ABSTRACT
Background
The technique to analyze circulating tumor DNA (ctDNA) in body fluid (so-called “liquid biopsy”) is recently developed.
Aims
Our aim was to assess the utility of liquid biopsy for predicting progression of pancreatic ductal adenocarcinoma (PDAC) after surgical resection or chemotherapy.
Methods
A total of 72 patients with PDAC were retrospectively enrolled for this study, 33 treated surgically and 39 given chemotherapy, either FOLFIRINOX (oxaliplatin/irinotecan/fluorouracil/leucovorin) or gemcitabine plus nab-paclitaxel. Prior to treatment, patients were screened for the presence of KRAS mutations (G12D and G12V) in plasma using droplet digital polymerase chain reaction, and outcomes were compared.
Results
KRAS mutations were identified in plasma samples of 12 patients (36%) underwent surgical resection. Patients with plasma KRAS mutations had significantly shorter disease-free survival (DFS) and overall survival (p < .01 and p = .01, respectively). Of 10 clinical variables analyzed, plasma KRAS mutation was the factor predictive of DFS in multivariate analysis (RR = 3.58, 95% CI: 1.36–9.60; p = .01). Although 12 patients (31%) given chemotherapy tested positive for plasma KRAS mutations, there was no demonstrable relation between plasma KRAS mutations and progression-free survival (PFS) or overall survival (OS) (p = .35 and p = .68, respectively).
Conclusions
In patients with PDAC, detection of KRAS mutations in plasma proved independently predictive of early recurrence after surgical resection but did not correlate with PFS following chemotherapy.
KEYWORDS: Chemotherapy, disease-free survival, droplet digital polymerase chain reaction, progression-fee survival, surgical resection
Introduction
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most aggressive types of cancer. The 5-year survival rate after diagnosis is a dismal 10%,1 with surgical resection offering the only chance of cure. Unfortunately, only 20% of patients are considered surgically resectable at the time of diagnosis;2 and despite curative intent at surgical resection, 60% of these patients develop tumor recurrences.3 In patients with advanced PDAC, the use of combination regimens, namely oxaliplatin, irinotecan, fluorouracil and leucovorin (FOLFIRINOX), or gemcitabine plus nab-paclitaxel, have boosted chemotherapeutic efficacy and currently are standard treatments, although the prognosis remains far from satisfactory.4–6 Biomarkers to predict effects of treatment are instrumental in improving the dire prognosis; yet there are few in routine clinical use, primarily carcinoembryonic antigen (CEA) and carbohydrate antigen (CA19-9), the available options having stagnated for several decades.7–14 Recently, endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) has become a popular means of procuring tissue for histologic examination and detection of malignancy. However, this procedure is invasive, carrying the risk of adverse effects.15–17 An alternative, a much less invasive method, called “liquid biopsy,” which analyzes circulating tumor DNA (ctDNA) in body fluids such as blood and urine, has been in development.18–22 This technique also addresses the issue of intra-tumor heterogeneity (ie, a single tumor with differing molecular characteristics), which may cause erroneous estimates of malignant potential based on limited biopsy specimens.23–26 Ostensibly, ctDNA is derived from the most aggressive parts of tumors, where turnover of cancer cells is the highest. These cells are prone to disintegration, releasing ctDNA into the blood stream. Thus, the information obtained via liquid biopsy provide more general representation of each tumor, unlike the restricted tumor sampling obtained through tissue biopsy. Adequate grading of tumor malignancy is critical in formulating treatment strategies.
Recently, we reported that KRAS mutations found in serum or plasma were viable prognosticators in patients with pancreatic cancer, even though their presence in tissue bears no association with disease progression.27,28 However, the utility of liquid biopsy in predicting individual therapeutic effects of surgical resection and chemotherapy has not been fully examined, particularly since the advent of the now standard FOLFIRINOX or gemcitabine/nab-paclitaxel regimens for advanced pancreatic cancer.4–6,29
In this study, we used a droplet digital polymerase chain reaction (ddPCR) test to evaluate the relation between plasma KRAS mutations and clinical progression of PDAC in patients after surgical resection or recent chemotherapeutic treatment.
