Significance
Despite advances in directed evolution and computational design, engineering of functional proteins remains challenging. Here we demonstrate that rearrangement of modular repeats from BRCA2 can yield chimeras with improved interaction properties, while deconstructing the relative energetic contributions of different modules across a large interaction interface and resolving the effects of shape complementary or physicochemical properties by empirical observation. This de- and reconstruction of binding interfaces supports a mix-and-match model for proteins in which repeat units can be manipulated and used to construct functional proteins that interfere with dsDNA repair. Our approach may aid in creating biochemical and therapeutic tools from natural modules, with minimal screening effort.
Keywords: protein evolution, modular protein engineering, synthetic biology, RAD51, BRC repeats
Abstract
Exchanges of protein sequence modules support leaps in function unavailable through point mutations during evolution. Here we study the role of the two RAD51-interacting modules within the eight binding BRC repeats of BRCA2. We created 64 chimeric repeats by shuffling these modules and measured their binding to RAD51. We found that certain shuffled module combinations were stronger binders than any of the module combinations in the natural repeats. Surprisingly, the contribution from the two modules was poorly correlated with affinities of natural repeats, with a weak BRC8 repeat containing the most effective N-terminal module. The binding of the strongest chimera, BRC8-2, to RAD51 was improved by −2.4 kCal/mol compared to the strongest natural repeat, BRC4. A crystal structure of RAD51:BRC8-2 complex shows an improved interface fit and an extended β-hairpin in this repeat. BRC8-2 was shown to function in human cells, preventing the formation of nuclear RAD51 foci after ionizing radiation.
The daunting combinatorial diversity arising from simultaneous mutation of all amino acid positions even in small proteins (leading, e.g., to 1039 variants of a 30-mer) renders exploration of such sequence space futile. It is therefore attractive to view proteins not only as combinations of variant amino acids, but as combinations of exchangeable segments, because the combinatorial diversity is dramatically reduced if such “modules” are instead recombined (1–6). Understanding protein modularity at different length scales, with building blocks of various sizes ranging from as few as 20 amino acids (7, 8) to entire domains (9) or beyond (10), is key to elucidating natural protein evolution and to achieving full control in protein design and engineering (6). Likewise de novo designed structures (11), consisting of stitched-together naturally occurring elements should provide more diverse starting points for computational protein engineering, as exemplified in the “SEWING” approach (12) or in novel repeat proteins (13–15). Designed ankyrin repeat proteins (DARPins, consisting of four to five repeats of a 33 amino acid motif) have already shown their potential as antibody-like therapeutics (16). Plausibly, addition of repeats of an original functional motif in evolution could enhance binding of a target through the increase in available binding surface area or by turning a monovalent interaction into a multivalent system, explaining why many extant proteins contain repeats as a result of duplication (17) and fusion events (e.g., in the TIM-barrel fold) (18, 19). The question then arises whether modular proteins can be deconstructed into their constituent parts and whether these parts can be reassembled to create functional proteins as chimeras of “standard parts,” realizing an ambition of synthetic biology. A reassembly reminiscent of this idea, shuffling, is often used in directed protein evolution (20–22) and has been rationalized by the automated determination of minimally folded domains for shuffling, for example using the SCHEMA algorithm (23, 24). Natural evolution may rely on rearrangement of protein modules through exon shuffling: the exon–intron architecture of eukaryotic genes may allow homologous recombination at introns, to arrange exons into novel combinations, and help bring about new protein functions (25–28), as illustrated by evolution of an alcohol dehydrogenase (29) that resulted in a shift from the progenitor enzyme’s substrate preference (26).
Here we address the relationship of modularity and function by probing whether binding motifs in linear peptides can be recombined to interact with a given target protein and by determining how the module order affects affinity. This analysis is related to systems where the modularity of the binding protein parallels the modularity of the target peptide, e.g., in natural and designed armadillo repeats (30, 31), where modules with a given specificity are assembled to interact with a linear epitope (32).
