ABSTRACT
Chemotherapy is currently the main treatment for unresectable or advanced postoperative gastric cancers. However, its efficacy is negatively affected by the occurrence of chemoresistance, which severely affects patient prognosis. Recently, dysregulation in autophagy has been suggested as a potential mechanism for chemoresistence, and long noncoding RNA (lncRNA) also shows its regulatory role in cancer drug resistance. Using RNA sequencing, we found that lncRNA EIF3J-DT was highly expressed in drug-resistant gastric cancer cells. In-vitro and in-vivo experiments showed that EIF3J-DT activated autophagy and induced drug resistance in gastric cancer cells by targeting ATG14. Bioinformatics and experimental results showed that EIF3J-DT regulated the expression of ATG14 through direct binding to enhance stabilization of ATG14 mRNA and via blocking the degradation of ATG14 mRNA through competitively binding with microRNA (miRNA) MIR188-3p. Therefore, EIF3J-DT increased the expression of ATG14, contributing to activation of autophagy and chemoresistance. Furthermore, it was confirmed that EIF3J-DT and ATG14 were highly expressed in gastric cancer patients resistant to chemotherapy, and this was closely associated with patient prognosis. In conclusion, EIF3J-DT is involved in the regulation of autophagy and chemoresistance in gastric cancer cells by targeting ATG14. It may be a suitable new target for enhancing chemosensitivity and improving prognosis.
Abbreviations: 3-MA: 3-methyladenine; 5-Fu: 5-fluorouracil; ATG: autophagy related; C-CASP3: cleaved caspase 3; C-CASP7: cleaved caspase 7; C-PARP: cleaved PARP; CQ: chloroquine; CR: complete response; DIG: digoxigenin; ESR1: estrogen receptor 1; FBS: fetal bovine serum; FISH: fluorescence in situ hybridization; IHC: immunohistochemistry; ISH: in situ hybridization; lncRNA: long noncoding RNA; miRNA: microRNA; MUT: mutant; NC: negative control; OXA: oxaliplatin; PBS: phosphate-buffered saline; PD: progressive disease; PFA: paraformaldehyde; PR: partial response; qPCR: quantitative polymerase chain reaction; RAPA: rapamycin; SD: stable disease; TEM: transmission electron microscopy; WT: wild type.
KEYWORDS: ATG14, autophagy, chemotherapy resistance, EIF3J-DT; gastric cancer
Introduction
Gastric cancer is a serious threat to public health with a 21% increase in incidence and a 1.4% increase in mortality compared to 2014. Its incidence is highest in East Asia, and it ranks the third in China, but occurs worldwide [1,2]. Compared to surgery and targeted therapy, chemotherapy remains the main treatment for postoperative or advanced gastric cancer, reducing postoperative recurrence, prolonging survival and improving quality of life [3]. However, the rise of chemotherapy resistance greatly restricts the therapeutic effect and might lead to the treatment failure in gastric cancer [4]. Therefore, there is a pressing need to study the specific mechanisms underlying drug resistance and find solutions in clinical settings.
Autophagy is a process in which cells protect themselves in an adverse environment, such as during nutrient deficiency, hypoxia, and drug toxicity [5,6], and is closely related to the occurrence of tumor cell drug resistance. Reports have shown that autophagy mediates drug resistance in tumor cells mainly by recycling energy via eliminating damaged organelles and maintaining homeostasis through the degradation of hazardous substances [6]. Mechanistically, autophagy can regulate drug resistance via affecting autophagy genes, microRNAs (miRNAs), or the PI3K-AKT-MTOR, TP53, and MAPK signaling pathways [7,8]. For example, inhibition of autophagy genes, such as ATG5 (autophagy related 5), ATG7, and BECN1, results in resensitization of tamoxifen-resistant breast cancer cells [9,10]. He et al. also clarify that the downregulation of ATG14 can sensitize ovarian cancer cells to cisplatin treatment [11]. However, the regulatory mechanism involving autophagy genes in gastric cancer still needs further study.
Long noncoding RNAs (lncRNAs) (> 200 bp) are emerging as key regulators of multiple essential biological behaviors in tumors and are involved in tumor physiology and pathology via regulating the expression of various genes [12], important to proliferation [13] and metastasis [14]. Recently, growing evidence has revealed that lncRNAs are also responsible for drug resistance, which is a major obstacle to effective cancer treatment. For example, the upregulated lncRNA NEAT1 can induce cisplatin and taxol resistance in triple-negative breast cancer [15]. Huang et al. report that lncRNA NR2F1-AS1 is upregulated in oxaliplatin (OXA)-resistant hepatocellular carcinoma and regulates OXA resistance by targeting ABCC1 [16]. In addition, recent studies have revealed the main regulatory mechanisms of lncRNAs participating in tumor resistance, including the cell cycle, apoptosis, drug efflux system, epithelial-mesenchymal transition, cancer stem cells, and resistance-related signaling pathways [17]. These findings indicate that targeting lncRNAs for cancer intervention has been a novel and promising treatment.
In this study, we investigated lncRNA EIF3J-DT, which was highly expressed in resistant gastric cancer cells. Through various in-vitro and in-vivo assays, we confirmed that EIF3J-DT was a drug resistance-related gene that upregulated the expression of ATG14 in gastric cancer cells resistant to OXA and 5-fluorouracil (5-Fu). EIF3J-DT stabilized ATG14 mRNA and alleviated its degradation via sequestering MIR188-3p, leading to autophagy activation and chemotherapy resistance. These findings may provide a clinical treatment target for reversing chemotherapy resistance.
Results
Chemotherapy activated autophagy in gastric cancer cells
First, we investigated the relationship between chemotherapy and autophagy. The gastric adenocarcinoma cell line, MGC803, was treated with OXA and 5-Fu. Western blotting was then performed at different time points (0, 3, 6, 9, 12, 15, 18, 21, and 24 h), and the expression levels of LC3-I, LC3-II, and SQSTM1 were detected to evaluate the extent of autophagy. When autophagy occurs, LC3-I transforms to LC3-II expressed on autophagosomes, and SQSTM1 is degraded to form complexes that are then degraded in lysosomes [18]. Therefore, increased LC3-II:LC3-I and decreased SQSTM1 indicate the activation of autophagy. The western blot analysis showed that LC3-II:LC3-I was increased while SQSTM1 was decreased in MGC803 cells treated with OXA and 5-Fu over time (Figures 1A and 1B). Next, we treated cells with OXA or 5-Fu combined with chloroquine (CQ), a classic autophagy inhibitor, to inhibit autophagic flux by decreasing the fusion of autophagosomes and lysosomes [19], leading to the accumulation of LC3-II. The results showed that with the combined treatment of CQ, activated autophagy in MGC803 cells following OXA and 5-Fu treatment was inhibited (Figures 1C and 1D).
Figure 1.

OXA and 5-Fu treatment activated autophagy in gastric cancer cells. (A and B) Western blotting performed on the MGC803 cells to evaluate the expression levels of LC3-I, LC3-II and SQSTM1 after being treated with OXA (10 μg/mL) (A) and 5-Fu (20 μg/mL) (B) over time. (C and D) Western blotting performed on the MGC803 cells to evaluate the expression levels of LC3-I and LC3-II after being treated with OXA (10 μg/mL) (C) and 5-Fu (20 μg/mL) (D) combined with or without CQ (10 μM) treatment over time. (E) Cell viability of the MGC803, MGC803/OXA R1 and MGC803/OXA R2 cells 24 h after being treated with OXA (left), and cell viability of the MGC803, MGC803/5Fu R1 and MGC803/5Fu R2 cells 24 h after being treated with 5-Fu (right) detected by MTT assay. (F) Western blotting performed on the MGC803, MGC803/OXA R1 and MGC803/OXA R2 cells (left), and the MGC803, MGC803/5Fu R1 and MGC803/5Fu R2 cells (right) against LC3-I, LC3-II and SQSTM1. (G) Representative TEM images of the MGC803, MGC803/OXA and MGC803/5Fu cells (left). The white arrows indicated the autophagosomes in the cytoplasm. Magnification: 4 × . The graph on the right summarized the numbers of autophagosomes in the cytoplasm of different cells. (H) Cell viability of the MGC803/OXA (left) and MGC803/5Fu (right) cells 24 h after treatment with OXA and 5-Fu combined with or without CQ (10 μM) detected by MTT assay. Data were quantified as mean ± SEM (n = 3). **P < 0.01, ***P < 0.001, vs the relative control
Next, we constructed MGC803 cells with secondary resistance to OXA, MGC803/OXA R1 and MGC803/OXA R2, and resistance to 5-Fu, MGC803/5Fu R1 and MGC803/5Fu R2. MTT assay was conducted to evaluate cell viability. Compared to the parental MGC803 cells, MGC803/OXA R1 and R2 both showed higher cell viability when being treated with increasing concentrations of OXA. Similar results were also shown in the MGC803/5Fu R1 and R2 cells (Figure 1E), confirming that the MGC803 OXA and 5-Fu resistant were successfully created. To further determine the relationship between autophagy and chemoresistance, we performed western blotting on the drug-resistant cells. The results showed that the MGC803/OXA R1 and R2, and the MGC803/5Fu R1 and R2 cells had higher LC3-II:LC3-I and decreased SQSTM1, indicating activated autophagy in the drug-resistant MGC803 cells (Figure 1F). For subsequent experiments, we chose the MGC803/OXA R1 (MGC803/OXA) and MGC803/5Fu R1 (MGC803/5Fu) cells for further exploration. The GFP-mCherry-LC3-transfected drug-resistant cells were constructed and observed by a fluorescence microscope. As green fluorescence is quenched in an acid environment, increased red fluorescence indicates activated autophagy. It was observed that there were more yellow puncta (G+R+) and red puncta (G−R+) shown in the MGC803/OXA and MGC803/5Fu cells compared to the parental MGC803 cells (Figure S1). Furthermore, transmission electron microscopy (TEM) images showed a greater number of autophagosomes, an important indication of the occurrence of autophagy, in the cytoplasm of the drug-resistant cells (Figure 1G). These results further supported that autophagy was activated in the drug-resistant MGC803 cells. After using CQ to inhibit autophagy, cell viability of the MGC803/OXA and MGC803/5Fu cells decreased significantly (Figure 1H). In conclusion, activated autophagy in gastric cancer cells was closely related to chemotherapy resistance.
