ABSTRACT
Non-fasting lipidemia (nFL), mainly contributed by postprandial lipidemia (PL), has recently been recognized as an important cardiovascular disease (CVD) risk as fasting lipidemia (FL). PL serves as a common feature of dyslipidemia in Type 2 Diabetes (T2D), albeit effective therapies targeting on PL were limited. In this study, we aimed to evaluate whether the therapy combining probiotics (Prob) and berberine (BBR), a proven antidiabetic and hypolipidemic regimen via altering gut microbiome, could effectively reduce PL in T2D and to explore the underlying mechanism. Blood PL (120 min after taking 100 g standard carbohydrate meal) was examined in 365 participants with T2D from the Probiotics and BBR on the Efficacy and Change of Gut Microbiota in Patients with Newly Diagnosed Type 2 Diabetes (PREMOTE study), a random, placebo-controlled, and multicenter clinical trial. Prob+BBR was superior to BBR or Prob alone in improving postprandial total cholesterol (pTC) and low-density lipoprotein cholesterol (pLDLc) levels with decrement of multiple species of postprandial lipidomic metabolites after 3 months follow-up. This effect was linked to the changes of fecal Bifidobacterium breve level responding to BBR alone or Prob+BBR treatment. Four fadD genes encoding long-chain acyl-CoA synthetase were identified in the genome of this B. breve strain, and transcriptionally activated by BBR. In vitro BBR treatment further decreased the concentration of FFA in the culture medium of B. breve compared to vehicle. Thus, the activation of fadD by BBR could enhance FFA import and mobilization in B. breve and diliminish the intraluminal lipids for absorption to mediate the effect of Prob+BBR on PL. Our study confirmed that BBR and Prob (B. breve) could exert a synergistic hypolipidemic effect on PL, acting as a gut lipid sink to achieve better lipidemia and CVD risk control in T2D.
KEYWORDS: Type 2 diabetes, probiotics, berberine, dyslipidemia, postprandial lipidemia, gut microbiome
Introduction
Hyperlipidemia is a major risk factor for atherosclerotic cardiovascular diseases (ASCVDs),1 particularly when combined with hyperglycemia and Type 2 Diabetes (T2D).2–4 Current diagnose criteria and treatment target are based on evaluating fasting lipidemia (FL). However, increasing evidence has supported that a high level of non-fasting lipidemia (nFL), mainly constituted by postprandial lipidemia (PL), is also an important CVD risk factor5–8 and multiple countries are currently changing their guidelines toward a consensus on measuring a lipid profile for cardiovascular risk prediction in the non-fasting state.7–11 Individuals are mainly in a non-fasting state during a regular 24-hour cycle, and non-fasting samples would simplify blood sampling and minimize the risk of hypoglycemia particularly for individuals with diabetes.12 Furthermore, postprandial hyperlipidemia is a common feature of insulin-resistant diabetes patients13–15 and has been recommended for evaluating T2D-related ASCVDs.16,17 Therefore, managing both FL and PL in T2D should be required to achieve better control for overall lipidemia and CVD risks, whereas, except of Niemann-Pick C1-like 1 (NPC1L1) inhibitor (Ezetimibe),18 there is few regimens developed for targeting PL or both.
Unlike FL that are mainly derived from liver-derived lipoproteins, PL alterations are constituted by intestinal lipid absorption, lipoprotein secretion and chylomicron production,15 and recently have been found to be tightly related with gut microbiota alterations,19 indicating that a different avenueto develop druggable targets for PL from that for FL. The intestinal microbiota is involved in host intestinal lipid absorption and lipid metabolism regulations in other metabolic organs contributing to host lipidomic profile alterations.20–28 However, in contrast to the piled-up evidence of associations, the intricate crosstalk between the microbiota and host circulation lipidomic alterations is far from fully elucidated.
Berberine (BBR), a plant alkaloid extracted from the Chinese herbal medicine Coptis chinensis (Huanglian), is known to elevate liver LDL uptake and adipose browning.29–33 Recently the impact of BBR on gut microbiota has been recognized and linked to its effect on metabolic disorders. Probiotics, such as strains from Bifidobacterium, improve dyslipidemia, via cholesterol binding, host intestinal absorption blocking, or altering host bile acid signaling.34–36 Both BBR and probiotics are defined as nutraceutical hypolipidemic agents, considering their effects on lowering FL.37–43 It is unknown if BBR or probiotics also lowering the nFL, whereastheir effects on gut microbiome suggested that they might be suitable candidate measures for treating PL. In “Probiotics and BBR on the Efficacy and Change of Gut Microbiota in Patients with Newly Diagnosed Type 2 Diabetes (PREMOTE) trial,”33 we confirmed that the antidiabetic effect of BBR could be mediated by its effect on gut microbial bile acid metabolism, and supplementation with probiotics can improve the hypoglycemic effect of BBR in participants older than 50. However, probiotics cannot improve the effect of BBR on lowering fasting lipidemia either in the whole cohort or in aged subgroup. This prompted us to ask how the combination treatment of BBR and probiotics, or either one could exert benefit on lowering PL, and whether their impact on gut microbiota could contribute to this effect .
To achieve this aim, in this study we compared lipidemia in postprandial blood samples collected from T2D patients assigned to the four treatment groups in the PREMOTE trial: placebo (Plac), probiotics alone (Prob), berberine alone (BBR), and probiotics combined with berberine (Prob+BBR). We further explored the potential underlying mechanism via multi-omics and comparative genomic analysis with in vitro culture experimental verification.
Methods
Clinical study
The PREMOTE study, a randomized, double-blind, placebo-controlled clinical trial in 20 medical centers in China (ClinicalTrials.gov number, NCT02861261), enrolled newly diagnosed T2D patients from August 18, 2016, to July 18, 2017. The PREMOTE trial evaluated glycemic control as the primary outcome and lipidaemia control as the secondary outcome.33 The primary outcome and its related microbiota mechanism have been previously published, showing that the combined treatment of BBR and probiotics showed superior hypoglycemic effect to BBR or probiotics alone in subjects aged ≥50 years. BBR alone and Prob+BBR showed similar effects in reducing fasting lipidemia.
This lipidomic study included 365 of the 409 enrolled participants from the PREMOTE trial based on the availability of postprandial lipid measurements before and after the 3-month intervention. Metagenomic sequencing data for the 1,192 fecal samples can be accessed from the National Center for Biotechnology Information BioProject Database with the dataset accession number PRJNA643353.
The detailed inclusion and exclusion criteria are listed in the online protocol and the previous study.33 In brief, drug-naïve participants were newly diagnosed with T2D according to the 1999 World Health Organization criteria, and included patients of both sexes, aged between 20 and 70 years, with a body mass index (BMI) between 19.0 and 35.0 kg/m2. All enrolled participants had HbA1c ≥6.5% and ≤10.0% and fasting plasma glucose ≥7.0 mmol L−1 (126.1 mg/dl) and ≤13.3 mmol L−1(239.6 mg/dl). Patients were excluded from the study if they had severe liver dysfunction, impaired renal function, severe organic heart diseases or heart failure (New York Heart Association class (NYHA) grade of heart function ≥ III), psychiatric disease, severe infection, severe anemia, neutropenia, or history of acute diabetic complications and are allergic to gentamicin, other amino glycoside antibiotics, berberine, or other probiotics.
All participants provided written informed consent. The study was approved by each institution’s human participant ethics committee at each participating center. The participants were randomly assigned into one of the following four groups in a 1:1:1:1 ratio as follows: BBR (0.6 g per 6 pills, twice daily before a meal) plus probiotics (4 g per 2 strips of powder, once daily at bedtime) (Prob+BBR), probiotics plus placebo (Prob), BBR plus placebo (BBR), or placebo plus placebo (Plac). Treatments were administered for 12 weeks, and patients visited the center every 4 weeks until the end of the study. The stratified randomization was achieved by utilizing a validated Interactive Web-based Response System (IWRS) as reported previously. The study personnel and participants were blinded to the assignment of treatment groups. BBR was manufactured by Northeast Pharmaceutical Group Co., Ltd., Shenyang, Liaoning, China. The multi-strain probiotic products containing nine proprietary strains of probiotics were produced by Shanghai Jiaoda Onlly Co., Ltd., Shanghai, China (Bifidobacterium longum CGMCC No. 2107; Bifidobacterium breve CGMCC No. 6402; Lactococcus gasseri CGMCC No. 10758; Lactobacillus rhamnosus CNCM I-4474; Lactobacillus salivariusCGMCC No. 6403; Lactobacillus crispatus CGMCC No. 6406; Lactobacillus plantarum; CGMCC No. 1258; Lactobacillus fermentum CGMCC No. 6407; and Lactobacillus caseiCNCM I-4458), and each sachet contains ≥50 billion CFU of live, freeze-dried bacteria. The placebos were provided along with the medications, details have been published in previous study.33
Dyslipidemia was defined according to the US National Cholesterol Education Program/Adult Treatment Panel III (NCEP/ATP III) criteria as total fasting levels of cholesterol ≥240 mg/dl, triglycerides ≥200 mg/dl, HDL-c ≤ 40 mg/dl, LDL-c ≥ 160 mg/dl and/or taking lipid-lowering medications,44and the others were defined as eulipidemia.
Postprandial lipid measures
In this study, we measured lipidemia of postprandial blood samples, which were drawn 120 min after taking 100 g standard carbohydrate meal provided by China Food Limited, COFCO (Beijing, China).45 The PL measurements were performed in the central laboratory of Ruijin Hospital. Postprandial plasma total cholesterol (pTC), triglycerides (pTGs), high-density lipoprotein cholesterol (pHDLc), and low-density lipoprotein cholesterol (pLDLc) were measured by the cholesterol oxidase method, glycerophosphate oxidase-peroxidase method, polyanion polymer/detergent method and solubilization method with an autoanalyzer (AU5800; Beckman Coulter, CA, USA). Other biochemical measures were taken as in our previous study.33
Metabolomic measures
1. Materials and Reagents
The organic solvents used in this experiment, including acetonitrile and methanol, were purchased from Merck (Darmstadt, Germany) at high-performance liquid chromatography (HPLC) grade. Formic acid (HPLC grade) was obtained from Sigma-Aldrich (St. Louis, MO, USA), and ammonium bicarbonate was obtained from Fluka (CH, Buchs, Switzerland) by liquid chromatography–mass spectrometry (LC-MS). Ultrapure water was obtained from a Milli-Q water system (Millipore, Billerica, USA).