The modular protein fragment used for the chimeragenesis in this work is involved in the interactions between RAD51 and BRCA2 that are critical for DNA double-strand break repair by homologous recombination. BRCA2 is a 3,418-amino-acid protein whose central part, exon 11, contains 8 conserved, approximately 35-residue repeats (33) (referred to as “BRC repeats” followed by the numbers 1 through 8; Fig. 1A). BRCA2 exerts a multitude of functions on RAD51 in the cell, such as localization, nucleofilament assembly and its depolymerization, and has been aptly termed the “custodian” of chromosomal numerical and structural integrity (34). The 8 BRC repeats, varying significantly in their affinity for RAD51:BRC repeats 1 to 4, are seen as high-affinity binders, while repeats 5 to 8 are generally weaker (35–37). The crystal structure of BRC4 in complex with RAD51 (38), together with structural modeling and biochemical experiments, revealed the existence of two distinct parts in the BRC repeats that interact with RAD51 (39). The first of these, known as the “FxxA module,” forms a β-hairpin structure and binds RAD51 with a Phe and Ala in two small binding pockets of the ATPase domain. The C-terminal part of the BRC repeat, with a conserved LFDE motif, interacts with the distal part of the ATPase domain in an α-helical conformation. In doing so, the BRC repeats directly compete with another FxxA module located in RAD51 itself (with the sequence FTTA), on an oligomerization epitope between RAD51’s C-terminal ATPase and N-terminal DNA-binding domains (40). The BRC4 repeat peptide has been shown to cause dissociation of RAD51 oligomers, and conditional expression of the repeat in breast cancer cells disrupts the RAD51:BRCA2 interaction and sensitizes them to radiation treatment (41). In isolation, the FxxA module makes a relatively weak contribution to the binding: a 4-residue FHTA peptide, representing the FxxA hotspot from the corresponding module in BRC4, bound RAD51-surrogate HumRadA2 with a Kd of 290 μM (42). Even the entire FxxA module—that is the FxxA hotspot and the surrounding residues—is not a strong binder of RAD51. About 500 μM of a 17-residue FHTA-containing peptide (the N-terminal half of the BRC4 repeat) was required to effect full disruption of the RAD51:BRC4 interaction in a competitive ELISA (39) and fluorescence polarization (FP) competition measurement using a monomeric surrogate for RAD51 determined a Kd of 36 μM for the FHTA module of BRC4 (43). It is the C-terminal LFDE module that ensures significantly enhanced affinities are achieved, even if its affinity on its own is too low to be determined biophysically (43). This second module binds to a groove on another surface of the RAD51 ATPase domain (38). Although the phylogeny of the BRC repeats remains to be fully elucidated, it is thought that the emergence of the BRCA2 repeats predates the emergence of the mammalian class (44) and perhaps even the divergence of birds and mammals 230 to 300 million years ago (45). As the eight repeats found in the BRCA2 protein all occur on the same exon (44), throughout their evolutionary history, these repeats would never have been subject to natural exon shuffling. Given the larger variation in the affinity of different BRC repeats for RAD51, the uneven contribution of FxxA and LFDE motifs for binding and distinctive structural features of these two building blocks in BRC4, led us to pose two questions. Firstly, what is the contribution of the different FxxA and LFDE motifs for RAD51 binding and do the relative affinities between repeats remains the same? Secondly, we wanted to see whether the BRC repeat building blocks can be reshuffled: are the resulting chimeras functional and how does the function of reassembled BRCA2 proteins depend on the motif order and identity?
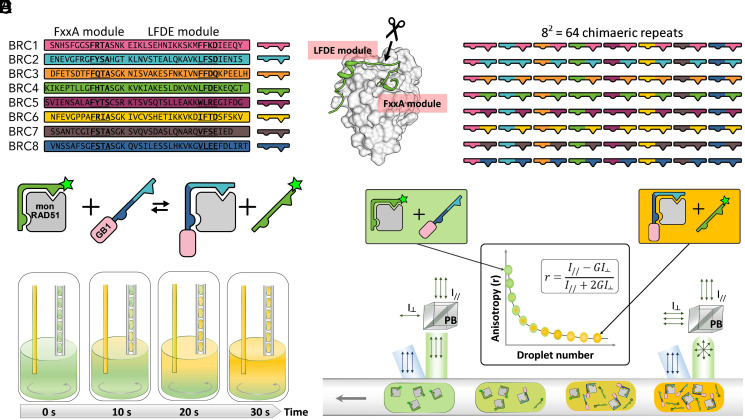
Fig. 1.
Shuffling of the binding modules comprising the eight RAD51-binding repeats in BRCA2. (A) The eight human BRC repeats with FxxA and LFDE motifs in bold. (B) The crystal structure of RAD51:BRC4 peptide complex (PDB ID: 1n0w). The arrow with scissors indicates the crossover point between the FxxA and LFDE modules used in this study. (C) Schematic representation of the 56 chimeric and eight natural repeats resulting from the shuffling around the crossover point described in B. (D) Schematic representation of the competition assay used for the affinity determination of the BRC-repeat shuffle set for monomeric RAD51 (mon RAD51) by fluorescence anisotropy. GB1-BRC recombinant peptide fusions were titrated into a complex of monomeric RAD51 and BRC4fl (a fluorescein-labeled BRC4 synthetic peptide), so that the Kd of the recombinant peptide could be calculated using the known Kd of BRC4fl 49,50. (E) Titration of GB1-BRC peptides using droplets on demand: microdroplets containing an increasing concentration of fusion peptides were produced from a well of a 384-well plate by a microcapillary aspiration technique. During 30 s, a stock solution of GB1-BRC peptide was injected in the well so that its concentration rose from 0 to 50% of stock concentration (left capillary). Simultaneously, droplets encapsulating the changing contents of the well were generated (right capillary). Flow directions are represented by gray arrows. Mixing was performed by a magnetic stir bar (gray circling arrow). The concentration of monomeric RAD51 and BRC4fl was kept constant throughout the titration. (F) The titration droplets were measured for their average fluorescence anisotropy using a fluorescence anisotropy imaging microscope. Upon traveling above the imaging area, linearly polarized light (in blue) excited the fluorescein tags of the BRC4fl peptides, either bound to monomeric RAD51 (larger hydrodynamic volume, resulting in high anisotropy) or freely tumbling after competition with GB1-BRC peptides (lower hydrodynamic volume, resulting in low anisotropy). Fluorescence emission intensity signals (in green) recorded for parallel and perpendicular polarizations enabled the quantification of average anisotropy for every droplet of the titration sequence. A G factor was calculated to correct for differences in detector sensitivities in the parallel and perpendicular channels. PB, polarizing beamsplitter.
Recognizing this modularity, we probe the idea that functional sophistication is brought about by combination of these relatively simple peptide building blocks, by testing the interactions of chimeras of rearranged BRC repeats with RAD51. Our objective was to explicitly explore the recombination of entire natural repeats in the creation of new functional proteins, in order to demonstrate the role of modularity in functional adaptation. The discovery that the natural, “parental,” combination of modules often turned out to be suboptimal for RAD51 binding, while novel BRC module combinations yielded more potent RAD51 binders, demonstrates the role that rearrangements of repeat building blocks can play in protein engineering.