EIF3J-DT was highly expressed in drug-resistant gastric cancer cells and promoted chemotherapy resistance
To identify the drug-resistant genes related to the occurrence of chemotherapy resistance, we performed RNA sequencing on the MGC803/OXA and MGC803/5Fu cells. The selected lncRNAs (fold change > 2) are shown in Figure 2A. We confirmed the expression levels of the five lncRNAs with the highest expression levels (PAX8-AS1, EIF3J-DT, AKT3-IT1, MALAT1, and TXNP6) via quantitative polymerase chain reaction (qPCR). The results showed that the expression levels of EIF3J-DT and MALAT1 significantly increased over time (0, 12, 24, and 36 h) after treatment with OXA and 5-Fu. PAX8-AS1 only showed significant increase following OXA treatment, and AKT3-IT1 was only increased by 5-Fu treatment, where there was no difference in the expression level of TXNP6 (Figure 2B). We then detected the expression levels of EIF3J-DT and MALAT1 in the drug-resistant cells. The results showed that EIF3J-DT and MALAT1 were highly expressed in both MGC803/OXA and MGC803/5Fu cells compared to the parental MGC803 cells (Figure 2C). These results suggested that EIF3J-DT and MALAT1 might be the lncRNAs related to OXA and 5-Fu resistance in gastric cancer cells.
Figure 2.

EIF3J-DT was highly expressed in the drug-resistant gastric tumor cells and EIF3J-DT silencing inhibited tumor growth. (A) Heatmap of differential expressed lncRNAs (fold change > 2) in the MGC803, MGC803/OXA, and MGC803/5Fu cells. (B) mRNA expression levels of PAX8-AS1, EIF3J-DT, AKT3-IT1, MALAT1, and TXNP6 with OXA (10 μg/mL) (upper) or 5-Fu (20 μg/mL) (lower) treatment over time detected by qPCR. (C) mRNA expression levels of EIF3J-DT (upper) and MALAT1 (lower) between the MGC803 and MGC803/OXA cells, or between the MGC803 and MGC803/5Fu cells detected by qPCR. (D) MTT assay comparing the cell viability between the NC and si-EIF3J-DT MGC803 cells 24 h after being treated with OXA (left) and 5-Fu (right). (E) MTT assay comparing the cell viability between the NC and si-EIF3J-DT MGC803/OXA cells or MGC803/5Fu cells 24 h after being treated with OXA (left) or 5-Fu (right). (F) Numbers of colony formation of the LV-NC and LV-Sh transfected MGC803 (left) and MKN45 (right) cells 14 d after being treated with PBS, OXA (10 μg/mL), and 5-Fu (20 μg/mL). LV-NC indicated the MGC803 and MKN45 cells with stable mock-vehicle transfection. LV-Sh indicated the MGC803 and MKN45 cells with sh-EIF3J-DT transfection. (G) Apoptosis rates of the LV-NC and LV-Sh MGC803 (left) and MKN45 (right) cells 24 h after being treated with PBS, OXA (10 μg/mL), and 5-Fu (20 μg/mL). (H) Western blotting performed on the NC, NC+OXA (20 and 40 μg/mL), Si, and Si+OXA (20 and 40 μg/mL) MGC803 cells (left), or the NC, NC+5-Fu (100 and 200 μg/mL), Si, and Si+5-Fu (100 and 200 μg/mL) MGC803 cells (right) with 24-h treatment against C-PARP, PARP, C-CASP3, and CASP3. Si indicated si-EIF3J-DT transfection. (I) Changes in tumor volumes (left) and tumor weight (right) of mice from the six groups (including NC, Sh, NC+OXA, Sh+OXA, NC+5-Fu, and Sh+5-Fu). Sh indicated sh-EIF3J-DT transfection. For OXA treatment, 0.1 mg/kg, intraperitoneal injection, three times per week; for 5-Fu treatment, 0.5 mg/kg, intraperitoneal injection, three times per week. (J) Representative images from the in vivo living imaging showing the subcutaneous tumors in the six groups (left) on day 28. The graph on the right calculated the total flux of tumors from different groups. Data were quantified as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, vs the relative control
Next, to confirm their effects on the MGC803 cells, we constructed the EIF3J-DT-silenced (si-EIF3J-DT) and MALAT1-silenced (si-MALAT1) MGC803 cells. The qPCR results showed that the expression of EIF3J-DT was significantly decreased by siRNA-1, siRNA-2, and siRNA-3, with the most significant decrease induced by siRNA-1 transfection. Also, the si-MALAT1-transfected MGC803 cells showed the decreased expression of MALAT1 (Figure S2A and S2B). Then, we performed MTT assays in the si-EIF3J-DT and si-MALAT1 MGC803 cells. Results showed that EIF3J-DT and MALAT1 silencing significantly sensitized the parental and drug-resistant MGC803 cells to OXA and 5-Fu treatment as evidenced by decreased cell viability (Figures 2D and 2E; Figures S2C and S2D). MALAT1 is reportedly an oncogene in multiple cancers, and our previous study also supports the promotion of tumorigenesis and metastasis by MALAT1 in gastric cancer [20]. Furthermore, MALAT1 has been shown to induce chemoresistance to vincristine in gastric cancer through autophagy regulation [21]. Therefore, we performed further study on EIF3J-DT. To clarify the relationship between EIF3J-DT and chemoresistance, we first analyzed three data sets in the GEO database. In melanoma (GDS5085/7,988,283), vemurafenib treatment induced the upregulation of EIF3J-DT. EIF3J-DT was also highly expressed in the DU-145 prostate cancer cell line, which is resistant to docetaxel (GDS3973/235,124) (Figure S3A and S3B). In the GSE69342 gastric cancer database, EIF3J-DT was significantly upregulated in vincristine- and adriamycin-resistant 7901 cells compared with the parental 7901 cells (Figure S3C). Furthermore, the Kaplan-Meier plotter analysis showed that high expression of EIF3J-DT indicated poor prognosis in patients receiving 5-Fu or other chemotherapy treatment (Figure S3D and S3E) [22]. These results suggested that EIF3J-DT might play an important role in the induction of chemotherapy resistance in cancer cells.
To further confirm our supposition regarding the function of EIF3J-DT, in-vitro and in-vivo experiments were conducted. First, we constructed the sh-EIF3J-DT MGC803 cells and sh-EIF3J-DT MKN45 cells (another human gastric cancer cell line) stably transfected with silenced EIF3J-DT. Clone formation assay results showed that the colony numbers of sh-EIF3J-DT were significantly reduced compared to the NC cells (the MGC803 and MKN45 cells transfected with negative control) when being treated with OXA or 5-Fu (Figure 2F; Figure S2E). As many chemotherapeutic drugs exert cytotoxicity in tumor cells via inducing apoptosis [23,24], we then studied whether apoptosis was also affected by EIF3J-DT. The flow cytometry results shown in Figure 2G and Figure S2F displayed that OXA and 5-Fu treatment significantly increased apoptosis rates in the sh-EIF3J-DT cells. Western blotting against cleaved PARP (C-PARP) and cleaved CASP3 (caspase 3; C-CASP3) was further performed to detect the effect of EIF3J-DT on cell apoptosis [25,26]. The results showed that C-PARP and C-CASP3 were significantly higher in the sh-EIF3J-DT MGC803 cells than in the NC cells after treatment with OXA or 5-Fu (Figure 2H), indicating EIF3J-DT silencing facilitated chemotherapy-induced apoptosis. Based on the in-vitro results, we concluded that EIF3J-DT was closely related to chemoresistance in gastric cancer cells and suppressed OXA- and 5-Fu-induced apoptosis.
Next, we subcutaneously inoculated the NC and sh-EIF3J-DT MGC803 cells into nude mice for in-vivo experiments. The mice were classified into six groups: NC, NC+OXA, NC+5-Fu, Sh, Sh+OXA, and Sh+5-Fu. The tumor volumes of the NC+OXA and NC+5-Fu groups were smaller than those in the NC group, while the Sh+OXA and Sh+5-Fu groups exhibited the slowest tumor growth and had the smallest tumor volumes (Figure 2I), suggesting that sh-EIF3J-DT cells showed greater chemosensitivity than NC cells in vivo. Furthermore, the changes in tumor weight and the results from in vivo imaging on day 28 supported the same conclusion (Figures 2I and 2J). In conclusion, highly expressed EIF3J-DT promoted tumor cell drug resistance in vitro and in vivo.
EIF3J-DT facilitated the autophagy in gastric cancer cells
To investigate the possible relationship between EIF3J-DT and autophagy, the expression levels of LC3-I and LC3-II were measured in the NC and si-EIF3J-DT-transfected MGC803 and MKN45 cells by western blotting. After being treated with CQ, higher expression of LC3-II was found in NC cells, indicating that EIF3J-DT silencing inhibited autophagy in gastric cancer cell (Figure 3A; Figure S2A). Furthermore, the increased LC3-II and decreased SQSTM1 were reversed by silenced EIF3J-DT following OXA and 5-Fu treatment (Figure 3B). These results suggested that EIF3J-DT contributed to the activated autophagy in gastric cancer cells.
Figure 3.
