Abstract
Transcriptional activation of the mouse mammary tumor virus (MMTV) promoter by ligand-bound glucocorticoid receptor (GR) is transient. Previously, we demonstrated that prolonged hormone exposure results in displacement of the transcription factor nuclear factor 1 (NF1) and the basal transcription complex from the promoter, the dephosphorylation of histone H1, and the establishment of a repressive chromatin structure. We have explored the mechanistic link between histone H1 dephosphorylation and silencing of the MMTV promoter by describing the putative kinase responsible for H1 phosphorylation. Both in vitro kinase assays and in vivo protein expression studies suggest that in hormone-treated cells the ability of cdk2 to phosphorylate histone H1 is decreased and the cdk2 inhibitory p21 protein level is increased. To address the role of cdk2 and histone H1 dephosphorylation in the silencing of the MMTV promoter, we used potent cdk2 inhibitors, Roscovitine and CVT-313, to generate an MMTV promoter which is associated predominantly with the dephosphorylated form of histone H1. Both Roscovitine and CVT-313 block phosphorylation of histone H1 and, under these conditions, the GR is unable to remodel chromatin, recruit transcription factors to the promoter, or stimulate MMTV mRNA accumulation. These results suggest a model where cdk2-directed histone H1 phosphorylation is a necessary condition to permit GR-mediated chromatin remodeling and activation of the MMTV promoter in vivo.
DNA in eukaryotic nuclei is highly packaged into chromatin by two molecules each of the core histone proteins H2A, H2B, H3, and H4 and one molecule of linker histone H1 (44). In addition, to the intrinsic stearic considerations of wrapping DNA around the histone octamer, the posttranslation modification of the core histones have come under increased scrutiny (22, 44). Numerous studies support a strong link between transcriptional regulation and the remodeling of chromatin structure through the acetylation of core histones H3 and H4 (20, 40, 46). The acetylation of core histones in vivo is presumed to play a role in increasing the accessibility of transcription factors to the promoters of target genes (23). More recently, the Mi-2 ATPase complex, which contains chromatin remodeling activity, has been linked to both DNA methylation and histone deacetylation (39, 47).
The role of histone H1 in the regulation of transcription is less clear, but there is evidence that histone H1 interacts differentially with transcriptionally active and inactive regions of chromatin (29). Indeed, studies in Xenopus and Tetrahymena thermophila have ruled out an exclusive role for histone H1 phosphorylation in chromatin condensation (31, 36). However, other studies in mammals and T. thermophila have found a correlation between transcriptional activation and decreased amounts of histone H1 (9, 12, 14). Thus, it is plausible, given the role of histone H1 in the packaging of the nucleosome, that posttranslational modifications of this protein may also be involved in transcriptional regulation.
Evidence to support a role for histone H1 phosphorylation in transcriptional regulation includes the correlation of increased histone H1 phosphorylation during mitosis, presumably by p34cdc2 kinase (8, 24). It has also been reported that ionizing radiation decreases phospho-H1 levels through kinase inactivation, which suggests that phosphorylation of histone H1 may be regulated in response to DNA damage (17). Moreover, recent studies in Tetrahymena have suggested that histone H1 phosphorylation mimics the removal or depletion of histone H1 and thus regulates the expression of specific genes (14). These studies suggest that the phosphorylation of histone H1 acts to create a “charge patch” or domain in H1 that is directly responsible for its ability to regulate gene expression (13). It has also been proposed that phosphorylated histone H1 has a decreased affinity for the nucleosome, thus leading to an open chromatin structure (19).
The mouse mammary tumor virus (MMTV) promoter represents a well-studied mammalian system in which chromatin structure and transcriptional activity have been intimately linked (5, 18). In the absence of glucocorticoid, the MMTV promoter is incorporated into six regularly positioned nucleosomes (33). This closed chromatin structure prevents the binding of ubiquitous trans-acting factors to the promoter and thus inhibits transcription (4). Glucocorticoid exposure rapidly disrupts the local chromatin structure, recruits transcription factors such as nuclear factor 1 (NF1), and induces maximal transcription within 1 h (25). In contrast, prolonged hormone treatment, in excess of 24 h, causes the promoter to enter a refractory state in which transcription is repressed. A striking feature of this refractory state is the simultaneous dephosphorylation of histone H1 that accompanies the silencing of the promoter (26).
We have explored the relationship between histone H1 dephosphorylation and inactivation of the MMTV promoter by investigating the mechanism(s) by which glucocorticoid mediates histone H1 dephosphorylation. We demonstrate that, in naive cells, cdk2 is able to phosphorylate histone H1 and that its activity is greatly decreased in cells exposed to glucocorticoid for 24 h. Furthermore, we demonstrate that cdk2 inhibitors block glucocorticoid receptor (GR)-mediated transcriptional activation of the MMTV promoter. Thus, the inhibitors mimic the repressive chromatin environment created by prolonged glucocorticoid exposure. These results are consistent with the hypothesis that histone H1 is an in vivo substrate of cdk2 and that the activity of this kinase plays a direct role in GR-mediated transcription within the context of the MMTV promoter.
MATERIALS AND METHODS
Cell culture and treatment with hormone and kinase inhibitors.
C127 mammary carcinoma cells (1471.1 cells) stably maintain ∼200 copies of a bovine papilloma virus vector with the MMTV promoter attached to the bacterial chloramphenicol acetyltransferase (CAT) gene (3). Cells were grown at 37°C with 5% CO2 in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum. Cells were treated with dexamethasone (10−7 M), Roscovitine (25 μM), and CVT-313 (25 μM) for the time periods indicated in the figure legends.
Immunoprecipitation and immunoblotting.
For immunoprecipitation assays, cells previously treated with or without hormone and/or inhibitors, were washed with cold phosphate-buffered saline (PBS) and pelleted. The cells were lysed in Buffer X plus bovine serum albumin (Buffer X+BSA; 100 mM Tris-HCl, pH 8.5; 250 mM NaCl; 1% [vol/vol] NP-40, 1 mM EDTA, 1 μg of aprotinin per ml; 2 mg of BSA per ml). The lysates were precleared once with protein A-Sepharose (3% [vol/vol]; Pharmacia) in Buffer X+BSA for 30 min with rotation at 4°C. Respective antibodies (Santa Cruz) were incubated with the supernatant for 1 h at 4°C and then with protein A-Sepharose for 1 h. Bound antibody-antigen complexes were washed three times in HEGND buffer (10 mM HEPES, pH 8.0; 1 mM EDTA; 10% glycerol; 50 mM NaCl; 2 mM dithiothreitol [DTT]) containing 0.1% Triton X-100 and once in HEGND buffer. Immunocomplexes then were suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and released by incubation at room temperature for 1 h. For immunoblot analysis, whole-cell protein lysate were solubilized in loading buffer, subjected to SDS-PAGE, and transferred to nitrocellulose, followed by incubation with the antibodies cited in the figure legends.
