Abstract
Affinity maturation is a key technique in protein engineering which is used to improve affinity and binding interactions in vitro, a process often required to fulfil the therapeutic potential of antibodies. There are many available display technologies and maturation methods developed over the years, which have been instrumental in the production of therapeutic antibodies. However, due to the inherent limitations in display capacity of these technologies, accommodation of expansive and complex library builds is still a challenge. In this article, we discuss our recent efforts in the affinity maturation of a difficult antibody lineage using an unbiased approach, which sought to explore a larger sequence space through the application of DNA recombination and shuffling techniques across the entire antibody region and selections using ribosome display. We also highlight the key features of several display technologies and diversification methods, and discuss the strategies devised by different groups in response to different challenges. Particular attention is drawn to examples which are aimed at the expansion of sequence, structural or experimental diversity through different means and approaches. Here, we provide our perspectives on these methodologies and the considerations involved in the design of effective strategies for the directed evolution of antibodies.
Keywords: affinity maturation, antibody engineering, display technologies
Antibodies are an increasingly important class of therapeutic molecules. Over the past decades we have seen a tremendous increase in the number of antibody drugs that were approved for clinical use in almost all disease areas, as we learnt to harness their unique properties to address different therapeutic needs [1,2]. There are many ways to generate antibodies in vivo and in vitro, the most common being immunisation and phage display; but the antibodies isolated from these methods often require extra steps to improve their affinities and/or drug-like properties, to fulfil the potency required in a therapeutic setting. In vitro affinity maturation usually involves a diversification of the antibody base sequence, followed by stringent selections to isolate higher-affinity binders, a directed evolution process much like the somatic hypermutation that naturally occurs in mammalian B cells [3]. With a wide choice of available technologies, there is a myriad of possible paths when it comes to affinity maturation, but how does one decide which approach to take?
In a recent report, we described the use of an unbiased approach to affinity maturation for the optimisation of an inhibitory antibody specific to Arginase 2 (ARG2) [4]. The premise of the project was to generate a therapeutic which neutralises the extracellular ARG2 secreted by cancer cells, thought to mediate an immunosuppressive response through the depletion of arginine [5]. In this context, high-affinity binding and a potent inhibitory effect are essential properties for the therapeutic, which is ideal for an antibody approach. An antibody candidate was first isolated from AstraZeneca's naïve phage display libraries [6], which showed specific binding to human ARG2 and inhibitory activity in an enzymatic assay in vitro [7]. This antibody was then taken through a comprehensive affinity maturation process, in which each of the six complementarity-determining regions (CDRs) were targeted for diversification and taken through selections in parallel. Surprisingly, little improvement in antibody affinity or potency were obtained. Moreover, a random error-prone mutagenesis approach was also unsuccessful, suggesting that this was a difficult antibody to affinity mature.
Given their ineffectiveness, we thought that perhaps the approaches taken, which only focused on small regions of the antibody at a time, were insufficient to produce appreciable improvements to this antibody lineage. Taking inspiration from the somatic recombination process in vivo, in which modular segments of immunoglobulin domains are rearranged to generate a diverse repertoire, we used antibody chain shuffling [8–10] and a staggered-extension process (StEP) [11,12] to recombine mutations sampled from all six CDRs (Figure 1). This recombination created fresh combinatorial diversity and produced antibody variants with mutations that span the length of the antibody construct in an unbiased way. Ribosome display was used for the selection of these libraries because it is capable of displaying such diverse library builds. Candidates with the most improved characteristics were identified and subsequently fine-tuned via a pool maturation technique to obtain further affinity gains. This resulted in a panel of high affinities antibodies, with over 50-fold improvements in affinity and potency estimated in the final leads.
Figure 1. Overview of the affinity maturation process in the optimisation of an ARG2-inhibitory antibody.
