ABSTRACT
Immunoglobulin G-based monoclonal antibodies (mAbs) have become a dominant class of biotherapeutics in recent decades. Approved antibodies are mainly of the subclasses IgG1, IgG2, and IgG4, as well as their derivatives. Over the decades, the selection of IgG subclass has frequently been based on the needs of Fc gamma receptor engagement and effector functions for the desired mechanism of action, while the effect on drug product developability has been less thoroughly characterized. One of the major reasons is the lack of systematic understanding of the impact of IgG subclass on the molecular properties. Several efforts have been made recently to compare molecular property differences among these IgG subclasses, but the conclusions from these studies are sometimes obscured by the interference from variable regions. To further establish mechanistic understandings, we conducted a systematic study by grafting three independent variable regions onto human IgG1, an IgG1 variant, IgG2, and an IgG4 variant constant domains and evaluating the impact of subclass and variable regions on their molecular properties. Structural and computational analysis revealed specific molecular features that potentially account for the differential behavior of the IgG subclasses observed experimentally. Our data indicate that IgG subclass plays a significant role on molecular properties, either through direct effects or via the interplay with the variable region, the IgG1 mAbs tend to have higher solubility than either IgG2 or IgG4 mAbs in a common pH 6 buffer matrix, and solution behavior relies heavily on the charge status of the antibody at the desirable pH.
KEYWORDS: Monoclonal antibodies, IgG subclass, viscosity, solubility, turbidity, isoelectric point, hydrophobicity, thermal unfolding, homology modeling
Introduction
Over the past 20 years, monoclonal antibodies (mAbs) have become the most dominant biotherapeutics in the pharmaceutical industry.1 Since the commercialization of the first antibody therapeutic (Orthoclone Okt3) in 1986, over 100 antibody-related therapeutics have been approved in the United States for various indications such as oncology, immunology, cardiovascular, neurology, and infectious diseases (www.antibodysociety.org/antibody-therapeutics-product-data/). The high specificity, long half-life, and generally safe profiles have made antibodies attractive as human therapeutics.
Although there are five naturally occurring classes of human immunoglobulins (IgA, IgD, IgE, IgG, and IgM), the majority of recombinant therapeutic antibodies under development to date belong to the IgG class. Among the antibody therapeutics marketed in the United States and Europe, ~90% (including antibody-drug conjugates) are IgG immunoglobulin and ~ 10% are either bispecific antibody, PEGylated antigen-binding fragments (Fabs), single-chain variable fragment (scFv), Fab, or nanobody. The IgG antibody family consists of IgG1, IgG2, IgG3, and IgG4 subclasses based on small differences in the constant region of the heavy chain (HC). These four IgG subclasses distinctly differ in their effector functions via interactions with Fc gamma receptors (FcγRs) and C1q, and only IgG1, IgG2, and IgG4 have been used for human therapeutics due to the favorable serum half-life. The most popular IgG subclass used as mAb therapeutics is IgG1, and its variants. Currently, ~74% of approved IgG antibody therapeutics are IgG1-based, while IgG2 and IgG4 comprise ~13% each. Inspired by the success of antibody therapeutics, there has been a tremendous interest in engineering antibodies to modulate effector functions such as antibody-dependent cell-mediated cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), and complement-dependent cytotoxicity (CDC) to derive superior antibody therapeutics, and some of these have also entered the clinical pipeline or have become human therapeutics.2,3
Due to their unique structures, the different IgG subclasses have distinct characteristics in product development. However, systematic physicochemical property studies across IgG1, IgG2, and IgG4 subclasses remain limited. This is compounded by the difficulty of separating the effect of the highly diversified variable region sequence from the IgG subclass structures. In 1997, Roux et al. found that IgG1 has greater Fab-Fab/Fab-Fc flexibility than IgG4 while IgG4 has greater flexibility than IgG2 with identical variable regions by electron microscopy.4 Later, Tian et al. used small-angle X-ray scattering technique to characterize the conformational flexibility of IgG1, IgG2, and IgG4 and confirmed the findings of Roux et al.5 These authors also suggested that the flexibility of IgG1 might shield the aggregation-prone motifs and contribute to IgG1’s stability against aggregation.6 In another study, Tian et al. found that antibodies of different IgG subclasses had distinctly different aggregation pathways under low pH condition, indicating the electrostatic charge of IgG subclasses plays a critical role of mAb aggregation.5 More recently, Heads et al. studied the electrostatic interactions and relative aggregation propensities for seven IgG1 and IgG4 pairs and concluded that the net charge state of the variable domain relative to that of the constant domain has a dominant effect on aggregation propensity over the global net charge status of the molecules.7 Despite these previous reports, the findings cannot be used to fully explain the relationship between IgG subclass and the physiochemical properties of IgG, such as hydrophobicity, viscosity, and solubility. The desire to understand the direct impact of IgG subclass on these physiochemical properties has recently become stronger due to the demand for faster drug discovery and development.
In this study, we conducted a systematic investigation of three independent series of mAbs. The molecules in each series share identical variable region sequences, bind to the same unique target, and are composed of IgG1, an IgG1 variant with diminished effector functions (IgG1EN), IgG2, and an IgG4 variant with reduced FcγR interaction and enhanced serum stability (IgG4PAA). By a detailed analysis of the biochemical and biophysical properties of these antibodies, we identified trends in molecular properties that are unique to each IgG subclass, as well as the interplay between the variable region and the IgG subclass structure. Homology modeling and structural analysis are leveraged to shed light on the mechanism of the observed differences. Our findings provide useful insight into the relationship between antibody physicochemical properties and IgG subclass, laying the foundation for subsequent investigation into how these properties manifest on developability of these antibodies.
Results
Description of the antibodies tested
Three unrelated humanized variable regions (Fv A, Fv B, and Fv C) for unique targets with distinct charge and hydrophobicity profiles were selected and cloned into respective IgG subclass series. Each series is comprised of IgG1, IgG1 L234A, L235E, G237A, A330S, P331S (IgG1EN),8 IgG2, and IgG4 S228P, L234A, L235A (IgG4PAA),9–11 resulting in a total of 12 antibodies (Table 1).
Table 1.
List of the antibodies used in the study
| mAb name | Constant region |
Variable region |
Mutations from germline | |
|---|---|---|---|---|
| CH | CL | Fv | ||
| IgG1-A | IgG1 | Kappa | A | None |
| IgG1EN-A | IgG1EN | Kappa | A | L234A, L235E, G237A, A330S, P331S |
| IgG2-A | IgG2 | Kappa | A | None |
| IgG4-A | IgG4PAA | Kappa | A | S228P, F234A, L235A |
| IgG1-B | IgG1 | Kappa | B | None |
| IgG1EN-B | IgG1EN | Kappa | B | L234A, L235E, G237A, A330S, P331S |
| IgG2-B | IgG2 | Kappa | B | None |
| IgG4-B | IgG4PAA | Kappa | B | S228P, F234A, L235A |
| IgG1-C | IgG1 | Kappa | C | None |
| IgG1EN-C | IgG1EN | Kappa | C | L234A, L235E, G237A, A330S, P331S |
| IgG2-C | IgG2 | Kappa | C | None |
| IgG4-C | IgG4PAA | Kappa | C | S228P, F234A, L235A |
All the antibodies were expressed in Chinese hamster ovary (CHO) cells and purified using standard antibody purification procedures. The purity of the mAbs was measured by analytical size-exclusion chromatography (SEC). All mAbs were >96% monomer by SEC-HPLC with the exception of IgG1EN-C and IgG2-C at 90% and 88%, respectively.
