Abstract
On average, men and women differ in brain structure and behavior, raising the possibility of a link between sex differences in brain and behavior. But women and men are also subject to different societal and cultural norms. We navigated this challenge by investigating variability of sex-differentiated brain structure within each sex. Using data from the Queensland Twin IMaging study (n = 1,040) and Human Connectome Project (n = 1,113), we obtained data-driven measures of individual differences along a male–female dimension for brain and behavior based on average sex differences in brain structure and behavior, respectively. We found a weak association between these brain and behavioral differences, driven by brain size. These brain and behavioral differences were moderately heritable. Our findings suggest that behavioral sex differences are, to some extent, related to sex differences in brain structure but that this is mainly driven by differences in brain size, and causality should be interpreted cautiously.
Keywords: masculinization, brain structure, neuroimaging, MRI, twin modeling, open data
Women and men differ, on average, in many ways. Obvious physical differences in measures such as height and strength are generally accepted to have a biological and evolutionary basis. But the basis of average differences in male and female behavior—for example, specific cognitive abilities (Gur & Gur, 2016) and personality traits (Archer, 2019)—is not well understood and is subject to controversy. On one hand, there is little doubt that historically and culturally ingrained social expectations and gender roles contribute to observed sex differences in behavior. On the other hand, there is strong resistance in some quarters to the idea that evolved predispositions—stemming from different selection pressures on our female and male ancestors—may also contribute to the observed behavioral sex differences (Eagly & Wood, 2013). Indeed, because many behavioral sex differences appear to fit with predictions from both evolutionary biology and social role theory, it is difficult to determine whether behavioral sex differences reflect evolved dispositions at all.
One clue is the observation of structural differences, on average, between female and male brains. In adulthood, male brains are, on average, 10% to 15% larger than female brains (Ruigrok et al., 2014) and remain larger even after body height is adjusted for (Ritchie et al., 2018). Also, several regional sex differences remain after adjusting for overall brain size: For instance, the largest single-sample study to date (N = 5,216; Ritchie et al., 2018) showed that after brain size was adjusted for, female UK Biobank participants had smaller volumes than male participants in the amygdala, pallidum, and putamen, whereas male participants had smaller volumes in the nucleus accumbens. A recent large voxel-wise study (N = 2,838; Lotze et al., 2019) also found sex differences in subcortical and cortical gray matter in adults. Other studies (Bruner et al., 2012; Kim et al., 2012) have reported sex differences in the shape of regional brain structures. Moreover, several studies have succeeded in predicting an individual’s biological sex on the basis of brain structure differences, showing an accuracy between 69% and 93% (Anderson et al., 2019; Chekroud et al., 2016; Del Giudice et al., 2016; Joel et al., 2018; Tunç et al., 2016; Xin et al., 2019), even after correction for height (Chekroud et al., 2016) or brain size (gray-matter volume; Anderson et al., 2019)—despite the substantial overlap on brain-structure measures between men and women (Ritchie et al., 2018). However, although these studies adjusted for global brain size, the findings may still be driven by differences in brain size because brain regions scale differently with brain size (de Jong et al., 2017).
Importantly, the well-established existence of sex differences in brain structure does not necessarily mean that these differences relate to behavioral sex differences. Indeed, some researchers have proposed that sex differences in brain structure may instead promote similarity in women’s and men’s behavior by compensating for scaling differences due to the sex difference in body and brain size (De Vries, 2004). A key obstacle to examining the association between sex differences in brain structure and behavior is that men and women, as well as having brains that differ on average, are also, on average, subject to different societal and cultural norms and expectations that might lead to behavioral sex differences. One way to eliminate sex-differentiated socialization as a confound is to examine brain differences among individuals of the same sex. Individuals vary in genetic predispositions as well as exposure and sensitivity to gonadal hormones: Some men will develop a more female-like brain, whereas other men an exaggeratedly male-like brain (and conversely, for women).
Such an approach has recently been applied successfully by predicting sex on the basis of differences in the structural connectome, that is, how the brain is wired. Using a large imaging data set of the Philadelphia Neurodevelopmental Cohort (N = 900), Tunç et al. (2016) found a weak but significant association between sex predictor scores based on the structural connectome and those based on motor and cognitive test performance. Using the same data set, Phillips et al. (2019) constructed a sex-differentiation score from several other brain-structure measures—surface area, volume, thickness, and diffusion—that correlated in the expected direction with externalizing symptoms within males but not within females; predicted correlations with internalizing symptoms were not significant in either sex. However, the question remains whether such an association between brain and behavioral sex predictor scores exists after we control for brain size on a regional level—that is, to take into account that different brain regions scale differently with brain size.
Statement of Relevance.
Women and men differ, on average, in brain structure and in behavior. A long-standing question is the extent to which these sex differences are related. The question is difficult to address because men and women are subject to different societal and cultural norms. We navigated this challenge in the present research by examining individual differences in brain structure along the male–female dimension separately for each gender group. We then determined whether the differences were associated with physical and behavioral measures such as endurance, body mass index, cognition, and personality traits. We found that brain differences on the male–female dimension were weakly associated with behavior, but this association was driven by differences in brain size. Importantly, the associations were small, suggesting that brain structure is only one of many factors explaining behavioral sex differences.
In this study, we obtained a measure of brain differences along a male–female dimension based on sex differences in brain shape and structure while adjusting for brain size on a regional level. Next, we derived a composite measure of behavioral differences along a male–female dimension from sex differences in behavior and tested whether individual differences along a male–female dimension for brain and behavior were correlated (within sex). Lastly, we used the classical twin design to estimate the extent to which these individual differences in brain and behavior can be explained by genetic and environmental influences.
Method
Participants
We analyzed two large independent imaging data sets to obtain a measure of brain differences along a male–female dimension and to test the relationship between individual differences along a male–female dimension for brain and behavior. Both data sets were drawn from the general population. The first consisted of 1,040 individuals from 616 families as part of the Queensland Twin IMaging (QTIM) study (64.81% female; age: M = 22.42 years, SD = 3.33, range = 15–30 years), including 157 identical (monozygotic [MZ]) twin pairs, 261 nonidentical (dizygotic [DZ]) twin pairs, and their siblings. Behavioral measures were collected as part of the Brisbane Longitudinal Twin Study (Gillespie et al., 2013), also known as the Brisbane Adolescent Twin Study (Wright & Martin, 2004). In addition, a subsample of 40 individuals (55% female; age: M = 23.36 years, SD = 2.27) was scanned a second time within 3 months. Diffusion-tensor imaging scans were available for 460 individuals (63.10% female; age: M = 22.20 years, SD = 2.71, range = 16.85–29.16 years) after we excluded 36 individuals, including 26 because of incidental findings of potential clinical relevance and 10 because of poor scan quality. Individuals with developmental, neurological, or psychiatric disorders; impaired intellectual functioning; or head trauma were excluded. Only right-handed twins were recruited in the study. All individuals gave written informed consent. Ethics approval for the study was given by the Human Research Ethics Committees of the QIMR Berghofer Medical Research Institute, University of Queensland, and UnitingCare Health.
The second data set was provided as part of the Human Connectome Project (HCP; Van Essen et al., 2012) and comprised 1,113 (left- and right-handed) individuals (54.40% female; age: M = 28.80 years, SD = 3.70, range = 22–37 years) from 428 families, including 129 MZ twin pairs, 72 DZ twin pairs, and their siblings. In addition, 46 individuals were scanned a second time (68.89% female; age: M = 30.29 years, SD = 3.34). Diffusion-tensor imaging scans were available for 972 individuals (53.60% female; age: M = 28.73 years, SD = 3.70, range = 22–37 years). Test-retest diffusion scans were available for 41 individuals (70.73% female; age: M = 30.46 years, SD = 3.15). Individuals with severe neurodevelopmental disorders, documented neuropsychiatric disorders, neurologic disorders, diabetes, or high blood pressure or those born prematurely were excluded. All individuals gave written informed consent. Ethics approval was given by the institutional review board.