2. Sample preparation
A 100 μL serum sample was mixed with 400 μL extraction solvent in 2 mL centrifuge tubes. This extraction solvent was made with methanol containing 0.1 µg/mL carnitine C2:0-d3, 0.1 µg/mL carnitine C10:0-d3, 0.15 µg/mL carnitine C16:0-d3, 0.75 µg/mL LPC 19:0, 2.5 µg/mL Fatty acid (FFA) C16:0-d3, 2.5 µg/mL FFA C18:0-d3, 4.25 µg/mL tryptophan-d5, 3.6 µg/mL phenylalanine-d5, 0.75 µg/mL SM (d18:1/12:0), 2 µg/mL choline-d4, and 0.1 µg/mL. After vertexing and centrifugation, two parts of 180 μL supernatant were freeze-dried and then stored at −80°C. Before analysis, 50 μL acetonitrile/water (1:4) solvent was added to each sample for reconstitution.
To assess the stability of the analysis process, quality control (QC) samples from the mixture of equal volumes of all serum samples were prepared using the same method as the serum samples and analyzed once after every ten serum samples.
3. Non-targeted LC-MS Methods
As described previously,46–48 a Vanquish UPLC-Q Exactive instrument (Thermo Fisher Scientific, Rockford, IL, USA) was used for LC-MS analysis.
In positive mode, a Waters BEH C8 column (50 mm × 2.1 mm, 1.7 μm, Waters, Milford, MA) was used for LC separation. The oven temperature was 60°C, and the flow rate was 0.4 mL/min. The autosampler temperature was set at 10°C, and the injection volume was 5 μL. Phase A was ultrapure water with 0.1% formic acid, and phase B was acetonitrile with 0.1% formic acid. The gradient programme was started from 5% B and maintained for 0.5 min, then increased to 40% B for 1.5 min, continued to 100% B for 6 min, maintained at 100% B for 2 min, returned to 5% B for 0.1 min and equilibrated for 2.5 min.
In negative mode, LC separation was achieved by an ACQUITY UPLC HSS T3 column (100 mm × 2.1 mm, 1.8 μm, Waters, Milford, MA). The oven temperature was 50°C, and the flow rate was 0.35 mL/min. Phase A was ultrapure water with 6.5 mM ammonium bicarbonate, and phase B was 95% methanol/water solvent with 6.5 mM ammonium bicarbonate. The gradient program was started from 0% B and maintained for 1 min, increased to 40% B for 2 min, further increased to 100% B for 13 min, maintained at 100% B for 5 min, returned to 0% B for 0.1 min and equilibrated for 2.9 min.
A 7.0E4 resolution MS full scan mode with a scan range of m/z 70–1050 was applied for analysis. The spray voltage was 3.5 kV for positive mode and 3.00 kV for negative mode. The capillary temperature was 300°C, and the aux gas heater temperature was 350°C. The sheath gas and aux gas were 45 and 10 (in arbitrary units).
4. Data processing
Nontargeted LC-MS data from multiple runs were extracted and aligned by TraceFinder 3.2 (Thermo Fisher Scientific, USA). The intensity of each retained peak was normalized using one of the internal standards. An intra-laboratory database including approximately 2000 metabolites was used to identify the metabolites in nontargeted metabolic profiling by retention time and MS1 and MS2 information.49 A total of 157 lipid-related metabolites were identified (details in Data Set 1 and Date Set 2).
QC samples were used to evaluate the reproducibility of the metabolomics analysis with the use of internal standard calibration (Figure S1). Furthermore, the reproducibility of the metabolite ions was also evaluated with relative standard deviation (RSD%) in the 125 QC samples: among the 157 identified metabolite ions, 98.7% of ions had an RSD% less than 30% (Figure S2).
Metagenomics study
1. Metagenomics sequencing
High-quality non-human metagenomic data (100 bp paired-end reads, BGISEQ-500 platform) and microbial profiling at the species and Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) level of all samples in this project were obtained from our previous study of the PREMOTE trial.33 Differentially enriched KEGG pathways between groups were identified according to the reporter Z scores of all detected KOs in the given pathway.50
2. Genome sequencing and de novo assembly of the genomes of the ingested probiotic strains
Bacterial genomic DNA was sheared randomly to construct three read libraries with lengths of 250 bp by a Bioruptor ultrasonicator (Diagenode, Denville, NJ, USA) and physicochemical methods. The paired-end fragment libraries with an insert size of 270 bp were sequenced according to the Illumina HiSeq 4000 system protocol at the Beijing Genomics Institute (Shenzhen, China). Low quality raw reads (>40% of the bases with Q value ≤20 or containing >10% ambiguous bases) were discarded. The remaining high-quality reads were assembled to form scaffolds using SOAP denovo v1.05 software. A summary of the genome assembly statistics for each genome is provided in Table S1.
3. Annotation of genes coding enzymes involved in lipid metabolism
The 9 assembled genomes of ingested probiotic strains contained in the probiotic formula and 1,520 high-quality genomes from cultivated human gut bacteria51 were functionally annotated to identify genes encoding lipid metabolism-related enzymes. All coding genes were translated into amino acid sequences to run BLASTX against the KEGG (version 76) pathway database (http://www.genome.jp/kegg) at an E-value threshold of 1.0E-5 and an identity threshold of 60%. All annotations were based on the best BLASTX hits. Among the 9 probiotic genomes, a total of 73 genes were annotated as functional genes encoding lipid metabolism-related enzymes, and most of the assigned sequences (94.5%, 69/73) had at least 90% sequence identity to reference sequences (Data Set 3).
In vitro growth experiment of B. breve and E. coli
The B. breve 6402 was derived from the China General Microbiological Culture Collection Center and cultured in a strain‐specific medium (seen in Table S2). The E. coli MG1655 was cultured in lysogeny broth medium (Sangon Biotech, Shanghai, China). Both strains were incubated in an anaerobic chamber (Whitley A35 anaerobic workstation, Don Whitley Scientific, UK) with 5% hydrogen, 10% carbon dioxide, and 85% nitrogen at 37°C. For the growth curve experiment, we seeded B. breve 6402 and E. coli MG1655 at 10% and 5%, respectively, in a volume of 5 ml media with different concentrations of BBR (0, 1.56, 3.125, 6.25, 12.5, and 25 μg ml−1) and cultured for 14 hours. The OD600 of the bacterial culture was measured every 2 hours with a plate reader (VarioskanFlash, Thermo Scientific, MA, USA).
For in vitro assaying bacterial fadD expression and non-esterified fatty acids (NEFAs) consumption, B. breve 6402 were seeded at 10% in media and cultured for 10 h, then treated with BBR (6.25 μg ml−1) for 4 h in either blank or LA (linolenic acid, at final concentration of 1 mg ml−1) containing conditioned media. The NEFAs content of the media was measured with colorimetric assays (LabAssay NEFA, Wako, Japan). Stock solutions of LA (#60-33-3, MedChemExpress NJ, USA) were prepared at 10 mg ml−1with 2% (w/v) Tween 80 after homogenization by vertexing 2100 rpm during 150 s (separated with three intervals of 30 s).
Real-time quantitative RT-PCR
RT-PCR was used to determine the relative levels of gene transcription in B. breve. Total B. breve RNA was extracted using an Eastep Super Total RNA Extraction Kit (Promega, Shanghai, China) and reverse transcribed to cDNA with a Reverse Transcription System Kit (Promega, Madison, USA) according to the manufacturer’s protocols. Real-time PCR amplification and detection were performed using SYBR Green II Master Mix (TaKaRa, Kusatsu, Japan) on a LightCycler 480 (Roche Applied Science, Indianapolis, USA). The sequences of primers used for four fadD genes are listed in Table S3. The gene expression levels were calculated and normalized to the levels of 16S rRNA.
Statistical analysis
Statistical analyses of clinical data were performed using SAS version 9.4 (SAS Institute, Cary, NC, USA). At baseline, analysis of variance (ANOVA) for continuous variables and a Chi-square test for categorical variables were performed for comparisons of the demographic and clinical characteristics of the treatment group. For lipidaemia analyses, the overall differences among treatment groups were compared with the use of a global test of unordered groups. If the difference was significant at a P value of <0.05, then pairwise comparisons were made with adjustment for multiple comparisons, statistical significance was defined as adjusted P < .05 after adjustment for multiple comparisons with Tukey correction, and ANCOVA was also used with stratified randomization factors as a covariate. The 95% confidence interval (CI) was constructed with the use of least-squares (LS) means. We also performed a sensitivity analysis for participants without hypolipidemic medication (Table S6).
Statistical analyses of metagenomic and metabolomic data were performed using R software (version 3.6.3; R Foundation for Statistical Computing). Kruskal–Wallis (KW) tests were applied to detect differences in the lipid metabolites among the four groups at baseline. Canonical analysis of principal coordinates (CAP) based on Bray–Curtis distance was conducted using alterations of lipid metabolites after treatment to determine the differences among the four treatment groups (capscale function, vegan package). Wilcoxon signed-rank tests were applied to detect differences in lipid metabolite levels between baseline and post-treatment measurements for each treatment group.
Partial least squares-discriminant analysis (PLS-DA) was performed to construct the variable importance of projection (VIP) scores. VIP scores ≥1 were considered significant for lipid metabolites altered by treatment (plsda and vip function, mixOmics package).52 Multivariate generalized estimating equation (GEE) model analysis53,54 was performed to assess the longitudinal associations between the two time-point measurements of lipid metabolites and clinical parameters or species abundances after adjustment for age, sex and BMI (geem function, geeM package), and the P value of each regression coefficient (β) was calculated. The Wilcoxon matched-pairs signed-rank tests were applied to detect differences in RA of gut microbial species between baseline and posttreatment measurements. The correlations between the RAs of microbial species and clinical parameters in the Prob+BBR treatment groups were assessed by partial Spearman’s correlation analysis after adjustment for age, sex and BMI (pcor function, ppcor package). The differences in RA of microbial species between participants with dyslipidemia and lipidaemia at baseline were assessed by using KW tests. The adjusted P-value (q) was calculated with Benjamini–Hochberg (BH) method to correct the multiple comparisons and correlations of the lipid metabolite levels and gut microbial species (p.adjust function, stats package). Two-way analysis of variance (ANOVA) and unpaired Student’s t-test were used to compare the growth curves of the in vitro culture experiment.