Materials and methods
Patients
We enrolled 72 patients with PDAC retrospectively, each admitted to Okayama University Hospital between February 2013 and December 2018. Eligibility criteria were as follows: (i) histopathologic diagnosis of PDAC, (ii) treatment of PDAC by surgical resection or by combination chemotherapy (FOLFIRINOX or gemcitabine plus nab-paclitaxel), and (iii) no medical history of other cancers. A diagnosis of PDAC with intraductal papillary mucinous neoplasm (IPMN) or surgical resection of PDAC following neoadjuvant chemotherapy was grounds for exclusion. Each patient underwent blood tests and dynamic contrast-enhanced CT every 2–6 months after onset of treatment. Disease-free survival (DFS) and progression-free survival (PFS) were defined from the date of diagnosis to the date of recurrence and disease progression identified by dynamic contrast-enhanced CT, respectively. The last follow-up date was in July 2020.
All patients provided written informed consent prior to enrollment. This study was approved by the ethics committee of Okayama University Hospital, conducted in accordance with the Declaration of Helsinki, and listed in the University Hospital Medical Information Network (UMIN) Clinical Trials Registry (UMIN000023529).
Sample preparation
Blood samples were collected from all patients prior to initial treatment, separating plasma within 3 hours after collection by centrifugation (1500 × g, 3000 rpm, 10 min, room temperature). The samples were then stored at −30°C for later DNA extraction. Cell-free DNA was subsequently extracted from 1 mL of plasma (QIAamp Circulating Nucleic Acid Kit; Qiagen, GmbH, Hilden, Germany) in accordance with kit instructions. All DNA eluents (50 uL) were again frozen (−30°C) pending droplet digital PCR. A Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) served for DNA quantitation.
Droplet digital PCR
The presence of KRAS mutations were detected via droplet digital PCR (QX200 system; Bio-Rad Laboratories, Hercules, CA, USA) as described previously,30 using the two customary probes (G12D and G12V) for KRAS mutations in pancreatic cancer.31,32 Plasma DNA eluent (5 μL) was combined with Droplet PCR Supermix (10 μL; Bio-Rad Laboratories), primer/probe mixture (2 μL), and sterile DNase- and RNase-free water (5 μL). This mixture (22 μL) was then added to Droplet Generation Oil (70 μL; Bio-Rad Laboratories) to produce droplets. The emulsion was thermal cycled as follows: (1) enzyme activation (10 min, 95°C); (2) a 40-cycle series (30 sec, 94°C); (3) a completion cycle (1 min, 60°C); and (4) enzymatic deactivation (10 min, 98°C). Thereafter, the fluorescence signal of each droplet was measured. Each sample was tested in duplicate to increase the sensitivity because the frequency of KRAS mutations derived from cancer is very low that may lead to false negative.
Data analysis
Fluorescence signal data were analyzed using proprietary software (Quanta v1.4.0; Bio-Rad Laboratories) as directed, to determine the number of droplets positive for wild-type and/or mutant KRAS. In this study, we set the threshold of fluorescence intensity as 2000 which we analyzed previously.27 Samples were deemed positive for KRAS mutation if there is one or more positive droplets for mutant KRAS in either one of the two analytic attempts.
Statistical analyses
Baseline characteristics were expressed as medians and ranges. All pertinent testing was two-sided, setting significance at p < .05. Survival curves were generated by Kaplan-Meier method and compared via log rank test. Uni- and multivariate Cox proportional hazards models were applied to assess factors linked with survival. Those yielding p-values <0.10 by univariate analysis qualified for multivariate analysis, as did CA19-9 (a reported risk factor in this setting). Above computations relied on standard software (JMP v13.0; SAS Institute, Cary, NC, USA).
Results
Patients characteristics
Median patient age was 70 years, and 46 patients (64%) were male. Median primary tumor size was 28 mm (Table 1). The distribution of patients by disease stage was as follows: I, 5 (7%); II, 30 (42%); III, 18 (25%); and IV, 19 (26%). As initial therapy, 33 patients (46%; stages I–III) underwent surgical resection, and the remaining 39 patients (54%; stages II–IV) received chemotherapeutic regimens (FOLFIRINOX, 14; gemcitabine plus nab-paclitaxel, 25). Median of cell-free DNA (cfDNA) concentration was 15.9 ng/mL (5–645 ng/mL). There was no significant difference between median cfDNA concentration of patients treated with surgery and those with chemotherapy (16.6 ng/mL and 15.5 ng/mL, p = .47).
Table 1.