Image acquisition
For the QTIM data set, structural MRI scans were obtained on a 4-tesla scanner (Siemens Bruker) that acquired a 3D structural T1-weighted image (longitudinal relaxation time [T1] = 700 ms, repetition time [TR] = 1,500 ms, echo time [TE] = 3.35 ms, flip angle = 8°, voxel size = 0.9375 × 0.9375 × 0.9 mm3); 81% had a coronal acquisition, and 19% had a sagittal acquisition. The test-retest sample included only participants scanned with a coronal acquisition on both occasions. Diffusion-weighted images were also collected (TR = 6,090 ms, TE = 91.7 ms, number of slices = 55, voxel size = 1.79 × 1.79 × 2 mm3, 94 directions with b = 1,159 s/mm2 and 11 b = 0 images).
For the HCP data set, structural MRI scans were obtained on a 3-tesla scanner (Siemens Connectome Skyra) that acquiring a 3D structural T1-weighted image (T1 = 1,000 ms, TR = 2,400 ms, TE = 2.14 ms, flip angle = 8°, slice thickness = 0.7 mm, voxel size = 0.70 × 0.70 × 0.70 mm3). Diffusion-weighted images were also collected (TR = 5,520 ms, TE = 89.5 ms, number of slices = 111, voxel size = 1.25 × 1.25 × 1.25 mm3, 90 directions with b = 1,000/2,000/3,000 s/mm2 and six b = 0 images).
Image preprocessing
All structural scans were preprocessed to remove signal inhomogeneity using the Statistical Parametric Mapping software package (Version 12; Friston et al., 1995) in MATLAB (Version R2018a; The MathWorks, Natick, MA). Scans were not registered to common template space to avoid distortions in the shape of the brain structures. Using the FMRIB Software Library (FSL; Jenkinson et al., 2012), we corrected diffusion-weighted images for eddy current distortions, applied a brain mask, and registered the images to the structural scan. For more details, see Jahanshad et al. (2011) for the QTIM data set and Glasser et al. (2013) for the HCP data set.
Obtaining a measure of brain differences along a male–female dimension
Using two different approaches, we obtained a measure of individual differences along a male–female dimension based on sex differences in brain shape (using the landmark approach) and structure (using the vertex-wise approach).
The landmark approach: a measure derived from brain shape
For the landmark approach, we developed and placed subcortical landmarks and placed existing cortical landmarks (called dense individualized and common connectivity-based cortical landmarks [DICCCOLs]; Zhu et al., 2013; Fig. 1). The initial landmark approach included landmarks placed in both subcortical and cortical regions using the T1-weighted scan only. Landmarks were placed on a mask on the standard template (MNI152 1 mm) in FSL’s FSLVIEW (Jenkinson et al., 2012) to serve as an example for automatic placement. Automatic placement to each individual scan was done using SPM’s “normalize” function. While visually inspecting the landmarks, we found that the placement of landmarks in cortical regions showed too much error using the method described above, so all cortical landmarks were excluded.
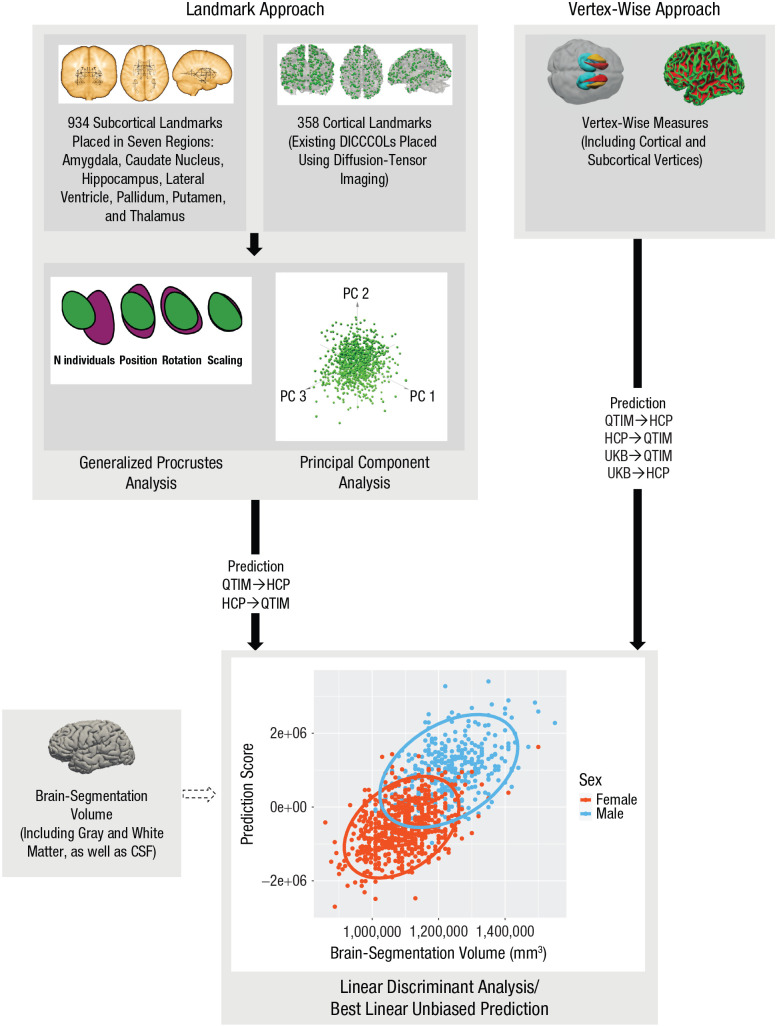
Fig. 1.
The different stages of obtaining a measure of brain differences along a male–female dimension, derived from the landmark and vertex-wise approaches. Brain size was used as a crude proxy for comparison. The landmark approach included landmarks placed in subcortical regions and dense individualized and common connectivity-based landmarks (DICCCOLs; see Ou et al., 2015) placed in cortical regions. Landmark coordinates from each individual were brought into standard space by applying a generalized Procrustes analysis, which removes variation in position, orientation, rotation, and brain size. During this process, a principal component (PC) analysis was also performed, which yields uncorrelated brain-shape variables. The vertex-wise approach included cortical and subcortical vertex-wise measures, derived from FreeSurfer (Fischl, 2012) and the ENIGMA Shape pipeline (see http://enigma.ini.usc.edu/ongoing/enigma-shape-analysis/), respectively. For each approach, the corresponding measures (landmarks/vertex-wise measures) were used in the model to predict an individual’s biological sex, using either a linear discriminant analysis or a best linear unbiased prediction, respectively, resulting in a linear combination of shape (or vertex-wise) variables that best discriminates males from females in the training sample. The same linear combination (i.e., algorithm) was then applied to predict sex in an independent sample, which assigned each individual a prediction score reflecting the position of their brain shape along this male–female dimension. As well as training the algorithm on the Queensland Twin IMaging (QTIM) sample and then applying this algorithm to predict sex in the Human Connectome Project (HCP) sample, and vice versa, for the vertex-wise measures, we also trained the algorithm on the UK Biobank (UKB) sample and then applied this algorithm to predict sex in both the QTIM and HCP samples. The scatterplot shows the association between these prediction scores and brain-segmentation volume (i.e., brain size), separately for men and women. The ovals display the 95% confidence ellipses for the corresponding means. CSF = cerebrospinal fluid.
This process resulted in 467 subcortical landmarks per hemisphere (934 in total) placed in seven subcortical regions: amygdala, caudate nucleus, hippocampus, lateral ventricle, pallidum, putamen, and thalamus (see Fig. S1 in the Supplemental Material available online). We visually inspected the placement of the 934 landmarks for 10 individuals each to confirm the accuracy of the placement method. Next, the 3D coordinates of the landmarks were extracted for each landmark. On a rare occasion, landmarks were not transformed to native space, which led to missing data. Missing data (0.035% of the data points) were imputed with the estimate-missing function (thin-plate spline setting) in the statistics package geomorph (Version 3; Adams & Otarola-Castillo, 2013) run in the R programming environment (Version 3.4.4; R Core Team, 2018).