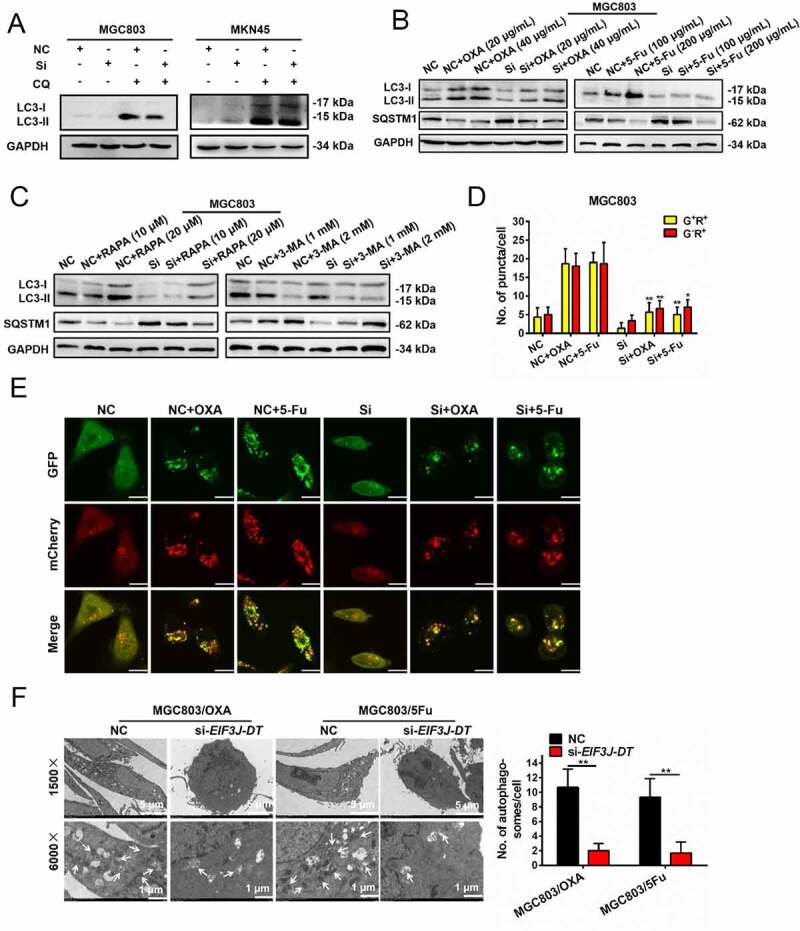
EIF3J-DT activated autophagy in the gastric cancer cells. (A) Expression levels of LC3-I and LC3-II in the NC and Si MGC803 (left) or MKN45 (right) cells treated with or without CQ (10 μM) detected by western blotting. (B) Expression levels of LC3-I, LC3-II and SQSTM1 in the NC and Si MGC803 cells treated with OXA (20 and 40 μg/mL, left) or 5-Fu (100 and 200 μg/mL, right) detected by western blotting. (C) Expression levels of LC3-I, LC3-II and SQSTM1 in the NC and Si MGC803 cells treated with RAPA (10 and 20 μM, left) or 3-MA (1 and 2 mM, right) detected by western blotting. (D and E) Representative immunofluorescence images of the GFP-mCherry-LC3 transfected NC and Si MGC803 cells 24 h after treatment with OXA (10 μg/mL) and 5-Fu (20 μg/mL) (E). Scale bar: 10 μm. Numbers of the G+R+ and G−R+ fluorescent puncta in different treated groups were calculated in D. (F) TEM performed on the MGC803/OXA and MGC803/5Fu cells transfected with NC or si-EIF3J-DT (left). The white arrows indicated the autophagosomes in the cytoplasm. Magnification: 4 × . The graph summarized the numbers of autophagosomes in different groups (right). Data were quantified as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, vs the relative control
Next, we treated the cells with the autophagy agonist rapamycin (RAPA) and the autophagy inhibitor 3-methyladenine (3-MA). Western blotting showed that RAPA enhanced, and 3-MA inhibited, autophagy in NC cells. While in EIF3J-DT-silenced cells, RAPA did not accurately function (Figure 3C), indicating that EIF3J-DT silencing blocked the activation of autophagy. Furthermore, less G+R+ and G−R+ puncta were found in the si-EIF3J-DT cells than in the NC cells following treatment with OXA and 5-Fu (Figures 3D and 3E). As observed by TEM, EIF3J-DT silencing also significantly decreased the numbers of autophagosomes in the cytoplasm of the MGC803/OXA and MGC803/5Fu cells (Figure 3F). Accordingly, it was concluded that the expression of EIF3J-DT facilitated autophagy in gastric cancer cells.
ATG14 enhanced chemotherapy resistance via apoptosis inhibition and autophagy activation
The specific mechanism that EIF3J-DT induced autophagy in gastric cancer cells required further study. Combined with TCGA Nature 2014 [27] and TCGA Provisional database analysis, we found that the expression of EIF3J-DT was most positively and significantly related to the mRNA expression of ATG14 (Spearmen’s correlation coefficient: 0.462 and 0.55 for TCGA Nature 2014 and TCGA Provisional database, respectively) (Figure 4A; Table S1). To further explore the correlation between EIF3J-DT and ATG14, we measured ATG14 mRNA expression in the MGC803 cells with overexpressed or silenced EIF3J-DT via qPCR. The results showed that the mRNA expression of ATG14 was increased by overexpressed EIF3J-DT, where it was decreased by silenced EIF3J-DT (Figure 4B). When the expression of EIF3J-DT in EIF3J-DT-silenced cells was recovered, the expression of ATG14 mRNA was also upregulated (Figure 4B). This indicated that the mRNA expression of ATG14 was positively correlated with the expression of EIF3J-DT. The western blotting results verified this conclusion (Figure S4A).
Figure 4.

The expression of ATG14 affected cell apoptosis and autophagy. (A) TCGA Nature 2014 and TCGA Provisional database analyses showing the correlation between EIF3J-DT and ATG14 mRNA expression levels. (B) mRNA expression level of ATG14 in the MGC803 cells with overexpressed (left), silenced (middle), or overexpressed+silenced (right) EIF3J-DT detected by qPCR. (C) Cell viability among the NC, siATG14 #1, and siATG14 #2 MGC803 cells 24 h after being treated with OXA (left) and 5-Fu (right) detected by MTT assay. (D) Cell viability among the NC, siATG14 #1, and siATG14 #2 drug-resistant MGC803 cells 24 h after being treated with OXA (left) and 5-Fu (right) detected by MTT assay. (E) Numbers of colony formation of the NC, siATG14 #1, and siATG14 #2 MGC803 (left) and MKN45 (right) cells 14 d after being treated with PBS, OXA (10 μg/mL) and 5-Fu (20 μg/mL). (F) Apoptosis rates of the NC, siATG14 #1, and siATG14 #2 MGC803 (left) and MKN45 (right) cells 24 h after being treated with PBS, OXA (10 μg/mL) and 5-Fu (20 μg/mL) detected by flow cytometry. (G) Western blotting performed on the NC, siATG14 #1, and siATG14 #2 MGC803 cells 24 h after being treated with OXA (20 μg/mL) (left) and 5-Fu (20 μg/mL) (right) against C-PARP, PARP, C-CASP3, CASP3, C- CASP7, and CASP7. (H) Expression levels of ATG14, LC3-I, LC3-II and SQSTM1 in the NC, siATG14 #1, and siATG14 #2 MGC803 (left) and MKN45 (right) cells detected by western blotting. (I) Expression levels of LC3-I and LC3-II in the NC, siATG14 #1, and siATG14 #2 MGC803 cells with or without CQ treatment (10 μM) detected by western blotting. (J) Representative fluorescence images of the GFP-mCherry-LC3 transfected NC and siATG14 (siRNA2) MGC803 cells 24 h after being treated with PBS, OXA (10 μg/mL) and 5-Fu (20 μg/mL) (left). Scale bar: 10 μm. The statistics was shown on the right. Data were quantified as mean ± SEM (n = 3). **P < 0.01, ***P < 0.001, vs the relative control
Further experiments focused on clarifying the relationship between ATG14 and chemotherapy resistance. First, western blotting results showed that ATG14 was upregulated in the drug-resistant cells (Figure S4B). ATG14-silenced MGC803 cells (ATG14 siRNA-1, siRNA-2, and siRNA-3) were then constructed, and siRNA-1 and siRNA-2 were selected for further experiments (Figure S4C). MTT assays showed that decreased expression of ATG14 sensitized the parental MGC803 cells to OXA and 5-Fu treatment (Figure 4C) and reversed the resistance of the MGC803/OXA and MGC803/5Fu cells to chemotherapy (Figure 4D). Clone formation assays showed that the colony numbers were significantly reduced in the MGC803 and MKN45 cells with ATG14 silencing (Figure 4E; Figure S4D and S4E). This indicated that ATG14 was important for chemoresistance in gastric cancer cells. Furthermore, the results from flow cytometry and western blotting showed that cell apoptosis was facilitated by ATG14 silencing (Figures 4F and 4G; Figure S4F).
Next, we studied the relationship between ATG14 and autophagy. Western blotting showed the inhibition of autophagy by ATG14 silencing both in MGC803 and MKN45 cells (Figures 4H and 4I). Furthermore, fluorescence imaging showed less G+R+ and G−R+ puncta in the GFP-mCherry-LC3 transfected cells with silenced ATG14 treated with OXA and 5-Fu (Figure 4J). In conclusion, ATG14 was closely related to chemotherapy resistance by regulating autophagy and cell apoptosis. As ATG14 was highly expressed in the drug-resistant cells, and its expression was positively correlated with the expression of EIF3J-DT, EIF3J-DT might modulate ATG14-mediated autophagy and chemotherapy resistance.
EIF3J-DT induced ATG14-mediated autophagy and chemotherapy resistance
To further identify interaction between EIF3J-DT and ATG14 mRNA, we constructed the EIF3J-DT+siATG14 #1- and EIF3J-DT+siATG14 #2-transfected MGC803 cells with overexpressed EIF3J-DT and silenced ATG14. MTT assays were conducted to measure cell viability in the four groups, including the pcDNA (MGC803 cells transfected with the control plasmid), EIF3J-DT (MGC803 cells with overexpressed EIF3J-DT), EIF3J-DT+siATG14 #1, and EIF3J-DT+siATG14 #2. The results showed that compared to the pcDNA cells, the EIF3J-DT group possessed the highest cell viability with OXA and 5-Fu treatment. After the silencing of ATG14, resistance to OXA and 5-Fu in the EIF3J-DT+siATG14 #1 and EIF3J-DT+siATG14 #2 groups was decreased (Figure 5A). Clone formation assays obtained similar results (Figure 5B). Next, we further detected the expression levels of autophagy related proteins in the MGC803 and MKN45 cells from the four groups. The results showed that compared to the EIF3J-DT group, the increased expression of LC3-II and decreased SQSTM1 were reversed by ATG14 silencing (Figure 5C). After being treated with CQ, the expression levels of LC3-II were both lower in the EIF3J-DT+siATG14 #1 and EIF3J-DT+siATG14 #2 groups than that in the EIF3J-DT group (Figure 5D), indicating that the EIF3J-DT-induced activation of autophagy would be inhibited by ATG14 silencing. Combined with the previous findings, we concluded that EIF3J-DT activated autophagy and induced chemoresistance in gastric cancer cells mediated by ATG14.
Figure 5.