Results
Systematic Shuffling of the Two Binding Modules of the Eight BRC Repeats and Evaluation of the 64 Resulting Chimeras in a Microfluidic Fluorescence Anisotropy Assay.
To be able to shuffle the two modules of the eight BRC repeats found in BRCA2 that bind RAD51 (Fig. 1A), a crossover point was defined immediately at the C terminus of the hairpin structure found in the FxxA module (Fig. 1B), as suggested by the RAD51:BRC4 crystal structure (38). The resulting 64 variant BRC peptides (Fig. 1C) were cloned through an oligonucleotide cassette method (SI Appendix, Fig. S1 and Table S1), as C-terminal fusions to the GB1 domain from protein G (SI Appendix, Fig. S2; we will refer to shuffled variants by two digits denoting the identity of the N-terminal FxxA and the C-terminal LFDE module, respectively, e.g., BRC2-4 is a peptide with the FxxA module from BRC2 and the LFDE module from BRC4). The GB1 domain does not interfere with the BRC-repeat interaction: an isothermal titration calorimetry (ITC) affinity measurement was carried out to confirm that the BRC4 peptide, upon fusion to the GB1 C terminus, maintained its ability to bind HumRadA22 [a monomeric yet faithful model of RAD51, and for simplicity we will refer to this as monomeric RAD51 (46); SI Appendix, Figs. S5 and S6 and accompanying SI Appendix, Supplementary Text 3) and matched affinities previously measured for BRC4 peptide (46–48)].
The 64 different variant peptide repeats were assayed in a competition assay with a fluorescently labeled BRC4 repeat (BRC4fl-peptide) as the tracer (Fig. 1D). We employed a microfluidic setup to facilitate measurement of affinities to minimize the sample quantities (49): dose–response curves were set up by coupling “droplet-on-demand” formation (Fig. 1E) and a microfluidic fluorescence anisotropy imaging platform (Fig. 1F), so that every measurement only required nanoliter volumes and minimal protein amounts.
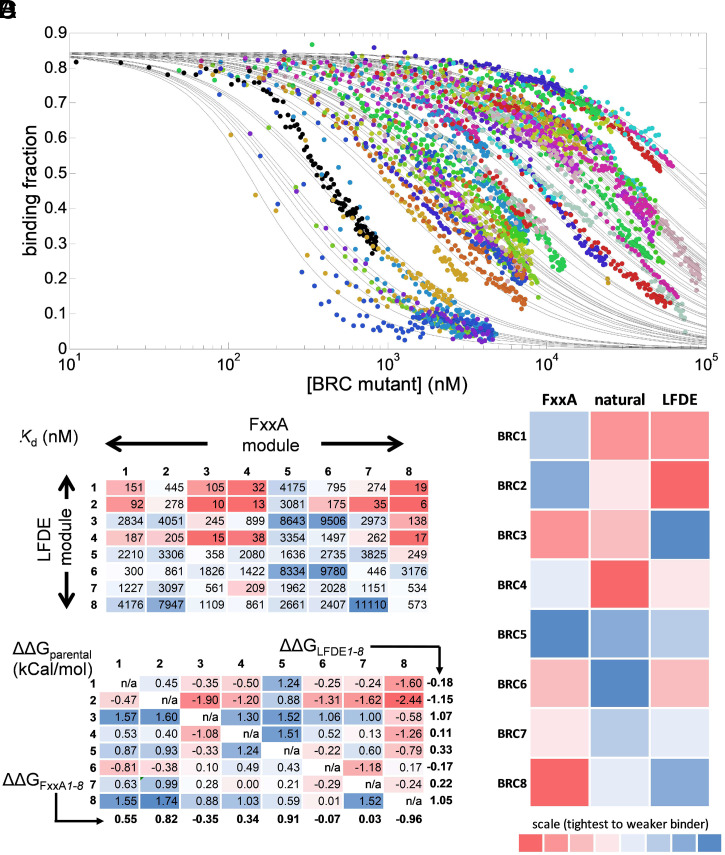
Although the droplet-on-demand method supported procurement of measurements over at least two orders of magnitude in concentration of titrant, we expected even larger differences in affinity between the different chimeras. Consequently, the BRC repeats were diluted to an appropriate concentration (as established by an initial, single-concentration point screen; SI Appendix, Table S3) for acquisition of a dose–response profile in microdroplets, representing individual titrations of the chimeras (SI Appendix, Fig. S7). The small amounts of peptides necessary to carry out droplet assays made it possible to screen close to saturating conditions (with the exception of the poorest binders), resulting in good-quality data, which could be fit to a competitive binding model (Fig. 2A and SI Appendix, Fig. S7) to derive dissociation constants for all 64 variants (Fig. 2B).
Fig. 2.