Isoelectric points
The isoelectric point (pI) of each mAb was measured by capillary isoelectric focusing (cIEF), and the results are summarized in Table 2 and included in Supplemental Material. With identical variable regions, IgG1 and IgG1EN mAbs demonstrated higher pI (~0.4–1.0 unit) compared to the corresponding IgG2 and IgG4PAA mAbs. The IgG2 mAb also showed ~0.3–0.5 units higher pI in comparison to the IgG4PAA mAb with the same variable region. The mutations introduced into IgG1EN mAbs resulted in a very minor pI reduction of ~0.1–0.2 units. In contrast to the IgG subclass, the variable regions had a more pronounced effect on the pI. The mAbs with Fv A exhibited the highest pIs, ranging from 8.2 to 9.1, regardless of IgG subclass. The mAbs with Fv C demonstrated pIs in the range of 6.3–7.4, while the mAbs with Fv B showed the lowest pI in the range of 6.1–7.1.
Table 2.
Isoelectric points of the antibodies measured by cIEF and calculated from primary sequences
| Antibodies | Measured pI | Calculated pI |
||
|---|---|---|---|---|
| IgG | Fab | Fc | ||
| IgG1-A | 9.1 | 8.0 | 8.4 | 6.7 |
| IgG1EN-A | 9.0 | 7.9 | 8.4 | 6.4 |
| IgG2-A | 8.7 | 7.6 | 7.9 | 6.7 |
| IgG4PAA-A | 8.2 | 7.4 | 8.2 | 5.7 |
| IgG1-B | 7.1 | 6.7 | 6.4 | 6.7 |
| IgG1EN-B | 6.9 | 6.5 | 6.4 | 6.4 |
| IgG2-B | 6.4 | 6.2 | 5.7 | 6.7 |
| IgG4PAA-B | 6.1 | 5.9 | 6.1 | 5.7 |
| IgG1-C | 7.4 | 7.0 | 6.9 | 6.7 |
| IgG1EN-C | 7.2 | 6.8 | 6.9 | 6.4 |
| IgG2-C | 6.7 | 6.4 | 5.8 | 6.7 |
| IgG4PAA-C | 6.3 | 6.0 | 6.5 | 5.7 |
To further understand the impact from IgG subclass constant regions, the pI of the IgG, Fab, and Fc were calculated from the primary sequences (Table 2). The discrepancy between the calculated and measured IgG pI is not surprising in light of the fact the calculated pI values are based on a fixed theoretical pKa value for each amino acid, whereas the measured pI is affected by the three-dimensional structure. Nevertheless, the calculated IgG pIs follow the same trend as the measured IgG pIs. Given the sequence differences in the CH1 domain between the IgG subclasses, the calculated pI of the Fab domain for a given variable region varies by ~0.5–1 unit and is only mildly affected by IgG1, IgG1EN, and IgG4PAA while exhibiting a more notable decrease for IgG2. The decrease for IgG2 Fab is due to multiple charged residue differences within its CH1 domain. There is also a subtle difference in the calculated pIs for the Fc domain between IgG1, IgG1EN, and IgG2, while a more significant pI decrease of 0.7–1.0 unit is found for IgG4PAA.
Thermal stability by differential scanning calorimetry
Differential scanning calorimetry (DSC) was used to evaluate the impact of IgG subclass and variable region on mAb stability against thermal denaturation. The thermal stability profiles of the 12 mAbs are shown in Figure 1. Both IgG subclass and variable region demonstrated significant effects on the thermal stability of the mAbs. The IgG1, IgG2, and IgG4PAA mAbs showed typical thermal stability profiles as reported previously,12–15 with an unfolding onset temperature (Tm-onset) of ~60°C . The thermogram of the IgG2 mAbs, especially IgG2-A, is more complex, presumably due to different disulfide isomer conformations.16,17 For a given variable region and with the assumption that the highest peak originates from the Fab transition, a trend emerges that the IgG1 mAbs demonstrate similar or slightly higher Fab transition temperature than that of the IgG2 mAb and higher Fab transition temperature than that of the IgG4PAA mAb. The mutations in IgG1EN shifted the Tm-onset and CH2 unfolding temperature to as low as ~54°C and ~64°C (Figure 1), respectively, without a significant effect on the thermal unfolding temperatures of the other domains.
Figure 1.
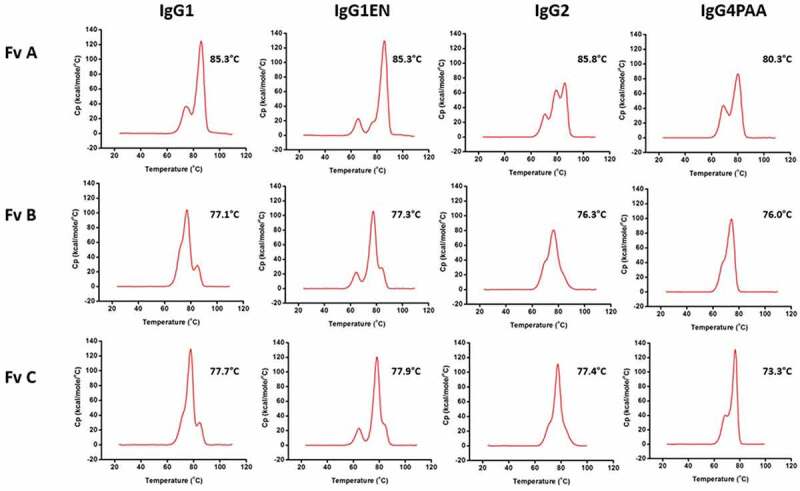
DSC profiles of the 12 antibodies with Fv A, Fv B, Fv C at pH 6. Temperatures listed represent the Tm of the highest peak
Beyond the IgG subclass, the variable regions dominate the Fab unfolding transition temperatures as expected. Antibodies with Fv A showed the highest Fab transition temperature of 80.3–86.0°C, whereas the mAbs with Fv B and Fv C displayed Fab transition temperature of 76.0–77.2°C and 76.4–77.7°C, respectively.
Hydrophobicity
In contrast to pIs and thermal stability, the apparent hydrophobicity measured by hydrophobic interaction chromatography (HIC) is almost solely driven by the variable regions with little impact from the IgG subclass except a small effect from IgG4PAA. The data is summarized as both retention time and hydrophobicity interaction potential (HIP%) in Table 3 and included in Supplemental Material. Longer retention time or higher HIP% indicate molecules with higher hydrophobicity. The mAbs with Fv A and Fv B exhibit retention times ranging from 13.0 to 14.0 minutes or HIP% ranging from 40.0–44.8%, while the mAbs with Fv C are more hydrophobic, with retention times of ~ 16 minutes or HIP% in a range 55.0–56.9%. For each series, a slight increase of hydrophobicity associated with the IgG4PAA subclass is observed.
Table 3.
Hydrophobicity interaction potential of the antibodies by hydrophobic interaction chromatography
| HIC-HPLC retention time (minutes) |
Hydrophobicity interaction potential (HIP%) |
|||||
|---|---|---|---|---|---|---|
| IgG subclass | Fv A | Fv B | Fv C | Fv A | Fv B | Fv C |
| IgG1 | 13.4 | 13.0 | 16.0 | 42.1 | 40.1 | 55.0 |
| IgG1EN | 13.4 | 13.0 | 16.0 | 42.1 | 40.0 | 55.2 |
| IgG2 | 13.5 | 13.0 | 16.1 | 42.3 | 40.1 | 55.7 |
| IgG4PAA | 14.0 | 13.6 | 16.4 | 44.8 | 43.0 | 56.9 |
Viscosity
The viscosity of each mAb except IgG2-A was measured at 125 mg/mL in a common pH 6 buffer matrix. The IgG2-A mAb was not measured due to significant precipitation during storage at 5°C in an intermediate buffer at a relative high protein concentration before buffer exchanging to pH 6. The mechanism of precipitation is pending further investigation. The viscosity results are summarized in Table 4. The effect from both variable region and the IgG subclass on viscosity is evident.
Table 4.
Viscosity of the antibodies
| Variable region | Viscosity (cP) |
|||
|---|---|---|---|---|
| IgG1 | IgG1EN | IgG2 | IgG4PAA | |
| A | 18.4 | 20.9 | ND* | 28.7 |
| B | 9.1 | 10.0 | 10.0 | 13.4 |
| C | 9.4 | 10.0 | 14.9 | 10.4 |
*Not determined due to insufficient material for measurement.
