Abstract
Id proteins are known to play important roles in the proliferation and differentiation of many cell types. The type 1 insulin-like growth factor receptor (IGF-IR), activated by its ligand, induces the differentiation of 32D IGF-IR cells, a murine hematopoietic cell line, expressing a human IGF-IR. Expression in 32D IGF-IR cells of a dominant negative mutant of Stat3 (DNStat3) inhibits IGF-I-mediated differentiation. DNStat3 causes a dramatic increase in Id2 gene expression. This increase, however, is IGF-I dependent and is abrogated by a mutation at tyrosine 950 of the IGF-IR. These results indicate that in 32D cells, the IGF-IR regulates the expression of the Id2 gene and that this regulation is modulated by both positive and negative signals. Our results also suggest that in this model, Id2 proteins influence the differentiation program of cells but are not sufficient for the full stimulation of their proliferation program.
The Id family of helix-loop-helix proteins are known to form heterodimers with similar proteins, mostly transcriptional activators, composed of a basic region and a helix-loop-helix region (57). Because the Id proteins lack a DNA binding region, these heterodimers cannot bind to DNA. The Id proteins therefore function as negative regulators of basic helix-loop-helix proteins through the formation of inactive heterodimers (6, 57). MyoD is the best-known transcription factor inhibited by Id proteins, but other genes important in neurogenic and hematopoietic differentiation are also inhibited (46). There are at least four Id proteins, but there is evidence in the literature the Id1 and Id3 are overlapping, while expression of Id4 is limited to specific tissues (32, 41). Id gene expression varies during mouse development (32) and is markedly increased in proliferating cells, in cycling cells, and in tumor cell lines (2, 6). Id gene expression has been implicated in the G1-to-S transition (28, 46). High levels of Id gene expression inhibit the differentiation of a variety of cell types (6, 36, 40), including mammary cell differentiation (16). Id expression is repressed in senescent cells (28), and the Id1 protein has been claimed to delay senescence of primary human keratinocytes (1, 45). The Id1 protein promotes mammary epithelial cell invasion (17) and increases the aggressive phenotype of human breast cancer cells (39). The Id2 protein has been reported to inhibit differentiation and enhance cellular proliferation by associating with the retinoblastoma protein (30). More recently, it has been reported that the Id2 promoter is the target of the proto-oncogene N-myc in neuroblastoma cells (38). Id proteins are also required for angiogenesis and vascularization of tumor xenografts (41).
The dual role of Id proteins in proliferation and differentiation has prompted us to examine their regulation by the type 1 insulin-like growth factor receptor (IGF-IR), which also sends a dual signal. The IGF-IR, activated by its ligands, sends an unambiguous mitogenic signal in many cell types, such as fibroblasts and epithelial cells (53). However, in other cell types, IGF-I (or IGF-II) can stimulate either proliferation or differentiation or both (3). We have studied these contradictory signals of the IGF-IR (mitogenesis versus differentiation) in 32D cells, a murine hematopoietic cell line, which undergo apoptosis within 24 h after withdrawal of interleukin-3 (IL-3) (51, 60, 61). 32D cells have low levels of IGF-I and insulin receptors and do not express IRS-1 or IRS-2 (60, 62), which are important substrates for both receptors. 32D cells expressing a human IGF-IR cDNA (32D IGF-IR cells) survive in the absence of IL-3 and, with the addition of IGF-I, grow exponentially for about 48 h (18, 48, 55, 59). After 48 h, the cells begin to differentiate along the granulocytic pathway and eventually decrease in number (60). This sequence of events is not unusual in hematopoietic cells, where induction of differentiation requires a short but intense period of cell proliferation (8, 61, 63). This dual response has been interpreted as indicating that differentiating growth factors send two signals, one for proliferation and one for differentiation, with the latter eventually prevailing.
We have asked whether signaling from the IGF-IR can regulate the expression of Id genes. If Id gene expression were to be regulated by the IGF-IR, it would be interesting to identify the domain(s) of the IGF-IR that regulates it. Specifically, we wished to investigate Id gene expression in 32D-derived cells, which either differentiate or grow indefinitely in the absence of IL-3. For differentiation, 32D IGF-IR cells (see above) are the obvious choice, since they differentiate under the control of IGF-I (60). To inhibit the differentiation of 32D IGF-IR cells in the absence of IRS-1, we introduced into these cells a dominant negative mutant of Stat3 (DNStat3), which has been reported to inhibit granulocyte colony-stimulating factor (G-CSF)-induced differentiation of 32D cells (15, 54).
We find that expression of DNStat3 in 32D IGF-IR cells abrogates IGF-I-mediated differentiation. In fact, the cells become transformed, by stringent criteria. Using this strategy, we report that Id2 RNA expression is regulated by the IGF-IR, with a dramatic up-regulation in cells expressing DNStat3. There are also changes in Id1 RNA levels, but these are not as clear as in the case of Id2 gene expression. A mutation at tyrosine 950 (Y950F) of the IGF-IR abrogates the up-regulation of Id2 gene expression, regardless of the presence or absence of DNStat3. IGF-I-mediated Id2 gene expression is also inhibited by inhibitors of the mitogen-activated protein kinase (MAPK) or phosphatidylinositide 3-kinase (PI3K) pathways. These results are compatible with a hypothesis in which Y950 of the IGF-IR, in 32D cells, sends a dual signal. One signal (through the MAPK and PI3K pathways) induces up-regulation of Id2 gene expression. Simultaneously, Y950 sends (through Stat3) a signal that represses Id2 gene expression. In this model, the role of Stat3 is to inhibit Id2 gene activation. The role of Id2 gene expression in proliferation versus differentiation cannot be defined at this point, especially since the data in the literature are sometimes contradictory. However, at least in our model system, up-regulation of Id2 gene expression is correlated with inhibition of the differentiation program. Overexpression of Id2 in 32D IGF-IR cells also inhibits the differentiation program of these cells (as determined by myeloperoxidase RNA levels). However, Id2 overexpression is not sufficient for transformation of 32D IGF-IR cells, indicating that DNStat3 sends additional signals, above and beyond the up-regulation of Id2 gene expression.
MATERIALS AND METHODS
Plasmids, cell lines, and retroviral infection.
The Stat3 Y705F cDNA with a FLAG tag at the 3′ end, kindly provided by J. E. Darnell, Jr. (The Rockefeller University, New York, N.Y.), was excised from pRcStat3Y705F and inserted into the pMSCVpac retroviral vector (29) to generate pMSCVpac DN Stat3 Y705F.
32D cells and 32D-derived cells were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS) and 10% WEHI cell conditioned medium as a source of IL-3. 32D, 32D IGF-IR, 32D IR, and 32D Y950F cells were previously described and characterized (18, 48, 60). These cell lines were transduced with pMSCVpac DN Stat3 Y705F to generate mixed populations of 32D DN Stat3, 32D IGF-IR/DN Stat3, 32D IR/DN Stat3, and 32D Y950F/DN Stat3 cells. Selection was carried out with 1.5 μg of puromycin per ml. The same strategy was used to generate from 32D IGF-IR cells cell lines stably expressing the Id2 cDNA (see “Northern blots” below). Selection was again with puromycin. The infection method was described by Prisco et al. (49). In one experiment, Id2 RNA levels were measured in quiescent or stimulated R600 mouse embryo fibroblasts, an NIH 3T3-like cell line (49).
Growth, survival, and differentiation.
Cells were washed three times with Hanks' balanced salt solution (HBSS) and seeded at a density of 5 × 104 cells/35-mm plate in 2 ml of RPMI 1640 medium supplemented with 10% FBS with or without 50 ng of IGF-I or insulin (GIBCO-BRL) per ml or 10% WEHI cell conditioned medium. The cells were counted by trypan blue exclusion (Life Technology) at the indicated times after IL-3 withdrawal. For analysis of differentiation, exponentially growing cells were collected, washed three times with HBSS, and seeded (5 × 104 cells/ml) in RPMI 1640 medium containing 10% FBS and 50 ng of IGF-I per ml. After 96 h, viable cells were counted by trypan blue exclusion (Life Technology) and cytospins were used for the morphological analysis as described by Valentinis et al. (60). Differentiation was expressed as the percentage of bands and polymorphonuclear cells in the total number of scored cells. Treatment with rapamycin was carried out with the concentrations and the modalities previously described (60).
Northern blots.
Cells were seeded under the same conditions used for growth analysis. At the indicated time points, the cells were collected and total RNA was extracted with an RNeasy mini kit (Qiagen). In some experiment, cells were washed, treated with the inhibitor PD98059 (Calbiochem) or LY94002 (Biomol) at 50 μM for 15 min in RPMI 1640 medium containing 10% FBS or left untreated, and, after being washed, seeded (5 × 104 cells/ml) in complete medium or in medium supplemented with 10% FBS and 50 ng of IGF-I per ml for the indicated times.
An 8-μg portion of total RNA for each sample was run on a 1% agarose–formaldehyde gel, blotted onto a nylon membrane, and hybridized with a 1.3-kb Id2 cDNA obtained from pLXSN Id2 plasmid or with a 1.1-kb Id1 sequence obtained from pEM Id1 (kind gifts of G. Condorelli, Kimmel Cancer Institute, Thomas Jefferson University, Philadelphia, Pa.).