Affinity determination of 64 chimeric BRC4 repeats by a microfluidic droplet-on-demand system interfaced with fluorescence anisotropy detection. (A) Fraction of BRC4fl peptide bound to monomeric RAD51 as a function of GB1-BRC peptide chimera concentration for nine examples of the dataset of 64 combinations, measured using the droplet-on-demand anisotropy competition assay. Measurement conditions were 100 nM BRC4fl, 150 nM monomeric RAD51 in a buffer of 20 mM CHES (pH 9.5), 100 mM NaCl, 1 mM EDTA, at 20 °C. Note the starting binding fraction of 0.85 that is calculated from the affinity of the BRC4fl peptide for monomeric RAD51 and the initial concentrations used (50). (All 64 binding curves are shown separately elsewhere (SI Appendix, Fig. S7). (B) Kd values (in nM) determined for all 64 BRCA2 peptide chimeras using data in A. (C) Analysis of the effect of recombination, expressed as the difference in ΔG (SI Appendix, Table S4) of each shuffled variant relative to the average of the two natural parental combinations (ΔΔGparental, in kCal/mol) for each variant. The parental combinations are depicted in gray (by definition, their ΔΔGparental is always zero). The values indicated below each column and next to each row represent the average ΔΔGparental value for FxxA and LFDE modules from each repeat, respectively, and are referred to as ΔΔGFxxA1–8 and ΔΔGLFDE1–8, respectively. (D) Binding rank order of the parental BRC repeats (Central column) and the individual FxxA (Left column) and LFDE (Right column) modules comprising the repeats, indicated by intensity (low intensity = top binder, high intensity = poor binder, as indicated by scale bar). The contributions to binding of the eight different repeat-derived modules are calculated using their ΔΔGLFDE1–8 and ΔΔGFxxA1–8 values.
Shuffling Leads to BRC Peptide Binders with Enhanced Affinity over Wild Type.
The 64 measured Kd values for BRC peptide binding to monomeric RAD51 spanned a range of three orders of magnitude from 11 μM (BRC7-8) to 6 nM (BRC8-2). To the best of our knowledge, the affinities of most of the shuffled variants had never been measured before (except BRC4-5 and BRC5-4) (39), so it is useful to compare the values we measured here for the natural repeats (which can be read as a diagonal from the Top Left corner to the Bottom Right corner in Fig. 2B) to previously reported values. First, using this competitive fluorescence anisotropy assay, BRC4 was found to have a Kd of 38 nM for monomeric RAD51, slightly higher than the value previously determined by direct titration by fluorescence anisotropy (12 nM) (49) or by ITC in this study (11 nM; SI Appendix, Fig. S5 B and G). These values fall within the range of previously measured affinities (6.2 to 64 nM) for this protein–protein interaction (46). Also, BRC4 was found in this assay to be the tightest binder of all the natural repeats, in agreement with previous studies (36, 51). The repeats 1 through 4 displayed higher affinity (median Kd 198 nM) than repeats 5 through 8 (median Kd 1,394 nM). This observation is also consistent with previous reports that BRC repeats 1 through 4 have higher affinity than repeats 5 through 8 for RAD51 (36). Beyond this broad analysis, more detailed comparisons (including the absolute Kd values) to previous studies are of limited value as there are important differences in the assays employed and the binders titrated against, e.g., full-length, oligomerization-capable RAD51 (36, 52) or truncated, monomeric RAD51 consisting of the catalytic domain only (51).
Having established that the affinity ranking for the natural repeats was consistent with previous reports, we next analyzed the affinities we determined for the novel recombinant peptides. The combinations with the FxxA module of BRC5 were found to be the weakest binders, easily rationalized by the fact that the conserved alanine in the FxxA module of BRC5 is replaced by a serine (FYTS), which has a hydrophilic side chain that would not form favorable steric and hydrophobic contacts within the small Ala pocket. Interestingly, the recombinant peptide BRC4-5 was also found to be a relatively weak binder (Kd 2.1 μM), despite previous findings that this chimera displayed a stronger affinity (39). This paradox is addressed below. Remarkably, a few chimeras containing the FxxA module from BRC8, from the “weak” group of repeats 5 to 8, were found to be the strongest binders from the entire set of 64 variants (Fig. 2B) with BRC8-2 being the peptide with highest affinity with a Kd of 6 nM. This demonstrates how shuffling can lead to the bringing together of elements that are naturally in “nonoptimal” combination for highest affinity—though likely optimized for endogenous function and fitness—and result in a significantly improved binder.
Discerning Module-Specific Contributions to Binding Affinity and the Effect of the Crossover Point Placement.
To allow comparisons across repeats and modules, the dissociation constants were expressed in units of Gibbs free energy (ΔG; SI Appendix, Table S4). The effect of shuffling was quantified by expressing each of the 56 novel, unexplored combinations in terms of relative change vis-à-vis their two parental repeats, e.g., the two parents of BRC1-2 are BRC1 and BRC2 (Fig. 2C and SI Appendix, Supplementary Text 5.1). A positive value for ΔΔGparental—indicating that the product of the shuffling was detrimental to the binding function—was observed in 33 out 56 peptides. Also, the average ΔΔGparental for all 56 shuffled repeats was 0.16 kCal/mol, indicating that shuffling had a net-detrimental effect on binding function. Nevertheless, 23 out of these 56 repeats had negative ΔΔGparental values and thus represented variants that were improved over the average of their parents. We asked whether the identity of the FxxA module affected binding to monomeric RAD51 more strongly than the identity of the LFDE module. At a first approximation, the FxxA module might appear to have dominated the interaction as proven by the fact that any combination with imperfect FxxA module of BRC5 (containing the stretch of residues Ser1662BRC5 to Arg1677BRC5 and in which the alanine of the FxxA motif is replaced by a serine) resulted in exceedingly weak interactions (ΔΔGFxxA5 = 0.91 kCal/mol) (Fig. 2C). The LFDE modules from BRC3 and BRC8 were found to cause the most significant reduction to binding in each of their respective seven recombinant peptides (ΔΔGLFDE3 = 1.07 kCal/mol; ΔΔGLFDE8 = 1.05 kCal/mol). As both the FxxA and LFDE modules in each repeat could thus make a significant contribution to binding, we considered whether the net contribution to binding was equally distributed within each repeat. In BRC repeat 5, both the FxxA and LFDE modules conspired to make a weak binder (ΔΔGFxxA5 = 0.91 kCal/mol; ΔΔGLFDE5 = 0.33 kCal/mol). By contrast, BRC repeat 8 could be considered “Janus-faced,” as it is composed of a net contributor (ΔΔGFxxA8 = −0.96 kCal/mol) and a net disruptor (ΔΔGLFDE8 = 1.05 kCal/mol) to binding. To a slightly lesser degree, BRC repeat 2 displayed the same contrast in intrarepeat properties, although in this natural repeat, the FxxA module was a net disruptor overall (ΔΔGFxxA2 = 0.82 kCal/mol), while the LFDE module was a net contributor (ΔΔGLFDE2 = −1.15 kCal/mol). This analysis is validated by the observation that BRC8-2, which combines the overall best FxxA module with overall best LFDE module, is the BRC peptide with the highest affinity of all, which we were able to cross-validate by ITC measurements (SI Appendix, Fig. S5) and is also the most improved over its parental sequences (ΔΔGparental = −2.44 kCal/mol). The depiction of the rank affinity ordering of individual modules by intensity, based on their ΔΔGFxxA1-8 or ΔΔGLFDE1-8 values, next to the rank order of the natural repeats’ affinities (Fig. 2D), highlighted that within the eight natural BRC repeats, binding function was not always equally distributed between modules. It is also interesting to note that the two modules of BRC4, the highest affinity natural repeat, are relatively poor contributors of affinity and highly dependent on which module they pair with. In contrast, BRC6, the natural repeat with the lowest affinity in our analysis, contains modules that, in combination with other modules, contribute positively to affinity.