The mAbs with Fv A display the highest viscosity (18.4–28.7 cP) compared to mAbs with Fv B (9.1–13.4 cP) and Fv C (9.4–14.9 cP). This observation illustrates a dominant impact of the variable domain on the viscosity. Interestingly, IgG subclass also shows a significant impact. For mAbs with Fv A, the viscosity rank order is IgG4PAA > IgG1EN > IgG1. Similarly, the rank order for the mAbs with Fv B is IgG4PAA > IgG2 ≈ IgG1EN > IgG1. In both cases the IgG4PAA mAbs show notably higher viscosity compared to the other IgG subclass mAbs. However, for mAbs containing Fv C the rank order is IgG2 > IgG4PAA ≈ IgG1EN ≈ IgG1. These results point to a likely interplay between the variable domain and the IgG subclass.
Solubility
The absolute maximal solubility of a protein is usually difficult to measure with confidence. During the concentration process, the solution viscosity normally dramatically increases, which prevents the protein solution from being further concentrated to its maximal solubility. Additionally, protein insolubility manifests through a wide variety of phenomenon, such as haze formation, high turbidity, visible precipitation, and phase separation, and may take days to months to be observed. The challenges in sample preparation and observation duration make the direct measurement of “solubility” difficult. Therefore, instead of measuring maximal soluble protein content, PEG-induced precipitation and microturbidity methods have been widely adopted to evaluate relative solubility at appropriate development stage.18,19
Solubility ranking by PEG-induced precipitation
The PEG-induced precipitation method was used to rank the solubility of the mAbs (Figure 2(a)). Both IgG subclass and variable region exert notable effects on apparent solubility as reported by %PEGonset. Within the series of identical variable region, the mAbs with IgG1 or IgG1EN required the highest %PEG to precipitate compared to the corresponding IgG2 or IgG4PAA mAbs, indicating higher solubility, which is more obvious for mAbs with Fv A and Fv C. No clear differentiation between IgG1 and IgG1EN mAbs was observed on solubility ranking by %PEG onset. No apparent difference was observed between IgG2 and IgG4PAA, when bearing either Fv B or Fv C.
Figure 2.
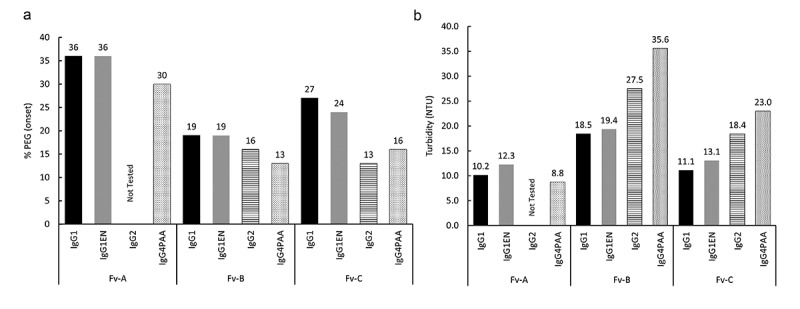
Solubility Ranking by PEG-induced precipitation assay (2a) and by microturbidity assay (2b). The %PEG onset required for protein precipitation and the turbidity (NTU) measured by microturbidity are plotted in the category of variable region and IgG subclass
Of the three series of mAbs assessed by PEG-induced precipitation, the impact of the variable region is more pronounced than the impact of IgG subclass. As shown in Figure 2(a), all mAbs with Fv A demonstrate higher %PEGonset in comparison to the mAbs with the other two variable regions. In general, the solubility ranking order is: mAbs with Fv A > mAbs with Fv C > mAbs with Fv B, while mAbs with Fv A having the highest solubility.
Solubility ranking by micro-turbidity assay
In the micro-turbidity assay, mAbs were concentrated to 90 mg/mL in the common pH 6.0 buffer matrix and the solution turbidity was measured using a microplate reader. The turbidity readout is taken as an indicator of protein association, which is a precursor of reaching maximal solubility.18 Higher turbidity values indicate a lower maximal solubility. Because of its reliability and ease of use, the microturbidity assay is frequently used in supporting formulation screening.19
As shown in Figure 2(b), both IgG subclass and variable regions demonstrate a significant effect on the mAb solubility ranking by microturbidity measurements. The overall ranking is mAbs with Fv A > mAbs with Fv C > mAbs with Fv B. Given the same variable region, the impact of IgG subclass on solubility ranking is also apparent. For mAbs with either Fv B or Fv C, the relative solubility ranking is IgG1 ≈ IgG1EN > IgG2 > IgG4PAA. The IgG subclass impact for mAbs with Fv A, however, is indistinguishable, as the turbidity for mAbs in this series is below the lower limit of assay quantification.
Computational interrogation on biophysical properties of variable and constant domains
In order to understand the potential mechanisms that drive the above observations, we performed computational analysis of the variable domains and the constant domains separately, focusing on their charge (Figures 3(a,b)) and hydrophobicity profiles (Figure 3(c)). On the surface electrostatic potential profile presented in Figure 3(a), the three Fv domains demonstrate distinctively different overall charge properties as well as charge distributions. At pH 6, the Fv A domain has a net charge of 4.1 and a predominantly charge-positive surface. The Fv B domain presents a pronounced charge-negative patch across its complementarity determining region (CDR) surface with a net charge of −3.3, while the Fv C domain presents a small charge-negative patch on its heavy chain CDR, with a net charge of −2.6. The charge profile of the constant domain of various IgG subclass is different from one another as well, especially in the CH3 surface (Figure 3(b)). The IgG1 constant domain has the most charge-positive surface at pH 6 with the highest net charge of 16.1, while the IgG4PAA constant domain has a strong charge-negative patch at the tail of CH3 and the least overall net charge of 8.9. The constant domain charge profile of IgG1EN and IgG2 are in between IgG1 and IgG4PAA with net charge of 14.7 and 12.3, respectively.
Figure 3.
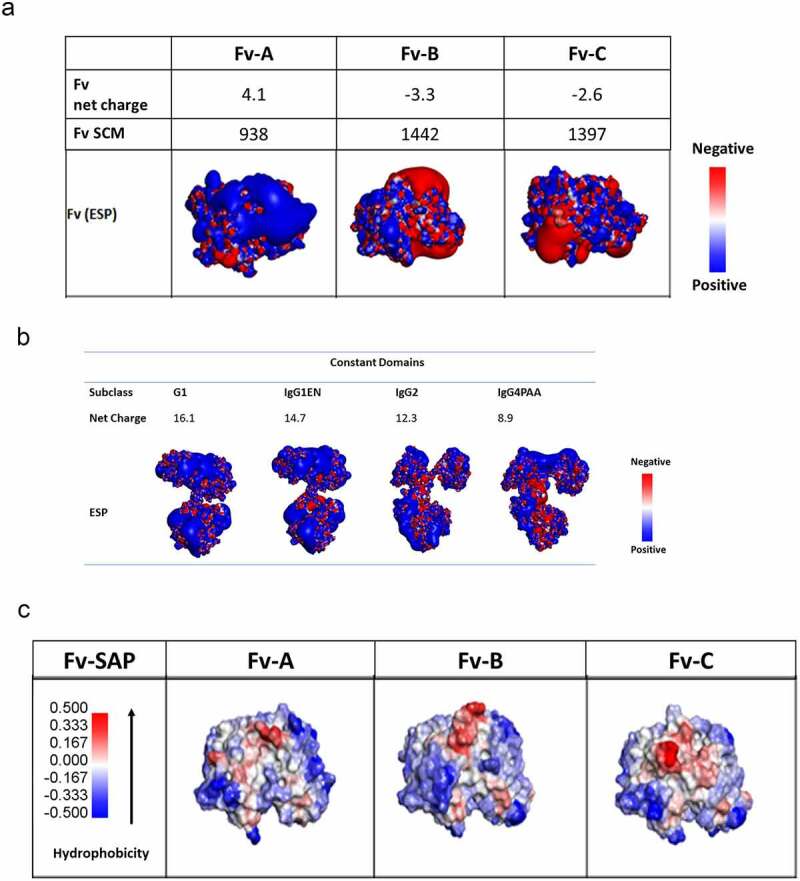
Surface charge analysis at pH 6 (3a and 3b) and hydrophobic patch analysis (3c) for the antibodies with variable region of Fv A, B, and C (3a) in the subclass structure of IgG1, IgG1EN, IgG2, and IgG4PAA by homology modeling
Similarly, we inspected the hydrophobicity profile of the three Fv domains and the corresponding constant domains by spatial aggregation potential (SAP). Hydrophobicity hot spots in Fv that potentially attribute to aggregation are shown in red in Figure 3(c). Among the Fv domains, Fv A has the least hydrophobic hotspots, Fv B has a protruding hotspot at the center of CDRs, and Fv C has the largest hydrophobic hotspot on its CDR surface. This is in contrast to the constant domain hydrophobicity profiles, where no significant difference is observed for various IgG subclasses (data not shown).