ATG14 silencing reversed the chemotherapy resistance induced by EIF3J-DT. (A) Cell viability of the four groups (including pcDNA, EIF3J-DT, EIF3J-DT+siATG14 #1 and EIF3J-DT+siATG14 #2) with OXA (left) and 5-Fu (right) treatment for 24 h detected by MTT assay. (B) Clone formation assay performed on the cells from the four groups 14 d after being treated with PBS, OXA (10 μg/mL), and 5-Fu (20 μg/mL) (left). The statistics was shown on the right. (C) Expression levels of ATG14, LC3-I, LC3-II and SQSTM1 in the cells from the four groups detected by western blotting. (D) Expression levels of LC3-I and LC3-II in the MGC803 (upper) and MKN45 (lower) cells from the four groups treated with or without CQ (10 μM) detected by western blotting. Data were quantified as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, vs the relative control
EIF3J-DT promoted ATG14 mRNA stability through direct interaction
LncRNAs mainly function through the endonuclear or intra-cytoplasmic pathways [28]. To clarify the specific mechanism of EIF3J-DT, it was important to first determine the subcellular localization of EIF3J-DT. Via fluorescence in situ hybridization (FISH) assay, we found that EIF3J-DT was mainly localized in the cytoplasm (Figure 6A). Therefore, we hypothesized that there may be three key pathways through which EIF3J-DT functions: regulation of mRNA translation, affecting mRNA stability, and acting as a sponge to sequester miRNAs [29]. The previous results showed that the mRNA expression of ATG14 was positively related to the expression of EIF3J-DT. Therefore, to clarify how EIF3J-DT affected the expression of ATG14, BLAST was used to predict the binding between EIF3J-DT and ATG14 mRNA, and found 211 binding sites (Figure 6B). This suggested that EIF3J-DT might directly bind with ATG14 mRNA to affect its expression. To further confirm this, we utilized α-amanitin to block the initiation of RNA synthesis and extension of the RNA strand, and actinomycin D to inhibit RNA synthesis by suppressing the activity of DNA-dependent RNA polymerase. qPCR assays showed that the mRNA expression level of ATG14 in the pcDNA MGC803 cells decreased gradually over time with α-amanitin and actinomycin D treatment. In the MGC803 cells overexpressing EIF3J-DT, ATG14 mRNA was also gradually degraded, but the degradation velocity was slower than in the pcDNA MGC803 cells (Figure 6C). Unlike the ATG14 mRNA, the mRNA level of GAPDH was not affected by the expression of EIF3J-DT (Figure 6D). This indicated that EIF3J-DT slowed the degradation of ATG14 mRNA.
Figure 6.

EIF3J-DT promoted the mRNA stability of ATG14 via direct binding to induce cell autophagy and chemotherapy resistance. (A) Representative images of FISH assay showing the localization of EIF3J-DT (red). The cell nuclei were stained with DAPI (blue). Scale bar: 30 μm. (B) Table showing the potential binding sites between EIF3J-DT and ATG14 mRNA. (C and D) Changes in mRNA expression levels of ATG14 (C) and GAPDH (D) in the pcDNA or EIF3J-DT transfected MGC803 cells with α-amanitin (50 mM, left) and actinomycin D (10 μg/mL, right) treatment over time detected by qPCR assay. (E) Changes in mRNA expression level of ATG14 in the pcDNA, EIF3J-DT WT, and EIF3J-DT MUT1 transfected MGC803 cells with α-amanitin (50 mM, left) and actinomycin D (10 μg/mL, right) treatment over time detected by qPCR assay. (F) MTT assay performed on the pcDNA, EIF3J-DT WT, and EIF3J-DT MUT1 transfected MGC803 cells showing the cell viability 24 h after OXA (left) and 5-Fu (right) treatment. (G) Numbers of colony formation of the pcDNA, EIF3J-DT WT, and EIF3J-DT MUT1 transfected MGC803 (left) and MKN45 (right) cells 14 d after PBS, OXA (10 μg/mL) and 5-Fu (20 μg/mL) treatment. (H) Western blotting performed on the pcDNA, EIF3J-DT WT, and EIF3J-DT MUT1 transfected MGC803 (left) and MKN45 (right) cells against ATG14, LC3-I, LC3-II and SQSTM1. (I) Relative luciferase activity of the indicated luciferase reporter vectors in MGC803 (left) and 293 T (right) cells co-transfected with EIF3J-DT or pcDNA plasmid. (J) Interaction between ATG14 mRNA and EIF3J-DT WT or EIF3J-DT MUT1 detected by RNA affinity-isolation assay (left). The statistics was shown on the right. Data were quantified as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, vs the relative control
To further confirm whether EIF3J-DT interacted with ATG14 mRNA, a plasmid vector carrying mutant EIF3J-DT was constructed (EIF3J-DT MUT1) and transfected into the MGC803 cells. The binding sites with ATG14 mRNA on EIF3J-DT was mutant in EIF3J-DT MUT1. mRNA detection of ATG14 at different time points revealed that both wild-type EIF3J-DT-transfected MGC803 cells (EIF3J-DT WT cells) and the EIF3J-DT MUT1 cells exhibited significantly decreased mRNA expression levels of ATG14 following α-amanitin and actinomycin D treatment, but a higher degradation velocity was observed in EIF3J-DT MUT1 cells (Figure 6E), indicating that only the EIF3J-DT WT could regulate the expression of ATG14. Results from the latter functional assays also showed that the resistance of the EIF3J-DT MUT1 cells to OXA and 5-Fu treatment was significantly reduced (Figures 6F and 6G; Figure S5A). Western blotting showed that compared to the EIF3J-DT WT cells, EIF3J-DT MUT1 cells showed decreased expression of ATG14 and LC3-II, and increased expression of SQSTM1 (Figure 6H). To further confirm these findings, we transfected EIF3J-DT WT and EIF3J-DT MUT1 into MKN45 cells and obtained similar results in the clone formation assay and western blotting (Figures 6G and 6H; Figure S5B). These results suggested that EIF3J-DT regulated the expression of ATG14, which was closely related to the induction of autophagy and chemoresistance in gastric cancer cells.
Next, we constructed luciferase reporter vectors (psi-CHECK2, ATG14 WT, and ATG14 MUT1) based on the binding sites, and the luciferase vectors were co-transfected into the MGC803 cells with pcDNA or EIF3J-DT plasmids. The results showed that EIF3J-DT led to the most significant increase in ATG14 WT luciferase activity (Figure 6I), indicating a strong affinity between EIF3J-DT and ATG14 mRNA. A similar result was also obtained in the 293 T human embryonic kidney cell line. In addition, affinity isolation of endogenous ATG14 mRNA using in vitro-transcribed biotin-labeled EIF3J-DT WT or EIF3J-DT MUT1 showed that only EIF3J-DT WT interacted with ATG14 mRNA (Figure 6J). Accordingly, we concluded that EIF3J-DT upregulated the expression of ATG14 via directly binding with ATG14 mRNA to increase its stability, contributing to activation of autophagy and chemotherapy resistance.
MIR188-3p accelerated the degradation of ATG14 mRNA and reversed autophagy and chemotherapy resistance induced by EIF3J-DT
Apart from direct binding with mRNA to affect its stability, lncRNAs also can play an important role in miRNAs sequestration [30]. Previous studies show that miRNAs can accelerate mRNA degradation or inhibit translation via specific binding with the targeted mRNA [31,32]. Based on the above results, we confirmed that EIF3J-DT regulated the expression of ATG14 through direct interaction with ATG14 mRNA. Whether other pathways participate in the regulation of ATG14 expression requires further research.
According to bioinformatics analysis of RegRNA2.0, we found that EIF3J-DT had seven potential binding sites for miRNA MIR188-3p (Figure 7A), which reportedly plays a crucial role in regulating chemoresistance and autophagy [33,34]. Interestingly, analysis of the Targetscan database revealed that MIR188-3p targeted the 3′ UTR of ATG14 mRNA (Figure 7A). Therefore, the interactions among MIR188-3p, EIF3J-DT, and ATG14 were investigated. First, to study whether MIR188-3p affected the expression of ATG14, we constructed MGC803 cells transfected with MIR188-3p AgomiR or MIR188-3p AntagomiR (anti-MIR188-3p). qPCR assays showed that the mRNA expression level of ATG14 was decreased in the MIR188-3p AgomiR-transfected MGC803 cells and increased in the anti-MIR188-3p-transfected MGC803 cells (Figure S6A). This indicated a negative relationship between the expression of MIR188-3p and ATG14 mRNA. In addition, western blotting showed similar changes in protein expression (Figure S6B). Therefore, we concluded that MIR188-3p inhibited the expression of ATG14. To further study how MIR188-3p affected the expression of ATG14, we co-transfected the psi-CHECK2, ATG14 WT, and ATG14 MUT2 luciferase reporter vectors with MIR188-3p AgomiR into the MGC803 and 293 T cells. The results showed that in both cell lines, MIR188-3p decreased ATG14 WT luciferase activity, but not ATG14 MUT2 (Figure 7B). This indicated the binding between MIR188-3p and ATG14 mRNA, which facilitated the degradation of ATG14 mRNA.
Figure 7.

MIR188-3p inhibited the expression of ATG14 and reversed the autophagy and chemotherapy resistance induced by EIF3J-DT. (A) RegRNA2.0 and Targetscan database analyzing the binding sites between MIR188-3p and EIF3J-DT, or ATG14 mRNA. (B) Relative luciferase activity of psi-CHECK2, ATG14 WT and ATG14 MUT2 in the MGC803 (left) and 293 T (right) cells co-transfected with MIR188-3p AgomiR. (C) Relative luciferase activity of EIF3J-DT WT and EIF3J-DT MUT2 in the MGC803 (left) and 293 T (left) cells co-transfected with MIR188-3p AgomiR or NC. (D) Expression of MIR188-3p and GAPDH isolated by EIF3J-DT WT or EIF3J-DT MUT2 from MGC803 (left) and MKN45 (right) cells detected by qPCR. (E) Representative fluorescence images of MGC803/OXA and MGC803/5Fu cells when NC or MIR188-3p AgomiR co-transfected with the GFP-mCherry-LC3. Scale bar: 10 μm. The statistics was shown on the right. (F) Western blotting performed on the NC and anti-MIR188-3p transfected MGC803 cells against LC3-I and LC3-II with treatment of OXA (10 μg/mL, left) and 5-Fu (20 μg/mL, right) with or without CQ (10 μM). (G) Cell viability of the MGC803 cells transfected with pcDNA, EIF3J-DT, EIF3J-DT+MIR188-3p AgomiR 24 h after OXA (left) and 5-Fu (right) treatment by MTT assay. (H) Western blotting performed on the pcDNA, EIF3J-DT WT, EIF3J-DT MUT2, and EIF3J-DT WT+MIR188-3p AgomiR transfected MGC803 (upper) and MKN45 (lower) cells against ATG14, LC3-I, LC3-II and SQSTM1. Data were quantified as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, vs the relative control
Next, to verify the interaction between EIF3J-DT and MIR188-3p, we cloned the binding sequence (EIF3J-DT WT) and the mutant binding sequence (EIF3J-DT MUT2) of EIF3J-DT into luciferase reporter vectors and co-transfected them with MIR188-3p AgomiR. The Luciferase reporter assay showed that luciferase activity of EIF3J-DT WT was significantly decreased by MIR188-3p in the MGC803 and 293 T cells, where EIF3J-DT MUT2 was not (Figure 7C). Then, we performed RNA affinity-isolation experiment and found that the amount of MIR188-3p was remarkably higher in cells transfected with EIF3J-DT WT than in cells transfected with EIF3J-DT MUT2 (Figure 7D). These results suggested that MIR188-3p directly bound to EIF3J-DT. As MIR188-3p decreased the mRNA level of ATG14, we therefore considered that EIF3J-DT might sequester MIR188-3p to suppress the ATG14 inhibition by MIR188-3p.