Results
Prob+BBR combined treatment significantly improves PL
Our previous study has shown that BBR and Prob+BBR exert similar effects in reducing fasting lipidemia,33 we thus sought to investigate how the two treatments affect PL. The baseline characteristics of postprandial plasma samples from 365 T2D participants in the PREMOTE study (NCT02861261), showed no significant difference in PL among the four treatment groups (Table 1). At the end of the follow-up period, participants in the Prob+BBR group had a greater reduction in pTC and pLDLc from baseline to week 13 than those in the Plac group (LS mean 95% [CI], −24.29 [−29.95, −18.64] vs −8.66 [−14.52, −2.79] mg/dl, P = .001 in pTC, and −16.54 [−21.30, −11.79] vs −7.35 [−12.29, −2.42] mg/dl, P = .043 in pLDLc, respectively, ANOVA, Table 2, Table S4). However, compared to the Plac group, neither the BBR alone group (pTC, P = .14 vs Plac; pLDLc, P = .91 vs Plac) nor the Prob alone group (pTC, P = .33 vs Plac; pLDLc, P = .34 vs Plac) showed significant changes in postprandial cholesterols. Furthermore, controlling for stratified randomization factors similar results were shown (Model 2, Table 2). No additional benefit in improving pTG was found in Prob+BBR group compared to BBR (Table 2, Table S5). Of note, the effects of Prob+BBR treatment in improving PL were still significant when those taking hypolipidemic medications were excluded (n = 360, see Methods, Table S6). Thus, our study demonstrated that BBR alone was effective in reducing fasting levels but not in postprandial levels of cholesterols, the latter of which might require a synergistic effect with probiotics.
Table 1.
Baseline characteristics of participants (n = 365)
| Plac (n = 91) | Prob (n = 92) | BBR (n = 84) | Prob+BBR (n = 98) | P value | |||
|---|---|---|---|---|---|---|---|
| Age, y | 52.56 ± 9.44 | 52.11 ± 8.74 | 52.07 ± 10.81 | 52.9 ± 9.1 | 0.92 | ||
| Male sex (%) | 53 (54.08) | 58 (63.04) | 51 (60.71) | 52 (57.14) | 0.61 | ||
| Body weight, kg | 72.04 ± 12.18 | 71.72 ± 11.45 | 71.62 ± 13.1 | 70.63 ± 11.15 | 0.86 | ||
| Waist circumference, cm | 91.9 ± 8.98 | 91.34 ± 8.35 | 90.65 ± 9.4 | 90.56 ± 8.23 | 0.7 | ||
| Hip circumference, cm | 98.63 ± 6.6 | 97.91 ± 6.28 | 97.58 ± 7.09 | 98.19 ± 5.95 | 0.74 | ||
| Systolic blood pressure, mmHg | 129.58 ± 14.72 | 127.9 ± 13.76 | 128.48 ± 14.6 | 125.71 ± 12.86 | 0.28 | ||
| Diastolic blood pressure, mmHg | 80.38 ± 9.16 | 79.65 ± 7.8 | 79.51 ± 9.1 | 78.69 ± 8.88 | 0.62 | ||
| Body mass index, kg/m2 | 26.26 ± 3.42 | 25.47 ± 2.91 | 25.78 ± 3.36 | 25.46 ± 2.85 | 0.27 | ||
| Fasting triglyceride (IQI), mg/dl | 109.77 (82.33, 143.61) | 113.53 (76.32, 181.01) | 116.92 (84.77, 159.21) | 124.44 (89.66, 177.82) | 0.32 | ||
| Fasting total cholesterol, mg/dl | 199.37 ± 37.99 | 203.08 ± 40.94 | 192.32 ± 40.61 | 203.34 ± 37.27 | 0.21 | ||
| Fasting LDL cholesterol, mg/dl | 128.51 ± 32.91 | 132.24 ± 33.74 | 124.3 ± 34.21 | 131.36 ± 31.71 | 0.38 | ||
| Fasting HDL cholesterol, mg/dl | 47.64 ± 10.38 | 46.94 ± 10.41 | 47.05 ± 10.77 | 46.2 ± 8.89 | 0.81 | ||
| Postprandial triglyceride (IQI), mg/dl | 112.03 (77.82, 150.38) | 113.53 (88.53, 169.55) | 112.03 (90.23, 145.68) | 119.55 (88.72, 158.08) | 0.3 | ||
| Postprandial total cholesterol, mg/dl | 186.96 ± 36.1 | 187.17 ± 37.07 | 181.6 ± 39.02 | 189.35 ± 36.65 | 0.56 | ||
| Postprandial LDL cholesterol, mg/dl | 108.11 ± 30.79 | 103.93 ± 34.39 | 101.24 ± 32.49 | 108.66 ± 28.47 | 0.34 | ||
| Postprandial HDL cholesterol, mg/dl | 39.92 ± 8.68 | 38.03 ± 8.34 | 38.73 ± 7.99 | 38 ± 6.71 | 0.32 | ||
Data are presented as the mean ± SD unless otherwise indicated. IQI, interquartile intervals.
Body mass index (BMI) is the weight in kilograms divided by the square of the height in meters.
The postprandial samples were drawn 120 min after taking 100 g standard noodle (a polysaccharide) provided by China Food Limited, COFCO (Beijing, China).45
Table 2.
Changes in postprandial lipidaemia after treatment
| Model 1 |
Model 2 |
||||||
|---|---|---|---|---|---|---|---|
| LS mean (95% CI) |
LS mean (95% CI) |
||||||
| Change from Baseline | Treatment Difference | Adjusted P value | Change from Baseline | Treatment Difference | Adjusted P value | ||
| pTC (mg/dl) | Plac (91) | −8.66 (−14.52, −2.79) | Reference | −8.65 (−14.52, −2.78) | Reference | ||
| Prob (92) | −1.55 (−7.39, 4.28) | 7.10 (−3.76, 17.96) | 0.33 | −1.52 (−7.37, 4.32) | 7.13 (−3.75, 18) | 0.33 | |
| BBR (84) | −17.89 (−24.00, −11.79) | −9.24 (−20.35, 1.87) | 0.14 | −17.87 (−23.99, −11.76) | −9.22 (−20.35, 1.91) | 0.14 | |
| Prob+BBR (98) | −24.29 (−29.95, −18.64) | −15.64 (−26.33, −4.94) | 0.001 | −24.34 (−30.01, −18.67) | −15.69 (−26.41, −4.97) | 0.001 | |
| pLDLc (mg/dl) | Plac (91) | −7.35 (−12.29, −2.42) | Reference | −7.36 (−12.3, −2.42) | Reference | ||
| Prob (92) | −1.43 (−6.34, 3.48) | 5.93 (−3.21, −15.06) | 0.34 | −1.46 (−6.38, 3.45) | 5.9 (−3.25, 15.04) | 0.34 | |
| BBR (84) | −9.81 (−14.95, −4.67) | −2.46 (−11.81, 6.90) | 0.91 | −9.84 (−14.98, −4.69) | −2.48 (−11.84, 6.89) | 0.90 | |
| Prob+BBR (98) | −16.54 (−21.30, −11.79) | −9.19 (−18.19, −0.19) | 0.043 | −16.48 (−21.25, −11.7) | −9.12 (−18.14, −0.1) | 0.046 | |
| pTG (mg/dl, log) | Plac (91) | −0.03 (−0.10, 0.04) | Reference | −0.03 (−0.1, 0.04) | Reference | ||
| Prob (92) | −0.05 (−0.12, 0.02) | −0.02 (−0.16, 0.12) | 0.98 | −0.05 (−0.12, 0.03) | −0.02 (−0.16, 0.12) | 0.98 | |
| BBR (84) | −0.18 (−0.26, −0.10) | −0.15 (−0.29, −0.01) | 0.037 | −0.18 (−0.25, −0.1) | −0.15 (−0.29, −0.01) | 0.038 | |
| Prob+BBR (98) | −0.17 (−0.24, −0.10) | −0.14 (−0.28, −0.01) | 0.035 | −0.18 (−0.25, −0.1) | −0.14 (−0.28, −0.01) | 0.031 | |
| pHDLc (mg/dl) | Plac (91) | 0.51 (−0.53, 1.56) | Reference | 0.51 (−0.54, 1.55) | Reference | ||
| Prob (92) | 0.55 (−0.49, 1.59) | 0.04 (−1.90, 1.97) | 1.00 | 0.53 (−0.51, 1.57) | 0.02 (−1.92, 1.95) | 1 | |
| BBR (84) | 1.42 (0.34, 2.51) | 0.91 (−1.07, 2.89) | 0.64 | 1.41 (0.32, 2.5) | 0.9 (−1.08, 2.88) | 0.65 | |
| Prob+BBR (98) | 1.16 (0.15, 2.17) | 0.65 (−1.26, 2.56) | 0.82 | 1.2 (0.19, 2.21) | 0.69 (−1.21, 2.6) | 0.78 | |
Model 1: Analysis of variance (ANOVA) was performed to compare the change in postprandial lipidaemia between groups using Tukey’s method for multiple pairwise comparisons.
Model 2: ANCOVA was performed to compare the postprandial change in lipidaemia between groups adjusted for prespecified age group using Tukey’s method for multiple pairwise comparisons. LS: least-squares means. All P values reported were two-sided, and statistical significance was defined as adjusted P < 0.05 after adjustment for multiple comparisons of Tukey correction. pTC: postprandial TC; pLDLc: postprandial LDL cholesterol; pTG (log): postprandial triglyceride.