Patient characteristics
| Surgical resection (n = 33) | Chemotherapy (n = 39) | Total (N = 72) | |
|---|---|---|---|
| Age, years | 73 (60–88) | 67 (52–78) | 70 (52–88) |
| Gender, male (%) | 20 (61%) | 26 (67%) | 46 (64%) |
| DM+ | 10 (30%) | 17 (44%) | 27 (38%) |
| CEA, ng/mL | 3.02 (0.2–35.7) | 4.03 (0.4–93.8) | 4 (0.2–93.8) |
| CA19-9, IU/mL | 63.5 (0.6–2718) | 208 (0.6–7121) | 126.8 (0.6–7121) |
| Tumor site (head) | 15 (45%) | 17 (44%) | 32 (44%) |
| Tumor size, mm | 23 (13–60) | 30 (15–58) | 28 (13–60) |
| Stage (UICC) | |||
| I | 5 (15%) | 0 (0%) | 5 (7%) |
| II | 25 (76%) | 5 (13%) | 30 (42%) |
| III | 3(9%) | 15 (38%) | 18 (25%) |
| IV | 0 (0%) | 19 (49%) | 19 (26%) |
| Positive margin | 6 (19%) | ||
| Adjuvant therapy | 23 (70%) | ||
| cfDNA amount (ng/mL) | 15.5 (5–331) | 16.6 (7.4–645) | 15.9 (5–645) |
DM, diabetes mellitus; CEA, carcinoembryonic antigen; UICC, Union for International Cancer Control; cfDNA, cell-free DNA.
Plasma KRAS mutation frequency
Overall Plasma KRAS mutations (G12D or G12V) were identified in 12 patients (36%) underwent surgical resection, and 12 patients (31%) with chemotherapy (Table 2). The frequency of G12D, which was 27% (n = 9) in instances of surgical resection and 21% (n = 8) in patients given chemotherapy, exceeded the frequency of G12V. One patient treated with chemotherapy had both of G12D and G12V. Eight of 23 patients (35%) who received adjuvant chemotherapy after surgical resection were positive for KRAS mutation. The positive rate was not statistically different from that of the patients without adjuvant chemotherapy (40%, p = .77).
Table 2.
Frequencies of KRAS mutations
| Surgical resection (n = 33) | Chemotherapy (n = 39) | Total (N = 72) | |
|---|---|---|---|
| G12D | 9 (27%) | 8 (21%) | 17 (24%) |
| G12V | 3 (9%) | 5 (13%) | 8 (11%) |
| KRAS (G12D or G12V) | 12 (36%) | 12 (31%) | 24 (33%) |
KRAS mutations, postsurgical recurrence and overall survival
In patients underwent surgical resection, the median follow-up period was 26.2 months (range: 5.3–74.2 months). During this time, recurrence developed in 20 patients (61%), presenting predominantly as distant metastasis (n = 16, 48%). Local recurrence and lymph node metastasis were observed in two patients, respectively. The ratio of patients with distant metastasis was not different between the patients with and without positive plasma KRAS mutations (p < .59). DFS time was significantly curtailed in patients with positive (vs negative) for plasma KRAS mutations (p < .01) (Figure 1(a)). Median DFS times in patients with and without plasma KRAS mutations were 7.7 months and 26.2 months, respectively. In this study, all DNA samples were examined in duplicate. The presence of plasma KRAS mutations was classified in three groups; no KRAS mutation (21 patients), once KRAS mutation positive (8 patients) or twice KRAS mutation positive (4 patients) in two tests. DFS in those three groups are clearly different (p = .01) (Figure 1(b)). Patients with KRAS mutation in both of two tests had clearly shorter DFS, that means all patients had recurrence within 6 months, while patients without KRAS mutation in neither of two tests had significantly longer DFS.
Figure 1.
