Abstract
Proteinase inhibitor 9 (PI-9) is a human serpin present in the cytoplasm of cytotoxic lymphocytes and epithelial cells. It inhibits the cytotoxic lymphocyte granule proteinase granzyme B (graB) and is thought to protect cytotoxic lymphocytes and bystander cells from graB-mediated apoptosis. Following uptake into cells, graB promotes DNA degradation, rapidly translocating to the nucleus, where it binds a nuclear component. PI-9 should therefore be found in cytotoxic lymphocyte and bystander cell nuclei to ensure complete protection against graB. Here we demonstrate by microscopy and subcellular fractionation experiments that PI-9 is present in the nuclei of human cytotoxic cells, endothelial cells, and epithelial cells. We also show that the related serpins, PI-6, monocyte neutrophil elastase inhibitor (MNEI), PI-8, plasminogen activator inhibitor 2 (PAI-2), and the viral serpin CrmA exhibit similar nucleocytoplasmic distributions. Because these serpins lack classical nuclear localization signals and are small enough to diffuse through nuclear pores, we investigated whether import occurs actively or passively. Large (∼70 kDa) chimeric proteins comprising PI-9, PI-6, PI-8, MNEI, or PAI-2 fused to green fluorescent protein (GFP) show similar nucleocytoplasmic distributions to the parent proteins, indicating that nuclear import is active. By contrast, CrmA-GFP is excluded from nuclei, indicating that CrmA is not actively imported. In vitro nuclear transport assays show that PI-9 accumulates at a rate above that of passive diffusion, that it requires cytosolic factors but not ATP, and that it does not bind an intranuclear component. Furthermore, PI-9 is exported from nuclei via a leptomycin B-sensitive pathway, implying involvement of the export factor Crm1p. We conclude that the nucleocytoplasmic distribution of PI-9 and related serpins involves a nonconventional nuclear import pathway and Crm1p.
Proteolysis mediated by serine proteinases is crucial to processes such as blood coagulation, fibrinolysis, complement activation, embryo implantation, extracellular matrix remodeling, and cell differentiation. Homeostatic regulation of serine proteinases is mainly achieved through interactions with inhibitors belonging to the large metazoan, plant, and virus serpin superfamily (44). Inhibitory serpins have a common structure and mode of action: each contains a variable C-terminal-reactive center loop resembling the substrate of its cognate proteinase. On proteinase binding, the serpin is cleaved between two residues in the loop designated P1 and P1′, and it undergoes a conformational change that distorts the proteinase and irreversibly locks the serpin-proteinase complex (27). The P1 residue is crucial and largely dictates the specificity of the serpin-proteinase interaction, while residues surrounding the cleavage site contribute to the affinity of the interaction (66).
The best-characterized serpins are involved in the regulation of extracellular proteolysis in vertebrates (reviewed in references 44 and 54); however, there is an emerging, more widespread subgroup resembling chicken ovalbumin (ov-serpins) which includes serpins that function intracellularly and some that target other proteinase classes, such as caspases and papains (14, 45, 48, 57). Ov-serpins display complex patterns of cellular distribution that probably reflect diverse physiological functions. Most are intracellular, with roles that are as yet unclear, but some are efficiently secreted to regulate cell-cell and cell-matrix interactions, and others exist in both intracellular and extracellular forms (5, 35, 71). Two of the intracellular ov-serpins, chicken MENT and human bomapin, accumulate efficiently in nuclei via classical nuclear localization sequences (NLSs) resembling the signal on the simian virus 40 large tumor antigen (T-ag) (13, 21). MENT is involved in chromatin condensation, but the nuclear role of bomapin is unknown (21).
Two other intracellular serpins, poxvirus CrmA and human proteinase inhibitor 9 (PI-9), are involved in the regulation of apoptosis (reviewed in reference 7). Cytotoxic lymphocytes (CLs) kill abnormal cells by using either one of two proapoptotic systems (37, 53). One system involves Fas ligand on the surface of the CL binding to Fas/Apo1/CD95 (Fas) on the target cell, resulting in receptor trimerization and recruitment of cytoplasmic adapter molecules to the receptor complex. The initiator caspase zymogen, procaspase 8, then binds to the complex, is activated, and in turn activates downstream effector caspases. CrmA efficiently inhibits caspase 8 (70) and potentially protects poxvirus-infected cells against CL Fas-mediated apoptosis (7).
The second cytotoxic system requires perforin to mediate entry of the granule serine proteinase granzyme B (graB) into the target cell, which then activates caspases, cleaves a variety of other proteins, and rapidly translocates to nuclei inducing DNA fragmentation (reviewed in reference 59). CLs do not commit fratricide or undergo autolysis as they sequentially engage and destroy target cells (19, 33, 38). This implies that their apoptotic machinery is precisely regulated. We have shown that PI-9 is a very efficient graB inhibitor produced by CLs (57), endothelial cells, and epithelial cells (11a). Cells expressing intracellular PI-9 resist apoptosis induced by graB and perforin but do not resist Fas-mediated apoptosis, since PI-9 does not efficiently inhibit caspases (3, 6). We have therefore proposed that the role of PI-9 is to protect CLs against autolysis directed by ectopic or misdirected graB and to protect bystander cells from graB released by activated CLs (6, 8). Importantly, the inability of PI-9 to inhibit caspases allows CLs to be deleted via death receptor-mediated apoptosis at the conclusion of the immune response and permits a response to stress-mediated apoptotic signals.
PI-9 is present in the CL cytoplasm outside the granules containing graB, which is consistent with a role in inactivating mislocalized graB (57). Since graB introduced into the cytoplasm of a cell rapidly translocates to the nucleus and binds to an intranuclear component (31, 52, 61, 62), it is likely that efficient protection of a CL by PI-9 requires its presence in the nucleus. Here we show that PI-9 is present in the nuclei of CLs, endothelial cells, and epithelial cells, that nuclear import occurs in the absence of a classical NLS, and that related ov-serpins exhibit a similar nucleocytoplasmic distribution. Although it is small enough to diffuse into the nucleus, uptake of PI-9 is an active process that depends on cytosolic factors but not ATP. Export of PI-9 from the nucleus occurs through a leptomycin B (LMB)-sensitive pathway. We conclude that nucleocytoplasmic distribution is a common feature of intracellular ov-serpins consistent with their predicted cytoprotective functions and appears to involve an unconventional nuclear import pathway as well as a nuclear export pathway requiring the export receptor Crm1p (exportin).
MATERIALS AND METHODS
Plasmids.
Plasmids used for expression of various proteins in COS-1 cells included pCMV/PI-9 (6) and pEUK/PAI-2 (50). A plasmid encoding PI-6 was constructed by subcloning the cDNA into the EcoRI site of pCMV2 (2). A plasmid encoding PI-8 was made by removing an internal EcoRI site in the PI-8 cDNA by site-directed mutagenesis and cloning the modified PI-8 cDNA into pSVTf (34). A monocyte neutrophil elastase inhibitor (MNEI) expression plasmid was constructed by removing the cDNA from pGEM-T/EI (41) using SacII (ends removed using T4 DNA polymerase) and PstI. This was cloned into pSVTf cut with SmaI and PstI. The plasmid pEGFP/PI-9 encodes a fusion protein comprising the human codon-enhanced green fluorescent protein (GFP) fused to the N terminus of PI-9. It was constructed by isolating a PI-9 cDNA on a 1-kb BamHI fragment from a yeast two-hybrid bait plasmid, pGTB/PI-9 (A. Calderone and P. Bird, unpublished data), and ligating it to BglII-digested pEGFP-c2 (Clontech). A similar fusion between neomycin 3′ phosphotransferase (neo) and PI-9 was also constructed. The neo gene was obtained by PCR amplification using pZeroBlunt (Invitrogen) as a template, the oligonucleotide primers 5′-GGGCTAGCCGCATGAATTGAACAAGATGGATTGCAC-3′ and 5′-CGCTCACCCGGGGAAGAACTCGTCAAGAAGCC-3′ (the latter introduces a SmaI site at the 3′ end), and Vent DNA polymerase (New England Biolabs) for 35 cycles of 95°C for 60 s, 50°C for 60 s, and 72°C for 60 s. The resulting 1-kb product was cloned into pZeroBlunt (Invitrogen) and then released and purified as a SmaI-EcoRI fragment. This was subcloned into pSVTf also digested with SmaI and EcoRI to generate pSVTf/neo. The plasmid pEGFP/PI-9 was digested with EagI and treated with T4 DNA polymerase. It was then cut with SalI, and the resulting 1.4-kb EagI-SalI fragment was ligated to pSVTf/neo digested with SmaI and SalI to generate pSVTf/neoPI-9. A plasmid encoding a GFP-CrmA fusion protein was constructed by digesting the plasmid pGEM7zf/CrmA (gift of D. Pickup, Duke University Medical Center, Durham, N.C.) with NcoI (ends filled in using T4 DNA polymerase) and ApaI. The resulting fragment was cloned into pEGFP-c2 cut with EcoRI (ends filled with T4 DNA polymerase) and ApaI. The plasmid pEGFP/PAI-2 was constructed by removing the plasminogen activator inhibitor 2 (PAI-2) cDNA from pSHT/PAI-2 (50) by BamHI (ends filled in using T4 DNA polymerase) and EcoRI digestion and cloning it into pEGFP-c2 cut with SacI (ends removed using T4 DNA polymerase) and EcoRI. The plasmids pEGFP/PI-8 and pEGFP/MNEI were constructed by removing the appropriate cDNA from pSHT/PI-8 or pSHT/MNEI (unpublished data) using BamHI and SpeI and cloning into pEGFP-c3 cut with BglII and XbaI. The plasmid pEGP/PI-6 was constructed by removing the PI-6 cDNA from the yeast two-hybrid bait vector pGTBK7/PI-6 (unpublished data) using EcoRI and SalI and cloning it into pEGFP-c2 cut with EcoRI and SalI.