In addition, we included 358 existing cortical landmarks (DICCCOLs) based on diffusion-weighted images (Zhu et al., 2013). These data-driven cortical landmarks are placed by using consistent white-matter fiber-connection patterns derived from diffusion-tensor imaging data. Fibers were extracted using the software package medINRIA (Version 1.9.0; Toussaint et al., 2007) for the QTIM data set (Zhu et al. 2015) and MRtrix (Version 3; Tournier et al., 2019) for the HCP data set, using a fractional anisotropy (FA) threshold of 0.2 and a minimum length of 20. We then placed the cortical landmarks by using the DICCCOL toolbox (Version 0.1; Zhu et al., 2013; see https://www.nitrc.org/projects/dicccol_0_1), and we extracted the 3D coordinates for each landmark.
Then, we brought the landmark coordinates from each individual into standard space by applying a generalized Procrustes analysis, which removes variation in size, position, orientation, and rotation of the brains (see Fig. S2 in the Supplemental Material). During this process, a principal component analysis was also performed (Fig. 1), rotating the data into uncorrelated components, using the R statistics package shapes (Version 1.2.3; Dryden, 2016). We ran this analysis separately for the cortical and subcortical landmarks because the cortical landmarks were extracted from diffusion space, whereas the subcortical landmarks were extracted in native (individual) T1 space. These analyses were performed while scaling for brain size in the Procrustes analysis to obtain a measure of brain shape independent of brain size.
Next, the first 52 principal components with an eigenvalue larger than or equal to one from both Procrustes analyses were used as predictors for the variable sex. We used the MASS package (Version 7.3; Venables & Ripley, 2002) in the R programming environment (Version 3.4.4; R Core Team, 2018) to perform a linear discriminant analysis (LDA; Fig. 1), which gives a linear combination of the shape variables that best discriminates men from women and assigns each individual a score reflecting the position of their brain shape along this male–female dimension.
The vertex-wise approach: a measure derived from brain structure
For the QTIM data set, the program FreeSurfer (Version 5.3; Fischl, 2012) was used to segment the brain from the structural T1-weighted scan and to extract the vertex-wise measures for thickness and surface area (Fig. 1). For the HCP data set, the processed images were downloaded. This segmentation also yielded a measure of brain size (i.e., brain-segmentation volume [BSV]), which includes gray and white matter and cerebrospinal fluid (see https://surfer.nmr.mgh.harvard.edu/fswiki/MorphometryStats). For processing in FreeSurfer, all individuals’ brain images were transformed to the FreeSurfer template. Then, the ENIGMA Shape pipeline (for details, see http://enigma.ini.usc.edu/ongoing/enigma-shape-analysis/) was run to extract vertex-wise measures for deep gray-matter volume as well (Fig. 1). For both the FreeSurfer and shape segmentation, we performed a detailed postprocessing quality check in line with procedures used by the ENIGMA consortium (see http://enigma.ini.usc.edu/protocols/imaging-protocols/). Next, FreeSurfer’s cortical and subcortical vertex-wise measures were included to predict sex to obtain a measure of brain differences along a male–female dimension derived from brain structure (Fig. 1).
Using the software package OSCA (a tool for omic-data-based complex trait analysis; Zhang et al., 2019; see http://cnsgenomics.com/software/osca/#Overview), we predicted the participants’ sex using best linear unbiased prediction (BLUP) scores, which allow handling the large number of vertex-wise measurements. BLUP scores are powerful and efficient predictors that do not require hyperparameter estimation (Robinson, 1991), unlike other machine-learning algorithms (e.g., support vector machine or penalized regression). In practice, BLUP scores constrain the weights given to the vertices to follow a normal distribution (Robinson, 1991). To improve prediction accuracy, we trained our BLUP scores on the first 9,888 participants of the UK Biobank who underwent MRI (Miller et al., 2016) and had usable cortical and subcortical data from processed T1-weighted and T2-FLAIR MRI images (Couvy-Duchesne et al., 2020). The UK Biobank participants were between the ages of 44.6 and 79.6 years (age: M = 62.60 years, SD = 7.5), and 52.40% of the sample was female (Couvy-Duchesne et al., 2020).
For the vertex-wise measure, we included three different approaches to obtain a measure of brain differences along a male–female dimension. Most importantly, because brain regions scale differently with brain size, we used an allometric scaling approach, adjusting the vertex-wise measures for brain size on a regional (vertex-by-vertex) level. For this, we applied a log-log regression—regressing out brain size (BSV) for each vertex by using the logs for brain size and the respective vertex and using the residuals of the vertices in the next analyses. For comparison, we regressed out brain size from the uncorrected prediction scores (instead of for each vertex as in the allometric approach). As another alternative, we regressed out only brain-size differences associated with sex from the vertex-wise measures before predicting sex to ensure that sex differences in brain size were not driving prediction accuracy.
Obtaining a measure of behavioral differences along a male–female dimension
Using a process similar to the one employed to derive our brain measures, we derived a measure of individual differences in behavior along a male–female dimension by using the behavioral variables to predict sex in an LDA. Behavioral data comprised a variety of measures including physical measures (e.g., body mass index, blood pressure), measures of intelligence (e.g., total, verbal, and performance intelligence), neurocognitive subtests (e.g., vocabulary, working memory, and visuospatial skills), and other measures (e.g., personality traits, anxiety, and depression symptoms).
Unlike the brain-imaging data, the behavioral variables were different in the QTIM and HCP samples (see Table S4 in the Supplemental Material); therefore, we divided each sample and trained the prediction in one half before predicting in the other half. For this, we used the MASS package in R (Version 3.4.4; R Core Team, 2018). Note that this approach excluded data for several behavioral measures and individuals to deal with missing values: We removed behavioral variables with scores for less than 75% of the individuals, resulting in 12 of 27 variables for QTIM and 26 of 26 measures for HCP retained in the prediction. Participants with missing values on one of the behavioral variables could not receive a prediction score (QTIM = 324, HCP = 69), resulting in the inclusion of 1,760 of the 2,153 individuals in the analyses.
Genetic analyses
For our genetic analysis, up to two siblings per family were included, and half siblings were excluded. We used a saturated univariate ACE model in the R package OpenMx (Boker et al., 2011) to examine how much of the variation in brain size, as well as the individual differences along a male–female dimension for brain and behavior, could be explained by genetic effects (A), common environmental effects (C), and residual effects including idiosyncratic environmental factors and measurement error (E), adjusting for sex and age. This model relies on the principle that MZ twins are genetically identical, whereas DZ twins share on average half of their segregating genes. Nontwin siblings were added to the classical twin design to improve statistical power.
We also tested the assumptions for twin modeling. These include (a) testing a mean and variance difference between the first twin and second twin, (b) testing a mean and variance difference between MZ and DZ (same-sex) twin pairs within women and within men, (c) testing a mean and variance difference between male MZ and DZ groups and female MZ and DZ groups, and (d) testing a mean and variance difference between women and men. We also examined whether we could identify sex limitation (which would indicate that the magnitude of the genetic effect differs between the sexes or that different genes in men and women affect the expression of the phenotype), while including sex and age as covariates in the model. All twin-modeling assumptions were met, and no significant sex limitation (i.e., different influences on men and women) was found (except for brain size, for which variances were greater in men than women). Therefore, only one mean and one variance were estimated in the ACE model (and two variances were estimated for brain size) as well as two covariances (MZ twins vs. DZ twins), whereas a sex effect was modeled to account for differences in means. We performed the above analyses for all measures of brain differences along a male–female dimension, as well as brain size and the measure of behavioral differences along a male–female dimension.
Next, using a bivariate Cholesky decomposition model (including sex and age as covariates), we examined the influence of genetic and environmental influences on the covariance between individual differences along a male–female dimension for brain and behavior as well as brain size and behavioral differences along a male–female dimension. Because we found robust associations between brain differences along a male–female dimension with both brain size and height, we also examined these variables for a common genetic and environmental factor. Because of the excellent prediction of sex when using the vertex-wise measure, the moderate-to-strong correlation between the brain measures with one another, and the similar heritability results for the different brain measures, the brain measure derived from only the vertex-wise approach (trained on the UK Biobank data set) was used for this analysis.