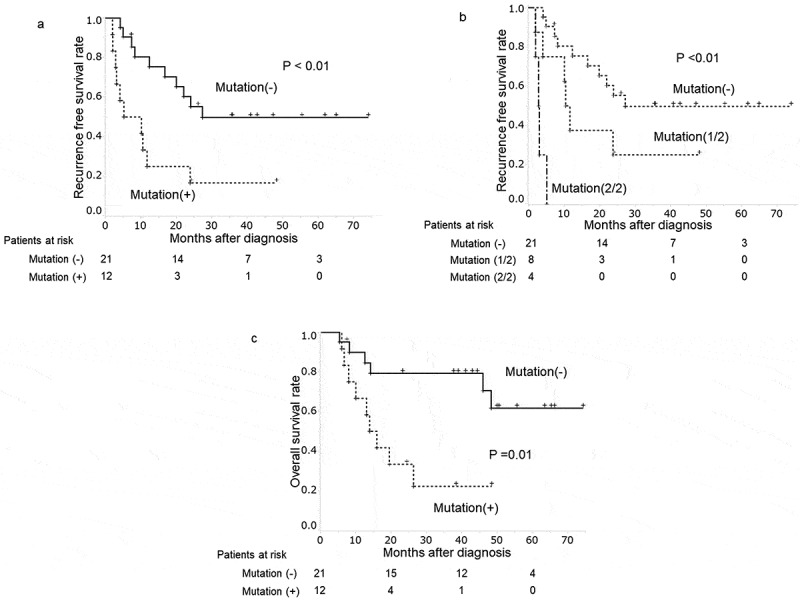
Plasma KRAS mutations and clinical progression of patients surgically treated for PDAC
(a) Kaplan Meier curve of patients with plasma KRAS mutations showed early recurrence (p < .01).(b) Disease-free survival time classified with the frequency of plasma KRAS mutations; No plasma KRAS mutation, once KRAS mutation positive or twice KRAS mutation positive in two tests.(c) Kaplan Meier curve of patients with plasma KRAS mutations showed significantly poor overall survival time after surgical resection (p = .01).
The univariate analysis showed plasma positivity for KRAS mutations, tumor size (≥30 mm) and CA19-9 (≥37mAU/mL) had significant associations with shortened DFS (p = .01, p = .01 and p = .02, respectively) (Table 3). The multivariate analysis revealed that the presence of plasma KRAS mutations and tumor size (≥30 mm) emerged as a significant variable (RR = 3.37, 95% CI: 1.36–9.60; p = .01 and RR = 6.36, 95% CI: 1.99–21.38; p = .01, respectively).
Table 3.
Analyses of disease-free survival (surgically treated patients)
| Univariate |
Multivariate |
|||||
|---|---|---|---|---|---|---|
| RR | 95% CI | P-value | RR | 95% CI | P-value | |
| Age (≤70 yrs) | 1.05 | 0.39–2.56 | .92 | 0.30 | 0.08–1.01 | .05 |
| Gender (male) | 0.84 | 0.37–2.09 | .70 | |||
| Tumor site (head) | 1.27 | 0.51–3.07 | .60 | |||
| Tumor size (≥30 mm) | 3.37 | 1.36–8.35 | .01 | 6.36 | 1.99–21.38 | .01 |
| Stage III (UICC) | 2.18 | 0.50–6.66 | .26 | |||
| CEA (≥5 ng/mL) | 1.75 | 0.61–4.39 | .28 | |||
| CA19-9 (≥37 mAU/mL) | 2.96 | 1.14–9.16 | .02 | 3.02 | 1.02–10.36 | .05 |
| Adjuvant chemotherapy | 0.71 | 0.22–2.16 | .54 | |||
| Positive margin | 1.25 | 0.36–3.45 | .70 | |||
| Plasma KRAS mutation | 3.37 | 1.36–8.35 | .01 | 3.58 | 1.36–9.60 | .01 |
CEA, carcinoembryonic antigen; RR, relative risk; UICC, Union for International Cancer Control.
During follow-up period, 15 out of 25 patients treated with surgery died with PDAC. Overall survival of patients with plasma KRAS mutations was significantly shorter than that of patients without plasma KRAS mutations (p = .01) (Figure 1(c)).
KRAS mutations and progression-free survival after chemotherapy
The median follow-up period of patients given chemotherapy was 13.6 months (range: 2.4–47.6 months). During this time, 32 patients (82%) changed chemotherapeutic regimens due to side effects (n = 12) or disease progression (n = 20). The PFS time between patients with and without plasma KRAS mutations was not significantly different (p = .35) (Figure 2(a)). Their median PFS time were 5.3 and 6.9 months respectively.
Figure 2.