Discussions
To date, the antibody therapeutics extensively used for the treatment of cancer, immunology, cardiovascular, and infectious diseases are primarily based on human IgG1, IgG2, or IgG4 and their variants. Choosing appropriate IgG subclass is not only crucial to the therapeutic efficacy, but also the mAb properties that can expedite and reduce the cost for product development. The frequent necessity for high concentration liquid formulations places limitations on the selection of mAb properties, which must be suitable for the manufacturing design space and allow for dosage form flexibility.
The emergence of many engineered Fc variants have provided options to tailor antibodies to specific FcγR interactions (for example, see reviews by Wang et al.2 and Saunders3) and to optimize molecular properties. The physicochemical properties of mAbs are generally viewed as mostly driven by CDR sequences, which usually have unique charge or hydrophobicity for antigen engagement. However, there has been a gradual realization that IgG subclass could also play an important role in mAb physiochemical properties, which can affect the subsequent drug product development.
In this study, three distinct series of antibodies with variable regions targeting three independent targets (Fv A, Fv B, and Fv C) were used to investigate the physiochemical properties that are associated with four representative constant domains: IgG1, IgG1EN, IgG2, and IgG4PAA. IgG1EN is an Fc variant engineered to eliminate the effector functions, while IgG4PAA is an Fc variant with enhanced interchain stability and reduced FcγR interactions. The impact both of variable region and of IgG subclass was observed in a number of molecular properties. Interestingly, this study not only shows that both the variable region and the IgG subclass can play a dominant role in particular molecular properties, but also highlights the interplay between them that contributes to the difficulty of developing a general guideline on the properties of IgG subclasses.
The overall hydrophobicity of the antibodies is almost solely determined by the variable region regardless of IgG subclass. Fv C introduced significant hydrophobicity to the antibodies while Fv A and Fv B antibodies were affected to a lesser extent. Within each mAb series, all antibodies demonstrated similar hydrophobicity interaction potential with a minor increase for IgG4PAA (Table 3). These observations are consistent with the homology modeling results, which identified the largest hydrophobic patch in the CDR surface of Fv C, but the least hydrophobic patch on Fv A and only a protruding hydrophobic area on Fv B. No significant difference in hydrophobic patches was observed across all IgG subclass constant domains from the structural analysis, in agreement with the experimental results from the HIC measurements. Therefore, switching IgG subclass is unlikely to affect overall hydrophobicity to a significant degree.
Unlike hydrophobicity, other molecular properties, including the transition temperature of thermal-induced unfolding, pI, viscosity, and solubility, can be significantly affected by both variable domain and IgG subclass. It is noted that, while the IgG subclass impact on thermal unfolding, pI, and solubility shows a clear trend, their effect on viscosity is more complicated, likely due to the interaction between the variable domain and the constant domains. It is also noted that, although the IgG2 antibodies are well known to have disulfide-mediated structural isoforms,17,20 they mainly manifest in the thermal-induced unfolding profiles with negligible impact on the other molecular attributes in our investigations.
We observed a significant impact of the variable region and IgG subclass on thermal stability (Figure 1). Consistent with previous reports,12,13,21,22 the first melting temperatures, presumably reflecting the CH2 domain unfolding, largely remain the same within each IgG subclass. The mutations in IgG1EN located in the CH2 domain led to a significantly lower CH2 melting temperature, analogous to some other IgG molecules with FcγR interactions modulated by either mutagenesis or deglycosylation.23–25 In this study, the individual domain melting temperature is difficult to determine unequivocally due to the overlapping transitions. Assuming that the highest peak in each thermogram represents Fab transition, the Fab melting temperature is affected by both variable domain and IgG subclass. Within each mAb series, the Tm of the Fab in IgG1 is virtually identical to that in IgG1EN, similar or slightly higher than that in IgG2 mAb, and higher than that in IgG4PAA. The observation of a decrease in Fab Tm changing from IgG1 to IgG4 is in agreement with a previous report by Heads et al.26 The mAbs with Fv A show higher Fab transition temperatures than the mAbs with either Fv B or Fv C. It has been reported that the unfolding temperature of antibodies could be correlated with the physical and chemical stability of the molecules at storage, accelerated degradation, and under other stress conditions.14,27,28 We are investigating the effect of the unfolding temperatures, especially the low CH2 unfolding temperature in IgG1EN, in another study.
The mAbs in this study have measured IgG pI in the range of 6–9, within the range reported by Kingbury et al.29 in their comprehensive studies of 59 therapeutic mAbs that included 43 FDA-approved products. The pI of a mAb is a molecular property highly affected by the variable region, but also moderately affected by the IgG subclass. The pI of mAbs with Fv A are in the basic pH range and are significantly higher than the pI of mAbs with either Fv B or Fv C. These high pIs are directly related to the charge contribution from the variable regions, as evidenced by the predominantly charge-positive surface located on Fv A by homology model (Figure 3(a)). The IgG subclass also makes noticeable contributions to the overall pI of the molecules. As shown in Table 2, the calculated pI of the Fc of IgG1, IgG1EN, and IgG2 are close to the neutral pH in the range of 6.4–6.7, but the pI of the Fc of IgG4 is significant lower (pI = 5.7), falling in the acidic range, which is attributed to the significant negative charge patches at the end of the CH3 domain of IgG4 shown by structural analysis of the homology model (Figure 3(b)).
The viscosity of protein solutions is complex, with elusive underlying principles. It has been proposed that multiple mechanisms could contribute to viscosity, such that several weak attractive interactions form network-like association. A surface charge map (SCM) was devised to quantify the sum of charge negative patch on the surface of the variable domains. Qualitatively, SCM has shown correlation with viscosity in a study of 19 mAbs by Agrawal et al.,30 which included only IgG1 subclass molecules. We calculated SCM scores for Fv A, Fv B, and Fv C to test the SCM and viscosity correlation. Our experimental viscosity data, however, contradict the previous finding that a variable region with higher SCM leads to higher viscosity, implicating contributing factors beyond the variable domain. In addition, the Fv charge symmetry parameter (Fv-CSP) was also computed. Yet there is no correlation of Fv-CSP to viscosity, as previously reported.29,31 We hypothesize that certain local hot spots on CDRs could make transient interactions with neighboring molecules in crowded environments, via Fv–Fv interaction, Fv–Fc interaction, or a mixture of both. In a recent report by Lai et al.,32 the role of net charge, hydrophobicity, and hydrophilicity on viscosity was extensively discussed. The authors reported a decision tree model to classify low- and high- viscosity mAbs, which could be very useful for hit screening at the early discovery stage. Yet it is still challenging to apply generalized rules describing complex high-concentration behavior when subtle changes are involved. Coarse-grained simulation, rational-based mutational studies, domain swapping, and salt influence experiments are worthy of further investigation.33,34 For the three antibody series studied, we found that the IgG1 and IgG1EN mAbs have either the lowest or similar viscosity compared with the corresponding IgG2 or IgG4 mAbs. Although the IgG4PAA mAbs with Fv A and Fv B show higher viscosity compared to the other IgG subclasses, the IgG4PAA mAb with Fv C exhibits viscosity similar to the corresponding IgG1EN mAb. This result underscores the importance of an interplay between Fv C and the constant domains. Among the IgG subclasses, IgG4PAA is the only one with a notable charge negative patch at the tail of the CH3 domain. We speculate that this CH3 tail in IgG4 creates additional attractive interactions and leads to more potential intermolecular association at high concentrations compared with other IgG subclasses.
