Abstract
Chemical carcinogenesis in mouse skin has been useful in delineating the molecular events that underlie squamous cell carcinoma progression. A late event in this progression, the epithelial-to-mesenchymal transition (EMT), is characterized by the loss of epithelial markers and the presence of mesenchymal markers. One mesenchymal marker associated with this transition is the matrix metalloproteinase stromelysin 1 (Str-1). To examine the molecular mechanisms regulating the expression of Str-1 during the EMT, genetically related mouse skin tumor cell lines representing the epithelial (B9SQ) and mesenchymal (A5SP) phenotypes were studied. As expected, B9SQ cells did not make Str-1, while A5SP cells did. B9SQ-A5SP somatic hybrids did not make Str-1, suggesting that a critical regulatory factor was a B9SQ-specific repressor. Str-1 promoter analysis revealed that a canonical AP-1 site was sufficient to maintain differential reporter gene activity. This result correlated with the observed loss of binding of the transcriptionally inactive JunB–Fra-2 AP-1 complex from B9SQ cells, being replaced primarily by the more active JunD–Fra-2 complex in A5SP cells. The higher level of JunB binding to both DNA and Fra-2 correlated with its hyperphosphorylation by Jun N-terminal kinase, an activity that was significantly higher in B9SQ cells. In the somatic hybrids, JunB gene expression was highly upregulated, a condition that also was sufficient to repress the expression of the endogenous Str-1 gene in A5SP cells. These data suggested that alterations in JunB activity, by changes in either phosphorylation or gene expression, contributed to the phenotypic differences that occur in this model of the EMT.
Chemical carcinogenesis of mouse skin has been investigated for decades and has contributed greatly to the understanding of tumor initiation and progression in general. Typically, this model involves treatment with the carcinogen 7,12-dimethylbenz[a]anthracene (DMBA), which acts as a tumor initiator, causing an activating mutation in the proto-oncogene H-ras (6). Treatment with a tumor promoter, such as 12-O-tetradecanoylphorbol-13-acetate (TPA), stimulates the growth of the initiated cells, resulting in benign papilloma formation. With continued application of the tumor promoter, the papilloma converts to a malignant squamous cell carcinoma, gaining the ability to invade surrounding connective tissue. During these stages, the chromosome carrying mutant H-ras is duplicated, the normal H-ras allele is lost, and p53 is mutated (5). Finally, in a process known as the epithelial-to-mesenchymal transition (EMT), the tumor cells undergo a fundamental loss of differentiation, changing to a metastatic spindle cell carcinoma. Besides the loss of expression of epithelial markers, such as E-cadherin and desmoplakins, little is known about the molecular events that control the EMT in skin tumor progression. In some tumor systems, the EMT is associated with the activity of growth factors, such as hepatocyte growth factor (HGF) (3) and transforming growth factor β (TGF-β) (9), as well as the activity of transcription factors, such as slug (43) and snail (2, 8).
The matrix metalloproteinases (MMPs) have been strongly implicated in all stages of tumor progression (10). Exogenous expression of MMPs in cell lines and in transgenic mice results in enhancement of tumor growth, invasion, and metastasis. For example, mice overexpressing human collagenase 1 in the skin experienced an increased papilloma incidence following DMBA-TPA treatment (14). During skin tumor progression, MMPs are initially expressed by connective tissue cells within and surrounding the tumor in a manner likened to a wound response (29). Coincident with the EMT, however, tumor cells begin expressing the stromal MMP stromelysin 1 (Str-1; MMP-3; EC 3.4.24.17) (49), potentially endowing them with the ability to invade and metastasize (28).
The expression of Str-1 by a majority of spindle cell tumors demonstrates that Str-1 is a mesenchymal marker for the EMT in skin. To better understand the molecular mechanisms underlying this step in tumor progression, we used a pair of genetically related skin tumor cell lines that represent the squamous and spindle cell stages of squamous cell tumor progression. B9SQ and A5SP are independent clones of the mouse skin tumor line MSC11, which originated from DMBA-TPA treatment of mouse skin (7). Although they arose from the same parent, as confirmed by the presence of identical allelic H-ras and p53 mutations (7), they differ significantly in morphology and behavior. B9SQ cells have a squamous cell morphology and express common epithelial markers. A5SP cells are spindle shaped, have lost epithelial markers, such as E-cadherin, and express mesenchyme-associated markers, such as vimentin (45). Both lines are tumorigenic and locally invasive; however, only A5SP successfully metastasizes (45). Finally, while B9SQ cells do not express Str-1, A5SP cells do (49). We therefore used the expression of Str-1 in this system as a tool to identify molecular alterations associated with the EMT. Our findings suggest that the loss of JunB activity is a major contributor to the onset of Str-1 expression during the EMT in this model system.
MATERIALS AND METHODS
Cell culturing and transfection.
B9SQ and A5SP cells and B9SQ-A5SP somatic hybrid cell clones were gifts from Allan Balmain (University of California, San Francisco). Both cell lines were grown in KGM or KGM-2 (Clonetics) without hydrocortisone but with 1% dialyzed calf serum (Gibco-BRL) at 37°C in 5% CO2.
Transient transfections were performed with N-[1-(2,3-dioleoyloxy)]-N,N,N-trimethylammonium propane methylsulfate (DOTAP) (Boehringer-Mannheim Biochemicals) according to the manufacturer's instructions. For stable transfection of pBABE-JunB into A5SP cells, TransFast (Promega) was used. Cells were selected in 200 μg of G418 (Gibco-BRL)/ml and cloned by ring cloning.
Str-1 promoter assays.
For chloramphenicol acetyltransferase (CAT) assays, 5 × 105 cells were plated on 60-mm dishes. One microgram of −2100TRCAT, −754TRCAT, −208TRCAT, and −116TRCAT (19), −208mAP-1, or 5XAP-1-CAT (a gift from Ronald Wisdom, Vanderbilt University) was cotransfected with 1 μg of a CMV–β-galactosidase plasmid (Promega) using DOTAP. At 48 h after transfection, cells were harvested by scraping, lysed by repeated freezing-thawing, and centrifuged to remove debris. Equal amounts of transfected extracts were determined for each sample by normalization to β-galactosidase activity and were incubated for 1 h at 37°C in 150-μl reaction mixtures containing 0.25 M Tris-HCl (pH 7.5), 0.5 mM acetyl coenzyme A (acetyl-CoA), and 2.7 μCi of [14C]chloramphenicol. Reactions were terminated by ethyl acetate extraction, and mixtures were spotted onto silica plates for thin-layer chromatography in methanol-chloroform. Samples on plates were visualized and quantitated with a PhosphorImager (Molecular Dynamics).
For luciferase assays, a transfection mixture of 1 μg of the 754TR-Luc construct and 100 ng of the Renilla luciferase plasmid (RL-tk) were cotransfected with 1, 2, or 3 μg of pCDNA3-JNK (18). Empty pCDNA3 vector (Invitrogen) was used to bring the total level of the cotransfected expression vector to 3 μg in each transfection mixture. TransFast reagent (Promega) was used to form transfection complexes, and each transfection mixture was divided into 3 wells of a 24-well plate, each well containing 5 × 105 A5SP cells. At 24 h after transfection, reporter activity was measured with a Dual Luciferase kit (Promega). Values were normalized to Renilla and plotted as a percentage relative to transfection with only pCDNA3.
Northern blotting.
Total RNA was harvested, and 10 μg of RNA was subjected to Northern blotting as previously described (49). The probes used were as follows. The Str-1 probe was an EcoRI isolate from pTRIIa, encoding a full-length mouse Str-1 cDNA (33). Human c-jun was an EcoRI fragment of c-jun cDNA (23). Full-length junB (42) and junD (41) cDNA plasmids were gifts from Daniel Nathans. Blot stripping was performed by incubating the previously hybridized blot in boiling 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer and allowing the solution to come to room temperature.
Cell extracts.