The observation of a ΔΔGparental of 1.24 kCal/mol (Fig. 2C) for BRC4-5 represented a notable discrepancy to previous data, obtained by competitive ELISA with synthetic chimeric peptides BRC4-5 and 5-4 (39). As expected, in agreement with our findings, BRC5-4 turned out to be a weak binder, due to the lack of conservation in repeat 5’s FxxA module. However, Rajendra and Venkitaraman (39) found BRC4-5 to be a stronger binder than the natural BRC4, whereas we found BRC4-5 to bind monomeric RAD51 with 55-fold lower affinity than BRC4. What could explain this? Apart from the obvious difference in the assays (heterogeneous ELISA-based assay vs. homogeneous polarization-based assay), the main remaining difference is the cutoff between the end of the FxxA module and the start of the LFDE module in the shuffled peptide. While Rajendra and Venkitaraman (39) defined Lys1533BRC4 as the last residue of the FxxA module and Ile1534BRC4 as the first of the LFDE module, our peptides were based on the cutoff point occurring between Lys1530BRC4 and Lys1531BRC4. The cutoff is arbitrary but resulted in our chimeric BRC4-5 repeat bearing two mutations compared to the other study, Val1532BRC4 → Thr1679BRC5 and Lys1533BRC4 → Ser1680BRC5. Retrospective analysis using SCHEMA computational algorithm to identify the optimal crossover points in a protein sequence (23) found a crossover point between Ile1534BRC4 and Ala1535BRC4 to be optimal (SI Appendix, Fig. S8 and SI Appendix, Supplementary Text 5.2). A cutoff point more distant from the FxxA hairpin results in fewer pairwise interactions (i.e., between residues from the N-terminal half with residues from the C-terminal half) being broken upon shuffling. Furthermore, we found that the deletion of Lys1530BRC4, located at our chosen crossover point, resulted in a significant loss of affinity (the Kd shifted almost 200-fold, from 21 nM to 4.1 µM; SI Appendix, Fig. S9 and SI Appendix, Supplementary Text 5.3). Thus, subtle differences in the linker region can lead to dramatic differences in affinity, explaining the effect of the exact placement of shuffle cutoff points to the relative contribution of the two modules.
BRC8-2 Forms a More Extensive β-Hairpin and Has Improved Helicity Compared to BRC4.
To gain structural insight into the increased affinity of BRC8-2 for RAD51, we determined the crystal structure of the monomeric RAD51:BRC8-2 complex at 1.95-Å resolution (Protein Data Bank [PDB] ID: 6HQU). There are eight complexes in the asymmetric unit of these crystals, all of which are very similar to each other with an average rmsd of 0.664 Å for 198-Cα atoms of RAD51. The bound BRC8-2 peptide is visible in seven of the eight RAD51 molecules, in essentially identical conformation in all complexes. Representative electron densities before the BRC8-2 have been modeled and after final refinement are provided in SI Appendix, Fig. S10. The best-defined complex (chains B and J) has been used in the subsequent analyses.