Interestingly, we found a trend between ΔpI (Fab – Fc) and mAb viscosity in the common pH 6 buffer matrix used in our studies. As shown in Figure 4, when ΔpI (Fab – Fc) is >1, such as for mAbs with Fv A, the larger ΔpI (Fab – Fc) leads to higher viscosity. Structural analysis of the homology models reveals that this observation can be potentially attributed to the electrostatic attractive interaction between the highly positively charged Fv A with the negatively charged patches on a neighboring Fc, particularly in the case of IgG4PAA, which has a strong negatively charged CH3 terminal, and warrants further investigations. We note that Heads et al. described similar electrostatic interactions when comparing IgG1 and IgG4 mAbs with identical variable regions, and found a correlation between the electrostatic interaction and aggregation propensity, which is believed to be the potential root cause of mAb viscosity behavior.7 More detailed modeling work and extensive data set is needed to further reveal the nature of the relationship between the overall pI, individual domain pI, hydrophobic patches, charge patches, and viscosity behavior.
Figure 4.
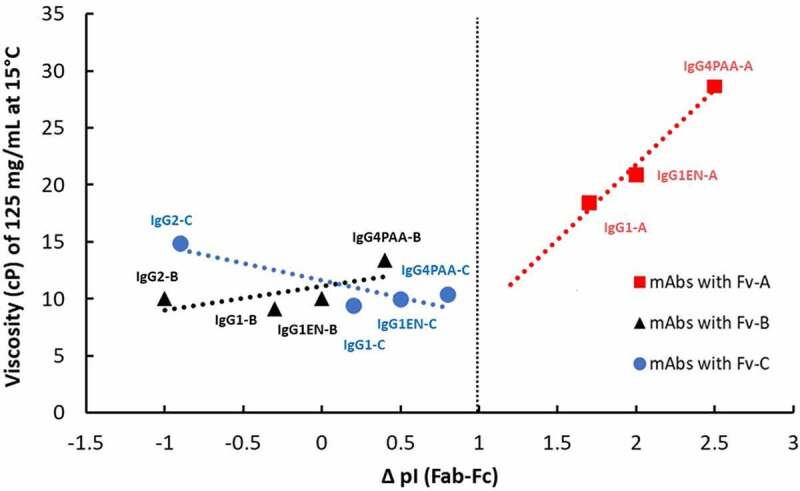
Correlation of the difference (Δ) of the calculated Fab isoelectric point (Fab pI) and the calculated Fc isoelectric point (Fc pI) with measured viscosity at 125 mg/mL in pH 6.0 L-histidine-based buffer at 15°C
Given that IgG subclass switching has been used as an engineering tool to improve mAb solubility,22,35 we are very interested in comparing the solubility of these antibodies. However, protein solubility is a complex phenomenon. Differing from precipitation or crystallization of small chemicals, the physical appearance of insoluble protein varies from protein to protein and in different buffer matrices. When protein concentration is beyond the solubility limit under a specific buffer condition, the insolubility phenomenon can be defined by terms such as precipitation, phase separation, haziness, high turbidity, and significant visible particulate formation. Additionally, the concentrated protein solution is prepared by centrifugation using appropriate molecular weight cut off membrane. The operational difficulties are significantly increased by increased viscosity when the protein is concentrated. Therefore, we decided to use the PEG-induced precipitation method and the micro-turbidity method to rank the solubility of the molecules instead of directly measuring the maximal solubility. Although the results are not quantitatively transferable between PEG-induced precipitation data (%PEG needed to precipitate mAb) and nephelometric turbidity unit (NTU) data, the overall trend is highly consistent between the two solubility ranking methods. Solubility can be highly pH/buffer-dependent and we chose to conduct these studies in a histidine buffer matrix, as such a buffer system is common for mAb-based drug products29 and has been shown to induce minimal injection site pain.36 Antibodies with Fv A require the highest %PEG to precipitate, which also show the lowest turbidity compared to the antibodies with the other variable regions. Similarly, the antibodies that require the lowest %PEG to precipitate also demonstrate the highest turbidity, such as the mAbs with Fv B. For the three series in this study, we found that the IgG1 and IgG1EN mAbs tend to display the highest %PEGonset and the lowest turbidity compared to the other IgG subclasses, indicating that the IgG1 and IgG1EN mAbs have higher solubility. This finding is consistent with the results from Kingsbury et al.,29 where a higher percentage of IgG1 mAbs exhibited good solution behavior than the percentage for the other two IgG subclasses.
Antibody solution behavior can be affected by both mAb charge status and hydrophobicity.35,37 We found that for the mAbs in our study, however, the solubility is predominantly driven by the overall charge property at pH 6, with little impact from hydrophobicity in the current range. As plotted in Figure 5(a), for antibodies that have a measured pI between 6.0 and 7.5, a strong linear correlation is observed between measured pI and solution turbidity at pH 6, where the increase of pI resulted in significant decrease of turbidity, such as the observation for mAbs with either Fv B and Fv C. This observation indicates that changing the overall net charge of the mAb can improve its solubility. However, when the measured pI is 1.5 units higher than the buffer pH, a plateau of solution turbidity is observed. Further increase of the pI does not result in further decrease of solution turbidity, which is the case for mAbs with Fv A.
Figure 5.
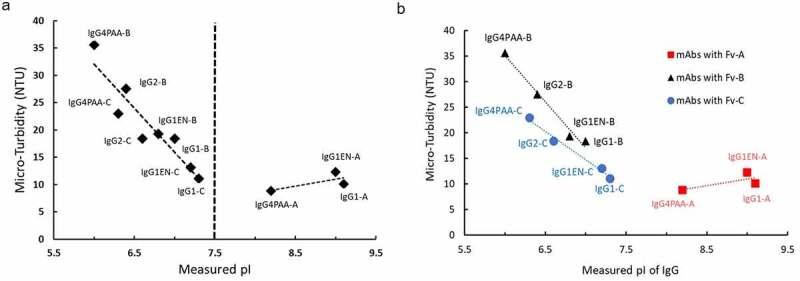
Correlation of mAb solution microturbidity (NTU) with measured isoelectric points (pI) of mAbs. (5a); Correlation of measured pI with solution microturbidity (NTU) of mAbs with Fv A, with Fv B, and with Fv C (5b)
Charge properties from IgG subclass constant regions (CH1, CH2, and CH3) contribute to overall pI of mAb molecules. To remove the impact from the variable region and investigate the role of charge property from IgG subclass on mAb solubility, the measured pI of the antibodies was compared against the turbidity data for a given variable region (Figure 5(b)). As expected, the antibodies with Fv A that have overall pI higher than 7.5 demonstrate insignificant differences on solution turbidity among IgG1, IgG1EN, and IgG4PAA. When the overall pI is below 7.5, however, a clear impact from IgG subclass is observed for mAbs with either Fv B or Fv C. Based on the turbidity trending, the solubility of IgG subclasses with Fv B and Fv C can be ranked as IgG4PAA < IgG2 < IgG1EN ≈ IgG1, following the increasing pI.