The antibody construct, in the single-chain variable fragment (scFv) format, was diversified and recombined in a comprehensive affinity maturation process. Mutations sampled from all six CDRs were recombined and shuffled in an unbiased way via chain-shuffling and StEP recombination, and selected using ribosome display. The most improved leads were then pooled and diversified by error-prone PCR, which introduced random mutations through the length of the construct. The resulting antibodies from these selections have a high number of mutations, which are scattered throughout the length of the constructs and across different CDRs.
Sequence and structural comparison of the parental and affinity-matured antibodies revealed that the antibody has gone through some very extensive changes during the affinity maturation process [4]. Substantial mutations across several regions of the antibody were translated into a large epitope shift that facilitated increases in the interface area and shape complementarity to the antigen, whilst preserving the key contacts of a hydrophobic cleft that is essential for its inhibitory mechanism [7]. Essentially, these changes have enabled the antibody to re-orient itself into a position allowing for superior binding without changing the observed mechanism of action, and in doing so seemed to have overcome some initial limitations of the parent. Indeed, there is evidence to suggest significant negative cooperativity in the binding mode of the parent antibody to trimeric ARG2, which was aptly resolved after affinity maturation. Such issues were unlikely to be overcome by small or focused changes, which might have been why the standard methods were ineffective. In contrast, the multiple changes applied simultaneously across the antibody provided sufficient scope to explore a larger region of combinatorial space, to escape the restrictions of the parental antibody and provide an improved binding solution.
It is extraordinary how such a solution would have presented itself through selections; nor could we have predicted or designed this. Based on the structural data, we might have attempted to improve the binding interactions at or around the key contacts of the hydrophobic cleft, which would have been unproductive or counterproductive. Indeed, our results suggested that mutations to CDRH3 of the antibody, which formed a large part of the hydrophobic cleft, were not tolerated and were rapidly eliminated during selections of the unbiased recombination libraries. There is existing evidence to suggest that feasible regions for affinity maturation are often not involved in key contacts, but lie in positions that provide indirect effects or establish fresh new interactions with its antigen [13–15]. Moreover, improved affinities can arise from multiple diverse mechanisms which are often unpredictable [15–17]. In vitro selections provide a way to probe the vast number of binding possibilities, with the potential to find the best available binding solution without requiring prior assumptions or dictation.
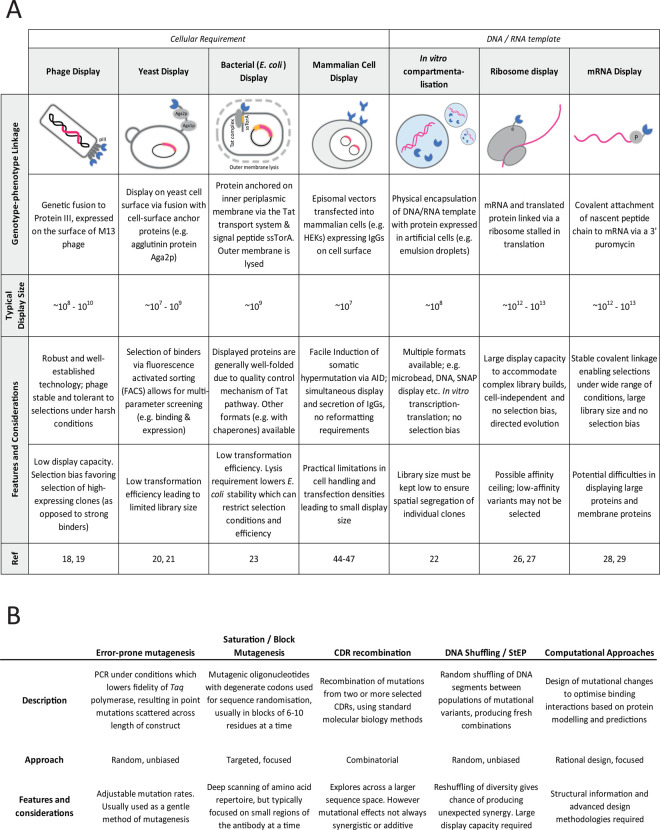
Given its utility, it is important that efforts are made to find the most suitable affinity maturation strategy for any given application. A diagram highlighting several features and considerations in the use of different technologies is shown in Figure 2. Generally, a key objective of affinity maturation is to maximise the sequence diversity of the initial library repertoire, as this would be translated to a higher structural diversity from which superior binders may be selected. However, we are often limited by the display capacity of the technology used. Methods with a cellular requirement, such as phage and yeast display, have a lower display size (typically 108–109) due to limitations in transformation efficiency [18–23]. It is not possible to sample every mutation at every position at the same time, which would require a theoretical capacity of ∼1078 for ∼60 positional variants across six CDRs. Besides the common compromise of limiting mutations to certain CDRs, there have been reports of alternative strategies to overcome this obstacle. Tiller et. al. recognised that combinations of mutations from different CDRs is important at an early stage [13]. To accommodate early re-combinatorial changes, they used alanine scanning to narrow down the permissive sites for mutagenesis, before generating libraries with restricted amino acid variants based on natural CDR diversity. Similarly, the ‘look-through mutagenesis’ method restricts the number of amino acid variants to nine representative residues based on side-chain chemistry, which allowed for the simultaneous mutation of positions in up to three CDRs [24]. Based on amino acid usage analyses, Gonzalez-Munoz et al. [25] selected a subset of seven amino acid variants that were typically over-presented from large datasets of affinity-matured antibodies. This tailored diversification approach allowed them to cover more positional ground with fewer library builds, with comparable effectiveness to full amino acid randomisation.