For detection of the myeloperoxidase mRNA level, cells were prepared and seeded under the same conditions used for growth analysis. At the indicated time points, the cells were collected and total RNA was extracted as above. An 8-μg portion of total RNA for each sample was run on a 1% agarose–formaldehyde gel, blotted onto a nitrocellulose membrane, and hybridized with a 1.45-kb myeloperoxidase cDNA fragment obtained from the pUC19-MMPO6 plasmid (a kind gift of Mauro Valtieri). The cDNA probe was labeled with [α-32P]dCTP by the random-primed DNA-labeling kit (Boehringer Mannheim) and purified using QuickSpinn G-50 Sephadex columns (Boehringer-Mannheim).
Western blots.
Cells were lysed with lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, 1% NP-40, 100 mM NaF, 10 mM sodium pyrophosphate, 0.2 mM sodium orthovanadate; 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml). For the detection of DN Stat3 Y705F, 300 μg of total extract was immunoprecipitated with an anti-FLAG antibody M2 (Sigma). After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS PAGE) (4 to 15% polyacrylamide) and transfer, the nitrocellulose membrane was probed with a monoclonal antibody against Stat3 (Transduction Lab.). Phosphorylated Stat3 was detected by immunoprecipitation or directly on whole lysates. For immunoprecipitation, cells were washed three times with HBSS and incubated in RPMI 1640 serum-free medium plus 0.1% bovine serum albumin for 3 h before stimulation with 50 ng of IGF-I 50 per ml (Gropep) for 2, 5, 30, or 60 min. The cells were collected, washed with cold phosphate-buffered saline, and lysed with lysis buffer. Portions (300 μg) of total extracts were immunoprecipitated with an anti-Stat3 monoclonal antibody (Transduction Lab.) and the precipitated proteins were separated by SDS-PAGE (4 to 15% polyacrylamide). After transfer, the nitrocellulose membrane was probed with a polyclonal antibody against antiphospho-Stat3 Y705 (New England Biolabs). After being stripped, the membrane was probed with a polyclonal antibody against Stat3 (Santa Cruz Biotechnology Inc.). For detection on whole lysates, cells were washed three times with HBSS and incubated in RPMI 1640 plus 10% FBS for 6 h before being stimulated with 50 ng of IGF-I per ml for 5 or 30 min or with G-CSF (Gibco, BRL) for 30 min. The cells were washed with phosphate-buffered saline, suspended in hypertonic buffer (50 mM HEPES [pH 7.5], 250 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10 mM NaF, 0.1 mM Na3VO4, 0.1% Tween 20, 10% glycerol, 1 mM dithiothreitol, 0.1 mM phenyl methylsulfonyl fluoride, 10 μg of Aprotinin per ml, 10 μg of leupeptin per ml, 10 μg of Pepstatin, per ml), and lysed by freezing and thawing. Then 100 μg of lysate was resolved by SDS-PAGE (4 to 15% polyacrylamide). After being blotted, the membrane was probed with a polyclonal antibody against antiphospho-Stat3 Y705 (New England Biolabs), stripped, and reprobed with a Stat3 monoclonal antibody (Transduction Lab.).
For the activation and detection of Shc and Akt proteins, we have used methods and antibodies described in previous papers (18, 44, 48). For extracellular signal regulated kinase (ERK) activation, cells were washed three times with HBSS and incubated in serum-free medium (SFM) for 3 h before being stimulated with 50 ng of IGF-I per ml at the indicated times. The cells were lysed with lysis buffer and 100 μg of total extracts was resolved by SDS-PAGE (4 to 15% polyacrylamide). ERK activation was detected with anti-phospho MAPK (Erk1/2) from UBI. The membrane was then stripped and probed with anti-Erk1 antibody, which recognizes both Erk1 and Erk2 (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). The same procedure was used for the detection of Id proteins, using antibodies from Santa Cruz. The Grb2 antibody was from Transduction Laboratories.
The phosphorylation of specific amino acids in p70S6K (phospho-Thr389) was detected with an antibody purchased from New England Biolabs. The total amount of p70S6K loaded was monitored after stripping of the filters by immunoblotting with an anti p70S6K antibody (C-18; Santa Cruz).
Tumor formation in nude mice.
The cell lines used are given in Table 1. The procedure used was exactly the same as the one described by Valentinis et al. (59).
TABLE 1.
Weights of liver and spleen in nude mice injected with 32D-derived cellsa
| Cell injected | Wt (mg) ofb:
|
||
|---|---|---|---|
| Liver | Spleen | Kidneys | |
| 32D IGF-IR | 1,270 (1,100–1,400) | 49 (48–50) | 50 (46–54) |
| 32D IGF-IR/DNStat3 | 2,285 (1,750–1,820) | 600 (400–800) | 535 (470–600) |
| 32D IGF-IR/IRS-1 | 4,000 (3,800–4,200) | 1,100 (1,000–1,200) | 430 (370–490) |
SCID mice were injected intraperitoneally with 107 cells and were examined after 6 weeks.
Results are shown as the mean and range of two experiments.
RESULTS
As anticipated, a dominant negative mutant of Stat3 inhibited IGF-I-mediated differentiation of 32D IGF-IR cells (see below). We therefore investigated Id gene expression in 32D IGF-IR and 32D IGF-IR/DNStat3 cells in the first 24 h after IL-3 withdrawal. We limited ourselves to Id1 and Id2, because Id3 and Id4 are not expressed in 32D cells (23).
Time course of Id gene expression after IGF-I stimulation.
After IL-3 withdrawal and IGF-I supplementation, Id2 mRNA levels were increased. In 32D IGF-IR cells, there was a modest but reproducible increase at 4 h after shifting to IGF-I. The increase in Id2 RNA levels was much more dramatic in 32D IGF-IR/DNStat3 cells than in 32D IGF-IR cells (Fig. 1A). There was another peak at 4 h, but Id2 RNA levels were still quite high at 24 h. An important point is that Id2 mRNA levels in both cell lines were higher when the cells were cultured in IGF-I than when they were growing in IL-3. These experiments have been repeated several times, with the same results (see also below). This observation is intriguing because in the first 48 h, 32D IGF-IR cells and 32D IGF-IR/DNStat3 cells doubled in number in each 24-h period, although their fates diverged soon after. The burst of cell proliferation that follows the addition of IGF-I to 32D IGF-IR cells has been repeatedly documented in previous papers (18, 48, 59) and was confirmed in the present experiments (data not shown). The growth of 32D IGF-IR/DNStat3 in IGF-I-supplemented medium will be documented below. Furthermore, in IL-3, all 32D-derived cell lines, even those with nonfunctional mutants of the IGF-IR, grow exponentially (18, 48, 59, 60). The results of Fig. 1A therefore indicate that the activated IGF-IR up-regulates Id2 gene expression and that the dramatic up-regulation in 32D IGF-IR/DNStat3 cells does not solely reflect the proliferative status of the cells. Finally, in the first 48 h, cell death in these cell lines is negligible (reference 60 and data not shown).
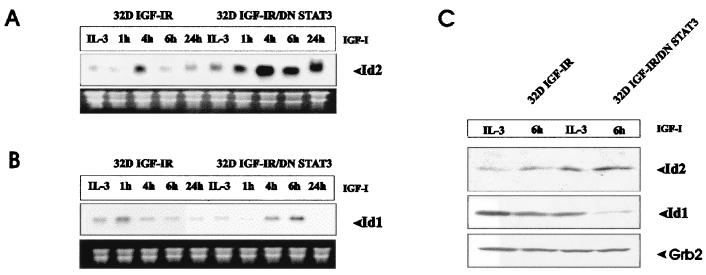
FIG. 1.
Time course of Id gene expression in 32D-derived cells. The cell lines used were 32D IGF-IR and 32D IGF-IR/DNStat3. The expression of the mRNAs for Id1 and Id2 was determined at the times indicated after IL-3 withdrawal and supplementation with IGF-I. The IL-3 lane refers to cells exponentially growing in IL-3. RNA and Northern blots were prepared as described in Materials and Methods. (A) mRNA levels of Id2. (B) mRNA levels of Id1. Levels of rRNA were used to monitor RNA amounts in each lane. (C) Levels of Id proteins in the same cell lines growing in IL-3 or 6 h after shifting to IGF-I. The antibodies are described in Materials and Methods. Grb2 levels were used to monitor the protein amounts in each lane.
The results with Id1 mRNA are inconclusive (Fig. 1B). There was a modest increase in Id1 RNA levels in 32D IGF-IR/DNStat3 cells when compared with the parental cell line, 32D IGF-IR cells. Although the increase was reproducible, it never was as impressive as with Id2. In addition, Id1 RNA levels were, in some experiments, as high in IL-3 as in IGF-I. The increase in Id2 RNA levels in 32D IGF-IR/DNStat3 cells was accompanied by an increase in the levels of Id2 protein (Fig. 1C). Again, the results with Id1 protein were inconclusive. These first experiments indicated a relationship between the IGF axis and the expression of Id genes. Id2 gene expression was dramatically increased by IGF-I in 32D IGF-IR cells expressing DNStat3. We next examined the dependence of these changes on signaling from the IGF-IR, focusing on the Id2 RNA levels.
Regulation of Id2 gene expression is IGF-I dependent.
