Abstract
The molecular mechanism of mRNA degradation in the chloroplast consists of sequential events including endonucleolytic cleavage, the addition of poly(A)-rich sequences to the endonucleolytic cleavage products, and exonucleolytic degradation by polynucleotide phosphorylase (PNPase). In Escherichia coli, polyadenylation is performed mainly by poly(A)-polymerase (PAP) I or by PNPase in its absence. While trying to purify the chloroplast PAP by following in vitro polyadenylation activity, it was found to copurify with PNPase and indeed could not be separated from it. Purified PNPase was able to polyadenylate RNA molecules with an activity similar to that of lysed chloroplasts. Both activities use ADP much more effectively than ATP and are inhibited by stem-loop structures. The activity of PNPase was directed to RNA degradation or polymerization by manipulating physiologically relevant concentrations of Pi and ADP. As expected of a phosphorylase, Pi enhanced degradation, whereas ADP inhibited degradation and enhanced polymerization. In addition, searching the complete Arabidopsis genome revealed several putative PAPs, none of which were preceded by a typical chloroplast transit peptide. These results suggest that there is no enzyme similar to E. coli PAP I in spinach chloroplasts and that polyadenylation and exonucleolytic degradation of RNA in spinach chloroplasts are performed by one enzyme, PNPase.
The chloroplast is the site of photosynthesis and other essential biosynthetic activities in plant cells. The chloroplast's structural proteins and enzymes are encoded by both nuclear and chloroplast genomes. During chloroplast development, chloroplast gene expression is tightly regulated at many levels, including mRNA accumulation (reviewed in references 3, 19, and 39). RNA metabolism involves a series of steps that are dependent on RNA secondary structures, nucleases, and regulatory RNA-binding proteins. A 100-kDa RNA-binding protein that is homologous to the bacterial exoribonuclease polynucleotide phosphorylase (PNPase) was isolated from a chloroplast protein extract and found to be the protein responsible for most exoribonucleolytic activity. The homology of the chloroplast and the bacterial enzymes was observed both in amino acid sequences and in biochemical characteristics (20).
PNPase is a phosphorolytic exonuclease involved in degrading prokaryotic and organelle RNAs (31, 40). It is a reversible enzyme that can degrade RNA by using inorganic phosphate (Pi) or synthesize RNA by using any nucleoside diphosphate. Until recently, it has always been assumed that due to the high concentration of Pi in bacteria and chloroplasts (about 10 mM), PNPase worked only degradatively (31). Indeed, PNPase has been shown to be an important component of the mRNA degradation system in bacteria and chloroplasts (6, 8, 17, 37). However, as described below, recent studies have shown that PNPase can also function in vivo as a polymerase in bacterial cells (33). PNPase is composed of several domains homologous to those in other RNA-binding proteins and ribonucleases. These include the KH and S1 domains, which are located at the C-terminal region, and two RNase PH domains that are also found in other phosphorolytic RNase enzymes (43). Structural and biochemical analyses have revealed a trimeric quaternary structure of PNPase (43). Part of the PNPase population in Escherichia coli is associated with the endoribonuclease RNase E, an RNA helicase, an enolase, and possibly several other proteins in a high-molecular-weight complex called a degradosome (6, 8, 17, 27, 37). In the chloroplast, PNPase was purified as a complex of about 600 kDa, composed only of this protein (20; Baginsky et al., submitted for publication). Therefore, a degradosome complex similar to that of E. coli does not exist in the chloroplast.
The molecular mechanism of RNA degradation in the chloroplast has been elucidated during the past few years and was found to be very similar to that of bacteria (19, 39). In both bacteria and chloroplasts, the first event is endoribonuclease cleavage of the RNA molecule. The endoribonuclease cleavage is followed by the addition of a poly(A) tail in the bacteria and poly(A) (23) or a poly(A)-rich tail in the chloroplasts (28–30). The polyadenylated cleavage products are then directed to rapid exonucleolytic degradation by PNPase and RNase II in E. coli and by PNPase and possibly other exoribonucleases in the chloroplasts (30). Therefore, polyadenylation is part of the RNA degradation machinery in bacteria, chloroplasts, and possibly also in plant mitochondria (6, 8, 15, 19, 24, 32, 37–39, 41). In contrast to what is observed with bacteria, evidences for activity of a 5′ to 3′ exoribonuclease in the chloroplasts of the green alga Chlamydomonas reinhardtii were obtained (10, 11, 34). Whether or not such an enzyme exists in the chloroplasts of higher plants and what its function is in the mechanism of RNA degradation are still open questions.
E. coli PAP I is responsible for 90 to 95% of the poly(A) tails, which are generally adenosine homopolymers. A smaller amount of nucleotides other than adenosine were reported in stationary-phase cells and when PNPase was overexpressed (5, 21, 33). It was recently reported that the second RNA polyadenylation activity in E. coli, which takes over in the absence of PAP I, is carried out by PNPase (33). In this situation, the elongated tails were found to consist not only of adenosines but also of the three other nucleotides. In spinach chloroplasts, the polyadenylated tails could be several hundred nucleotides long and were found to consist of about 70% adenosines, 25% guanosines, and 5% cytosines and uridines (28). Heterologous poly(A)-rich tails were obtained when several chloroplast transcripts including mRNAs and rRNA were analyzed (28; Anbussi-AbuToami and Schuster, unpublished results). Therefore, the heteropolymeric tails obtained by reverse transcription-PCR from spinach chloroplast transcripts resemble the tails obtained from E. coli in the absence of PAP I when PNPase is the polyadenylation enzyme. In contrast to the situation in spinach chloroplasts, only polyadenylated tails were obtained when five chloroplast transcripts, including mRNAs, rRNAs, and tRNAs of C. reinhardtii, were analyzed (23). Therefore, the poly(A) tails of chloroplast transcripts in C. reinhardtii resemble those in E. coli when PAP I is active.