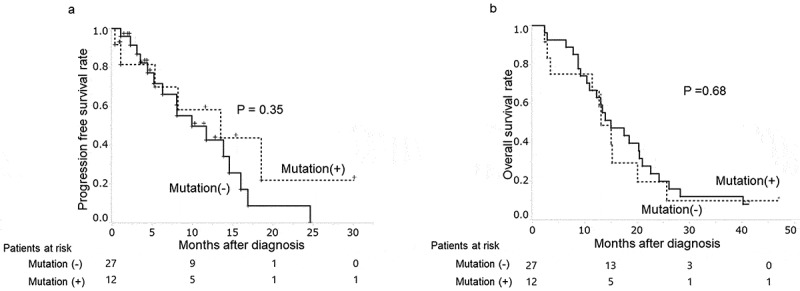
Plasma KRAS mutations and clinical progression patients given chemotherapy for PDAC
(a) Progression-free survival time in pancreatic cancer patients with and without plasma KRAS mutations.(b) Overall survival time in pancreatic cancer patients with and without plasma KRAS mutations.
In univariate analysis of PFS-related variables following chemotherapy (including the presence of plasma KRAS mutations), no significant factors were identified (Table 4). Multivariate analysis was conducted using age (≥70 years) and disease stage (IV), weakly correlating with PFS in univariate analysis (p < .10), as well as CA19-9 and plasma KRAS mutation positivity, as test parameters. The positivity of plasma KRAS mutations was not the significant factor for PFS time, but age (≥70 years) and disease stage (IV) proved to be significant (p < .01 each).
Table 4.
Analyses of progression-free survival (chemotherapy recipients)
| Univariate |
Multivariate |
|||||
|---|---|---|---|---|---|---|
| RR | 95% CI | P-value | RR | 95% CI | P-value | |
| Age (≤70 yrs) | 2.83 | 1.05–9.05 | .04 | 3.90 | 1.38–13.1 | <.01 |
| Gender (male) | 1.43 | 0.58–4.06 | .45 | |||
| Tumor site (head) | 0.90 | 0.36–2.14 | .81 | |||
| Tumor size (≥30 mm) | 1.37 | 0.57–3.62 | .50 | |||
| Stage IV (UICC) | 2.30 | 0.95–5.76 | .06 | 4.00 | 1.44–12.2 | <.01 |
| CEA (≥5 ng/mL) | 1.49 | 0.63–3.48 | .36 | |||
| CA19-9 (≥37 mAU/mL) | 1.81 | 0.74–5.10 | .20 | 1.93 | 0.78–5.48 | .16 |
| Plasma KRAS mutation | 0.63 | 0.22–1.57 | .34 | 0.51 | 0.17–1.33 | .17 |
CEA, carcinoembryonic antigen; RR, relative risk; UICC, Union for International Cancer Control.
Among them, 34 patients died with PDAC. Ten patients had plasma KRAS mutations and other 15 patients were without plasma KRAS mutation. The presence of plasma KRAS mutations also did not affect their overall survival (p = .68) (Figure 2(b)).
Discussion
It is readily acknowledged that KRAS mutations are detectable even in early stages of PDAC and pancreatic intraepithelial neoplasia (PanIN), so their presence in cancerous tissue carries no weight in predicting outcomes of patients with PDAC.27 However, some researchers and our previous study have confirmed that overall survival is poor in patients whose blood samples harbor KRAS mutations.27,28,33 Herein, we mainly focused on disease-free survival or progression-free survival of patients recently treated by surgical resection or combination chemotherapy (FOLFIRINOX or gemcitabine plus nab-paclitaxel, the current standard regimens for advanced pancreatic cancer),4–6,29 assessing the clinical utility of detecting plasma KRAS mutations.
Our study showed even droplet digital PCR could detect plasma KRAS mutations in 36% of operable patients that was similar to the rates reported in previous studies (31–48%).27,34,35 Droplet digital PCR is one of the most beneficial methods to detect rare mutations in cfDNA. The high sensitivity relies on technology which partition cfDNA into around 10000 droplets and perform PCR in each independent droplet.36,37In this study, around 500 droplets were filled with cfDNA that includes genomic DNA contamination, and others were blank because of the low amount of plasma cfDNA. So, every single cfDNA fragment could be distributed to each droplet, that makes the analysis accurate.
Also, we did not use mutant allele frequencies (MAF) generally used for cutoff value.
Aside from the finding that DFS time was shorter in patients with (vs without) plasma KRAS mutations, results of multivariate analysis underscored the superiority of plasma KRAS mutations as a biomarker of early recurrence (p < .01), surpassing CA19-9 level as customary measures in this regard.38,39 Also, the analysis classified by the frequency of plasma KRAS mutations showed patients with plasma KRAS mutations in both of two tests had clearly shorter DFS time. The background of this result is that the tumor with more aggressive turnover flows out more tumor cells and tumor DNA to the blood, called circulating tumor cells (CTC) and circulating tumor DNA (ctDNA). That might lead to micrometasitasis before surgical resection. Also, one research said ctDNA might originate from micrometastasis.40 So, our study can indicate multiple tests in one sample can catch a higher risk group of recurrence before surgical treatment.