For electrophoretic mobility shift assays (EMSA), nuclear extracts were prepared by growing cells to confluence, washing them three times in cold phosphate-buffered saline (PBS), and harvesting them by scraping. Cells were precipitated in a microcentrifuge, rinsed with hypotonic buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl), and resuspended in two pellet volumes of hypotonic buffer. After 5 min on ice, the suspension was lysed by adding 1 μl of 5% Nonidet P-40 and agitated by pipetting through a Pasteur pipette. Plasma membrane lysis was monitored by trypan blue exclusion of a portion of the sample. The nuclear suspension was then layered onto 300 μl of a sucrose pad (60 mM KCl, 15 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, 15 mM HEPES [pH 7.9], 0.876 M sucrose) in an Eppendorf tube and spun at 16,110 × g in a microcentrifuge for 20 min at 4°C. The pellet was washed in hypotonic buffer, pulsed in a microcentrifuge for 5 s, and aspirated. Nuclei were then gently resuspended in a one-half packed nuclear volume of low-salt buffer (20 mM HEPES [pH 7.9], 25% glycerol, 1.5 mM MgCl2, 0.02 M KCl, 0.2 mM EDTA). A one-half packed nuclear volume of high-salt buffer (20 mM HEPES [pH 7.9], 25% glycerol, 1.5 mM MgCl2, 1.2 M KCl, 0.2 mM EDTA) was added dropwise to lyse the nuclei. Nuclei were then rocked for 30 min at 4°C to ensure complete lysis. Nuclear debris was then spun out by centrifugation at full speed in a microcentrifuge, and the supernatant was dialyzed against buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 100 mM KCl, 0.2 mM EDTA). Dithiothreitol (DTT) (0.5 mM), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg of leupeptin/ml, 1 μg of aprotinin/ml, 1 mM sodium vanadate, and 20 mM sodium fluoride were added to each solution just prior to use.
Nuclear extracts used in glutathione S-transferase (GST) pulldown assays (see Fig. 8B and C) were prepared as described by Schreiber et al. (44), except that 1 mM sodium vanadate, 20 mM sodium fluoride, 1 μg of leupeptin/ml, and 1 μg of aprotinin/ml were added.
FIG. 8.
The GST–Fra-2 interaction with JunB in nuclear extracts shows a preference for phosphorylated JunB. (A) GST–Fra-2 binds preferentially to phosphorylated JunB. JunB immunoblot of 5 μg of B9SQ cell nuclear extracts before and after pulldown with the GST–Fra-2 leucine zipper domain (LZD) attached to glutathione-agarose beads. (B) Fra-2 binds significantly more JunB from B9SQ nuclear extracts than from A5SP nuclear extracts. Shown are JunB immunoblots of B9SQ or A5SP nuclear extracts either before (in) and after (pd) pulldown with the Fra-2 LZD. The difference in the JunB–Fra-2 association was consistent with the difference in the amounts of the JunB phosphoforms. (C) Fra-2 pulldown samples from panel B were immunoblotted for JunD and c-Jun. No significant difference was noted in the Fra-2 LZD association for JunD or c-Jun.
Total extracts for the in vitro kinase assays and kinase antibody immunoblots were prepared by scraping cells into lysis buffer (0.5% Nonidet P-40, 50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM sodium vanadate, 20 mM sodium fluoride, 1 μg of leupeptin/ml, 1 μg of aprotinin/ml, 1 mM PMSF, 5 mM DTT) and clarifying them by centrifugation.
EMSA.
For EMSA, 5 μg of nuclear extract was incubated with 5 × 105 cpm of the end-labeled double-stranded oligonucleotide 5′-GGAAGCAATTATGAGTCAGTTTG-3′ (the AP-1 site is shown in bold) in buffer D for 45 min at 15°C followed by 30 min at room temperature. For supershift reactions, the optimal concentration of each antibody was determined by titration into the EMSA reaction mixture. All antibodies were added at the same concentration (12.5 ng/μl) to correspond to the optimal concentration of the least potent antibody that showed a discernible supershift. Antibodies were obtained from Santa Cruz Biotechnology, except for the c-Fos antibody (Upstate Biotechnology; 06-340). Samples were resolved on a nondenaturing 4% polyacrylamide–2% glycerol gel, dried, and visualized and quantitated with a PhosphorImager. Images shown are autoradiographs of the same gels.
Immunoprecipitation, phosphatase treatment, and immunoblotting.
Antibodies and working concentrations were as follows: for JunB, PC28 (Calbiochem) at 1 μg/ml; for JunD, sc-074X at 0.1 μg/ml; for c-Jun, sc-1694 at 0.7 μg/ml; for extracellular signal-regulated kinase 1 (ERK-1) or ERK-2, sc-154G at 1 μg/ml; for ERK-1/2-PTyr204 (active), sc-7383 at 1 μg/ml; for Jun N-terminal kinase 1 (JNK-1) (agarose conjugated), sc-571, which recognizes JNK-1, JNK-2, and JNK-3, at 0.4 μg/ml; for JNK-2, sc-7345, which recognizes JNK-1, JNK-2, and JNK-3, at 0.4 μg/ml; for JNK-PThr183/PTyr185, sc-6254 (Santa Cruz Biotechnology), which recognizes JNK-1, JNK-2, and JNK-3, at 0.2 μg/ml; for glycogen synthase kinase 3β (GSK-3β), G22320 (Transduction Laboratories) at 0.1 μg/ml; for GSK-3β-Ptyr216, 44-604 (Quality Controlled Biochemicals) at 1 μg/ml; for GSK-3α, 06-391 at 0.7 μg/ml; and for GSK-3α-PSer21, 06-733-MN (Upstate Biotechnology) at 2 μg/ml.
Immunoblotting was performed with 5 μg of nuclear extract (for Jun antibodies) or total extract (for GSK-3 antibodies). Samples were resolved in sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to Immobilon P (Millipore) or nitrocellulose (Micron Systems Inc.) membranes. The membranes were blocked in Tris-buffered saline–Tween 20 (TBST; 150 mM NaCl, 10 mM Tris-HCl [pH 8], 0.05% Tween 20) with 10% milk, incubated with the appropriate antibody in TBST with 1% milk overnight at 4°C, washed in TBST, incubated with secondary antibody for 1 h at room temperature, washed, and visualized by enhanced chemiluminescence. Band intensities were determined by densitometry.
Immunoprecipitation for JNK and active JNK was done as follows. Ten microliters of anti-JNK-1 beads was mixed with 500 μg of total extract in 1 ml of lysis buffer plus protease and phosphatase inhibitors at 4°C overnight. On the following day, beads were washed four times with lysis buffer plus inhibitors and boiled in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (50 mM Tris Hcl [pH 6.8], 5% 2-mercaptoethanol, 10% glycerol, 1% SDS) for loading onto a gel for SDS–10% PAGE. For phosphatase treatment, 5 μg of nuclear extract was incubated at 30°C for 1 h with 3 U of calf intestinal phosphatase (CIP; Promega) in 50 mM Tris-HCl (pH 7.4)–1 mM MgCl2. For phosphatase-kinase treatment, 10 μg of nuclear extract was incubated at 30°C for 30 min without (input) or with (shrimp alkaline phosphatase [SAP] and SAP-JNK samples) 1 U of SAP (Boehringer) in phosphatase buffer (50 mM Tris [pH 7.4], 1 mM MgCl2) and heat inactivated at 65°C for 10 min. To this mixture were added ATP to 0.25 mM 10× kinase buffer (Stratagene), and 0 (input and SAP samples), 0.25, or 0.75 μg of JNK (Stratagene); incubation was carried out for 30 min at 30°C.
Purification of GST fusion proteins.
GST-JunB and GST–ATF-2 plasmids were kind gifts from K. E. Paulson (Tufts University). GST plasmids were transformed into Escherichia coli BL21 and grown and induced with isopropyl-β-d-galactopyranoside (IPTG) following standard protocols (Pharmacia Biotech). Bacterial sonicates were bound to glutathione-agarose (Sigma) at room temperature for 30 min, washed, loaded onto columns, and eluted in batches with 20 mM reduced glutathione (Sigma).
Kinase assays.
The in vitro kinase assay was done with GST–c-Jun, recombinant rat JNK-1, and 10× reaction buffer from a c-Jun N-terminal kinase assay kit (Stratagene). Total cell extracts were prepared from confluent flasks of cells. One microgram of total cell extract was mixed with 2 μCi of [γ-32P]ATP and 1 μg of GST–c-Jun, 1 μg of GST–ATF-2, or 10 μg of GST-JunB in a 50-μl reaction mixture containing 1× reaction buffer and 1 mM sodium vanadate. When JNK was used in place of cell extract, 0.25 μg of enzyme was used with 1 μg of substrate. Mixtures were placed at 30°C for 30 min, and reactions were stopped with SDS-PAGE sample buffer. Reaction mixtures were resolved in SDS–10% polyacrylamide gels, washed in double-distilled H2O, dried, and autoradiographed. Quantitative results were obtained using a PhosphorImager. Values represent the mean for three different sets of extracts and were analyzed with the paired t test.
