Abstract
Over the past decade, artificial intelligence (AI) and machine learning (ML) have become the breakthrough technology most anticipated to have a transformative effect on pharmaceutical research and development (R&D). This is partially driven by revolutionary advances in computational technology and the parallel dissipation of previous constraints to the collection/processing of large volumes of data. Meanwhile, the cost of bringing new drugs to market and to patients has become prohibitively expensive. Recognizing these headwinds, AI/ML techniques are appealing to the pharmaceutical industry due to their automated nature, predictive capabilities, and the consequent expected increase in efficiency. ML approaches have been used in drug discovery over the past 15–20 years with increasing sophistication. The most recent aspect of drug development where positive disruption from AI/ML is starting to occur, is in clinical trial design, conduct, and analysis. The COVID-19 pandemic may further accelerate utilization of AI/ML in clinical trials due to an increased reliance on digital technology in clinical trial conduct. As we move towards a world where there is a growing integration of AI/ML into R&D, it is critical to get past the related buzz-words and noise. It is equally important to recognize that the scientific method is not obsolete when making inferences about data. Doing so will help in separating hope from hype and lead to informed decision-making on the optimal use of AI/ML in drug development. This manuscript aims to demystify key concepts, present use-cases and finally offer insights and a balanced view on the optimal use of AI/ML methods in R&D.
Graphical abstract
KEY WORDS: Artificial intelligence, Machine learning, Drug development, Precision medicine, Probability of success, Clinical trial design, Risk-based monitoring, Predictive modeling
Introduction
Artificial intelligence (AI) and machine learning (ML) have flourished in the past decade, driven by revolutionary advances in computational technology. This has led to transformative improvements in the ability to collect and process large volumes of data. Meanwhile, the cost of bringing new drugs to market and to patients has become prohibitively expensive. In the remainder of this paper, we use “R&D” to generally describe the research, science, and processes associated with drug development, starting with drug discovery to clinical development and conduct, and finally the life-cycle management stage.
Developing a new drug is a long and expensive process with a low success rate as evidenced by the following estimates: average R&D investment is $1.3 billion per drug [1]; median development time for each drug ranges from 5.9 to 7.2 years for non-oncology and 13.1 years for oncology; and proportion of all drug-development programs that eventually lead to approval is 13.8% [2]. Recognizing these headwinds, AI/ML techniques are appealing to the drug-development industry, due to their automated nature, predictive capabilities, and the consequent expected increase in efficiency. There is clearly a need, from a patient and a business perspective, to make drug development more efficient and thereby reduce cost, shorten the development time and increase the probability of success (POS). ML methods have been used in drug discovery for the past 15–20 years with increasing sophistication. The most recent aspect of drug development where a positive disruption from AI/ML is starting to occur, is in clinical trial design, operations, and analysis. The COVID-19 pandemic may further accelerate utilization of AI/ML in clinical trials due to increased reliance on digital technology in patient data collection. With this paper, we attempt a general review of the current status of AI/ML in drug development and also present new areas where there might be potential for a significant impact. We hope that this paper will offer a balanced perspective, help in separating hope from hype, and finally inform and promote the optimal use of AI/ML.
We begin with an overview of the basic concepts and terminology related to AI/ML. We then attempt to provide insights on when, where, and how AI/ML techniques can be optimally used in R&D, highlighting clinical trial data analysis where we compare it to traditional inference-based statistical approaches. This is followed by a summary of the current status of AI/ML in R&D with use-case illustrations including ongoing efforts in clinical trial operations. Finally, we present future perspectives and challenges.
AI And ML: Key Concepts And Terminology
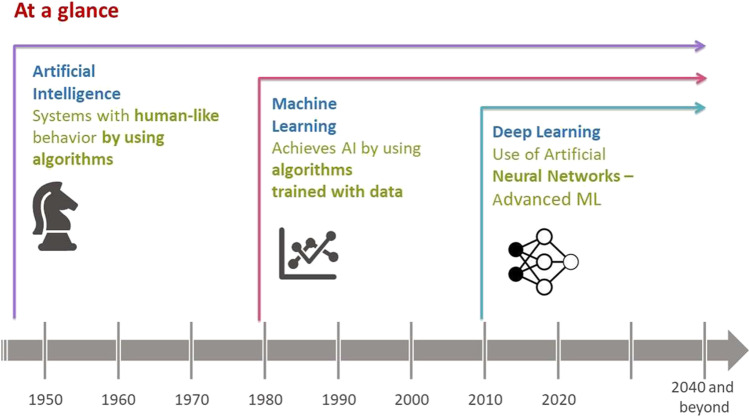
In this section, we present an overview of key concepts and terminology related to AI and ML and their interdependency (see Fig. 1 and Table I). AI is a technique used to create systems with human-like behavior. ML is an application of AI, where AI is achieved by using algorithms that are trained with data. Deep learning (DL) is a type of ML vaguely inspired by the structure of the human brain, referred to as artificial neural networks.