To further study the effect of MIR188-3p on chemotherapy resistance, the parental and drug-resistant MGC803 cells with MIR188-3p AgomiR were constructed. MTT assay results showed that chemoresistance was reduced in the MIR188-3p overexpressing drug-resistant cells compared to the NC cells treated with OXA and 5-Fu (Figure S6C). In addition, the MIR188-3p overexpressing MGC803 cells were also more sensitive to chemotherapy with decreased colony formation (Figure S6D). According to the fluorescence images and western blotting, MIR188-3p negatively regulated autophagy in MGC803 cells (Figures 7E and 7F). This indicated that the expression of MIR188-3p reduced autophagy and chemoresistance in gastric cancer cells.
We confirmed that EIF3J-DT and MIR188-3p showed antagonistic regulation on cell autophagy and chemotherapy resistance. As shown in Figure 7G, EIF3J-DT+MIR188-3p overexpressing MGC803 cells showed decreased cell viability compared to the EIF3J-DT overexpressing cells. Furthermore, it was found that the expression of ATG14 and LC3-II was downregulated in the MGC803 and MKN45 cells co-transfected with EIF3J-DT WT and MIR188-3p (Figure 7H), further supporting the antagonistic function of EIF3J-DT and MIR188-3p on the regulation of autophagy and chemoresistance in gastric cancer cells.
EIF3J-DT and ATG14 were correlated with gastric cancer patient prognosis
Based on the above experiments, we confirmed that chemotherapy resistance in gastric cancer cells was closely related to high expression of EIF3J-DT and ATG14, leading to activation of autophagy. Therefore, we further confirmed the expression of EIF3J-DT and ATG14 and explored their effects on prognosis in gastric cancer patients. Fresh tumor samples were collected from 43 gastric cancer patients, including 24 patients sensitive to chemotherapy (CR, PR, or SD after treatment) and 19 patients resistant to chemotherapy (PD after treatment) (detailed clinicopathological features of these patients were listed in Table S2). qPCR assays showed that the mRNA expression of EIF3J-DT and ATG14 was significantly increased in tumors resistant to chemotherapy treatment (Figures 8A and 8B). Following further analysis, the expression of EIF3J-DT was positively correlated with the expression of ATG14 (Figure 8C, R = 0.550, P < 0.001). Furthermore, we performed in situ hybridization (ISH) and immunohistochemistry (IHC) on 107 paraffin-embedded gastric tumor samples, and observed the higher expression of EIF3J-DT and ATG14 in tumor tissues from resistant patients compared with sensitive patients (Figure 8D). According to the ISH and IHC results, we classified the paraffin-embedded gastric tumor samples by the expression levels of EIF3J-DT and ATG14, including EIF3J-DT high (56 cases, 52.3%) and EIF3J-DT low (51 cases, 47.7%), and ATG14 high (59 cases, 55%) and ATG14 low (48 cases, 45%) (Figure 8E). There were 41 cases together with high expression of EIF3J-DT and ATG14, and 33 cases together with low expression of EIF3J-DT and ATG14. Next, prognostic analysis showed that high expression of EIF3J-DT or ATG14 was associated with shorter disease-free time for patients (Figures 8F and 8G, P < 0.001). Notably, patients together with high expression of EIF3J-DT and ATG14 had the worst prognosis with the shortest disease-free time (Figure 8H, P < 0.001). In conclusion, we confirmed that EIF3J-DT and ATG14 were upregulated in gastric cancer patients resistant to chemotherapy, consistent with the findings obtained from in-vitro cellular experiments. It was worth noting that high expression of EIF3J-DT and ATG14 significantly damaged patient prognosis.
Figure 8.

The expression of EIF3J-DT and ATG14 related to the prognosis of gastric cancer patients with chemotherapy treatment. (A and B) Expression of EIF3J-DT (A) and ATG14 (B) in fresh tumor tissues collected from gastric cancer patients sensitive (n = 24) or resistant (n = 19) to chemotherapy detected by qPCR. *P < 0.05, **P < 0.01 (C) Correlation between EIF3J-DT and ATG14 mRNA expression levels. Two-tailed Spearman’s correlation analysis (n = 43). P < 0.001. (D) Representative images showing the expression of EIF3J-DT (detected by ISH) and ATG14 (detected by IHC) in paraffin-embedded gastric tumor tissues (n = 107). Magnification: 2 × . Scale bar: 50 μm. (E) Table showing the classification of 107 paraffin-embedded gastric tumor tissues by different expression levels of EIF3J-DT (high or low) and ATG14 (high or low) according to ISH and IHC. (F-H) Retrospective analysis of Kaplan-Meier plots for EIF3J-DT (F), ATG14 (G), and the two (H) expression in association with disease-free time
Discussion
Chemotherapy has been applied in patients with unresectable gastric tumors to decrease the risk of recurrence and metastasis [35], with a five-year overall survival of 20%–35% [36]. Moreover, perioperative chemotherapy is able to significantly improve patient prognosis with resectable tumors [37]. However, 70%–90% of gastric cancer patients still relapse because they develop chemotherapy resistance [38], which remains a major challenge in gastric cancer therapy. Further investigation of chemoresistance mechanisms is urgently warranted. In this study, we investigated a novel model in which upregulation of lncRNA EIF3J-DT expression enhanced autophagy via targeting ATG14 resulting in OXA and 5-Fu resistance in gastric cancer cells.
LncRNAs are involved in almost all cellular functions, such as proliferation, migration, epithelial-mesenchymal transition, and metastasis [39]. In gastric cancer, a variety of abnormally expressed lncRNAs were identified to promote tumor progression [40], targeted therapy sensitivity [41,42], and chemoresistance [43]. In the present study, we performed RNA sequencing in OXA- and 5-Fu-resistant MGC803 cells and their parental cells. Combined with qPCR results, they showed EIF3J-DT to be significantly upregulated in MGC803/OXA and MGC803/5Fu cells. Silencing EIF3J-DT resensitized MGC803/OXA and MGC803/5Fu cells to OXA and 5-Fu treatment both in vitro and in vivo. Recent studies have shown that EIF3J-DT is involved in the biological behaviors of cancer. For example, EIF3J-DT can increase proliferation and impede apoptosis in colorectal cancer through the MIR3163-YAP1 axis [44] and accelerate hepatocellular carcinoma progression via targeting the MIR122-5p-CTNND2 axis [45]. However, there are still no studies focused on its effect on chemoresistance in gastric cancer.
Tumors enhance autophagy activity to survive microenvironmental stress and to facilitate proliferation and aggressiveness [46,47]. Recently, dysregulations in autophagy function have been proven to be a potential mechanism of developing chemoresistance [48,49]. Our results confirmed that autophagy was activated in the MGC803/OXA and MGC803/5Fu cells compared with their parental cells. LncRNAs have also been found to regulate autophagy. For instance, Wang et al. find that H19 induces autophagy activation, contributing to tamoxifen resistance in breast cancer [50]. In this study, silencing EIF3J-DT decreased autophagy in MGC803/OXA and MGC803/5Fu cells. Dysregulated expression of lncRNA provides a valuable clue for the discovery of functional molecular that modulates chemotherapy resistance [51]. According to bioinformatics analysis, the expression of ATG14 mRNA was positively related to the expression of EIF3J-DT. ATG14 is one of the known ATGs, acting as an effective activator for autophagy [52–54]. Our results showed that EIF3J-DT upregulated ATG14 in drug-resistant gastric cancer cells. Therefore, ATG14 might function as a direct function protein of EIF3J-DT to regulate the autophagy and chemoresistance.
By means of BLAST, we found 211 binding sites between EIF3J-DT and ATG14. Combined with luciferase reporter and RNA affinity-isolation assays, we confirmed that EIF3J-DT directly interacted with ATG14 mRNA to increase its stability, leading to increased expression of ATG14. The lncRNA-mRNA network has been verified in various cancers, such as colorectal cancer [55], pancreatic ductal adenocarcinoma [56], and breast cancer [57]. This plays an important regulatory role in tumorigenesis and tumor progression. It has been reported that lncRNA TMPO-AS1 is upregulated in endocrine therapy-resistant MCF-7 cells. By stabilizing ESR1 (estrogen receptor 1) mRNA through interaction with ESR1 mRNA, TMPO-AS1 positively regulates ESR1 mRNA expression, promoting the progression of estrogen receptor-positive breast cancer [57]. In our study, the interaction between EIF3J-DT and ATG14 mRNA activated autophagy, and induced chemoresistance in gastric cancer cells.
Increasing evidence has shown that lncRNAs exported to the cytoplasm can affect gene expression in multiple ways, including directly binding to mRNA to affect mRNA translation [58,59], and acting as decoys for miRNAs [60] and proteins [61]. According to bioinformatics analysis of RegRNA2.0, we found that EIF3J-DT had seven potential binding sites for MIR188-3p. Fortunately, MIR188-3p also targeted ATG14 mRNA 3′ UTR. Thus, we assumed that EIF3J-DT also modulated the OXA and 5-Fu resistance of gastric cancer cells by targeting ATG14 via sponging MIR188-3p. Our results confirmed that MIR188-3p and EIF3J-DT played the antagonistic function on the regulation of ATG14. Therefore, in addition to stabilizing ATG14 mRNA, EIF3J-AS also sequestered MIR188-3p to block the degradation of ATG14 mRNA, contributing to the activation of autophagy and induction of chemoresistance in gastric cancer cells. Increasing evidence has illustrated the important role of lncRNAs in cancer drug resistance via acting as a miRNA sponge. For example, lncRNA NEAT1 is upregulated in renal cell carcinoma tissue and NEAT1 knockdown increases the sensitivity of tumor cells to sorafenib by acting as a competitive sponge for MIR34A [62].