The in vivo kinase assay was done with a PathDetect c-Jun trans-Reporting system (Stratagene). Cells were plated in 12-well tissue culture plates, grown to 90% confluence, and transfected using TransFast at 3 μl/μg of DNA. One liposome-forming reaction was prepared for three wells of cells to obtain luciferase readings in triplicate. pFR-Luc is the reporter plasmid, containing GAL4 binding sites upstream of firefly luciferase. pFA2-cJun encodes a fusion protein of the amino terminus of c-Jun and the DNA binding domain of GAL4. pFC2-dbd encodes only the GAL4 DNA binding domain with no c-Jun sequences. RL-CMV (Promega) was cotransfected for normalization of transfection efficiency. One microgram of reporter plasmid, 75 ng of RL-CMV, and 50 ng of the effector plasmids were used in each reaction. Empty expression vector was used to maintain a constant total amount of DNA for each transfection. Cell media were changed 12 h after the addition of liposomes, and lysates were taken for analysis 12 h later. Lysates were analyzed for luciferase activity using a Dual Luciferase reporter assay system (Promega). Results represent the mean for four independent experiments.
GST–Fra-2 pulldown assay.
Human Fra-2 cDNA was obtained from Ronald Wisdom, and the sequence encoding amino acids 99 to 216 was subcloned by PCR and then cloned in frame with GST in plasmid pGEX-2T (Pharmacia) to yield GST–Fra-2(aa99–216). BL21 lysates containing GST–Fra-2(aa99–216) were divided into aliquots and frozen at −70°C. GST–Fra-2(aa99–216) lysates were incubated with glutathione beads for 1 h at room temperature with rocking. The beads were washed four times with PBS containing protease and phosphatase inhibitors and resuspended 1:1 (vol/vol) in PBS with inhibitors. For each sample, 15 μl of GST–Fra-2(aa99–216)-bound beads was diluted with 25 μl of 1:1 unbound beads to minimize nonspecific interactions with nuclear extract components. In a 100-μl total volume, 40 μg of nuclear extract was combined with the bead mixture and 20 μl of 5× binding buffer (50 mM HEPES [pH 7.4], 5 mM EDTA, 250 mM NaCl, 40% glycerol, 5 mM DTT) containing protease and phosphatase inhibitors. Reaction mixtures were rocked at room temperature for 2 h, and the beads were washed three times with lysis buffer. The beads were then resuspended in SDS-PAGE sample buffer, boiled, and subjected to SDS-PAGE for immunoblotting.
RESULTS
Role of AP-1 factors in Str-1 expression in B9SQ and A5SP cells.
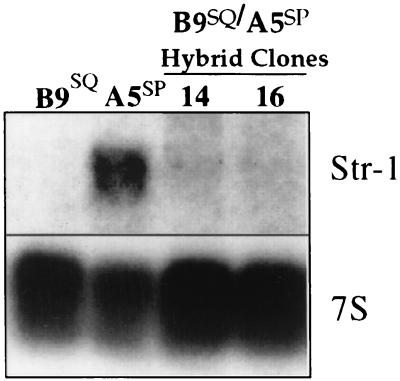
In order to determine whether the epithelial or mesenchymal phenotype was dominant, somatic hybrids of B9SQ and A5SP cells were generated (37). The majority of clones derived from this study have a squamous cell morphology, express epithelial markers, and have an invasive but nonmetastatic phenotype in vivo (37). The dominance of the B9SQ phenotype suggests that the EMT in this system is due to the loss of a dominant epithelial factor. Using Northern blot analysis, we confirmed that the neomycin-resistant B9SQ parental cell line did not express Str-1 and that the hygromycin-resistant A5SP parental cell line did (Fig. 1). Somatic hybrid cell clones that maintained the squamous phenotype did not express Str-1 mRNA. This pattern of Str-1 expression suggested that B9SQ cells contain a repressive factor that is capable of turning off Str-1 expression.
FIG. 1.
B9SQ-A5SP somatic hybrids do not express Str-1. Northern blot for Str-1 using 10 μg of total RNA from neomycin-resistant B9SQ and hygromycin-resistant A5SP parental cell lines and two somatic hybrid clones, both with a squamous phenotype. 7S rRNA hybridization is shown as a loading control.
Many potential mechanisms have been proposed for the repression of gene expression, including DNA methylation, histone acetylation, and trans-acting factors. Neither inhibition of methyltransferases with 5-azacytidine nor reversal of histone acetylation with trichostatin A induced production of the Str-1 message (data not shown). We next addressed the possibility that the Str-1 promoter was being repressed in B9SQ cells by specific trans-acting factors.
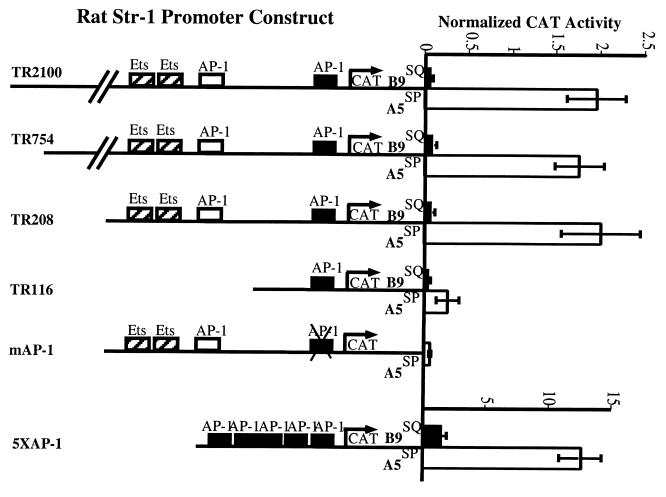
Using reporter constructs in which the rat Str-1 promoter was cloned upstream of a CAT reporter, we determined the relative Str-1 promoter activities in these cell lines. Str-1 promoter activity was high in A5SP cells and undetectable above the background in B9SQ cells (Fig. 2), consistent with the expression of the endogenous gene. Constructs containing 2,100, 754, and 208 bases of upstream promoter sequence were equally active in A5SP cells. When sequences between −208 and −116 were deleted, there was a substantial decrease in the overall activity observed in A5SP cells, probably due to the elimination in the intervening region of two Ets sites and an AP-1 site, each of which is important for Str-1 promoter activity (19, 24). Despite this decrease in activity, differential activity of the −116 promoter construct in the B9SQ and A5SP cells remained. This differential activity of the short Str-1 reporter suggested that the proximal AP-1 site, already described as being important in basal promoter activity for a number of similar MMP promoters, including Str-1 (12), was also a potential site of differential regulation. Mutation of this site in the TR208 construct (mAP-1) revealed that it was essential for reporter expression in A5SP cells. To test if AP-1 activity was different in B9SQ and A5SP cells, an artificial promoter containing five copies of a canonical collagenase AP-1 site (5XAP-1) was used. 5XAP-1 activity was considerably higher in A5SP cells than in B9SQ cells, supporting the hypothesis that the differential regulation of the Str-1 promoter was due, at least in part, to differential AP-1 activity.
FIG. 2.
The Str-1 promoter is active in A5SP cells but not in B9SQ cells. CAT reporter constructs under the control of the rat Str-1 (formerly called transin) promoter were transiently cotransfected into B9SQ and A5SP cells along with a CMV–β-galactosidase plasmid as a normalization control. Extracts were collected 48 h after transfection, and the amount of extract used was normalized according to β-galactosidase activity prior to the CAT assay. The abscissa represents CAT activity relative to that of an empty SV0-CAT reporter plasmid. Data bars represent the mean of four independent experiments, performed in triplicate. Schematics of promoter deletion constructs with pertinent transcription factor binding sites are shown. Numbers in the construct names refer the numbers of nucleotides included upstream of the transcription start site. mAP-1, TR208 construct with an inactive mutant proximal AP-1 site; 5XAP-1, artificial promoter containing five copies of the collagenase AP-1 site driving the CAT reporter gene.