Fig. 1.
Chronology of AI and ML
Table I.
below provides simple descriptors of the basic terminology related to AI, ML, and related techniques
| General AI | Systems that have human-like behavior in thought and action |
|---|---|
| ML | The practice of using algorithms to parse data, learn from data, and then make predictions about unseen data without being explicitly programmed to do so |
| Neural Network (NN) | A highly parameterized model, inspired by the biological neural networks that constitute the human brain |
| Deep learning (DL) | A subfield of ML where a multi-layered (deep) architecture is used to map the relationships between inputs or observed features and outcomes |
| Supervised learning | A subfield of ML that uses labeled datasets to train algorithms that classify data or predict outcomes accurately |
| Unsupervised learning | A subfield of ML that uses unlabeled data to discover patterns that help solve clustering or association problems |
| Semi-supervised learning | A subfield of ML that combines a small amount of labeled data with a large amount of unlabeled data during training |
| Reinforcement learning | A subfield of ML that is concerned with taking a sequence of actions in a previously unknown environment in order to maximize some form of cumulative reward |
| Bayesian probabilistic programming | A field in which Bayesian models are represented as programs, and inference, learning, and querying are operations that can be represented by programs as well |
| Bayesian nonparametric learning | The field of models and related Bayesian inference routines where the number of parameters grows with the data |
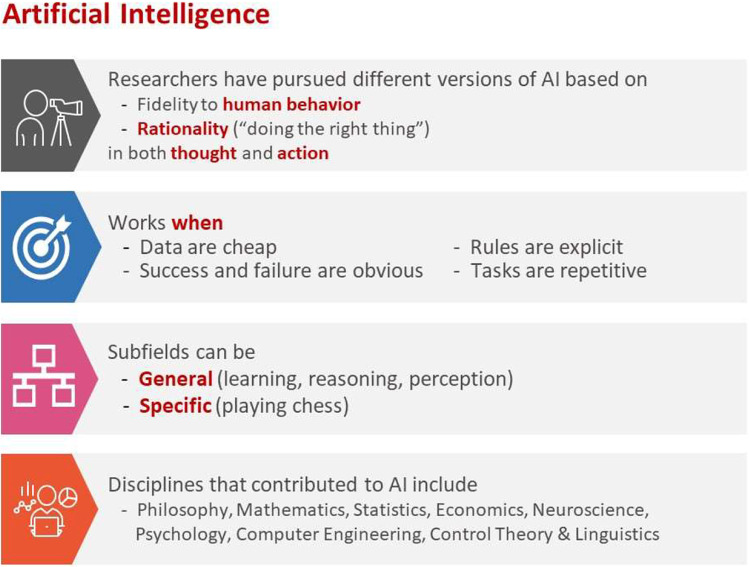
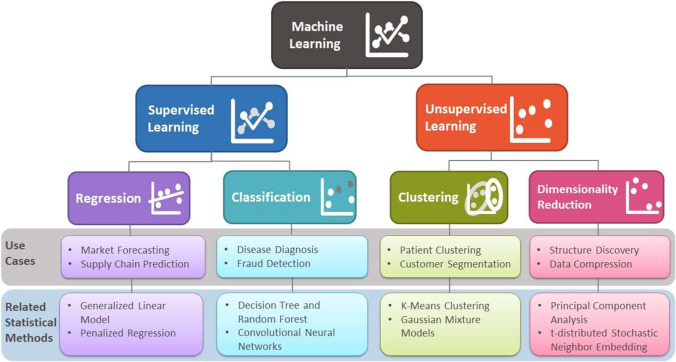
Human intelligence is related to the ability of the human brain to observe, understand, and react to an ever-changing external environment. The field of AI not only tries to understand how the human brain works but also tries to build intelligent systems that can react to an ever-changing external environment in a safe and effective way (see Fig. 2 for a brief overview of AI [3]). Researchers have pursued different versions of AI by focusing on either fidelity to human behavior or rationality (doing the right thing) in both thought and action. Subfields of AI can be either general focusing on perception, learning, reasoning, or specific such as playing chess. A multitude of disciplines have contributed to the creation of AI technology, including philosophy, mathematics, and neuroscience. ML, an application of AI, uses statistical methods to find patterns in data, where data can be text, images, or anything that is digitally stored. ML methods are typically classified as supervised learning, unsupervised learning, and reinforcement learning. (See Fig. 3 for a brief overview of supervised and unsupervised learning.)