In contrast to what occurs in bacteria, RNA polyadenylation in plants is expected to be of both eukaryotic and prokaryotic types. In the eukaryotic process, nucleus-derived RNA polymerase II transcripts are polyadenylated and this polyadenylated tract is important for translation initiation. The prokaryotic process occurring in the chloroplasts and possibly also in the mitochondria includes the polyadenylation of organelle-encoded transcripts as part of the RNA-degradation mechanism (39). In order to understand the molecular mechanism and the elements that modulate and control the RNA polyadenylation reaction in the chloroplast, we sought to identify the chloroplast polyadenylation enzyme(s). Since the mechanism of mRNA polyadenylation and degradation in the chloroplast is so similar to that in E. coli, the default hypothesis was that our target was a chloroplast homologue of PAP I. Surprisingly, however, a PAP I homologue could not be detected in chloroplasts from either spinach or pea. When in vitro polyadenylation activity from pea leaves was purified previously, a fraction containing two proteins was purified (26). Since one of these was identified as PNPase, it was proposed to be a component of the polyadenylation machinery in the chloroplast whose role was to bind the RNA to be polyadenylated but not the catalytic polyadenylating enzyme by itself (26). Here, we suggest that chloroplast PNPase performs both polyadenylation and exonucleolytic degradation. The observation that no putative PAP protein carrying a typical chloroplast transit peptide could be identified in the completely sequenced Arabidopsis genome is in agreement with this hypothesis.
MATERIALS AND METHODS
Chloroplast isolation, preparation of soluble protein extract, and lysed-chloroplast system.
Chloroplasts were purified on Percoll gradients from young leaves of hydroponically grown spinach plants (Spinacia oleracea cv. Viroflay) under a 10.5-h light–13.5-h dark cycle, as previously described (28). To prepare lysed chloroplasts, intact chloroplasts were resuspended in a volume equal to that of the pellet with buffer E (20 mM HEPES [pH 7.9]–60 mM KCl–12.5 mM MgCl2–0.1 mM EDTA–2 mM dithiothreitol–17% glycerol) so that each constituent was diluted to one-half the concentration in the chloroplasts (22, 29). The chloroplasts were lysed by freezing and thawing (22), and [32P]RNA was added in the amount of 10,000 cpm (1.3 fmol) and incubated at 25°C for the times indicated in the figures. Reactions were terminated as described previously (22), and RNA was extracted and analyzed by denaturing gels and autoradiography. A soluble protein extract was prepared from isolated intact chloroplasts, as previously described (16).
Purification of PNPase.
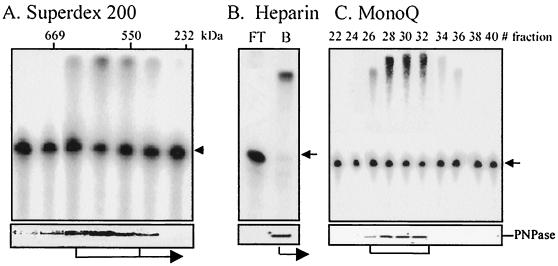
The soluble chloroplast protein extract was fractionated on a size-exclusion Superdex 200 (Pharmacia) column. Fractions containing protein complexes of 550 to 650 kDa were pooled and applied to a 1-ml heparin column (Hi-Trap; Pharmacia) that was developed with a linear gradient of KCl in buffer E. Proteins eluted at 0.2 M KCl were dialyzed against buffer E and applied to a Mono-Q column (Pharmacia). The column was developed with a linear gradient of KCl in buffer E, and the PNPase was eluted at 0.3 M as a single silver-stained polypeptide (30) (see Fig. 3).
FIG. 3.
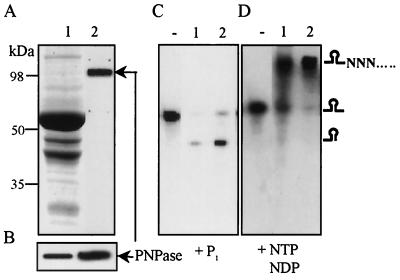
Purified PNPase is active in vitro both as a polymerase and as an exoribonuclease. (A) Protein profile (silver-stained gel electrophoresis) of chloroplast soluble protein extract (1) and purified PNPase (2). The identification of PNPase by an immunoblot assay is presented in panel B. (C) RNase activity of PNPase. [32P]RNA representing the psbA gene was incubated without (-) or with soluble protein extract (1) or purified PNPase (2) for 35 min in the presence of 10 mM Pi. (D) Polymerization activity of PNPase. Conditions were the same as for panel B except that 1 mM ADP was added to the reaction mixture and no Pi ions were present. A schematic representation of the RNA molecules is presented on the right.
Synthetic RNAs.
The plasmid used for the in vitro transcription of part of the spinach chloroplast gene psbA (encoding the D1 protein of photosystem II) has been previously described (28). The stem-loop structure of the malE-malF intergenic region (GenBank accession no. M19202), with the single mutation replacing A to C at the fifth nucleotide of the stem in order to strengthen the stem structure (4), was PCR amplified using the corresponding oligonucleotide primers and genomic DNA (45). The T7 promoter was included in the first oligonucleotide to drive in vitro transcription. Six cytosine residues were added to the second oligonucleotide to generate RNA molecules with the addition of six residues 3′ to the stem-loop structure, as shown in Fig. 7. RNAs were transcribed using T7 RNA polymerase and radioactively labeled with [α-32P]UTP to a specific activity of 8 × 103 to 10 × 103 cpm/fmol (45). The full-length transcription products were purified from 5% denaturing polyacrylamide gels.
FIG. 7.
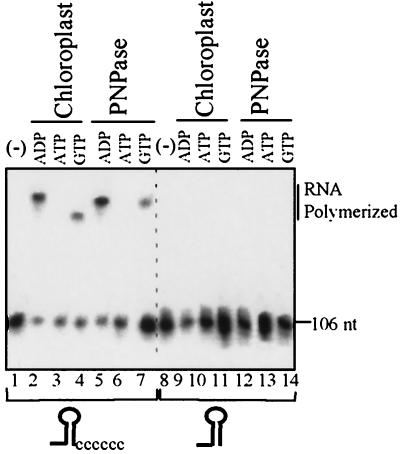
A stem-loop structure inhibits the polymerization activities of both lysed chloroplasts and purified PNPase. A 106-nucleotide [32P]RNA fragment representing the malE-malF intergenic region of E. coli with or without the addition of six cytosine residues, as shown schematically at the bottom, was incubated with lysed chloroplasts or purified PNPase with the addition of ADP (0.5 mM), ATP (1 mM), or GTP (1 mM) as indicated. Lane (-), input RNA. Following an incubation of 5 min for the lysed chloroplasts and 25 min for the purified PNPase, RNA was purified and analyzed.
In vitro degradation and polyadenylation activity assays.