Results
Obtaining a measure of brain differences along a male–female dimension
Using data from either the landmark or the vertex-wise approach, we trained the algorithm to predict sex on the basis of brain shape or structure, and predicted sex in an independent imaging sample, to derive a score for each individual reflecting the position of their brain shape or structure along a male–female dimension (Fig. 1). Both the landmark and vertex-wise approaches yielded scores that differed substantially (although with considerable overlap) between the sexes, as expected (see Figs. S3b–S3h in the Supplemental Material). Brain size also showed a comparable difference in female and male distributions (see Fig. S3a in the Supplemental Material). Although the landmark approach already scaled the brains to the same size, it is possible that brain shape covaries with brain size. If this is the case, then the brain measures based only on shape (i.e., brain size controlled) may still contain brain-size information. To derive a brain measure that is independent of brain size, we also used an allometric scaling approach to adjust for brain size (BSV) on a regional (vertex-wise) level. Specifically, we regressed out brain size for each vertex and used the residuals of the vertices in the prediction. This adjustment for brain size yielded a brain measure that showed more overlap between men and women than the vertex-wise measure in which only brain-size differences associated with sex were removed (see Fig. S3h), but the measure could still accurately discriminate between the sexes (d = 1.01, 95% confidence interval [CI] = [0.88, 1.14]; red lines in Fig. 2). We found similar results when regressing out brain size from the uncorrected prediction scores (orange lines in Fig. 2; see Fig. S3g).
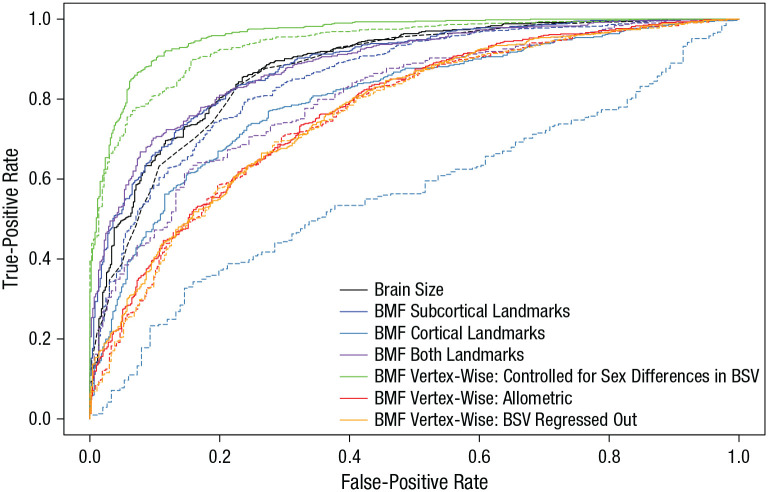
Fig. 2.
Receiver operating characteristic (ROC) curve (the true-positive rate as a function of the false-positive rate) predicting biological sex from the brain data in the Queensland Twin IMaging (QTIM) and Human Connectome Project (HCP) data sets. Results are shown for each of the two approaches (landmark and vertex-wise), as well as for brain size as a crude proxy for comparison. The landmark approach included the subcortical, cortical, and combined subcortical and cortical landmarks. The vertex-wise approach included scores controlled for brain size (brain-segmentation volume [BSV]) by regressing out brain-size differences associated with sex, applying allometric scaling, and regressing out brain size from the uncorrected scores. Predictions of sex in the QTIM data set are displayed with a dashed line, and predictions of sex in the HCP data set are displayed with a solid line.
Validity and reliability of the brain measures
Brain measures based on both approaches showed good to excellent test-retest reliability (see Table S1 in the Supplemental Material), defined as the correlation between the brain scores from the two time points. The validity (i.e., to what extent the measure could predict sex in an independent sample) was measured with the area under the curve (AUC)—defined by the true-positive rate against the false-positive rate—using the pROC package (Version 1.11 and above; Robin et al., 2011). The AUC is, unlike accuracy, insensitive to class imbalance. Both approaches predicted sex well (Fig. 2; see Table S1)—as reflected in the good to excellent AUC—and the prediction was often better when brain size was not filtered out (see Table S1).
Comparing the two approaches, we found that the landmark approach (including the subcortical landmark or both subcortical and cortical landmarks) resulted in a more accurate prediction than the vertex-wise approach after controlling for brain size (cf. rows B2 and B4 with row B5 in Table S1). However, when we trained the vertex-wise model on the large UK Biobank data set (N = 9,888), the prediction improved markedly (green lines in Fig. 2; see row A2 in Table S1). However, in both data sets, after we applied an allometric scaling approach to adjust for brain size (red lines in Fig. 2; see row A3 in Table S1), the prediction worsened and was no longer better than when using the landmark approach—results were similar when we regressed out brain size from the uncorrected vertex-wise scores (orange lines in Fig. 2).
Further, the prediction based only on cortical landmarks was not significantly better than chance when we predicted from the QTIM (n = 1,040) to the HCP (n = 1,113) data set and vice versa (Fig. 2; see row B3 in Table S1) or when we divided the QTIM data set in two halves (see row C3 in Table S1). In contrast, when we divided the HCP data set in two halves, the cortical landmarks were predictive of sex (see row D3 in Table S1). Because of the poor predictive power of the QTIM cortical landmarks in comparison with the HCP cortical landmarks, the prediction scores derived from the cortical landmarks for the QTIM data set were excluded from further analysis. The poor performance of the prediction based on the QTIM cortical landmarks may be explained by the poorer resolution and lower signal-to-noise ratio of the diffusion scans of the QTIM data set compared with the HCP data set, which may have led to more error in landmark placement. For all further analyses, outliers (z ± 3.29) were winsorized within each sex.
Correlations among brain measures
Brain measures derived from the landmark and vertex-wise approaches were associated with one another (Table 1) after we adjusted for sex, age, and scanning acquisition in the total sample. The vertex-wise scores for which brain size was regressed out of the uncorrected prediction scores showed high overlap with the allometric scores (r = .999, p < .001) and were therefore excluded from further analyses. As expected, brain measures were associated with brain size across samples (ps ≤ .001; Table 1), even after we adjusted the brain measures for brain size (by scaling brains to the same size or by regressing out BSV). This association raised the question of whether sex differences in brain size may still have been confounding the brain measures (i.e., the prediction of sex). To further examine this possibility, we used two subsamples in which female and male brains were matched for brain size (maximum of 10-ml difference in BSV; QTIM: n = 262; HCP: n = 372; for more details, see van Eijk et al., 2020). The association between brain measures and brain size remained in both subsamples in which men and women were matched for brain size (see Table S2 in the Supplemental Material). This finding shows that our prediction of sex (and resulting brain measures) was not driven by potential confounding sex differences in brain size and provides additional evidence for the scaling relationship between brain differences along a male–female dimension and brain size.
Table 1.
Correlation Matrix for the Measures of Brain Differences Along a Male–Female Dimension (BMF) Controlled for Covariates (Sex, Age, and Scan Acquisition), Displaying Correlations for the QTIM Data Set (Below the Diagonal) and for the HCP Data Set (Above the Diagonal)
| Measure | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1. BMF (subcortical landmarks) | — | .242** | .686** | .227** | .116** | .110** |
| 2. BMF (cortical landmarks) | .114* | — | .278** | .208** | −.097* | .322** |
| 3. BMF (subcortical + cortical landmarks) | .535** | .271** | — | .210** | .045 | .178** |
| 4. BMF (vertex-wise score controlled for sex differences in BSV) | .222** | .041 | .084 | — | .531** | .491** |
| 5. BMF (vertex-wise allometric score) | .072* | −.062 | .039 | .664** | — | −.452** |
| 6. Brain size | .173** | .149** | .065 | .370** | −.389** | — |
Note: Measures were derived from the landmark approach (subcortical, cortical, and both landmarks) and vertex-wise approach (a correction adjusting for sex differences in brain size and an allometric brain-size correction) as well as from brain size as a crude proxy for comparison. Brain-segmentation volume was used as a measure of brain size. Because the prediction of sex using brain measures derived from cortical landmarks in the Queensland Twin IMaging (QTIM) data set was no better than chance, these prediction scores were excluded from further analysis. HCP = Human Connectome Project.
p ≤ .05. **p ≤ .001.
Association between sex differences in brain and behavior
We tested for a link between sex differences in brain and behavior by computing a composite score of brain differences along a male–female dimension and testing its association with a score of behavioral differences along a male–female dimension. This followed a similar approach to that of Tunç et al. (2016). Further, we examined the associations between the brain scores with specific behavioral measures.