Fig. 2.
Brief overview of AI
Fig. 3.
Brief overview of supervised and unsupervised learning
Current Status
AI/ML techniques have the potential to increase the likelihood of success in drug development by bringing significant improvements in multiple areas of R&D including: novel target identification, understanding of target-disease associations, drug candidate selection, protein structure predictions, molecular compound design and optimization, understanding of disease mechanisms, development of new prognostic and predictive biomarkers, biometrics data analysis from wearable devices, imaging, precision medicine, and more recently clinical trial design, conduct, and analysis. The impact of the COVID-19 pandemic on clinical trial execution will potentially accelerate the use of AI and ML in clinical trial execution due to an increased reliance on digital technology for data collection and site monitoring.
In the pre-clinical space, natural language processing (NLP) is used to help extract scientific insights from biomedical literature, unstructured electronic medical records (EMR) and insurance claims to ultimately help identify novel targets; predictive modeling is used to predict protein structures and facilitate molecular compound design and optimization for enabling selection of drug candidates with a higher probability of success. The increasing volume of high-dimensional data from genomics, imaging, and the use of digital wearable devices, has led to rapid advancements in ML methods to handle the “Large p, Small n” problem where the number of variables (“p”) is greater than the number of samples (“n”). Such methods also offer benefits to research in the post-marketing stage with the use of “big data” from real-world data sources to (i) enrich the understanding of a drug’s benefit-risk profile; (ii) better understand treatment sequence patterns; and (iii) identify subgroups of patients who may benefit more from one treatment compared with others (precision medicine).
While AI/ML have been widely used in drug discovery, translational research and the pre-clinical phase with increasing sophistication over the past two decades, their utilization in clinical trial operations and data analysis has been slower. We use “clinical trial operations” to refer to the processes involved in the execution and conduct of the clinical trials, including site selection, patient recruitment, trial monitoring, and data collection. Clinical trial data analysis refers to data management, statistical programming, and statistical analysis of participant clinical data collected from a trial. On the trial operations end, patient recruitment has been particularly challenging with an estimated 80% of trials not meeting enrollment timelines and approximately 30% of phase 3 trials terminating early due to enrollment challenges [4]. Trial site monitoring (involving in-person travel to sites) is an important and expensive quality control step mandated by regulators. Furthermore, with multi-center global trials, clinical trial monitoring has become labor-intensive, time-consuming, and costly. In addition, the duration from the “last subject last visit” trial milestone for the last phase 3 trial to the submission of the data package for regulatory approval, has been largely unchanged over the past two decades and presents a huge opportunity for positive disruption by AI/ML. Shortening this duration will have a dramatic impact on our ability to get drugs to patients faster while reducing cost. The steps in-between include cleaning and locking the trial database, generating the last phase 3 trial analysis results (frequently involving hundreds of summary tables, data listings, and figures), writing the clinical study report, completing the integrated summary of efficacy and safety, and finally creation of the data submission package. The impact of COVID-19 may further accelerate the push to integrate AI/ML into clinical trial operations due to an increased push toward 100% or partially virtual (or “decentralized”) trials and the increased use of digital technology to collect patient data. AI/ML methods can be used to enhance patient recruitment and project enrollment and also to allow real-time automated and “smart” monitoring for clinical data quality and trial site performance monitoring. We believe AI/ML hold potential to have a transformative effect on clinical trial operations and clinical trial data analyses particularly in the areas of trial data analysis, creation of clinical study reports, and regulatory submission data packages.
Case Studies
Below, we offer a few use cases to illustrate how AI/ML methods have been used or are in the process of improving existing approaches in R&D.