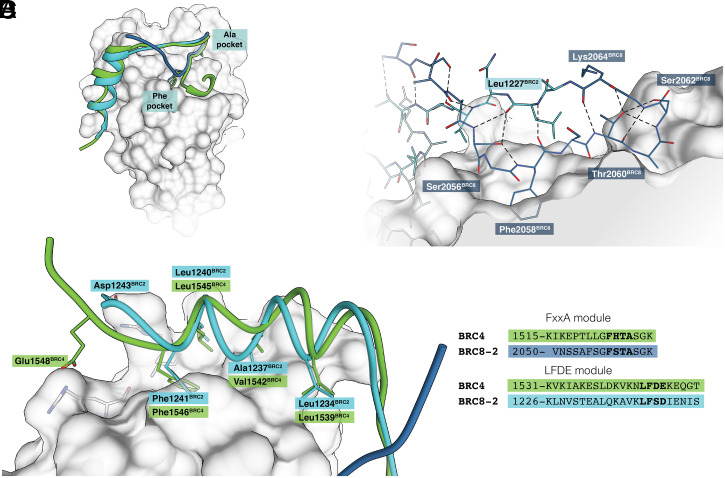
Comparison of the refined structure for monomeric RAD51:BRC8-2 with that of the RAD51:BRC4 complex (PDB: 1n0w) (38) shows a similar overall topology (Fig. 3A). Phe2058BRC8 and Ala2061BRC8 of the BRC8 FxxA module form identical contacts to those seen between BRC4 and RAD51. C-terminal to Ala2061BRC8, the peptide forms a β-hairpin that extends the central β-sheet of monomeric RAD51 in an intermolecular fashion, reminiscent of the RAD51:BRC4 complex (37, 38). Five residues at the C-terminal ends of the BRC8 and BRC4 FxxA modules are identical in sequence (TASGK) and both hairpins are stabilized by the hydroxyl groups of Thr2060BRC8/Thr1526BRC4 forming hydrogen bonds with the backbone amine of Lys2064BRC8/Lys1530BRC4 and the hydroxyl of Ser2062BRC8/Ser1528BRC4 (Fig. 2B). In the RAD51:BRC4 complex, the C-terminal LFDE module forms a 10-residue α-helix that interacts with RAD51 through a shallow interface using a mixture of hydrophobic and polar contacts. In the BRC2 LFDE module (with sequence LFSD) Leu1240BRC2 and Phe1241BRC2 bind the same hydrophobic interface that BRC4 interacts with and Asp1243BRC2 interacts with a nearby Arg270monomeric RAD51 as seen in BRC4 (Fig. 3C). Contact areas calculated for the BRC4 and BRC8-2 complexes are very similar, 1,042 and 942 Å2, respectively, consistent with the previously noted weak correlation between buried surface area and binding affinity (53).
Fig. 3.
Crystal structure of the monomeric RAD51:BRC8-2 complex compared with RAD51:BRC4. BRC8-2 is depicted in dark and light blue, corresponding to BRC8 and BRC2 sequences, respectively. BRC4 is shown in green. Peptides were superimposed by aligning the structures of their respective protein-binding partners. Monomeric RAD51 is represented by a gray surface. Selected residues of the monomeric RAD51 are depicted in gray. (A) Overall topologies of the two peptides, with the Phe and Ala pockets of the FxxA site shown. (B) Hydrogen-bonding network of the BRC8-2 β-hairpin. (C) LFDE interface with side chains of crucial residues depicted. (D) Sequence alignment of BRC4 and BRC8-2 FxxA and LFDE modules.
The most significant difference between the BRC4 and BRC8-2 peptides is the extent of the intramolecular hydrogen-bonding network that forms the β-hairpin in the FxxA module. In BRC8-2, the β-hairpin is significantly extended, with its N-terminal end, before Phe2058BRC8, folding back toward the rest of the peptide (Fig. 2B). The hairpin extends a total of 19 amino acids, from Ser2053BRC8 to Thr1231BRC2 and is significantly longer than the nine-residue hairpin in BRC4. Formation of the extended hairpin is enabled by Ser2056BRC8, whose side chain fits tightly between the two antiparallel strands of the peptide and the surface of RAD51, hydrogen bonding with the carbonyl of Leu1227BRC2, and the backbone amide of Phe2058BRC8. This allows the peptide to fold back on itself and to form an extended intramolecular H-bonding network (Fig. 3B). BRC4 has a larger, hydrophobic Leu1522 in the equivalent position of Ser2056BRC8, which cannot satisfy the steric and electrostatic requirements of the topology we observe in BRC8-2, forcing the N terminus of the peptide to point away from the β-hairpin and the rest of the peptide. Interestingly, the FxxA module of BRC3 contains a threonine in the position equivalent to Ser2056 and has the second highest ΔΔGFxxA1-8 (ΔΔGFxxA3 = −0.35 kCal/mol). BRC3 is likely to form a similar extended hairpin to BRC8, with its threonine forming equivalent hydrogen bonds to Ser2056. To examine the contribution of Ser2056 to binding, we designed a mutant repeat BRC8-2(S2056A). Its affinity for monomeric RAD51 was measured by ITC as a Kd of 5 nM, i.e., fivefold lower affinity than that measured for BRC8-2 by ITC, confirming the significance of Ser2056 for binding (SI Appendix, Fig. S5 E and F).
The binding modes of the LFDE module of BRC8-2 also shows subtle differences compared to BRC4 (Fig. 2C). In BRC4, the bulky side chain of interface-forming Val1542BRC4 pushes the peptide away from RAD51, forming an outward-facing bulge and disrupting the optimal helical geometry of the peptide. The equivalent residue in BRC8-2, Ala1237BRC2, is smaller and allows the α-helix to form a closer interaction with monomeric RAD51 and to retain a more regular helical geometry. Thus, the increased binding affinity of the BRC8-2 repeat for monomeric RAD51 appears to result from the extended hydrogen-bonding network of the FxxA module of BRC8 and the improved packing and helical geometry of the LFDE module from BRC2.
BRC8-2 Can Disrupt RAD51 Function.
To validate the utility of improved binding of BRC8-2 binding to RAD51 for biological intervention, we used two functional tests. We first evaluated whether BRC8-2 can modulate RAD51 interaction with single-stranded DNA (ssDNA), which is dependent on oligomerization of RAD51. Using an electrophoretic mobility shift assay (EMSA) with the fluorescein-labeled dT60 oligonucleotide as a substrate, we can show breakdown of nucleofilaments when BRC8-2 or BRC4 disrupts the RAD51:ssDNA complex (SI Appendix, Fig. S11). Due to high concentrations of protein needed in this assay, it is not possible to evaluate differences between BRC4 and BRC8-2 quantitatively, yet similar concentration dependencies confirm that the shuffled repeat has retained its ability to disrupt the RAD51:ssDNA filament.