Association between brain and behavioral scores
Prediction of individuals’ sex on the basis of behavioral measures (see Table S3 in the Supplemental Material) yielded a good AUC (74.94%–78.89%). After we combined both samples and adjusted for sex, age, and a dummy variable for study (QTIM/HCP), the resulting behavioral score correlated significantly with the brain scores derived from the landmark and vertex-wise approaches (with the exception of the measure based only on cortical landmarks; Table 2). We also tested the same correlations within each sex—these tests have lower power (because of the split sample) but would reveal whether the brain–behavior association was markedly different in each sex (Table 2). Statistical significance was inconsistent across methods, but the point estimates were small and positive. For the vertex-wise measure, we found a significant correlation within both sexes: Effect sizes were similar to those found by Tunç et al. (2016; within women: r = .129, 95% CI = [.068, .188], p < .001; within men: r = .137, 95% CI = [.065, .207], p < .001). Note that brain size itself showed a stronger association with the behavioral score than any of the shape-based brain scores (r = .162, 95% CI = [.116, .207], p < .001). After we controlled for brain size, the association between brain and behavioral scores was no longer significant (Table 2), whereas when we adjusted for body size (height) instead of brain size, the association remained significant in the total sample and within men (although the effect became smaller; r = .066, 95% CI = [.018, .114], p = .007; women: r = .034, 95% CI = [−.029, .096], p = .294; men: r = .105, 95% CI = [.031, .178], p = .006).
Table 2.
Association Between Measures of Brain and Behavioral Differences Along a Male–Female Dimension, Adjusted for Sex, Age, and Study and for Sex, Age, Study, and Brain Size
| Measure | Total sample | Within women | Within men | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | 95% CI | t | df | p | r | 95% CI | t | df | p | r | 95% CI | t | df | p | |
| Adjusted for sex, age, and study | |||||||||||||||
| Brain size | .162 | [0.116, 0.207] | 6.869 | 1758 | < .001 | .145 | [0.085, 0.204] | 4.718 | 1033 | < .001 | .181 | [0.109, 0.250] | 4.943 | 723 | < .001 |
| Subcortical landmarks | .060 | [0.013, 0.106] | 2.510 | 1758 | .012 | .014 | [−0.047, 0.075] | 0.453 | 1033 | .651 | .119 | [0.046, 0.190] | 3.218 | 723 | .001 |
| Cortical landmarks a | .044 | [−0.019, 0.107] | 1.361 | 953 | .174 | .017 | [−0.070, 0.104] | 0.383 | 506 | .702 | .077 | [−0.016, 0.168] | 1.6020 | 445 | .106 |
| Both subcortical and cortical landmarks a | .073 | [0.009, 0.135] | 2.247 | 953 | .025 | .044 | [−0.043, 0.131] | 0.993 | 506 | .321 | .108 | [0.016, 0.199] | 2.300 | 445 | .022 |
| Vertex-wise score controlled for sex differences in BSV | .132 | [0.090, 0.178] | 5.580 | 1758 | < .001 | .129 | [0.068, 0.188] | 4.165 | 1033 | < .001 | .137 | [0.065, 0.207] | 3.711 | 723 | < .001 |
| Adjusted for sex, age, study, and brain size | |||||||||||||||
| Subcortical landmarks b | .040 | [−0.007, 0.086] | 1.652 | 1758 | .100 | −.005 | [−0.066, 0.056] | −0.159 | 1033 | .873 | .097 | [0.024, 0.169] | 2.618 | 723 | .009 |
| Cortical landmarksa,b | −.017 | [−0.081, 0.046] | −0.549 | 953 | .583 | −.027 | [−0.114, 0.060] | −0.605 | 506 | .545 | −.001 | [−0.094, 0.092] | −0.020 | 445 | .984 |
| Both subcortical and cortical landmarksa,b | .037 | [−0.026, 0.1000] | 1.144 | 953 | .253 | .022 | [−0.065, 0.109] | 0.500 | 506 | .618 | .057 | [−0.036, 0.149] | 1.200 | 445 | .231 |
| Vertex-wise allometric score c | −.004 | [−0.051, 0.043] | −0.165 | 1736 | .869 | .009 | [−0.052, 0.070] | 0.288 | 1023 | .773 | −.020 | [−0.094, 0.053] | −0.544 | 711 | .586 |
Note: Brain-segmentation volume (BSV) was used as a measure of brain size. r = Pearson correlation; CI = confidence interval. aThese values were based on the Human Connectome Project (HCP) cohort only, including 972 of 1,113 individuals of the HCP cohort because diffusion data were not available for 89 participants with a behavioral score. bBSV was regressed out of the brain scores before the correlation with the behavioral score was tested. cBSV was regressed out of the brain data before a brain score was derived on a (vertex) regional level using an allometric scaling approach.
Associations of brain scores with physical and behavioral measures
Next, we aimed to gain more insight into whether and how the brain scores were associated with specific physical and behavioral measures. We also examined associations within each sex, under the hypothesis that we would find a similar correlation within each sex. Because we found an association between the brain scores and brain size (Table 1), brain size may possibly confound the correlations between the brain scores and behavioral traits, which is why we adjusted correlations for brain size as well as sex and age.
The brain scores showed only very weak associations with physical and behavioral measures regardless of the approach used (see Tables S6–S14 in the Supplemental Material) and not always in the direction of the sex effect found for these measures (see Table S5 in the Supplemental Material). One association that remained across samples and across the different brain measures (except the allometric approach) was the association between the brain scores and height (r = .064–.203; see Tables S6–S14). However, no association showed a trend (p ≤ .05) in both the total sample and within-sex analyses and was consistent across the different brain measures (see Tables S6–S14). As a comparison, brain size showed more and stronger associations with behavioral measures (rs = .059–.243, ps ≤ .05; see Tables S17 and S18 in the Supplemental Material) than did any of the brain scores (rs = .059 to −.207, ps ≤ .05; see Tables S6–S14). After we adjusted the brain scores for body size (height) instead of brain size (see Tables S12 and S13), several associations remained for the brain measures with physical and behavioral measures (see Tables S15 and S16 in the Supplemental Material)—suggesting that the associations were driven by brain size more so than body size.
Genetic analyses
In both data sets and all brain measures, intraclass correlation in MZ twin pairs was greater than in DZ twin pairs or nontwin siblings, which suggests the influence of genetic effects (see Tables S19–S21 in the Supplemental Material). Consistent with previous work (Rentería et al., 2014), results showed that brain size was highly heritable (see Table S22 in the Supplemental Material): 86% to 92% of the variation in brain size could be explained by genetic influences (A), 0% to 7% by shared environmental influences (C), and 7% to 8% by residual effects (E), which include idiosyncratic environmental factors and measurement error. In contrast, the other brain measures showed more modest heritability: Depending on the measure used, 33% to 50% of the variance could be explained by genetic influences, 0% to 10% by shared environmental influences, and 40% to 67% by residual effects (see Table S22). The behavioral measure was also moderately heritable: 32% to 51% of the variation in the behavioral measure could be explained by genetic influences, 0% to 6% by shared environmental influences, and 43% to 68% by residual effects (see Table S22). Results were similar when we excluded opposite-sex twin or sibling pairs (see Table S23 in the Supplemental Material).
Next, we examined the extent to which common genetic, shared environmental, or residual factors underlie the association of brain and behavioral differences along a male–female dimension, of brain differences along a male–female dimension with brain size and height, and of brain size with height. Because there was no evidence for shared environmental influence, we used a bivariate model with an AE model. To improve the power of our analyses, we combined the two samples. Our analyses showed a genetic correlation (rg) between brain and behavioral measures (combined rg = .296; within women: rg = .220; within men: rg = .409) when we derived the brain measure from the vertex-wise scores (for which brain-size differences associated with sex were removed). However, this correlation was no longer significant when we used the vertex-wise allometric scores, suggesting that brain size may be driving this correlation. In line with this possibility, we found a similar genetic correlation between brain size and the behavioral measure (combined rg = .261; within women: rg = .208; within men: rg = .333), and we found a genetic correlation between brain size and the brain measure (combined rg = .566; within women: rg = .526; within men: rg = .602) when we derived the brain score from the vertex-wise scores (for which brain-size differences associated with sex were removed) and also when we used the allometric approach (removing all brain-size differences), although the association became negative (combined rg = −.571; within women: rg = −.640; within men: rg = −.455).