Cells and transfections.
Activated primary CLs were prepared from human peripheral blood as previously described (6). YT is a human NK leukemia cell line (68) and was maintained in RPMI 1640 containing 10% heat-inactivated fetal bovine serum, 50 U of penicillin/ml, 50 μg of streptomycin/ml, 2 mM glutamine, 0.1 mM β-mercaptoethanol, and 1 mM sodium pyruvate. The placental choriocarcinoma cell line BeWo was obtained from the American Type Culture Collection and was maintained in Ham's F12 containing 10% heat-inactivated fetal bovine serum, 50 U of penicillin/ml, 50 μg of streptomycin/ml, 2 mM glutamine. Human umbilical vein endothelial cells (HUVEC) were isolated and maintained as described (58). COS-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, antibiotics, and 2 mM glutamine. Cells of the rat hepatoma tissue culture line (HTC; a derivative of Morris hepatoma 7288C) used for the in vitro transport studies were cultured in DMEM supplemented with 10% fetal bovine serum as described previously (26, 29, 30). COS-1 cells were transiently transfected by the dextran-chloroquine method (12).
Antibodies.
Rabbit antibodies to recombinant PI-9 and recombinant PI-6 have been described earlier (50, 57). A rabbit polyclonal antiserum to PI-8 was raised in a similar fashion and affinity purified on immobilized recombinant PI-8. Rabbit antibodies raised to native MNEI, CrmA, and lactate dehydrogenase (LDH) were provided by E. Remold O'Donnell (Harvard University, Boston, Mass.), D. Pickup (Duke University Medical Center), and J. Wilson (Michigan State University, East Lansing, Mich.), respectively. A rat monoclonal antibody against human Apaf-1 (23) was a gift of D. Huang (Walter and Eliza Hall Institute, Melbourne, Australia). Mouse monoclonal antibodies to cytochrome c and PAI-2 were purchased from Research Diagnostics Inc. (clone 7H8.2C12; Flanders, N.J.) and American Diagnostica Inc. (Greenwich, Conn.), respectively. Anti-GFP monoclonal antibody was purchased from Boehringer Mannheim. The nuclear protein B23 was detected using a mouse monoclonal antibody (40). Rabbit pan-cytokeratin antibodies were purchased from DAKO.
Cell fractionation.
Methods for the digitonin-based fractionation of YT, BeWo, and transfected COS-1 cells were based on previously published methods (23, 63, 65). A range of digitonin concentrations was tested in each experiment, and the results shown are those from cells treated with the lowest concentration that efficiently extracted the cytosolic marker protein (either LDH or Apaf-1) such that little or none was apparent in the subsequent nuclear fraction. Monolayers were removed from dishes by trypsinization and washed twice with ice-cold medium containing fetal calf serum and then washed once with ice-cold phosphate-buffered saline (PBS). Cells were counted and resuspended at 106/ml in HMKE buffer (20 mM HEPES [pH 7.2], 10 mM KCl, 5 mM MgCl2, 1 mM EDTA, 250 mM sucrose). One-milliliter aliquots were pelleted at approximately 200 × g and resuspended in HMKE buffer containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin/ml, 1 μg of pepstatin/ml, 1 μg of leupeptin/ml) and 50 to 200 μg of digitonin/ml (Sigma; 50-mg/ml stock in dimethyl sulfoxide). Cells were left on ice for 10 min and then centrifuged at 500 × g to separate cytosol from membranes, organelles, and cytoskeleton. The supernatant (cytosol fraction C) was carefully removed, and the pellet was washed in HMKE buffer. To extract proteins from membranes and organelles, the pellet was solubilized in extraction buffer containing 0.1 M Tris HCl (pH 9), 0.1 M NaCl, 5 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 0.5% Nonidet P-40, and protease inhibitors for 20 min on ice. Samples were transferred to a 1.5-ml microcentrifuge tube and centrifuged at 15,000 × g, the supernatant was carefully removed (fraction N), and a solution containing 20 mM Tris HCl (pH 6.8) and 2% sodium dodecyl sulfate (SDS) was added to the pellet to solubilize cytoskeletal proteins. The resulting lysate was collected and passed through a 26G needle until no longer viscous (fraction R).
The protein content of fraction C was determined, and typically 10 μg of protein was used for immunoblotting. The detergent in fractions N and R precluded direct protein estimation. To analyze and compare protein complement and distribution in the fractions, all were made up to the same final volume so that a sample taken from one fraction and compared to an equal volume from another represents the same number of starting cells. In each experiment, equal volumes of fractions C, N, and R were reduced and subjected to electrophoresis on SDS–10% polyacrylamide gels. Fractionation of COS cells producing CrmA was performed according to the methods of Schikendanz et al. (49).
Rabbit antisera (PI-6, PI-9, and MNEI) were used at 1:2,000 to 1:5,000 dilutions for immunoblotting, and the B23 monoclonal antibody was used at a dilution of 1:4,000. The CrmA and PI-8 antisera were used at dilutions of 1:100. The Apaf-1, cytochrome c, PAI-2, LDH, and GFP antibodies were used at dilutions of 1:1,000. Immunoblots were developed with an enhanced chemiluminescence detection kit (DuPont).
Indirect immunofluorescence microscopy and in situ cell extractions.
Cell monolayers grown on 12-well microscope slides were washed in PBS containing 0.1 mM CaCl2 and 1.0 mM MgCl2 (PBS+), fixed in 3.7% formaldehyde in PBS+ for 20 min, quenched with 20 mM ammonium chloride, and permeabilized by incubation in 0.5% Triton X-100 in PBS+ for 5 min. Alternatively, monolayers were fixed and permeabilized in 50% acetone–50% methanol for 2 min at room temperature. Glass slides used for nonadherent cells were first treated with poly-l-lysine (0.1 mg/ml) for 15 min at room temperature. Antigens were detected by incubation of the cells for 30 min with the appropriate dilution of primary antibody (typically 1:1,000 for PI-6 and PI-9 antisera, 1:50 for Apaf-1 antibodies, and 1:200 for all others). After being washed with PBS+ the cells were incubated with 1:200 dilutions of fluorescein isothiocyanate (FITC)- or rhodamine isothiocyanate-conjugated secondary antibodies. After 30 min cells were washed in PBS+ and, in some experiments, were stained with propidium iodide (1-μg/ml concentration in PBS) for 5 min at room temperature. Cells were washed, mounted in phenlyenediamine-buffered glycerol, and examined using either epifluorescence microscopy or confocal laser scanning microscopy (CLSM). In some indicated instances CLSM using 2-photon excitation was employed.
For in situ extractions, 104 BeWo cells per well were grown on 12-well microscope slides. Untreated cells were fixed and permeabilized as described above. Cells were stained with rabbit anti-PI-9 (1:1,000 dilution), rabbit anti-cytokeratin (1:200), and anti-Apaf-1 (1:50) followed by a 1:200 dilution of the appropriate secondary antibody conjugated to FITC. To remove proteins from the cytoplasm, cells were placed on ice, washed twice with ice-cold HMKE buffer, and then exposed to digitonin (25, 50, or 75 μg/ml) in 20 μl of HMKE buffer containing protease inhibitors for 10 min. At this point cells were either fixed for staining as above (digitonin treated) or washed with HMKE buffer and then incubated in 20 μl of extraction buffer for 10 min to remove detergent-soluble and salt-extractable (including most nuclear) proteins. The remaining monolayers were washed very gently and then fixed with formaldehyde and stained as above to visualize detergent-insoluble proteins.
Nuclear transport.
Analysis of nuclear import kinetics at the single-cell level in vitro using mechanically perforated HTC cells in conjunction with CLSM (MRC-600; Bio-Rad) was performed as described previously (10, 16, 29). NLS-dependent nuclear protein import can be reconstituted in this system through the exogenous addition of cytosolic extract (untreated reticulocyte lysate [Promega]), an ATP regenerating system (0.125 mg of creatine kinase/ml, 30 mM creatine-phosphate, 2 mM ATP), and transport substrate. In some experiments 0.025% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) was used to permeabilize the nuclear envelope; under these conditions nuclear accumulation can only occur through binding to intranuclear components such as lamins or chromatin (16, 17). Image analysis of CLSM files was performed using the MacIntosh NIH Image 1.60 public domain software. Each point shown in Results (Table 1 and Fig. 5) represents the average of 6 to 10 separate measurements (the standard error of the mean [SEM] was <10.2% of the value of the mean) for each of nuclear (Fn) and cytoplasmic (Fc) fluorescence, respectively, with autofluorescence subtracted. Data were fitted for the function Fn/c(t) = Fn/cmax × (1 − e−kt), where Fn/c is the ratio of nuclear to cytoplasmic fluorescence intensity, t is time in minutes, Fn/cmax is the maximal level of nuclear accumulation, and k is the first-order rate constant (10, 16, 29). Recombinant PI-9 was produced in a yeast expression system and purified as described previously (57) and was conjugated to FITC using standard procedures (22). The T-ag–CcN–β-Gal fusion protein used as a control for nuclear import studies contains T-ag amino acids 111 to 135, including the NLS, fused N terminal to β-galactosidase amino acids 9 to 1,023 (29, 46). It was expressed in Escherichia coli, purified by affinity chromatography, and labeled with 5-iodoacetamidofluorescein as described previously (29, 46).
TABLE 1.