On the other hand, the presence of plasma KRAS mutations (31% of patients given chemotherapy) in patients treated with chemotherapy had no relation to their efficacy. This study enrolled only the patients treated with FOLFIRINOX or gemcitabine plus nab-paclitaxel, those are the current first line option for chemotherapy. Those regimens have been using for several years, but it has been unclear how the presence of plasma KRAS mutations affect the clinical outcome of this current chemotherapy. The current result newly shows that the current regimens (FOLFIRINOX or gemcitabine plus nab-paclitaxel) should thus exert anti-tumor effects in even highly proliferative tumor associated with plasma KRAS mutations.
The reports about ctDNA clinical progression generally have been using three (G12D, G12V, and G12R) or more kinds of KRAS mutations.41,42 Our prior attempts at identifying all three mutations in serum and plasma showed rates of G12R detection that were consistently quite low; and similar results have been reported by another group.34 Therefore, we restricted our analysis to G12D and G12V. Indeed, detection rates of overall KRAS mutations in this study did not differ from those of previous reports, implying that just two KRAS mutations (G12D and G12V) are sufficient in analyzing liquid biopsies of PDAC.
This study has several limitations. First, the number of patients enrolled was relatively small. But, the sufficient follow-up period clearly revealed the relation between the presence of plasma KRAS mutations and the disease progression. Second, 21 patients treated with surgical resection and 17 patients received chemotherapy were the same as those enrolled in our previous study.27 However, this study focused on different points with longer follow-up period; the disease-free survival and progression-free-survival in each treatment method, though previous study analyzed only the overall survival without regard to the treatment methods. Third, we excluded the patients who received neoadjuvant chemotherapy, which was becoming a standard option for resectable and boderline-resectable PDAC. This current study can give the suggestions how to decide the treatment method especially surgical resection.
In conclusion, we have confirmed the feasibility of detecting plasma KRAS mutations in patients with PDAC. Their presence can be a good biomarker for early recurrence after surgical resection. Plasm KRAS mutations and the efficacies of chemotherapeutic regimens (FOLFIRINOX or gemcitabine plus nab-paclitaxel) were unrelated.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A.. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Barugola G, Partelli S, Marcucci S, Sartori N, Capelli P, Bassi C, Pederzoli P, Falconi M. Resectable pancreatic cancer: who really benefits from resection? Ann Surg Oncol. 2009;16:3316–3322. doi: 10.1245/s10434-009-0670-7. [DOI] [PubMed] [Google Scholar]
- 3.La Torre M, Nigri G, Lo Conte A, Mazzuca F, Tierno SM, Salaj A, Marchetti P, Ziparo V, Ramacciato G. Is a preoperative assessment of the early recurrence of pancreatic cancer possible after complete surgical resection? Gut Liver. 2014;8(1):102–108. doi: 10.5009/gnl.2014.8.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein D, El-Maraghi RH, Hammel P, Heinemann V, Kunzmann V, Sastre J, Scheithauer W, Siena S, Tabernero J, Teixeira L, et al. nab-Paclitaxel plus gemcitabine for metastatic pancreatic cancer: long-term survival from a phase III trial. J Natl Cancer Inst. 2015;107:dju413–dju413. doi: 10.1093/jnci/dju413. [DOI] [PubMed] [Google Scholar]
- 5.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, Adenis A, Raoul J-L, Gourgou-Bourgade S, de la Fouchardière C, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 7.Haas M, Heinemann V, Kullmann F, Laubender RP, Klose C, Bruns CJ, Holdenrieder S, Modest DP, Schulz C, Boeck S, et al. Prognostic value of CA 19-9, CEA, CRP, LDH and bilirubin levels in locally advanced and metastatic pancreatic cancer: results from a multicenter, pooled analysis of patients receiving palliative chemotherapy. J Cancer Res Clin Oncol. 2013;139(4):681–689. doi: 10.1007/s00432-012-1371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tas F, Karabulut S, Ciftci R, Sen F, Sakar B, Disci R, Duranyildiz D. Serum levels of LDH, CEA, and CA19-9 have prognostic roles on survival in patients with metastatic pancreatic cancer receiving gemcitabine-based chemotherapy. Cancer Chemother Pharmacol. 2014;73(6):1163–1171. doi: 10.1007/s00280-014-2450-8. [DOI] [PubMed] [Google Scholar]