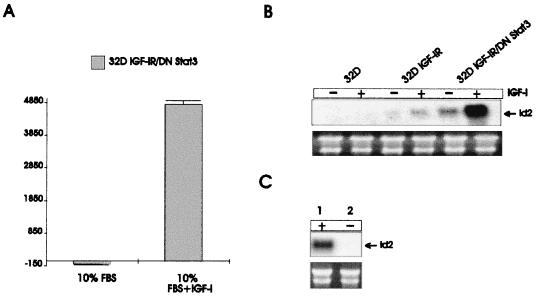
32D IGF-IR/DNStat3 cells were grown either in 10% serum or in serum supplemented with IGF-I. It is clear that supplementation with IGF-I is obligatory for the sustained growth of these cells in the absence of IL-3 (Fig. 2A). We then determined Id gene expression under the same conditions. A typical experiment is shown in Fig. 2B, where we compared Id2 RNA levels in three different cell lines: parental 32D cells, 32D IGF-IR cells, and 32D IGF-IR/DNStat3 cells. The cells were incubated either in IL-3 or in IGF-I (50 ng/ml) for 6 h. Id2 RNA was barely detectable in parental 32D cells, and its level increased modestly in 32D IGF-IR cells stimulated with IGF-I. It was markedly increased in 32D IGF-IR/DNStat3 cells, especially when incubated with IGF-I. These experiments indicate in 32D IGF-IR cells, the dramatic up-regulation of Id2 mRNA requires both a dominant negative mutant of Stat3 and an IGF-IR activated by its ligand. The IGF-IR requirement for the proliferation of 32D IGF-IR/DNStat3 cells will be further documented below. We asked at this point whether Id2 RNA up-regulation also occurred in another cell line. For this experiment, we chose an NIH 3T3-like cell line of mouse embryo fibroblasts called R600 cells (49). Figure 2C shows that the levels of Id2 mRNA were very high in stimulated cells (lane 1) but undetectable in quiescent cells (lane 2). Since activation of the IGF-IR is required for Id2 up-regulation in the 32D model, we next attempted to identify the domain(s) of the receptor sending this signal.
FIG. 2.
IGF-I is required for up-regulation of Id2 RNA and inhibition of differentiation by DNStat3. (A) Growth of 32D IGF-IR/DN Stat3 cells was determined in 10% serum in the presence or absence of IGF-I (50 ng/ml). The results given were obtained at 96 h after IL-3 withdrawal. (B) Expression of Id2 mRNA in parental 32D cells, 32D IGF-IR cells, and 32D IGF-IR/DNStat3. The cells were exponentially growing either in IL-3 or in IGF-I, as indicated above the lanes. IGF-I was added for 6 h. (C) Id2 mRNA levels in mouse embryo fibroblasts (R600 cells). The cells were either quiescent (lane 2) or stimulated (lane 1). RNA amounts in the last two panels were monitored with rRNA.
Effect of a mutation at Y950 of the IGF-IR on IGF-I regulation of Id2 RNA expression.
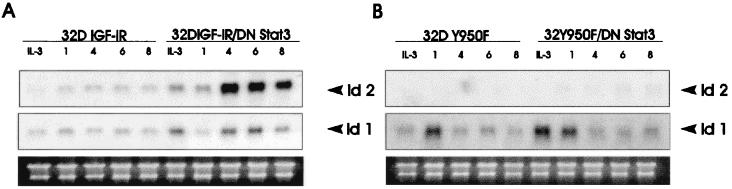
It is generally agreed that the main mitogenic and antiapoptotic pathways of the IGF-IR largely depend on the activation of one of its major substrates, IRS-1 (7, 64). IRS-1, in turn, activates the PI3K/Akt pathway (19, 33, 34, 37). In 32D cells, which do not express IRS-1 (or IRS-2), the mitogenic and antiapoptotic signals originating from the IGF-IR depend on two other pathways (48). One of these pathways originates from Y950 (18, 44), which is also involved in IGF-I-mediated differentiation (60). Accordingly, we have investigated the expression of Id mRNAs in four cell lines: 32D IGF-IR, 32D IGF-IR/DNStat3, 32D Y950F, and 32D Y950F/DNStat3. The 32D IGF-IR/Y950F cell line has already been described (18, 44, 60). DNStat3 was transduced into these cells (see below for expression levels). The cells were shifted from IL-3 to IGF-I, and the mRNA levels were determined at the times indicated in Fig. 3. The results of a typical experiment are again different for the two Id mRNAs. We confirmed that the levels of Id2 RNA were markedly increased in 32D IGF-IR/DNStat3 cells, compared with the parental 32D IGF-IR cell line (Fig. 3A). However, the results with the cells expressing the Y950F mutant receptor were very different. No Id2 mRNA was detectable in these two cell lines, whether expressing DNStat3 or not (Fig. 3B). This experiment was repeated three times, and always yielded the same results. Id1 mRNA levels were again somewhat higher in 32D IGF-IR/DNStat3 cells than in 32D IGF-IR cells (Fig. 3A). However, Id1 gene expression was not really affected by a mutation at Y950 (Fig. 3B). If anything, a mutation at Y950 actually increased Id1 gene expression. This is an important control, because it shows that the mutation at Y950 selectively abrogates Id2 gene expression and that 32D Y950F/DNStat3 cells, at these times, are still viable and in satisfactory condition.
FIG. 3.
Effect of a mutation at Y950 of the IGF-I receptor on Id gene expression. The same experiments described in the legend to Fig. 1 were carried out with four cell lines: 32D IGF-IR (A), 32D IGF-IR/DNStat3 (A), 32D Y950F (B), and 32D Y950F/DNStat3 (B). Y950F refers to a mutation in tyrosine 950 of the IGF-IR (see text). The levels of Id1 and Id2 mRNA were determined at the indicated times (in hours after shifting from IL-3 to IGF-I) as described in Materials and Methods. The cell lines are indicated above the lanes (the amounts of RNA in each lane are in the lower rows).
Mechanism of Y950 activation of Id2 gene expression.
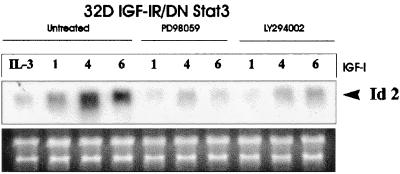
Y950 of the IGF-IR binds Shc proteins (13). Shc proteins are known to activate the Ras-Raf-MAPK pathway (5, 56). We have repeatedly shown that 32D IGF-IR cells with a mutation at Y950F have a decreased MAPK activity (18, 44). However, it has recently been reported that Shc phosphorylation may also activate the PI3K pathway (26). In addition, IGF-I causes a modest but reproducible increase in PI3K activity in 32D IGF-IR cells, even in the absence of IRS-1, an increase that is translated into an increase in Akt activation (44, 60). To distinguish between these two possibilities, we have used inhibitors of MAPK and PI3K to study their effect on Id2 gene expression in 32D IGF-IR/DNStat3 cells. The results are shown in Fig. 4. Again, there was a sharp increase in Id2 RNA levels when 32D IGF-IR/DNStat3 cells were shifted from IL-3 to IGF-I. Both a MEK inhibitor (PD98059) and a PI3K inhibitor (LY294002) effectively inhibited Id2 gene expression, indicating that both pathways are important for Id2 up-regulation.
FIG. 4.
Inhibitors of PI3K and ERK pathways inhibit IGF-I-mediated up-regulation of Id2 RNA. The cell line examined was the 32D IGF-IR/DNStat3 cell line. The treatment and the times (in hours) after IGF-I stimulation are indicated above the lanes. The first four lanes refer to untreated cells. The other lanes refer to cells treated with either PD98059 (MEK inhibitor) or LY294002 (PI3K inhibitor). RNA and Northern blot analyses were carried out as for previous figures.
Shc phosphorylation and MAPK activation in 32D-derived cells.
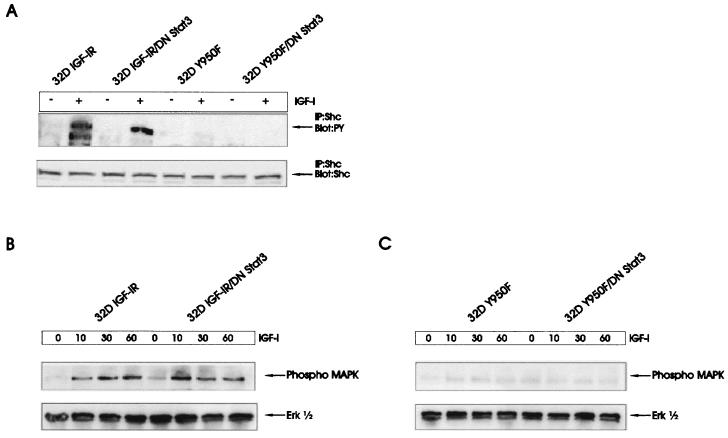
The experiments in Fig. 3 indicate that in this model, Id2 gene expression requires an intact Y950 residue. The literature (see Discussion) strongly supports the notion that Y950 sends a signal through Shc to the MAPK pathway. The effect of a MAPK inhibitor on Id2 gene expression suggests an involvement of MAPK in Id activation (Fig. 4). We have already reported that a mutation at Y950 of the IGF-IR markedly decreases or even completely abrogates the phosphorylation of the 52-kDa isoform of the Shc proteins in 32D IGF-IR cells (18, 44, 60). MAPK activity is also markedly decreased in 32D cells expressing the Y950F receptor (18, 44, 48). We have repeated these experiments and extended them to cells expressing DNStat3. Not surprisingly, DNStat3 did not restore the inhibition of Shc and MAPK activation in cells expressing the mutant receptor (Fig. 5).
FIG. 5.