In vitro degradation and polyadenylation experiments were carried out as previously described (28). Briefly, in vitro-synthesized RNA (1 fmol) was incubated with lysed chloroplasts (≈10 mg/ml), a chloroplast soluble protein extract (5 mg/ml), or isolated PNPase (1.5 μg/ml) for the times indicated in the figure legends. Following incubation, the RNA was analyzed by gel electrophoresis and autoradiography.
Determination of the PNPase concentration in the lysed-chloroplast system.
To determine the amount of PNPase in lysed chloroplasts, different volumes of lysed chloroplasts and purified PNPase were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Coomassie blue staining and immunoblotting with PNPase-specific antibodies (30), and the intensities of the immunological signals were compared. The amount of purified PNPase was determined by comparison to a bovine serum albumin dilution series in the same gel. Protein concentrations were determined using the Bio-Rad protein assay kit.
Determination of ATP, Pi, and chlorophyll concentrations.
To analyze the ATP and Pi concentrations, lysed chloroplasts were heated to 90°C for 3 min followed by centrifugation to remove insoluble material. ATP and Pi concentrations were determined using the Roche ATP Bioluminescence CLS II kit and the method of Chen et al. (7), respectively. Chlorophyll concentration was determined in 80% acetone (29).
Sequence analyses.
The BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and TIGR (http://www.tigr.org/tdb/ath1/htmls/index.html) sites were used to identify putative PAP and PNPase genes in the genome of Arabidopsis. ChloroP (http: //www.cbs.dtu.dk/services/ChloroP/) (14) and TargetP (http://www.cbs.dtu.dk /services/TargetP/) (13) were used to analyze whether or not these proteins were likely to contain a chloroplast transit peptide and therefore be localized in the chloroplast.
RESULTS
Copurification of PNPase and polyadenylation activity.
In recent years, polyadenylation has been found to be an integral part of RNA degradation in E. coli and chloroplasts. To further our understanding of the process in chloroplasts, we set out to identify and isolate the enzyme that polyadenylates RNA in this organelle. We first searched for a chloroplast homologue to E. coli PAP I using antibodies raised against it (35); however, no reaction was observed with chloroplast proteins of Arabidopsis or Chlamydomonas (data not shown). Only a small amount of cross-reactivity was observed with a spinach protein that was purified and identified as the nucleotidyl transferase enzyme which is a distant relative of PAP I (35).
We next used the in vitro polyadenylation assay to follow the enzyme during purification. Since we knew that GTP could be used as a substrate for polymerization activity, it was also used during the purification steps (28). When GTP was added to the in vitro polyadenylation assay, the activity was much more robust than when ATP was used (see below), most likely because polyadenylated RNA is very rapidly degraded in the extract, whereas polyguanylidated RNA is stable because of a tertiary structure formed by poly(G) that is resistant to exonucleolytic degradation (10, 12, 34, 44). In addition, we found that ADP gave much better activity than ATP (see below). This observation already suggested the possibility that PNPase was the enzyme responsible for this activity, since it is a phosphorylase enzyme that uses nucleoside diphosphates rather than triphosphates as for PAP I. Therefore, we decided to analyze each fraction using ATP, ADP, and GTP for polymerization activity and using immunoblotting for PNPase. First, it was necessary to determine whether polymerization activity was exclusively present in the soluble fraction of the chloroplast. Purified chloroplasts were gently lysed by osmotic disruption and separated into soluble and insoluble fractions by centrifugation. The membrane pellet was extensively washed of residual soluble proteins, and the three fractions were analyzed for polymerization activity using a synthetic [32P]RNA as a substrate. Polymerization activity using nucleoside triphosphates was found exclusively in the soluble fraction (Fig. 1). No activity was found in the membrane fraction even when the autoradiogram was overexposed (not shown).
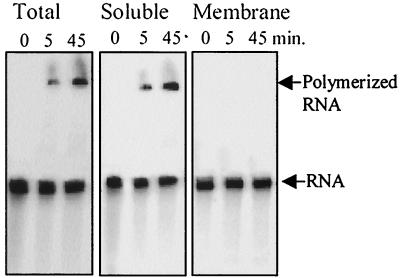
FIG. 1.
RNA polyadenylation activity is carried out exclusively in the soluble fraction of the chloroplast. Lysed chloroplasts (Total) and soluble and membrane fractions were incubated with [32P]RNA (representing the psbA gene) and 5 mM GTP. Following incubation for the times indicated in the figure, RNA was purified and analyzed by denaturing gel electrophoresis and phosphorimaging. Equivalent results were obtained when ADP or ATP was used instead of GTP.
We then further fractionated the soluble proteins using a size-exclusion Superdex 200 column and observed that the ADP, ATP, and GTP polymerization activities, as well as PNPase, were found at about 600 kDa and that no activity or PNPase signal was obtained in any other fraction of the column (Fig. 2A). The Superdex 200 fractions containing the polymerization activity were pooled and applied to a heparin column. Both the activity and PNPase bound to the column (Fig. 2B). The next step involved applying the bound fraction to an anion exchange Mono-Q column. Again, a perfect coelution was obtained for the polymerization activity and the PNPase protein detected by the antibodies (Fig. 2C). This purification profile was obtained with each of the three nucleotides used. In addition, we could not detect activity in any other fraction during the purification process, even when a large amount of protein was used and the autoradiograms were overexposed (not shown). Similar results of cofractionation of PNPase and polymerization activity were previously obtained when an extract of total leaf proteins from pea was fractionated (25, 26). These results suggested that the only polymerization activity in the chloroplast could be attributed to PNPase. In order to see that a putative PAP, possibly present in the chloroplast, is not inhibited by a protein or another component of the chloroplast extract, we performed an experiment in which E. coli PAP was added to the chloroplast soluble proteins and its polyadenylation activity was analyzed throughout the purification procedure. It was found that the polyadenylation activity was not inhibited (data not shown). Nevertheless, we could not completely exclude the possibility that in addition to PNPase, another polymerase, such as PAP I, is present in the chloroplast and its activity is inhibited in the in vitro polymerization assay or even that this protein fortuitously copurified with PNPase, for example as part of a complex. Therefore, we decided to determine whether purified PNPase could perform RNA polymerization under physiological conditions.
FIG. 2.