In vitro nuclear import kinetics of PI-9 compared to those of control molecules
| Molecule | Import conditions | Nuclear import parametera
|
||
|---|---|---|---|---|
| Fn/cmax | t1/2 (min) | n | ||
| PI-9 | + ATP, + cytosol | 2.26 ± 0.08 | 1.68 ± 0.52 | 2 |
| − ATP, + cytosol | 2.46 ± 0.33 | 1.28 ± 0.41 | 3 | |
| + ATP, − cytosol | 0.80 ± 0.04 | NDb | 2 | |
| − ATP, − cytosol | 1.12 ± 0.10 | ND | 2 | |
| − ATP, + cytosol, + CHAPS | 1.16 ± 0.01 | ND | 1 | |
| − ATP, − cytosol, + CHAPS | 0.96 ± 0.03 | ND | 1 | |
| T-ag-CcN-β-Gal | + ATP, + cytosol | 5.51 ± 0.21 | 12.4 ± 2.0 | 2 |
| − ATP, + cytosol | 0.82 ± 0.13 | ND | 2 | |
| + ATP, − cytosol | 1.10 ± 0.04 | ND | 2 | |
| − ATP, + cytosol, + CHAPS | 1.15 ± 0.15 | ND | 2 | |
| Dextran (70 kDa) | + ATP, + cytosol | 0.16 ± 0.03 | ND | 3 |
| − ATP, + cytosol | 0.19 ± 0.02 | ND | 1 | |
| − ATP, − cytosol | 0.27 ± 0.06 | ND | 2 | |
| − ATP, + cytosol, + CHAPS | 0.98 ± 0.06 | ND | 2 | |
| − ATP, − cytosol, + CHAPS | 0.89 ± 0.09 | ND | 1 | |
| Dextran (20 kDa) | + ATP, + cytosol | 0.87 ± 0.04 | ND | 3 |
Raw data were fitted for the function Fn/c(t) = Fn/cmax × (1 − e−kt), where t is time in minutes (10, 16, 26, 29, 30). Results represent the mean ± SEM; where n = 1, the standard error was determined from the curve fit.
ND, not able to be determined due to low nuclear accumulation.
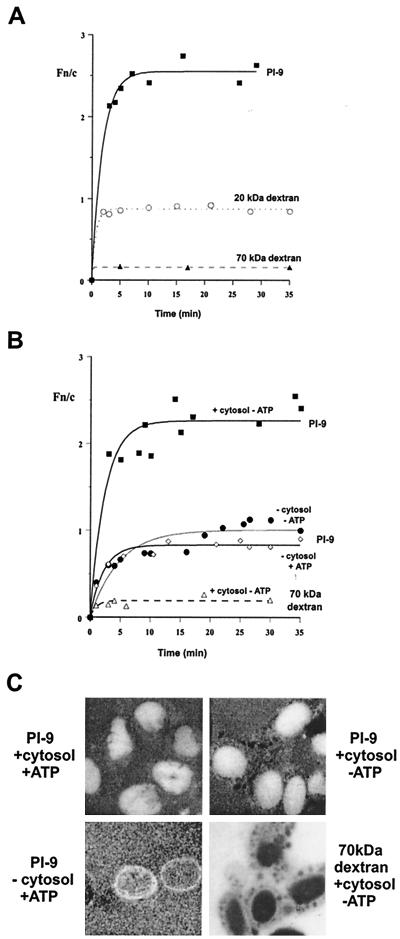
FIG. 5.
Nuclear import kinetics of PI-9 in vitro. Uptake of FITC-conjugated PI-9 and dextran was examined in mechanically perforated HTC cells at room temperature in the presence (A) or absence (B) of exogenous cytosol and/or an ATP regenerating system as indicated. (C) CLSM images of nuclei 20 min after the addition of the indicated components. Fn/c is the ratio of nuclear to cytoplasmic fluorescence. Table 1 shows pooled data.
Estimation of the proportion of nuclear PI-9.
Estimation of the nuclear and cytoplasmic volume for BeWo cells, YT cells, and HUVECs was performed using conventional as well as 2-photon CLSM (Bio-Rad) and standard cell measurement procedures. The percent nuclear PI-9 was calculated by multiplying the percent nuclear volume (>11 separate estimations) by the Fn/c for each cell type.
Treatment of cells with LMB.
COS-1 cells were transfected with pCMV/PI-9 or RevMAPKK-GFP (24). Cells grown on 12-well microscope slides were exposed to 4 ng of LMB/ml in complete medium for 0, 3, and 6 h and then fixed with formaldehyde and permeabilized as described above. RevMAPKK/GFP in transfected cells was followed by GFP fluorescence, whereas pCMV/PI-9-transfected cells were stained with rabbit anti-PI-9 diluted 1:500 and then with FITC-conjugated anti-rabbit immunoglobulins. Samples were mounted in Permafluor (Immunotech, Marseille, France). Quantitative analysis for nucleocytoplasmic distribution was performed as for the nuclear transport studies, where nontransfected cells were used to quantify background fluorescence due to autofluorescence or nonspecific staining by antibodies. Measurements of nucleolar fluorescence of cells producing RevMAPKK/GFP were carried out in a similar fashion.
RESULTS
PI-9 is present in CL, endothelial cell, and epithelial cell nuclei.
We have proposed that intracellular PI-9 protects CLs and bystander cells against mislocalized graB resulting from either granule leakage or misdirection during the immune response (6). Since free graB in the cell rapidly translocates from the cytoplasm to the nucleus (31, 52, 61, 62), the model predicts that PI-9 should also be present in nuclei to deal with any graB that evades the cytoplasmic pool of PI-9. To test this prediction we used indirect immunofluorescence and CLSM to examine primary human CLs prepared from peripheral blood, HUVECs, and the human cell lines YT (NK leukemia) and BeWo (choriocarcinoma). We have previously demonstrated that all of these cell types produce PI-9.
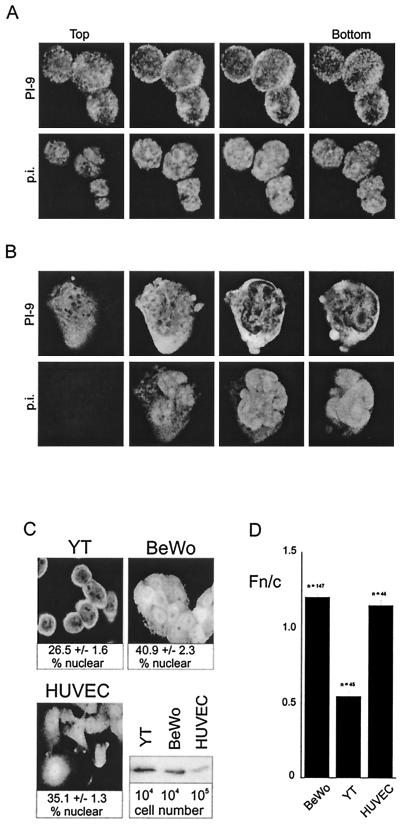
Shown in Fig. 1A is a series of confocal sections through a cluster of primary CLs that have been fixed, permeabilized, and stained for PI-9 (upper panel). In these cells the nucleus occupies most of the interior (shown by propidium iodide staining in the lower panel), and the cytoplasm is evident as a thin halo surrounding it. It is clear from these images that PI-9 is present in the cytoplasm and nucleus, though at lower concentration in the nucleus. Similar experiments on YT cells confirmed these observations (Fig. 1B and C). Again PI-9 was evident in both cytoplasm and nucleus, with less material in the nucleus.
FIG. 1.
Nucleocytoplasmic distribution of PI-9. (A) Human primary CLs. Interleukin-2-activated CLs were prepared from peripheral blood. Cells on glass slides were fixed and permeabilized with acetone-methanol and were incubated with rabbit antiserum against PI-9 and then incubated with FITC-conjugated anti-rabbit immunoglobulin and propidium iodide (p.i.). A series of CLSM sections through a cluster of cells shows PI-9 in both the nucleus and cytoplasm. (B) YT cells. Cells were fixed and stained as above. Shown is a series of CLSM sections of a single YT cell demonstrating PI-9 in the cytoplasm and nucleus. (C) Relative expression level and proportion of PI-9 in the nuclei of BeWo cells, YT cells, and HUVECs. Estimations of the nuclear and cytoplasmic volumes were performed using conventional and 2-photon CLSM and standard cell measurement procedures. The percent nuclear PI-9 was calculated by multiplying the percent nuclear volume by the ratio of nuclear to cytoplasmic fluorescence intensity (Fn/c). For comparison of PI-9 levels, 106 cells were lysed directly in SDS sample buffer, separated by SDS-polyacrylamide gel electrophoresis, and immunoblotted using PI-9 antibodies. Ten times more HUVEC than BeWo or YT lysate was run on the gel. (D) Concentration of PI-9 in the cytoplasm and nuclei of YT cells, BeWo cells, and HUVECs as measured by relative fluorescence intensity under CLSM. Results shown for the Fn/c ratio represent the mean ± SEM, with n being the number of cells analyzed.
As shown in Fig. 1C, we also observed PI-9 in the cytoplasm and nucleus of primary endothelial cells (HUVECs) and BeWo cells (which have epithelial characteristics of placental cytotrophoblasts). PI-9 levels in BeWo cells were comparable to those in YT cells, but levels in HUVECs were 10- to 100-fold lower (Fig. 1C, bottom left panel). Interestingly, the amount of PI-9 in the nucleus differed in the various cell types but did not seem to be related to the overall expression level of PI-9. To investigate this further we used CLSM and image analysis to measure the proportion of PI-9 in the nuclei of YT cells, HUVECs, and BeWo cells (Fig. 1C). This indicated that YT cells have the least PI-9 in the nucleus (26.5%) and that BeWo cells have the most (40.9%). Since the size of the nucleus varies from cell type to cell type, the concentration of PI-9 in the nucleus compared to the concentration in the cytoplasm may also vary. To investigate this we used image analysis to derive a value for the nuclear-to-cytoplasmic ratio (Fn/c) of PI-9 in the three cell types. As shown in Fig. 1D, YT cells have about twice the concentration of PI-9 in the cytoplasm as the nucleus, whereas HUVECs and BeWo cells have an equal concentration of PI-9 in the cytoplasm and nucleus.