Further, we found a genetic correlation between the brain measures and height (combined rg = .162; within women: rg = .128; within men: rg = .145) but only for the vertex-wise scores (from which brain-size differences associated with sex were removed) and not for the vertex-wise allometric scores. In comparison, brain size showed a similar genetic correlation with height (combined rg = .195; within women: rg = .147; within men: rg = .205), as found for the other brain measures.
Discussion
We investigated whether sex differences in brain structure are associated with sex differences in behavior within sex, thereby circumventing the confound of different socialization of women and men. We obtained a data-driven measure of brain differences along a male–female dimension (derived from sex differences in brain shape and structure) and behavioral differences along a male–female dimension (derived from sex differences in behavior) while adjusting brain measures for brain size using an allometric scaling approach. Our key finding is that there is a small positive association between sex differences in brain and behavior, but that association disappears when we take into account differences in brain size.
Previous research (Phillips et al., 2019; Tunç et al., 2016) showed some (mixed) evidence of an association between brain and behavioral differences along a male–female dimension. However, our research used two independent samples (total sample size more than double that in the studies by Tunç et al., 2016, and Phillips et al., 2019) and two different methods for deriving the brain measures, and we carefully considered whether brain size may drive the brain–behavior association. It is possible that brain size could drive the association between brain and behavior found previously because the previous studies either did not adjust for brain size (Tunç et al., 2016) or adjusted only some brain measures for brain size (Phillips et al, 2019), and neither applied an allometric approach to consider that different brain regions scale differently from brain size. As a consequence, their brain data could still contain shape differences that are associated with the original size differences, and their score reflecting brain differences along a male–female dimension could be driven by these size differences.
Our findings are consistent with this possibility. First, we showed that the brain measures were substantially correlated with brain size in the total sample and within each sex, even though all brains were scaled to the same size from the start. This is consistent with the concept of allometry, that is, that a structure’s shape is not independent of its size. Larger brains tend to have a different shape from those of smaller brains, for example, showing more folding on average. Second, we found an association between individual differences in brain and behavior similar to that previously reported by Tunç et al. (2016), but after we applied an allometric approach, adjusting for brain size on a regional (vertex-by-vertex) level, the correlation between brain and behavior disappeared. Our results suggest that any previous findings of a relationship between sex differences in brain structure and behavior may have been driven by brain size.
The brain measures were associated with both physical and behavioral variables (although possibly driven by brain-size differences), which implies that brain and behavioral sex differences may be subject to the same underlying processes of masculinization without being directly causally related. This possibility is strengthened by the correlation of the brain measures with height because there is no obvious reason to suspect that brain differences along a male–female dimension and height are causally related. Further, although it is well established that brain size is functionally relevant—for example, it is correlated at around .24 to .33 with IQ after adjustment for differences in body size (Goriounova & Mansvelder, 2019)—its relation to the nexus of brain and behavioral sex differences is less clear. In the HPC sample (which also formed part of our sample), van der Linden et al. (2017) found that brain size partially mediated the small sex difference in IQ—but many studies have found a negligible sex difference in IQ—and when van der Linden et al. (2017) used male and female samples matched on IQ, men still had larger brains. This finding raises the possibility that there are sex differences in brain structure that compensate for size differences between the sexes. On the one hand, it could be that some sex differences in brain structure are compensatory and make female and male behavior more similar despite different average brain sizes. On the other hand, other sex differences in brain structure may result in adaptive behavioral sex differences and, because of joint hormonal mediation, also covary with brain size. We are not able to resolve these complexities here. Also, the weakness of the associations suggests that sex differences in brain structure are among many other factors related to sex differences in behavior.
We also estimated the heritability of brain and behavioral differences along a male–female dimension. Using twin modeling, we estimated that variance in the brain and behavioral measures can be attributed in roughly similar proportions to genetic (32%–50%) and unshared environmental (40%–68%) influences. Phillips et al. (2019) estimated the heritability of sex-differentiated brain structure at 0% to 1.5% using single-nucleotide polymorphism (SNP) data. SNP heritability estimates are extremely imprecise in samples of that size (N = 900), and in any case, SNPs typically do not capture most of the total heritability of complex traits (Wainschtein et al., 2021). Twin studies such as ours estimate a trait’s total heritability. As for behavior, our heritability estimates were in line with those of a previous twin study using a different method with different data (Verweij et al., 2016).
This project has some limitations. Most importantly, our research does not imply that no association could exist between behavior and sex differences in regional (as opposed to global) brain structure, microstructure, or brain function, all of which our study is silent on. Second, the range of sexually dimorphic behaviors that we analyzed was limited by the measures that happened to have been collected in the QTIM and HCP studies, and they may not be the most sensitive to detect sex differences in behavior compared with more sexually differentiated behavioral traits. However, our prediction performance was similar to that previously reported by Tunç et al. (2016). In addition, several behavioral measures in the QTIM data set were obtained at a different time from when the imaging scans were acquired. Further, it is unclear to what degree the sex differences from which our measures are derived are influenced by genetic factors (e.g., number of X chromosomes, the presence of a Y chromosome, and mitochondrial DNA inheritance; Pearse & Young-Pearse, 2019) as well as sex hormone levels. In addition, despite our efforts to remove the confound of socialization between women and men by looking at within-sex differences, our measures may capture environmental differences among women and among men beyond those based on biology.
Future research with even larger samples and richer brain and behavioral measures, as well as a longitudinal study design, will further elucidate the biological and social influences on brain and behavioral sex differences. Such an approach will help to answer questions such as at what stage (or stages) across the life span sex hormones play the most prominent role in influencing brain and behavior and whether specific sex hormones have distinct influences on brain and behavior. It will also provide insights into the directionality of the association between sex differences in brain and behavior and shed light on the distinction between biological sex and gender differences.
Supplemental Material
Supplemental material, sj-docx-1-pss-10.1177_0956797621996664 for Are Sex Differences in Human Brain Structure Associated With Sex Differences in Behavior? by Liza van Eijk, Dajiang Zhu, Baptiste Couvy-Duchesne, Lachlan T. Strike, Anthony J. Lee, Narelle K. Hansell, Paul M. Thompson, Greig I. de Zubicaray, Katie L. McMahon, Margaret J. Wright and Brendan P. Zietsch in Psychological Science
Acknowledgments
We thank the participants, Marlene Grace and Ann Eldridge for twin recruitment, the radiographers for scanning, and Kerrie McAloney and Daniel Park for research support. We thank Bill von Hippel for his thoughtful comments.
Footnotes
ORCID iDs: Liza van Eijk  https://orcid.org/0000-0001-8254-1019
https://orcid.org/0000-0001-8254-1019
Anthony J. Lee  https://orcid.org/0000-0001-8466-2594
https://orcid.org/0000-0001-8466-2594
Greig I. de Zubicaray  https://orcid.org/0000-0003-4506-0579
https://orcid.org/0000-0003-4506-0579
Katie L. McMahon  https://orcid.org/0000-0002-6357-615X
https://orcid.org/0000-0002-6357-615X
Supplemental Material: Additional supporting information can be found at http://journals.sagepub.com/doi/suppl/10.1177/0956797621996664
Transparency
Action Editor: Steven W. Gangestad
Editor: D. Stephen Lindsay
Author Contributions
B. P. Zietsch developed the study concept. L. van Eijk and B. P. Zietsch contributed to the study design. M. J. Wright, K. L. McMahon, G. I. de Zubicaray, and P. M. Thompson designed the Queensland Twin IMaging study. L. van Eijk, D. Zhu, B. Couvy-Duchesne, L. T. Strike, A. J. Lee, N. K. Hansell, and K. L. McMahon analyzed the data. L. van Eijk and B. P. Zietsch drafted the manuscript and interpreted the results. All the authors revised the manuscript and approved the final manuscript for submission.