Copurification of PNPase and the RNA polymerization activity. (A) A chloroplast soluble protein extract was fractionated on a Superdex 200 size-exclusion column. Fractions were assayed for polymerization activity using [32P]RNA representing the psbA gene and GTP. Equivalent results were obtained when ATP, GTP, or ADP was used. Following incubation, the RNA was analyzed using denaturing gel electrophoresis (upper panel). The presence of PNPase in each fraction was analyzed using immunoblotting (lower panel). The migration of molecular weight markers thyroglobulin (669 kDa), RubP-carboxylase (550 kDa), and catalase (232 kDa) fractionated on the same column is shown at the top of the left panel. Only the column fractions having molecular masses exceeding 232 kDa are shown, since no activity whatsoever was detected in any other fraction. (B) The three fractions of the Superdex 200 column containing polymerization activity, as shown at the bottom of panel A, were pooled and loaded on a heparin column. The unbound (FT) and bound (B) fractions were analyzed as described in the legend to panel A for the Superdex 200 column. (C) The bound fraction of the heparin column was dialyzed and applied to a Mono-Q column that was developed with a KCl gradient. Fractions 26 to 32 were found to possess all of the polymerization activity and the PNPase polypeptide (see Fig. 3).
Characterization of the PNPase polymerization and exoribonuclease activities.
In order to explore the possibility of PNPase being responsible for RNA chloroplast polyadenylation, we first purified the PNPase to homogeneity (Fig. 3, panels A and B) and compared the degradation and polymerization activities of a chloroplast soluble protein extract to those of purified PNPase. As shown in Fig. 3, their activities were very similar. RNA degradation was observed without the addition of nucleotides, and the degradation activity paused at the stem-loop structure located in the middle of the RNA molecule (Fig. 3C). The addition of Pi dramatically enhanced the degradation activity of both the soluble protein extract and the purified PNPase. However, when Pi was absent and nucleotides were added to the in vitro assay, no degradation was observed and the RNA was elongated by several hundred nucleotides (Fig. 3D). When different nucleotides were analyzed in a concentration-dependent assay for polymerization activity using purified PNPase, the order (best substrate first) was ADP (0.05) > GDP (0.075) > ATP (0.25) > GTP (0.5) ≈ CTP (0.5) > UTP (2) (the numbers in parentheses are the millimolar concentrations at which one-half of the maximal activity was obtained). These results suggested that the Pi and ADP concentrations could significantly modulate the direction of PNPase activity for degradation or polymerization.
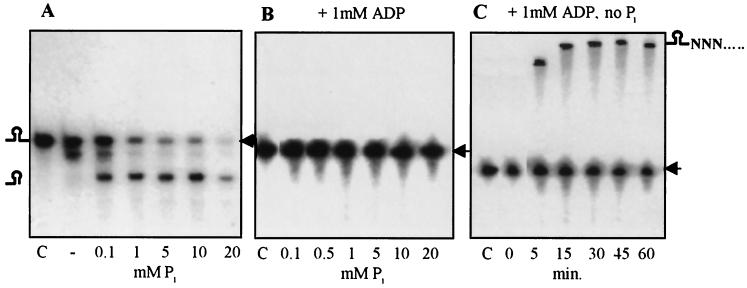
In order to better characterize the modulation of PNPase activity by Pi and ADP, we assayed the effects of different concentrations of each one separately as well as in combination. Figure 4A shows that Pi strongly enhanced RNA degradation activity in a concentration-dependent manner. This result was previously obtained for the chloroplast and E. coli PNPases and was expected since Pi is a substrate, in addition to RNA, for the degradation activity (31). At low Pi concentrations, purified PNPase paused at the stem-loop structure, as did the E. coli PNPase (4). However, at a concentration of 20 mM, where the activity is strongly enhanced, the enzyme can degrade the RNA including the stem-loop structure (Fig. 4A). When ADP was included in the reaction at a concentration of 1 mM, the degradation activity of the purified enzyme was completely blocked, and the RNA molecules remained stable during the incubation time even at a concentration of Pi as high as 20 mM (Fig. 4B). However, when only ADP was present, a time-dependent elongation of the RNA molecule was observed (Fig. 4C). These results demonstrated that PNPase activity is modulated between degradation and polyadenylation by concentrations of Pi and ADP known to be physiologically relevant (see below). When purified PNPase was incubated with RNA and ADP for longer times or when much more protein or ADP was present in the reaction mixture, polyadenylation was transient and RNA degradation followed (data not shown, but see Fig. 6).
FIG. 4.
Effects of Pi and ADP on PNPase activity. Purified PNPase (150 ng) was incubated with [32P]RNA representing the psbA gene (marked by an arrowhead) for 45 min with the addition of Pi and ADP in the concentrations shown. The first lane in each panel (marked C) represents the input RNA. In panel C, the RNA was incubated with purified PNPase and 1 mM ADP. The RNA was purified and analyzed at the time points indicated at the bottom. Schematic representations of the RNA molecules are presented on the right and left.
FIG. 6.
Transient polyadenylation and degradation of RNA in lysed chloroplasts and by purified PNPase. Lysed chloroplasts (Chloroplast) or 150 ng of purified PNPase (PNPase) were incubated with a 296-nucleotide [32P]RNA corresponding to the 3′ end of the psbA gene, with the addition of 1 and 5 mM ADP for the lysed chloroplasts and purified PNPase, respectively. At the time points indicated at the top, the RNA was purified and analyzed. For time point zero the sample was taken several seconds following the initiation of the reaction. In the case of the lysed chloroplast a degradation of the input [32P]RNA was already observed due to the high protein concentration of this system.
Comparison of polyadenylation and degradation activities in lysed chloroplasts and with purified PNPase.