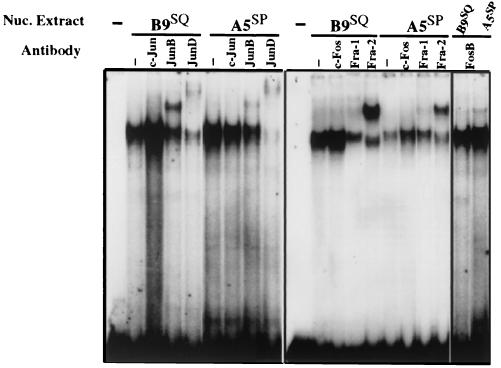
AP-1 is a dimeric complex, consisting of many different potential combinations of proteins from the Fos and Jun families encoded by nuclear proto-oncogenes. In order to identify which members are involved in the regulation of Str-1 in B9SQ and A5SP cells, nuclear extracts were taken from both cell lines and EMSAs were performed using the Str-1 proximal AP-1 site as an oligonucleotide probe.
Total AP-1 binding was approximately equivalent in the two extracts. However, the use of antibodies for supershift analyses revealed differences in the AP-1 components within the two extracts (Fig. 3). Of the Jun proteins, c-Jun binding was not observed in either B9SQ or A5SP cell nuclear extracts. B9SQ cell extracts showed binding by both JunB and JunD at approximately the same levels, whereas A5SP cell AP-1 complexes consisted primarily of JunD, with relatively little JunB activity. In both cell lines, Fra-2 appeared to be the major Fos family member that bound the AP-1 site, with Fra-1 and FosB representing relatively minor components of the complexes. Taken together, these data indicated that the AP-1 activity in B9SQ cells consisted largely of JunB or JunD with Fra-2, while in A5SP cells, JunB activity was greatly downregulated, leaving JunD and Fra-2 as the predominant heterodimeric complex components.
FIG. 3.
B9SQ and A5SP have different AP-1 binding activities. EMSA were performed using 5 μg of nuclear (Nuc.) extracts isolated from B9SQ or A5SP cells and an oligonucleotide probe representing the downstream AP-1 site of the Str-1 promoter. Supershift antibodies are indicated above each lane, with Jun family members in the left panel and Fos family members in the right panel. −, no nuclear extract or antibody addition.
JunB inhibits endogenous Str-1 gene expression.
The Jun proteins are not equivalent in their transactivation abilities. JunB, particularly in combination with Fra-2, is not a strong transcriptional activator, while JunD–Fra-2 transactivates well (11, 46). Consequently, for example, in dermal fibroblasts, JunB overexpression represses collagenase expression (11). Considering the relatedness of the promoter regions of collagenase and Str-1, we hypothesized that JunB may act as a repressor of Str-1 in B9SQ cells. As the EMSA experiments indicated a difference in the Jun components that make up the actively bound AP-1 complexes in B9SQ and A5SP cells, the mechanism controlling this differential activity was examined.
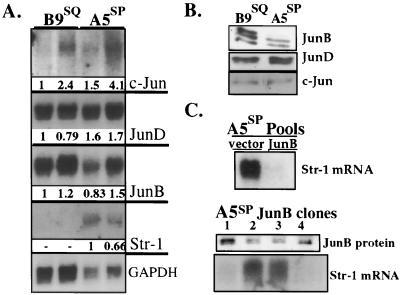
Northern blotting showed variable but low c-Jun transcript levels in both cell lines, while transcript levels for JunB and JunD were both high and did not correlate with Str-1 expression (Fig. 4A). Immunoblotting revealed that the JunD and c-Jun proteins were present in equal amounts, but B9SQ cells produced approximately 1.8-fold more JunB protein than A5SP cells (Fig. 4B).
FIG. 4.
JunB expression in B9SQ and A5SP cells. (A) Northern blot of 10 μg of two separate RNA preparations from B9SQ and A5SP cells. Probes are indicated to the right. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a loading control. Probes had approximately the same specific activities. c-Jun exposure time was 96 h. JunB and JunD exposure times were approximately 12 h. Numbers beneath panels show the relative quantity of the transcript after normalization to the GAPDH loading control. (B) Immunoblot for the Jun family using 5 μg of nuclear extracts from B9SQ and A5SP cells. Jun antibodies are indicated to the right. (C) Overexpression of JunB in A5SP cells. (Top panel) Str-1 Northern blot of 10 μg of total RNA from A5SP populations stably transfected with empty vector or JunB expression vector. (Middle panel) JunB immunoblot of 5 μg of nuclear extracts prepared from A5SP JunB stable clones demonstrating various levels of JunB expression. (Bottom panel) Str-1 Northern blot of 10 μg of total RNA from the same clones as those shown in the middle panel.
To determine if altering the ratio of JunB to JunD was sufficient to suppress Str-1 expression in A5SP cells, a JunB expression plasmid was stably transfected into these cells. Compared to the population transfected with the empty plasmid control, the JunB-transfected population produced considerably less endogenous Str-1 message (Fig. 4C, top panel). Examination of individual clones revealed that increased JunB protein production (Fig. 4C, middle panel) correlated with low levels of Str-1 mRNA (Fig. 4C, bottom panel). These results were consistent with the interpretation that JunB could repress Str-1 in A5SP cells.
JunB and JunD are differentially phosphorylated in B9SQ and A5SP cells.
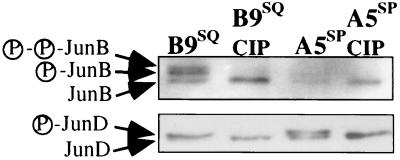
Although Str-1 expression in B9SQ and A5SP cells correlated inversely with the levels of JunB protein, it was not clear that the slightly higher JunB levels in B9SQ cells were sufficient to account for the differential binding activity observed in the supershift assays. Further examination of immunoblots of JunB and JunD (as in Fig. 4B) revealed multiple species for both JunB and JunD that differed between B9SQ and A5SP cells. For JunB, there was a consistent dominance of the uppermost band in B9SQ cell extracts compared to A5SP cell extracts. Because multiple bands seen by immunoblotting are often indicative of phosphorylation differences, we dephosphorylated nuclear extracts by incubation with CIP. CIP treatment resulted in a complete collapse of the slower-migrating JunB species into the fastest-migrating species, indicating that the slower-migrating species represented phosphorylated forms of JunB and JunD (Fig. 5). Therefore, JunB and JunD were hyperphosphorylated in B9SQ cells compared to A5SP cells.
FIG. 5.
Phosphorylation patterns for JunB and JunD differ in B9SQ and A5SP cells. Immunoblots of 10 μg of nuclear extracts incubated at 30°C alone (first and third lanes) or with CIP (second and fourth lanes) are shown. The JunB and JunD phosphoforms are indicated to the left. Data shown are representative of four independent experiments.
JNK activity is higher in B9SQ cells than in A5SP cells.
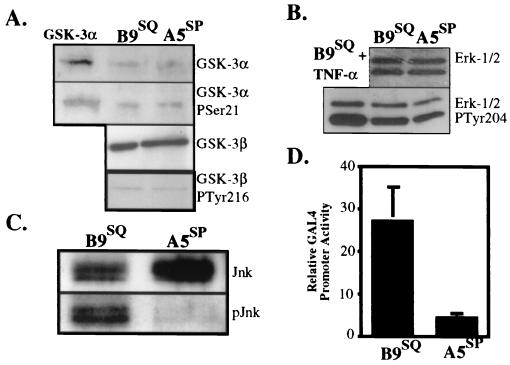
Phosphorylation of JunB and JunD can alter their transactivation abilities and suggests how the expression of Str-1 might be regulated differentially in B9SQ and A5SP cells in an AP-1-dependent manner. In order to identify the kinase responsible for the differential phosphorylation, we investigated the status of several enzymes that are known to phosphorylate Jun proteins. GSK-3 is a ubiquitously expressed enzyme that phosphorylates all three Jun proteins at the carboxy terminus, resulting in strong inhibition of DNA binding and transactivation (15, 32). Immunoblotting with GSK-3α- and GSK-3β-specific antibodies (Fig. 6A) demonstrated equivalent levels of these kinases in B9SQ and A5SP cells. In addition, phospho-specific antibodies revealed that the relative amounts of inactive GSK-3α (phosphorylated at Ser 21) and active GSK-3β (phosphorylated at Tyr 216) (Fig. 6A) were also equivalent between the cell lines.