In vitro kinase reaction.
Commercially purified histone H1 (Gibco-BRL) was incubated with cdk2 immunoprecipitated from whole-cell extracts equivalent to 100 μg of total protein in 50 μl of kinase reaction buffer (50 mM Tris, pH 7.2; 10 mM MgCl2; 1mM DTT; 5 μCi of [γ-32P]ATP) for 10 min at 25°C. Samples were analyzed by SDS-PAGE, followed by autoradiography.
Isolation of total histones and separation of phosphorylated H1 by acid-urea gel electrophoresis.
Nuclei were isolated from cells as previously described and acid-soluble proteins were prepared by resuspending the nuclei in 0.4 N H2SO4 at 4°C for 1 h (25). The suspension was centrifuged, and basic proteins were precipitated from the supernatant overnight at −20°C by the addition of 1 ml of acetone. Proteins were collected by centrifugation, air dried, and dissolved in solution contained 0.9 M acetic acid and 25 μl of 75% sucrose. A total 30-μg equivalent of total proteins was separated on a 16% acrylamide acid-urea gel (0.75 mm thick by 12 cm long) as previously described (26, 32) and transferred to nitrocellulose membrane (Amersham) for 24 h at 50 V and 4°C in a transfer buffer (25 mM Tris-HCl, 0.19 M glycine, 20% methanol). Membranes were probed with antibodies (Upstate Biotechnology) specific for either phosphorylated or unphosphorylated histone H1 protein.
In vivo restriction enzyme hypersensitivity analysis.
Nuclei were isolated and digested with restriction endonucleases as previously described (25). Genomic DNA was purified by phenol-chloroform extraction and then digested to completion with HaeIII in vitro. This provided an internal standard for accessing the extent of in vivo cleavage and confirmed that equivalent amounts of DNA were used in each reaction. Then, 10 μg of each sample was analyzed by reiterative primer extension with Taq polymerase and a 32P-labeled single-stranded primer (MMTV#22, 5′-CTGGAAAGTGAAGGATAAGTGACGA-3′) corresponding to the +60 to +84 portion of the MMTV promoter. The purified extension products were separated on 7% polyacrylamide denaturing gels and exposed to film.
In vivo analysis of transcription factor loading.
Isolated nuclei were digested by HaeIII (1,000 U/ml) and Exonuclease III (625 U/ml) to detect the 5′ boundaries of transcription factors on the MMTV promoter (25). DNA was purified, and single-stranded overhangs were removed with mung bean nuclease. All samples were digested to completion with AlwnI prior to analysis by reiterative primer extension with the 32P-labeled primer, MMTV#22. The purified extension products were analyzed on 7% polyacrylamide denaturing gels.
RNA isolation and primer extension.
Total RNA was prepared using Trizol reagent (Gibco-BRL) as described by the manufacturer. Oligonucleotide primers MMTV#22 and 18S (5′-ACCAAAGGAACCATAACTG-3′) were labeled with [32P]ATP by using T4 polynucleotide kinase. Total RNA (20 μg) and labeled oligonucleotide were dissolved in 15 μl of hybridization buffer (0.15 M KCl; 10 mM Tris, pH 8.3; 1 mM EDTA), heated to 65°C for 90 min, and allowed to cool slowly to 42°C. Extension of the primer was carried out at 42°C for 1 h after the addition of 30 μl of reverse transcriptase buffer (30 mM Tris-HCl pH 8.3; 15 mM MgCl2; 8.3 mM DTT: 75 μg of actinomycin D per ml; 0.22 mM deoxy nucleoside triphosphate [dNTP] mix) and 20 U of Superscript reverse transcriptase (Gibco-BRL). RNase reaction mix (100 μg of salmon sperm DNA per ml; 20 μg of RNase A per ml; 100 mM NaCl; 10 mM Tris-HCl, pH 7.5; 1 mM EDTA) was added to each tube, and RNase digestion was carried out at 37°C for 15 min. The reaction was terminated by the addition of 3 M sodium acetate. DNA was extracted with phenol-chloroform-isoamyl alcohol and precipitated by the addition of 100% ethanol. The cDNA pellet was washed with 70% ethanol, air dried, and resuspended in loading buffer. Products were analyzed on a 7% polyacrylamide denaturing gel.
Reverse transcriptase-PCR.
cDNA was synthesized from 5 μg of total RNA and 750 ng of oligo(dT)12–18 primer (Pharmacia) in a solution containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 500 μM concentrations of each dNTP, and 20 U of Superscript reverse transcriptase (Gibco-BRL). After synthesis for 1 h at 37°C and 10 min at 75°C, cDNA was stored at −80°C until use. The PCR reaction contained cDNA derived from 20 ng of RNA, 5 pmol of each primer, and 5 U of Taq DNA polymerase in 50 μl of solution containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, and 100 μM concentrations of each dNTP. PCR assays for the MT gene used primer MT5-p (5′-CGGATCCCGGAATGGACCCCAACTGCT-3′) and primer MT3-p (5′-CGGATCCAGACTCAAACAGGCTTTTAT-3′). PCR assays for the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene used primers GAP5-p (5′-TATTGGGCGCCTGGTCACCA-3′) and GAP3-p (5′-CCACCTTCTTGATGTCATCA-3′).
Transient transfections.
One day before transfection, 1471.1 cells were seeded into six-well plates at 350,000 cells/well. Transient transfections were carried out with 4 μg of pLTR.luc using LipofectAMINE Plus Reagent (Life Technologies, Inc.) according to manufacturer's protocol. Cells were allowed to recover for 16 h in DMEM containing 10% fetal bovine serum before the start of hormone treatment.
Luciferase assays.
Transiently transfected 1471.1 cells were treated with dexamethasone (100 nM), CVT-313 (25 μM), or a combination of both compounds for the times indicated in the figure legends. Following treatment, cells were washed twice with sterile filtered PBS and lysed directly on the plate with 400 μl of Passive Lysis Buffer (Promega, Madison, Wis.) per well. The plates were scraped, and lysates were vortexed at high speed for 5 s and then pelleted by centrifugation at 20,800 × g for 1 min at room temperature. Next, 20 μl of each lysate was added to 100 μl of luciferase substrate, and the activity was monitored for 5 s. Relative light units were normalized to the total protein measured.
Histone H1-MMTV DNA cross-linking in vivo.