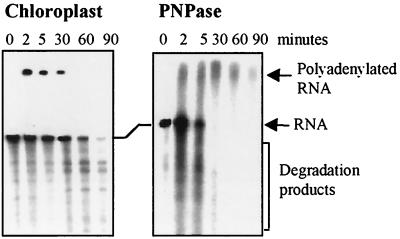
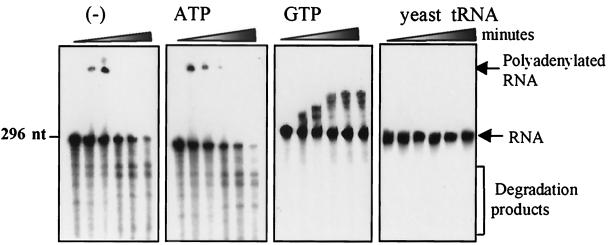
Since PNPase activity could be modulated between polyadenylation and degradation according to Pi and ADP concentrations, we wanted to analyze and compare these activities to those occurring in a situation that is as similar as possible to the in vivo situation. The lysed-chloroplast system has been previously used to analyze the degradation pathway of psbA mRNA (22, 28). In this system, chloroplasts are isolated and gently lysed by freezing and thawing in a buffer amount equivalent to the chloroplast pellet volume. In this way, every constituent is diluted twofold in comparison to intact chloroplasts. We first determined the internal concentrations of ATP, Pi, and PNPase in that system as described in Materials and Methods. The ATP and Pi concentrations were measured as 2 and 18 mM, respectively, which would correspond to 4 and 36 mM in the chloroplast; PNPase concentration was determined to be 0.0665 mg/ml, while chlorophyll and protein concentrations were 4 and 70 mg/ml, respectively. Although these concentrations are similar to those measured previously (18, 42), some changes could have occurred following the breakage of the chloroplasts. The ADP concentration in the chloroplast was previously reported to be about 0.5 mM (18, 42). Together, the ADP, ATP, and Pi concentrations are in agreement with those used for the PNPase assays shown in Fig. 3 and 4. The amount of PNPase was determined to be 0.1% of total chloroplast proteins. We then examined polyadenylation and degradation activities of lysed chloroplasts in the presence of different nucleotides. When no addition other than the [32P]RNA was made, transient polyadenylation followed by degradation of the RNA was observed (Fig. 5, left panel). This polymerization activity probably used the internal chloroplast nucleotides as substrates. The transient polymerization of the RNA added externally followed by its degradation is in agreement with what was predicted from the model of polyadenylation-dependent RNA degradation in the chloroplast (19, 39). According to this model, RNA polyadenylation precedes the exoribonucleolytic degradation. In the lysed-chloroplast system this process occurs very rapidly, as could be observed with the samples of time zero. These samples were taken several seconds following the initiation of the reaction, and degradation products of the input RNA were already observed (Fig. 5; see also Fig. 6). The addition of 1 mM ADP or 5 mM ATP to the system produced similar results, but the polyadenylation activity occurred more extensively and rapidly, probably due to the addition of these substrates to the internal chloroplast concentrations (Fig. 5; see also Fig. 6).
FIG. 5.
Transient polyadenylation and degradation of RNA in lysed chloroplasts. Lysed chloroplasts (3.5 mg of protein) were incubated with a 296-nucleotide [32P]RNA corresponding to the 3′ end of the psbA gene without (-) or with the addition of 5 mM ATP, 5 mM GTP, or 0.5 mg of yeast tRNA per ml, as indicated at the top. At time points 0, 2, 5, 30, 60, and 90 min, RNA was purified and analyzed. For time point zero the sample was taken several seconds following the initiation of the reaction. In the case of the lysed chloroplast, a degradation of the input [32P]RNA was already observed due to the high protein concentration of this system.
Another interpretation of these results could be that the RNA added externally undergoes either polyadenylation or direct degradation. Transient polyadenylation would therefore not be a prerequisite for the degradation that follows. Poly(G) is an effective inhibitor of exoribonucleases due to its formation of a strong tertiary structure (9, 10). Indeed, the addition of GTP resulted exclusively in polymerization activity (Fig. 5). Polyadenylation is thought to precede exonucleolytic degradation (19, 39), and this result offers conclusive evidence that this is the case in the lysed-chloroplast system. Finally, we note that the addition of a large amount of Saccharomyces cerevisiae tRNA to lysed chloroplasts completely blocked both polyadenylation and degradation, possibly due to competition for PNPase (Fig. 5). These results are consistent with the possibility that PNPase performs both polymerization and exonucleolytic degradation. In order to further explore this hypothesis, we compared the polymerization activity of purified PNPase and lysed chloroplasts. When [32P]RNA was added to lysed chloroplasts or purified PNPase in the presence of ADP, polyadenylation occurred in both cases within 2 min and was followed by degradation of the polyadenylated molecules (Fig. 6). Therefore, the activities of RNA polyadenylation and degradation in the lysed chloroplasts could be mimicked by purified PNPase. It should be noted that the ADP concentration was 5 mM in this experiment, in comparison to the 1 mM used in the experiment illustrated in Fig. 4C. This explains why only polyadenylation is observed in Fig. 4C, whereas both polyadenylation and degradation are seen in Fig. 6. In addition, although the activities of the purified PNPase and the lysed chloroplasts were generally similar, several differences could be detected even when the protein and ADP concentrations were carefully tuned. For example, the input [32P]RNA disappeared much more rapidly when incubated with the purified PNPase (Fig. 6). The small differences between the polyadenylation and degradation of RNA in the purified PNPase compared to the lysed chloroplast fractions could be attributed to the presence of other proteins and RNA molecules at high concentrations in the lysed chloroplast system and not with the purified PNPase. The lysed chloroplast fraction contains, in addition to the PNPase, many other RNA-binding proteins and chloroplast RNA molecules that compete with the PNPase for the externally added [32P]RNA. Taken together, these results suggest that PNPase could be responsible for both polyadenylation and degradation activities in the chloroplast.
The polymerization activities of both lysed chloroplasts and PNPase are inhibited by a stem-loop structure and prefer ADP over ATP.
We have previously shown that polyadenylation activity in chloroplast extracts is inhibited by a 3′ end stem-loop structure (28). It was suggested that this inhibition prevents RNA degradation since the degradation process is initiated by an endonucleolytic cleavage removing the stem-loop, producing a cleavage product containing a 3′ end that is not protected by a stem-loop structure. This unprotected 3′ end can then undergo polyadenylation and exonucleolytic degradation. If PNPase were responsible for polyadenylation activity in the chloroplast, we would expect its polymerization activity to be inhibited by stem-loop structures. To test this, we compared the polymerization activity of lysed chloroplasts to that of purified PNPase using RNA substrates terminating either with a stable stem-loop structure or with the addition of a platform of six cytosines (45) (Fig. 7). Polymerization activity was observed both for the lysed chloroplast and for the purified PNPase exclusively with the RNA that was extended by six nucleotides (Fig. 7). Similar to the results presented in Fig. 5 and 6, activity was observed for both systems with ADP and GTP (lanes 2, 4, 5, and 7). Under these experimental conditions, only degradation was observed with ATP (lanes 3 and 6). This is because polyadenylation occurred rapidly and is transient in both systems. Polyadenylation was observed in experiments where shorter time points were taken (not shown; Fig. 5).