Prob+BBR treatment induces substantial changes in the postprandial lipidomic profile
To explore how lipid metabolism regulation might be affected by Prob+BBR, we further performed LC/MS-based lipidomic analysis on postprandial blood samples. The pre-treatment postprandial lipidomic composition between groups was similar (Data Set 1). CAP ordination analysis demonstrated that the treatment induced changes in lipidomic composition in the Prob+BBR group significantly differed from those in the Plac, Prob, and BBR groups (Figure 1a, p < .001 at CAP1). Thirty-one lipidomic metabolites were uniquely and significantly altered by the Prob+BBR treatment (Figure 1b, Wilcoxon signed-rank test, Data Set 2, q < 0.01). However, only eight in the BBR group and two in the Prob group of postprandial lipid metabolites were changed more than what were changed in the Plac group after follow-up. PLS-DA analysis showed that the 20 of the 31 postprandial lipid metabolites altered in the Prob+BBR group significantly contributed to the separation of baseline and post-treatment samples (VIP score > 1, Figure 1c, Figure S3), which we then designated as the key combined treatment responding lipid metabolites, including long to medium chain fatty acids (FFAs), acyl-carnitines and multiple glycerophospholipids: lysoglycerophosphatidylcholine (LPC), lysoglycerophatidylethanolamine (LPE), glycerophosphatidylcholine (PC), glycerophatidylethanolamine (PE) with alkyl and alkenyl substituents. The alterations in these key lipid metabolites were strongly associated with the improvement of fasting and/or the postprandial levels of LDLc and TC and, to a lesser extent, with those of TG and glycemia indices in the Prob+BBR group (Figure 1d, GEE, q < 0.05, Data Set 5). Thus, the decreases in multiple postprandial FFAs and phospholipids after Prob+BBR treatment might contribute to the overall reduced PL levels.
Figure 1.
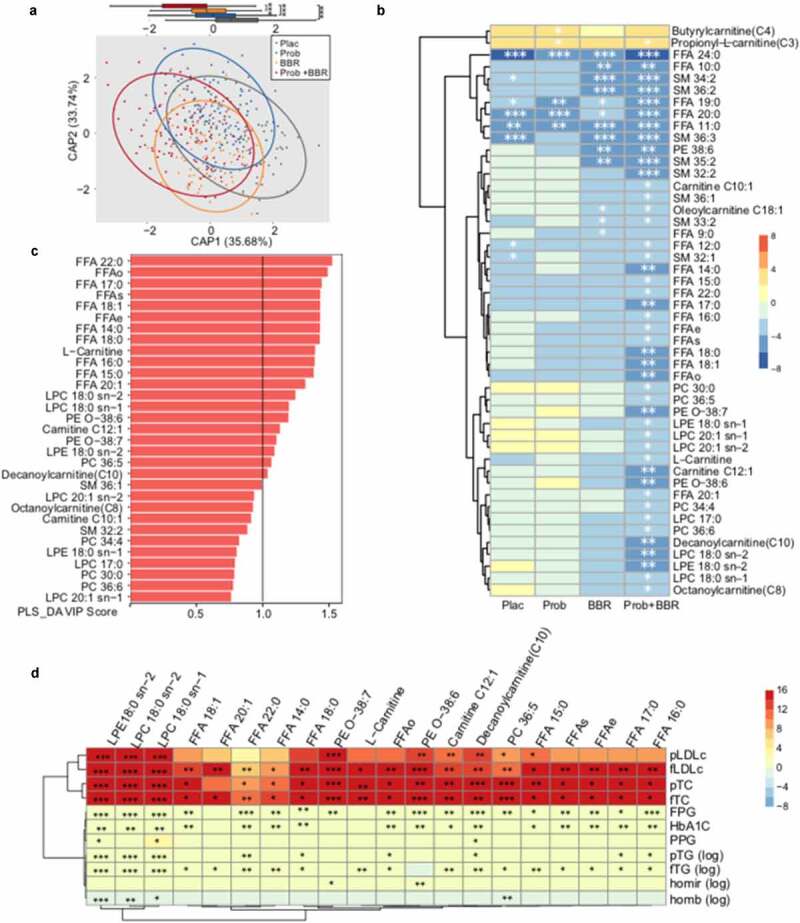
Lipidomic study on postprandial blood samples
(a) Canonical analysis of principal coordinates (CAP) ordination plot (Bray-Curtis) of the changes in pre-post-treatment postprandial blood lipidomic metabolites in four treatment groups; with the two primary axes (CAP 1 and CAP 2) accounting for 69.42% of the total variance. The Tukey-style box plot shows the distribution of the first canonical axis (CAP 1) of lipid profiles. ***, P < .001. Samples beyond the end of the whiskers are called “outlying” points. Plac, placebo, n = 91; Prob, probiotic treatment, n = 92; BBR, berberine treatment, n = 84; Prob+BBR: berberine plus probiotic treatment, n = 98. (b) Heatmap of significant changes in postprandial lipidomic metabolites by four different treatments. The Z-score was calculated with the Wilcoxon signed-rank test. A Z-score >0 (red) indicated an increase after treatment, and a Z-score <0 (blue) indicated a decrease after treatment. *, q < 0.01, **, q < 0.001, ***, q < 0.0001. FFA: fatty acid; FFAo: the sum of odd-carbon-chain fatty acids; FFAe: the sum of even-carbon-chain fatty acids; FFAs: the sum of all fatty acids; LPC: lysoglycerophosphatidylethanolamine; LPE: lysoglycerophartylethanolamine; PC: glycerophosphosphatidylcholine; PE: glycerophatidyethanolamine; SM: sphingomyelin; PE O-:PE with alkyl and alkenyl substituents. (c) Bar plots showing the variable importance in projection (VIP) scores of postprandial lipidomic metabolites from the partial least squares-discriminant analysis (PLS-DA) in the Prob+BBR group. A total of 31 metabolites that were uniquely and significantly altered in the Prob+BBR group (see panel B) are presented. (d) Heatmap of longitudinal associations between the changes in twenty important lipidomic metabolites (VIP score >1, see panel B) and clinical characteristics response to Prob+BBR treatment. Multivariate Generalized Estimating Equation (GEE) analysis, controlling for age, sex and BMI. The color key represents the β value from GEE models. *, q < 0.05, **, q < 0.01, ***, q < 0.001. fTC: the fasting level of total cholesterol; fLDLc: low-density lipoprotein cholesterol; fTG (log): log of the fasting level of triglyceride; pTC: postprandial TC; pLDLc: postprandial LDL cholesterol; pTG (log): log of postprandial triglyceride; HbA1c: glycated hemoglobin; homair (log): log of (fasting serum insulin * fasting plasma glucose)/22.5, homeostasis model assessment index for assessing insulin resistance; homab (log): log of (20 * fasting serum insulin)/(fasting plasma glucose – 3.5), homeostasis model assessment index for assessing ß cell function; Prob+BBR: berberine plus probiotics treatment. n = 98.
Recovering fecal enrichment of B. breve could be responsible for Prob+BBR induced PL improvement
As shown in our previous study,33 we found significant changes in gut microbiota in the Prob+BBR group. We then sought to ask whether the gut microbial alterations could underlie the benefit of Prob+BBR on PL compared to BBR alone. Twenty-four species in the gut microbiome were found to only respond to Prob+BBR treatment, including nine ingested probiotic strains33 (Figure 2a, Wilcoxon signed-rank test, q < 0.05). B. breve was the only taxon that was significantly reduced by BBR and increased in the Prob+BBR group. RAs of B. breve and Eggerthella lenta were correlated with the level of PL after Prob+BBR treatment (Figure 2b, Spearman, P < .05, Data Set 6). Higher fecal levels of B. breve and lower levels of E. lenta after treatment were associated with better control of PL in Prob+BBR group. Multivariate GEE analysis further suggested that increment of B. breve after treatment were significantly associated with the reductions of pTC (Figure 3a, β [SE] – 5.25 [0.95], P = 4.00 E-08) and pLDLc (Figure 3a, β [SE] – 4.18 [0.76], P = 3.00 E-08). In addition, GEE analysis of alterations in lipid metabolites and microbial taxa relating with PL improvements (listed in Figure 2b) showed that the changes in RAs of probiotic species including B. breve were negatively correlated with the changes in carnitines and phospholipids such as LPC 18:0 sn-2, PE-O 38:7 and PE O-38:6 (Figure 3b, q < 0.05, Data Set 7). These metabolites were also positively correlated with PL changes (Figure 1d). Furthermore, we found that baseline fecal RAs of B. breve, B. longum and the genus Bifidobacterium were significantly lower in participants with dyslipidemia than those with eulipidemia (Figure 3c, KW, P < .05, Data Set 8). Hence, considering the different treatment induced changes of B. breve between BBR alone and Prob+BBR groups, its correlations with PL and lipidomic metabolites, and its enrichment in participants with eulipidemia, B. breve might be an effective component in the probiotic formula to improve PL with BBR. In in vitro culture, B. breve growth was significantly inhibited by BBR in a dose-dependent manner, whereas the growth of E. coli was not affected by BBR even at the highest concentration applied for B. breve (Figure 3d). These results further consolidated our hypothesis that the superior effects of the combined treatment in improving PL and postprandial lipidomic profile might be related to recovering the reduced biomass of B. breve with BBR treatment.
Figure 2.
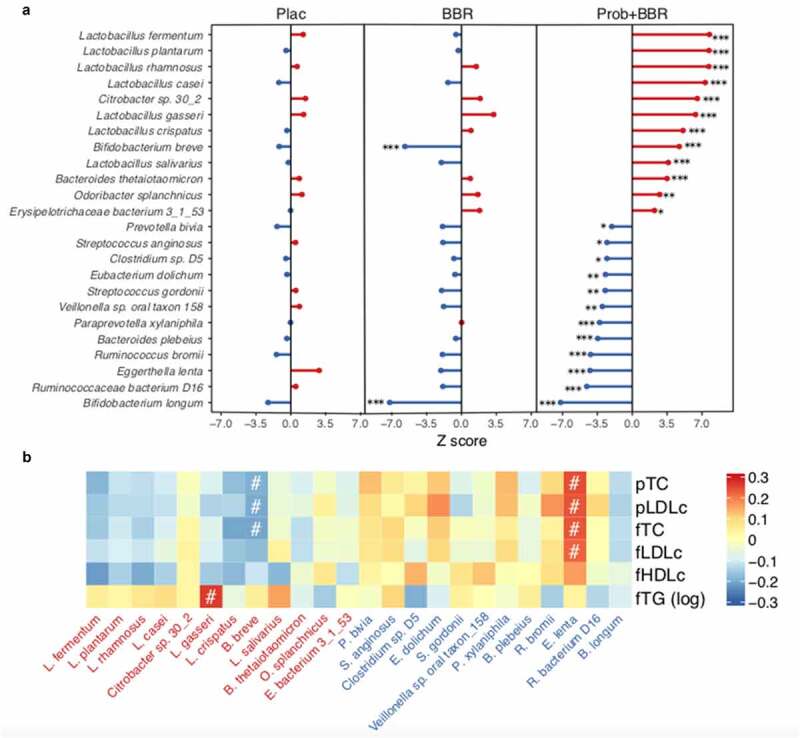
Gut microbial species correlate with blood lipidemia profiles
(a) The changes in RAs of 24 selected microbial species uniquely altered in the Prob+BBR (right) compared to the Plac group (left), including 15 species altered uniquely and significantly by Prob+BBR and the 9 species from the probiotic mixture. Z-scores were calculated with the two-sided Wilcoxon signed rank tests. A Z-score >0 (red) indicated an increase after treatment and a Z-score <0 (blue) indicated a decrease after treatment. *, q < 0.05, **, q < 0.01, ***, q < 0.001. Plac, Placebo, n = 91; BBR: berberine alone treatment, n = 84; Prob+BBR: berberine plus probiotic treatment, n = 98. (b) Correlations between post-treatment abundances of species in (A) and serum levels of cholesterol and triglycerides in the Prob+BBR group. Spearman correlation, #, q < 0.1. The color key represents the rho value. Red and blue represent the enriched and depleted species (ranked as same as panel A) after Prob+BBR treatment, respectively.