Phosphorylation of Shc and ERKs in 32D-derived cells. (A) The indicated cell lines were stimulated with IGF-I (50 ng/ml) for 10 min. Phosphorylated Shc proteins and Shc protein amounts were determined as described in Materials and Methods. (B) MAPK activation in 32D IGF-IR and 32D IGF-IR/DNStat3 cells. Above the lanes are the times (in minutes) after stimulation with IGF-I. (C) Same experiment as in panel B, but in the cell lines expressing the IGF-IR with a mutation at Y950.
According to the experiments in Fig. 4, the PI3K pathway may also send a signal for the activation of Id2 gene expression. IGF-I activates Akt in 32D IGF-IR cells, albeit at much decreased levels compared to those in cells expressing IRS-1 (44, 60). A mutation at Y950, however, completely abrogates the modest activation of Akt in these cells (44). Again, the expression of DNStat3 did not restore the activation of Akt in 32D/Y950F cells (data not shown). It seems, therefore, that the failure of 32D cells expressing the mutant receptor to up-regulate Id2 gene expression depends on the inability of Y950F to activate the MAPK and PI3K pathways (Fig. 4).
Activation of Stat3 by the IGF-IR.
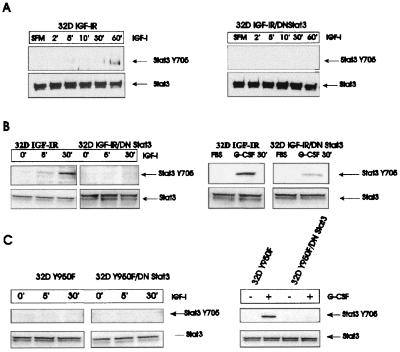
The previous experiments indicate that both Y950 and Stat3 play an important role in the up-regulation of Id2 gene expression. Since DNStat3 inhibits differentiation of 32D IGF-IR cells, it is reasonable to assume that the IGF-IR activates Stat3 and that its activation is necessary for their differentiation. We asked whether Stat3 would be activated in 32D cells expressing the IGF-IR with a mutation at Y950. There are already reports in the literature indicating that the IGF-IR activates Stat3 (68, 69), although usually in association with transformation rather than differentiation. We have investigated the phosphorylation of tyrosine 705 (Y705) of Stat3 in 32D IGF-IR cells and in 32D IGF-IR/DNStat3 cells (Fig. 6). In the first experiment (Fig. 6A), lysates were immunoprecipitated with an antibody to Stat3 and the gels were blotted with an antibody recognizing the phosphorylated Y705 residue. Y705 phosphorylation was detectable in 32D IGF-IR cells between 30 and 60 min after stimulation with IGF-I. Under the conditions used, Y705 phosphorylation was not detectable in 32D IGF-IR/DNStat3 cells, presumably because the dominant negative mutant interferes with its detection. In a second experiment, Western blotting was done directly on lysates from the same cells (Fig. 6B). A Y705 phosphorylated Stat3 was again detectable in 32D IGF-IR cells, with a slight increase already visible 5 min after IGF-I stimulation. This band was not clearly detectable in 32D IGF-IR/DNStat3 cells, although there was more Stat3 protein in the lysates from these cells (as expected). As a control, we used stimulation with G-CSF (Fig. 6B, right). In the presence of G-CSF, which is a strong activator of Stat3, a band was also visible in 32D IGF-IR/DNStat3 cells, although its intensity was decreased in comparison to that in 32D IGF-IR cells.
FIG. 6.
Phosphorylation of tyrosine 705 of Stat3 by IGF-I. The cell lines are indicated above the panels. (A) The times (in minutes) are times after placing the cells in serum-free medium (SFM) and stimulation with IGF-I. The lysates were immunoprecipitated with an antibody to Stat3, and the blots were developed with a phosphoantibody to tyrosine 705 of Stat3. The blots were then reprobed with an antibody to Stat3. (B) Western blots on lysates of the same cell lines. The blots were probed directly with an antibody to Y705 (upper row) and then reprobed with anti-Stat3 antibody (lower row). In the experiment on the right, G-CSF was used as control for the phosphorylation of Y705. (C) A mutation at Y950 of the IGF-IR abrogates the detection of Y705 phosphorylation of Stat3, regardless of the presence or absence of DNStat3 (left). The right panel shows that Y705 phosphorylation of Stat3 is not abrogated in the same cell lines stimulated with G-CSF.
We have tested the activation of Stat3 in 32D cells expressing the receptor mutated at Y950. The results are shown in Fig. 6C. Phosphorylation of Stat3 at Y705 was no longer detectable in 32D cells expressing the mutant IGF-IR, regardless of IGF-I stimulation or the presence of DNStat3. The parental cells expressing the Y950F receptor, though, were still capable of phosphorylating Y705 when stimulated with G-CSF (Fig. 6C). These results are compatible with a model in which Y950 is required for the phosphorylation of Stat3 at Y705. Failure to activate Stat3 (for instance, with DNStat3) resulted in up-regulation of Id2 gene expression and inhibition of differentiation.
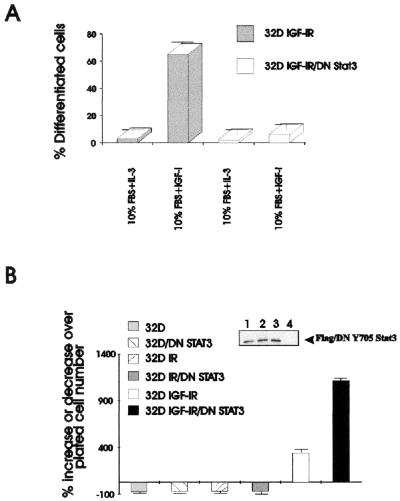
A dominant negative mutant of Stat3 inhibits IGF-I-mediated differentiation and causes transformation of 32D IGF-IR cells.
Id proteins play a role in differentiation and tumor development (see Introduction), and DNStat3 has a dramatic effect on their expression, especially of Id2. We have asked whether this strong up-regulation of Id2 in 32D IGF-IR/DNStat3 cells correlates with their growth and differentiation. As already reported, 32D IGF-IR cells differentiate along the granulocytic pathway, provided that the medium is supplemented with IGF-I (59, 60). This was confirmed in the experiment in Fig. 7A. Expression of a dominant negative mutant of Stat3 (DN/Stat3) in 32D IGF-IR cells caused inhibition of differentiation (Fig. 7A). For clarity, we show in Fig. 7A only the extent of differentiation on day 4 after shifting the cells from IL-3 to IGF-I. Later days, however, were also monitored, and differentiation in 32D IGF-IR/DNStat3 was essentially abrogated. The inhibition of differentiation was accompanied by increased growth rates, IL-3 independence (Fig. 7B), and malignant transformation. The difference in growth between the 32D IGF-IR cells and 32D IGF-IR/DNStat3 cells was already evident at 48 h, and it became more pronounced at later times (Fig. 2A), when 32D IGF-IR cells began to differentiate (60). Figure 7B also shows that the transformation of 32D IGF-IR cells by DNStat3 (IL-3 independence) is dependent on a functional IGF-IR. When DNStat3 was expressed in parental 32D cells or in 32D cells overexpressing the insulin receptor, the cells remained IL-3 dependent and died rapidly in its absence. The failure of DNStat3 to transform 32D cells overexpressing the insulin receptor confirms previous results indicating that in the absence of IRS-1, the insulin receptor cannot protect 32D cells from apoptosis induced by IL-3 withdrawal (18, 48, 66). The expression of DNStat3 in these cell lines is shown in the inset of Fig. 7B, lanes 1 to 3. Since DN/Stat3 carried a FLAG tag, it was immunoprecipitated with an anti-FLAG antibody, and the protein was detected with an antibody to Stat3. Stat3 was not detectable in parental 32D cells by this method (lane 4) but was present (see above).
FIG. 7.
A dominant negative mutant of Stat3 inhibits IGF-I-mediated differentiation and causes transformation of 32D IGF-IR cells. 32D cells were transduced with the appropriate retroviral vectors, and mixed populations were selected. (A) IGF-IR cells and 32D IGF-IR/DN Stat3 cells were grown in medium supplemented with 10% serum and either IL-3 or IGF-I. The percentage of differentiated cells is indicated on the ordinate. In this experiment, the cells were fixed and stained after 4 days in the indicated medium. The percentage of differentiated cells was determined by standard methods (60). (B) After withdrawal of IL-3, the cells were grown in medium supplemented with 10% serum and either IGF-I or insulin at a concentration of 50 ng/ml. The cells were counted 48 h after IL-3 withdrawal. The cell lines are indicated on the left of the figure. 32D, parental cells; 32D IR, cells overexpressing the insulin receptor; 32D IGF-IR, cells expressing increased levels of IGF-IR. DN/STAT3 indicates the same cell lines stably transduced with the dominant negative mutant of Stat3. The inset shows levels of expression of DN/Stat3, after immunoprecipitation with a FLAG antibody and blotting with an anti-Stat3 antibody. Lanes of inset: 1, 32D/DN Stat3; 2, 32D IR/DN Stat3; 3, 32D IGF-IR/DN Stat3; 4, parental 32D cells.