Although mAbs with Fv A exhibit higher viscosity than mAbs with Fv B and Fv C, they display the highest %PEGonset and the lowest turbidity. Such an inverse correlation between viscosity and solubility was also observed for some mAbs by Kingsbury et al.29 and warrants further investigations. It is interesting to note that, although in our studies IgG1 and IgG1EN mAbs display higher solubility than either IgG2 or IgG4 mAbs in each series, Pepinsky et al.22 found IgG2 and IgG4 mAb more soluble than the corresponding IgG1 mAb, and Bethea et al.35 found IgG4 mAb more soluble than the corresponding IgG1 mAb. These findings, together with our viscosity data, point to the potential for a significant amount of interplay between the variable domain and the constant domains.
In summary, we found that both variable domain and IgG subclass structure can contribute significantly to the molecular properties of an antibody. Some properties are unique to each IgG subclass, some are dominated by the variable region, while others are influenced by both the variable region and the IgG subclass. There can also be a significant interplay between the variable region and the IgG subclass for some antibodies. Among the mAbs studied in this report, the IgG1 and IgG1EN mAbs tend to have higher solubility and lower or similar viscosity compared with either IgG2 or IgG4 mAbs of identical variable regions. The solubility ranking, whether by PEG-induced precipitation or by microturbidity, relies heavily on the charge status of the antibody at a fixed pH, which is attributed to charge status of both variable domain and IgG subclass. In addition to mechanistic considerations for subclass selection, our data indicate that IgG subclass choice can have a significant impact on the molecular properties, which may affect the success of drug product development. Therefore, consideration of subclass selection should be explored early in each case to enable a balanced decision between biology and product development. Our findings establish a foundation for the subsequent investigation of how the IgG subclass properties influence the developability of these antibodies.
Materials and methods
Antibody preparation
Three antibodies (denoted mAb A, mAb B, and mAb C) with discrete targets were obtained from rabbit immunizations and subsequently humanized and affinity matured. Humanized variable domains were cloned into respective constant domains; each VL was cloned into a human kappa constant domain while each VH was cloned into IgG1, IgG1EN, IgG2, and IgG4PAA constant domains.9,38,39
The antibodies used in this study were expressed in CHO by either transient transfection or stable bulk cell lines using methods adopted from Barnard, Rajendra and coworkers.40,41 Each antibody was purified using conventional antibody purification processes.
Isoelectric point measurement and calculation
Isoelectric points were measured by capillary isoelectric focusing (cIEF) for each mAb using Protein Simple iCE3 with autosampler on a FC-coated cIEF cartridge (Protein Simple Part # 101701). Antibody samples were prepared at 0.25 mg/ml in 3 M urea/0.35% methylcellulose with Pharmalytes 3–10 (Sigma-Aldrich Cat.# 17–0456-01) and pI markers (ProteinSimple). Samples were focused at 1500 V for 1-minute followed by a second focusing period of 3000 V for 8 minutes. MAb pIs were calibrated to low and high internal pI makers. The pI of each Fab and Fc were calculated from the primary sequence using algorithm in Molecular Operating Environment (MOE2019) (Chemical Computing Group, Montreal, Canada) and outlined in Sillero and Ribeiro.42
Differential scanning calorimetry
Thermal stability of the antibody samples was assessed by DSC. Samples were prepared at 1 mg/mL in 5 mM histidine, 280 mM mannitol, pH6.0. Thermal transition of each antibody was measured using an automated MicroCal PEAQ-DSC (Malvern Panalytical, UK) in a scan rate of 1°C/min for the temperature range of 20–110°C, after equilibration at 20°C for 3 minutes. Thermograms for each sample were buffer referenced, baseline subtracted, and fit to three peaks using Origin 7. The CH2 and CH3 domain peaks were assigned based on convention.12,43
Hydrophobic interaction chromatography
Relative hydrophobicity was assessed by HIC-HPLC. Samples were diluted to 1 mg/ml in phosphate-buffered saline and injected onto a MAbPac HIC-10 1000 Å, 5um, 4.6 × 100 mm (Thermo) HIC column preequilibrated in 25 mM potassium phosphate, 1.5 M ammonium sulfate, pH 6.8. Samples were eluted with a 20-minute linear gradient into 25 mM potassium phosphate, pH 6.8, 20% isopropanol. Hydrophobicity interaction potential (HIP%) was calculated based on % retention time over gradient as reported by Datta-Mannan et al.44
Viscosity measurement
All mAbs were concentrated to 125 mg/ml in 5 mM histidine buffer at pH 6.0, with 280 mM mannitol and 0.02%(w/v) polysorbate 80. Viscosity of each antibody was measured at 15°C on VROC Initium (RheoSense, San Ramon, CA) viscometer at a shear rate of 1550 1/s with 8–10 measurements per sample.
PEG-induced precipitation for mAb solubility ranking
Stock solutions of 0.4 M L-histidine, pH 6.0 and 20% (w/v) D-mannitol, were used to prepare a matrix containing varying final Polyethylene glycol 3350 (PEG 3350) levels (from 4 to 36%) in a deep well 2× Master Block plate using a Formulator (Formulatrix, Bedford, Ma), which was mixed overnight at 1200 RPM at room temperature on a BioShakeIQ plate shaker (QInstruments, Jena, Germany). MAb samples were buffer-exchanged into and diluted with water to 1 mg/mL and plated (50 μL/well) onto the wells of four rows of a 96-well polystyrene, V-bottomed assay plates (Greiner Cat # REF651101). A Biomek i7 (Beckman Coulter, Indianapolis, IN) liquid handler was used to add 50 μL/well of the stock solutions from the 2x Master Block plate into the assay plates containing mAb samples. The assay plates were sealed with Microseal ‘F’ Foil (Cat# MSF1001, Bio-Rad, Hercules, CA) and then incubated at room temperature on a titer plate shaker (Lab-Line Instruments, Melrose Park, IL) for three hours on gentle shaking (setting 3). After incubation 80% of the contents of the 96-well plates were transferred to a 384-well optical bottom plate (Thermo Scientific, Cat# 242764) using a Biomek i7. The optical bottom plate was read on a SpectraMax Pro plate reader (Molecular Devices, San Jose, CA) at 350 nm. The absorbance data was de-convoluted and plotted in Excel (Microsoft, Redmond, WA), where onset of precipitation (abrupt increase in A350) was determined visually. After reading, the assay plates were sealed with Microseal ‘F’ Foil and then incubated at room temperature on the bench for a total of 24 hours. After incubation the assay plate was centrifuged at 3500 RPM for 15 minutes at 25°C on an Allegra X12R benchtop centrifuge (Beckman Coulter, Brea, CA). Without disturbing the pelleted precipitants, 50 μL per well was transferred from each well of the 384-well optical bottom plate into a new UVStar 384-well plate (Greiner Cat # 781801) using a Biomek i7. After sealing the UV Star plate with Microseal ‘F’ Foil, the plate was centrifuged for two minutes at 3000 RPM to remove air bubbles. Finally, the plate was read on a Tecan Infinite M1000 Pro UV/Vis Spectrophotometer (Männedorf, Switzerland) at 280 nm (with background subtraction at 320 nm), using iControl software. The absorbance data was de-convoluted and plotted in Excel (Microsoft, Redmond, WA), where the point of abrupt decrease in absorbance was determined visually.
Turbidity measurement by micro-turbidity method
Turbidity was assessed by microplate spectrofluorometer (SpectraMax M5, San Jose, CA). 100 μL aliquot samples were plated in 96-well Special Optics Black Plates (Corning, Glendale, AZ) and read at ambient temperatures of 20–25°C. Plate based small-volume turbidity analysis (Microturbidity) is a noncompendial method, developed in-house that affords a numerical value to the opalescence and turbidity of mAb formulations. Analysis is based on using absorbance, to measure the amount of light transmission through a sample at a wavelength of 540 nm. The increase or decrease in absorbance can be converted to NTU by a linear regression means using a calibration curve generated from Formazin calibration standards (Millipore Sigma, St. Louis, MO) at various ranges of turbidity. All mAb samples were measured in formulation condition of 90 mg/mL in buffer matrix consisting of 5 mM histidine buffer pH6.0, 280 mM mannitol, and 0.02% (w/v) polysorbate 80. All prepared samples were equilibrated at 5°C over 24 hours prior to measurement.