FIG. 6.
B9SQ cells possess significantly more active JNK than do A5SP cells. (A) Immunoblots for total GSK-3α (top panel) and its inactive Ser 21 phosphoform (second panel from top) and for total GSK-3β (third panel from top) and its active Tyr 216 phosphoform (bottom panel) in 5 μg of whole-cell extracts from B9SQ and A5SP cells. (B) Immunoblots for ERK-1 and ERK-2 (top panel) and their active forms (bottom panel). Nuclear extracts (20 μg) from B9SQ cells stimulated with tumor necrosis factor alpha (TNF-α) were used as a positive control for the ERK active forms. (C) Immunoprecipitation-immunoblot for JNK protein using 10 μl of anti-JNK beads and 500 μl of total cell extracts. Immunoblotting was performed using antibodies recognizing total (Jnk) or active (phosphorylated at Thr 183 and Tyr 185) (pJnk) JNK. (D) In vivo assay for JNK activity in B9SQ and A5SP cells. B9SQ and A5SP cells were transiently transfected with a GAL4 UAS reporter construct and an expression construct encoding either a fusion protein containing the amino-terminal transactivation domain (JNK substrate) of c-Jun fused to GAL4dbd or GAL4dbd alone. Luciferase activity was measured and is shown as the activity of the c-Jun–GAL4dbd fusion normalized to that of GAL4dbd alone (n = 4) for each cell line.
Next, we examined two mitogen-activated protein kinases, ERK and JNK, both of which are capable of phosphorylating Jun proteins at the amino terminus in vitro and in vivo (4, 21, 25, 31, 38). There were equivalent levels of ERK-1 and ERK-2 in the two cell lines, as determined by immunoblotting with an anti-ERK-1/2 antibody (Fig. 6B). B9SQ cells possessed slightly more active ERK (approximately 1.4-fold), as detected by an antibody specific for ERK-1/2 phosphorylated at Tyr 204 (Fig. 6B). Overall, JNK levels appeared to be slightly lower in B9SQ cells (approximately 1.3-fold) (Fig. 6C). However, a phospho-specific antibody that recognizes all isoforms of active JNK showed that active p46 JNK levels were over fourfold higher in B9SQ cells than in A5SP cells (Fig. 6C). Due to interference from the immunoprecipitation antibody, we were unable to ascertain the status of the p55 JNK isoforms.
To compare overall JNK activities in B9SQ and A5SP cells, an in vivo assay that takes advantage of the fact that c-Jun phosphorylation by JNK strongly enhances its ability to activate transcription (21) was used. A plasmid expressing a fusion protein consisting of the amino terminus of c-Jun and the DNA binding domain of the yeast transcription factor GAL4 (GAL4dbd) was transfected, along with a reporter consisting of the GAL4 binding sequence (upstream activation sequence [UAS]) preceding a luciferase reporter, into the B9SQ and A5SP cell lines. The fusion protein binds to the UAS via GAL4dbd, and if JNK activity is present in the cells, phosphorylation of the c-Jun portion enhances its ability to transactivate the luciferase reporter gene. As depicted in Fig. 6D, when normalized to the reporter plus GAL4dbd alone, reporter activity with the cotransfected fusion protein was nearly sixfold higher in B9SQ cells than in A5SP cells, supporting the conclusion that B9SQ cells have significantly more constitutive JNK activity than do A5SP cells.
Phosphorylation of JunB by JNK.
JNK phosphorylation of c-Jun enhances not only the transactivation ability of c-Jun but also its capacity for binding DNA, albeit in an indirect manner (21, 34). In order to understand how higher JNK activity in B9SQ cells might lead to an inactive AP-1 complex on the Str-1 AP-1 site, we considered the facts that JunB was a major component of this complex in B9SQ cells and that JunB also was more phosphorylated in B9SQ cells. Because JunB is also a target of JNK (26, 31), the constitutive JNK activity in B9SQ cells might be responsible for the observed JunB hyperphosphorylation which could then enhance the capacity of JunB for DNA binding, as is the case for c-Jun (21, 34).
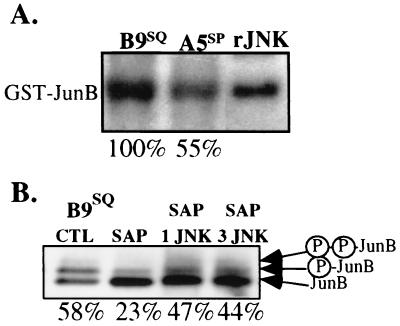
In order to examine JunB N-terminal phosphorylation in these cell lines, a GST-JunB fusion protein containing JunB amino acids 1 to 138 was used as the substrate in in vitro kinase assays. The reaction mixtures were incubated for 30 min at 30°C and then subjected to SDS-PAGE. JunB N-terminal phosphorylation activity present in the cell extracts was indicated by labeling of GST-JunB with 32P. Recombinant rat JNK phosphorylated GST-JunB (Fig. 7A), although not as efficiently as it did GST–c-Jun or GST–ATF-2 (data not shown). GST alone was not phosphorylated (data not shown). To test intracellular levels of activity, multiple sets of B9SQ and A5SP cell extracts were assayed, and results from each experiment were quantitated. Both cell lines had JunB N-terminal phosphorylation activity, but A5SP extracts possessed 63% ± 8% (n = 9) of the level found in B9SQ extracts. This result was consistent with the degree of difference in the phosphorylation of endogenous JunB in B9SQ and A5SP cells (Fig. 4B). There were similar differences in the phosphorylation of GST–c-Jun and GST–ATF-2 substrates by B9SQ and A5SP extracts (data not shown).
FIG. 7.
JNK can phosphorylate JunB in vitro. (A) In vitro phosphorylation of GST-JunB by B9SQ and A5SP cell extracts. GST-JunB was prepared, purified, and incubated with 1 μg of B9SQ or A5SP total extracts or recombinant JNK (rJNK) in the presence of [γ-32P]ATP. Values below the lanes reflect percent activity relative to that in the B9SQ extract. Three independently isolated sets of B9SQ and A5SP extracts were assayed in triplicate. The A5SP densitometric value was divided by the B9SQ value from the same experiment. The value shown is that for the autoradiogram in the figure and is representative of an overall average value of A5SP activity relative to B9SQ activity (63% ± 8%) examined in parallel. (B) JNK activity can contribute to both JunB phosphoforms. B9SQ cell extracts were incubated with SAP and then with 0.25 or 0.75 μg of rJNK (SAP 1 JNK and SAP 3 JNK, respectively). Treated extracts were then immunoblotted for JunB. Arrows indicate the migrations of the different phosphoforms of JunB. Values below the lanes reflect the relative amount of phosphorylated JunB species relative to the amount of total JunB, as determined by densitometry. CTL, control.
To determine if JNK phosphorylation of JunB could contribute to the formation of the slower-migrating species observed by immunoblotting, nuclear extracts were treated with recombinant JNK. First, JunB was dephosphorylated by treating B9SQ cell nuclear extracts with SAP. The reaction mixture was then heat inactivated, and recombinant JNK was added for further incubation. The addition of JNK after SAP treatment resulted in a twofold enhancement of the two upper JunB immunoblot bands compared to the results obtained with SAP treatment alone (Fig. 7B). Because SAP was not as capable as CIP in fully dephosphorylating JunB, we cannot conclude that JNK alone can lead to the formation of both phosphorylated species. However, these data suggested that the increased JNK activity in B9 cells could contribute to the formation of both phosphorylated species by phosphorylating either unphosphorylated or partially phosphorylated JunB.
JNK activity affects JunB protein-protein interactions and suppresses the Str-1 promoter.
We have observed that B9SQ cells produce more JunB than A5SP cells and that JunB in B9SQ cells exists in a hyperphosphorylated state that could be attributed to enhanced JNK activity. In addition, EMSA experiments showed that JunB–Fra-2 binding activity was substantially more prominent in B9SQ cells than in A5SP cells. The interaction of Fra-2 with JunB and JunD in B9SQ and A5SP cells was verified by immunoprecipitation of the Jun proteins followed by Western blotting for Fra-2 (data not shown). Given that phosphorylation is often found to affect protein-protein interactions (4), we tested the effects of the phosphorylation of JunB on its binding with Fra-2.