Declaration of Conflicting Interests: The author(s) declared that there were no conflicts of interest with respect to the authorship or the publication of this article.
Funding: The Queensland Twin IMaging study was funded by the National Institutes of Health (NIH; Project HD050735; Award No. U54 EB020403, Subaward No. 56929223) and National Health and Medical Research Council (1009064, 496682). Data from the Human Connectome Project were provided in part by the HCP, WU-Minn Consortium (principal investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657), funded by the 16 NIH institutes and centers that support the NIH Blueprint for Neuroscience Research, and by the McDonnell Center for Systems Neuroscience at Washington University. L. van Eijk was supported in part by the Australian Government Research Training Program Scholarship and the Imaging Genomics Laboratory, Queensland Brain Institute, as well as by the Hendrik Muller Vaderlandsch Fund, the Vrijvrouwe van Renswoude Fund, and the Prince Bernhard Culture Fund. P. M. Thompson was also supported in part by NIH (U54 EB20403, R01MH116147, R56AG058854, and P41 EB015922). B. P. Zietsch was also supported in part by ZonMw Grant 849200011 from The Netherlands Organisation for Health Research and Development.
Open Practices: The Queensland Twin IMaging study data set used in this study has been made publicly available via UQ eSpace and can be accessed at https://doi.org/10.14264/38dde15. Researchers can request access to the Human Connectome Project data set through the Human Connectome Project team (see https://www.humanconnectome.org/contact-us). The design and analysis plan for this study were not preregistered. This article has received the badge for Open Data. More information about the Open Practices badges can be found at http://www.psychologicalscience.org/publications/badges.

References
- Adams D. C., Otarola-Castillo E. (2013). geomorph: an R package for the collection and analysis of geometric morphometric shape data. Methods in Ecology and Evolution, 4(4), 393–399. 10.1111/2041-210X.12035 [DOI] [Google Scholar]
- Anderson N. E., Harenski K. A., Harenski C. L., Koenigs M. R., Decety J., Calhoun V. D., Kiehl K. A. (2019). Machine learning of brain gray matter differentiates sex in a large forensic sample. Human Brain Mapping, 40(5), 1496–1506. 10.1002/hbm.24462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer J. (2019). The reality and evolutionary significance of human psychological sex differences. Biological Reviews, 94, 1381–1415. 10.1111/brv.12507 [DOI] [PubMed] [Google Scholar]
- Boker S., Neale M., Maes H., Wilde M., Spiegel M., Brick T., Spies J., Estabrook R., Kenny S., Bates T., Mehta P., Fox J. (2011). OpenMx: An open source extended structural equation modeling framework. Psychometrika, 76(2), 306–317. 10.1007/s11336-010-9200-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner E., de la Cuétara J. M., Colom R., Martin-Loeches M. (2012). Gender-based differences in the shape of the human corpus callosum are associated with allometric variations. Journal of Anatomy, 220(4), 417–421. 10.1111/j.1469-7580.2012.01476.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekroud A. M., Ward E. J., Rosenberg M. D., Holmes A. J. (2016). Patterns in the human brain mosaic discriminate males from females. Proceedings of the National Academy of Sciences, USA, 113(14), Article E1968. 10.1073/pnas.1523888113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvy-Duchesne B., Strike L. T., Zhang F., Holtz Y., Zheng Z., Kemper K. E., Yengo L., Colliot O., Wright M. J., Wray N. R., Yang J., Visscher P. M. (2020). A unified framework for association and prediction from vertex-wise grey-matter structure. Human Brain Mapping, 41(14), 4062–4076. 10.1002/hbm.25109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong L. W., Vidal J.-S., Forsberg L. E., Zijdenbos A. P., Haight T., Alzheimer’s Disease Neuroimaging Initiative, Sigurdsson S., Gudnason V., van Buchem M. A., Launer L. J. (2017). Allometric scaling of brain regions to intra-cranial volume: An epidemiological MRI study. Human Brain Mapping, 38(1), 151–164. 10.1002/hbm.23351 [DOI] [PMC free article] [PubMed]
- Del Giudice M., Lippa R. A., Puts D. A., Bailey D. H., Bailey J. M., Schmitt D. P. (2016). Joel et al.’s method systematically fails to detect large, consistent sex differences. Proceedings of the National Academy of Sciences, USA, 113(14), Article E1965. 10.1073/pnas.1525534113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries G. J. (2004). Minireview: Sex differences in adult and developing brains: Compensation, compensation, compensation. Endocrinology, 145(3), 1063–1068. 10.1210/en.2003-1504 [DOI] [PubMed] [Google Scholar]
- Dryden I. L. (2016). shapes: Statistical shape analysis. https://CRAN.R-project.org/package=shapes
- Eagly A. H., Wood W. (2013). The nature–nurture debates: 25 years of challenges in understanding the psychology of gender. Perspectives on Psychological Science, 8(3), 340–357. 10.1177/1745691613484767 [DOI] [PubMed] [Google Scholar]
- Fischl B. (2012). FreeSurfer. NeuroImage, 62(2), 774–781. 10.1016/j.neuroimage.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K. J., Holmes A. P., Poline J. B., Grasby P. J., Williams S. C. R., Frackowiak R. S. J., Turner R. (1995). Analysis of fMRI time-series revisited. NeuroImage, 2(1), 45–53. 10.1006/nimg.1995.1007 [DOI] [PubMed] [Google Scholar]
- Gillespie N. A., Henders A. K., Davenport T. A., Hermens D. F., Wright M. J., Martin N. G., Hickie I. B. (2013). The Brisbane Longitudinal Twin Study: Pathways to Cannabis Use, Abuse, and Dependence project—current status, preliminary results, and future directions. Twin Research and Human Genetics, 16(1), 21–33. 10.1017/thg.2012.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser M. F., Sotiropoulos S. N., Wilson J. A., Coalson T. S., Fischl B., Andersson J. L., Xu J., Jbabdi S., Webster M., Polimeni J. R., Van Essen D. C., Jenkinson M., & WU-Minn HCP Consortium. (2013). The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage, 80, 105–124. 10.1016/j.neuroimage.2013.04.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriounova N. A., Mansvelder H. D. (2019). Genes, cells and brain areas of intelligence. Frontiers in Human Neuroscience, 13, Article 44. 10.3389/fnhum.2019.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur R. E., Gur R. C. (2016). Sex differences in brain and behavior in adolescence: Findings from the Philadelphia Neurodevelopmental Cohort. Neuroscience & Biobehavioral Reviews, 70, 159–170. 10.1016/j.neubiorev.2016.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N., Aganj I., Lenglet C., Joshi A., Jin Y., Barysheva M., McMahon K. L., de Zubicaray G. I., Martin N. G., Wright M. J., Toga A. W., Sapiro G., Thompson P. M. (2011, March 30–April 2). Sex differences in the human connectome: 4-tesla high angular resolution diffusion imaging (HARDI) tractography in 234 young adult twins [Paper presentation]. 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, United States. [Google Scholar]
- Jenkinson M., Beckmann C. F., Behrens T. E. J., Woolrich M. W., Smith S. M. (2012). FSL. NeuroImage, 62(2), 782–790. 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Joel D., Persico A., Salhov M., Berman Z., Oligschläger S., Meilijson I., Averbuch A. (2018). Analysis of human brain structure reveals that the brain “types” typical of males are also typical of females, and vice versa. Frontiers in Human Neuroscience, 12, Article 399. 10.3389/fnhum.2018.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Kim N., Kim S., Hong S., Park K., Lim S., Park J.-M., Na B., Chae Y., Lee J., Yeo S., Choe I.-H., Cho S.-Y., Cho G. (2012). Sex differences in amygdala subregions: Evidence from subregional shape analysis. NeuroImage, 60(4), 2054–2061. 10.1016/j.neuroimage.2012.02.025 [DOI] [PubMed] [Google Scholar]