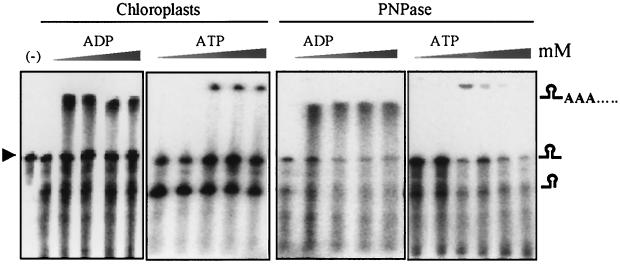
PNPase as a phosphorolytic enzyme uses ADP much more efficiently than ATP for polymerization, whereas in the case of E. coli PAP the opposite is true. If PNPase were the only polyadenylation enzyme in the chloroplast, then the preference for ADP should also be observed in the lysed-chloroplast system. To explore this possibility, we incubated [32P]RNA with lysed chloroplasts or purified PNPase in the presence of increasing concentrations of either ADP or ATP. The results of this experiment, presented in Fig. 8, show that polymerization activity in lysed chloroplasts is very similar, if not identical, to the activity of purified PNPase. ADP was found to be more effective, with a concentration of <50 μM for half saturation compared to approximately 150 μM for ATP. Taken together, the biochemical characteristics of polymerization activities of lysed chloroplasts and the purified PNPase strongly argue that PNPase is responsible for polyadenylation in the chloroplast.
FIG. 8.
Similar polymerization activities of lysed chloroplasts and purified PNPase. Lysed chloroplasts and purified PNPase (150 ng of protein) were incubated with [32P]RNA and ADP or ATP at concentrations of 0, 0.1, 0.25, 0.5, and 1.5 mM. The incubation times were 5 min for the lysed chloroplasts and 25 min for the purified PNPase. The input RNA is marked with an arrowhead in lane (-). A schematic representation of the RNA molecules is shown on the right.
Putative Arabidopsis PAPs do not contain typical chloroplast transit peptides.
Most of the polyadenylation activity in E. coli and eukaryotic cells is carried out by PAP. Since no chloroplast open reading frames encode proteins related to PAP, any such protein must be encoded in the nucleus. Using the complete Arabidopsis genome sequence, we found six putative PAP encoding sequences. Using ChloroP and TargetP (see Materials and Methods) for the identification of chloroplast transit peptides, none of these were predicted to be chloroplast or mitochondrion targeted (Table 1). In contrast, at least two different PNPases were detected, of which one, located on chromosome 3, is predicted to contain a chloroplast transit peptide. The second, located on chromosome 5, is predicted to be located at the mitochondria (Table 1). Together with our biochemical data, these results suggest that there is no PAP similar to that of E. coli or the nucleus in higher-plant chloroplasts. However, it should be noted that an argument based on the ChloroP and TargetP prediction is tentative at best and should be backed by experimental evidence.
TABLE 1.
PAP and PNPase sequences in the Arabidopsis genome
| Protein no. | Gene | Putative function | Chromosome | Length (no. of amino acids) | ChloroP scorea | Protein location in the cellb |
|---|---|---|---|---|---|---|
| 1 | AAF97277 | PAP | 1 | 713 | 0.44 | NPc |
| 2 | AAD32915 | PAP | 2 | 241 | 0.42 | NP |
| 3 | AAF66438 | PAP | 2 | 795 | 0.44 | NP |
| 4 | AAF63637 | PAP | 3 | 482 | 0.43 | NP |
| 5 | T10692 | PAP | 4 | 729 | 0.47 | NP |
| CAB80002 | ||||||
| 6 | BAA97232.1 | PAP | 5 | 499 | 0.47 | NP |
| 1 | CAB43864 | PNPase | 5 | 991 | 0.54 | Mitochondria |
| CAB87625 | ||||||
| CAB43865 | ||||||
| T48631 | ||||||
| 2 | AAF00646 | PNPase | 3 | 922 | 0.57 | Chloroplast |
| AAF03462 | ||||||
| CAB85703 |
Score according to ChloroP for a chloroplast transit peptide. Usually a protein with a score below 0.5 has a low probability of containing a chloroplast transit peptide.
As predicted by TargetP.
NP, none predicted.
DISCUSSION
In order to study the molecular mechanism of RNA polyadenylation in the chloroplast, we sought to purify and characterize the enzyme responsible for this activity. The most likely candidate was a putative chloroplast homologue of E. coli PAP, which is its major polyadenylating enzyme. Nevertheless, our attempts, as well as others', to find such a homologue in the chloroplasts failed (25, 26). In the absence of PAP, the most likely candidate became PNPase, due to the highly reversible nature of its reaction mechanism (31). Indeed, several attempts to biochemically purify chloroplast polyadenylation activity yielded PNPase (25, 26), which has been assumed for a long time to be active only as an exoribonuclease in bacteria and chloroplasts due to their high concentrations of Pi (31). However, Mohanty and Kushner recently showed that PNPase could be active in vivo as a polymerase in bacteria (33). This polymerization activity is mostly masked by PAP but becomes the major one in PAP deletion strains. In this situation, RNA tails are not the adenosine homopolymers resulting from PAP activity but rather are comprised of all four nucleotides, which can be utilized by PNPase (33). In other words, the nature of the tails reveals the nature of the enzyme. In this respect, we note that heterogeneous mRNA tails were found in spinach chloroplasts, consistent with PNPase activity (28) and also with the lack of a PAP-encoding gene preceded by a typical chloroplast transit peptide in the Arabidopsis genome. In contrast, reverse transcription-PCR experiments in Chlamydomonas revealed only homopolymeric adenosine tails in the chloroplast (23). This raises the possibility that a chloroplast PAP might be active in this organism (see below).
Our findings raise the question of how the opposing activities of PNPase might be regulated in time and space. One possibility is that there are two distinct populations of PNPase in the chloroplast, in terms of both activity and perhaps a posttranslational modification such as phosphorylation. Indeed, spinach chloroplast PNPase is a good substrate for phosphorylation when incubated with a protein kinase and ATP (Shtieman-Kotler and Schuster, unpublished results). However, any role for phosphorylation seems unclear since at least in vitro, PNPase can be directed towards either exclusive degradation or polyadenylation by varying the Pi and ADP concentrations (Fig. 3 and 4). A second possibility is that the nature of its activity is determined by supramolecular structure. Recently, the crystal structure of PNPase from the bacterium Streptomyces antibioticus was determined (43). Three polypeptides were found to be associated in a trimeric structure establishing an RNA channel, suggesting a possible modulation of processivity and of degradation and polyadenylation activities by structural changes. Third, one might speculate that chloroplast PNPase activity might be influenced by association with additional proteins, for example in the context of a chloroplast equivalent to the E. coli degradosome (6, 8, 27, 37). However, in contrast to earlier reports (20), it was recently found that an E. coli-like degradosome is not present in spinach chloroplasts and thus, PNPase is not associated with other proteins (Baginski et al., submitted).