Living cells were fixed by adding formaldehyde (37%) directly to the growth medium (1:100) and incubated for 10 min. Nuclei were isolated and digested completely with restriction enzyme HaeIII. Subsequent chromatin immunoprecipitations with anti-H1 or anti-phosphorylated H1 antibodies were carried out as described previously (26). A PCR-based approach was used with forward and reverse primers (5′-TTAGCTTCCTTAGCTCCTGAAAAT-3′ and 5′-TTAAAGTAAGTTTTTGGTTACAAACT-3′, respectively) that amplify a 325-bp fragment that encompassed nucleosome B within the HaeIII-digested MMTV promoter.
RESULTS
Prolonged hormone treatment suppresses the ability of cdk2 to phosphorylate histone H1.
For the MMTV promoter, prolonged hormone exposure leads to the eviction of transcription factors from the promoter and the reconstitution of a repressive chromatin environment that is refactory to further stimulation by the GR (25). In addition, prolonged hormone treatment leads to the global dephosphorylation of histone H1. This raises the question as to whether the hormone dependent refractory period is, in part, due to suppression of the activity of a kinase(s) responsible for histone H1 phosphorylation in vivo.
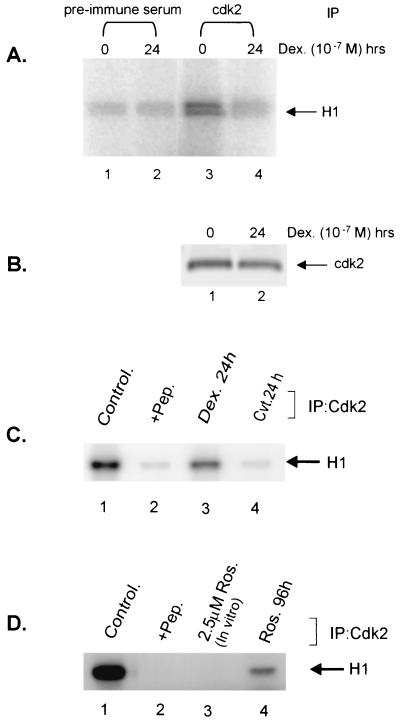
To begin to investigate the kinase(s) that might be involved in histone H1 phosphorylation, we tested the ability of a cell cycle-dependent kinase (cdk2) to phosphorylate histone H1 in vitro. Whole-cell lysates from C127 mammary carcinoma cells (1471.1 cells) were prepared from untreated cells (Fig. 1A, lanes 1 and 3) and cells treated for 24 h with dexamethasone (Fig. 1A, lanes 2 and 4). Following immunoprecipitation with a cdk2 specific antibody, cdk2 kinase activity was determined in a kinase assay with histone H1 as a substrate (Fig. 1A, lanes 3 and 4). These results demonstrate that cdk2, immunoprecipitated from mouse mammary cells, can efficiently phosphorylate histone H1 in vitro and that this activity is severely reduced in cells treated with dexamethasone for 24 h (Fig. 1A, compare lanes 3 and 4).
FIG. 1.
Glucocorticoids and CVT-313 block phosphorylation of histone H1 in Vitro. (A) Prolonged glucocorticoid treatment inhibits cdk2 activity. Mouse mammary cells were untreated (lanes 1 and 3) or treated with dexamethasone (10−7 M) for 24 h (lanes 2 and 4). cdk2 (lanes 3 and 4) was immunoprecipitated from 100 μg of total proteins in whole-cell lysate. As a control, an identical reaction was carried out with a preimmune serum (lanes 1 and 2). Immunocomplexes were then incubated with purified histone H1 in presence of [γ-32P]ATP for 10 min at 25°C. Samples were analyzed by SDS-PAGE, followed by autoradiography. (B) The Western blot below the kinase gel indicates the level of cdk2 kinase immunoprecipitated by the anti-CDK2 antibody from the same cellular lysates used for the kinase reaction in panel A. (C) CVT-313 inhibits H1 phosphorylation in vitro. Cells were untreated (lanes 1 and 2) or treated with dexamethasone (lane 3) or CVT-313 25 μM (lane 4) for 24 h. cdk2 was immunoprecipitated from cell lysate with affinity-purified anti-cdk2 antibodies (lanes 1, 3, and 4) or anti-cdk2 antibodies preabsorbed with an excess of peptide antigen (lane 2). The kinase reactions were performed as described above. (D) Roscovitine inhibits H1 phosphorylation in vitro. Cells were untreated (lanes 1, 2, and 3) or treated with Roscovitine at 25 μM (lane 4) for 96 h. cdk2 was immunopurified with purified anti-cdk2 antibodies (lanes 1, 3, and 4) or anti-cdk2 antibodies preabsorbed with an excess of peptide antigen (lane 2). Immunopurified cdk2 was then incubated in vitro with 2.5 μM Roscovitine (lane 3) prior to the kinase reaction. The samples were analyzed as in panel A.
cdk2 inhibitors prevent the phosphorylation of histone H1 in vitro.
To examine a potential relationship between cdk2 phosphorylation of histone H1 and MMTV activation, we examined the effects of two cdk2 inhibitors, Roscovitine and CVT-313, on histone H1 phosphorylation (10, 28). Previous characterization of these inhibitors revealed that they are most effective at inhibiting cdk2 and, in the case of CVT-313, the 50% inhibitory concentration (IC50) is 0.5 μM for cdk2, while for cdk4 the IC50 is 215μM and greater than 1,250 μM for protein kinase C (10). Mouse mammary cells were cultured in the presence of cdk2 inhibitors for various time periods, and the kinase activity of cdk2 was monitored by an in vitro kinase assay. Both CVT-313 and Roscovitine were found to be potent inhibitors of cdk2 in our cells (Fig. 1C and D). For example, under similar experimental conditions, cdk2-associated incorporation of [32P]phosphate to histone H1 was found to be decreased (70 to 80%) when CVT-313 and Roscovitine were incubated for 24 and 96 h, respectively (Fig. 1C, compare lanes 1 and 4; Fig. 1D, compare lanes 1 and 4). The kinase reactions were performed following immunoprecipitation of cdk2 from both treated and untreated cells. As a control, phosphorylation of histone H1 was not detected when anti-cdk2 antibodies had been preabsorbed with cdk2 antigenic peptide (Fig. 1C and D, lane 2) prior to immunoprecipitation. Furthermore, the addition of as little as 2.5 μM Roscovitine to the immunocomplexes was also found to completely block cdk2 activity (Fig. 1D, lane 3). These and subsequent experiments revealed that both inhibitors were effective at inhibiting cdk2 in these cells and suggest that this is accomplished at least in part by the reduction in cdk2 protein levels (see Fig. 7 and also below).
FIG. 7.