Figure 3.
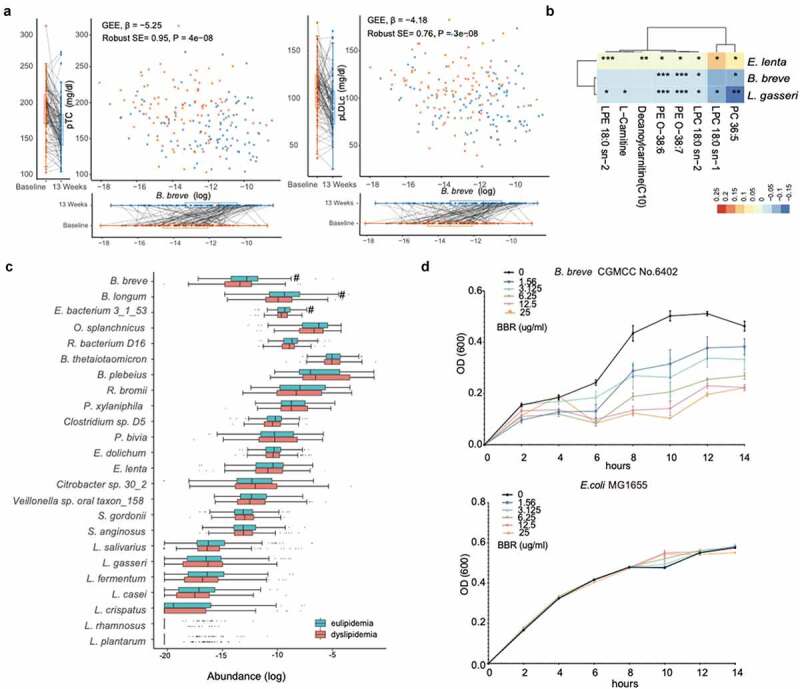
B. breve correlates with lipid metabolites changes and is depleted by BBR
(a) Scatter plot of the levels of postprandial total cholesterol (pTC, left panel) and LDL cholesterol (pLDLc, right panel) levels against the relative abundances of B. breve in each subject at baseline and after 13 weeks treatment in the Prob+BBR group. β value and P-value for their longitudinal associations are calculated by GEE model. Tukey-style box plots showing the levels of pTC (left), pLDLc (right), and relative abundances of B. breve (at the bottom of the scatter plot) in the corresponding subjects, respectively (orange: baseline, n = 98; blue: 13 weeks, n = 98). (b) Heatmap of longitudinal associations between the changes in important lipidomic metabolites (VIP score >1, Figure 1b) and microbial species response to Prob+BBR treatment. Multivariate GEE analysis, controlling for age, sex, and BMI. The color key represents the β value from GEE models. *, q < 0.05, **, q < 0.01, ***, q < 0.001. Only three species significantly correlated with blood lipid profiles (Figure 2b) are shown in the panel. (c) Tukey-style bar plot showing the differences in RAs of 24 microbial species between participants with dyslipidaemia (n = 171) and eulipidaemia (n = 194) at baseline. Only the nine probiotic species and those uniquely responses to the Prob+BBR treatment (Figure 2a) are shown. Kruskal–Wallis tests, #, q < 0.1. (d) The growth curve of B. breve CGMCC No. 6402 (left) and E. coli MG1655 (right) with different concentrations of BBR (0–25 ug/mL) in the in vitro culture experiment. n = 4, P < .001, determined by two-way repeated measures ANOVA. Data are shown as the mean ± SE.
BBR induces the expression of genes regulating FFA simulation in B. breve
We then asked whether there were factors other than recovering the decreased intraluminal biomass of B. breve after BBR treatment could improve PL lowering effect in Prob+ BBR. KEGG functional analysis of the fecal sample showed that Prob+BBR exhibited higher fatty acid metabolism and lower fatty acid synthesis potential than BBR alone treatment group (Figure S4). Using the draft genomes of the 9 probiotic strains in our formula33 and 1,520 high-quality genomes from cultivated human gut bacteria,51 we illustrated the distribution of genes regulating bacterial lipid metabolism in all 24 species responding to Prob+BBR treatment (Data Set 4, Figure 4a). Notably, the 9 probiotic strains all contained phospholipid biosynthesis genes including gspA (glycerol-3-phosphate dehydrogenase (NAD(P)+), K00057), plsC (1-acyl-sn-glycerol-3-phosphate acyltransferase, K00655) and cdsA (phosphatide cytidylyltransferase, K00981). However, only two Bifidobacterium strains exclusively expressed fadD (long-chain acyl-CoA synthetase, K01897, EC 6.2.1.3 (Figure 4a), a key bacterial enzyme that involved in the import and mobilization of exogenous FFA.55 Four fadD genes were annotated in the B. breve and B. longum genomes, indicating that both strains might possess more active FFA import and mobilization capacity compared to those have none (Data Set 4). Interestingly, when B. breve was cultured in vitro with BBR, the RNA expression of all its four fadD genes was significantly elevated compared to control medium with vehicle (Figure 4b). Consistently, the non-esterified FFA levels in culture media with adding Linolenic acid at final concentration of 1 mg ml−1, were significantly reduced by B. breve and could be enhanced by BBR treatment (Figure 4c). Thus, in addition to maintaining intraluminal abundancy of B. breve via supplementing probiotics formula with BBR treatment, activating the capacities of B. breve lipid simulation by BBR also might contribute to the synergetic hypolipidemic benefit of the combined treatment.
Figure 4.
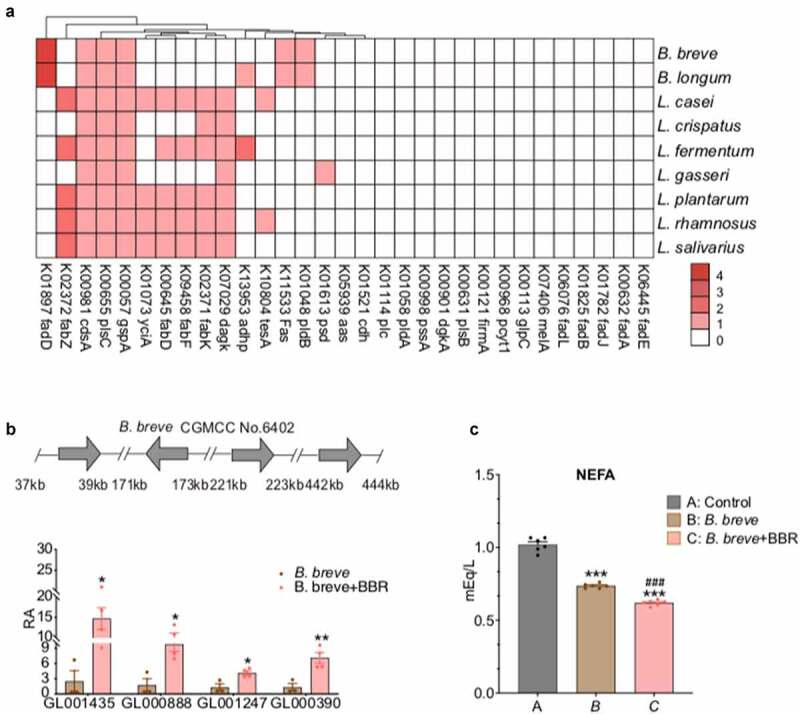
BBR activates lipid metabolism in B. breve.
(a) The distribution of genes encoding enzymes regulating bacterial glycerophospholipid metabolism, fatty acid biosynthesis and degradation pathways predicted in genomes of the nine bacterial strains in probiotics. Color key indicates gene numbers under KO in an individual strain.(b) The expression of fadD increased in B. breve after treatment with BBR (6.25 μg ml−1). The mRNA levels of the four fadD genes in B. breve were determined by quantitative PCR. n = 4, *, P < .05, **, P < .01 compared to vehicle, determined by unpaired two-tailed student’s t test.(c) In vitro FFA consumption of B. breve, non-esterified fatty acids (NEFAs) were measured in the medium supplemented with LA. n = 6, ***, P < .001 relative to control, ###, P < .001 relative to B. breve after treatment with BBR, unpaired two-tailed student’s t test. BBR, berberine; fadD, long-chain acyl-CoA synthetase, LA, linolenic acid.
Discussion
This study based on a randomized clinical trial confirmed that Prob+BBR combined therapy exerted a similar effect on reducing fasting lipidaemia with BBR alone but a superior effect on the levels of pTC and pLDLc compared to either BBR or probiotics alone. A pseudo-target lipidomic study revealed a substantial decrease in various lipid species after Prob+BBR treatment, implying decreased intestinal lipid uptake. Further we found that the hypolipidemic effect of combined treatment could be gained from the recovery of BBR-induced B. breve depletion by ingested probiotics and the induction of microbial lipid import and mobilization by BBR. Thus, our study provided both clinical and experimental evidence to support the synergistic effect of supplemental empirical probiotics containing Bifidobacteria such as B. breve with BBR in lowering PL, which could serve as an effective remedy for managing T2D PL and general dyslipidemia with its effect in lowering FL.