JunB–Fra-2 interactions were analyzed using a GST–Fra-2 construct that contains the basic leucine zipper and surrounding residues of Fra-2 necessary for interactions with its cofactors (30). GST–Fra-2 beads were incubated with B9SQ or A5SP cell nuclear extracts, washed under stringent conditions, and loaded onto an SDS-polyacrylamide gel for immunoblotting. GST–Fra-2 bound the hyperphosphorylated but not the unphosphorylated form of JunB (Fig. 8A). The intermediate phosphoform of JunB was faintly present after Fra-2 pulldown (data not shown). In addition, GST–Fra-2 bound more JunB (twofold) from B9SQ extracts than from A5SP extracts (Fig. 8B, lanes 2 and 3). GST–Fra-2 interacted exclusively with the phosphorylated form of JunD (Fig. 8C, top panel) and pulled down approximately equal amounts of JunD and c-Jun in B9SQ and A5SP extracts (Fig. 8C). These results indicated that Fra-2 bound preferentially to phosphorylated JunB and JunD, and the degree of differential phosphorylation of JunB was sufficient to significantly alter the overall levels of Fra-2 interactions.
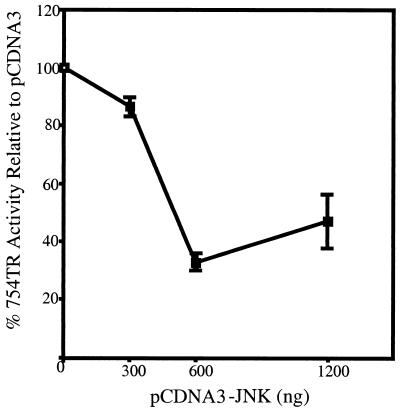
Together, these data suggested that AP-1 contributes to the differential expression of Str-1 in these cell lines and that JNK activity is a major determinant in this differential expression, given that there is little difference in the expression of the AP-1 factors themselves. If this hypothesis is accurate, then supplying A5SP cells with abundant JNK activity should be capable of inhibiting Str-1 promoter activity. To test this notion, an expression vector for JNK was cotransfected into A5SP cells with a −754 Str-1 promoter–luciferase reporter vector. Compared to the results obtained with the empty vector control, JNK overexpression inhibited the Str-1 promoter in a dose-dependent manner to a maximum of approximately threefold (Fig. 9). Therefore, restoration of JNK activity by overexpression of exogenous JNK can inhibit Str-1 promoter-driven transcription, supporting the hypothesis that the loss of JNK activity was a key event in releasing the inhibition of Str-1 expression in this skin tumor model system.
FIG. 9.
Overexpression of JNK in A5SP cells represses the Str-1 promoter. Three hundred nanograms of the rat Str-1 promoter–luciferase reporter was cotransfected with 300, 600, or 1,200 ng of pCDNA3-JNK-1 expression vector into 3 × 105 A5SP cells. Twenty nanograms of the thymidine kinase (tk) promoter driving Renilla luciferase was cotransfected as a normalization control. Firefly luciferase (Str-1 promoter) values were normalized to Renilla activity (tk promoter) and plotted as percent activity relative to the activity obtained with the empty pCDNA3 control. Error bars represent standard errors (n = 3).
Increased expression of JunB in B9SQ-A5SP somatic hybrid cell lines.
Our study of the regulators of Str-1 expression in the skin cancer EMT began with an analysis of somatic hybrid B9SQ-A5SP cell lines; data suggested the presence of a squamous cell repressor. Subsequent analysis of the parental B9SQ and A5SP cell lines led to the hypothesis that it is the increase in JunB activity, as regulated by JNK phosphorylation, that constitutes this squamous cell repressor. To confirm that this was the case, the somatic hybrid cell lines were examined once again.
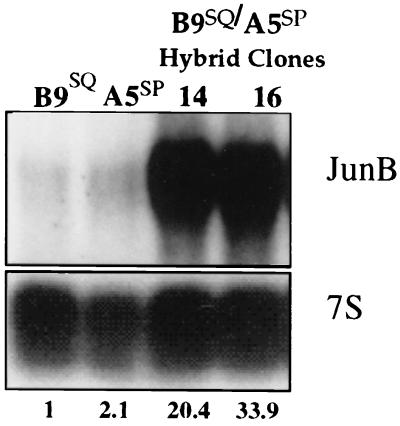
By all criteria examined, JNK activity and the degree of JunB phosphorylation were slightly elevated in the somatic hybrid lines compared to A5SP cells but were lower than in B9SQ cells (data not shown). Surprisingly, however, JunB expression itself was upregulated 20- to 30-fold above that in B9SQ parental cells and 10- to 15-fold above that in A5SP parental cells (Fig. 10). As we showed that the overexpression of JunB was sufficient to reduce Str-1 expression in A5SP cells (Fig. 4C), these data strongly suggested that it is the higher level of expression of JunB itself that is responsible for the lack of Str-1 expression in these hybrid lines. Thus, JunB activity fit the criteria for being the hypothetical squamous cell repressor in this model system.
FIG. 10.
JunB expression is highly upregulated in B9SQ-A5SP somatic cell hybrids. The Northern blot shown in Fig. 1 was stripped and rehybridized with a JunB-specific probe. Numbers beneath the 7S rRNA panel represent the quantitation of JunB expression normalized to the 7S loading control and shown as values relative to the B9SQ value.
DISCUSSION
In an effort to further understand the molecular mechanisms of the EMT in skin tumor progression, we examined the expression of the EMT marker Str-1 in two genetically related mouse skin tumor cell lines, B9SQ and A5SP. B9SQ, which does not express Str-1, is invasive in vivo but relatively well differentiated, being squamous in morphology. The A5SP spindle cell line, which has lost many of the hallmarks of epithelial differentiation and is metastatic, expresses Str-1 (49). Analysis of B9SQ-A5SP somatic hybrids strongly suggested that the critical factor governing the differential expression of Str-1 was a repressor in B9SQ cells. In this study, we used the rat Str-1 promoter, which shares 92% identity with the mouse Str-1 promoter within the −208 minimal promoter region. Promoter analysis led us to focus on components interacting with the proximal AP-1 site, a site identical in the rat and mouse promoters and one necessary for constitutive and differential Str-1 promoter activities in this system. EMSA experiments indicated that the AP-1 complexes primarily contained combinations of Fra-2, JunD, and JunB proteins in B9SQ cells but almost exclusively Fra-2 and JunD in A5SP cells. Thus, a major difference between the AP-1 complexes in these cells was the binding activity of JunB in B9SQ cells.
JunB has a long history of being considered an inhibitory AP-1 factor. More accurately, JunB is a transactivator whose level of activity is much lower than those of the other Jun family members, c-Jun and JunD. Therefore, when JunB effectively competes with c-Jun or JunD, overall AP-1 activity on many promoters is reduced. This mode of activity has been shown for promoters controlling genes as diverse as those for collagenase (11) and cyclin D1 (1) and has been substantiated by examination of the expression of AP-1-regulated growth factors in a JunB-null genetic background (47).
Recent evidence has suggested that the inhibitory nature of JunB activity is oversimplified. JunB can be an activator of some AP-1-responsive promoters, as powerful as or more powerful than c-Jun or JunD (26). The difference between the effective inhibitor and powerful activator functions of JunB is apparently determined by its interactions with other proteins in the target promoters. For instance, the inhibitory nature of JunB is most evident when it is complexed with the AP-1 protein c-Jun (16) or Fra-2 (46), while the stimulatory role is evident in the interleukin 4 promoter upon interaction with c-Maf (26). JNK phosphorylation of JunB at threonines 102 and 104, equivalent to the JNK phosphorylation sites at threonines 91 and 93 in c-Jun (26, 34), has been shown to enhance its binding to the interleukin 4 promoter and its interaction with c-Maf (26). These results are strikingly similar to those obtained with the B9SQ-A5SP system, where hyperphosphorylated JunB bound better to its inhibitory partner, Fra-2, as well as to the Str-1 AP-1 site. Thus, JNK phosphorylation of JunB appears to be a major regulatory mechanism that can result in the activation of particular promoters and the repression of others, with both activities being dependent upon AP-1 sites in the target promoters and the cellular context, as defined by the expression of its protein binding partners. It is important to note that in this study, we used a concatemerized collagenase AP-1 site reporter construct that has been shown to be inhibited by JunB (11). Thus, our data relating the onset of the mesenchymal phenotype to the loss of JunB repression at the Str-1 AP-1 site does not contradict the possibility that the higher level of JunB activity in B9SQ cells activated another group of genes that is important for maintaining the epithelial phenotype of these cells.