Figure 2. Comparison of (A) different display technologies and (B) in vitro diversification and maturation methods.
A variety of technologies and methods with different strengths and limitations; features which may guide the design of an effective affinity maturation strategy.
Cell-free systems such as ribosome and mRNA display have a much higher display capacity in the range of 1012–1013 [26–29], and are favoured methods for the exploration of larger sequence space. Direct comparisons of phage and ribosome display methods have suggested higher diversity and affinity gains in the outputs of the latter [30,31], which lends itself to a broader range of applications. A recent study described the use of insertion and deletion (InDel) mutagenesis to create large diversified libraries in the affinity maturation of an anti-IL-13 antibody [32]. Random in-frame InDels were introduced using a transposon-based system and selected using ribosome display, which uncovered positions of tolerance and allowed for the exploration of loop length variation on maturation outcomes. This is a particularly interesting application as increasing evidence have shown the importance of InDels in antibody maturation, both in vivo and in vitro [33–35]. Lengths of CDRs, particularly that of CDR3, vary considerably in nature [36–39], yet much of our focus during antibody engineering have tended to remain on point substitutions. There is evidence that unconventional loop lengths may confer advantages for antibodies against challenging antigens, such as G-protein-coupled receptors (GPCRs) [40,41] and rapidly evolving pathogens (influenza and HIV-1) [35,42]. The likely expansion of structural or conformational diversity that may be attained through length diversification makes it an attractive strategy for affinity maturation.
Ribosome display is also ideal for the selection of libraries diversified through DNA shuffling. In one example, this was achieved through the random digestion by DNAse I followed by enzymatic ligation to recombine point mutations accumulated from error-prone selections [43], with interesting parallels to our approach. Such recombination and shuffling methods have the potential to eliminate deleterious mutations, as a result of backcrossing with original template DNA segments in the sequence pool. Moreover, spontaneous mutations which occur through the numerous amplification steps through the selection and recovery cycles further adds to the diversity, promoting the simultaneous evolution of non-targeted regions, with favourable implications for directed evolution.
Progressive or continuous diversification at the same time as selections is a clear advantage for in vitro maturation, as it allows for the gradual emergence of epistatic and synergistic mutations, lessening the demand on the diversity of the initial library repertoire. This is exemplified in the case of mammalian cell display, which, despite having a small display size, can be induced to diversify in vitro through the addition of activation-induced cytidine deaminase (AID) [44–47]. Full-length IgG or Fabs are typically expressed on the mammalian cell surface, and between rounds of selections and sorting, AID can be introduced to induce somatic hypermutation in situ, without further requirements for reformatting or library builds. This can be viewed as an evolving library; with diversification concurrent with selections. Advancements in the use of gene editing techniques have also allowed for a more directed approach. A recent report describes the use of TALE nucleases and CRISPR-Cas9 to promote site-specific integration of antibody gene populations; allowing for the creation of large diversified libraries which were successfully used to affinity mature a PD1-blocking antibody using mammalian cell display [48].