Hyperglycemia and hyperlipidemia commonly coexist in patients with T2D, and both are main risk factors for ASCVDs events.56 Statins, the current main hypolipidemia medication to reduce major adverse cardiovascular events (MACEs) in T2D patients,3 exhibits unfavorable effects, such as increasing intestinal cholesterol absorption, blood glucose level and diabetes incidence, as well as the hepatoxicity and myotoxicity. These side effects (SE) have brought concerns to the clinical practice ,57,58 particularly in East Asia population, including the Chinese, who is more susceptible to the SEs of statins.59,60 A cross-sectional survey in a nationally representative sample of 15,540 Chinese adults reports only 3.5% and 3.4% of men and women with a total cholesterol ≥200 mg/dL has been treated with any antilipidemic medicine.61 Thus, our regimen of combined therapy with Prob+BBR, targeting the PL with antidiabetic effect, could provide an alternative treatment for managing hyperlipidemia in patients with T2D, particularly those who are intolerant to statins.
In most studies on the role of BBR in lipid metabolism regulation,29–33,62,63 only fasting lipid levels have been evaluated. Here, we first reported that BBR was less potent in lowering the PL in participants with T2D compared to its effect in lowering fasting lipidaemia. We attributed this diverse effect partly to the suppression of commensal gut Bifidobacteria by BBR, either in our data (Table S7) or in previous study.64 Supplementation with a probiotic strain of B. breve recovered the loss of its indigenous counterpart and significantly improved PL in BBR-treated participants with T2D. Apparently, the BBR induced gut microbiota alterations might not all be beneficial to host metabolism. The negative effect of BBR on Bifidobacterium taxa might compromise its effect in reducing PL and raised caution that the effect of the gut microbiome should not be neglected when developing new treatment strategies for metabolic diseases.
The effect of Bifidobacterium on lowering blood lipids, particularly cholesterol levels, has been well recognized and made it recognized as a nature hypolipidemic agent like BBR,37–43 albeit the underlying mechanism has not yet been clarified. In our study, we found that B. breve and B. longum as well as the genus Bifidobacterium were enriched in T2D participants with better lipidemia. Further we found the enrichment of fadD genes might mediate the distinguish lipid lowering effect of B. breve. FadD is a fatty acyl-CoA synthetase that facilitates bacterial exogenous FFA uptake,55,65 mobilizes medium-chain and long-chain fatty acids (for FFA elongation, degradation, phospholipid biosynthesis,66,67 and represses expression of genes for FFA biosynthesis.55,68 The significantly enhanced transcription of all 4 fadD genes and FFA consumption in B. breve by BBR could underlie the effect of Prob+BBR in reducing host intestinal lipid uptake and blood cholesterol levels. The activated gut microbial lipid metabolism brought by the combined treatment could hijack host intraluminal lipids so as to significantly improve postprandial lipidomic profile. This might explain why Prob alone was neutral in lowering lipidemia and became effective only when it was combined with BBR for treating T2D patients. Thus, we thought it could be necessary to supplement with probiotics containing Bifidobacterium spp. strains, when BBR is clinically used as a hypolipidemic agent.
However, though both B. breve and B. longum were supplemented in the probiotic formula and both contained multiple fadD genes, metagenomics analysis showed that only B.breve was significantly recovered in Prob+BBR group33 and only its RA alterations was correlated with those in pTC and pLDLc. It might be explained that the amount of B.longum applied in this study was less than what was required. In addition, we also found that some commensal gut species other than those were supplemented by probiotics, such as E. lenta, altered uniquely after Prob+BBR treatment, and positively correlated either with PLs or postprandial lipid metabolites (Figure 1d, Figure 3b), indicating its relevance to the effect of Prob+BBR on lowering PL. E. lenta has been reported to decrease in fecal samples from T2D subjects treated with Acarbose, in which multiple Bifidobacterium and Lactobacillus are elevated.69 Future work is warranted to evaluate if E. lenta could elevate the PL autonomously to serve as a potential new target for treating postprandial dyslipidemia, or just be a marker of lactic bacteria flourish in gut.
Our study has several limitations. The predesigned 13-week multicenter randomized, double-blind, placebo-controlled study can avoid bias and obtain powerful evidence but does not allow for the assessment of long-term efficacy on MACEs in the combined treatment of Prob+BBR. The PL were tested after a 100 g carbon meal with post-load blood glucose and might be constituted by hepatobiliary secretion or transintestinal cholesterol excretions.70,71 How the PL responses to mix or fat meal could be affected by the combined treatment requires further study. Considering that the gut microbiota could directly stimulate atherosclerosis via bacterial-host co-metabolism of the choline/betaine diet to produce TMA,72–74 the potential of our remedy to target this metabolic pathway will be of interest for future investigations. In addition, the findings derived from this investigation may not be generalized to those on statin therapy populations without caution.
In conclusion, this clinical trial-based study proved the therapeutic effect of a combined treatment of oral administration of probiotics with berberine on improving PL in patients newly diagnosed with T2D and proposed a new gut microbiome related remedy for managing dyslipidemia, covering both PL and FL, in patients with T2D.
Supplementary Material
Acknowledgments
We thank all the patients enrolled in the PREMOTE study. We thank Drs. Dalong Zhu (Nanjin Drum Tower Hospital, Nanjing University Medical School, Jiangsu Province, China), Tao Yang (Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing University Medical School, Nanjing Jiangsu, China), Yanbing Li (The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, China), Li Yan (Sun Yat-sen Memory Hospital, Sun Yat-sen University, Guangzhou, Guangdong, China), Wei Gu (The Second Affiliated hospital of Zhejiang University School of Medicine, Hangzhou, Zhejiang, China), Lulu Chen (Wuhan Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, China), Fan Zhang (Peking University Shenzhen Hospital, Shenzhen, Guangdong, China), Qian Zhang (Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong, China), Lili Xia (Tong Ren Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China), Yujuan Fan (Central Hospital of Minhang district, Shanghai, China), Junping Wen (Fujian Provincial Hospital, Fuzhou, Fujian, China), Wei Xu (Xuzhou Central Hospital, Xu Zhou, Jiangsu, China), Yanyan Hu (Chang Hai Hospital, Second Military Medical University, Shanghai, China), Yufan Wang (Shanghai First People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China), Xi Xia (Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China) for their collection of data and blood samples and for taking care of the patients. We thank Zhiyun Zhao, Min Xu (Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China), and Karsten Kristiansen (Department of Biology, University of Copenhagen, Denmark, BGI-Shenzhen, Shenzhen, China) for all discussions of clinical and metagenomics data and multi-omics analysis. We thank Northeast Pharmaceutical Group Co., Ltd., Shenyang, Liaoning, China, and Shanghai Jiaoda Onlly Co., Ltd., Shanghai, China for providing treatment agents and placebo as a courtesy.
Funding Statement
This study was funded by grants from the National Key R&D Program of China (2016YFC0901200); Fund of the Prevention and Control of Major Chronic Noncommunicable Diseases Research of China (2018YFC1313804); the Program for Shanghai Outstanding Medical Academic Leader (2019LJ07); the National Nature Science Foundation of China (91857205, 81870555); and the Fund of the Shenzhen Municipal Government of China (No. JCYJ20170817145809215).
Disclosure statement
No potential conflict of interest was reported by the authors.
Author Contributions
Y.G., W.W., Y.Z. and J.L. had full access to all the data in the study and accept responsibility for the integrity of the data and the accuracy of the data analyses. W.W., J.L., S.W., Y.Z., Y.G. designed the study. Y.G., Y.Z., S.W., J.M., X.G., Y.X., S.H., J.Y., L.C., G.C., S.Q., J.L., L.Q., Q.H., Y.P., G.N., and W.W. conducted the clinical trial, enrolled and managed the patients. Y.Z., S.W. and Y.G. performed quality control and analysed the clinical data. H.R., Z.S., H.Z. and J.L. performed the metagenomics sequencing and bioinformatics analyses. C.L., S.W., Y.G. and H.Z. designed and performed the in vitro experiments. X.Z., X.W., Q.L., and G.X. performed the LC/MS experiments for assaying blood BAs. Y.G., H.R. S.W., H.Z., and J.L. performed the multi-omics data analysis. Y.G., W.W., J.L., Y.Z., H.R., S.W., H.Z., J.W. and H.Y. discussed the data and wrote the manuscript.
Data statement
The data that support the findings of this study are available in https://www.ncbi.nlm.nih.gov/bioproject/PRJNA643353, reference number PRJNA643353, and within the article and its supplementary materials.
Abbreviations
- Nonstandard Abbreviations and Acronyms
- ASCVD
Atherosclerotic cardiovascular disease
- ANOVA
Analysis of variance
- BBR
Berberine
- B. breve
Bifidobacterium breve
- BH
Benjamini–Hochberg
- CAP
Canonical analysis of principal coordinates analysis
- FFA
Fatty acid
- FL
Fasting lipidemia
- fLDLc
Fasting levels of low-density lipoprotein cholesterol
- fTC
Fasting levels of total cholesterol
- fTG
Fasting levels of triglycerides
- GEE
Generalized estimating equation
- HPLC
High-performance liquid chromatography
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KO
KEGG Orthology
- KW
Kruskal–Wallis
- LC-MS
Liquid chromatography–mass spectrometry
- LPC
Lyso-glycerophosphatidylcholine
- LPE
Lyso-glycerophatidylethanolamine
- LS
Least-squares
- MACE
Major adverse cardiovascular events
- NCEP/ATP III National Cholesterol Education Program/Adult Treatment Panel III
- NPC1L1
Niemann-Pick C1-like 1
- nFL
Non-fasting lipidaemia
- PC
Glycerophosphatidylcholine
- PE
Glycerophatidylethanolamine
- pHDLc
Postprandial high-density lipoprotein cholesterol
- pLDLc
Postprandial low-density lipoprotein cholesterol
- PLS-DA
Partial least squares-discriminant analysis
- Plac
Placebo
- Prob
Probiotics
- Prob+BBR
Probiotics combined with Berberine
- PREMOTE
Probiotics and BBR on the Efficacy and Change of Gut Microbiota in Patients with Newly Diagnosed Type 2 Diabetes trial.