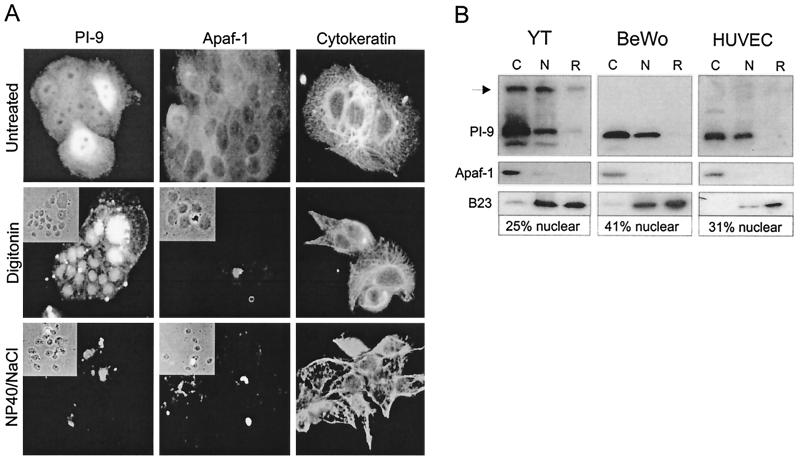
To confirm that PI-9 is present in the nuclei of these cells we carried out two digitonin-based fractionation procedures. One involved sequential extraction of protein from cells in situ with changes in protein content followed at the single-cell level using indirect immunofluorescence microscopy, and the other entailed fractionation of larger numbers of cells with analysis of the fractions by immunoblotting. Digitonin is extensively used in in vitro nuclear import studies to separate nuclei with intact envelopes and functional pore complexes from cytosolic components because it selectively permeabilizes the plasma membrane, leaving the nuclear membrane intact (23, 63, 65). Following digitonin treatment and washing to remove cytosolic protein, nuclei can be lysed in a high-salt buffer containing detergent to release nuclear proteins. Any remaining insoluble material contains mostly cytoskeletal and some nuclear or nucleolar protein that can be solubilized in SDS. In both procedures we expected to extract cytosolic, nuclear, and cytoskeletal protein sequentially. To verify this we followed the release of the cytosolic protein Apaf-1 (23), the nuclear or nucleolar protein B23 (40), and cytoskeletal cytokeratins.
As shown in Fig 2A, in situ extractions demonstrated PI-9 in the nuclei of BeWo cells. Untreated cells showed PI-9 in the cytoplasm and nucleus (upper left panel), Apaf-1 in the cytoplasm (upper center panel), and cytokeratin throughout the cell (upper right panel). Treatment of cells with digitonin completely removed Apaf-1 from the cytoplasm (middle center panel) but did not fully release PI-9 from cells, as the protein is clearly still evident in nuclei (middle left panel). Cytokeratin was not removed by digitonin treatment (right middle panel). Subsequent extraction with high salt and detergent completely removed PI-9 from nuclei (lower left panel) but did not remove cytokeratin from the cells (lower right panel), indicating that PI-9 is not associated with the cytoskeleton. In situ extraction experiments on HUVECs and YT cells yielded similar results (data not shown).
FIG. 2.
In situ extraction and subcellular fractionation of PI-9 expressing cells. (A) BeWo cells growing on microscope slides were sequentially extracted with digitonin-containing and high salt–detergent-containing buffers. After each treatment, cells were fixed and permeabilized with acetone-methanol and incubated with antibodies against PI-9, Apaf-1, or cytokeratins. After being stained with FITC-conjugated anti-rabbit immunoglobulins, the cells were examined by phase-contrast (inset panels) and fluorescence microscopy. (B) YT cells, BeWo cells, and HUVECs were harvested and sequentially treated with digitonin, high salt-detergent, and SDS to generate cytosolic (C), nuclear (N), and remnant (R) fractions. Equal amounts of each fraction were separated by SDS–10% polyacrylamide gel electrophoresis and analyzed by immunoblotting with PI-9, Apaf-1, or B23 antibodies. The percentage of PI-9 in the nuclear fraction was estimated by densitometry. The PI-9–graB complex is indicated with an arrow.
Immunoblotting analysis of digitonin-treated HUVECs, YT cells, and BeWo cells separated into cytoplasmic (C), nuclear (N), and remnant (R) fractions confirmed the results of the single-cell extraction procedure (Fig. 2B). PI-9 was evident in both the cytoplasmic and nuclear fractions of all the cell types and was not associated with cytoskeletal material. Densitometry was used to estimate the amount of nuclear PI-9 in these cells (Fig. 2B), and the resulting values were in very good agreement with the results of the confocal analysis (Fig. 1C).
The higher-molecular-weight species seen in both the cytosolic and nuclear fractions of YT cells represents PI-9 complexed with graB. Complex formation is a postlysis phenomenon, as graB is released from granules by detergents and rapidly equilibrates between the cytosol and nucleus via an unknown mechanism (61). We have shown previously that if cells are broken mechanically and intact granules are separated from the soluble components, little or no complex is detected in the cytosol (57). Furthermore, if whole cells are lysed rapidly in SDS sample buffer, no complexes are observed (data not shown).
Taken together, the above results clearly show that PI-9 has a nucleocytoplasmic distribution in accordance with our cytoprotective model and that the proportion of PI-9 in the nucleus is not related to the expression level. To see if the nuclear localization of PI-9 requires a pathway or factors peculiar to HUVECs, YT cells, or BeWo cells, we also examined a number of other human cell lines of lymphoid or epithelial origin (SKW6, MCF7, HeLa) that are normally PI-9 negative but have been stably transfected with PI-9 cDNAs. In every case PI-9 was present in both the cell cytoplasm and nucleus (data not shown). We also expressed PI-9 in transiently transfected COS-1 cells (Fig. 3A). Fractionation experiments and epifluorescence microscopy of cells stained with rabbit anti-PI-9 antibodies showed PI-9 in both the cytoplasm and nucleus in these cells (Fig. 3A), and the concentration of nuclear PI-9 in transfected COS cells was similar to that of YT cells (Fn/c, ∼0.5; see Fig. 6B). Thus, nuclear localization of PI-9 is unlikely to require cell type-specific machinery.
FIG. 3.
Nucleocytoplasmic distribution of PI-9-related ov-serpins. COS-1 cells transiently transfected with appropriate expression vectors were grown either on glass slides for examination by indirect immunofluorescence or in dishes for cytosolic and nuclear fractionation experiments. Cells expressing either PI-9 (A), PI-6 (B), PI-8 (C), MNEI (D), PAI-2 (E), or CrmA (F) were fixed and stained with appropriate primary antibodies and then with FITC-conjugated secondary antibodies. In each case the images clearly show the serpin in the cytoplasm and nucleus. The distribution of protein within cytosolic (C), nuclear (N), or remnant (R) fractions in each transfected line was determined by immunoblotting. Also shown are the LDH and B23 controls for each experiment. CrmA-transfected cells were fractionated according to previously published methods (49), and cytochrome c (cyt) was used to monitor release of cytoplasmic protein.
FIG. 6.
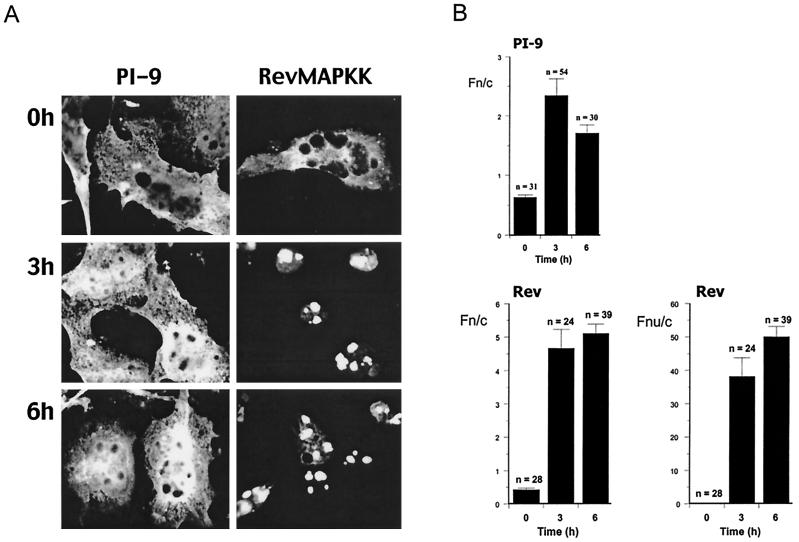
Nuclear export of PI-9 is blocked by LMB. COS-1 cells transfected with the expression vectors encoding PI-9 or RevMAPKK/GFP were exposed to 4 ng of LMB/ml for the indicated times. Cells were prepared for visualization (A) and quantitation (B) of nuclear/cytoplasmic ratios (Fn/c) or nucleolar/cytoplasmic ratios (Fnu/c by CLSM as described in Materials and Methods). Results are the mean ± SEM, with n being the number of cells analyzed.
Related ov-serpins also exhibit a nucleocytoplasmic distribution.