Homology modeling and surface property analysis
Homology models of Fab region and Fc portion are built by the Molecular Operating Environment (MOE2019) (Chemical Computing Group, Montreal, Canada), based on crystal structures of Fab obtained in house and closest matching templates. Specifically, structure modeling of Fv-B, and Fv-C is based on crystal structures obtained in house. Fv-A homology model is built using MOE with the top scoring homology model among 5 models selected for further analysis. Solvent-accessible surface area, surface exposure (%) for each residue, and net charge and hydrophobicity score for each CDR were calculated based on the modeled structure using the protein properties analysis module (MOE2019). Specifically, hydrophobicity and net charge of each CDR segment are computed and reported in supplemental material. IMGT definition is applied to CDR segment and standard calculation by MOE protein descriptor calculation was used, which include pro_patch_cdr related calculation, Fv-CSP, and parsed score for each CDR segment. In addition, electrostatic potential calculation, spatial aggregation propensity (SAP), and spatial-charge map (SCM) were computed according to the method in the BIOVIA Discovery Studio 2020 software (San Diego, CA).
Supplementary Material
Acknowledgments
The authors kindly acknowledge Maria Hougland and Rob Peery for assisting antibody production, and Dr. Michael R De Felippis for providing technical guidance.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
List of Abbreviations
| ADCC | Antibody-dependent cell-mediated cytotoxicity |
| ADCP | Antibody dependent phagocytosis |
| CDC | Complement dependent cytotoxicity |
| CDR | Complementarity-determining region |
| cIEF | Capillary isoelectric focusing |
| DSC | Differential scanning calorimetry |
| Fab | Antigen-binding fragment |
| Fc | Crystallizable fragment |
| FcγR | Fc gamma receptor |
| HC | Heavy chain |
| HIC | Hydrophobic-Interaction-Chromatography |
| IgG1EN | Human IgG1 with mutations consisting of L234A, L235E, G237A, A330S, and P331S |
| IgG4PAA | Human IgG4 variant with mutations consisting of S228P, L234A, and L235A |
| LC | Light chain |
| mAb | Monoclonal antibody |
| NTU | Nephelometric Turbidity Units |
| pI | Isoelectric point |
| SAP | Spatial aggregation potential |
| SCM | Surface charge map |
| SEC | Size-exclusion-chromatography |
Disclosure statement
All authors in this report are employees of Eli Lilly and Company. The authors do not have any conflict of interest or financial disclosure to report.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Kaplon H, Reichert JM.. Antibodies to watch in 2021. mAbs. 2021;13(1):1860476. doi: 10.1080/19420862.2020.1860476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Mathieu M, Brezski RJ. IgG Fc engineering to modulate antibody effector functions. Protein Cell. 2018;9(1):63–12. doi: 10.1007/s13238-017-0473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saunders KO. Conceptual approaches to modulating antibody effector functions and circulation half-life. Front Immunol. 2019;10:1296. doi: 10.3389/fimmu.2019.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roux KH, Strelets L, Michaelsen TE. Flexibility of human IgG subclasses. J Immunol. 1997;159(7):3372–82. PMID: 9317136 [PubMed] [Google Scholar]
- 5.Tian X, Langkilde AE, Thorolfsson M, Rasmussen HB, Vestergaard B. Small-angle x-ray scattering screening complements conventional biophysical analysis: comparative structural and biophysical analysis of monoclonal antibodies IgG1, IgG2, and IgG4. J Pharm Sci. 2014;103(6):1701–10. doi: 10.1002/jps.23964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian X, Vestergaard B, Thorolfsson M, Yang Z, Rasmussen HB, Langkilde AE. In-depth analysis of subclass-specific conformational preferences of IgG antibodies. IUCrJ. 2015;2(1):9–18. doi: 10.1107/S205225251402209X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heads JT, Lamb R, Kelm S, Adams R, Elliott P, Tyson K, Topia S, West S, Nan R, Turner A, et al. Electrostatic interactions modulate the differential aggregation propensities of IgG1 and IgG4P antibodies and inform charged residue substitutions for improved developability. Protein Engi Design Sel PEDS. 2019;32(6):277–88. doi: 10.1093/protein/gzz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross JA, Dillon SR, Mudri S, Johnston J, Littau A, Roque R, Rixon M, Schou O, Foley KP, Haugen H, et al. TACI-Ig neutralizes molecules critical for B cell development and autoimmune disease. impaired B cell maturation in mice lacking BLyS. Immunity. 2001;15(2):289–302. doi: 10.1016/S1074-7613(01)00183-2. [DOI] [PubMed] [Google Scholar]
- 9.Angal S, King DJ, Bodmer MW, Turner A, Lawson AD, Roberts G, Pedley B, Adair JR. A single amino acid substitution abolishes the heterogeneity of chimeric mouse/human (IgG4) antibody. Mol Immunol. 1993;30(1):105–08. doi: 10.1016/0161-5890(93)90432-B. [DOI] [PubMed] [Google Scholar]
- 10.Bloom JW, Madanat MS, Marriott D, Wong T, Chan SY. Intrachain disulfide bond in the core hinge region of human IgG4. Protein Sci Publ Protein Soc. 1997;6(2):407–15. doi: 10.1002/pro.5560060217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuurman J, Perdok GJ, Gorter AD, Aalberse RC. The inter-heavy chain disulfide bonds of IgG4 are in equilibrium with intra-chain disulfide bonds. Mol Immunol. 2001;38(1):1–8. doi: 10.1016/S0161-5890(01)00050-5. [DOI] [PubMed] [Google Scholar]
- 12.Garber E, Demarest SJ. A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem Biophys Res Commun. 2007;355(3):751–57. doi: 10.1016/j.bbrc.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 13.Ito T, Tsumoto K. Effects of subclass change on the structural stability of chimeric, humanized, and human antibodies under thermal stress. Protein Sci Publ Protein Soc. 2013;22(11):1542–51. doi: 10.1002/pro.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavakoli-Keshe R, Phillips JJ, Turner R, Bracewell DG. Understanding the relationship between biotherapeutic protein stability and solid-liquid interfacial shear in constant region mutants of IgG1 and IgG4. J Pharm Sci. 2014;103(2):437–44. doi: 10.1002/jps.23822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garidel P, Eiperle A, Blech M, Seelig J. Thermal and chemical unfolding of a monoclonal IgG1 antibody: application of the multistate Zimm-Bragg theory. Biophys J. 2020;118(5):1067–75. doi: 10.1016/j.bpj.2019.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryazantsev S, Tischenko V, Nguyen C, Abramov V, Zav’yalov V, Nikolaidis N. Three-dimensional structure of the human myeloma IgG2. PloS One. 2013;8(6):e64076. doi: 10.1371/journal.pone.0064076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dillon TM, Ricci MS, Vezina C, Flynn GC, Liu YD, Rehder DS, Plant M, Henkle B, Li Y, Deechongkit S, et al. Structural and functional characterization of disulfide isoforms of the human IgG2 subclass. J Biol Chem. 2008;283(23):16206–15. doi: 10.1074/jbc.M709988200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chai Q, Shih J, Weldon C, Phan S, Jones BE. Development of a high-throughput solubility screening assay for use in antibody discovery. mAbs. 2019;11(4):747–56. doi: 10.1080/19420862.2019.1589851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann M, Winzer M, Weber C, Gieseler H. Low-volume solubility assessment during high-concentration protein formulation development. J Pharm Pharmacol. 2018;70(5):636–47. doi: 10.1111/jphp.12621. [DOI] [PubMed] [Google Scholar]