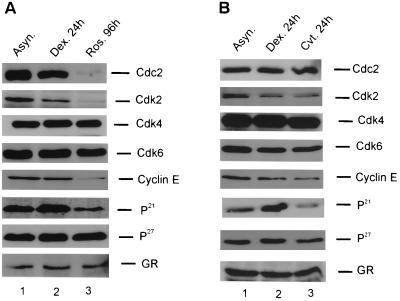
Western analysis of cell cycle proteins after exposure to dexamethasone or cdk2 inhibitors. (A) Effects of Roscovitine or prolonged dexamethasone treatment on cell cycle proteins. Cells were untreated (lane 1), treated with dexamethasone (10−7 M) for 24 h (lane 2), or treated with Roscovitine for 96 h (lane 3). Whole-cell extract proteins were solubilized in loading buffer, boiled for 3 min at 100°C, subjected to SDS-PAGE, and transferred to nitrocellulose membrane before immunoblotting with primary antibodies as indicated in the figure. The various kinases and inhibitors were evaluated a minimum of three times, and the figure shown is a representative data set. (B) Effects of CVT-313 or prolonged dexamethasone treatment on cell cycle proteins. Cells were untreated (lane 1), treated with dexamethasone for 24 h (lane 2), or treated with CVT-313 for 24 h (lane 3). Analysis was as described in panel A.
Treatment with cdk2 inhibitors leads to the dephosphorylation of histone H1 in vivo.
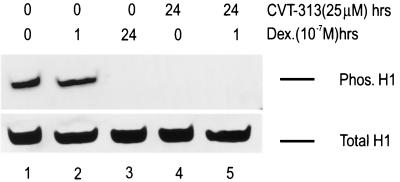
The previous in vitro kinase experiments indicate that the cdk2 kinase may be involved in phosphorylation of histone H1 in vivo. To assess the in vivo relevance of cdk2-mediated histone H1 phosphorylation, we incubated mouse mammary cells with the cdk2 inhibitors, CVT-313 and Roscovitine, to mimic the dephosphorylation of histone H1 observed in hormone-treated cells. Cells grown without hormone or CVT-313 contained the highest levels of phospho-H1 protein (Fig. 2, lane 1). Cells treated with hormone for an hour, which maximizes transcriptional activity from the MMTV promoter, had levels of phosphorylated histone H1 protein similar to those of nontreated cells (Fig. 2, lanes 1 and 2). In contrast, histone H1 from cells exposed to hormone for 24 h was completely dephosphorylated (Fig. 2, lane 3). Consistent with its in vitro effects, treatment with CVT-313, both in the absence and in the presence of hormone, leads to a decrease in the phosphorylation of histone H1 in vivo (Fig. 2, lanes 4 and 5). Similar results were seen with Roscovitine (data not shown), and the levels of total histone H1 in the cells were unchanged under all conditions (Fig. 2, lower panel).
FIG. 2.
Prolonged treatment with dexamethasone and CVT-313 dephosphorylates histone H1 in vivo. Mouse cells were untreated (lane 1), treated with dexamethasone (10−7 M) for 1 h (lane 2), treated with dexamethasone for 24 h (lane 3), treated with CVT-313 (25 μM) for 24 h (lane 4), or pretreated with CVT-313 (25 μM) for 23 h prior to dexamethasone addition for 1 h (lane 5). Total histones were prepared from the nuclei by H2SO4 extraction as described in Materials and Methods. Histones (30 μg) were separated on a 16% acrylamide acid-urea gel, transferred to a nitrocellulose membrane, and analyzed by Western blot analysis using a polyclonal anti-phosphorylated H1 antibody. Equal loading of protein was confirmed by staining the blot with amido black dye to reveal the total H1 present (lower panel).
cdk2 inhibitors mirror the effects of prolonged hormone treatment on MMTV promoter.
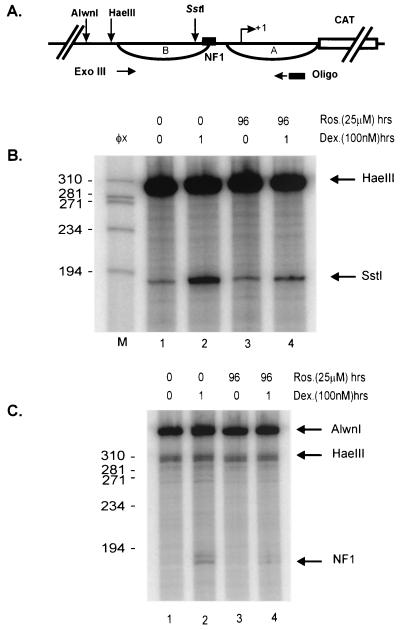
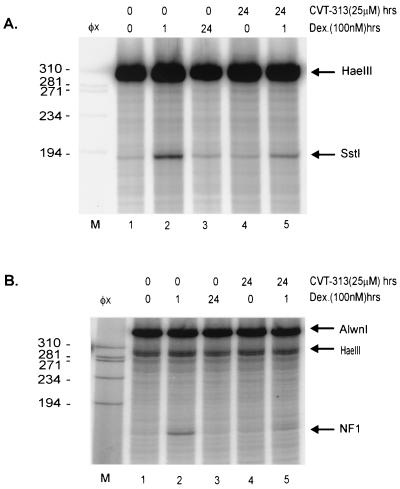
Within the MMTV promoter, the second nucleosome (nuc-B) encompasses the binding site for the transcription factor NF1, as well as the binding sites for the GR (33). The glucocorticoid-induced disruption of nuc-B, which allows access of transcription factors, makes the promoter hypersensitive to digestion by restriction endonucleases in this region (3). If histone H1 phosphorylation was linked to GR-mediated chromatin remodeling of the MMTV promoter, one would predict that blocking histone H1 phosphorylation might prevent chromatin remodeling. Therefore, we examined the effects of Roscovitine and CVT-313 on the chromatin organization of the MMTV promoter. As shown in Fig. 3 and 4, Roscovitine and CVT-313 both inhibited GR-mediated hypersensitivity and NF1 loading on the MMTV promoter. Compared to untreated cells, dexamethasone treatment for 1 h resulted in substantially elevated cleavage (six- to sevenfold) by the restriction enzyme SstI (Fig. 3B, compare lanes 1 and 2), which cleaves within the region of nuc-B. Concomitant treatment of cells with Roscovitine partially blocked (60 to 70%) the induction of restriction enzyme hypersensitivity induced by dexamethasone (Fig. 3B, lane 4). In addition to restriction enzyme hypersensitivity, treatment with dexamethasone for 1 h increased the level of bound NF1 (Fig. 3C, compare lanes 1 and 2). However, NF1 binding was reduced (60 to 70%) in cells pretreated with Roscovitine and subsequently induced by dexamethasone (Fig. 3C, compare lanes 2 and 4). CVT-313 displayed a pattern similar to that of Roscovitine in that it also inhibited the ability of the GR to remodel MMTV chromatin, as measured by restriction enzyme hypersensitivity (Fig. 4A, compare lanes 2 and 5). Consistent with its repression of the GR chromatin remodeling activity, CVT-313 treatment also blocked NF1 binding to the promoter in the presence of dexamethasone (Fig. 4B, compare lanes 2 and 5). Taken together, these experiments demonstrate that when H1 phosphorylation is significantly decreased in cells exposed to cdk2 inhibitors, GR is unable to remodel the MMTV promoter.