- 9.Hammad N, Heilbrun LK, Philip PA, Shields AF, Zalupski MM, Venkatramanamoorthy R, El-Rayes BF. CA19-9 as a predictor of tumor response and survival in patients with advanced pancreatic cancer treated with gemcitabine based chemotherapy. Asia Pac J Clin Oncol. 2010;6(2):98–105. doi: 10.1111/j.1743-7563.2010.01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An X, Li YH, Lin XB, Wang FH, Feng F, Xu RH, Jiang WQ, He YJ.. Prognostic value of serum CA19-9 in patients with advanced pancreatic cancer receiving gemcitabine based chemotherapy. Ai Zheng. 2009;28:286–291. [PubMed] [Google Scholar]
- 11.Zamcheck N. The present status of carcinoembryonic antigen (CEA) in diagnosis, detection of recurrence, prognosis and evaluation of therapy of colonic and pancreatic cancer. Clin Gastroenterol. 1976;5:625–638. doi: 10.1016/S0300-5089(21)00311-4. [DOI] [PubMed] [Google Scholar]
- 12.Ni XG, Bai XF, Mao YL, Shao YF, Wu JX, Shan Y, Wang CF, Wang J, Tian YT, Liu Q, et al. The clinical value of serum CEA, CA19-9, and CA242 in the diagnosis and prognosis of pancreatic cancer. Eur J Surg Oncol. 2005;31:164–169. doi: 10.1016/j.ejso.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Kilickap S, Arslan C. The effect on prognosis of postoperative CA 19-9 level in patients with pancreatic cancer. Am J Clin Oncol. 2010;33:320; author reply. doi: 10.1097/COC.0b013e31819fe1ff. [DOI] [PubMed] [Google Scholar]
- 14.Wu L, Huang P, Wang F, Li D, Xie E, Zhang Y, Pan S. Relationship between serum CA19-9 and CEA levels and prognosis of pancreatic cancer. Ann Transl Med. 2015;3:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamada T, Yasunaga H, Nakai Y, Isayama H, Horiguchi H, Matsuda S, Fushimi K, Koike K. Severe bleeding and perforation are rare complications of endoscopic ultrasound-guided fine needle aspiration for pancreatic masses: an analysis of 3,090 patients from 212 hospitals. Gut Liver. 2014;8:215–218. doi: 10.5009/gnl.2014.8.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada T, Yasunaga H, Nakai Y, Isayama H, Horiguchi H, Matsuda S, Fushimi K, Koike K. Rarity of severe bleeding and perforation in endoscopic ultrasound-guided fine needle aspiration for submucosal tumors. Dig Dis Sci. 2013;58:2634–2638. doi: 10.1007/s10620-013-2717-7. [DOI] [PubMed] [Google Scholar]
- 17.Carrara S, Arcidiacono PG, Mezzi G, Petrone MC, Boemo C, Testoni PA. Pancreatic endoscopic ultrasound-guided fine needle aspiration: complication rate and clinical course in a single centre. Dig Liver Dis. 2010;42:520–523. doi: 10.1016/j.dld.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Ma M, Zhu H, Zhang C, Sun X, Gao X, Chen G. “Liquid biopsy”-ctDNA detection with great potential and challenges. Ann Transl Med. 2015;3:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, Lax S, Waldispuehl-Geigl J, Mauermann O, Lackner C, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 20.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4:650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 22.De Mattos-Arruda L, Cortes J, Santarpia L, Vivancos A, Tabernero J, Reis-Filho JS, Seoane J. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat Rev Clin Oncol. 2013;10:377–389. doi: 10.1038/nrclinonc.2013.80. [DOI] [PubMed] [Google Scholar]
- 23.Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61:112–123. doi: 10.1373/clinchem.2014.222679. [DOI] [PubMed] [Google Scholar]