AP-1 activity has been shown to play a critical role in mouse skin tumor progression (39, 50). In another skin tumor model system, AP-1 factors act to inhibit the expression of some of the MMP genes (17). It also has been noted previously that JunB activity is reduced in an advanced-stage mouse skin tumor cell line relative to earlier-stage skin tumor cells (22). JNK activity has also been noted to change during skin tumor progression (51), although that study focused primarily on progression from papilloma to squamous cell carcinoma. Here we have shown how these properties of regulated AP-1 activity can converge to alter gene expression in a manner relevant to the EMT, a process marked by a loss of cellular differentiation. Interestingly, the absence of JunB activity appears to relate to the state of differentiation of keratinocytes in the differentiation program of normal epidermis as well as in the tumor progression program. During differentiation, the level of JunB expression is high in differentiated superbasal keratinocytes but is absent in stem cell basal keratinocytes in both in vivo (48) and in vitro (20, 38, 40) models. These observations strongly suggest that JunB activity is important in maintaining the differentiation of keratinocytes in both normal and pathological processes. It will be of interest to determine how JunB may regulate other aspects of the epithelial phenotype.
In the scheme of skin tumor progression, the importance of losing JunB activity offers some intriguing mechanistic possibilities. It has been shown that the progression from squamous to spindle cells is under the control of TGF-β activity both in vitro (9) and in transgenic model systems (13). Initially, TGF-β acts to inhibit papilloma outgrowth (13), but once squamous cell conversion takes place, the tumor cells selectively escape the inhibitory effects of TGF-β. Instead, chronic exposure to TGF-β enhances the EMT (9) and is thought to be required for this transition (36). TGF-β, in turn, has been shown to upregulate both JNK activity (18) and JunB expression (35). Our results suggest that one mechanism of escape from TGF-β-dependent negative pressure is the loss of JunB activity which, in turn, leads to Str-1 expression and potentially to spindle cell conversion itself. That is, B9SQ cells are largely under the inhibitory control of the combination of a high level of JunB expression and a high level of JNK activity. A5SP cells appear to have adapted to and escaped from this inhibitory pressure by downregulating JNK activity. This interpretation presumes that it is the loss of JunB activity that is being selected for and not the loss of JNK activity per se. This notion is supported by the observation that despite the lower level of activity of JunB in a model of skin tumor progression examined by Joseloff and Bowden, this loss of activity did not correlate with a decrease in JunB phosphorylation (22). Furthermore, the extremely high level of expression of JunB in B9SQ-A5SP somatic hybrid cells relative to either parental cell line supports the notion that it is JunB activity that is being selected against during the EMT. While these data strongly suggest that the specific mechanism of JunB downregulation during the EMT is less important than the end result, we cannot dismiss additional consequences of JNK downregulation pertinent to the EMT, given that JNK substrates are not limited to the Jun proteins on which this study focuses.
The onset of Str-1 expression has been shown to be sufficient to induce the EMT in mammary cell lines (27), a phenomenon that may be equally valid in the skin tumor system, particularly given the correlation of Str-1 expression with the progression from squamous to spindle cells. Whether Str-1 is a cause or a symptom of the EMT in the skin remains a question for the future. However, in this study, we have shown that Str-1 expression provides a powerful tool with which relevant changes in signaling molecules that occur during the transition can be revealed.
ACKNOWLEDGMENTS
We thank Sheelagh Frame for helpful discussions. We also thank Eric Paulson and Ron Wisdom for gifts of plasmids. We are especially grateful to Allan Balmain for the kind gifts of B9SQ, A5SP, and B9SQ-A5SP somatic hybrid cells.
This work was supported by grants NIH R01 CA46843 (to L.M.M.) and NIH CA67429 (to H.C.C.).
REFERENCES
- 1.Bakiri L, Lallemand D, Bossy-Wetzel E, Yaniv M. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 2000;19:2056–2068. doi: 10.1093/emboj/19.9.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia D H. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 3.Birchmeier C, Birchmeier W, Brand-Saberi B. Epithelial-mesenchymal transitions in cancer progression. Acta Anat (Basel) 1996;156:217–226. doi: 10.1159/000147848. [DOI] [PubMed] [Google Scholar]
- 4.Boulikas T. Phosphorylation of transcription factors and control of the cell cycle. Crit Rev Eukaryot Gene Expr. 1995;5:1–77. [PubMed] [Google Scholar]
- 5.Bremner R, Balmain A. Genetic changes in skin tumor progression: correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell. 1990;61:407–417. doi: 10.1016/0092-8674(90)90523-h. [DOI] [PubMed] [Google Scholar]
- 6.Brown K, Balmain A. Transgenic mice and squamous multistage skin carcinogenesis. Cancer Metastasis Rev. 1995;14:113–124. doi: 10.1007/BF00665795. [DOI] [PubMed] [Google Scholar]
- 7.Burns P A, Kemp C J, Gannon J V, Lane D P, Bremner R, Balmain A. Loss of heterozygosity and mutational alterations of the p53 gene in skin tumors of interspecific hybrid mice. Oncogene. 1991;6:2363–2369. [PubMed] [Google Scholar]
- 8.Cano A, Perez-Moreno M A, Rodrigo I, Locascio A, Blanco M J, del Barrio M G, Portillo F, Nieto M A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 9.Caulin C, Scholl F G, Frontelo P, Gamallo C, Quintanilla M. Chronic exposure of cultured transformed mouse epidermal cells to transforming growth factor-beta 1 induces an epithelial-mesenchymal transdifferentiation and a spindle tumoral phenotype. Cell Growth Differ. 1995;6:1027–1035. [PubMed] [Google Scholar]
- 10.Chambers A F, Matrisian L M. Changing views of the role of matrix metalloproteinases in metastasis. J Natl Cancer Inst. 1997;89:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- 11.Chiu R, Angel P, Karin M. Jun-B differs in its biological properties from, and is a negative regulator of, c-Jun. Cell. 1989;59:979–986. doi: 10.1016/0092-8674(89)90754-x. [DOI] [PubMed] [Google Scholar]
- 12.Crawford H C, Matrisian L M. Mechanisms controlling the transcription of matrix metalloproteinase genes in normal and neoplastic cells. Enzyme Protein. 1996;49:20–37. doi: 10.1159/000468614. [DOI] [PubMed] [Google Scholar]
- 13.Cui W, Fowlis D J, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst R J. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 14.D'Armiento J, DiColandrea T, Dalal S S, Okada Y, Huang M T, Conney A H, Chada K. Collagenase expression in transgenic mouse skin causes hyperkeratosis and acanthosis and increases susceptibility to tumorigenesis. Mol Cell Biol. 1995;15:5732–5739. doi: 10.1128/mcb.15.10.5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Groot R P, Auwerx J, Bourouis M, Sassone-Corsi P. Negative regulation of Jun/AP-1: conserved function of glycogen synthase kinase 3 and the Drosophila kinase shaggy. Oncogene. 1993;8:841–847. [PubMed] [Google Scholar]
- 16.Deng T, Karin M. JunB differs from c-Jun in its DNA-binding and dimerization domains, and represses c-Jun by formation of inactive heterodimers. Genes Dev. 1993;7:479–490. doi: 10.1101/gad.7.3.479. [DOI] [PubMed] [Google Scholar]