In light of the above considerations, we propose that anabolic versus catabolic activity of chloroplast PNPase is most likely modulated by transient and localized changes in the Pi and nucleotide diphosphate concentrations. Since Pi is a substrate of PNPase, it is possible that extensive processive degradation activity could lead to a transient reduction in the phosphate concentration within a hypothetical microenvironment. In this situation and in the presence of nucleoside diphosphates, the enzyme begins polymerizing. In this direction, the reaction would eventually restore the Pi concentration while depleting nucleoside diphosphates, bringing the cycle full circle. Such a dynamic situation could explain the existence of long poly(A)-rich tails of several hundred nucleotides on the one hand and the very small steady-state amount of polyadenylated RNA due to rapid degradation on the other hand (28). A dynamic regulation of PNPase activity has also been suggested to occur in E. coli (33, 37). In this situation, RNA structural elements are critical for regulating enzyme activity, since both degradation and polymerization activities are tremendously sensitive to a stem-loop structure (28, 45) (Fig. 7). In addition to inhibiting processive degradation, a stem-loop structure inhibits the binding of the PNPase to the RNA and thus prevents polymerization. A minimal platform of 7 to 10 unpaired nucleotides is required for the binding of PNPase (43). Therefore, a 3′ end stem-loop structure protects it from degradation by preventing polyadenylation and/or direct exonucleolytic degradation.
Despite the close similarity of the molecular mechanisms of RNA polyadenylation and degradation in bacteria and in chloroplasts, the results of this work suggesting that there is no PAP in the chloroplasts of higher plants point to a major difference between the two systems. Assuming a common evolutionary origin, it is an interesting question whether bacteria have acquired PAP or whether it was lost from the higher plants' chloroplast lineage. The observation that the cyanobacterium Synechocystis sp., believed to be related to the chloroplast ancestor, contains two putative PAP genes (BAA18159 and BAA10528 in BLAST) homologous to PAP I of E. coli suggests that PAP was lost from the chloroplasts of higher plants. However, it should be noted that the homology of PAP from bacteria to the tRNA nucleotidyl transferase family (36) makes it difficult to predict which protein belongs to each function. In that respect, an interesting question can be posed regarding the situation in the chloroplasts of green algae. On one hand, the analysis of polyadenylated transcripts in the chloroplast of C. reinhardtii revealed only homogenous poly(A) tails (23). On the other hand, analyzing the expressed sequence tags of this alga revealed only a PAP homologous to the nuclear enzymes and none homologous for the PAP of E. coli (AV630342 and seven other expressed sequence tags related to the same protein) (1, 2). Therefore, the question whether or not there is a PAP in the chloroplasts of green algae remains open. For either possibility, the evolutionary importance of this event is an open and interesting question. Analyzing the situation in the other organelle of prokaryotic origin, the mitochondrion, and in other algae and higher plants, as well as revealing whether cyanobacteria really have the PAP enzyme, would help to answer the questions as to when and why in the evolutionary process PAP was omitted from the chloroplasts of higher plants and whether it was acquired by bacteria. In addition, experiments in which E. coli or nucleus PAPs are introduced into the chloroplast and changes in RNA metabolism are analyzed are in progress.
ACKNOWLEDGMENTS
We thank A. J. Carpousis for the PAP I antibodies and David Stern, Takahiro Nakamura, Ruth Rott, and Varda Liveanu for providing helpful comments on the manuscript. Special thanks to David Stern for help in editing the manuscript.
This work was supported by grants from the Israel Science Foundation, the Israel-Japan Corporation Foundation, and the Israel-USA Binational Agriculture Research and Development Foundation (BARD).
REFERENCES
- 1.Asamizu E, Miura K, Kucho K, Inoue Y, Fukuzawa H, Ohyama K, Nakamura Y, Tabata S. Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 2000;7:305–307. doi: 10.1093/dnares/7.5.305. [DOI] [PubMed] [Google Scholar]
- 2.Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S. A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res. 1999;6:369–373. doi: 10.1093/dnares/6.6.369. [DOI] [PubMed] [Google Scholar]
- 3.Barkan A, Stern D B. Chloroplast mRNA processing: intron splicing and 3′-end metabolism. In: Bailey-Serres J, Gallie D R, editors. A look beyond transcription: mechanisms determining mRNA stability and translation in plants. Rockville, Md: American Society of Plant Physiologists; 1998. pp. 162–173. [Google Scholar]
- 4.Blum E, Carpousis A J, Higgins C F. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J Biol Chem. 1999;274:4009–4016. doi: 10.1074/jbc.274.7.4009. [DOI] [PubMed] [Google Scholar]
- 5.Cao G, Sarkar N. Stationary phase-specific mRNAs in Escherichia coliare adenylated. Biochem Biophys Res Commun. 1997;239:46–50. doi: 10.1006/bbrc.1997.7421. [DOI] [PubMed] [Google Scholar]
- 6.Carpousis A J, Vanzo N F, Raynal L C. mRNA degradation, a tale of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 7.Chen P S, Toribara T Y, Warner H. Microdetermination of phosphorus. Anal Chem. 1956;28:1756–1758. [Google Scholar]
- 8.Coburn G A, Mackie G A. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog Nucleic Acid Res. 1999;62:55–108. doi: 10.1016/s0079-6603(08)60505-x. [DOI] [PubMed] [Google Scholar]
- 9.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 10.Drager R G, Girard-Bascou J, Choquet Y, Kindle K L, Stern D B. In vivoevidence for 5′–>3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 1998;13:85–96. doi: 10.1046/j.1365-313x.1998.00016.x. [DOI] [PubMed] [Google Scholar]
- 11.Drager R G, Higgs D C, Kindle K L, Stern D B. 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J. 1999;19:521–531. doi: 10.1046/j.1365-313x.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- 12.Drager R G, Zeidler M, Simpson C L, Stern D B. A chloroplast transcript lacking the 3′ inverted repeat is degraded by 3′ to 5′ exoribonuclease activity. RNA. 1996;2:652–663. [PMC free article] [PubMed] [Google Scholar]