The increased capacity for diversification is particularly advantageous for optimisation strategies seeking to cover a broader ground. While it is widely established that CDRs are major determinants of antigen recognition, there is evidence to suggest that framework (FR) regions may also play an important role. In particular, a loop identified in the FR-3 region has been known to exhibit CDR-like characteristics, in terms of sequence and structural diversity as well as antigen binding, and is sometimes referred to as the ‘CDR4’ [49–52]. Other FR regions in the VH and VL domains may also contribute to antigen binding, through direct antigen contact or distal effects [53–55]. Pairings of different VH and VL frameworks has been shown to affect antigen and Fc receptor (FcR) engagement [56], and conversely isotype selection for the constant region can also influence antigen binding [57–59]. Such observations remind us to think of an antibody as a whole protein during engineering, with interconnected domains that can exert influence on each other [60].
The numerous display and maturation strategies, each with their own unique features, provide us with an extensive toolkit to fulfil our antibody engineering needs. Exploration of a larger experimental and sequence space has the clear ability to provide a wider range of conceivable binding solutions, with the potential to deliver greater improvements. On the other hand it can also lead to increased functional divergence or thermostability trade-offs, which may require additional screening steps or compensatory mechanisms [61]. As exemplified by the many different approaches devised in response to different challenges, it is about choosing the right strategy for the antibody lineage under examination. Assessment of antigenic properties, choice of an optimal diversification strategy paired with appropriate display and screening methods, adaptation of selection strategy as the protein evolves; these are all important considerations which are key to the success of affinity maturation.
Acknowledgements
The Cancer Research UK-AstraZeneca Antibody Alliance Laboratory is a long-term strategic alliance jointly supported by Cancer Research UK and AstraZeneca.
Abbreviations
- AID
activation-induced cytidine deaminase
- ARG2
Arginase 2
- CDRs
complementarity-determining regions
- Fab
antigen-binding fragment
- FcR
Fc receptor
- FR
Framework
- GPCR
G-protein-coupled receptors
- IgG
immunoglobulin
- InDels
insertions and deletions
- scFv
single-chain variable fragment
- StEP
staggered-extension process
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
Both authors contributed to the design, writing and revision of the manuscript.
References
- 1.Kaplon, H., Muralidharan, M., Schneider, Z. and Reichert, J.M. (2020) Antibodies to watch in 2020. MAbs 12, 1703531 10.1080/19420862.2019.1703531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu, R.M., Hwang, Y.C., Liu, I.J., Lee, C.C., Tsai, H.Z., Li, H.J.et al. (2020) Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 27, 1 10.1186/s12929-019-0592-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Noia, J.M. and Neuberger, M.S. (2007) Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76, 1–22 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- 4.Chan, D.T.Y., Jenkinson, L., Haynes, S.W., Austin, M., Diamandakis, A., Burschowsky, D.et al. (2020) Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation. Proc. Natl Acad. Sci. U.S.A. 117, 16949–16960 10.1073/pnas.1919565117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mussai, F., De Santo, C., Abu-Dayyeh, I., Booth, S., Quek, L., McEwen-Smith, R.M.et al. (2013) Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood 122, 749–758 10.1182/blood-2013-01-480129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaughan, T.J., Williams, A.J., Pritchard, K., Osbourn, J.K., Pope, A.R., Earnshaw, J.C.et al. (1996) Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat. Biotechnol. 14, 309–314 10.1038/nbt0396-309 [DOI] [PubMed] [Google Scholar]