In the clinical setting, we confirmed that EIF3J-DT and ATG14 were upregulated in gastric cancer patients resistant to chemotherapy, relating to poor patient prognosis. Researchers suggest that lncRNAs have great potential as prognostic or diagnostic indicators as their cancer-restricted expression characteristics and excellent stability in biological fluids [63,64]. Gupta et al. find that lncRNA HOTAIR can serve as a promising predictor of metastasis and death of breast cancer patients [63]. Moreover, antisense oligonucleotides have been reported to potently decrease lncRNA function by cleaving complementary target lncRNA [65]. Using antisense oligonucleotides to downregulate MALAT1 expression in mouse mammary tumor virus-PyMT mice decreases tumor cell proliferation and metastasis [66]. In this study, results showed that the overexpression of EIF3J-DT served as a promising prognostic biomarker for clinical gastric cancer patients. Further research on the inhibition of EIF3J-DT could provide opportunities for EIF3J-DT-targeted therapies in chemoresistant gastric cancer.
In summary, our study and data showed the role of EIF3J-DT in OXA- and 5-Fu-resistant gastric cancer cells. Further experiments revealed the regulatory mechanism of the EIF3J-DT-MIR188-3p-ATG14 pathway on gastric cancer chemoresistance, suggesting the vital role of EIF3J-DT in providing the novel therapeutic target for gastric cancer.
Materials and Methods
Patient tissue samples
43 fresh gastric cancer specimens were collected from gastric cancer patients sensitive (24 cases) or resistant (19 cases) to chemotherapy from Nanfang Hospital, Southern Medical University. The fresh tumor samples were preserved in liquid nitrogen immediately after resection. 107 paraffin-embedded gastric cancer specimens were obtained from Nanfang Hospital as well. Each specimen was attached to a confirmed pathological diagnosis. All experiments in this study were endorsed by the Ethics Committee of Southern Medical University and complied with the Declaration of Helsinki. The Ethics Committee of Southern Medical University specifically approved that no informed consent was required, because data were analyzed anonymously.
Cell lines and culture
Human gastric cancer cell lines-MGC803 and MKN45, and human embryonic kidney cell line-293 T, were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). MGC803 and MKN45 cells were maintained in RPMI-1640 medium (Hyclone, SH30809.01) supplemented with 10% fetal bovine serum (FBS) (Gibco, 10,270–106). 293 T cells were maintained in DMEM (Gibco, 12,800,017) supplemented with 10% FBS. All cells were incubated in a humidified atmosphere of 5% CO2 at 37°C.
RNA sequencing
The expression profiling data of differential lncRNAs in MGC803, MGC803/OXA and MGC803/5Fu cells was analyzed by LongSee lncRNA microarray. Detailed information was listed in Table S3. LncRNAs with the fold change () above 2 in both MGC803/OXA and MGC803/5Fu were selected. Statistical significance was established at P < 0.001. Among them, to search the drug-resistant associated lncRNAs, five lncRNAs (PAX8-AS1, EIF3J-DT, AKT3-IT1, MALAT1, and TXNP6) with the highest gene expression were selected for further verification.
Construction of the OXA- and 5-Fu-resistant MGC803 cells
MGC803 cells were seeded into the 6-well plates. After growing to 70%–80% confluence, the media contained OXA or 5-Fu (10 mg/mL for OXA; 0.2 mg/mL for 5-Fu) were added into the wells. At 1 h after drug treatment, the drug-contained media was replaced by fresh media (without OXA and 5-Fu) after the cells were washed by phosphate-buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) twice. The drug-contained media was again added at the second cell passage and the treatment time was prolonged gradually (for 2, 3, 6, 12, 24, 36, 48, 60, and 72 h, respectively). The following process was the same as aforementioned. Until the cell sensitivity to OXA and 5-Fu treatment was decreased obviously and became stable, MTT assays were performed to confirm the successful construction of the OXA- and 5-Fu-resistant MGC803 cells.
Cell transfection
Before transfection, the MGC803 or MKN45 cells (/mL) were seeded in the 6-well plates. When growing to 30%–40% confluence, siRNAs against EIF3J-DT (siRNA-1, siRNA-2, and siRNA-3), MALAT1, ATG14 (siRNA-1, siRNA-2, and siRNA-3), and a NC siRNA (GEMA, Shanghai, China); EIF3J-DT WT, EIF3J-DT MUT1 and EIF3J-DT MUT2 expression plasmids and pcDNA (GEMA, Shanghai, China); MIR188-3p AntagomiR, MIR188-3p AgomiR and matched MIR-NC (GEMA, Shanghai, China), were transfected into the MGC803 cells using Lipofectamine 2000 Transfection Reagent (Invitrogen, 11,668–019) according to manufacturer’s instructions. siRNAs against EIF3J-DT (siRNA-1, siRNA-2, and siRNA-3) and ATG14 (siRNA-1, siRNA-2, and siRNA-3); EIF3J-DT WT, EIF3J-DT MUT1 and EIF3J-DT MUT2 expression plasmids; and MIR188-3p AgomiR, were transfected into the MKN45 cells. After being incubated for 4–6 h, the cells were washed by PBS twice and the fresh media (supplemented with 10% FBS) were added for another 24-h to 48-h incubation. Then the cells were collected and used for qPCR assay, western blotting, MTT assay, clone formation assay and so on. All sequences were listed in Table S4.
For EIF3J-DT knockdown, sh-EIF3J-DT-Puro lentiviruses (LV-EIF3J-DT) and sh-negative control-Puro lentiviruses (LV-NC) were constructed (GEMA, Shanghai, China). MGC803 and MKN45 cells were seeded into the 12-well plates. After growing to 30%–40% confluence, 1 mL fresh media supplemented with 10 μl polybrene (Obio technology, OGTR(C)20,181,002) and 5 μl lentiviruses (LV-EIF3J-DT or LV-NC) were added into each well. The cells were observed by microscopy per 4 h to evaluate the cell states. If the cell morphology changed or the cells were in poor states, the media would be replaced by the fresh media for another 3-d incubation. Puromycin (1 μg/mL) (Medchem Express, HY-B1743A) was used to selected the successfully transfected cells.
MTT assay
In brief, /mL suspended cells were plated per well in 96-well plates. After incubation for 24 h at 37°C with 5% CO2, 100 μl drug-contained media were added into each well by certain concentration gradients (0, 2.5, 5, 10, 20, 40, and 80 μg/mL for OXA; 0, 12.5, 25, 50, 100, 200, and 300 μg/mL for 5-Fu). Twenty-four h later, add 10 μl MTT reagent (5 mg/mL, without RPMI 1640) (Sigma-Aldrich, M2128) to per well and incubate for another 2–4 h. Then 150 μl/well dimethyl sulfoxides were used to terminate the reaction. After concussion for 10 min in a low speed, the absorbance values were recorded at 570 nm using an enzyme-linked immunometric meter (SpectraMax M5, USA).
Colony formation assay
The suspended cells ( cells/well) were seeded into the 6-well plates. After incubation overnight, OXA (10 μg/mL) and 5-Fu (20 μg/mL) was added and the cells were continuously cultured until the colonies were visible (14 d). After aspirating the supernatant, the cells were washed with PBS three times, fixed with 4% paraformaldehyde (PFA) for 20 min, and stained with crystal violet for 30 min. Finally, the staining reagent was washed away gently with PBS, and the plates were dried in air for photographs. The numbers of colonies containing more than 50 cells were recorded by microscopy Bx51 (Japan).
Apoptosis assay
To assess the cell apoptosis in vitro, the right amounts of cells were inoculated into the 6-well plates to ensure 70% confluence at the next day. PBS, OXA (10 μg/mL), and 5-Fu (20 μg/mL) were, respectively, added for stimulation for 24 h. After digested by trypsin without EDTA, centrifuged at 1000 g for 5 min, and washed with PBS twice, the cells in each group were resuspended in 100 μl dilute Binding Buffer (Deionized Water:Binding Buffer = 10:1), and 5 μl ANXA5/annexinV-FITC and 5 μl PI dye (Solarbio, CA1020) were successively added for 10-min incubation protecting from light. After being supplemented with 390 μl cold PBS, the cells were tested in the fluorescence-activated cell sorter (LSRFortessa X-20, USA).
TEM
For the observation of autophagyosomes, the suspended cells were collected, fixed with 2.5% glutaraldehyde at 4°C for 2 h followed by 0.1 M Na-cacodylate buffer (pH 7.4) washing thrice for 15 min, and post-fixed with 1% osmium tetroxide in 0.1 M Na-cacodylate buffer for 2 h at room temperature. For dehydration, the cells were soaked in a gradient (50%, 70%, 80%, 90%, 95%, and 100%) of ethyl alcohol and 100% acetone, respectively, for 15 min. Next, they were fixed with SPI-Pon 812 for 5–8 h after being permeabilized with SPI-Pon 812 epoxy resin monomer (SPI Supplies, 90,529–77-4) and acetone at a ratio of 1:1 for 4 h and a ratio of 1:2 overnight. The cells were then placed into the embedded plates containing SPI-Pon 812, incubated at 37°C overnight and at 68°C for 48 h. They were then stained using uranyl acetate and lead citrate for 15 min and dried at room temperature overnight. Finally, these were imaged by TEM HT7700 (Japan).
Confocal assay
GFP-mCherry-LC3 (GEMA, Shanghai, China) transfected cells ( cells) were inoculated in the confocal dish and then incubated overnight. The next day OXA (10 μg/mL) and 5-Fu (20 μg/mL) were, respectively, added into each dish for a 24-h stimulation. After discarding cell media and washing with PBS three times, the cells were added with 1 mL 4% PFA for fixation for 20 min, and then washed again with PBS three times. The images were obtained by confocal microscopy FV10i (Japan).