- 24.Diaz LA Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J clin oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Emburgh BO, Arena S, Siravegna G, Lazzari L, Crisafulli G, Corti G, Mussolin B, Baldi F, Buscarino M, Bartolini A, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun. 2016;7(1):13665. doi: 10.1038/ncomms13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ako S, Nouso K, Kinugasa H, Dohi C, Matushita H, Mizukawa S, Muro S, Akimoto Y, Uchida D, Tomoda T, et al. Utility of serum DNA as a marker for KRAS mutations in pancreatic cancer tissue. Pancreatology. 2017;17(2):285–290. doi: 10.1016/j.pan.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Kinugasa H, Nouso K, Miyahara K, Morimoto Y, Dohi C, Tsutsumi K, Kato H, Matsubara T, Okada H, Yamamoto K, et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121(13):2271–2280. doi: 10.1002/cncr.29364. [DOI] [PubMed] [Google Scholar]
- 29.Tjensvoll K, Lapin M, Buhl T, Oltedal S, Steen-Ottosen Berry K, Gilje B, Søreide JA, Javle M, Nordgård O, Smaaland R, et al. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol Oncol. 2016;10(4):635–643. doi: 10.1016/j.molonc.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berz D, Raymond VM, Garst JH, Erlander MG. Non-invasive urine testing of EGFR activating mutation and T790M resistance mutation in non-small cell lung cancer. Exp Hematol Oncol. 2015;5:24. doi: 10.1186/s40164-016-0052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reckamp KL, Melnikova VO, Karlovich C, Sequist LV, Camidge DR, Wakelee H, Perol M, Oxnard GR, Kosco K, Croucher P, et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol. 2016;11:1690–1700. doi: 10.1016/j.jtho.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Meng Q, Wang C, Li F, Zhu Z, Liu S, Shi Y, Huang J, Chen S, Li C. Investigation of transrenal KRAS mutation in late stage NSCLC patients correlates to disease progression. Biomarkers. 2017;22(7):654–660 [DOI] [PubMed] [Google Scholar]
- 33.Nakano Y, Kitago M, Matsuda S, Nakamura Y, Fujita Y, Imai S, Shinoda M, Yagi H, Abe Y, Hibi T, et al. KRAS mutations in cell-free DNA from preoperative and postoperative sera as a pancreatic cancer marker: a retrospective study. Br J Cancer. 2018;118(5):662–669. doi: 10.1038/bjc.2017.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadano N, Murakami Y, Uemura K, Hashimoto Y, Kondo N, Nakagawa N, Sueda T, Hiyama E. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br J Cancer. 2016;115(1):59–65. doi: 10.1038/bjc.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takai E, Totoki Y, Nakamura H, Morizane C, Nara S, Hama N, Suzuki M, Furukawa E, Kato M, Hayashi H, et al. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci Rep. 2015;5(1):18425. doi: 10.1038/srep18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishio K, Kimura K, Amano R, Yamazoe S, Ohrira G, Nakata B, Hirakawa K, Ohira M. Preoperative predictors for early recurrence of resectable pancreatic cancer. World J Surg Oncol. 2017;15(1):16. doi: 10.1186/s12957-016-1078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hata S, Sakamoto Y, Yamamoto Y, Nara S, Esaki M, Shimada K, Kosuge T. Prognostic impact of postoperative serum CA 19-9 levels in patients with resectable pancreatic cancer. Ann Surg Oncol. 2012;19(2):636–641. doi: 10.1245/s10434-011-2020-9. [DOI] [PubMed] [Google Scholar]
- 40.Schwarzenbach H, Chun FK, Lange I, Carpenter S, Gottberg M, Erbersdobler A, Friedrich MG, Huland H, Pantel K. Detection of tumor-specific DNA in blood and bone marrow plasma from patients with prostate cancer. Int J Cancer. 2007;120(7):1465–1471. doi: 10.1002/ijc.22470. [DOI] [PubMed] [Google Scholar]
- 41.Miglio U, Oldani A, Mezzapelle R, Veggiani C, Paganotti A, Garavoglia M, Boldorini R. KRAS mutational analysis in ductal adenocarcinoma of the pancreas and its clinical significance. Pathol Res Pract. 2014;210(5):307–311. doi: 10.1016/j.prp.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 42.Visani M, De Biase D, Baccarini P, Fabbri C, Polifemo AM, Zanini N, Pession A, Tallini G. Multiple KRAS mutations in pancreatic adenocarcinoma: molecular features of neoplastic clones indicate the selection of divergent populations of tumor cells. Int J Surg Pathol. 2013;21:546–552. doi: 10.1177/1066896912475073. [DOI] [PubMed] [Google Scholar]