- 7.Austin, M., Burschowsky, D., Chan, D.T.Y., Jenkinson, L., Haynes, S., Diamandakis, A.et al. (2020) Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action. MAbs 12, 1801230 10.1080/19420862.2020.1801230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockmann, E.C., Akter, S., Savukoski, T., Huovinen, T., Lehmusvuori, A., Leivo, J.et al. (2011) Synthetic single-framework antibody library integrated with rapid affinity maturation by VL shuffling. Protein Eng. Des. Sel. 24, 691–700 10.1093/protein/gzr023 [DOI] [PubMed] [Google Scholar]
- 9.Kang, A.S., Jones, T.M. and Burton, D.R. (1991) Antibody redesign by chain shuffling from random combinatorial immunoglobulin libraries. Proc. Natl Acad. Sci. U.S.A. 88, 11120–3 10.1073/pnas.88.24.11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshinaga, K., Matsumoto, M., Torikai, M., Sugyo, K., Kuroki, S., Nogami, K.et al. (2008) Ig L-chain shuffling for affinity maturation of phage library-derived human anti-human MCP-1 antibody blocking its chemotactic activity. J. Biochem. 143, 593–601 10.1093/jb/mvn009 [DOI] [PubMed] [Google Scholar]
- 11.Zhao, H. (2004) Staggered extension process in vitro DNA recombination. Methods Enzymol. 388, 42–49 10.1016/S0076-6879(04)88005-4 [DOI] [PubMed] [Google Scholar]
- 12.Zhao, H. and Zha, W. (2006) In vitro ‘sexual’ evolution through the PCR-based staggered extension process (StEP). Nat. Protoc. 1, 1865–1871 10.1038/nprot.2006.309 [DOI] [PubMed] [Google Scholar]
- 13.Tiller, K.E., Chowdhury, R., Li, T., Ludwig, S.D., Sen, S., Maranas, C.D.et al. (2017) Facile affinity maturation of antibody variable domains using natural diversity mutagenesis. Front. Immunol. 8, 986 10.3389/fimmu.2017.00986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zahnd, C., Spinelli, S., Luginbuhl, B., Amstutz, P., Cambillau, C. and Pluckthun, A. (2004) Directed in vitro evolution and crystallographic analysis of a peptide-binding single chain antibody fragment (scFv) with low picomolar affinity. J. Biol. Chem. 279, 18870–7 10.1074/jbc.M309169200 [DOI] [PubMed] [Google Scholar]
- 15.Midelfort, K.S. and Wittrup, K.D. (2006) Context-dependent mutations predominate in an engineered high-affinity single chain antibody fragment. Protein Sci. 15, 324–334 10.1110/ps.051842406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra, A.K. and Mariuzza, R.A. (2018) Insights into the structural basis of antibody affinity maturation from next-generation sequencing. Front. Immunol. 9, 117 10.3389/fimmu.2018.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeliazkov, J.R., Sljoka, A., Kuroda, D., Tsuchimura, N., Katoh, N., Tsumoto, K.et al. (2018) Repertoire analysis of antibody CDR-H3 loops suggests affinity maturation does not typically result in rigidification. Front. Immunol. 9, 413 10.3389/fimmu.2018.00413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter, G., Griffiths, A.D., Hawkins, R.E. and Hoogenboom, H.R. (1994) Making antibodies by phage display technology. Annu. Rev. Immunol. 12, 433–455 10.1146/annurev.iy.12.040194.002245 [DOI] [PubMed] [Google Scholar]
- 19.Dufner, P., Jermutus, L. and Minter, R.R. (2006) Harnessing phage and ribosome display for antibody optimisation. Trends Biotechnol. 24, 523–529 10.1016/j.tibtech.2006.09.004 [DOI] [PubMed] [Google Scholar]
- 20.Chao, G., Lau, W.L., Hackel, B.J., Sazinsky, S.L., Lippow, S.M. and Wittrup, K.D. (2006) Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1, 755–768 10.1038/nprot.2006.94 [DOI] [PubMed] [Google Scholar]