32D IGF-IR/DNStat3 cells are fully transformed. They can be passaged indefinitely in the absence of IL-3, and they can form tumors in nude mice (Table 1). As already reported, the parental cells, 32D IGF-IR cells, cannot form tumors in nude mice (59). 32D IGF-IR/DNStat3 cells form tumors in SCID mice, as evidenced by the increase in the weights of livers and spleens. The spleen is especially enlarged, to about 10 times the weight of a normal spleen. This is also what we found previously with 32D IGF-IR/IRS1 cells (59), which were repeated in Table 1 for comparison. It may be argued that 32D IGF-IR/IRS1 cells form larger tumors than 32D IGF-IR/DNStat3 cells. However, these latter cells also colonize the livers and spleens of injected mice. The pathology is that of a leukemia, infiltrating both liver and spleen, which has been previously documented histologically (59).
The results therefore indicate that in 32D IGF-IR cells, expression of DNStat3 inhibits IGF-I-mediated differentiation and causes IL-3-independence and malignant transformation. These effects of DNStat3, however, are dependent on the presence of a functional IGF-IR. This was confirmed in the following experiments.
A mutation at tyrosine 950 of IGF-IR abrogates the proliferative effect of DNStat3.
Given that Y950 is necessary for the up-regulation of Id2 gene expression by the IGF-IR (Fig. 3), one would expect that a mutation at Y950 should abrogate the ability of DNStat3 to transform 32D IGF-IR cells. As mentioned above, we transduced DNStat3 into 32D cells expressing the Y950F mutant of the IGF-I receptor. The expression of the transduced DNStat3 in this cell line is shown in the inset of Fig. 8. Stat3 was again indicated by using an antibody to the FLAG epitope, and therefore the parental cells are negative. Both cell lines, parental 32D/Y950F and 32D/Y950F/DNStat3 cells, grew very well in IL-3, as expected. However, neither of these cell lines survived after IL-3 withdrawal, not even when the medium was supplemented with IGF-I (Fig. 8). In fact, the cells expressing DNStat3 and the mutant receptor were indistinguishable from the parental cells expressing only the mutant receptor (Fig. 8). This is dramatically different from the effect of DNStat3 on 32D cells expressing the wild-type IGF-IR.
FIG. 8.
Effect of a mutation at Y950 on the growth of 32D IGF-IR cells expressing DNStat3. The cell lines are indicated to the right of the figure or above the lanes. Y950F is a mutation at tyrosine 950 of the IGF-I receptor. This mutation has been previously described (see the text). The cell number was determined after 48 h of incubation in the medium indicated on the abscissa. The inset shows the level of expression of DNStat3 in the new cell line.
These experiments indicate that expression of DNStat3 is not sufficient for the transformation of 32D IGF-IR cells (and the up-regulation of Id2 gene expression). The proliferative stimulus originates from the IGF-IR, and the function of DNStat3 is to extinguish the differentiation program, which is simultaneously implemented by the IGF-IR. When Y950 is mutated, DNStat3 cannot stimulate the proliferation of 32D IGF-IR cells and cannot up-regulate Id2 gene expression.
Functional significance of Id2 gene expression in 32D cells.
Our results clearly indicate that the IGF-IR and Stat3 cooperate in regulating the expression of Id2 RNA and proteins in 32D cells. We therefore wanted to know the functional significance of this regulation. Since Id proteins are involved in both proliferation and differentiation of cells (see Introduction), we have investigated the effect that blockade or overexpression of Id2 may have on either proliferation or differentiation of 32D-derived cells.
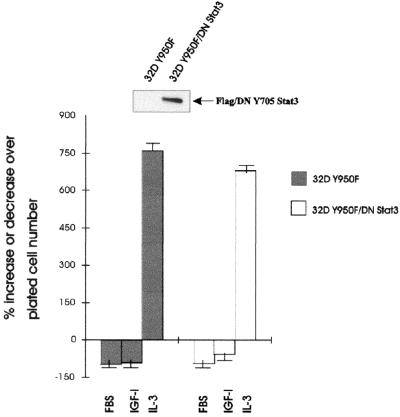
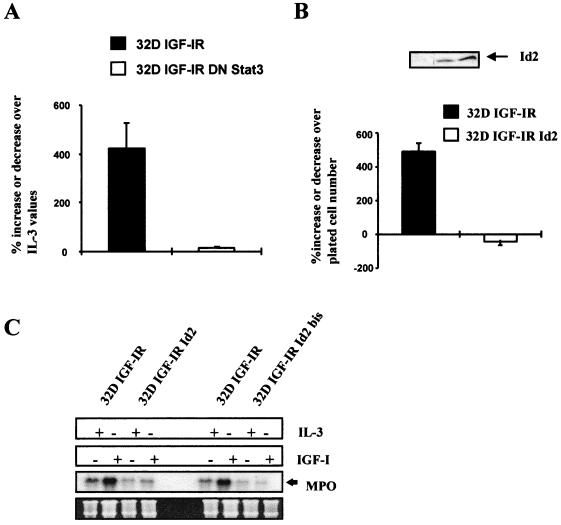
Differentiation of 32D IGF-IR cells is a slow process that becomes clearly evident only on day 4 after shifting from IL-3 to IGF-I (60). It is therefore not always possible to score for morphological differentiation the cell lines that die in the first 24 to 48 h after IL-3 withdrawal and IGF-I supplementation. However, we and others (59–61) have shown that cells programmed for differentiation have increased levels of myeloperoxidase (MPO) RNA in the first 24 h, although the cells at that time are actively proliferating (62). Therefore, MPO RNA levels are an early marker of differentiation that indicates whether the cells have started a differentiation program (60, 61). We first confirmed this finding in experiments with 32D IGF-IR and 32D IGF-IR/DNStat3 cells (Fig. 9A). We measured MPO RNA levels in these two cell lines at 72 h after the shifting of cells from IL-3 to IGF-I. As expected, MPO RNA levels increased sharply in differentiating 32D IGF-IR cells but remained low in 32D IGF-IR/DNStat3 cells. These experiments suggest that Id2 gene expression plays a role in the inhibition of differentiation (see also below). We then asked whether overexpression of Id2 in 32D IGF-IR cells would affect either the proliferation or the differentiation programs or both. For this purpose, we transduced a retroviral vector expressing the human Id2 cDNA (see Materials and Methods) into 32D IGF-IR cells. Its effect on the growth of these cells is shown in Fig. 9B. 32D IGF-IR cells stably overexpressing Id2 (see inset in Fig. 9C) do not transform. Actually, they underwent apoptosis, beginning at about 48 h after withdrawal of IL-3. These results are in agreement with those of Florio et al. (23), who reported that overexpression of Id2 in parental 32D cells resulted in an acceleration of apoptosis caused by IL-3 withdrawal. However, overexpression of Id2 protein affects the differentiation program of 32D IGF-IR cells. Since the cells expressing Id2 die more quickly, we determined the levels of expression of MPO RNA at 24 h after shifting from IL-3 to IGF-I. The results are shown in Fig. 9C. Again, IGF-I increased MPO RNA levels in 32D IGF-IR cells but failed to do so in 32D IGF-IR Id2 cells. It seems, therefore, that overexpression of Id2 protein can inhibit the differentiation program of 32D IGF-IR cells but has no effect on their survival.
FIG. 9.
MPO RNA levels in 32D-derived cells. The levels of MPO RNA, a marker of differentiation, were measured in the indicated cell lines. The MPO RNA levels are expressed as percentage of values in the same cell lines growing in IL-3-supplemented medium. The cell lines designated 32D IGF-IR and 32D IGF-IR/DNStat3 have been described in the previous figures. 32D IGF-IR Id2 cells are two mixed populations of 32D IGF-IR cells stably overexpressing Id2 (see Materials and Methods for the retroviral vector used). (A) MPO RNA levels in 32D IGF-IR and 32D IGF-IR/DNStat3 cells at 72 h after shifting from IL-3 to IGF-I. (B) Survival of 32D IGF-IR and 32D IGF-IR Id2 cell lines at 72 h after shifting from IL-3 to IGF-I (expressed as the percent decrease or increase over the plated number). The inset shows expression of Id2 protein in the parental cell lines (lane 1) and in the two mixed populations transduced with the Id2 retroviral vector (lanes 2 and 3). (C) MPO RNA levels in 32D IGF-IR and 32D IGF-IR Id2 cells, growing for 24 h in either IL-3 or IGF-I. The Northern blots give two distinct populations of 32D IGF-IR Id2 cells, selected separately at different times. Amounts of RNA in each lane were monitored with rRNA.
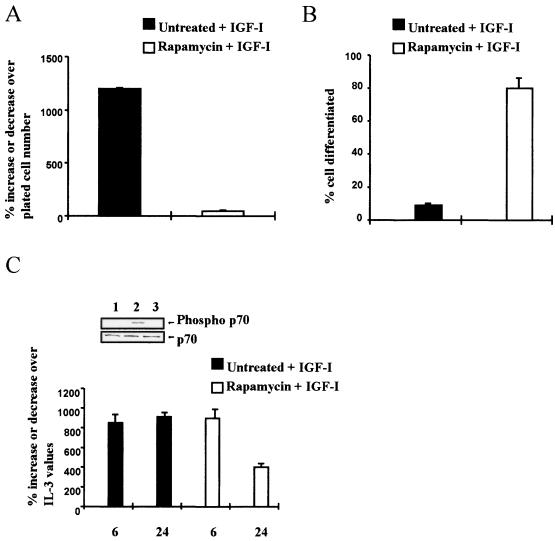
To confirm the effect of Id2 gene expression on differentiation, we have used a second approach, based on a previous paper, in which we showed that the mTOR-specific inhibitor, rapamycin (20), induces the differentiation of 32D IGF-IR/IRS-1 cells (60). If rapamycin induces differentiation in these cells, it may also do so in 32D IGF-IR/DNStat3 cells, which behave like 32D IGF-IR/IRS-1 cells, forming tumors in nude animals (reference 60 and results cited above). Indeed, rapamycin treatment of 32D IGF-IR/DNStat3 cells inhibited their growth (Fig. 10A) and induced differentiation (Fig. 10B). Rapamycin inhibits the activation of p70S6K (20), which was confirmed (inset in Fig. 10C). If Id2 down-regulation is necessary for differentiation, then Id2 gene expression should decrease in 32D IGF-IR/DNStat3 cells treated with rapamycin. This is what happened (Fig. 10C) when Id2 gene expression was compared in these cells with or without rapamycin treatment. The levels of Id2 RNA were essentially the same as in 32D IGF-IR cells treated with IGF-I (compare the IL-3 and IGF-I treatments in Fig. 2B).