- pTC
Postprandial plasma total cholesterol
- pTG
Postprandial plasma triglycerides
- QC
Quality control
- RA
Relative abundancy
- RSD
Relative standard deviation
- SM
Sphingomyelin
- VIP
Variable importance in projection
Supplementary Material
Supplemental data for this article can be accessed on Publisher’s website.
References
- 1.Goff DC Jr., Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB, Gibbons R, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129(25 Suppl 2):S49–19. doi: 10.1161/01.cir.0000437741.48606.98. [DOI] [PubMed] [Google Scholar]
- 2.Emerging Risk Factors C, Sarwar N, Gao P, Seshasai SR, Gobin R, Kaptoge S, et al. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet. 2010;375(9733):2215–2222. doi: 10.1016/S0140-6736(10)60484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Low Wang CC, Hess CN, Hiatt WR, Goldfine AB.. Clinical Update: cardiovascular Disease in Diabetes Mellitus: atherosclerotic Cardiovascular Disease and Heart Failure in Type 2 Diabetes Mellitus - Mechanisms, Management, and Clinical Considerations. Circulation. 2016;133(24):2459–2502. doi: 10.1161/CIRCULATIONAHA.116.022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baddour LM, Wilson WR, Bayer AS, Fowler VG Jr., Tleyjeh IM, Rybak MJ, et al. Infective Endocarditis in Adults: diagnosis, Antimicrobial Therapy, and Management of Complications: a Scientific Statement for Healthcare Professionals From the American Heart Association. Circulation. 2015;132(15):1435–1486. doi: 10.1161/CIR.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 5.Nordestgaard BG, Benn M, Schnohr P, Tybjaerg-Hansen A. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA. 2007;298(3):299–308. doi: 10.1001/jama.298.3.299. [DOI] [PubMed] [Google Scholar]
- 6.Katsanos CS. Clinical considerations and mechanistic determinants of postprandial lipemia in older adults. Adv Nutr. 2014;5(3):226–234. doi: 10.3945/an.113.004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welsh C, Celis-Morales CA, Brown R, Mackay DF, Lewsey J, Mark PB, et al. Comparison of Conventional Lipoprotein Tests and Apolipoproteins in the Prediction of Cardiovascular Disease. Circulation. 2019;140(7):542–552. doi: 10.1161/CIRCULATIONAHA.119.041149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warnick GR, Nakajima K. Fasting versus nonfasting triglycerides: implications for laboratory measurements. Clin Chem. 2008;54(1):14–16. doi: 10.1373/clinchem.2007.098863. [DOI] [PubMed] [Google Scholar]
- 9.Eckel RH. LDL cholesterol as a predictor of mortality, and beyond: to fast or not to fast, that is the question? Circulation. 2014;130(7):528–529. doi: 10.1161/CIRCULATIONAHA.114.011512. [DOI] [PubMed] [Google Scholar]
- 10.Nordestgaard BG, Langsted A, Mora S, Kolovou G, Baum H, Bruckert E, et al. Fasting is not routinely required for determination of a lipid profile: clinical and laboratory implications including flagging at desirable concentration cut-points-a joint consensus statement from the European Atherosclerosis Society and European Federation of Clinical Chemistry and Laboratory Medicine. Eur Heart J. 2016;37(25):1944–1958. doi: 10.1093/eurheartj/ehw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varbo A, Nordestgaard BG. Nonfasting Triglycerides, Low-Density Lipoprotein Cholesterol, and Heart Failure Risk: two Cohort Studies of 113 554 Individuals. Arterioscler Thromb Vasc Biol. 2018;38(2):464–472. doi: 10.1161/ATVBAHA.117.310269. [DOI] [PubMed] [Google Scholar]
- 12.Langsted A, Nordestgaard BG. Nonfasting versus fasting lipid profile for cardiovascular risk prediction. Pathology. 2019;51(2):131–141. doi: 10.1016/j.pathol.2018.09.062. [DOI] [PubMed] [Google Scholar]
- 13.Adeli K, Lewis GF. Intestinal lipoprotein overproduction in insulin-resistant states. Curr Opin Lipidol. 2008;19(3):221–228. doi: 10.1097/MOL.0b013e3282ffaf82. [DOI] [PubMed] [Google Scholar]
- 14.Haidari M, Leung N, Mahbub F, Uffelman KD, Kohen-Avramoglu R, Lewis GF, et al. Fasting and postprandial overproduction of intestinally derived lipoproteins in an animal model of insulin resistance. Evidence that chronic fructose feeding in the hamster is accompanied by enhanced intestinal de novo lipogenesis and ApoB48-containing lipoprotein overproduction. J Biol Chem. 2002;277(35):31646–31655. doi: 10.1074/jbc.M200544200. [DOI] [PubMed] [Google Scholar]
- 15.Xiao C, Dash S, Morgantini C, Lewis GF. New and emerging regulators of intestinal lipoprotein secretion. Atherosclerosis. 2014;233(2):608–615. doi: 10.1016/j.atherosclerosis.2013.12.047. [DOI] [PubMed] [Google Scholar]
- 16.Doran B, Guo Y, Xu J, Weintraub H, Mora S, Maron DJ, et al. Prognostic value of fasting versus nonfasting low-density lipoprotein cholesterol levels on long-term mortality: insight from the National Health and Nutrition Examination Survey III (NHANES-III). Circulation. 2014;130(7):546–553. doi: 10.1161/CIRCULATIONAHA.114.010001. [DOI] [PubMed] [Google Scholar]
- 17.Doran B, Guo Y, Xu J, Weintraub H, Mora S, Maron DJ, et al. Reply to Letters Regarding Article, “Prognostic Value of Fasting Versus Nonfasting Low-Density Lipoprotein Cholesterol Levels on Long-Term Mortality: insight From the National Health and Nutrition Examination Survey III (NHANES-III)”. Circulation. 2015;131(19):e473. doi: 10.1161/CIRCULATIONAHA.114.014177. [DOI] [PubMed] [Google Scholar]
- 18.Bozzetto L, Annuzzi G, Corte GD, Patti L, Cipriano P, Mangione A, et al. Ezetimibe beneficially influences fasting and postprandial triglyceride-rich lipoproteins in type 2 diabetes. Atherosclerosis. 2011;217(1):142–148. doi: 10.1016/j.atherosclerosis.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021. doi: 10.1038/s41591-020-01183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Guryn K, Hubert N, Frazier K, Urlass S, Musch MW, Ojeda P, et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe. 2018;23(4):458–69 e5. doi: 10.1016/j.chom.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molinaro A, Wahlstrom A, Marschall HU. Role of Bile Acids in Metabolic Control. Trends Endocrinol Metab. 2018;29(1):31–41. doi: 10.1016/j.tem.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Kuang Z, Wang Y, Li Y, Ye C, Ruhn KA, Behrendt CL, et al. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science. 2019;365(6460):1428–1434. doi: 10.1126/science.aaw3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Koonen D, Hofker M, Fu J. Gut microbiome and lipid metabolism: from associations to mechanisms. Curr Opin Lipidol. 2016;27(3):216–224. doi: 10.1097/MOL.0000000000000308. [DOI] [PubMed] [Google Scholar]
- 24.Araujo JR, Tazi A, Burlen-Defranoux O, Vichier-Guerre S, Nigro G, Licandro H, et al. Fermentation Products of Commensal Bacteria Alter Enterocyte Lipid Metabolism. Cell Host Microbe. 2020;27(3):358–75 e7. doi: 10.1016/j.chom.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 25.Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010;51(5):1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, et al. The Gut Microbiome Contributes to a Substantial Proportion of the Variation in Blood Lipids. Circ Res. 2015;117(9):817–824. doi: 10.1161/CIRCRESAHA.115.306807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witkowski M, Weeks TL, Hazen SL. Gut Microbiota and Cardiovascular Disease. Circ Res. 2020;127(4):553–570. doi: 10.1161/CIRCRESAHA.120.316242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenny DJ, Plichta DR, Shungin D, Koppel N, Hall AB, and Fu B, et al. Cholesterol Metabolism by Uncultured Human Gut Bacteria Influences Host Cholesterol Level Cell Host Microbe . 2020;28(2):245–57 e6. doi: 10.1016/j.chom.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong W, Wei J, Abidi P, Lin M, Inaba S, Li C, et al. Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat Med. 2004;10(12):1344–1351. doi: 10.1038/nm1135. [DOI] [PubMed] [Google Scholar]
- 30.Yao S, Yuan Y, Zhang H, Meng X, Jin L, Yang J, et al. Berberine attenuates the abnormal ectopic lipid deposition in skeletal muscle. Free Radic Biol Med. 2020;159:66–75. doi: 10.1016/j.freeradbiomed.2020.07.028. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Zhang H, Li B, Meng X, Wang J, Zhang Y, et al. Berberine activates thermogenesis in white and brown adipose tissue. Nat Commun. 2014;5:5493. doi: 10.1038/ncomms6493. [DOI] [PubMed] [Google Scholar]
- 32.Pirillo A, Catapano AL. Berberine, a plant alkaloid with lipid- and glucose-lowering properties: from in vitro evidence to clinical studies. Atherosclerosis. 2015;243(2):449–461. doi: 10.1016/j.atherosclerosis.2015.09.032. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Gu Y, Ren H, Wang S, Zhong H, and Zhao X, et al. Gut microbiome-related effects of berberine and probiotics on type 2 diabetes (the PREMOTE study). Nat Commun. 2020;11(1):5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallianou N, Stratigou T, Christodoulatos GS, Dalamaga M. Understanding the Role of the Gut Microbiome and Microbial Metabolites in Obesity and Obesity-Associated Metabolic Disorders: current Evidence and Perspectives. Curr Obes Rep. 2019;8(3):317–332. doi: 10.1007/s13679-019-00352-2. [DOI] [PubMed] [Google Scholar]
- 35.Jang HR, Park HJ, Kang D, Chung H, Nam MH, Lee Y, et al. A protective mechanism of probiotic Lactobacillus against hepatic steatosis via reducing host intestinal fatty acid absorption. Exp Mol Med. 2019;51(8):1–14. doi: 10.1038/s12276-019-0352-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi SB, Lew LC, Yeo SK, Nair Parvathy S, Liong MT. Probiotics and the BSH-related cholesterol lowering mechanism: a Jekyll and Hyde scenario. Crit Rev Biotechnol. 2015;35(3):392–401. doi: 10.3109/07388551.2014.889077. [DOI] [PubMed] [Google Scholar]