FISH
FISH was performed to identify the subcellular localization of EIF3J-DT. The experiment was conducted according to the instructions of RiboTM lncRNA FISH Probe Mix (Red) (RiboBio, C10920). The cover glasses were placed into a 24-well plate and MGC803 cells were inoculated at the density of cells/well. When reaching 80% confluence, the cells were washed by PBS and fixed with 1 mL 4% PFA at room temperature. After being treated with protease K, glycine and acetylation reagent, 250 μl prehybridization solution was added and the cells were incubated at 42°C for 1 h. Discard prehybridization solution, and add 250 μl hybridization solution containing EIF3J-DT probes for overnight hybridization at 42°C. Next day, the cells were washed by PBS supplemented with tween-20 three times, and stained with DAPI (Sigma-Aldrich, D8417) for 5 min. After being washed with PBS for 5 min three times, cells were photographed by confocal microscopy FV10i (Japan).
Luciferase reporter assay
The wild-type and mutant plasmids of ATG14 mRNA binding EIF3J-DT or MIR188-3p (ATG14 WT/MUT) were sub-cloned into a psiCHECK2 dual-luciferase plasmid and cotransfected with EIF3J-DT plasmid or MIR188-3p AgomiR; the wild-type and mutant plasmids of EIF3J-DT binding MIR188-3p (EIF3J-DT WT/MUT2) were sub-cloned into a psiCHECK2 dual-luciferase plasmid and cotransfected with MIR188-3p AgomiR, into the MGC803 or 293 T cells via Lipofectamine 2000. After 48-h transfection, luciferase activities were detected using the dual-luciferase reporter assay system (Promega, E2920).
RNA affinity-isolation assay
Biotin-labeled EIF3J-DT WT, EIF3J-DT MUT1, and negative control (Beads), and biotin-labeled EIF3J-DT WT, EIF3J-DT MUT2, and Beads, obtained from Thermo Fisher Scientific, were transfected into the MGC803 and MKN45 cells, respectively. Cell lysates were harvested 48 h after transfection and incubated with DynabeadsTM M-280 Streptavidin (Invitrogen, 11206D) for 3 h at 4°C according to the manufacturer’s protocol. Then, the beads were washed three times with ice-cold lysis buffer and once with high salt buffer (0.1% SDS, 1% Triton X-100 [Invitrogen, HFH10], 2 mM EDTA, 20 mM Tris-HCl, and 500 mM NaCl, pH 8.0). The bound RNAs were purified for the qPCR analysis.
RNA degradation assay
Twenty-four h after cell seeding, α-amanitin (Apexbio, A4548) and actinomycin D (Apexbio, A4452) were, respectively, treated at a final concentration of 50 mM and 10 μg/mL. Cells were collected at indicated times (0, 2, 4, 6 and 8 h after α-amanitin and actinomycin D treatment) and the expression of ATG14 and GAPDH mRNA were detected by qPCR.
Animal study
Animal experiments were carried out according to the policy of the animal welfare and care of Nanfang Hospital, and Nanfang Hospital Ethics Review Board approved the study. To construct the subcutaneous gastric tumor model, NC MGC803 and Sh-EIF3J-DT MGC803 cells were, respectively, injected into the left thighs of 4- to 5-week-old nude mice after being diluted with PBS to 100 μl (the mice were bought from the Animal Center of Southern Medical University, China). The drug was injected intraperitoneally (100 μl/d PBS for the control group, 0.1 mg/kg/d OXA, or 0.5 mg/kg/d 5-Fu for the treatment group) three times per week. Tumor nodule volumes were measured every three days using the formula: V = π × (d2 × D)/6, where d was the minor tumor axis and D was the long diameter, until day 27. After euthanatized on day 28, tumors were collected and weighted.
Bioluminescent in vivo imaging
On day 28 after the construction of the aforementioned subcutaneous gastric tumor model, bioluminescent in vivo imaging was performed to evaluate tumors. Briefly, the tumor-bearing mice were intraperitoneally injected with the VivoGloTM Luciferin (Promega, P1041) in vivo Grade solution (150 mg/kg or 10 μl/g). Bioluminescence positive signals were then photographed via in vivo imaging system (FX Pro, USA) within 10–12 min of injection.
ISH
ISH was performed to detect the expression of EIF3J-DT in paraffin-embedded tumor tissues collected from gastric cancer patients. The digoxigenin (DIG)-EIF3J-DT probes (probes 1–3) were purchased from Guangzhou MssBio, China. In brief, after being deparaffinized and blocked with 3% H2O2, slices were incubated with proteinase K (2 μg/mL) for 30 min at 37°C and prehybridized for 2 h at 37°C. Then, the slices were hybridized with DIG-EIF3J-DT probes overnight. After being washed for 10 min the next day, the slices were incubated with anti-DIG-horseradish peroxidase antibody for 1 h at room temperature. Staining was visualized using 3ʹ3-diaminobenzidine tetrahydrochloride. The slices were photographed by microscopy Bx51 (Japan) after being sealed by neutral balsam. Sequences of DIG-EIF3J-DT probes were listed in Table S5.
IHC
IHC was performed to detect the expression of ATG14 in paraffin-embedded tumor tissues collected from gastric cancer patients. In brief, paraffin-embedded slices (4 μm) were dried at 62°C for 30 min to 1 h. They were deparaffinized by xylene for 20 min twice and then soaked in 100%, 95%, 90%, 80%, and 70% ethyl alcohol for 3–5 min in order. After antigen retrieval at 120°C in citrate buffer (pH 6.0) for 5 min, the slices were cooled at room temperature. Being washed with PBS twice, incubated in 3% H2O2 and washed with PBS three times again, the slices were blocked in 5% bovine serum albumin for 1 h and incubated overnight at 4°C with primary antibodies against ATG14 (Cell Signaling Technology, 96,752). The next day, the slices were rewarmed for 1 h and washed for 10 min three times. Afterward, 50 μl secondary antibody was added to the slices for 1 h incubation at room temperature. Then color development was performed using 3ʹ3-diaminobenzidine tetrahydrochloride as a chromogen and the slices were counterstained using hematoxylin for 10 min. After staining, they were dehydrated by increasing concentrations of ethanol and xylene. The slices were photographed by microscopy Bx51 (Japan) after being sealed by neutral balsam.
Western blotting
Protein lysates from cultured cells were extracted in lysis buffer (KeyGEN BioTECH, KGP10100) and their concentration was measured with a BCA Protein Assay Kit (KeyGEN BioTECH, KGP902). Equal amounts of denatured protein lysates were subjected to SDS-PAGE electrophoresis, transferred to PVDF membranes (Millipore, IPFL00010), and blocked in 5% bovine serum albumin for 1 h at room temperature. Next, membranes were incubated overnight at 4°C with primary antibodies against LC3 (Novus, NB600-1384), SQSTM1 (Cell Signaling Technology, 16,177), C-PARP (Cell Signaling Technology, 9185), PARP (Cell Signaling Technology, 9532), C-CASP3 (Cell Signaling Technology, 9664), CASP3/caspase 3 (Cell Signaling Technology, 9662), C-CASP7 (Cell Signaling Technology, 8438), CASP7/caspase 7 (Cell Signaling Technology, 12,827), and ATG14 (Cell Signaling Technology, 96,752). After rewarming for 1 h at room temperature and washing with TBS-Tween 20 (0.1%) (0.1 M TBS: 3 g Tris, 0.2 g KCl, 8 g NaCl, pH 7.4) for 10 min three times, the membranes were incubated with IRDye® 680RD goat anti-mouse (Li-Cor Bioscience, P/N 926–68,070) or IRDye® 800CW goat anti-rabbit (Li-Cor Bioscience, P/N 926–32,211) secondary antibodies for 1 h at room temperature. They were then washed for 10 min three times. Bands were visualized on Odyssey Infrared Imaging System (Li-Cor Bioscience, USA). The antibody against GAPDH (Proteintech, 60,004-1-Ig) was used as a loading control.
qPCR
The total RNA was extracted from cultured cells using RNAiso Plus reagent (TaKaRa, 9108) according to manufacturer’s instruction, and then cDNA was synthesized using PrimeScriptTM RT Master Mix (TaKaRa, RR036A). The amplification of cDNA was performed in 10 μl reactions on LightCycler 480 system (Roche Diagnostics, USA) via SYBR Premix Ex Taq II (TaKaRa, RR820A). The expression of MIR188-3p was quantified by TaqMan miRNA assays (Applied Biosystems, 4,440,885). The primers used in our experiments are listed in Table S6. The mean cycle threshold (Ct) was determined by triplicate PCR runs, and the relative expression was normalized to that of internal control (GAPDH or RNU6) via the 2–ΔΔCt method.
Bioinformatics
The correlation between the expression of EIF3J-DT and various autophagy related genes was analyzed by TCGA Nature 2014 and TCGA Provisional Databases. The expression of EIF3J-DT in melanoma, prostate cancer and gastric cancer was analyzed by GEO databases. BLAST was used to predict the potential binding sites between EIF3J-DT and ATG14 mRNA. RegRNA2.0 and Targetscan were used to predict the potential MIR188-3p targets on EIF3J-DT and ATG14 mRNA. Furthermore, the expression of EIF3J-DT and ATG14 and their prognosis roles in gastric cancer were confirmed by Kaplan-Meier plotter analysis. All the databases are listed in Table S7.
Statistical analysis
Sample sizes were denoted in the figure legends. All experiments were performed in triplicate. Samples included in the analyses surely met proper experimental conditions. Results were quantified as mean ± SEM. All statistical analyses were conducted with SPSS v20.0 software (Abbott Laboratories, USA) and GraphPad Prism software 6.0 (GraphPad Software, USA), and performed using the Student’s t test, one-way ANOVA, or a Mann-Whitney U test for nonparametric data. Kaplan-Meier plotter analysis were used to estimate the prognostic relevance of EIF3J-DT and ATG14 in univariate analysis. Two-tailed Spearman’s correlation analysis was used to analyze the associations between EIF3J-DT and ATG14 mRNA expression. P < 0.05 was considered statistically significant.
Supplementary Material
Funding Statement
This work was supported by Guangzhou Municipal Science and Technology Project [201803010070]; National Natural Science Foundation of China [81811530024]; National Natural Science Foundation of China [81772580].
Acknowledgments
This work was supported by Guangzhou Municipal Science and Technology Project (No. 201,803,010,070, WL), and the National Natural Science Foundation of China (No. 81,772,580, WL; No. 81,811,530,024, WL).
Disclosure statement
The authors declare no competing interests.
Supplemental material
Supplemental data for this article can be accessed here.
