ABSTRACT
Macroautophagy/autophagy is special because the double-layer lipid-formed autophagosome is formed by de novo generation. Phosphatidylinositol-3-phosphate (PtdIns3P) produced by class III phosphatidylinositol 3-kinase complex I (PtdIns3K-CI) is an essential source lipid for the formation of autophagosomes. However, how autophagy is initiated is unknown. In other words, the mechanism by which PtdIns3K-CI is recruited to the phagophore assembly site (PAS) to initiate autophagosome formation is unclear. We recently uncovered the pivotal role of yeast Vac8 in autophagy initiation through the recruitment of PtdIns3K-CI to the PAS. N-terminal palmitoylation of Vac8 anchors it to the vacuole membrane, and the middle ARM domains bind PtdIns3K-CI, leading to the generation of PtdIns3P at the PAS and subsequent autophagosome formation. We found that mouse ARMC3 is the homolog of yeast Vac8 and that its autophagic roles are conserved. Interestingly, spermatids from mice with Armc3 deletion showed blocked ribophagy, low energy levels of mitochondria and motionless flagella, which caused male infertility. These findings revealed a germ tissue-specific autophagic function of ARMC3 in complex eukaryotic species.
KEYWORDS: ARM, ARMC3, flagella, mitochondria, palmitoylation, PtdIns3K-CI, PtdIns3P, ribosome, sperm, Vac8
Main text
Lipid-formed organelles are essential for the compartmentalization of intracellular materials in eukaryotes. In addition to existing organelles, such as the endoplasmic reticulum (ER), Golgi, and mitochondria, autophagosomes are de novo-generated double-layered lipid vesicles that can transport sequestered cytosolic materials into vacuoles (in yeast and plants) or lysosomes (in mammalian cells) for eventual degradation. New organelles, such as the ER and mitochondria, are formed based on the original source organelles. Autophagosomes, however, are formed at certain locations called phagophore assembly sites (PASs) using source lipids from the ER, Golgi, plasma membrane, and mitochondria. This unique de novo generation of autophagosomes emphasizes the spatial and temporal regulation of autophagosome formation initiation.
Very early autophagic stages involve class III phosphatidylinositol 3-kinase complex I (PtdIns3K-CI), which is composed of the lipid kinase PIK3C3/Vps34, the regulatory kinase PIK3R4/Vps15, the regulatory subunits BECN1/Vps30 and ATG14. The function of the PtdIns3K-CI complex is to generate phosphatidylinositol-3-phosphate (PtdIns3P), whose lipids are essential for autophagosome formation. Thus, targeting PtdIns3K-CI to the phagophore assembly site (PAS) is essential for its activity in autophagosome formation.
We recently uncovered a conserved role of the autophagic factor Vac8 in recruiting PtdIns3K-CI to vacuole membranes where autophagosome formation is initiated [1]. In yeast, Vac8 was shown to be involved in autophagy by Dr. Daniel Klionsky’s laboratory, but the mechanism by which it affects autophagy is unclear. Recently, we found that Vac8 was a candidate essential autophagic factor in a genome-wide screen in yeast, and our immunoprecipitation-mass spectrometry results showed that Vac8 could bind the four subunits of the PtdIns3K-CI complex. We first confirmed the essential role of Vac8 in autophagy, and the results showed that vac8 deletion blocks autophagy. Based on the observation that no autophagosomes are formed in Vac8-deficient yeast cells, we hypothesized that Vac8 regulates upstream autophagosome formation instead of autophagosome fusion with vacuoles.
What is the mechanism by which Vac8 regulates autophagosome formation? As we mentioned above, Vac8 was thought to bind the PtdIns3K-CI complex, and this binding was confirmed. Further analysis showed that the middle ARM domains of Vac8 and the Vps34 subunit of the PtdIns3K-CI complex mediate the Vac8-PtdIns3K-CI interaction. The binding of PtdIns3K-CI was thought to be necessary for the function of Vac8 in autophagy because the Vac8 mutant with deleted ARM domains loses its effect on autophagy.
Another clue suggesting the role of Vac8 in autophagy is that Vac8 has been shown to be N-terminally palmitoylated at cystines 4, 5, and 7. Therefore, we examined whether N-terminal palmitoylation of Vac8 anchors it at the vacuole membrane and whether Vac8 then binds and recruits PtdIns3K-CI. In wild-type (WT) yeast cells, the four subunits of PtdIns3K-CI are mainly located at the vacuole membrane. Yeast cells expressing Vac8 with palmitoylation site mutations or with ARM deletion exhibit disturbed localization of PtdIns3K-CI. Furthermore, the lipid product of PtdIns3K-CI, PtdIns3P, is also abnormally distributed in these Vac8-mutated yeast cells.