FIG. 10.
Effect of rapamycin on 32D IGF-IR/DNStat3 cells. 32D IGF-IR/DNStat3 cells were incubated in 10% FBS plus IGF-I only or plus IGF-I and rapamycin (10 ng/ml). (A) Growth of cells, expressed as the percent increase over the number of plated cells, at 72 h after IL-3 withdrawal and IGF-I supplementation (plus or minus rapamycin). (B) Percentage of differentiated cells under the same conditions. (C) Id2 RNA levels in 32D IGF-IR/DNStat3 cells, plus or minus rapamycin, at the indicated times after IL-3 withdrawal and IGF-I supplementation. The inset shows activation of p70S6K Lanes: 1, unstimulated; 2, stimulated with IGF-I (20 ng/ml) for 20 min; 3, same in the presence of rapamycin. Total protein is shown in the lower row.
DISCUSSION
The novel findings in this communication can be summarized as follows. (i) the IGF-IR, activated by its ligand, regulates Id2 gene expression. When Stat3 activation is inhibited by a dominant negative mutant of Stat3, IGF-I causes a dramatic increase in Id2 gene expression. (ii) in 32D cells, which do not express IRS-1 (48, 62), the tyrosine residue at 950 of the IGF-IR plays an important role in the regulation of Id2 gene expression. (iii) regulation of Id2 gene expression by Y950 is both positive (through MAPK and PI3K) and negative (through Stat3). (iv) Up-regulation by IGF-I of Id2 gene expression inhibits the differentiation program of 32D IGF-IR cells. (v) the differentiation program of 32D IGF-IR cells is also inhibited by overexpression of Id2. (vi) 32D IGF-IR/DNStat3 cells are actually transformed (tumor formation in nude mice). However, the up-regulation of Id2 gene expression, by itself, is not sufficient for transformation, since 32D IGF-IR Id2 cells do not survive withdrawal of IL-3. It seems, therefore, that the Id2 protein can regulate the differentiation program of 32D cells but requires other factors for their transformation, factors that are provided by the inhibition of Stat3.
As a corollary to these findings, we would like to formulate a hypothesis that will be discussed in detail below. In 32D IGF-IR cells, which do not express IRS-1 or IRS-2 (60, 62), the tyrosine 950 residue of the IGF-IR simultaneously sends two signals. One signal, through the activation of Shc and the MAPK and PI3K pathways (22, 25, 64), induces Id2 gene expression and a proliferative program. At the same time, Y950 sends (through the activation of Stat3) a signal which represses Id2 gene activation and promotes cell differentiation. Inhibition of Stat3 by DNStat3 causes a marked increase in Id2 gene expression and inhibition of the differentiation program, resulting in continuous proliferation and transformation. The dual signal from Y950 is seemingly contradictory, but it has a reasonable explanation. As already pointed out, 32D IGF-IR cells proliferate actively for the first 48 h after shifting from IL-3 to IGF-I. As is the case with other growth factors of hematopoietic cells (see Introduction), IGF-I sends two signals, with the differentiation signal eventually prevailing.
While there is substantial information about the functions of the Id proteins, little is known about the growth factors and the pathways of signal transduction that regulate the expression of the Id genes. There are reports that Id gene expression is up-regulated by serum (28) and platelet-derived growth factor (10). Id2 expression is induced by serum (38) and by cytokines that drive granulocytic differentiation (11). Our data provide evidence that the activated IGF-IR regulates Id2 gene expression. Addition of IGF-I is necessary for the increase in Id2 gene expression, whether in 32D IGF-IR cells or 32D IGF-IR/DNStat3 cells. Id2 RNA levels are actually increased, in the presence IGF-I, over the levels in IL-3-cultured cells, indicating that the up-regulation is not simply the consequence of cell proliferation (see also below). A mutation at Y950 of the IGF-IR abrogates the up-regulation of Id2 RNA, even in 32D IGF-IR/DNStat3 cells, indicating that Y950 is required in these cells for the up-regulation of Id2 gene expression. It also confirms that the signal originates from the IGF-IR.
The expression of DNStat3 in 32D-derived cells is accompanied by dramatic changes in their biological behavior. DNStat3 inhibits differentiation and causes malignant transformation of cells. DNStat3 is known to interfere with Stat3 signaling (9, 24, 42, 43). Stat3 is involved in cell differentiation (42, 43, 65), and the same dominant negative mutant of Stat3 we have used in these experiments is known to effectively inhibit differentiation by other growth factors (47, 50, 54, 56). Another interesting biological change is the effect of a mutation at Y950 on the survival of 32D cells. We have already reported that a Y950F mutant receptor no longer protects 32D cells from apoptosis induced by IL-3 withdrawal (18, 60). DNStat3, although inhibiting IGF-I-mediated differentiation in 32D IGF-IR cells, cannot rescue 32D Y950F cells from apoptosis. Clearly, in this model, both the differentiation and the mitogenic and survival signals originate from the IGF-IR and, more precisely, from Y950. The function of DNStat3 is to inhibit the differentiation program while leaving intact the proliferation program of the IGF-IR (59).
The role of Id2 in transformation and differentiation of 32D cells is complex. Decreased Id2 gene expression correlates with cell differentiation (see Introduction), and this has been confirmed in our experiments in several ways. Blockade of Id2 gene expression by rapamycin induces the activation of the differentiation program of 32D-derived cells, as evidenced by morphological differentiation. Conversely, the overexpression of Id2 protein in 32D IGF-IR cells inhibits the activation of the differentiation program (MPO RNA levels). However, for transformation (growth in IL-3-free medium), Id2 up-regulation is not sufficient. 32D IGF-IR cells stably overexpressing Id2 undergo apoptosis after IL-3 withdrawal, even when supplemented with IGF-I. Florio et al. (23) have reported that Id2 overexpression accelerates apoptosis in 32D cells. Although our 32D IGF-IR/Id2 cells do not die as quickly (probably because of the protective effect of the IGF-IR), they certainly are not IL-3 independent. Therefore, Id2 down-regulation seems to be important for differentiation but Id2 up-regulation is not sufficient for transformation. These findings clearly dissociate the effects of Id2 on differentiation and transformation.
More uncertain is the identification of the pathways originating from Y950 of the IGF-IR in the absence of IRS-1. Especially puzzling, as already mentioned, are the apparently contradictory signals originating from Y950. The Y950 residue is the main binding site for Shc proteins (13, 58), but it also binds IRS-1 (13, 58) and the Crk proteins (35), all of which could send positive signals. We can rule out IRS-1 in our case, since it is not expressed in 32D cells, but we cannot rule out the Crk proteins (35). The fact that Shc phosphorylation is impaired in cells expressing the Y950 mutant indicates a role of Shc proteins, without excluding a possible participation of other transducing molecules. Interestingly, both PD98059 (an inhibitor of MEK) and LY294002 (an inhibitor of P13K) suppress Id2 activation in 32D IGF-IR/DNStat3 cells. Apparently, both pathways are involved in the up-regulation of Id2 gene expression. It is not the first time that both pathways have been found to be required in 32D cells for transformation. Neither overexpression of Ras nor overexpression of IRS-1 can transform 32D cells, but the two combined cause malignant transformation with tumor formation in mice (14). While the MAPK and PI3K pathways seem to be involved in the up-regulation of Id2 gene expression (Fig. 4), the IGF-IR must also send a signal to repress Id2 gene expression. It is reasonable to connect the inhibitory signal again to Y950 and its ability to activate Stat3. The IGF-IR is known to activate Stat3 by phosphorylation of Stat3 Y705 (68, 69), and this was confirmed to occur in our cells. A mutation at Y950 of the IGF-IR inhibits phosphorylation of Stat3 at Y705 and abrogates the ability of the IGF-IR to up-regulate Id2 gene expression in 32D IGF-IR/DNStat3 cells. The fact that a dominant negative mutant of Stat3 markedly increases IGF-IR-mediated activation of Id2 gene expression clearly indicates that the inhibitory signal goes through Stat3. Possible candidates for this inhibition are SOCS-1 or SOCS-3, both of which have been related to IGF-IR signaling and Stat3 (68). SOCS proteins bind the insulin receptor at Y960, which is the homologue of Y950 of the IGF-IR (21). At this point, it would be too speculative to discuss the respective merits of Shc, Crk, SOCS, PI3K, and MAPK in activating the two programs.