- 37.Bordoni A, Amaretti A, Leonardi A, Boschetti E, Danesi F, Matteuzzi D, et al. Cholesterol-lowering probiotics: in vitro selection and in vivo testing of bifidobacteria. Appl Microbiol Biotechnol. 2013;97(18):8273–8281. doi: 10.1007/s00253-013-5088-2. [DOI] [PubMed] [Google Scholar]
- 38.Ruscica M, Pavanello C, Gandini S, Macchi C, Botta M, Dall’Orto D, et al. Nutraceutical approach for the management of cardiovascular risk - a combination containing the probiotic Bifidobacterium longum BB536 and red yeast rice extract: results from a randomized, double-blind, placebo-controlled study. Nutr J. 2019;18(1):13. doi: 10.1186/s12937-019-0438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivey KL, Hodgson JM, Kerr DA, Thompson PL, Stojceski B, Prince RL. The effect of yoghurt and its probiotics on blood pressure and serum lipid profile; a randomised controlled trial. Nutr Metab Cardiovasc Dis. 2015;25(1):46–51. doi: 10.1016/j.numecd.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 40.Guardamagna O, Amaretti A, Puddu PE, Raimondi S, Abello F, Cagliero P, et al. Bifidobacteria supplementation: effects on plasma lipid profiles in dyslipidemic children. Nutrition. 2014;30(7–8):831–836. doi: 10.1016/j.nut.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Cicero AFG, Colletti A, Bajraktari G, Descamps O, Djuric DM, Ezhov M, et al. Lipid-lowering nutraceuticals in clinical practice: position paper from an International Lipid Expert Panel. Nutr Rev. 2017;75(9):731–767. doi: 10.1093/nutrit/nux047. [DOI] [PubMed] [Google Scholar]
- 42.Barrios V, Escobar C, Cicero AF, Burke D, Fasching P, Banach M, et al. A nutraceutical approach (Armolipid Plus) to reduce total and LDL cholesterol in individuals with mild to moderate dyslipidemia: review of the clinical evidence. Atheroscler Suppl. 2017;24:1–15. doi: 10.1016/j.atherosclerosissup.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Sahebkar A, Serban MC, Gluba-Brzozka A, Mikhailidis DP, Cicero AF, Rysz J, et al. Lipid-modifying effects of nutraceuticals: an evidence-based approach. Nutrition. 2016;32(11–12):1179–1192. doi: 10.1016/j.nut.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 44.National Cholesterol Education Program Expert Panel on Detection E, Treatment of High Blood Cholesterol in A . Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106(25):3143–3421. doi: 10.1161/circ.106.25.3143. [DOI] [PubMed] [Google Scholar]
- 45.Deng Y, Zhang Y, Zheng S, Hong J, Wang C, Liu T, et al. Postprandial glucose, insulin and incretin responses to different carbohydrate tolerance tests. J Diabetes. 2015;7(6):820–829. doi: 10.1111/1753-0407.12245. [DOI] [PubMed] [Google Scholar]
- 46.Luo P, Yin P, Hua R, Tan Y, Li Z, Qiu G, et al. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2018;67(2):662–675. doi: 10.1002/hep.29561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Su B, Zeng Z, Li C, Zhao X, Lv W, et al. Ion-Pair Selection Method for Pseudotargeted Metabolomics Based on SWATH MS Acquisition and Its Application in Differential Metabolite Discovery of Type 2 Diabetes. Anal Chem. 2018;90(19):11401–11408. doi: 10.1021/acs.analchem.8b02377. [DOI] [PubMed] [Google Scholar]
- 48.Shao Y, Zhu B, Zheng R, Zhao X, Yin P, Lu X, et al. Development of urinary pseudotargeted LC-MS-based metabolomics method and its application in hepatocellular carcinoma biomarker discovery. J Proteome Res. 2015;14(2):906–916. doi: 10.1021/pr500973d. [DOI] [PubMed] [Google Scholar]
- 49.Zhao X, Zeng Z, Chen A, Lu X, Zhao C, Hu C, et al. Comprehensive Strategy to Construct In-House Database for Accurate and Batch Identification of Small Molecular Metabolites. Anal Chem. 2018;90(12):7635–7643. doi: 10.1021/acs.analchem.8b01482. [DOI] [PubMed] [Google Scholar]
- 50.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Zou Y, Xue W, Luo G, Deng Z, Qin P, Guo R, et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol. 2019;37(2):179–185. doi: 10.1038/s41587-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westerhuis JA, van Velzen EJ, Hoefsloot HC, Smilde AK. Multivariate paired data analysis: multilevel PLSDA versus OPLSDA. Metabolomics. 2010;6(1):119–128. doi: 10.1007/s11306-009-0185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23(7):859–868. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 54.Wang B, Li M, Zhao Z, Lu J, Chen Y, Xu Y, et al. Urinary bisphenol A concentration and glucose homeostasis in non-diabetic adults: a repeated-measures, longitudinal study. Diabetologia. 2019;62(9):1591–1600. doi: 10.1007/s00125-019-4898-x. [DOI] [PubMed] [Google Scholar]
- 55.Weimar JD, DiRusso CC, Delio R, Black PN. Functional role of fatty acyl-coenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids. Amino acid residues within the ATP/AMP signature motif of Escherichia coli FadD are required for enzyme activity and fatty acid transport. J Biol Chem. 2002;277(33):29369–29376. doi: 10.1074/jbc.M107022200. [DOI] [PubMed] [Google Scholar]
- 56.Eckel RH, Bornfeldt KE, Goldberg IJ. Cardiovascular disease in diabetes, beyond glucose. Cell Metab. 2021;33(8):1519–1545. doi: 10.1016/j.cmet.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ridker PM, Danielson E, Fonseca FA, Genest J, Gotto AM Jr., Kastelein JJ, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med. 2008;359(21):2195–2207. doi: 10.1056/NEJMoa0807646. [DOI] [PubMed] [Google Scholar]
- 58.Shen L, Gu Y, Qiu Y, Cheng T, Nie A, Cui C, et al. Atorvastatin Targets the Islet Mevalonate Pathway to Dysregulate mTOR Signaling and Reduce beta-Cell Functional Mass. Diabetes. 2020;69(1):48–59. doi: 10.2337/db19-0178. [DOI] [PubMed] [Google Scholar]
- 59.Haynes R, Jiang L, Hopewell JC, Li J, Chen F, Parish S, Landray MJ, Collins R, Armitage J, Collins R; Group HTC . HPS2-THRIVE randomized placebo-controlled trial in 25 673 high-risk patients of ER niacin/laropiprant: trial design, pre-specified muscle and liver outcomes, and reasons for stopping study treatment. Eur Heart J. 2013;34(17):1279–1291. doi: 10.1093/eurheartj/eht055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Handelsman Y, Jellinger PS, Guerin CK, Bloomgarden ZT, Brinton EA, Budoff MJ, et al. Consensus Statement by the American Association of Clinical Endocrinologists and American College of Endocrinology on the Management of Dyslipidemia and Prevention of Cardiovascular Disease Algorithm – 2020 Executive Summary. Endocrine Pract 2020;26(10):1196–1224. doi: 10.4158/CS-2020-0490. [DOI] [PubMed] [Google Scholar]
- 61.He J, Gu D, Reynolds K, Wu X, Muntner P, Zhao J, et al. Serum total and lipoprotein cholesterol levels and awareness, treatment, and control of hypercholesterolemia in China. Circulation. 2004;110(4):405–411. doi: 10.1161/01.CIR.0000136583.52681.0D. [DOI] [PubMed] [Google Scholar]
- 62.Liu YT, Hao HP, Xie HG, Lai L, Wang Q, Liu CX, et al. Extensive intestinal first-pass elimination and predominant hepatic distribution of berberine explain its low plasma levels in rats. Drug Metab Dispos. 2010;38(10):1779–1784. doi: 10.1124/dmd.110.033936. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Li X, Zou D, Liu W, Yang J, Zhu N, et al. Treatment of type 2 diabetes and dyslipidemia with the natural plant alkaloid berberine. J Clin Endocrinol Metab. 2008;93(7):2559–2565. doi: 10.1210/jc.2007-2404. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PloS One. 2012;7(8):e42529. doi: 10.1371/journal.pone.0042529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Black PN, DiRusso CC. Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiol Mol Biol Rev. 2003;67(3):454–472. doi: 10.1128/MMBR.67.3.454-472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Groot PH, Scholte HR, Hulsmann WC. Fatty acid activation: specificity, localization, and function. Adv Lipid Res. 1976;14:75–126. [DOI] [PubMed] [Google Scholar]
- 67.Yao J, Rock CO. Exogenous fatty acid metabolism in bacteria. Biochimie. 2017;141:30–39. doi: 10.1016/j.biochi.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dirusso CC, Black PN. Bacterial long chain fatty acid transport: gateway to a fatty acid-responsive signaling system. J Biol Chem. 2004;279(48):49563–49566. doi: 10.1074/jbc.R400026200. [DOI] [PubMed] [Google Scholar]
- 69.Gu Y, Wang X, Li J, Zhang Y, Zhong H, Liu R, et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun. 2017;8(1):1785. doi: 10.1038/s41467-017-01682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Temel RE, Brown JM. A new model of reverse cholesterol transport: enTICEing strategies to stimulate intestinal cholesterol excretion. Trends Pharmacol Sci. 2015;36(7):440–451. doi: 10.1016/j.tips.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Le May C, Berger JM, Lespine A, Pillot B, Prieur X, Letessier E, et al. Transintestinal cholesterol excretion is an active metabolic process modulated by PCSK9 and statin involving ABCB1. Arterioscler Thromb Vasc Biol. 2013;33(7):1484–1493. doi: 10.1161/ATVBAHA.112.300263. [DOI] [PubMed] [Google Scholar]
- 72.Marques FZ, Nelson E, Chu PY, Horlock D, Fiedler A, Ziemann M, et al. High-Fiber Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in Hypertensive Mice. Circulation. 2017;135(10):964–977. doi: 10.1161/CIRCULATIONAHA.116.024545. [DOI] [PubMed] [Google Scholar]
- 73.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nemet I, Saha PP, Gupta N, Zhu W, Romano KA, Skye SM, et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell. 2020;180(5):862–77 e22. doi: 10.1016/j.cell.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