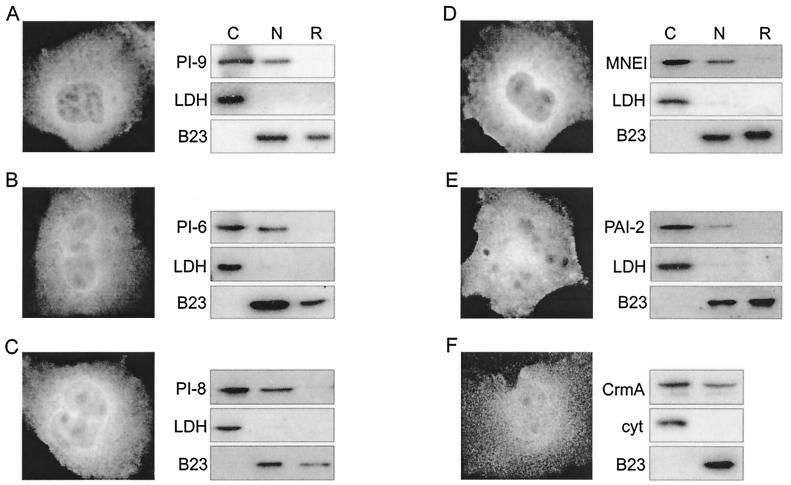
PI-9 belongs to the ov-serpin family that comprises intracellular proteins very similar in structure and size. Two ov-serpins (MENT and bomapin) possess conventional nuclear import signals located in a region of the molecule known as the interhelical loop (13, 21). PI-9 lacks this loop and has no identifiable NLS, implying that its uptake into the nucleus occurs via a different mechanism. To determine if this type of nuclear accumulation is unique to PI-9, we transiently transfected COS-1 cells with expression vectors encoding other ov-serpins lacking the interhelical loop and an identifiable NLS (PI-6, PI-8, MNEI) with an ov-serpin containing the loop but no obvious import signal (PAI-2) and with the viral intracellular serpin, CrmA. Cells were either fractionated for analysis by immunoblotting or plated on glass slides and prepared for examination by indirect immunofluorescence microscopy (Fig. 3). In every case, expression in the nucleus was detected by both methods, although the cytoplasmic-to-nuclear ratio varied from protein to protein. With the exception of PAI-2 (Fig. 3E), there was generally good correlation between the proportion of protein observed in the nuclei by microscopy and the amount evident by subcellular fractionation. For PAI-2 there appeared to be more material in the nucleus by microscopy than was indicated by analysis of fractions. The reason for the discrepancy is unknown but may reflect postlysis modification in the nuclear fractions of the epitope for the PAI-2 monoclonal antibody so that the protein is no longer recognized efficiently. This explanation is supported by the experiments on the GFP–PAI-2 fusion protein (Fig. 4D) in which a different monoclonal antibody was used (anti-GFP) and no such discrepancy between the two methods was evident.
FIG. 4.
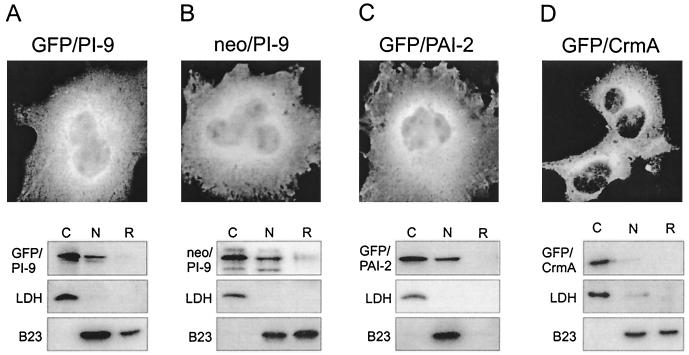
PI-9 is imported into the nucleus through a facilitated mechanism. COS-1 cells were transfected with expression vectors encoding GFP–PI-9 (A), neo–PI-9 (B), GFP–PAI-2 (C), or GFP-CrmA (D), examined by indirect immunofluorescence microscopy, or fractionated into cytosolic and nuclear components. Cells were fixed and permeabilized using acetone-methanol and then incubated with primary antibodies (GFP-CrmA and GFP–PAI-2 were monitored using a monoclonal antibody against GFP) and the appropriate FITC-conjugated secondary antibodies. The distribution of protein within cytosolic (C), nuclear (N), or remnant (R) fractions in each transfected line was determined by immunoblotting with the same primary antibodies. Also shown are the LDH and B23 controls for each experiment.
Finally, examination of primary human monocytes and mesothelial cells, which endogenously produce PAI-2, PI-6, and MNEI (51 and unpublished results), showed nucleocytoplasmic distributions of the three proteins (data not shown). Taken together, these results indicate that nucleocytoplasmic localization is a common feature of ov-serpins.
Nuclear accumulation of PI-9 is an active process.
Proteins up to 40 to 60 kDa can pass freely through the nuclear pore complex. However, even very small proteins with specific nuclear functions carry NLSs, ensuring efficient nuclear targeting. Examination of the amino acid sequence of PI-9 and related serpins failed to reveal known classical, bipartite, or hnRNP A1 M9-like import signals (39). The lack of an identifiable NLS together with its relatively small size (42 kDa) suggested that PI-9 enters the nucleus by passive diffusion. To test this we generated a chimeric protein (GFP–PI-9) consisting of GFP (27 kDa) fused to the N terminus of PI-9. If PI-9 diffuses into the nucleus and does not require a dedicated NLS, the 69-kDa fusion protein should be excluded from the nucleus because it is too large to move through the pores.
By microscopy the fusion protein produced in transfected COS-1 cells was evident in both cytoplasm and nuclei (Fig. 4A). Fractionation experiments showed the 69-kDa fusion protein in the cytoplasm and nucleus, and densitometry indicated that the same proportion of GFP–PI-9 as PI-9 is found in nuclear fractions (approximately 20 to 30%). Pulse-chase experiments of metabolically labeled cells over 5 h showed that the fusion protein is stable and that no degradation into smaller (potentially diffusible) fragments occurred (data not shown).
To rule out the generation of a cryptic NLS during the construction of GFP–PI-9, we made a similar fusion (neo–PI-9) between the 28-kDa aminoglycoside 3′-phosphotransferase (neo) protein and PI-9. Like GFP, neo is small enough to passively enter the nucleus, but the chimeric protein should be too large to do so. Fractionation experiments and microscopy revealed that neo–PI-9 also enters the nucleus (Fig. 4B). Taken together these results suggest that although PI-9 is under the nominal nuclear pore cutoff, it enters the nucleus in a facilitated process.
To determine whether facilitated nuclear import is unique to PI-9, we also constructed similar fusion proteins consisting of GFP linked to PAI-2, PI-6, MNEI, PI-8, or CrmA. As judged by microscopy and fractionation of transfected COS cells, the GFP–PAI-2 fusion protein accumulated in the nucleus as efficiently as PAI-2 alone (Fig. 4C). By contrast, the GFP-CrmA fusion protein was essentially excluded from the nucleus (Fig. 4D), indicating that CrmA is not actively imported and that CrmA observed in the nucleus results from diffusion only (see Fig. 3F). Fusions between GFP and PI-6, MNEI, or PI-8 showed distributions similar to those of the parent proteins (data not shown). These results demonstrate that PI-9 and related ov-serpins can be imported into nuclei via a facilitated mechanism that does not depend on a classical NLS.
Nuclear uptake of PI-9 requires cytosolic factors but not ATP.
Nuclear accumulation of PI-9 was also examined in vitro. As shown in Fig. 5, the nuclear import properties of FITC-labeled recombinant PI-9 were compared to those of control molecules at the single-cell level in mechanically perforated HTC cells (29). Addition of exogenous cytosol and an ATP-generating system to the HTC cells is sufficient to reconstitute nuclear transport, as shown by the uptake of the well-characterized chimeric protein (T-ag–CcN–β-Gal) which comprises a conventional NLS derived from T-Ag fused to the N terminus of E. coli β-galactosidase (46). This protein accumulated in the nucleus to levels over fivefold greater than those in the cytoplasm, with half-maximal accumulation achieved within 12 min (Table 1). By contrast, 70-kDa dextran was excluded from the nucleus (Fn/cmax of about 0.2), while 20-kDa dextran equilibrated between nuclear and cytoplasmic compartments but did not accumulate in the nucleus (Fn/cmax of about 1). In this system, nuclear accumulation of PI-9 in HTC cell nuclei was observed, occurring to a maximum of 2.5 times that of the cytoplasmic levels, with transport half maximal within 2 min (Table 1 and Fig. 5A).
Conventional signal-mediated nuclear protein import in vitro is dependent on energy in the form of ATP and exogenous cytosol (20), the latter containing the NLS-recognizing importin heterodimer, the monomeric guanine nucleotide-binding protein Ran, and other interacting proteins essential for nuclear accumulation (20). The conventional NLS-containing fusion protein T-ag–CcN–β-Gal requires both ATP and cytosol for nuclear accumulation (see Table 1 and reference 17). By contrast, nuclear import of PI-9 required cytosol but not ATP (Fig. 5B and Table 1).
The detergent CHAPS can be used to perforate the nuclear envelope to enable molecules to diffuse freely between cytoplasm and nucleoplasm (16, 17). For example, 70-kDa dextran is no longer excluded from nuclei after CHAPS treatment (Table 1). In the presence of CHAPS, nuclear accumulation can occur only through binding to nuclear components (16, 17). As demonstrated by T-ag–CcN–β-gal (Table 1), most proteins containing classical nuclear import signals do not exhibit nuclear accumulation in the presence of CHAPS; instead they equilibrate between the nuclear and cytoplasmic compartments. Likewise, in the presence of CHAPS PI-9 did not accumulate in the nucleus in either the absence or presence of cytosol (Table 1) and is thus clearly unable to bind to nuclear components and/or accumulate in the nucleus under these conditions. This contrasts with graB, which accumulates in nuclei in the presence of CHAPS by binding a nuclear component (28). In summary, PI-9 appears to accumulate in the nucleus through a novel nuclear import pathway which requires cytosolic factors, does not require ATP, and does not involve intranuclear binding.
Export of PI-9 from the nucleus occurs via a LMB-sensitive pathway.