FIG. 3.
Roscovitine inhibits GR activation of the MMTV promoter. (A) Schematic of the MMTV promoter indicating the sites of cleavage by restriction enzymes and the NF1 binding site. (B) Roscovitine inhibits restriction enzyme hypersensitivity. Mouse mammary cells were untreated (lane 1), treated with dexamethasone for 1 h (lane 2), treated with Roscovitine for 96 h (lane 3), or pretreated with Roscovitine for 95 h prior to dexamethasone addition for 1 h (lane 4). Nuclei were isolated and digested with restriction endonuclease SstI (in vivo). After purification, genomic DNA was digested with HaeIII in vitro to provide an internal standard. Reiterative primer extension analysis of this digested DNA was performed using a 32P-labeled primer. The purified extended products were separated on a polyacrylamide denaturing gel before autoradiography. Lane M is the size standard φX174 cut with HaeIII. The arrows indicate HaeIII and SstI cleavage products. (C) Roscovitine inhibits NF1 binding to the MMTV promoter. Cells were treated as described in Fig. 3A. Isolated nuclei were digested with HaeIII and Exonuclease III to detect stops corresponding to the 5′ boundary of NF1. Purified DNA was digested to completion with AlwnI to provide an internal standard and loading control. Single-stranded DNA was removed with mung bean nuclease digestion and then analyzed by primer extension with Taq polymerase using a 32P-labeled primer specific for the MMTV promoter.
FIG. 4.
CVT-313 inhibits GR-mediated hypersensitivity and NF1 loading on the MMTV promoter. Cells were untreated (lane 1), treated with dexamethasone for 1 h (lane 2), treated with dexamethasone for 24 h (lane 3), treated with CVT-313 for 24 h (lane 4), or pretreated with CVT-313 for 23 h prior to dexamethasone addition for 1 h (lane 5). Details of the experiment are as described in Fig. 3.
Selective inhibition of GR-dependent transcriptional activation of MMTV by CVT-313.
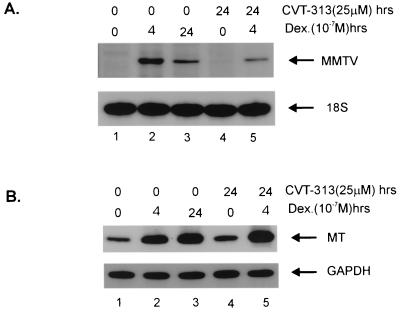
In vivo dephosphorylation of histone H1 due to dexamethasone or CVT-313 treatment is a global phenomenon and is not restricted to the MMTV promoter (Fig. 2). We therefore investigated whether the reduced transcription from the MMTV promoter was part of a more global response to the widespread dephosphorylation of histone H1. This was accomplished by assessing, in parallel, the accumulation of MMTV mRNA and mRNA from two genes which are either independent of or induced by glucocorticoid treatment (GAPDH, glucocorticoid independent; metallothionein [MT], glucocorticoid inducible). For this experiment, cells were treated with hormone for 4 h instead of 1 h because mRNA accumulation is maximal at 4 h, as demonstrated previously (26). Cells were also treated for 24 h with hormone and CVT-313 in order to assess the effect of the dephosphorylation of histone H1 on transcription from the three promoters (Fig. 5). In the absence of hormone the cells maintain very low levels of mRNA indicative of little transcription from the MMTV promoter (Fig. 5A, lane 1). However, there is a marked elevation in mRNA at 4 h post-hormone treatment (Fig. 5A, lane 2). Prolonged hormone treatment results in greater than 70% reduction in MMTV mRNA (Fig. 5A, lane 3). Similarly, CVT-313 treatment also reduced MMTV mRNA levels to roughly the same point, 60 to 70%, as had the glucocorticoid treatment (Fig. 5A, compare lanes 2 and 5). Neither hormone nor CVT-313 had an appreciable effect on 18S rRNA, which was run as a loading control (Fig. 5A, lower panel). These results suggest that prolonged hormone treatment and treatment with CVT-313 both effectively reduce transcription from the MMTV promoter. Analysis of the MT and the GAPDH gene transcription from the same RNA preparations gave different results for both glucocorticoid and CVT-313 treatment (Fig. 5B). The glucocorticoid-inducible MT gene exhibited an eightfold induction of mRNA within 4 h of hormone treatment (Fig. 5B, lane 2). In contrast to MMTV, transcription from the MT promoter was not inhibited after 24 h of hormone exposure (Fig. 5B, lane 3). Consistently, no reduction in MT transcription was observed in cells pretreated with CVT-313 and subsequently induced by hormone (Fig. 5B, lane 5). Transcription from the GAPDH gene, which is not regulated by glucocorticoid, was not affected by either prolonged glucocorticoid treatment or exposure to CVT-313. These results confirm that, while both the hormone and the cdk2 inhibitor block global histone H1 phosphorylation, they can mediate gene specific effects, as seen for the GR-activated MMTV and MT promoters.
FIG. 5.
Effects of CVT-313 on activation of the MMTV, MT, and GAPDH promoters. (A) Prolonged hormone and CVT-313 exposure inhibit MMTV transcription from chromatin. Cells were untreated (lane 1), treated with dexamethasone for 4 h (lane 2), treated with dexamethasone for 24 h (lane 3), treated with CVT-313 for 24 h (lane 4), or pretreated with CVT-313 for 20 h prior to dexamethasone addition for 4 h (lane 5). Total RNA was isolated as described in Materials and Methods. MMTV RNA and 18S rRNA levels were analyzed by primer extension from 20 and 0.2 μg of total RNA, respectively, using gene-specific primers as described in the text. (B) Prolonged hormone and CVT-313 exposure do not affect transcription from MT and GAPDH promoters. Cells were treated as in panel A. A total of 5 μg of total RNA was reverse transcribed to cDNA, and 20 ng of the cDNA was analyzed by PCR with 32P-labeled primers specific for the MT or GAPDH.
Silencing of the MMTV promoter occurs with dephosphorylated histone H1 at the promoter.