Case Study 1 (Drug Discovery)—DL for Protein Structure Prediction and Drug Repurposing
A protein’s biological mechanism is determined by its three-dimensional (3D) structure that is encoded in its one-dimensional (1D) string of amino acid sequence. Knowledge about protein structures is applied to understand their biological mechanisms and help discover new therapies that can inhibit or activate the proteins to treat target diseases. Protein misfolding has been known to be important in many diseases, including type II diabetes, as well as neurodegenerative diseases such as Alzheimer’s, Parkinson’s, Huntington’s, and amyotrophic lateral sclerosis [5]. Given the knowledge gap between a proteins’1D string of amino acid sequence and its 3D structure, there is significant value in developing methods that can accurately predict 3D protein structures to assist new drug discovery and an understanding of protein-folding diseases. AlphaFold [6, 7] developed by DeepMind (Google) is an AI network used to determine a protein’s 3D shape based on its amino acid sequence. It applied a DL approach to predict the structure of the protein using its sequence. The central component of AlphaFold is a convolutional neural network that was trained on the Protein Data Bank structures to predict the distances between every pair of residues in a protein sequence, giving a probabilistic estimate of a 64 × 64 region of the distance map. These regions are then tiled together to produce distance predictions for the entire protein for generating the protein structure that conforms to the distance predictions. In 2020, AlphaFold released the structure predictions of five understudied SARS-CoV-2 targets including SARS-CoV-2 membrane protein, Nsp2, Nsp4, Nsp6, and Papain-like proteinase (C terminal domain), which will hopefully deepen the understanding of under-studied biological systems [8].
Beck et al. [9] developed a deep learning–based drug-target interaction prediction model, called Molecule Transformer-Drug Target Interaction (MT-DTI), to predict binding affinities based on chemical sequences and amino acid sequences of a target protein, without their structural information, which can be used to identify potent FDA-approved drugs that may inhibit the functions of SARS-CoV-2’s core proteins. Beck et al. computationally identified several known antiviral drugs, such as atazanavir, remdesivir, efavirenz, ritonavir, and dolutegravir, which are predicted to show an inhibitory potency against SARS-CoV-2 3C–like proteinase and can be potentially repurposed as candidate treatments of SARS-CoV-2 infection in clinical trials.
Case Study 2 (Translational Research/Precision Medicine)—Machine Learning for Developing Predictive Biomarkers
Several successful case studies have now been published to show that the biomarkers derived by the ML predictive models were used to stratify patients in clinical development. Predictive models were developed [10] to test whether the models derived from cell line screen data could be used to predict patient response to erlotinib (treatment for non-small cell lung cancer and pancreatic cancer) and sorafenib (treatment for kidney, liver, and thyroid cancer), respectively. The predictive models have IC50 values as dependent variables and gene expression data from untreated cells as independent variables. The whole-cell line panel was used as the training dataset and the gene expression data generated from tumor samples of patients treated with the same drug was used as the testing dataset. No information from the testing dataset was used in training the drug sensitivity predictive models. The BATTLE clinical trial data was used as an independent testing dataset to evaluate the performance of the drug sensitivity predictive models trained by cell line data. The best models were selected and used to predict IC50s that define the model-predicted drug-sensitive and drug-resistant groups.
Li et al. [10] applied the predictive model to stratify patients in the erlotinib arm from the BATTLE trial. The median progression-free survival (PFS) for the model-predicted erlotinib-sensitive patient group was 3.84 months while the PFS for model-predicted erlotinib-resistant patients was 1.84 months, which suggests that the erlotinib-sensitive patients predicted by the model had more than doubled PFS benefit relative to erlotinib-resistant patients. Similarly, the model-predicted sorafenib-sensitive group had a median PFS benefit of 2.66 months over the sorafenib-resistant group with a p-value of 0.006 and a hazard ratio of 0.32 (95%CI, 0.15 to 0.72). The median PFS was 4.53 and 1.87 months, for model-predicted sorafenib-sensitive and model-predicted sorafenib-resistant groups, respectively.
Case Study 3—Nonparametric Bayesian Learning for Clinical Trial Design and Analysis
Many of the existing ML methods are focused on learning a set of parameters within a class of models using the appropriate training data, which is often referred to as model selection. However, an important issue encountered in practice is the potential model over-fitting or under-fitting, as well as the discovery of an underlying data structure and related causes [11]. Examples include but are not limited to the following: selecting the number of clusters in clustering problem, the number of hidden states in a hidden Markov model, the number of latent variables in a latent variable model, or the complexity of features used in nonlinear regression. Thus, it is important to appropriately train ML methods to perform reliably under real-world conditions with trustworthy predictions. Cross-validation is commonly used as an efficient way to evaluate how well the ML methods perform in the selection of tuning parameters.
Nonparametric Bayesian learning has emerged as a powerful tool in modern ML framework due to its flexibility, providing a Bayesian framework for model selection using a nonparametric approach. More specifically, a Bayesian nonparametric model allows us to use an infinite-dimensional parameter space and involve only a finite subset of the available parameters on the given sample set. Among them, the Dirichlet process is currently a commonly used Bayesian nonparametric model, particularly in Dirichlet process mixture models (also known as infinite mixture models). Dirichlet process mixtures provide a nonparametric approach to model densities and identify latent clusters within the observed variables without pre-specification of the number of components in a mixture model. With advances in Markov Chain Monte Carlo (MCMC) techniques, sampling from infinite mixtures can be done directly or using finite truncations.