PI-9 is detectable in both the nucleus and the cytoplasm of the cell. Since PI-9 is able to localize strongly in the nucleus via a facilitated mechanism, it is likely that active export of PI-9 from the nucleus to the cytoplasm occurs in order to maintain the correct nucleocytoplasmic distribution. Most proteins exported from the nucleus travel on a pathway that can be blocked by the compound LMB (20). We therefore tested the ability of LMB to inhibit the export of PI-9, as indicated by increased nuclear accumulation of PI-9 in LMB-treated cells. As a control for LMB activity we obtained a plasmid encoding a mutant form of nucleolar human immunodeficiency virus (HIV) Rev linked to GFP (RevMAPKK/GFP) (24). This fusion protein also carries a strong, heterologous nuclear export signal (NES) from mitogen-activated protein kinase kinase. Under normal conditions, in the absence of LMB the vast majority of this protein is present in the cytoplasm, since the rate of nuclear export directed by this NES is far greater than that of nuclear import (24). However, in the presence of LMB nuclear export is prevented and there is a dramatic shift from predominantly cytoplasmic to predominantly nuclear compartmentalization of RevMAPKK/GFP, with most accumulation in the nucleoli, as expected for Rev (43).
COS-1 cells expressing PI-9 or RevMAPKK/GFP were exposed to LMB for 0, 3, or 6 h and then were examined by indirect immunofluorescence using CLSM to measure the ratio of fluorescence intensities in the cytosol and nucleus at the single-cell level (Fig. 6). As expected, RevMAPKK/GFP rapidly accumulated in the nucleolus, peaking at 3 h with a concentration 40- to 50-fold higher in the nucleolus than in the cytoplasm. Some RevMAPKK/GFP accumulated in the nucleus outside the nucleolus, and this concentration was fivefold higher than that in the cytoplasm. Nuclear accumulation of PI-9 was also observed, peaking at 3 h with two to three times the concentration in the nucleus than that in the cytoplasm. This represented a significant shift from untreated cells, which show about twice the concentration of PI-9 in the cytoplasm as that in the nucleus. These results clearly show that PI-9 is actively exported from the nucleus through a mechanism dependent on Crm1p (exportin).
DISCUSSION
The ov-serpins are an emerging subgroup of the serpin superfamily distinguished by their largely intracellular localization. Indeed, most of the new serpins recently identified through whole-genome analysis of Caenorhabditis elegans and Drosophila melanogaster have the characteristics of intracellular proteins (67 and A. Lesk, P. Bird, and J. Whisstock, unpublished results). Although 1 of the 12 human ov-serpins (bomapin) is imported into the nucleus via a classical NLS (13), we show here for the first time that PI-9 and at least four related human ov-serpins (PAI-2, MNEI, PI-6, and PI-8) enter nuclei without possessing obvious classical nuclear import signals. On the basis of the behavior of GFP fusion proteins, it is clear that import of these serpins depends almost entirely on a facilitated (active) pathway. This contrasts with the viral intracellular serpin CrmA, which has functional similarities to PI-9 but apparently enters the nucleus entirely by diffusion. On the basis of our results, it is reasonable to suggest that other ov-serpins—and perhaps unrelated intracellular serpins—will exhibit similar subcellular localization.
A nucleocytoplasmic distribution pattern is consistent with the proposed cytoprotective roles of several of these serpins. For example, it is thought that PI-9 protects cytotoxic and bystander cells against misdirected graB, which is known to translocate efficiently from the cytoplasm to the nucleus of cytotoxic and target cells and to degrade cytoplasmic and nuclear substrates (60). Obviously, the presence of PI-9 in both the cytoplasm and nucleus of a cell would provide efficient protection against graB-mediated damage. Likewise, PI-6 and MNEI inhibit the monocyte and granulocyte granule proteinases, elastase, cathepsin G, and proteinase 3, and may also protect cells against protease-directed autolysis (51, 56). These proteinases are all small enough to enter nuclei by diffusion, so that if introduced into the host cell cytoplasm they have the potential to threaten viability in a manner similar to that of misdirected graB in cytotoxic cells. Indeed, cathepsin G can activate caspase 7 via cleavage at a noncanonical site (69) and cleaves the nuclear protein brm (5a), suggesting it is proapoptotic if released into the interior of the cell. The presence of PI-6 and MNEI in the nucleus as well as the cytoplasm therefore offers the cell an additional level of protection against misdirected granule proteinases. A similar protective role can be invoked for two other ov-serpins, SCCA-1 and SCCA-2, that were not investigated in this study. These proteins interact with lysosomal and mast cell proteinases, respectively (47, 48). Given that loss of lysosomal membrane integrity and the release of contents into the cytoplasm are known to occur under stress (11) and that lysosomal proteinases are also small enough to diffuse into the nucleus, it is possible that the SCCAs will also exhibit a nucleocytoplasmic distribution.
The role of PAI-2 in cells is probably different from that of other ov-serpins, although still cytoprotective in scope. PAI-2 protects cells against tumor necrosis factor alpha-mediated apoptosis (15) and virus infection (4). Protection against virus infection occurs through PAI-2-mediated induction of autocrine alpha and beta interferon, and it has been suggested that PAI-2 acts on a transcription factor pathway (4). Although the intracellular targets of PAI-2 are unknown, the presence of PAI-2 in the nucleus is certainly consistent with a direct or indirect impact on the transcription of cytoprotective factors.
An interesting question not addressed in our study is whether the nucleocytoplasmic distribution of PI-9 and other intracellular ov-serpins is regulated, in that levels of nuclear import or export alter in response to specific signals. Clearly different cell types show different proportions of nuclear PI-9 (compare HUVECs, YT cells, and BeWo cells), and this is not related to the expression level of PI-9, ruling out saturation of the nuclear import machinery. While different cells may possess different levels of key mediators of PI-9 import or export, an alternative possibility is that the nucleocytoplasmic distribution of PI-9 is actively regulated. There are several specific mechanisms by which nuclear transport can be regulated, perhaps the best known being cytoplasmic retention, in which a nuclear-targeted protein is held in the cytoplasm by binding either to an anchored structure or to a partner that sequesters its NLS (25). Signal-mediated phosphorylation or proteolysis then releases the protein by disrupting binding or revealing its NLS, allowing import to occur. It remains to be seen if the nucleocytoplasmic distribution of PI-9 and related ov-serpins alters in response to cell activation, differentiation, stress, or other stimuli and whether they have intracellular binding partners.
The structure of the nuclear pore and the main players in the import machinery are reasonably well understood (for reviews, see references 1 and 20). Import of proteins carrying a classical NLS involves the formation of a complex between the cargo, importin-α (adapter), and importin-β. In a process requiring energy, a G protein (Ran), Ran-binding proteins, guanine nucleotide exchange factors, and GTPase activators the complex docks at the nuclear pore, is translocated through it, and then dissociates within the nucleoplasm. Several lines of evidence suggest that alternative import routes and components are used by proteins lacking classical import signals. For example, importin-β family members can mediate the import of ribosomal proteins independently of importin-α, and other Ran-binding proteins have also been implicated in adapter-free transport. In this study we have shown that PI-9 accumulates in the nucleus via an atypical import pathway that requires cytosolic factors but not ATP, which may also hold true for other ov-serpins. Although it lacks an apparent classical NLS, PI-9 is imported via a facilitated process, as indicated by its ability to mediate uptake of large fusion proteins. Preliminary evidence suggests that PI-9 does indeed possess an NLS, but it is conformational, comprising a number of noncontiguous residues (unpublished results). Like many nuclear proteins, PI-9 is exported from the nucleus via an LMB-sensitive pathway, strongly implying that it possesses an NES and that the nuclear export receptor Crm1p (exportin) is involved.
What is the import pathway followed by PI-9? Proteins bearing a classical NLS bind to importin-α, and translocation requires ATP in vitro (1, 20). Since PI-9 lacks a classical NLS, it may not bind importin-α. This is supported by preliminary experiments using established assays in which PI-9 failed to bind to mouse importin-α2 in either the presence or absence of importin-β (unpublished results). However, this conclusion should be qualified by noting that importin-α now appears to be a member of a larger family of proteins with similar functions and that there is some diversity in the sequence of signals recognized by importin-α (reviewed in references 1 and 55). Hence, the formal possibility that the PI-9 NLS binds a different importin-α family member cannot be excluded, but it is more likely that PI-9 is imported in a process that does not require importin-α. Perhaps it binds an importin-β family member directly, as is the case for import of ribosomal proteins and certain transcription factors (20, 42, 55).
PI-9 nuclear import is clearly distinguishable from conventional import on the basis of its ATP independence in vitro. This nonconventional mechanism is probably not unique or restricted to particular cell types, because nuclear import of PI-9 and related ov-serpins occurs in epithelial, lymphoid, and fibroblast lines. PI-9 does not accumulate in the nucleus in the presence of CHAPS, so its lower energy requirement for translocation is not due to retention or anchorage in the nucleus, as is the case for the unconventional import of nucleoplasmin, granzymes A and B, and HIV Vpr and Tat (16, 28, 32, 52, 64). Further distinctions between the nonconventional import pathways utilized by the latter proteins and that utilized by PI-9 are that import of HIV Vpr does not require cytosolic factors, such as Ran or importins (32), while the import of Tat requires ATP hydrolysis but not cytosolic factors (16).