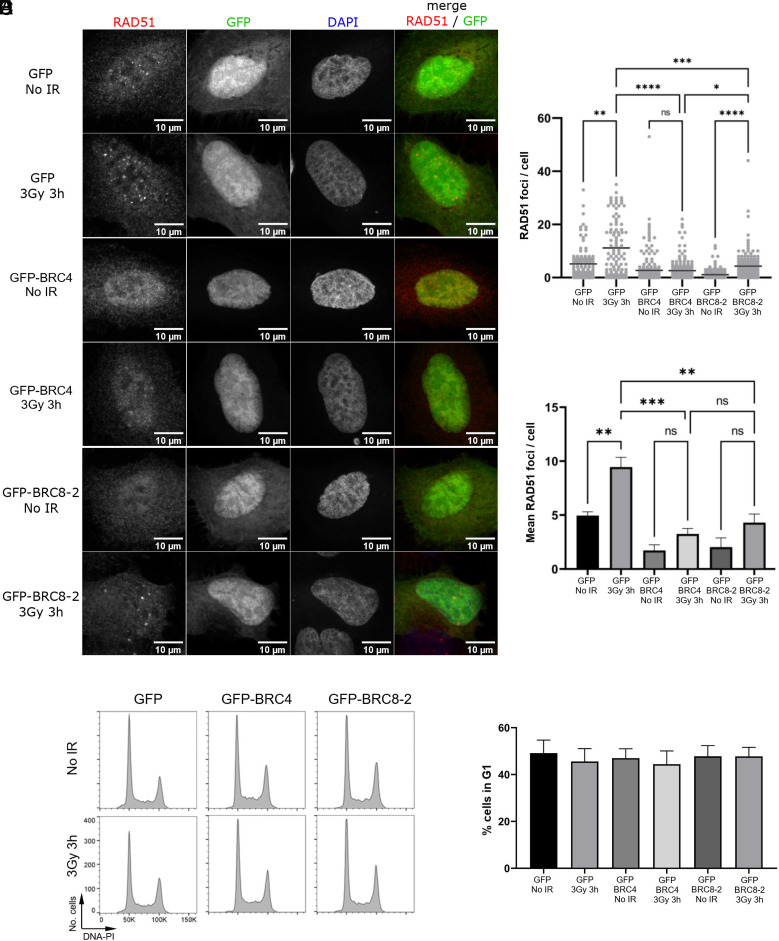
Having confirmed this, we investigated the ability of this peptide to disrupt RAD51 function in human cells. Following treatment with ionizing radiation (IR), RAD51 translocates to the sites of DNA damage and forms foci that are visible by immunofluorescence. The formation of such foci is dependent on BRCA2 (54), and it has previously been shown that foci formation can be disrupted by expression of native BRC repeats (55, 56).
To determine whether BRC8-2 impaired RAD51 foci formation, we generated construct GFP-NLS-BRC8-2, in which the peptide was fused to a green fluorescent protein (GFP) containing a nuclear localization signal (NLS) and transfected this construct alongside the negative control GFP-NLS construct into U2OS osteosarcoma cells. As a positive control, we also transfected cells with a construct expressing a GFP-NLS-BRC4 peptide. RAD51 foci formation was monitored in GFP-positive cells after exposure to IR or in the absence of any treatment (Fig. 4A). The control GFP-NLS cells showed the expected increase in the mean number of RAD51 foci after irradiation (IR, 3 Gy) (Fig. 4 B and C). In addition, a small number of foci were present in the absence of IR, most likely reflecting homologous recombination events associated with replicative stress. In contrast, cells expressing either the GFP-NLS-BRC8-2 or the GFP-NLS-BRC4 constructs had fewer RAD51 foci both prior to irradiation and following IR exposure (Fig. 4 B and C), suggesting that the BRC8-2 peptide is able to interfere with RAD51 foci formation in cells. Because RAD51 foci formation is limited to the S and G2 phases of the cell cycle, we wanted to determine whether the reduction in foci formation was due to an indirect effect via increased G1 phase cells in the GFP-NLS-BRC8-2–expressing cell population. We therefore monitored the cell cycle profile of GFP-expressing cells by flow cytometry and found that there were no significant differences between the GFP-, GFP-BRC4, and GFP-BRC8-2 cell populations, indicating that the effect on RAD51 foci formation is not due to cell cycle alterations (Fig. 4 D and E). While it is difficult to compare the extent of interference with RAD51 foci formation due to variability in expression levels and transfection efficiency, these data demonstrate that BRC8-2, like BRC4, is capable of disrupting RAD51 foci formation in human cells through binding and sequestering RAD51 away from sites of DNA damage. In support of this, we also noted that the pan-nuclear signal of RAD51 in both GFP-NLS-BRC8-2– and GFP-NLS-BRC4–expressing cells is greater than in the GFP-NLS control cells (SI Appendix, Fig. S12). Taken together, these data support the conclusion that the BRC8-2 chimera is functional in a physiological context.
Fig. 4.