There are many applications of such Bayesian nonparametric models in clinical trial design. For example, in oncology dose-finding clinical trials, nonparametric Bayesian learning can offer efficient and effective dose selection. In oncology first in human trials, it is common to enroll patients with multiple types of cancers which causes heterogeneity. Such issues can be more prominent in immuno-oncology and cell therapies. Designs that ignore the heterogeneity of safety or efficacy profiles across various tumor types could lead to imprecise dose selection and inefficient identification of future target populations. Li et al. [12] proposed nonparametric Bayesian learning–based designs for adaptive dose finding with multiple populations. These designs based on the Bayesian logistic regression model (BLRM) allow data-driven borrowing of information, across multiple populations, while accounting for heterogeneity, thus improving the efficiency of the dose search and also the accuracy of estimation of the optimal dose level. Liu et al. [13] extended another commonly used dose-finding design, modified toxicity probability interval (mTPI) designs to BNP-mTPI and fBNP-mTPI, by utilizing Bayesian nonparametric learning across different indications. These designs use the Dirichlet process, which is more flexible in prior approximation, and can automatically group patients into similar clusters based on the learning from the emerging data.
Nonparametric Bayesian learning can also be applied in master protocols including basket, umbrella, and platform trials, which allow investigation of multiple therapies, multiple diseases, or both within a single trial [14–16]. With the use of nonparametric Bayesian learning, these trials have an enhanced potential to accelerate the generation of efficacy and safety data through adaptive decision-making. This can affect a reduction in the drug development timeline in an area of significant unmet medical need. For example, in the evaluation of potential COVID-19 therapies, adaptive platform trials have quickly emerged as a critical tool, e.g., the clinical benefits of remdesivir and dexamethasone have been demonstrated using such approaches in the Adaptive COVID-19 Treatment Trial (ACTT) and the RECOVERY [17] trial.
One of the key questions in master protocols is whether borrowing across various treatments or indications is appropriate. For example, ideally, each tumor subtype in a basket trial should be tested separately; however, it is often infeasible given the rare genetic mutations. There is potential bias due to the small sample size and variability as well as the inflated type I error if there is a naïve pooling of subgroup information. Different Bayesian hierarchical models (BHMs) have been developed to overcome the limitation of using either independent testing or naïve pooling approaches, e.g., Bayesian hierarchical mixture model (BHMM) and exchangeability-non-exchangeability (EXNEX) model. However, all these models are highly dependent on the pre-specified mixture parameters. When there is limited prior information on the heterogeneity across different disease subtypes, the misspecification of parameters can be a concern. To overcome the potential limitation of existing parametric borrowing methods, Bayesian nonparametric learning is emerging as a powerful tool to allow flexible shrinkage modeling for heterogeneity between individual subgroups and for automatically capturing the additional clustering. Bunn et al. [18] show that such models require fewer assumptions than other more commonly used methods and allow more reliable data-driven decision-making in basket trials. Hupf et al. [19] further extend these flexible Bayesian borrowing strategies to incorporate historical or real-world data.
Case Study 4—Precision Medicine with Machine Learning
Based on recent estimates, among phase 3 trials with novel therapeutics, 54% failed in clinical development, with 57% of those failures due to inadequate efficacy [20]. A major contributing factor is failure in identification of the appropriate target patient population with the right dose regimen including the right dose levels and combination partners. Thus, precision medicine has become a priority in pharmaceutical industry for drug development. One approach could be a systematic model utilizing ML applied to (a) build a probabilistic model to predict probability of success; and (b) identify subgroups of patients with a higher probability of therapeutic benefit. This will enable the optimal match of patients with the right therapy and maximize the resources and patient benefit. The training datasets can include all ongoing early-phase data, published data, and real-world evidence but are limited to the same class of drugs.
One major challenge to establish the probabilistic model is defining endpoints that can best measure therapeutic effect. Early-phase clinical trials (particularly in oncology) frequently adopt different primary efficacy endpoints compared with confirmatory pivotal trials due to a relatively shorter follow-up time and need for faster decision-making. For example, common oncology endpoints are overall response rate or complete response rate in phase I/II and progression-free survival (PFS) and/or overall survival (both measure long-term benefit) in pivotal phase III trials. In oncology, it is also common that phase I/II trials use single-arm settings to establish the proof of concept and generate the hypothesis of treatment benefit, while in pivotal trials, especially in randomized phase III trial with a control arm, the purpose is to demonstrate superior treatment benefit over available therapy. This change in the targeted endpoints from the early phase to late phase makes the prediction of POS in the pivotal trial, using early-phase data, quite challenging. Training datasets using previous trials for drugs with a similar mechanism and/or indications can help establish the relationship between the short-term endpoints and long-term endpoints, which ultimately determines the success of drug development.