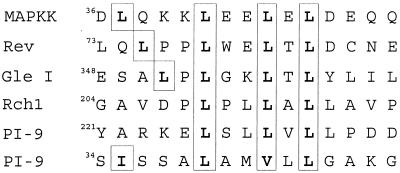
PI-9 apparently exits the nucleus via a conventional process. Its sensitivity to LMB indicates that egress depends on the factor Crm1p (exportin), which conventionally recognizes Leu-rich NESs (reviewed in reference 20). Studies on a number of exported proteins have led to the definition of a consensus NES, which comprises LeuXxx(1-3)LeuXxx(2-3)LeuXxxLeu, but not all sequences conforming to the consensus are functional, and avid interaction with Crm1p can occur in the absence of a consensus sequence (20, 24). As shown in Fig. 7, PI-9 has at least two sequences matching the consensus NES motif. However, the large number of available serpin crystal structures (66) and the overall sequence similarity between serpins have allowed us to build a model of PI-9 with a high degree of certainty based on the crystal structure of the related serpin, antithrombin (J. Whisstock and P. Bird, unpublished results). This clearly shows that both sequences are buried within the body of the molecule and would not be available for a protein-protein interaction unless PI-9 is unfolded. As there is no evidence at present that nuclear export requires unfolding of the passenger protein, this burying of a potential NES may explain why some sequences that match the consensus motif are not functional. We suggest that export of PI-9 either involves a linear NES that does not resemble the proposed consensus sequence or involves a conformational rather than linear NES. Alternatively, PI-9 may lack an NES altogether but can exit the nucleus by binding to a protein that possesses a classical NES.
FIG. 7.
Potential Leu-rich NESs in PI-9. Shown are two sequences on PI-9 with similarity to known NESs on Rev (18), Gle1 (36), Rch1 (9), and mitogen-activated protein kinase kinase (MAPKK) (24). Conserved residues of the NES are boxed.
ACKNOWLEDGMENTS
We thank B. Henderson (Westmead Hospital, Sydney, Australia) for advice and the RevMAPKK/GFP plasmid, A. Calderone for construction of the GFP–PAI-2 plasmid, and J. Whisstock (Monash University) for molecular modeling. We also thank D. Huang (Walter and Eliza Hall Institute) for Apaf-1 antibodies, E. Remold-O'Donnell (Center for Blood Research, Boston, Mass.) for MNEI antibodies, J. Wilson and J. Wang (Michigan State University) for LDH antiserum, and D. Pickup (Duke University Medical Center) for the CrmA cDNA and antiserum.
This work was supported by the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Adam S A. Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr Opin Cell Biol. 1999;11:402–406. doi: 10.1016/S0955-0674(99)80056-8. [DOI] [PubMed] [Google Scholar]
- 2.Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 3.Annand R R, Dahlen J R, Sprecher C A, de Dreu P, Foster D C, Mankovich J A, Talanian R V, Kisiel W, Giegel D A. Caspase-1(interleukin-1b-converting enzyme) is inhibited by the human serpin analogue proteinase inhibitor 9. Biochem J. 1999;342:655–665. [PMC free article] [PubMed] [Google Scholar]
- 4.Antalis T M, La Linn M, Donnan K, Mateo L, Gardner J, Dickinson J L, Buttigieg K, Suhrbier A. The serine proteinase inhibitor (serpin) plasminogen activation inhibitor type 2 protects against viral cytopathic effects by constitutive interferon alpha/beta priming. J Exp Med. 1998;187:1799–1811. doi: 10.1084/jem.187.11.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belin D. Biology and facultative secretion of plasminogen activator inhibitor-2. Thromb Haemostasis. 1993;70:144–147. [PubMed] [Google Scholar]
- 5a.Biggs J R, Yang J, Gullberg U, Muchardt C, Yaniv M, Kraft A S. The human brm protein is cleaved during apoptosis: the role of cathepsin-G. Proc Natl Acad Sci USA. 2001;98:3814–3819. doi: 10.1073/pnas.071057398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird C H, Sutton V R, Sun J, Hirst C E, Novak A, Trapani J A, Bird P I. Selective regulation of apoptosis: the cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol Cell Biol. 1998;18:6387–6398. doi: 10.1128/mcb.18.11.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird P I. Serpins and regulation of cell death. Results Probl Cell Differ. 1998;24:63–89. doi: 10.1007/978-3-540-69185-3_4. [DOI] [PubMed] [Google Scholar]
- 8.Bird P I. Regulation of pro-apoptotic leukocyte granule serine proteinases by intracellular serpins. Immunol Cell Biol. 1999;77:47–57. doi: 10.1046/j.1440-1711.1999.00787.x. [DOI] [PubMed] [Google Scholar]
- 9.Boche I, Fanning E. Nucleocytoplasmic recycling of the nuclear localization signal receptor alpha subunit in vivo is dependent on a nuclear export signal, energy and RCC1. J Cell Biol. 1997;139:313–325. doi: 10.1083/jcb.139.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs L J, Stein D, Goltz J, Corrigan V C, Efthymiadis A, Hubner S, Jans D A. The cAMP-dependent protein kinase site (Ser312) enhances dorsal nuclear import through facilitating nuclear localization sequence/importin interaction. J Biol Chem. 1998;273:22745–22752. doi: 10.1074/jbc.273.35.22745. [DOI] [PubMed] [Google Scholar]
- 11.Brunk U T, Dalen H, Roberg K, Hellquist H B. Photo-oxidative disruption of lysosomal membranes causes apoptosis of cultured human fibroblasts. Free Radic Biol Med. 1997;23:616–626. doi: 10.1016/s0891-5849(97)00007-5. [DOI] [PubMed] [Google Scholar]
- 11a.Buzza, M. S., C. E. Hirst, C. H. Bird, P. Hosking, J. McKendrick, and P. I. Bird. The granzyme B inhibitor, PI-9, is present in endothelial and mesothelial cells, suggesting it protects bystander cells during immune responses. Cell Immunol., in press. [DOI] [PubMed]
- 12.Chu M, Bird C H, Teasdale M S, Bird P I. Turnover of thrombomodulin at the cell surface occurs at a similar rate to receptors that are not actively internalized. Thromb Haemostasis. 1998;80:119–127. [PubMed] [Google Scholar]
- 13.Chuang T L, Schleef R R. Identification of a nuclear targeting domain in the insertion between helices C and D in protease inhibitor-10. J Biol Chem. 1999;274:11194–11198. doi: 10.1074/jbc.274.16.11194. [DOI] [PubMed] [Google Scholar]
- 14.Coughlin P, Sun J, Cerruti L, Salem H H, Bird P. Cloning and molecular characterization of a human intracellular proteinase inhibitor. Proc Natl Acad Sci USA. 1993;90:9417–9421. doi: 10.1073/pnas.90.20.9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickinson J L, Bates E J, Ferrante A, Antalis T M. Plasminogen activator inhibitor 2 inhibits tumor necrosis factor alpha-induced apoptosis. Evidence for an alternate biological function. J Biol Chem. 1995;270:27894–27904. doi: 10.1074/jbc.270.46.27894. [DOI] [PubMed] [Google Scholar]
- 16.Efthymiadis A, Briggs L J, Jans D A. The HIV-1 Tat nuclear localization sequence confers novel nuclear import properties. J Biol Chem. 1998;273:1623–1628. doi: 10.1074/jbc.273.3.1623. [DOI] [PubMed] [Google Scholar]
- 17.Efthymiadis A, Shao H, Hubner S, Jans D A. Kinetic characterization of the human retinoblastoma protein bipartite nuclear localization sequence (NLS) in vivo and in vitro. A comparison with the SV40 large T-antigen NLS. J Biol Chem. 1997;272:22134–22139. doi: 10.1074/jbc.272.35.22134. [DOI] [PubMed] [Google Scholar]
- 18.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 19.Golstein P. Sensitivity of cytotoxic T cells to T-cell mediated cytotoxicity. Nature. 1974;252:81–83. doi: 10.1038/252081a0. [DOI] [PubMed] [Google Scholar]
- 20.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 21.Grigoryev S A, Bednar J, Woodcock C L. MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member. J Biol Chem. 1999;274:5626–5636. doi: 10.1074/jbc.274.9.5626. [DOI] [PubMed] [Google Scholar]
- 22.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 23.Hausmann G, O'Reilly L A, van Driel R, Beaumont J G, Strasser A, Adams J A, Haung D C S. Pro-apoptotic apoptosis protease-activating factor 1 (Apaf-1) has a cytoplasmic localization distinct from Bcl-2 or Bcl-xL. J Cell Biol. 2000;149:623–633. doi: 10.1083/jcb.149.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson B R, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res. 2000;256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- 25.Hood J K, Silver P A. In or out? Regulating nuclear transport. Curr Opin Cell Biol. 1999;11:241–247. doi: 10.1016/s0955-0674(99)80032-5. [DOI] [PubMed] [Google Scholar]
- 26.Hubner S, Xiao C Y, Jans D A. The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T-antigen nuclear localization sequence by importin. J Biol Chem. 1997;272:17191–17195. doi: 10.1074/jbc.272.27.17191. [DOI] [PubMed] [Google Scholar]
- 27.Huntington J A, Read R A, Carrell R W. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 28.Jans D A, Briggs L J, Jans P, Froelich C J, Parasivam G, Kumar S, Sutton V R, Trapani J A. Nuclear targeting of the serine proteinase granzyme A (fragmentin-1) J Cell Sci. 1998;111:2645–2654. doi: 10.1242/jcs.111.17.2645. [DOI] [PubMed] [Google Scholar]