The results with Id1 RNA expression are much less dramatic. More significantly, the Y950F mutation has no effect on Id1 gene expression. It confirms the importance of Y950 in Id2 gene expression. This discrepancy on the effect of IGF-I on Id1 and Id2 expression is not unique (32, 57), especially in hematopoietic cells (12). Another slight discrepancy occurs with the results of Ishiguro et al.(31), who correlated the levels of Id2 with differentiation of myeloid cells. The discrepancy may be more apparent than real, since Id2 RNA levels do increase in differentiating 32D IGF-IR cells. The difference is that this increase is much more pronounced when differentiation is inhibited by DN Stat3. We did not address in this paper the link between the IGF-IR and Stat3, apparently through the JAK proteins, which has already been considered by other investigators (27, 67, 68).
In conclusion, the present results show that Id2 gene expression is regulated by the activated IGF-IR. In the absence of IRS-1, Y950 of the IGF-IR sends contradictory signals, one for stimulation and one for repression of Id2 gene expression. The negative effect on Id2 expression signals through Stat3 and correlates with differentiation of cells. The positive signal uses both the PI3K and MAPK pathways and correlates with proliferation. When the Stat3 signal is inhibited by a dominant negative mutant of Stat3, only the proliferative stimulus remains, Id2 RNA levels are very high, and the cells become permanently IL-3 independent. However, Id2 up-regulation is not sufficient to confer IL-3 independence to 32D cells. The implications of these results are important. Evidence is accumulating that Id1 plays a role in the growth of breast cancer cells (16, 17, 39) while Id2 binds the retinoblastoma protein (30) and is activated by the proto-oncogene N-myc in neuroblastoma cells (38). Although our results do not directly correlate Id2 gene expression with transformation, given the importance of the IGF-IR in the establishment and maintenance of the transformed phenotype (3, 4), the link between the IGF-IR and the Id genes could offer new insights into the processes of differentiation and malignant transformation.
ACKNOWLEDGMENTS
This work is supported by grants CA 56309 and CA 78890 from the National Institutes of Health.
REFERENCES
- 1.Alani R M, Hasskari J, Grace M, Hernandez M C, Israel M A, Munger K. Immortalization of primary human keratinocytes by the helix-loop-helix protein. Id1. Proc Natl Acad Sci USA. 1999;96:9637–9641. doi: 10.1073/pnas.96.17.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barone M V, Pepperkok R, Peverali F A, Philipson L. Id proteins control growth induction in mammalian cells. Proc Natl Acad Sci USA. 1994;91:4985–4988. doi: 10.1073/pnas.91.11.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baserga R, Morrione A. Differentiation and malignant transformation: two roads diverged in a wood. J Cell Biochem. 1999;32/33:68–75. doi: 10.1002/(sici)1097-4644(1999)75:32+<68::aid-jcb9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 4.Baserga R, Prisco M, Hongo A. IGFs and cell growth. In: Rosenfeld R G, Roberts C T Jr, editors. The IGF system. Totowa. N.J: Humana Press; 1999. pp. 329–353. [Google Scholar]
- 5.Basu T, Warne P H, Downward J. Role of Shc in the activation of Ras in response to epidermal growth factor and nerve growth factor. Oncogene. 1994;9:3483–3491. [PubMed] [Google Scholar]
- 6.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 7.Blakesley V A, Butler A A, Koval A P, Okubo Y, LeRoith D. IGF-I receptor function: transducing the IGF-I signal into intracellular events. In: Rosenfeld R G, Roberts C T Jr, editors. The IGF system. Totowa, N.J: Humana Press; 1999. pp. 143–163. [Google Scholar]
- 8.Brown G, Choudhry M A, Durham J, Drayson M T, Michell R H. Monocytically differentiating HL60 cells proliferate rapidly before they mature. Exp Cell Res. 1999;253:511–518. doi: 10.1006/excr.1999.4660. [DOI] [PubMed] [Google Scholar]
- 9.Chaturvedi P, Sharma S, Reddy P. Abrogation of interleukin-3 dependence of myeloid cells by the v-src oncogene requires SH2 and SH3 domains which specify activation of STATs. Mol Cell Biol. 1997;17:3295–3304. doi: 10.1128/mcb.17.6.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christy B A, Saunders L K, Lau L F, Copeland N G, Jenkins N A, Mathans D. An Id-related helix-loop-helix protein encoded by a growth factor inducible gene. Proc Natl Acad Sci USA. 1991;88:1815–1819. doi: 10.1073/pnas.88.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper C L, Newburger P E. Differential expression of Id genes in multipotent myeloid progenitor cells: Id1 is induced by early- and late-acting cytokines while Id2 is selectively induced by cytokines that drive terminal granulocytic differentiation. J Cell Biochem. 1998;71:277–285. doi: 10.1002/(sici)1097-4644(19981101)71:2<277::aid-jcb12>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 12.Cooper C L, Brady G, Bilia F, Iscove N N, Quesenberry P J. Expression of the Id family helix-loop-helix regulators during growth and development in the hematopoietic system. Blood. 1997;89:3155–3165. [PubMed] [Google Scholar]
- 13.Craparo A, O'Neill T J, Gustafson T A. Non-SH2 domains within insulin receptor substrate-1 and SHC mediate their phosphotyrosine-dependent interaction with the NPEY motif of the insulin-like growth factor-I receptor. J Biol Chem. 1995;270:15639–15643. doi: 10.1074/jbc.270.26.15639. [DOI] [PubMed] [Google Scholar]
- 14.Cristofanelli B, Valentinis B, Soddu S, Rizzo M G, Marchetti A, Bossi G, Morena A R, Dews M, Baserga R, Sacchi A. Co-operative transformation of 32D cells by the combined expression of IRS-1 and v-Ha-Ras. Oncogene. 2000;19:3245–3255. doi: 10.1038/sj.onc.1203664. [DOI] [PubMed] [Google Scholar]
- 15.De Koning J P, Soede-Bobok A A, Ward A C, Schelen A M, Antonissen C, van Leeuwen D, Lowenberg B, Touw I P. Stat-3-mediated differentiation and survival of myeloid cells in response to granulocyte colony-stimulating factor: role for the cyclin-dependent kinase inhibitor p27Kip1. Oncogene. 2000;19:3290–3298. doi: 10.1038/sj.onc.1203627. [DOI] [PubMed] [Google Scholar]
- 16.Desprez P Y, Hara E, Bissell M J, Campisi J. Suppression of mammary epithelial cell differentiation by the helix-loop-helix protein Id-1. Mol Cell Biol. 1995;15:3398–3404. doi: 10.1128/mcb.15.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desprez P Y, Lin C Q, Thomasset N, Simpson C J, Bissell M J, Campisi J. A novel pathway for mammary epithelial cell invasion by the helix-loop-helix protein Id-1. Mol Cell Biol. 1998;18:4577–4588. doi: 10.1128/mcb.18.8.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dews M, Prisco M, Peruzzi F, Romano G, Morrione A, Baserga R. Domains of the IGF-I receptor required for the activation of extracellular signal-regulated kinases. Endocrinology. 2000;141:1289–1300. doi: 10.1210/endo.141.4.7414. [DOI] [PubMed] [Google Scholar]
- 19.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 20.Dufner A, Thomas G. Ribosomal S6 kinase signaling and the control of translation. Exp Cell Res. 1999;253:100–109. doi: 10.1006/excr.1999.4683. [DOI] [PubMed] [Google Scholar]
- 21.Emanuelli B, Peraldi P, Filloux C, Sawka-Verhelle D, Hilton D, van Obberghen E. SOCS-3 is an insulin-induced negative regulator of insulin signaling. J Biol Chem. 2000;275:15985–15991. doi: 10.1074/jbc.275.21.15985. [DOI] [PubMed] [Google Scholar]
- 22.English J, Pearson G, Wilsbacher J, Swantek J, Karandikar M, Xu S, Cobb M H. New insights into the control of MAP kinase pathways. Exp Cell Res. 1999;253:255–270. doi: 10.1006/excr.1999.4687. [DOI] [PubMed] [Google Scholar]
- 23.Florio M, Hernandez M C, Yang H, Shu H K, Cleveland J L, Israel M A. Id2 promotes apoptosis by a novel mechanism independent of dimerization to basic helix-loop-helix factors. Mol Cell Biol. 1998;18:5435–5444. doi: 10.1128/mcb.18.9.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia R, Yu C L, Hudnall A, Catlett R, Nelson K L, Smithgall T, Fujita D J, Ethier S P, Jove R. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differentiation. 1997;8:1267–1276. [PubMed] [Google Scholar]
- 25.Garrington T P, Johnson G L. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 26.Gu H, Maeda H, Moon J J, Lord J D, Yoakim M, Nelson B H, Neel B G. New role for Shc in activation of the phosphatidylinositol 3-kinase/Akt pathway. Mol Cell Biol. 2000;20:7109–7120. doi: 10.1128/mcb.20.19.7109-7120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gual P, Baron V, Lequoy V, van Obberghen E. Interaction of Janus kinases JAK-1 and JAK-2 with the insulin receptor and the insulin-like growth factor 1 receptor. Endocrinology. 1998;139:884–893. doi: 10.1210/endo.139.3.5829. [DOI] [PubMed] [Google Scholar]
- 28.Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. Id related genes encoding HLH proteins are required for G1 progression and are repressed in senescent human fibroblasts. J Biol Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]