- 21.Cherf, G.M. and Cochran, J.R. (2015) Applications of yeast surface display for protein engineering. Methods Mol. Biol. 1319, 155–175 10.1007/978-1-4939-2748-7_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, O.J., Bernath, K., Agresti, J.J., Amitai, G., Kelly, B.T., Mastrobattista, E.et al. (2006) Directed evolution by in vitro compartmentalization. Nat. Methods 3, 561–570 10.1038/nmeth897 [DOI] [PubMed] [Google Scholar]
- 23.Moghaddam-Taaheri, P., Ikonomova, S.P., Gong, Z., Wisniewski, J.Q. and Karlsson, A.J. (2016) Bacterial inner-membrane display for screening a library of antibody fragments. J. Vis. Exp. 116, 54583 10.3791/54583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajpal, A., Beyaz, N., Haber, L., Cappuccilli, G., Yee, H., Bhatt, R.R.et al. (2005) A general method for greatly improving the affinity of antibodies by using combinatorial libraries. Proc. Natl Acad. Sci. U.S.A. 102, 8466–8471 10.1073/pnas.0503543102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Munoz, A., Bokma, E., O'Shea, D., Minton, K., Strain, M., Vousden, K.et al. (2012) Tailored amino acid diversity for the evolution of antibody affinity. MAbs 4, 664–672 10.4161/mabs.21728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis, L. and Lloyd, C. (2012) Optimisation of antibody affinity by ribosome display using error-prone or site-directed mutagenesis. Methods Mol. Biol. 805, 139–161 10.1007/978-1-61779-379-0_9 [DOI] [PubMed] [Google Scholar]
- 27.Pluckthun, A. (2012) Ribosome display: a perspective. Methods Mol. Biol. 805, 3–28 10.1007/978-1-61779-379-0_1 [DOI] [PubMed] [Google Scholar]
- 28.Seelig, B. (2011) mRNA display for the selection and evolution of enzymes from in vitro-translated protein libraries. Nat. Protoc. 6, 540–552 10.1038/nprot.2011.312 [DOI] [PubMed] [Google Scholar]
- 29.Wang, H. and Liu, R. (2011) Advantages of mRNA display selections over other selection techniques for investigation of protein-protein interactions. Expert Rev. Proteomics 8, 335–346 10.1586/epr.11.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groves, M., Lane, S., Douthwaite, J., Lowne, D., Rees, D.G., Edwards, B.et al. (2006) Affinity maturation of phage display antibody populations using ribosome display. J. Immunol. Methods 313, 129–139 10.1016/j.jim.2006.04.002 [DOI] [PubMed] [Google Scholar]
- 31.Groves, M.A., Amanuel, L., Campbell, J.I., Rees, D.G., Sridharan, S., Finch, D.K.et al. (2014) Antibody VH and VL recombination using phage and ribosome display technologies reveals distinct structural routes to affinity improvements with VH-VL interface residues providing important structural diversity. MAbs 6, 236–245 10.4161/mabs.27261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skamaki, K., Emond, S., Chodorge, M., Andrews, J., Rees, D.G., Cannon, D.et al. (2020) In vitro evolution of antibody affinity via insertional scanning mutagenesis of an entire antibody variable region. Proc. Natl Acad. Sci. U.S.A. 117, 27307–27318 10.1073/pnas.2002954117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowers, P.M., Verdino, P., Wang, Z., da Silva Correia, J., Chhoa, M., Macondray, G.et al. (2014) Nucleotide insertions and deletions complement point mutations to massively expand the diversity created by somatic hypermutation of antibodies. J. Biol. Chem. 289, 33557–33567 10.1074/jbc.M114.607176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brockmann, E.C., Pyykko, M., Hannula, H., Khan, K., Lamminmaki, U. and Huovinen, T. (2021) Combinatorial mutagenesis with alternative CDR-L1 and -H2 loop lengths contributes to affinity maturation of antibodies. N. Biotechnol. 60, 173–182 10.1016/j.nbt.2020.09.002 [DOI] [PubMed] [Google Scholar]