- 29.Jans D A, Ackermann M J, Bischoff J R, Beach D H, Peters R. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV-40 T antigen proteins. J Cell Biol. 1991;115:1203–1212. doi: 10.1083/jcb.115.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jans D A, Jans P. Negative charge at the casein kinase II site flanking the nuclear localization signal of the SV40 large T-antigen is mechanistically important for enhanced nuclear transport. Oncogene. 1994;9:2961–2968. [PubMed] [Google Scholar]
- 31.Jans D A, Jans P, Briggs L J, Sutton V, Trapani J A. Nuclear transport of granzyme B (fragmentin 2) J Biol Chem. 1996;271:30781–30789. doi: 10.1074/jbc.271.48.30781. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins Y, McEntee M, Weis K, Greene W C. Characterization of HIV-1 Vpr nuclear import: analysis of signals and pathways. J Cell Biol. 1998;143:875–885. doi: 10.1083/jcb.143.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kranz D M, Eisen H N. Resistance of cytotoxic lymphocytes to lysis by a clone of cytotoxic T lymphocytes. Proc Natl Acad Sci USA. 1987;84:3375–3379. doi: 10.1073/pnas.84.10.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madison E L, Bird P. A vector, pSHT, for the expression and secretion of protein domains in mammalian cells. Gene. 1992;121:179–180. doi: 10.1016/0378-1119(92)90179-s. [DOI] [PubMed] [Google Scholar]
- 35.Miyata T, Nangaku M, Suzuki D, Inagi R, Uragami K, Sakai H, Okubo K, Kurokawa K. A mesangium-predominant gene, megsin, is a new serpin upregulated in IgA nephropathy. J Clin Investig. 1998;120:828–836. doi: 10.1172/JCI2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy R, Wente S R. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 37.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 38.Nagler-Anderson C, Verret C R, Firmenich A A, Berne M, Eisen H N. Resistance of primary CD8+ cytotoxic T lymphocytes to lysis by cytotoxic granules from cloned T cell lines. J Immunol. 1988;141:3299–3305. [PubMed] [Google Scholar]
- 39.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 40.Ochs R, Lischwe M, O'Leary P, Busch H. Localization of nucleolar phosphoproteins B23 and C23 during mitosis. Exp Cell Res. 1983;46:139–149. doi: 10.1016/0014-4827(83)90332-4. [DOI] [PubMed] [Google Scholar]
- 41.Ooms L, Nicholl J, Bird P, Sutherland G R. Localization of the human monocyte/neutrophil elastase inhibitor gene to chromosome 6p25. Chromosome Res. 1995;3:447. [Google Scholar]
- 42.Pemberton L F, Rosenblum J S, Blobel G. Nuclear import of the TATa-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J Cell Biol. 1999;145:1407–1417. doi: 10.1083/jcb.145.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perkins A, Cochrane A W, Ruben S M, Rosen C A. Structural and functional characterization of the human immunodeficiency virus rev protein. J Acquir Immune Defic Syndr. 1989;2:256–263. [PubMed] [Google Scholar]
- 44.Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function and regulation. J Biol Chem. 1994;269:15957–15960. [PubMed] [Google Scholar]
- 45.Remold-O'Donnell E. The ovalbumin family of serpin proteins. FEBS Lett. 1993;315:105–108. doi: 10.1016/0014-5793(93)81143-n. [DOI] [PubMed] [Google Scholar]
- 46.Rihs H P, Peters R. Nuclear transport kinetics depend on phosphorylation-site-containing sequences flanking the karyophilic signal of the simian virus 40 T-antigen. EMBO J. 1989;8:1479–1484. doi: 10.1002/j.1460-2075.1989.tb03531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schick C, Kamachi Y, Bartuski A J, Cataltepe S, Schechter N M, Pemberton P A, Silverman G A. Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase. J Biol Chem. 1997;272:1849–1855. doi: 10.1074/jbc.272.3.1849. [DOI] [PubMed] [Google Scholar]
- 48.Schick C, Pemberton P A, Shi G-P, Kamachi Y, Cataltepe S, Bartuski A J, Gornstein E R, Bromme D, Chapman H A, Silverman G A. Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry. 1998;37:5258–5266. doi: 10.1021/bi972521d. [DOI] [PubMed] [Google Scholar]
- 49.Schikendanz J, Scheidtmann K H, Walter G. Kinetics of nuclear transport and oligomerization of simian virus 40 large T antigen. Virology. 1986;148:47–57. doi: 10.1016/0042-6822(86)90402-2. [DOI] [PubMed] [Google Scholar]
- 50.Scott F L, Coughlin P B, Bird C, Cerruti L, Hayman J A, Bird P. Proteinase inhibitor 6 cannot be secreted, which suggests it is a new type of cellular serpin. J Biol Chem. 1996;271:1605–1612. doi: 10.1074/jbc.271.3.1605. [DOI] [PubMed] [Google Scholar]
- 51.Scott F L, Hirst C E, Sun J, Bottomley S P, Bird C H, Bird P I. The intracellular serpin proteinase inhibitor 6 (PI-6) is expressed in monocytes and granulocytes and is a potent inhibitor of the azurophilic granule proteinase, cathepsin G. Blood. 1999;93:2089–2097. [PubMed] [Google Scholar]
- 52.Shi L, Mai S, Israels S, Browne K A, Trapani J A, Greenberg A H. Granzyme B (GraB) autonomously crosses the cell membrane and perforin initiates apoptosis and GraB nuclear localization. J Exp Med. 1997;185:855–866. doi: 10.1084/jem.185.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smyth M J, Trapani J A. Granzymes: exogenous proteinases that induce target cell apoptosis. Immunol Today. 1995;16:202–206. doi: 10.1016/0167-5699(95)80122-7. [DOI] [PubMed] [Google Scholar]
- 54.Stein P E, Carrell R W. What do dysfunctional serpins tell us about molecular mobility and disease. Struct Biol. 1995;2:96–113. doi: 10.1038/nsb0295-96. [DOI] [PubMed] [Google Scholar]
- 55.Stochaj U, Rother K L. Nucleocytoplasmic trafficking of proteins: with or without Ran? Bioessays. 1999;21:579–589. [Google Scholar]
- 56.Sugimori T, Cooley J, Hoidal J R, Remold-O'Donnell E. Inhibitory properties of recombinant monocyte/neutrophil elastase inhibitor. Am J Respir Cell Mol Biol. 1995;13:314–322. doi: 10.1165/ajrcmb.13.3.7654387. [DOI] [PubMed] [Google Scholar]
- 57.Sun J, Bird C H, Sutton V, McDonald L, Coughlin P B, De Jong T A, Trapani J A, Bird P I. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J Biol Chem. 1996;271:27802–27809. doi: 10.1074/jbc.271.44.27802. [DOI] [PubMed] [Google Scholar]
- 58.Teasdale M S, Bird C H, Bird P. Internalization of the anticoagulant thrombomodulin is constitutive and does not require a signal in the cytoplasmic domain. Immunol Cell Biol. 1994;72:480–488. doi: 10.1038/icb.1994.72. [DOI] [PubMed] [Google Scholar]
- 59.Trapani J A. Dual mechanisms of apoptosis induction by cytotoxic lymphocytes. Int Rev Cytol. 1998;182:111–192. doi: 10.1016/s0074-7696(08)62169-5. [DOI] [PubMed] [Google Scholar]
- 60.Trapani J A, Bird P I. Granzymes: mediation of pro-apoptotic and nonapoptotic functions of cytolytic lymphocytes and regulation by endogenous and circulating inhibitors. In: Sitkovsky M V, Henkart P A, editors. Cytotoxic cells: basic mechanisms and medical applications. Philadelphia, Pa: Lippincott, Williams & Wilkins; 2000. pp. 179–196. [Google Scholar]
- 61.Trapani J A, Browne K A, Smyth M J, Jans D A. Localization of granzyme B in the nucleus. J Biol Chem. 1996;271:4127–4133. doi: 10.1074/jbc.271.8.4127. [DOI] [PubMed] [Google Scholar]
- 62.Trapani J A, Smyth M J, Apostolidis V A, Dawson M, Browne K. Granule serine proteinases are normal nuclear constituents of natural killer cells. J Biol Chem. 1994;269:18359–18365. [PubMed] [Google Scholar]
- 63.Tsay Y-G, Lin N Y, Voss P G, Patterson R J, Wang J L. Export of galactin-3 from nuclei of digitonin-permeabilized mouse 3T3 fibroblasts. Exp Cell Res. 1999;252:250–261. doi: 10.1006/excr.1999.4643. [DOI] [PubMed] [Google Scholar]
- 64.Vancurova I, Lou W, Paine T M, Paine P L. Nucleoplasmin uptake by facilitated transport and intranuclear binding. Eur J Cell Biol. 1993;62:22–33. [PubMed] [Google Scholar]
- 65.Vyakarnam A, Lenneman A J, Lakkides K M, Patterson R J, Wang J L. A comparative nuclear localization study of galectin-1 with other splicing components. Exp Cell Res. 1998;242:419–428. doi: 10.1006/excr.1998.4111. [DOI] [PubMed] [Google Scholar]
- 66.Whisstock J, Skinner R, Lesk A M. An atlas of serpin conformations. Trends Biochem Sci. 1998;23:63–67. doi: 10.1016/s0968-0004(97)01172-9. [DOI] [PubMed] [Google Scholar]
- 67.Whisstock J C, Irving J A, Bottomley S P, Pike R N, Lesk A M. Serpins in the Caenorhabditis elegans genome. Proteins. 1999;36:31–41. doi: 10.1002/(sici)1097-0134(19990701)36:1<31::aid-prot3>3.3.co;2-h. [DOI] [PubMed] [Google Scholar]
- 68.Yodoi J, Teshigawara K, Nikaido T, Fukui K, Noma T, Honjo T, Takigawa M, Sasaki M, Minato N, Tsudo M, et al. TCGF (IL 2)-receptor inducing factor(s). I. Regulation of IL 2 receptor on a natural killer-like cell line (YT cells) J Immunol. 1985;134:1623–1630. [PubMed] [Google Scholar]
- 69.Zhou Q, Salvesen G S. Activation of pro-caspase-7 by serine proteases includes a non-canonical specificity. Biochem J. 1997;324:361–364. doi: 10.1042/bj3240361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Q, Snipas S, Orth K, Muzio M, Dixit V, Salvesen G. Target protease specificity of the viral serpin CrmA. J Biol Chem. 1997;272:7797–7800. doi: 10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]
- 71.Zou Z, Anisowicz A, Hendrix M J C, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]