The function of yeast Vac8 in autophagy was suggested to be conserved, as its mammalian homolog ARMC3 is pivotal for autophagosome formation and binds to mammalian PtdIns3K-CI in mouse testes, which is consistent with the specific testicular expression pattern of ARMC3. Proteomics analysis of spermatids from WT and ARMC3-deficient mice show that ribosomal proteins accumulate in the absence of ARMC3, which is consistent with previous findings showing that ribosomes are cargoes for autophagic degradation. Further analysis showed that ATP levels, mitochondrial activity, and sperm motility are also reduced in ARMC3-deficient spermatids. Thus, we proposed a model showing that palmitoylated Vac8 recruits PtdIns3K-CI to the PAS, leading to the generation of PtdIns3P and subsequent autophagosome formation (Figure 1A).
Figure 1.
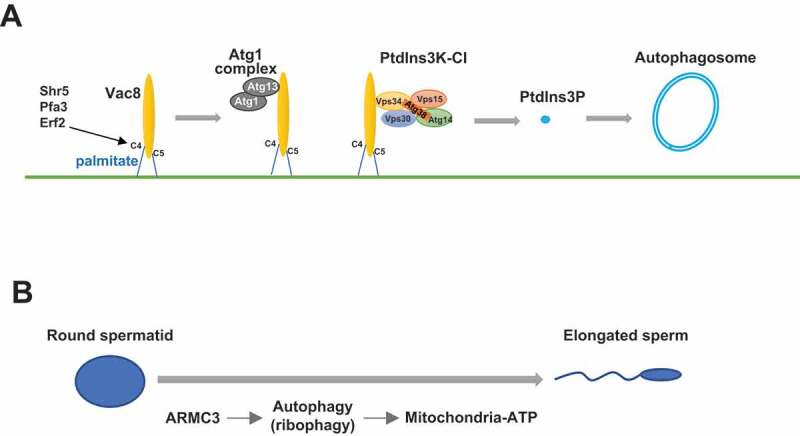
Illustration of Vac8 functions in autophagy and ARMC3-mediated ribophagy in spermiogenesis. (A) The N-terminal cysteines of Vac8 are palmitoylated, which enables Vac8 localization to the vacuole membrane. Vac8 then recruits the Atg1 complex and the PtdIns3K-CI complex to the vacuole membrane, which leads to the generation of PtdIns3P and initiates autophagosome formation. (B) During spermatogenesis, ARMC3 is specifically expressed and promotes the autophagic degradation of excessive ribosomes, providing energy resources for mitochondria and thus ensuring sperm flagellar motility
In addition, our study uncovered an interesting function of autophagy in spermiogenesis (Figure 1B). After two rounds of meiosis, diploid spermatogonia produce four round-shaped haploid spermatids that undergo a series of morphological and functional changes to form elongated mature spermatozoa. During this spermiogenesis process, excessive cytoplasmic organelles are thought to be removed and discarded by the formation of residual bodies. However, our study showed that there is ribosome recycling (by autophagic degradation), in addition to simple discarding during spermiogenesis. This finding is highly consistent with the functional nature of autophagy, which is the recycling of intracellular materials through degradation. Furthermore, this recycling provides energy for mitochondria in sperm to ensure motility and thus is more economic than simply discarding material.
In complex eukaryotic species, the activity levels of autophagy are different in various organs/tissues. It is thought that there are organ/tissue-specific regulators of autophagy that maintain certain levels of autophagy and either upregulate or downregulate autophagy according to spatial and temporal conditions. Identifying such organ/tissue-specific regulators of autophagy might help researchers understand the multiple functions of autophagy in complex multicellular eukaryotes, which can provide insights into the physiological roles of autophagy.
There are some questions that remain unanswered in our study. Because the Atg1 complex is recruited to the PAS by the Atg13-Vac8 interaction, how Vac8 coordinates the recruitment of the Atg1 complex and PtdIns3K-CI is unclear. It is thought that Atg13 and Vps34 bind to different ARM domains in Vac8; alternatively, one Vac8 protein binds Atg13, while the other Vac8 protein binds Vps34, as Vac8 is a dimer. ARMC3 is specifically expressed in the testis, and this expression is also specifically triggered when spermatogenesis begins. Thus, it would be interesting to clarify the temporal control of ARMC3 expression. Furthermore, considering that large autophagic cargoes such as organelles are usually targeted to phagophores by selective receptors, it would also be interesting to discover whether or which receptor can mediate the specific bundling of excessive ribosomes within autophagosomes during spermiogenesis.
Funding Statement
This work was supported by the National Key R&D Program of China [2018YFC1003603]; National Natural Science Foundation [81902997]; National Natural Science Foundation [31970693]; National Key R&D Program of China [2017YFA0506300].
Disclosure statement
All authors have no conflicts of interest to declare.
Reference
- [1].Lei Y, Zhang X, Xu Q, et al. Autophagic elimination of ribosomes during spermiogenesis provides energy for flagellar motility. Dev Cell. 2021. Aug 23;56(16):2313–2328.e7. DOI: 10.1016/j.devcel.2021.07.015. PMID: 34428398. [DOI] [PubMed] [Google Scholar]