Additionally, the clustering of patients can be done using unsupervised learning. For example, nonparametric Bayesian hierarchical models using the Dirichlet process enables patient grouping (without pre-specified number of clusters) with key predictive or prognostic factors, to represent various levels of treatment benefit. This DL approach will bring efficiency in patient selection for precision medicine clinical development.
Case Study 5—AI/ML-assisted Tool for Clinical Trial Oversight
Monitoring of trials by a sponsor is a critical quality control measure mandated by regulators to ensure the scientific integrity of trials and safety of subjects. With increasing complexity of data collection (increased volume, variety, and velocity), and the use of contract research organizations (CROs)/vendors, sponsor oversight of trial site performance and trial clinical data has become challenging, time-consuming, and extremely expensive. Across all study phases (excluding estimated site overhead costs and costs for sponsors to monitor the study), trial site monitoring is among the top three cost drivers of clinical trial expenditures (9–14% of total cost) [21].
For monitoring of trial site performance, risk-based monitoring (RBM) has recently emerged as a potential cost-saving and efficient alternative to traditional monitoring (where sponsors sent study monitors to visit sites for 100% source-data verification (SDV) according to a pre-specified schedule). While RBM improves on traditional monitoring, inconsistent RBM approaches used by CROs and the current prospective nature of the operational/clinical trial data reviews—has meant that sponsor’s ability to detect critical issues with site performance, may be delayed or compromised (particularly in lean organizations where CRO oversight is challenging due to limited resources).
For monitoring of trial data quality, current commonly used approaches largely rely on review of traditional subject and/or aggregate data listings and summary statistics based on known risk factors. The lack of real-time data and widely available automated tools limit the sponsor’s ability for prospective risk mitigation. This delayed review can have a significant impact on the outcome of a trial, e.g., in an acute setting where the primary endpoint uses ePRO data—monthly transfers may be too late to prevent incomplete or incorrect data entry. The larger impact is a systemic gap in study team oversight that could result in critical data quality issues.
One potential solution is the use of AI/ML-assisted tools for monitoring trial site performance and trial data quality. Such a tool could offer an umbrella framework, overlaid on top of the CRO systems, for monitoring trial data quality and sites. With the assistance of AI/ML, study teams may be able to use an advanced form of RBM (improved prediction of risk and thresholds for signal detection) and real-time clinical data monitoring with increased efficiency/quality and reduced cost in a lean resourced environment. Such a tool could apply ML and predictive analytics to current RBM and data quality monitoring—effectively moving current study monitoring to the next generation of RBM. The use of accumulating data from the ongoing trial and available data from similar trials, to continuously improve on the data quality and site performance checks, could have a transformative effect on sponsor’s ability to protect patient safety, reduce trial duration, and trial cost.
In terms of data quality reviews, data fields, and components contributing to the key endpoints that impact the outcome of the trial would be identified by the study team. For trial data monitoring, an AI/ML-assisted tool can make use of predictive analytics and R Shiny visualization for cross-database checks and real-time “smart monitoring” of clinical data quality. By “smart monitoring,” we mean the use of AI/ML techniques that continuously learn from accumulating trial data and improve on the data quality checks, including edit checks. Similarly, for trial site performance, monitoring an AI/ML tool could begin with the Transcelerate (a non-profit cross-pharma consortium) library of key risk indicators (KRIs) and team-specified thresholds to identify problem sites based on operational data. In addition, the “smart” feature of an AI/ML tool could use accumulating data to continuously improve on the precision of the targeted site monitoring that makes up RBM. The authors of this manuscript are currently collaborating with a research team at MIT to advance research in Bayesian probabilistic programming approaches that could aid the development of an AI/ML tool with the features described above for clinical trial oversight of trial data quality and trial site performance.
Summary
AI/ML as a field has tremendous growth potential in R&D. As with most technological advances, this presents both challenges and hope. With modern-day data collection, the magnitude and dimensionality of data will continue to increase dramatically because of the use of digital technology. This will increase the opportunities for AI/ML techniques to deepen understanding of biological systems, to repurpose drugs for new indications, and also to inform study design and analysis of clinical trials in drug development.