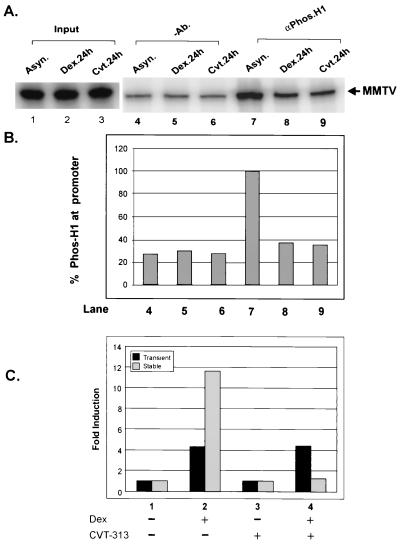
In order to determine the in vivo phosphorylation status of histone H1 both in silenced and in inducible MMTV chromatin, we used protein-DNA cross-linking followed by chromatin immunoprecipitation (CHIP) assays as described previously (26). The cells were treated with dexamethasone and CVT-313 for 24 h. DNA was then cross-linked with protein by the addition of formaldehyde to the cells. Nuclei were isolated and cleaved by restriction endonuclease, and fragments of chromatin were then immunoprecipitated with an antibody specific to phosphorylated histone H1. The total amount of MMTV DNA associated with phosphorylated H1 was then detected and quantified by PCR amplification using specific primers encompassing the nucleosomes A and B of the MMTV promoter. The input of MMTV DNA used in the immunoprecipitation experiment was detected by PCR (Fig. 6A, lanes 1 to 3). A small amount of MMTV DNA was nonspecifically bound to the protein A-Sepharose (Fig. 6A and B, lanes 4 to 6). However, the MMTV promoter in transcriptionally competent naive cells was found to be associated with higher levels of phosphorylated histone H1 (60 to 70%) compared to cells exposed to prolonged hormone or CVT-313 treatment (Fig. 6A and B, compare lane 7 to lanes 8 and 9). Thus, the phosphorylation status of histone H1 on the MMTV promoter is quantitatively different in transcriptionally competent cells and refractory cells, a result consistent with a role for phosphorylated histone H1 in GR-mediated chromatin remodeling.
FIG. 6.
CVT-313 reduces phosphorylation of histone H1 at the MMTV promoter in vivo but does not inhibit GR activation of a transiently transfected MMTV reporter. (A) Loss of phosphorylated H1 at the MMTV promoter upon prolonged treatment with hormone and CVT-313. Cells were untreated (lanes 1, 4, and 7), treated with dexamethasone (10−7 M) for 24 h (lanes 2, 5, and 8), or treated with CVT-313 (25μM) for 24 h (lanes 3, 6, and 9). The cells were fixed by adding formaldehyde (37%) directly to the medium (1:100) for 10 min. Nuclei were then isolated and digested completely with HaeIII. Digested nuclei were immunoprecipitated without (lanes 4 to 6) or with (lanes 7 to 9) an antibody specific for phosphorylated histone H1. Input (lanes 1 to 3) and immunoprecipitated DNA (lanes 4 to 9) was purified as described previously, and nucleosome B sequences within the MMTV promoter were detected by PCR amplification (26). (B) PCR products were analyzed using 8% polyacrylamide denaturing gels and quantified by use of a PhosphorImager using ImageQuant software. (C) CVT-313 does not block GR activation from a transiently transfected reporter. Cells were left untreated (column 1), treated with dexamethasone (Dex) for 8 h (column 2), treated with CVT-313 for 24 h (column 3), or treated with CVT-313 for 24 h followed by the addition of dexamethasone for 8 h (column 4). Lysates were prepared and assayed for luciferase and CAT activity from transient and stable constructs, respectively. Induction of MMTV luciferase and CAT activity is indicated as the fold induction relative to untreated cells.
CVT-313 treatment does not block GR activation from a transient MMTV template.
We have shown previously that the effects of prolonged glucocorticoid exposure are unique to the MMTV promoter assembled as an ordered array of nucleosomes (25). Consequently, for transiently transfected DNA the dephosphorylation of H1 fails to inhibit the GR. In the next series of experiments we examined the impact of CVT-313 mediated H1 dephosphorylation on transiently transfected MMTV reporter. Consistent with previous observations, GR-activated transcription of the transiently transfected reporter was not inhibited by CVT-313 exposure (Fig. 6C, compare dark columns 2 and 4). In contrast, the stable MMTV reporter was fully inhibited within the same 1471.1 cells (Fig. 6C, compare light columns 2 and 4). Given that the MMTV regulatory sequences are identical between the transient and stable templates within the cells, these results suggest that H1 interactions with MMTV chromatin are critical for the ability of the cdk2 inhibitor to repress GR activation of the promoter.
cdk2 inhibitors downregulate cdk2, cyclin E, and p21 protein levels.
To further investigate the mechanism(s) of cdk2 regulation in cells treated with hormone, Roscovitine, or CVT-313, we determined the expression levels of various cell cycle regulatory proteins by Western blotting (Fig. 7). The protein levels of cdk4, cdk6, p27, and GR were not significantly altered as a result of prolonged hormone treatment (Fig. 7A and B, column 2) or treatment with Roscovitine and CVT-313 (Fig. 7A and B, column 3). Both CVT-313 and Roscovitine decreased the expression of cdk2, the cdk2 inhibitor p21, and cyclin E protein but did not affect the expression of the cdk2 inhibitor p27. Interestingly, Roscovitine suppressed the expression of both cdc2 and cdk2 protein, whereas CVT-313 only inhibits cdk2 expression (Fig. 7, compare panel A, column 3, to panel B, column 3). Glucocorticoid treatment also reduced the expression of the cdk2 protein; however, in contrast to both Roscovitine and CVT-313, glucocorticoid treatment increased the levels of p21 protein (Fig. 7, cdk2 and p21 panels). Therefore, the inhibition of cdk2 activity by glucocorticoid may occur by simultaneously increasing the inhibitor protein p21 and decreasing the expression of cdk2. In contrast, the cdk inhibitors decreased both cdk2 and cyclin E levels, along with p21 levels, to effectively reduce cdk2 activity (Fig. 7, cdk2, cyclin E, and p21 panels). These data are consistent with the hypothesis that cdk2 may be an in vivo kinase for histone H1, and inactivation of this kinase blocks GR-mediated chromatin remodeling and transactivation of the MMTV promoter.