- 13.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 14.Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagliardi D, Leaver C J. Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 1999;18:3757–3766. doi: 10.1093/emboj/18.13.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruissem W, Greenberg B M, Zurawski G, Hallick R B. Chloroplast gene expression and promoter identification in chloroplast extracts. Methods Enzymol. 1986;118:253–270. doi: 10.1016/0076-6879(86)18077-3. [DOI] [PubMed] [Google Scholar]
- 17.Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu Rev Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 18.Hampp R, Goller M, Ziegler H. Adenylate levels, energy charge and phosphorylation potential during dark-light and light-dark transition in chloroplast, mitochondria and cytosol of mesophyll protoplasts form Avena sativa L. Plant Physiol. 1982;69:448–455. doi: 10.1104/pp.69.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes R, Kudla J, Gruissem W. Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem Sci. 1999;24:199–202. doi: 10.1016/s0968-0004(99)01388-2. [DOI] [PubMed] [Google Scholar]
- 20.Hayes R, Kudla J, Schuster G, Gabay L, Maliga P, Gruissem W. Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J. 1996;15:1132–1141. [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson M D, Popowski J, Cao G J, Shen P, Sarkar N. Bacteriophage T7 mRNA is polyadenylated. Mol Microbiol. 1998;27:23–30. doi: 10.1046/j.1365-2958.1998.00649.x. [DOI] [PubMed] [Google Scholar]
- 22.Klaff P. mRNA decay in spinach chloroplasts: psbAmRNA degradation is initiated by endonucleolytic cleavages within the coding region. Nucleic Acids Res. 1995;23:4885–4892. doi: 10.1093/nar/23.23.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komine Y, Kwong L, Anguera M, Schuster S, Stern D B. Polyadenylation of three classes of chloroplast RNA in Chlamydomonas reinhardtii. RNA. 2000;6:598–607. doi: 10.1017/s1355838200992252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn J, Tengler U, Binder S. Transcript lifetime is balanced between stabilizing stem-loop structures and degradation-promoting polyadenylation in plant mitochondria. Mol Cell Biol. 2001;21:731–742. doi: 10.1128/MCB.21.3.731-742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q S, Gupta J D, Hunt A G. A plant poly(A) polymerase requires a novel RNA-binding protein for activity. J Biol Chem. 1996;271:19831–19835. doi: 10.1074/jbc.271.33.19831. [DOI] [PubMed] [Google Scholar]
- 26.Li Q S, Gupta J D, Hunt A G. Polynucleotide phosphorylase is a component of a novel plant poly(A) polymerase. J Biol Chem. 1998;273:17539–17543. doi: 10.1074/jbc.273.28.17539. [DOI] [PubMed] [Google Scholar]
- 27.Liou G G, Jane W N, Cohen S N, Lin N S, Lin-Chao S. RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc Natl Acad Sci USA. 2001;98:63–68. doi: 10.1073/pnas.011535498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lisitsky I, Klaff P, Schuster G. Addition of poly(A)-rich sequences to endonucleolytic cleavage sites in the degradation of spinach chloroplast mRNA. Proc Natl Acad Sci USA. 1996;93:13398–13403. doi: 10.1073/pnas.93.23.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lisitsky I, Klaff P, Schuster G. Blocking polyadenylation of mRNA in the chloroplast inhibits its degradation. Plant J. 1997;12:1173–1178. [Google Scholar]
- 30.Lisitsky I, Kotler A, Schuster G. The mechanism of preferential degradation of polyadenylated RNA in the chloroplast: the exoribonuclease 100RNP/PNPase displays high binding affinity for poly(A) sequence. J Biol Chem. 1997;272:17648–17653. doi: 10.1074/jbc.272.28.17648. [DOI] [PubMed] [Google Scholar]
- 31.Littauer U Z, Grunberg-Manago M. Polynucleotide phosphorylase. In: Creighton T E, editor. The encyclopedia of molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1999. pp. 1911–1918. [Google Scholar]
- 32.Lupold D S, Caoile A G, Stern D B. Polyadenylation occurs at multiple sites in maize mitochondrial cox2mRNA and is independent of editing status. Plant Cell. 1999;11:1565–1578. doi: 10.1105/tpc.11.8.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohanty B K, Kushner S R. Polynucleotide phosphorylase functions both as a 3′ to 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nickelsen J, Fleischmann M, Boudreau E, Rahire M, Rochaix J-D. Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell. 1999;11:957–970. doi: 10.1105/tpc.11.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raynal L C, Carpousis A J. Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol Microbiol. 1999;32:765–775. doi: 10.1046/j.1365-2958.1999.01394.x. [DOI] [PubMed] [Google Scholar]
- 36.Raynal L C, Krisch H M, Carpousis A J. The Bacillus subtilisnucleotidyltransferase is a tRNA CCA-adding enzyme. J Bacteriol. 1998;180:6276–6282. doi: 10.1128/jb.180.23.6276-6282.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regnier P, Arraiano C M. Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 38.Schuster, G., and R. Bock. Editing, polyadenylation and degradation of mRNA in the chloroplast. Adv. Photosynth., in press. [DOI] [PMC free article] [PubMed]
- 39.Schuster G, Lisitsky I, Klaff P. Update on chloroplast molecular biology: polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol. 1999;120:937–944. doi: 10.1104/pp.120.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soreq H, Littauer U Z. Polynucleotide phosphorylase. Vol. 15. New York, N.Y: Academic Press; 1982. [Google Scholar]
- 41.Steege D A. Emerging features of mRNA decay in bacteria. RNA. 2000;6:1079–1090. doi: 10.1017/s1355838200001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stitt M, Wirtz W, Heldt H W. Metabolite levels during induction in the chloroplast and extrachloroplast compartments of spinach protoplasts. Biochim Biophys Acta. 1980;593:85–102. doi: 10.1016/0005-2728(80)90010-9. [DOI] [PubMed] [Google Scholar]
- 43.Symmons M F, Jones G H, Luisi B F. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 44.Williamson J R, Raghuraman M K, Cech T R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 45.Yehudai-Resheff S, Schuster G. Characterization of the E. colipoly(A)-polymerase: specificity to nucleotides, RNA-binding affinities and RNA-structure dependence activity. Nucleic Acids Res. 2000;28:1139–1144. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]