Although the development of recent ML/AI methods represents major technological advances, the conclusions made could be misleading if we are not able to tease out the confounding factors, use reliable algorithms, look at the right data, and fully understand the clinical questions behind the endpoints and data collection. It is imperative to train ML algorithms properly to have trustworthy performance in practice using various data scenarios. Additionally, not every research question can be answered utilizing AI/ML, particularly if there is high variability, limited data, poor quality of the data collection, under-represented patient populations, or flawed trial design. The issue of under-represented patient populations is particularly concerning as it could lead to a systematic bias. Furthermore, in line with the emerging concerns in other spaces where AI/ML have been used, care and caution needs to be exercised to address patient privacy and bioethical considerations.
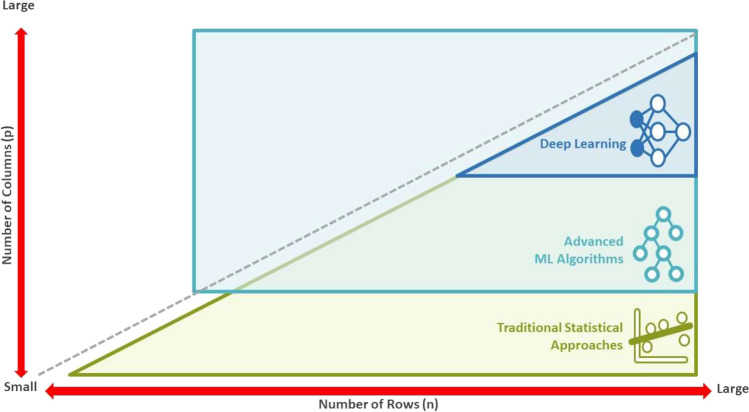
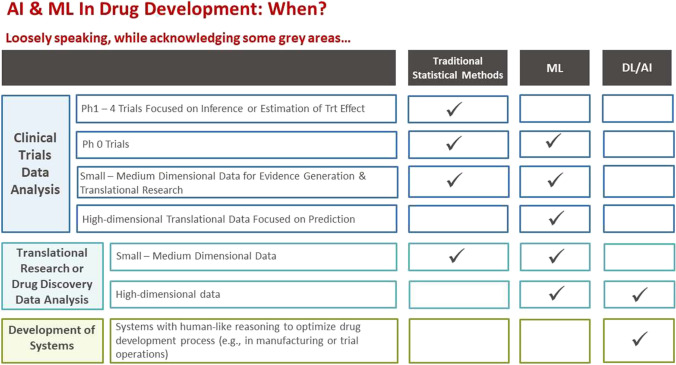
It is also important to be aware when DL/AI vs. ML vs. traditional inference-based statistical methods are most effective in R&D. In Fig. 4 below, we attempt to provide a recommendation based on the dimensionality of the dataset. In Fig. 5, we attempt to provide a similar recommendation, this time based on different aspects of drug development. Although many ML algorithms are able to handle high-dimensional data with the “Large p, Small n” problem, the increased number of variables/predictors, especially those not related to the response, continues to be a challenge. As the number of irrelevant variables/predictors increases, the volume of the noise becomes greater, resulting in the reduced predictive performance of most ML algorithms.
Fig. 4.
Application of ML/AI based on the dimensionality of the data
Fig. 5.
Application of ML/AI based on various aspects of drug development
In summary, a combination of appropriate understanding of both R&D and advanced ML/AI techniques can offer huge benefits to drug development and patients. The implementation and visualization of AI/ML tools can offer user-friendly platforms to maximize efficiency and promote the use of breakthrough techniques in R&D. However, a sound understanding of the difference between causation and correlation is vital, as is the recognition that the evolution of sophisticated prediction capabilities does not render the scientific method to be obsolete. Credible inference still requires sound statistical judgment and this is particularly critical in drug development, given the direct impact on patient health and safety. This further underscores that a well-rounded understanding of ML/AI techniques along with adequate domain-specific knowledge in R&D is paramount for their optimal use in drug development.
Author Contribution
SK, JL, RL, YZ, and WZ contributed to the ideas, implementation, and interpretation of the research topic, and to the writing of the manuscript.
Declarations
Conflict of Interest
Sheela K. was previously employed by Takeda Pharmaceuticals and is currently employed by Teva Pharmaceuticals (West Chester PA USA) during the development and revision of this manuscript. All other authors are employed by Takeda Pharmaceuticals during the development and revision of this manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.DiMasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Wong CH, Siah KW, Lo AW. Estimation of clinical trial success rates and related parameters. Biostatistics. 2019;20(2):273–286. doi: 10.1093/biostatistics/kxx069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell S, Norvig P. Artificial intelligence: a modern approach (4th edition), 2021; Pearson Series in Artificial Intelligence.