- 17.Dong Z G, Crawford H C, Lavrovsky V, Taub D, Watts R, Matrisian L M, Colburn N H. A dominant negative mutant of jun blocking 12-o-tetradecanoylphorbol-13-acetate-induced invasion in mouse keratinocytes. Mol Carcinog. 1997;19:204–212. doi: 10.1002/(sici)1098-2744(199707)19:3<204::aid-mc8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 18.Engel M E, McDonnell M A, Law B K, Moses H L. Interdependent SMAD and JNK signaling in transforming growth factor-beta-mediated transcription. J Biol Chem. 1999;274:37413–37420. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- 19.Gaire M, Magbanua Z, McDonnell S, McNeil L, Lovett D H, Matrisian L M. Structure and expression of the human gene for the matrix metalloproteinase matrilysin. J Biol Chem. 1994;269:2032–2040. [PubMed] [Google Scholar]
- 20.Gandarillas A, Watt F M. Changes in expression of members of the fos and jun families and myc network during terminal differentiation of human keratinocytes. Oncogene. 1995;11:1403–1407. [PubMed] [Google Scholar]
- 21.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 22.Joseloff E, Bowden G T. Regulation of the transcription factor AP-1 in benign and malignant mouse keratinocyte cells. Mol Carcinog. 1997;18:26–36. doi: 10.1002/(sici)1098-2744(199701)18:1<26::aid-mc4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 23.Kerr L D, Miller D B, Matrisian L M. TGF-beta 1 inhibition of transin-stromelysin gene expression is mediated through a fos binding sequence. Cell. 1990;61:267–278. doi: 10.1016/0092-8674(90)90807-q. [DOI] [PubMed] [Google Scholar]
- 24.Kirstein M, Sanz L, Quinones S, Moscat J, Diaz-Meco M T, Saus J. Cross-talk between different enhancer elements during mitogenic induction of the human stromelysin-1 gene. J Biol Chem. 1996;271:18231–18236. doi: 10.1074/jbc.271.30.18231. [DOI] [PubMed] [Google Scholar]
- 25.Leppa S, Saffrich R, Ansorge W, Bohmann D. Differential regulation of c-Jun by ERK and JNK during PC12 cell differentiation. EMBO J. 1998;17:4404–4413. doi: 10.1093/emboj/17.15.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Tournier C, Davis R J, Flavell R A. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lochter A, Galosy S, Muschler J, Freedman N, Werb Z, Bissell M J. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J Cell Biol. 1997;139:1861–1872. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majmudar G, Nelson B R, Jensen T C, Voorhees J J, Johnson T M. Increased expression of stromelysin-3 in basal cell carcinomas. Mol Carcinog. 1994;9:17–23. doi: 10.1002/mc.2940090105. [DOI] [PubMed] [Google Scholar]
- 29.Matrisian L M, Bowden G T. Stromelysin/transin and tumor progression. Semin Cancer Biol. 1990;1:107–115. [PubMed] [Google Scholar]
- 30.Matsui M, Tokuhara M, Konuma Y, Nomura N, Ishizaki R. Isolation of human fos-related genes and their expression during monocyte-macrophage differentiation. Oncogene. 1990;5:249–255. [PubMed] [Google Scholar]
- 31.Mendelson K G, Contois L R, Tevosian S G, Davis R J, Paulson K E. Independent regulation of JNK/p38 mitogen-activated protein kinases by metabolic oxidative stress in the liver. Proc Natl Acad Sci USA. 1996;93:12908–12913. doi: 10.1073/pnas.93.23.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikolakaki E, Coffer P J, Hemelsoet R, Woodgett J R, Defize L H. Glycogen synthase kinase 3 phosphorylates Jun family members in vitro and negatively regulates their transactivating potential in intact cells. Oncogene. 1993;8:833–840. [PubMed] [Google Scholar]
- 33.Ostrowski L E, Finch J, Krieg P, Matrisian L M, Patskan G, O'Connell J F, Philips J, Slaga T J, Breathnach R, Bowden G T. Expression pattern of a gene for a secreted metalloproteinase during late stages of tumor progression. Mol Carcinog. 1988;1:13–19. doi: 10.1002/mc.2940010106. [DOI] [PubMed] [Google Scholar]
- 34.Papavassiliou A G, Treier M, Bohmann D. Intramolecular signal transduction in c-Jun. EMBO J. 1995;14:2014–2019. doi: 10.1002/j.1460-2075.1995.tb07193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pertovaara L, Sistonen L, Bos T J, Vogt P K, Keski-Oja J, Alitalo K. Enhanced jun gene expression is an early genomic response to transforming growth factor beta stimulation. Mol Cell Biol. 1989;9:1255–1262. doi: 10.1128/mcb.9.3.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portella G, Cumming S A, Liddell J, Cui W, Ireland H, Akhurst R J, Balmain A. Transforming growth factor beta is essential for spindle cell conversion of mouse skin carcinoma in vivo: implications for tumor invasion. Cell Growth Differ. 1998;9:393–404. [PubMed] [Google Scholar]
- 37.Portella G, Liddell J, Crombie R, Haddow S, Clarke M, Stoler A B, Balmain A. Molecular mechanisms of invasion and metastasis during mouse skin tumour progression. Invasion Metastasis. 1994;14:7–16. [PubMed] [Google Scholar]
- 38.Rosenberger S F, Finch J S, Gupta A, Bowden G T. Extracellular signal-regulated kinase 1/2-mediated phosphorylation of JunD and FosB is required for okadaic acid-induced activator protein 1 activation. J Biol Chem. 1999;274:1124–1130. doi: 10.1074/jbc.274.2.1124. [DOI] [PubMed] [Google Scholar]
- 39.Rutberg S E, Adams T L, Glick A, Bonovich M T, Vinson C, Yuspa S H. Activator protein 1 transcription factors are fundamental to v-rasHa-induced changes in gene expression in neoplastic keratinocytes. Cancer Res. 2000;60:6332–6338. [PubMed] [Google Scholar]
- 40.Rutberg S E, Saez E, Glick A, Dlugosz A A, Spiegelman B M, Yuspa S H. Differentiation of mouse keratinocytes is accompanied by PKC-dependent changes in AP-1 proteins. Oncogene. 1996;13:167–176. [PubMed] [Google Scholar]
- 41.Ryder K, Lanahan A, Perez-Albuerne E, Nathans D. jun-D: a third member of the jun gene family. Proc Natl Acad Sci USA. 1989;86:1500–1503. doi: 10.1073/pnas.86.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryder K, Lau L F, Nathans D. A gene activated by growth factors is related to the oncogene v-jun. Proc Natl Acad Sci USA. 1988;85:1487–1491. doi: 10.1073/pnas.85.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savagner P, Yamada K M, Thiery J P. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts,’ prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoler A B, Stenback F, Balmain A. The conversion of mouse skin squamous cell carcinomas to spindle cell carcinomas is a recessive event. J Cell Biol. 1993;122:1103–1117. doi: 10.1083/jcb.122.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki T, Okuno H, Yoshida T, Endo T, Nishina H, Iba H. Difference in transcriptional regulatory function between c-Fos and Fra-2. Nucleic Acids Res. 1991;19:5537–5542. doi: 10.1093/nar/19.20.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szabowski A, Maas-Szabowski N, Andrecht S, Kolbus A, Schorpp-Kistner M, Fusenig N E, Angel P. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal-epidermal interaction in skin. Cell. 2000;103:745–755. doi: 10.1016/s0092-8674(00)00178-1. [DOI] [PubMed] [Google Scholar]
- 48.Wilkinson D G, Bhatt S, Ryseck R P, Bravo R. Tissue-specific expression of c-jun and junB during organogenesis in the mouse. Development. 1989;106:465–471. doi: 10.1242/dev.106.3.465. [DOI] [PubMed] [Google Scholar]
- 49.Wright J H, McDonnell S, Portella G, Bowden G T, Balmain A, Matrisian L M. A switch from stromal to tumor cell expression of stromelysin-1 mRNA associated with the conversion of squamous to spindle carcinomas during mouse skin tumor progression. Mol Carcinog. 1994;10:207–215. doi: 10.1002/mc.2940100405. [DOI] [PubMed] [Google Scholar]
- 50.Young M R, Li J J, Rincon M, Flavell R A, Sathyanarayana B K, Hunziker R, Colburn N. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci USA. 1999;96:9827–9832. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zoumpourlis V, Papassava P, Linardopoulos S, Gillespie D, Balmain A, Pintzas A. High levels of phosphorylated c-Jun, Fra-1, Fra-2 and ATF-2 proteins correlate with malignant phenotypes in the multistage mouse skin carcinogenesis model. Oncogene. 2000;19:4011–4021. doi: 10.1038/sj.onc.1203732. [DOI] [PubMed] [Google Scholar]