DISCUSSION
Within cells, glucocorticoid-mediated transcriptional activation of the MMTV promoter requires multistep changes in chromatin architecture that include the transitory removal of histone H1 from the nucleosomes (9) and binding of the transcription factors NF1 and OTF (5). This activation is initiated by the recruitment of BRG1-BAF or SWI-SNF complex to the promoter and is short-lived (15). After 24 h of continuous hormone treatment, transcription factors and the basal transcription complex are displaced from the MMTV promoter. This in turn results in the establishment of a repressive chromatin structure that is refractory to subsequent hormone activation (25). The dephosphorylation of histone H1 in response to ligand-bound GR is highly correlated with MMTV promoter chromatin dynamics and the repression of transcription (26). The work presented here extends earlier observations by demonstrating that the closing of the chromatin structure, repression of the MMTV promoter, and histone H1 dephosphorylation are concomitant with cdk2 inactivation, suggesting that these events are functionally linked. We observed that in naive cells, which respond to hormone with rapid chromatin remodeling and transactivation of the MMTV promoter, cdk2 kinase activity is maintained (Fig. 1). In contrast, prolonged hormone treatment and inactivation of the MMTV promoter was associated with a loss of cdk2 kinase activity (Fig. 1).
To provide further evidence linking cdk2 phosphorylation of histone H1 and MMTV promoter regulation, we examined the effects of two cdk2 inhibitors on histone H1 phosphorylation and transcription from the MMTV promoter (16). We report that both cdk2 inhibitors blocked histone H1 phosphorylation in vitro (Fig. 1C and D) and in vivo (Fig. 2), as well as simultaneously inhibited the expression of cdk2 and cyclin E proteins in vivo (Fig. 7). As a consequence, the cdk2 inhibitors lead to growth arrest, and for CVT-313 the arrest is distributed fairly equally among the G0/G1, S, and G2/M phases of the cell cycle (data not shown). Furthermore, consistent with their effects on H1 phosphorylation, the cdk2 inhibitors also closed the MMTV promoter chromatin structure (Fig. 3 and 4). The consequence of this architectural transition was an approximately 60 to 70% decrease in both hypersensitivity and binding of NF1 to the promoter, facilitating a reduced transcriptional response. Thus, the downregulation of cdk2 activity may represent a key component of the mechanism by which this promoter becomes refractory upon prolonged hormone treatment.
The elevated expression of the cdk2 inhibitor protein p21 in dexamethasone-dependent refractory cells (Fig. 7) suggests an additional mechanism by which cdk2 activity is reduced. CHIP assays revealed that an active promoter is associated with substantially more phosphorylated histone H1 than was detected at the promoter made refractory by cdk inhibitors (Fig. 6). However, the dephosphorylation of histone H1 by prolonged hormone treatment or treatment with cdk2 inhibitors is a global effect and, as such, would not be restricted to the MMTV promoter. Therefore, it was of interest to investigate the expression of other endogenous genes in the context of these treatments. In contrast to MMTV, expression of neither the glucocorticoid-inducible MT gene nor the glucocorticoid-neutral GAPDH gene was substantially modulated by the phosphorylation status of histone H1 (Fig. 5). This finding is reminiscent of studies of Xenopus and T. thermophila in which the absence of histone H1 or potential phosphorylation sites on histone H1 does not initiate a global effect on transcription (7, 14, 35, 42). Instead, it was found that specific genes in Tetrahymena were either activated (ngoA) or repressed (Cyp1), depending on the growth conditions employed (14, 35). Similarly, experiments in Xenopus oocytes revealed that histone H1 incorporation in the nucleosome where the TRE resides in the TrβA gene was critical to the ability of the TR-RXR heterodimer to activate transcription (45). Finally, a compelling role for histone H1 in the gene-specific regulation of transcription has been seen in the in vivo regulation of the Xenopus MyoD gene, suggesting that H1 has selective functions in transcriptional regulation (37).
The linker histone H1 is believed in some cases to act as a repressor of transcription due to its role in chromatin condensation (11). Although the level of histone H1 phosphorylation has been shown to be modulated with respect to the cell cycle (34), the precise role of this modification in chromatin remodeling and transcriptional regulation is less well understood. It has been postulated that phosphorylation of histone H1 decreases its net positive charge and repels it from negatively charged DNA. This depletion or removal of histone H1 from nucleosomes may then lead to a structural reorganization of chromatin to provide access for transcription factors involved in replication and transcription (27, 34).
More recent observations have provided insight into the role of histone H1 phosphorylation on chromatin structure and transcription regulation. For example, cdk2 phosphorylates histone H1 in late G1, and the deregulation of this activity was found to correlate with a relaxed chromatin structure in retinoblastoma protein (Rb)-deficient fibroblasts (19). The binding and displacement of histone H1 has been found to exert regulatory effects on transcription from certain genes. For example, when histone H1 is bound to the beta interferon promoter transcription is repressed, and this effect can be reversed by the displacement of histone H1 by HMG1 protein (6). Similarly for the MMTV promoter, a transient displacement of histone H1 has been seen in concert with steroid-dependent chromatin remodeling (9). Furthermore, incorporation of mutations in potential histone H1 phosphorylation sites in Tetrahymena suggest that the phosphorylation of H1 may regulate gene expression in vivo (14).
CBP is a histone acetyltransferase (30), and the acetylation of core histones by a CBP containing complex (38) has been shown to be a crucial determinant in the transcriptional activation of many genes (22). Interestingly, it has also been reported that phosphorylation of CBP by cdk2 modulates the intrinsic acetyltransferase activity of CBP (also known as p300) (1). cdk2 also phosphorylates Rb in vivo and the regulates the phosphorylation status of Rb during the cell cycle (2). Indeed, inhibition of cdk2 activity with the cdk2 inhibitors not only blocks histone H1 phosphorylation but also blocks phosphorylation of other cellular transcription factors such as Rb (41) and CBP. It is clear that cdk2 activity is crucial in a broad range of cellular functions, including the control of cell cycle progression, chromatin remodeling, and the regulation of transcription. To this list we would now add the glucocorticoid-dependent activation of the MMTV promoter. The inhibition of cdk2 activity in either hormone-refractory mouse cells or cells treated with cdk2 inhibitors promotes the dephosphorylation of histone H1 which modifies MMTV chromatin structure in such a way that the GR is neither able to remodel chromatin nor recruit transcription factors to the promoter.
ACKNOWLEDGMENTS
We thank Laurent Meijer (Roscoff) and Dov Schiffman (CV Therapeutics) for Roscovitine and CVT-313, respectively. We thank H. K Kinyamu for technical help. We are grateful to K. Brown, B. Deroo, H. K. Kinyamu, C. Weinberger, J. O'Bryan, and J. Cidlowski for critical review of the manuscript. We also thank Bonnie Deroo for help with the figures.
This work was supported in its initial stages by grants to T.K.A. from the National Cancer Institute of Canada and the Canadian Breast Cancer Research Initiative of Canada.