References
- [1].Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond). 2019;39(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chandra R, Balachandar N, Wang S, et al. The changing face of gastric cancer: epidemiologic trends and advances in novel therapies. Cancer Gene Ther. 2020. DOI: 10.1038/s41417-020-00234-z. [DOI] [PubMed] [Google Scholar]
- [3].Shen L, Shan Y-S, Hu H-M, et al. Management of gastric cancer in Asia: resource-stratified guidelines. Lancet Oncol. 2013;14(12):e535–47. [DOI] [PubMed] [Google Scholar]
- [4].Ruan T, Liu W, Tao K, et al. A review of research progress in multidrug-resistance mechanisms in gastric cancer. Onco Targets Ther. 2020;13:1797–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Singletary K, Milner J.. Diet, autophagy, and cancer: a review. Cancer Epidemiol Biomarkers Prev. 2008;17(7):1596–1610. [DOI] [PubMed] [Google Scholar]
- [6].Ouyang L, Shi Z, Zhao S, et al. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45(6):487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kumar P, Zhang D-M, Degenhardt K, et al. Autophagy and transporter-based multi-drug resistance. Cells. 2012;1(3):558–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wu Q, Yang Z, Nie Y, et al. Multi-drug resistance in cancer chemotherapeutics: mechanisms and lab approaches. Cancer Lett. 2014;347(2):159–166. [DOI] [PubMed] [Google Scholar]
- [9].Qadir MA, Kwok B, Dragowska WH, et al. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112(3):389–403. [DOI] [PubMed] [Google Scholar]
- [10].Cook KL, Shajahan AN, Clarke R.. Autophagy and endocrine resistance in breast cancer. Expert Rev Anticancer Ther. 2011;11(8):1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].He J, Yu -J-J, Xu Q, et al. Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy. 2015;11(2):373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jiang S, Cheng S-J, Ren L-C, et al. An expanded landscape of human long noncoding RNA. Nucleic Acids Res. 2019;47(15):7842–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang G, Zhang Z-J, Jian W-G, et al. Novel long noncoding RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell renal cell carcinoma proliferation via the Wnt/β-catenin signaling pathway. Mol Cancer. 2019;18(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhuo W, Liu Y, Li S, et al. Long Noncoding RNA GMAN, up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of ephrin A1 by competitively binding GMAN-AS. Gastroenterology. 2019;156(3):676–691.e11. [DOI] [PubMed] [Google Scholar]
- [15].Shin VY, Chen J, Cheuk IW-Y, et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10(4):270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Huang H, Chen J, Ding C-M, et al. LncRNA NR2F1-AS1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC1 via miR-363. J Cell Mol Med. 2018;22(6):3238–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Qu Y, Tan H-Y, Chan Y-T, et al. The functional role of long noncoding RNA in resistance to anticancer treatment. Ther Adv Med Oncol. 2020;12:1758835920927850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jian X, Xiao-yan Z, Bin H, et al. MiR-204 regulate cardiomyocyte autophagy induced by hypoxia-reoxygenation through LC3-II. Int J Cardiol. 2011;148(1):110–112. [DOI] [PubMed] [Google Scholar]
- [19].Pellegrini P, Strambi A, Zipoli C, et al. Acidic extracellular pH neutralizes the autophagy-inhibiting activity of chloroquine: implications for cancer therapies. Autophagy. 2014;10(4):562–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li Y, Wu Z, Yuan J, et al. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31–44. [DOI] [PubMed] [Google Scholar]
- [21].YiRen H, YingCong Y, Sunwu Y, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Szasz AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gray C, Campbell K, Regional Chemotherapy for the Treatment of Breast Cancer: a Review of Clinical Effectiveness, Cost-Effectiveness and Guidelines. CADTH Rapid Response Reports. 2018, Ottawa (ON): Canadian Agency for Drugs and Technologies in Health. [PubMed] [Google Scholar]
- [24].Koumenis C, Giaccia A. Transformed cells require continuous activity of RNA polymerase II to resist oncogene-induced apoptosis. Mol Cell Biol. 1997;17(12):7306–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Frenzel A, Grespi F, Chmelewskij W, et al. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis. 2009;14(4):584–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].D’Orsi B, Mateyka J, Prehn J. Control of mitochondrial physiology and cell death by the Bcl-2 family proteins Bax and Bok. Neurochem Int. 2017;109:162–170. [DOI] [PubMed] [Google Scholar]
- [27].Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang FF, Luo YH, Wang H, et al. Metastasis-associated long noncoding RNAs in gastrointestinal cancer: implications for novel biomarkers and therapeutic targets. World J Gastroenterol. 2016;22(39):8735–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen L, Zhou Y, Li H. LncRNA, miRNA and lncRNA-miRNA interaction in viral infection. Virus Res. 2018;257:25–32. [DOI] [PubMed] [Google Scholar]
- [30].Panzarini E, Dini L. Nanomaterial-induced autophagy: a new reversal MDR tool in cancer therapy? Mol Pharm. 2014;11(8):2527–2538. [DOI] [PubMed] [Google Scholar]
- [31].Yu Y, Nangia-Makker P, Farhana L, et al. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol Cancer. 2017;16(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liang B, Liu X, Liu Y, et al. Inhibition of autophagy sensitizes MDR-phenotype ovarian cancer SKVCR cells to chemotherapy. Biomed Pharmacother. 2016;82:98–105. [DOI] [PubMed] [Google Scholar]
- [33].Wang K, Liu C-Y, Zhou L-Y, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6(1):6779. [DOI] [PubMed] [Google Scholar]
- [34].Shi W, Zhang C, Ning Z, et al. Long non-coding RNA LINC00346 promotes pancreatic cancer growth and gemcitabine resistance by sponging miR-188-3p to derepress BRD4 expression. J Exp Clin Cancer Res. 2019;38(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu D, Lu M, Li J, et al. The patterns and timing of recurrence after curative resection for gastric cancer in China. World J Surg Oncol. 2016;14(1):305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chon SH, Berlth F, Plum PS, et al. Gastric cancer treatment in the world: germany. Transl Gastroenterol Hepatol. 2017;2(5):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yang Y, Yin X, Sheng L, et al. Perioperative chemotherapy more of a benefit for overall survival than adjuvant chemotherapy for operable gastric cancer: an updated Meta-analysis. Sci Rep. 2015;5(1):12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Russi S, Verma HK, Laurino S, et al. Adapting and Surviving: intra and Extra-Cellular Remodeling in Drug-Resistant Gastric Cancer Cells. Int J Mol Sci. 2019;20(15):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer. 2016;139(2):269–280. [DOI] [PubMed] [Google Scholar]
- [40].Zhang F, Li Y, Xu W, et al. Long non-coding RNA ZFAS1 regulates the malignant progression of gastric cancer via the microRNA-200b-3p/Wnt1 axis. Biosci Biotechnol Biochem. 2019;83(7):1289–1299. [DOI] [PubMed] [Google Scholar]
- [41].Liu XH, Sun M, Nie F-Q, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wei Y, Liu Z, Fang J. H19 functions as a competing endogenous RNA to regulate human epidermal growth factor receptor expression by sequestering let7c in gastric cancer. Mol Med Rep. 2018;17(2):2600–2606. [DOI] [PubMed] [Google Scholar]
- [43].Xi Z, Si J, Nan J. LncRNA MALAT1 potentiates autophagyassociated cisplatin resistance by regulating the microRNA30b/autophagyrelated gene 5 axis in gastric cancer. Int J Oncol. 2019;54(1):239–248. [DOI] [PubMed] [Google Scholar]
- [44].Liu D, Zhang H, Cong J, et al. H3K27 acetylation-induced lncRNA EIF3J-AS1 improved proliferation and impeded apoptosis of colorectal cancer through miR-3163/YAP1 axis. J Cell Biochem. 2020;121(2):1923–1933. [DOI] [PubMed] [Google Scholar]
- [45].Yang X, Yao B, Niu Y, et al. Hypoxia-induced lncRNA EIF3J-AS1 accelerates hepatocellular carcinoma progression via targeting miR-122-5p/CTNND2 axis. Biochem Biophys Res Commun. 2019;518(2):239–245. [DOI] [PubMed] [Google Scholar]
- [46].Guo JY, Chen H-Y, Mathew R, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25(5):460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].White E. The role for autophagy in cancer. J Clin Invest. 2015;125(1):42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lefort S, Joffre C, Kieffer Y, et al. Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers. Autophagy. 2014;10(12):2122–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Corazzari M, Rapino F, Ciccosanti F, et al. Oncogenic BRAF induces chronic ER stress condition resulting in increased basal autophagy and apoptotic resistance of cutaneous melanoma. Cell Death Differ. 2015;22(6):946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang J, Xie S, Yang J, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol. 2019;12(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chen G, Guo Z, Liu M, et al. Clinical Value of Capecitabine-Based Combination Adjuvant Chemotherapy in Early Breast Cancer: a Meta-Analysis of Randomized Controlled Trials. Oncol Res. 2017;25(9):1567–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Obara K, Ohsumi Y. Atg14: a key player in orchestrating autophagy. Int J Cell Biol. 2011;2011:713435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fogel AI, Dlouhy BJ, Wang C, et al. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol. 2013;33(18):3675–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Diao J, Liu R, Rong Y, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520(7548):563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhang Z, Jia H, Gu T, et al. RNA sequencing and bioinformatics analysis of the long noncoding RNA-mRNA network in colorectal cancer. J Cell Biochem. 2018;119(12):9957–9966. [DOI] [PubMed] [Google Scholar]
- [56].Hao S, Yao L, Huang J, et al. Genome-Wide Analysis Identified a Number of Dysregulated Long Noncoding RNA (lncRNA) in Human Pancreatic Ductal Adenocarcinoma. Technol Cancer Res Treat. 2018;17:1533034617748429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mitobe Y, Ikeda K, Suzuki T, et al. ESR1-Stabilizing Long Noncoding RNA TMPO-AS1 Promotes Hormone-Refractory Breast Cancer Progression. Mol Cell Biol. 2019;39(23):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3ʹ UTRs via Alu elements. Nature. 2011;470(7333):284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lee S, Kopp F, Chang T-C, et al. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164(1–2):69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Liu F, Chen N, Gong Y, et al. The long non-coding RNA NEAT1 enhances epithelial-to-mesenchymal transition and chemoresistance via the miR-34a/c-Met axis in renal cell carcinoma. Oncotarget. 2017;8(38):62927–62938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tan D, Chong FT, Leong HS, et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med. 2017;23(10):1167–1175. [DOI] [PubMed] [Google Scholar]
- [65].Ideue T, Hino K, Kitao S, et al. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15(8):1578–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30(1):34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


