics/131.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McVey M, Kaeberlein M, Tissenbaum H A, Guarente L. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics. 2001;157:1531–1542. doi: 10.1093/genetics/157.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mills K D, Sinclair D A, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 48.Moore S P, Rinckel L A, Garfinkel D J. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol. 1998;18:1105–1114. doi: 10.1128/mcb.18.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puranam K L, Blackshear P J. Cloning and characterization of RecQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 50.Rattray A J, Shafer B K, Garfinkel D J. The Saccharomyces cerevisiae DNA recombination and repair functions of the RAD52 epistasis group inhibit Ty1 transposition. Genetics. 2000;154:543–556. doi: 10.1093/genetics/154.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 52.Roy N, Runge K W. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr Biol. 2000;10:111–114. doi: 10.1016/s0960-9822(00)00298-0. [DOI] [PubMed] [Google Scholar]
- 53.Sharon G, Burkett T J, Garfinkel D J. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14:6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinclair D A, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 56.Sinclair D A, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 57.Smith J S, Boeke J D. An unusual form of transcriptional silencing in the yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 58.Smith J S, Caputo E, Boeke J D. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swanson M S, Malone E A, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 61.Watt P M, Hickson I D, Borts R H, Louis E J. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watt P M, Louis E J, Borts R H, Hickson I D. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 63.Weinstock K G, Mastrangelo M F, Burkett T J, Garfinkel D J, Strathern J N. Multimeric arrays of the yeast retrotransposon Ty. Mol Cell Biol. 1990;10:2882–2892. doi: 10.1128/mcb.10.6.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu H, Boeke J D. Host genes that influence transposition in yeast: the abundance of a rare tRNA regulates Ty1 transposition frequency. Proc Natl Acad Sci USA. 1990;87:8360–8364. doi: 10.1073/pnas.87.21.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamagata K, Kato J-I, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc Natl Acad Sci USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Youngren S D, Boeke J D, Sanders N J, Garfinkel D J. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol Cell Biol. 1988;8:1421–1431. doi: 10.1128/mcb.8.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu C E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]