- 20.Wypych J, Li M, Guo A, Zhang Z, Martinez T, Allen MJ, Fodor S, Kelner DN, Flynn GC, Liu YD, et al. Human IgG2 antibodies display disulfide-mediated structural isoforms. J Biol Chem. 2008;283(23):16194–205. doi: 10.1074/jbc.M709987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa T, Ito T, Endo R, Nakagawa K, Sawa E, Wakamatsu K. Influence of pH on heat-induced aggregation and degradation of therapeutic monoclonal antibodies. Biol Pharm Bull. 2010;33(8):1413–17. doi: 10.1248/bpb.33.1413. [DOI] [PubMed] [Google Scholar]
- 22.Pepinsky RB, Silvian L, Berkowitz SA, Farrington G, Lugovskoy A, Walus L, Eldredge J, Capili A, Mi S, Graff C, et al. Improving the solubility of anti-LINGO-1 monoclonal antibody Li33 by isotype switching and targeted mutagenesis. Protein Sci Publ Protein Soc. 2010;19(5):954–66. doi: 10.1002/pro.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oganesyan V, Damschroder MM, Leach W, Wu H, Dall’Acqua WF. Structural characterization of a mutated, ADCC-enhanced human Fc fragment. Mol Immunol. 2008;45(7):1872–82. doi: 10.1016/j.molimm.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 24.Zheng K, Bantog C, Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. mAbs. 2011;3(6):568–76. doi: 10.4161/mabs.3.6.17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam SH, McCarthy SG, Armstrong AA, Somani S, Wu SJ, Liu X, Gervais A, Ernst R, Saro D, Decker R, et al. Functional, biophysical, and structural characterization of human IgG1 and IgG4 Fc variants with ablated immune functionality. Antibodies (Basel, Switzerland). 2017;6(3):12. doi: 10.3390/antib6030012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heads JT, Adams R, D’Hooghe LE, Page MJ, Humphreys DP, Popplewell AG, Lawson AD, Henry AJ. Relative stabilities of IgG1 and IgG4 Fab domains: influence of the light-heavy interchain disulfide bond architecture. Protein Sci Publ Protein Soc. 2012;21(9):1315–22. doi: 10.1002/pro.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson CM. Differential scanning calorimetry as a tool for protein folding and stability. Arch Biochem Biophys. 2013;531(1–2):100–09. doi: 10.1016/j.abb.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Sasahara K, Goto Y. Application and use of differential scanning calorimetry in studies of thermal fluctuation associated with amyloid fibril formation. Biophys Rev. 2013;5(3):259–69. doi: 10.1007/s12551-012-0098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kingsbury JS, Saini A, Auclair SM, Fu L, Lantz MM, Halloran KT, Calero-Rubio C, Schwenger W, Airiau CY, Zhang J, et al. A single molecular descriptor to predict solution behavior of therapeutic antibodies. Sci Adv. 2020;6(32):eabb0372. doi: 10.1126/sciadv.abb0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agrawal NJ, Helk B, Kumar S, Mody N, Sathish HA, Samra HS, Buck PM, Li L, Trout BL. Computational tool for the early screening of monoclonal antibodies for their viscosities. mAbs. 2016;8(1):43–48. doi: 10.1080/19420862.2015.1099773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma VK, Patapoff TW, Kabakoff B, Pai S, Hilario E, Zhang B, Li C, Borisov O, Kelley RF, Chorny I, et al. In silico selection of therapeutic antibodies for development: viscosity, clearance, and chemical stability. Proc Natl Acad Sci U S A. 2014;111(52):18601–06. doi: 10.1073/pnas.1421779112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai PK, Fernando A, Cloutier TK, Gokarn Y, Zhang J, Schwenger W, Chari R, Calero-Rubio C, Trout BL. Machine learning applied to determine the molecular descriptors responsible for the viscosity behavior of concentrated therapeutic antibodies. Mol Pharm. 2021;18(3):1167–75. doi: 10.1021/acs.molpharmaceut.0c01073. [DOI] [PubMed] [Google Scholar]
- 33.Chaudhri A, Zarraga IE, Kamerzell TJ, Brandt JP, Patapoff TW, Shire SJ, Voth GA. Coarse-grained modeling of the self-association of therapeutic monoclonal antibodies. J Phys Chem B. 2012;116(28):8045–57. doi: 10.1021/jp301140u. [DOI] [PubMed] [Google Scholar]
- 34.Arora J, Hu Y, Esfandiary R, Sathish HA, Bishop SM, Joshi SB, Middaugh CR, Volkin DB, Weis DD. Charge-mediated Fab-Fc interactions in an IgG1 antibody induce reversible self-association, cluster formation, and elevated viscosity. mAbs. 2016;8(8):1561–74. doi: 10.1080/19420862.2016.1222342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bethea D, Wu SJ, Luo J, Hyun L, Lacy ER, Teplyakov A, Jacobs SA, O’Neil KT, Gilliland GL, Feng Y, et al. Mechanisms of self-association of a human monoclonal antibody CNTO607. Protein Engi Design Sel PEDS. 2012;25(10):531–37. doi: 10.1093/protein/gzs047. [DOI] [PubMed] [Google Scholar]
- 36.Shi GH, Pisupati K, Parker JG, Corvari VJ, Payne CD, Xu W, Collins DS, De Felippis MR. Subcutaneous injection site pain of formulation matrices. Pharm Res. 2021;38(5):779–93. doi: 10.1007/s11095-021-03047-3. [DOI] [PubMed] [Google Scholar]
- 37.Pindrus M, Shire SJ, Kelley RF, Demeule B, Wong R, Xu Y, Yadav S. Solubility challenges in high concentration monoclonal antibody formulations: relationship with amino acid sequence and intermolecular interactions. Mol Pharm. 2015;12(11):3896–907. doi: 10.1021/acs.molpharmaceut.5b00336. [DOI] [PubMed] [Google Scholar]
- 38.Tamm A, Schmidt RE. IgG binding sites on human Fc gamma receptors. Int Rev Immunol. 1997;16(1–2):57–85. doi: 10.3109/08830189709045703. [DOI] [PubMed] [Google Scholar]
- 39.Lund J, Pound JD, Jones PT, Duncan AR, Bentley T, Goodall M, Levine BA, Jefferis R, Winter G. Multiple binding sites on the CH2 domain of IgG for mouse Fc gamma R11. Mol Immunol. 1992;29(1):53–59. doi: 10.1016/0161-5890(92)90156-R. [DOI] [PubMed] [Google Scholar]
- 40.Barnard GC, Hougland MD, Rajendra Y. High-throughput mAb expression and purification platform based on transient CHO. Biotechnol Prog. 2015;31(1):239–47. doi: 10.1002/btpr.2012. [DOI] [PubMed] [Google Scholar]
- 41.Rajendra Y, Balasubramanian S, Peery RB, Swartling JR, McCracken NA, Norris DL, Frye CC, Barnard GC. Bioreactor scale up and protein product quality characterization of piggyBac transposon derived CHO pools. Biotechnol Prog. 2017;33(2):534–40. doi: 10.1002/btpr.2447. [DOI] [PubMed] [Google Scholar]
- 42.Sillero A, Ribeiro JM. Isoelectric points of proteins: theoretical determination. Anal Biochem. 1989;179(2):319–25. doi: 10.1016/0003-2697(89)90136-X. [DOI] [PubMed] [Google Scholar]
- 43.Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, Li L, Brems DN, Remmele RL. Structure and stability changes of human IgG1 Fc as a consequence of methionine oxidation. Biochemistry. 2008;47(18):5088–100. doi: 10.1021/bi702238b. [DOI] [PubMed] [Google Scholar]
- 44.Datta-Mannan A, Estwick S, Zhou C, Choi H, Douglass NE, Witcher DR, Lu J, Beidler C, Millican R. Influence of physiochemical properties on the subcutaneous absorption and bioavailability of monoclonal antibodies. mAbs. 2020;12(1):1770028. doi: 10.1080/19420862.2020.1770028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