- 4.Mitchell A, Sharma Y, Ramanathan S, Sethuraman V. Is data science the treatment for inefficiencies in clinical trial operations? White paper. https://www.zs.com/insights/is-data-science-the-treatment-for-inefficiencies-in-clinical-trial-operations.
- 5.Dill KA and MacCallum JL. The protein-folding problem, 50 years on. Science. 2012;338(6110):1042–6. 10.1126/science.1219021. [DOI] [PubMed]
- 6.Senior A, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AWR, Bridgland A, Penedones H, Petersen S, Simonyan K, Crossan S, Kohli P, Jones DT, Silver D, Kavukcuoglu K, Hassabis D. Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13) Proteins. 2019;87:1141–1148. doi: 10.1002/prot.25834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senior A, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AWR, Bridgland A, Penedones H, Petersen S, Simonyan K, Crossan S, Kohli P, Jones D. T, Silver D, Kavukcuoglu K, Hassabis. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577. 10.1038/s41586-019-1923-7. [DOI] [PubMed]
- 8.John Jumper, Kathryn Tunyasuvunakool, Pushmeet Kohli, Demis Hassabis, and the AlphaFold Team, Computational predictions of protein structures associated with COVID-19, Version 3, DeepMind website, 4 August 2020, https://deepmind.com/research/open-source/computational-predictions-of-protein-structures-associated-with-COVID-19.
- 9.Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Shin H, Gulbekyan G, Pustovalova O, Nikolsky Y, Hope A, Bessarabova M, Schu M, Kolpakova-Hart E, Merberg D, Dorner A, Trepicchio WL. Development of a drug-response modeling framework to identify cell line derived translational biomarkers that can predict treatment outcome to erlotinib or sorafenib. PLoS ONE. 2015;10(6):e0130700. doi: 10.1371/journal.pone.0130700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teh YW. Dirichlet process. In Sammut C, Webb GI (Eds) Encyclopedia of Machine Learning. 2011; pp. 280–287, Springer. 10.1007/978-0-387-30164-8_219.
- 12.Li M, Liu R, Lin J, Bunn V, Zhao H. Bayesian semi-parametric design (BSD) for adaptive dose-finding with multiple strata. J Biopharm Stat. 2020;30(5):806–820. doi: 10.1080/10543406.2020.1730870. [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Lin J, Li P. Design considerations for phase I/II dose finding clinical trials in immuno-oncology and cell therapy. Contemporary Clinical Trials. 2020;96:106083. doi: 10.1016/j.cct.2020.106083. [DOI] [PubMed] [Google Scholar]
- 14.Saville BR, Berry SM. Efficiencies of platform clinical trials: a vision of the future. Clin Trials. 2016;13(3):358–366. doi: 10.1177/1740774515626362. [DOI] [PubMed] [Google Scholar]
- 15.Woodcock J, LaVange L. Master protocols to study multiple therapies, multiple diseases, or both. N Engl J Med. 2017;377:62–70. doi: 10.1056/NEJMra1510062. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Lin L, Bunn V, and Liu R. Adaptive randomization for master protocols in precision medicine. In Contemporary Biostatistics with Biopharmaceutical Application, 2019; Springer, 251–270.
- 17.NIAID (2020). Adaptive COVID-19 treatment trial (ACTT). https://clinicaltrials.gov/ct2/show/study/NCT04280705.
- 18.Bunn V, Liu R, Lin J, Lin J. Flexible Bayesian subgroup analysis in early and confirmatory trials. Contemp Clin Trials. 2020;98:106149. doi: 10.1016/j.cct.2020.106149. [DOI] [PubMed] [Google Scholar]
- 19.Hupf B, Bunn V, Lin J, Dong C. Bayesian semiparametric meta-analytic-predictive prior for historical control borrowing in clinical trials. Stat Med (accepted) 2021 doi: 10.1002/sim.8970. [DOI] [PubMed] [Google Scholar]
- 20.Hwang TJ, Carpenter D, Lauffenburger JC. Failure of investigational drugs in late-stage clinical development and publication of trial results. JAMA Intern Med. 2016;176(12):1826–1833. doi: 10.1001/jamainternmed.2016.6008. [DOI] [PubMed] [Google Scholar]
- 21.Sertkaya A, Wong H, Jessup A, Beleche T. Key cost drivers of pharmaceutical clinical trials in the United States. Clin Trials. 2016;13(2):117–126. doi: 10.1177/1740774515625964. [DOI] [PubMed] [Google Scholar]