REFERENCES
- 1.Ait-Si-Ali S, Ramirez S, Barre F-X, Dkhissi F, Magnaghi-jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama T, Ohuchi T, Sumida S, Matsumoto K, Toyoshima K. Phosphorylation of the retinoblastoma protein by cdk2. Proc Natl Acad Sci USA. 1992;89:7900–7904. doi: 10.1073/pnas.89.17.7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer T K, Cordingley M G, Wolford R G, Hager G L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11:688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 5.Archer T K, Fryer C J, Lee H-L, Zaniewski E, Liang T, Mymryk J S. Steroid hormone receptor status defines the MMTV promoter chromatin structure in vivo. J Steroid Biochem. 1995;53:421–429. doi: 10.1016/0960-0760(95)00088-h. [DOI] [PubMed] [Google Scholar]
- 6.Bonnefoy E, Bandu M-T, Doly J. Specific binding of high-mobility-group 1 (HMG1) protein and histone H1 to the upstream AT-rich region of the murine beta interferon promoter: HMG1 protein acts as a potential antirepressor of the promoter. Mol Cell Biol. 1999;19:2803–2816. doi: 10.1128/mcb.19.4.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouvet P, Dimitrov S I, Wolffe A P. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 1994;8:1147–1159. doi: 10.1101/gad.8.10.1147. [DOI] [PubMed] [Google Scholar]
- 8.Bradbury E M. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 9.Bresnick E H, Bustin M, Marsaud V, Richard-Foy H, Hager G L. The transcriptionally active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992;20:273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks E E, Gray N S, Joly A, Kerwar S S, Lum R, Mackman R L, Norman T C, Rosete J, Rowe M, Schow S R, Schultz P G, Wang X, Wick M M, Shiffman D. CVT-313, a specific and potent inhibitor of CDK2 that prevents neointimal proliferation. J Biol Chem. 1997;272:29207–29211. doi: 10.1074/jbc.272.46.29207. [DOI] [PubMed] [Google Scholar]
- 11.Croston G E, Kerrigan L A, Lira L M, Marshak D R, Kagonaga J T. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 12.Dedon P C, Soults J A, Allis C D, Gorovsky M A. Formaldehyde cross-linking and immunoprecipitation demonstrate developmental changes in H1 association with transcriptionally active genes. Mol Cell Biol. 1991;11:1729–1733. doi: 10.1128/mcb.11.3.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dou Y, Gorovsky M A. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol Cell. 2000;2:225–231. doi: 10.1016/s1097-2765(00)00024-1. [DOI] [PubMed] [Google Scholar]
- 14.Dou Y, Mizzen C A, Abrams M, Allis C D, Gorovsky M A. Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol Cell. 1999;4:641–647. doi: 10.1016/s1097-2765(00)80215-4. [DOI] [PubMed] [Google Scholar]
- 15.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 16.Gray N, Detivaud L, Doerig C, Meijer L. ATP-site directed inhibitors of cyclin-dependent kinases. Curr Med Chem. 1999;9:859–875. [PubMed] [Google Scholar]
- 17.Guo C Y, Wang Y, Brautigan D L, Larner J M. Histone H1 dephosphorylation is mediated through a radiation-induced signal transduction pathway dependent on ATM. J Biol Chem. 1999;274:18715–18720. doi: 10.1074/jbc.274.26.18715. [DOI] [PubMed] [Google Scholar]
- 18.Hager G L, Archer T K, Fragoso G, Bresnick E H, Tsukagoshi Y, John S, Smith C L. Influence of chromatin structure on the binding of transcription factors to DNA. Cold Spring Harbor Symp Quant Biol. 1993;58:63–71. doi: 10.1101/sqb.1993.058.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Herrera R E, Chen F, Weinberg R A. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc Natl Acad Sci USA. 1996;93:11510–11515. doi: 10.1073/pnas.93.21.11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 21.Koff A, Giordano A, Desai D, Yamashita K, Harper J W, Elledge S, Nishimoto T, Morgan D O, Franza B R, Roberts J M. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 22.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 23.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Histone acetyltransferase activity of yeast Gen5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langan T A, Gautier J, Lohka M, Hollingsworth R, Moreno S, Nurse P, Maller J, Sclafani R A. Mammalian growth associated H1 histone kinase: a homolog of cdc2+/CDC28 protein kinases controlling mitotic entry in yeast and frog cells. Mol Cell Biol. 1989;9:3860–3868. doi: 10.1128/mcb.9.9.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H-L, Archer T K. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994;14:32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee H-L, Archer T K. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 1998;17:1454–1466. doi: 10.1093/emboj/17.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu M J, Mpoke S S, Dadd C A, Allis C D. Phosphorylated and dephosphorylated linker histone H1 reside in distinct chromatin domains in tetrahymena macronuclei. Mol Biol Cell. 1995;6:1077–1087. doi: 10.1091/mbc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meijer L, Borgne A, Mulner O, Chong J P, Blow J J, Inagaki N, Inagaki M, Delcros J G, Moulinoux J P. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur J Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- 29.Nacheva G A, Guschin D Y, Preobrazhenskaya O V, Karpov V L, Ebralidse K K, Mirzabekov A D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989;58:27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- 30.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 31.Ohsumi K, Katagiri C, Kishimoto T. Chromosome condensation in Xenopus mitotic extracts without histone H1. Science. 1993;262:2033–2035. doi: 10.1126/science.8266099. [DOI] [PubMed] [Google Scholar]
- 32.Panyim S, Chalkley R. High-resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969;130:337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- 33.Richard-Foy H, Hager G L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth S Y, Allis C D. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem Sci. 1992;17:93–98. doi: 10.1016/0968-0004(92)90243-3. [DOI] [PubMed] [Google Scholar]
- 35.Shen X, Gorovsky M A. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 36.Shen X, Yu L, Weir J W, Gorovsky M A. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 37.Steinbach O C, Wolffe A P, Rupp R A. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 1997;389:395–399. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- 38.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 39.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;1:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 40.Wade P A, Wolffe A P. Histone acetyletransferases in control. Curr Biol. 1997;7:R82–R84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 41.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 42.Wolffe A P. The role of transcription factors, chromatin structure and DNA replication in 5S RNA gene regulation. J Cell Sci. 1994;107:2055–2063. doi: 10.1242/jcs.107.8.2055. [DOI] [PubMed] [Google Scholar]
- 43.Wolffe A P. Histone H1. Int J Biochem Cell Biol. 1997;12:1463–1476. doi: 10.1016/s1357-2725(97)00026-5. [DOI] [PubMed] [Google Scholar]
- 44.Wolffe A P. Chromatin. London, England: Academic Press, Ltd.; 1998. [Google Scholar]
- 45.Wong J, Li Q, Levi B Z, Shi Y B, Wolffe A P. Structural and functional features of a specific nucleosome containing a recognition element for the thyroid hormone receptor. EMBO J. 1997;16:7130–7145. doi: 10.1093/emboj/16.23.7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;15:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]