- 35.Kepler, T.B., Liao, H.X., Alam, S.M., Bhaskarabhatla, R., Zhang, R., Yandava, C.et al. (2014) Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell Host Microbe 16, 304–313 10.1016/j.chom.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, T.T., Johnson, G. and Kabat, E.A. (1993) Length distribution of CDRH3 in antibodies. Proteins 16, 1–7 10.1002/prot.340160102 [DOI] [PubMed] [Google Scholar]
- 37.Kabat, E.A., Foeller, C., Perry, H.M. and Gottesman, K.S. (1992) Sequences of Proteins of Immunological Interest, Diane Publishing Company [Google Scholar]
- 38.Dondelinger, M., Filee, P., Sauvage, E., Quinting, B., Muyldermans, S., Galleni, M.et al. (2018) Understanding the significance and implications of antibody numbering and antigen-binding surface/residue definition. Front. Immunol. 9, 2278 10.3389/fimmu.2018.02278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Wildt, R.M., van Venrooij, W.J., Winter, G., Hoet, R.M. and Tomlinson, I.M. (1999) Somatic insertions and deletions shape the human antibody repertoire. J. Mol. Biol. 294, 701–710 10.1006/jmbi.1999.3289 [DOI] [PubMed] [Google Scholar]
- 40.Douthwaite, J.A., Sridharan, S., Huntington, C., Hammersley, J., Marwood, R., Hakulinen, J.K.et al. (2015) Affinity maturation of a novel antagonistic human monoclonal antibody with a long VH CDR3 targeting the Class A GPCR formyl-peptide receptor 1. MAbs 7, 152–166 10.4161/19420862.2014.985158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma, Y., Ding, Y., Song, X., Ma, X., Li, X., Zhang, N.et al. (2020) Structure-guided discovery of a single-domain antibody agonist against human apelin receptor. Sci. Adv. 6, eaax7379 10.1126/sciadv.aax7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krause, J.C., Ekiert, D.C., Tumpey, T.M., Smith, P.B., Wilson, I.A. and Crowe, Jr, J.E. (2011) An insertion mutation that distorts antibody binding site architecture enhances function of a human antibody. mBio 2, e00345-10 10.1128/mBio.00345-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chodorge, M., Fourage, L., Ravot, G., Jermutus, L. and Minter, R. (2008) In vitro DNA recombination by L-Shuffling during ribosome display affinity maturation of an anti-Fas antibody increases the population of improved variants. Protein Eng. Des. Sel. 21, 343–351 10.1093/protein/gzn013 [DOI] [PubMed] [Google Scholar]
- 44.Bowers, P.M., Horlick, R.A., Neben, T.Y., Toobian, R.M., Tomlinson, G.L., Dalton, J.L.et al. (2011) Coupling mammalian cell surface display with somatic hypermutation for the discovery and maturation of human antibodies. Proc. Natl Acad. Sci. U.S.A. 108, 20455–20460 10.1073/pnas.1114010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowers, P.M., Boyle, W.J. and Damoiseaux, R. (2018) The use of somatic hypermutation for the affinity maturation of therapeutic antibodies. Methods Mol. Biol. 1827, 479–489 10.1007/978-1-4939-8648-4_24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McConnell, A.D., Do, M., Neben, T.Y., Spasojevic, V., MacLaren, J., Chen, A.P.et al. (2012) High affinity humanized antibodies without making hybridomas; immunization paired with mammalian cell display and in vitro somatic hypermutation. PLoS ONE 7, e49458 10.1371/journal.pone.0049458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horlick, R.A., Macomber, J.L., Bowers, P.M., Neben, T.Y., Tomlinson, G.L., Krapf, I.P.et al. (2013) Simultaneous surface display and secretion of proteins from mammalian cells facilitate efficient in vitro selection and maturation of antibodies. J. Biol. Chem. 288, 19861–9 10.1074/jbc.M113.452482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parthiban, K., Perera, R.L., Sattar, M., Huang, Y., Mayle, S., Masters, E.et al. (2019) A comprehensive search of functional sequence space using large mammalian display libraries created by gene