- Lotze M., Domin M., Gerlach F. H., Gaser C., Lueders E., Schmidt C. O., Neumann N. (2019). Novel findings from 2,838 adult brains on sex differences in gray matter brain volume. Scientific Reports, 9(1), Article 1671. 10.1038/s41598-018-38239-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. L., Alfaro-Almagro F., Bangerter N. K., Thomas D. L., Yacoub E., Xu J., Bartsch A. J., Jbabdi S., Sotiropoulos S. N., Andersson J. L. R., Griffanti L., Douaud G., Okell T. W., Weale P., Dragonu I., Garratt S., Hudson S., Collins R., Jenkinson M., . . . Smith S. M. (2016). Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nature Neuroscience, 19(11), 1523–1536. 10.1038/nn.4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J., Xie L., Li X., Zhu D., Terry D. P., Puente A. N., Jiang R., Chen Y., Wang L., Shen D., Zhang J., Miller L. S., Liu T. (2015). Atomic connectomics signatures for characterization and differentiation of mild cognitive impairment. Brain Imaging and Behavior, 9(4), 663–677. 10.1007/s11682-014-9320-1 [DOI] [PubMed] [Google Scholar]
- Pearse R. V., Young-Pearse T. L. (2019). Lost in translational biology: Understanding sex differences to inform studies of diseases of the nervous system. Brain Research, 1722, Article 146352. 10.1016/j.brainres.2019.146352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips O. R., Onopa A. K., Hsu V., Ollila H. M., Hillary R. P., Hallmayer J., Gotlib I. H., Taylor J., Mackey L., Singh M. K. (2019). Beyond a binary classification of sex: An examination of brain sex differentiation, psychopathology, and genotype. Journal of the American Academy of Child & Adolescent Psychiatry, 58(8), 787–798. 10.1016/j.jaac.2018.09.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2018). R: A language and environment for statistical computing (Version 3.4.4) [Computer software]. http://www.r-project.org/
- Rentería M. E., Hansell N. K., Strike L. T., McMahon K. L., de Zubicaray G. I., Hickie I. B., Thompson P. M., Martin N. G., Medland S. E., Wright M. J. (2014). Genetic architecture of subcortical brain regions: Common and region-specific genetic contributions. Genes, Brain, and Behavior, 13(8), 821–830. 10.1111/gbb.12177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie S. J., Cox S. R., Shen X., Lombardo M. V., Reus L. M., Alloza C., Harris M. A., Alderson H. L., Hunter S., Neilson E., Liewald D. C. M., Auyeung B., Whalley H. C., Lawrie S. M., Gale C. R., Bastin M. E., McIntosh A. M., Deary I. J. (2018). Sex differences in the adult human brain: Evidence from 5216 UK Biobank participants. Cerebral Cortex, 28(8), 2959–2975. 10.1093/cercor/bhy109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M. (2011). pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics, 12, Article 77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G. K. (1991). That BLUP is a good thing: The estimation of random effects. Statistical Science, 6(1), 15–32. 10.1214/ss/1177011926 [DOI] [Google Scholar]
- Ruigrok A. N., Salimi-Khorshidi G., Lai M.-C., Baron-Cohen S., Lombardo M. V., Tait R. J., Suckling J. (2014). A meta-analysis of sex differences in human brain structure. Neuroscience & Biobehavioral Reviews, 39, 34–50. 10.1016/j.neubiorev.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier J.-D., Smith R. E., Raffelt D., Tabbara R., Dhollander T., Pietsch M., Christiaens D., Jeurissen B., Yeh C.-H., Connelly A. (2019). MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage, 202, Article 116137. 10.1016/j.neuroimage.2019.116137 [DOI] [PubMed] [Google Scholar]
- Toussaint N., Souplet J. C., Fillard P. (2007). MedINRIA: Medical image navigation and research tool by INRIA. In Proceedings of MICCAI’07 Workshop on interaction in medical image analysis and visualization. The Medical Image Computing and Computer Assisted Intervention Society. [Google Scholar]
- Tunç B., Solmaz B., Parker D., Satterthwaite T. D., Elliott M. A., Calkins M. E., Ruparel K., Gur R. E., Gur R. C., Verma R. (2016). Establishing a link between sex-related differences in the structural connectome and behaviour. Philosophical Transactions of the Royal Society B: Biological Sciences, 371(1688), Article 20150111. 10.1098/rstb.2015.0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden D., Dunkel C. S., Madison G. (2017). Sex differences in brain size and general intelligence (g). Intelligence, 63, 78–88. 10.1016/j.intell.2017.04.007 [DOI] [Google Scholar]
- van Eijk L., Hansell N. K., Strike L. T., Couvy-Duchesne B., de Zubicaray G. I., Thompson P. M., McMahon K. L., Zietsch B. P., Wright M. J. (2020). Region-specific sex differences in the hippocampus. NeuroImage, 215, Article 116781. 10.1016/j.neuroimage.2020.116781 [DOI] [PubMed] [Google Scholar]
- Van Essen D. C., Ugurbil K., Auerbach E., Barch D., Behrens T. E. J., Bucholz R., Chang A., Chen L., Corbetta M., Curtiss S. W., Della Penna S., Feinberg D., Glasser M. F., Harel N., Heath A. C., Larson-Prior L., Marcus D., Michalareas G., Moeller S., . . . WU-Minn HCP Consortium. (2012). The Human Connectome Project: A data acquisition perspective. NeuroImage, 62(4), 2222–2231. 10.1016/j.neuroimage.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W. N., Ripley B. D. (2002). Modern applied statistics with S (4th ed). Springer. [Google Scholar]
- Verweij K. J., Mosing M. A., Ullen F., Madison G. (2016). Individual differences in personality masculinity-femininity: Examining the effects of genes, environment, and prenatal hormone transfer. Twin Research and Human Genetics, 19(2), 87–96. 10.1017/thg.2016.8 [DOI] [PubMed] [Google Scholar]
- Wainschtein P., Jain D. P., Yengo L., Zheng Z., Cupples L. A., Shadyab A. H., McKnight B., Shoemaker B. M., Mitchell B. D., Psaty B. M., Kooperberg C., Roden D., Darbar D., Arnett D. K., Regan E. A., Boerwinkle E., Rotter J. I., Allison M. A., McDonald M.-L. N., . . . Visscher P. M. (2021). Recovery of trait heritability from whole genome sequence data. bioRxiv. 10.1101/588020 [DOI]
- Wright M. J., Martin N. G. (2004). Brisbane Adolescent Twin Study: Outline of study methods and research projects. Australian Journal of Psychology, 56(2), 65–78. 10.1080/00049530410001734865 [DOI] [Google Scholar]
- Xin J., Zhang Y., Tang Y., Yang Y. (2019). Brain differences between men and women: Evidence from deep learning. Frontiers in Neuroscience, 13, Article 185. 10.3389/fnins.2019.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Chen W., Zhu Z., Zhang Q., Nabais M. F., Qi T., Deary I. J., Wray N. R., Visscher P. M., McRae A. F., Yang J. (2019). OSCA: A tool for omic-data-based complex trait analysis. Genome Biology, 20(1), Article 107. 10.1186/s13059-019-1718-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Li K., Guo L., Jiang X., Zhang T., Zhang D., Chen H., Deng F., Faraco C., Jin C., Wee C.-Y., Yuan Y., Lv P., Yin Y., Hu X., Duan L., Hu X., Han J., Wang L., . . . Liu T. (2013). DICCCOL: Dense individualized and common connectivity-based cortical landmarks. Cerebral Cortex, 23(4), 786–800. 10.1093/cercor/bhs072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Zhan L., Faskowitz J., Daianu M., Jahanshad N., de Zubicaray G. I., McMahon K. L., Martin N. G., Wright M. J., Thompson P. M. (2015). Genetic analysis of structural brain connectivity using DICCCOL models of diffusion MRI in 522 twins. In Proceedings of the IEEE 12th International Symposium on Biomedical Imaging (pp. 1167–1171). 10.1109/ISBI.2015.7164080 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-pss-10.1177_0956797621996664 for Are Sex Differences in Human Brain Structure Associated With Sex Differences in Behavior? by Liza van Eijk, Dajiang Zhu, Baptiste Couvy-Duchesne, Lachlan T. Strike, Anthony J. Lee, Narelle K. Hansell, Paul M. Thompson, Greig I. de Zubicaray, Katie L. McMahon, Margaret J. Wright and Brendan P. Zietsch in Psychological Science