- 29.Hawley R G, Lieu F H, Fong A Z, Hawley T S. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 30.Iavarone A, Garg P, Lasorella A, Hsu J, Israel M A. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 31.Ishiguro A, Spirin K S, Shioara M, Tobler A, Gombart A F, Israel M A, Norton J D, Koeffler H P. Id2 expression increases with differentiation of human myeloid cells. Blood. 1996;87:5225–5231. [PubMed] [Google Scholar]
- 32.Jen Y, Manova K, Benezra R. Expression patterns of Id1, Id2, and Id3 are highly related but distinct from that of Id4 during mouse embryogenesis. Dev Dyn. 1996;207:235–252. doi: 10.1002/(SICI)1097-0177(199611)207:3<235::AID-AJA1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 33.Kennedy S G, Wagner A J, Conzen S D, Jordan J, Bellacosa A, Tsichlis P N, Hay N. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 34.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne P H, Downward J. Matrix adhesion and ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/AAkt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koval A P, Blakesley V A, Roberts C T, Jr, Zick V, LeRoith D. Interaction in vitro of the product of the c-Crk-II proto-oncogene with the insulin-like growth factor 1 receptor. Biochem J. 1998;330:923–932. doi: 10.1042/bj3300923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreider B L, Benezra R, Rovera G, Kadesch T. Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science. 1992;255:1700–1702. doi: 10.1126/science.1372755. [DOI] [PubMed] [Google Scholar]
- 37.Kulik G, Weber M J. Akt-dependent and -independent survival signaling pathways utilized by insulin-like growth factor 1. Mol Cell Biol. 1998;18:6711–6718. doi: 10.1128/mcb.18.11.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasorella A, Noseda M, Beyna M, Iavarone A. Id2 is a retinoblasoma protein target and mediates signaling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 39.Lin C Q, Singh J, Murata K, Itahana Y, Parrinello S, Liang S H, Gillett C E, Campisi J, Desprez P Y. A role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res. 2000;60:1332–1340. [PubMed] [Google Scholar]
- 40.Lister J, Forrester W C, Baron M H. Inhibition of an erythroid differentiation switch by the helix-loop-helix protein Id1. J Biol Chem. 1995;270:17939–17946. doi: 10.1074/jbc.270.30.17939. [DOI] [PubMed] [Google Scholar]
- 41.Lyden D, Young A Z, Zagzag D, Yan W, O'Reilly R, Bader B L, Hynes R O, Zhuang Y, Manova K, Benezra R. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 42.Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto D, Akira S. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. A central role of Stat3 on IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 44.Navarro M, Baserga R. Limited redundancy of survival signals from the type 1 insulin-like growth factor receptor. Endocrinology. 2001;142:1073–1081. doi: 10.1210/endo.142.3.7991. [DOI] [PubMed] [Google Scholar]
- 45.Nickoloff B J, Chaturvedi V, Bacon P, Qin J Z, Denning M F, Diaz M O. Id-1 delays senescence but does not immortalize keratinocytes. J Biol Chem. 2000;275:27501–27504. doi: 10.1074/jbc.C000311200. [DOI] [PubMed] [Google Scholar]
- 46.Norton J D, Deed R W, Craggs G, Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- 47.Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui A L F, Kitamura T. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 1999;18:4754–4765. doi: 10.1093/emboj/18.17.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peruzzi F, Prisco M, Dews M, Salomoni P, Grassilli E, Romano G, Calabretta B, Baserga R. Multiple signaling pathways of the IGF-I receptor in protection from apoptosis. Mol Cell Biol. 1999;19:7203–7215. doi: 10.1128/mcb.19.10.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prisco M, Romano G, Peruzzi F, Valentinis B, Baserga R. Insulin and IGF-I receptor signaling in protection from apoptosis. Horm Metab Res. 1999;31:80–89. doi: 10.1055/s-2007-978703. [DOI] [PubMed] [Google Scholar]
- 50.Raz R, Lee C K, Cannizzaro L A, D'Eustachio P, Levy D E. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci USA. 1999;96:2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez-Tarduchy G, Collins M K L, Garcia I, Lopez-Rivas A. Insulin-like growth factor I inhibits apoptosis in IL-3 dependent hemopoietic cells. J Immunol. 1992;149:535–540. [PubMed] [Google Scholar]
- 52.Sasaoka T, Draznin B, Leitner J W, Langlois W J, Olefsky J M. Shc is the predominant signaling molecule coupling insulin receptors to activation of guanine nucleotide releasing factor and p21ras-GTP formation. J Biol Chem. 1994;269:10734–10738. [PubMed] [Google Scholar]
- 53.Scher C D, Shephard R C, Antoniades H N, Stiles C D. Platelet derived growth factor and the regulation of the mammalian fibroblast cell cycle. Biochim Biophys Acta. 1979;560:217–241. doi: 10.1016/0304-419x(79)90020-9. [DOI] [PubMed] [Google Scholar]
- 54.Shimozaki K, Nakajima K, Hirano T, Nagata S. Involvement of STAT3 in the granulocyte colony-stimulating factor-induced differentiation of myeloid cells. J Biol Chem. 1997;272:25184–25189. doi: 10.1074/jbc.272.40.25184. [DOI] [PubMed] [Google Scholar]
- 55.Soon L, Flechner L, Gutkind J S, Wang L H, Baserga R, Pierce J H, Li W. Insulin-like growth factor 1 synergizes with interleukin 4 for hematopoietic cell proliferation independent of insulin receptor substrate expression. Mol Cell Biol. 1999;19:3816–3828. doi: 10.1128/mcb.19.5.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinman R A, Iro A. Suppression of G-CSF-mediated Stat signalling by IL-3. Leukemia. 1999;13:54–61. doi: 10.1038/sj.leu.2401253. [DOI] [PubMed] [Google Scholar]
- 57.Sun X H, Copeland N G, Jenkins N A, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tartare-Deckert S, Sawka-Verhelle D, Murdaca J, Obberghen van E. Evidence for a differential interaction of SHC and the insulin receptor substrate-1 (IRS-1) with the insulin-like growth factor-I (IGF-I) receptor in the yeast two-hybrid system. J Biol Chem. 1996;271:23456–23460. doi: 10.1074/jbc.270.40.23456. [DOI] [PubMed] [Google Scholar]
- 59.Valentinis B, Navarro M, Zanocco-Marani T, Edmonds P, McCormick J, Morrione A, Sacchi A, Romano G, Reiss K, Baserga R. Insulin receptor substrate-1, p70S6K and cell size in transformation and differentiation of hemopoietic cells. J Biol Chem. 2000;275:25451–25459. doi: 10.1074/jbc.M002271200. [DOI] [PubMed] [Google Scholar]
- 60.Valentinis B, Romano G, Peruzzi F, Morrione A, Prisco M, Soddu S, Cristofanelli B, Sacchi A, Baserga R. Growth and differentiation signals by the insulin-like growth factor 1 receptor in hemopoietic cells are mediated through different pathways. J Biol Chem. 1999;274:12423–12430. doi: 10.1074/jbc.274.18.12423. [DOI] [PubMed] [Google Scholar]
- 61.Valtieri M, Tweardy D J, Caracciolo D, Johnson K, Mavilio F, Altmann S, Santoli D, Rovera G. Cytokine dependent granulocytic differentiation. J Immunol. 1987;138:3829–3835. [PubMed] [Google Scholar]
- 62.Wang L M, Myers M G, Jr, Sun X J, Aaronson S A, White M, Pierce J H. IRS-1: essential for insulin- and IL-4-stimulated mitogenesis in hemopoietic cells. Science. 1993;261:1591–1594. doi: 10.1126/science.8372354. [DOI] [PubMed] [Google Scholar]
- 63.Ward A C, Smith L, de Koning J P, van Aesch Y, Touw I P. Multiple signals mediate proliferation, differentiation and survival from the granulocyte-colony stimulating factor receptor in myeloid 32D cells. J Biol Chem. 1999;274:14956–14962. doi: 10.1074/jbc.274.21.14956. [DOI] [PubMed] [Google Scholar]
- 64.White M F. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol Cell Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
- 65.Wu Y Y, Bradshaw R A. Activation of Stat3 signaling pathway is required for differentiation by interleukin-6 in OC12–E2 cells. J Biol Chem. 2000;275:2147–2156. doi: 10.1074/jbc.275.3.2147. [DOI] [PubMed] [Google Scholar]
- 66.Yenush L, Zanella C, Uchida T, Bernal D, White M F. The pleckstrin homology and phosphotyrosine binding domains of insulin receptor substrate 1 mediate inhibition of apoptosis by insulin. Mol Cell Biol. 1998;18:6784–6794. doi: 10.1128/mcb.18.11.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang V, Turkson J, Carter-Su C, Smithgall T, Levitzki A, Kraker A, Krolewski J J, Medveczky P, Jove R. Activation of Stat3 in v-src transformed fibroblasts requires cooperation of Jak1 kinase activity. J Biol Chem. 2000;275:24935–24944. doi: 10.1074/jbc.M002383200. [DOI] [PubMed] [Google Scholar]
- 68.Zong C S, Chan J, Levy D E, Horvath C, Sadowski H B, Wang L H. Mechanism of STAT3 activation by insulin-like growth factor I receptor. J Biol Chem. 2000;275:15099–15105. doi: 10.1074/jbc.M000089200. [DOI] [PubMed] [Google Scholar]
- 69.Zong C S, Zong L, Jiang Y, Sadowski H B, Wang L H. Stat3 plays an important role in oncogenic ros- and insulin-like growth factor 1 receptor-induced anchorage-independent growth. J Biol Chem. 1998;273:28065–28072. doi: 10.1074/jbc.273.43.28065. [DOI] [PubMed] [Google Scholar]