editing. MAbs 11, 884–898 10.1080/19420862.2019.1618673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelow, S.P., Adolf-Bryfogle, J. and Dunbrack, R.L. (2020) Hiding in plain sight: structure and sequence analysis reveals the importance of the antibody DE loop for antibody-antigen binding. MAbs 12, 1840005 10.1080/19420862.2020.1840005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirik, U., Persson, H., Levander, F., Greiff, L. and Ohlin, M. (2017) Antibody heavy chain variable domains of different germline gene origins diversify through different paths. Front. Immunol. 8, 1433 10.3389/fimmu.2017.01433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burkovitz, A., Sela-Culang, I. and Ofran, Y. (2014) Large-scale analysis of somatic hypermutations in antibodies reveals which structural regions, positions and amino acids are modified to improve affinity. FEBS J. 281, 306–319 10.1111/febs.12597 [DOI] [PubMed] [Google Scholar]
- 52.Fanning, S.W. and Horn, J.R. (2011) An anti-hapten camelid antibody reveals a cryptic binding site with significant energetic contributions from a nonhypervariable loop. Protein Sci. 20, 1196–1207 10.1002/pro.648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koenig, P., Lee, C.V., Walters, B.T., Janakiraman, V., Stinson, J., Patapoff, T.W.et al. (2017) Mutational landscape of antibody variable domains reveals a switch modulating the interdomain conformational dynamics and antigen binding. Proc. Natl Acad. Sci. U.S.A. 114, E486–EE95 10.1073/pnas.1613231114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su, C.T., Ling, W.L., Lua, W.H., Poh, J.J. and Gan, S.K. (2017) The role of antibody Vkappa Framework 3 region towards antigen binding: effects on recombinant production and protein L binding. Sci. Rep. 7, 3766 10.1038/s41598-017-02756-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sela-Culang, I., Kunik, V. and Ofran, Y. (2013) The structural basis of antibody-antigen recognition. Front. Immunol. 4, 302 10.3389/fimmu.2013.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ling, W.L., Lua, W.H., Poh, J.J., Yeo, J.Y., Lane, D.P. and Gan, S.K. (2018) Effect of VH-VL families in pertuzumab and trastuzumab recombinant production, Her2 and FcgammaIIA binding. Front. Immunol. 9, 469 10.3389/fimmu.2018.00469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janda, A., Bowen, A., Greenspan, N.S. and Casadevall, A. (2016) Ig constant region effects on variable region structure and function. Front. Microbiol. 7, 22 10.3389/fmicb.2016.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lua, W.H., Ling, W.L., Yeo, J.Y., Poh, J.J., Lane, D.P. and Gan, S.K. (2018) The effects of antibody engineering CH and CL in trastuzumab and pertuzumab recombinant models: Impact on antibody production and antigen-binding. Sci. Rep. 8, 718 10.1038/s41598-017-18892-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torres, M., Fernandez-Fuentes, N., Fiser, A. and Casadevall, A. (2007) The immunoglobulin heavy chain constant region affects kinetic and thermodynamic parameters of antibody variable region interactions with antigen. J. Biol. Chem. 282, 13917–13927 10.1074/jbc.M700661200 [DOI] [PubMed] [Google Scholar]
- 60.Phua, S.X., Chan, K.F., Su, C.T., Poh, J.J. and Gan, S.K. (2019) Perspective: the promises of a holistic view of proteins-impact on antibody engineering and drug discovery. Biosci. Rep. 39, BSR20181958 10.1042/BSR20181958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Julian, M.C., Li, L., Garde, S., Wilen, R. and Tessier, P.M. (2017) Efficient affinity maturation of antibody variable domains requires co-selection of compensatory mutations to maintain thermodynamic stability. Sci. Rep. 7, 45259 10.1038/srep45259 [DOI] [PMC free article] [PubMed] [Google Scholar]