BRC8-2 impairs RAD51 foci formation in human U2OS cells. (A) Representative images of U2OS cells expressing GFP-NLS, that is GFP with a nuclear location signal only (GFP), a GFP-NLS-BRC8-2 peptide (GFP-BRC8-2), or a GFP-NLS-BRC4 peptide (GFP-BRC4). Cells were monitored 3 h after no treatment (No IR) or irradiation with 3 Gy (3Gy 3h) for GFP fluorescence or stained with RAD51 or DAPI as indicated. (B) Dot plot graph from one biological replicate plotting the number of RAD51 foci per GFP-positive cell. Mean values for each population are indicated with a bar. More than 50 GFP-positive cells were analyzed for each condition. Statistical analysis was done using Kruskal–Wallis rank sum test followed by Dunn’s procedure for pairwise comparison (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant). (C) Bar graph showing the average of the mean RAD51 foci per GFP-positive cell from four independent biological experiments. Data are presented as mean ± SEM, n = 4 biological repeats. **P < 0.01, ***P < 0.001; ns, not significant, using ANOVA test [F(5,15) = 15.29, P < 0.0001], assuming sphericity) followed by Tukey’s method. (D) Representative cell cycle profiles from GFP-positive cells transfected with GFP-NLS (GFP) or GFP-NLS-BRC4 (GFP-BRC4) or GFP-NLS-BRC8-2 (GFP-BRC8-2). Cells were analyzed by flow cytometry 3 h after no treatment (No IR) or irradiation with 3 Gy (3Gy 3h). PI, propidium iodide. (E) Bar graph showing the percentage of cells in G1 phase. Data are the mean values from three independent biological experiments ± SD.
Discussion
Shuffling of BRC Modules: Anciently Diverged Parts Meet.
In this work we successfully exploited a droplet-on-demand platform to comprehensively survey the effect of shuffling of the BRC modules. This approach revealed BRC-repeat combinations with unprecedented affinity for monomeric RAD51 and demonstrated in cellula activity. Due to our systematic analysis of the contribution of each module to binding, as well as our crystallographic study of the monomeric RAD51:BRC8-2 complex, we shed light on structure–function relationships within the BRCA2:RAD51 complex, a highly dynamic and context-specific protein–protein interaction: although repeats 1 to 4 were found to bind free RAD51 more strongly than repeats 6 to 8, by us and others, repeats 5 to 8 were reported to have higher binding affinity for the RAD51:ssDNA complex than repeats 1 to 4 (36, 57). Repeats 5 to 8 may also bind in concert to stimulate certain RAD51 functions (58).
If combinations of modules achieve a wide range of affinities, their assembly context may matter (in addition to intrinsic effects of each module), pointing to cooperative effects of the different modules. We found that both the FxxA and LFDE modules of the eight BRC repeats make relevant contributions to the binding of monomeric RAD51, as evidenced by both FxxA and LFDE being associated with net disruption of binding function upon shuffling with other modules. This is also consistent with the finding by previous studies that the FxxA module in isolation (i.e., lacking the LFDE module) has only a modest affinity for RAD51 (39, 48, 56). Interestingly, we found that both modules often failed to act in concert to give high-affinity binders within the natural repeats.
Our shuffling approach helps to pick apart the role played by both modules in their various repeats, allowing us to discern module-specific effects that are otherwise obscured when measured in their parental combinations. The BRC repeats provide a fascinating example of how Nature can exploit modularity to fine tune function. By mixing variant FxxA β-hairpins and LFDE α-helices, a range of affinities (spanning beyond the magnitude of the natural BRC repeats) was achieved, without the need to resort to entirely novel sequences.
Structural Insight into Beneficial Effect of Shuffling and Scope for Future Work.
The crystal structure of the RAD51:BRC8-2 complex has provided further insight into the BRC-repeat binding to RAD51, in particular in identification of the extended β-hairpin formed by the FxxA module and the critical role of Ser2056BRC8 in facilitating the formation of this structure. The enhanced affinity of BRC8-2 may render it an attractive tool in studies that seek to investigate the effect of disrupting the RAD51:BRCA2 interaction using cell penetrating peptide derivatives of BRC repeats (56, 59). We were able to demonstrate the utility of BRC8-2 in a functional cellular assay for disruption of radiation-induced RAD51 foci formation. Strategies to further stabilize this β-hairpin may involve the use of tryptophan–tryptophan cross-strand pairs (60), or even artificial crosslinks such as triazole (61), to achieve an additional enhancement of binding through a reduction in conformational heterogeneity prior to complex formation (as well as improving resistance to proteolytic degradation). Our work revealed that the linker between the FxxA module and LFDE module is also likely to play a role in determining the affinity of the peptide. Our data on the importance of the linker region is further corroborated by a study showing that mutating the wild-type Val1532BRC4 to Ile or Phe results in respectively enhanced and diminished binding of BRC4 peptide to RAD51 (62). Several studies have investigated the effect of point mutations within the modules, that future studies may be able to combine with our shuffling approach. Nomme et al. succeeded, through docking BRC motifs in complex with RAD51, in generating a BRC4 repeat peptide mutant that was 10 times more efficient in inhibiting the RAD51:ssDNA complex than the original BRC4-repeat peptide itself (48). Similarly, Cole et al. calculated molecular mechanics energies combined with the Poisson–Boltzmann surface area continuum solvation (MM-PBSA) to successfully identify the BRC-repeat binding hotspots as well as enabling an accurate prediction of relative binding-free energies of the natural BRC repeats (51). Scott et al. probed the contribution of individual residues in the FxxA epitope identifying changes that resulted in increased affinity toward RAD51 (63) (by up to ΔΔG = −0.64 kCal/mol).
In this study we exploited a shuffling library of natural repeats only, successfully obtaining both insight into relative contributions of the different modules and identification of affinity-enhanced variants. Equally importantly, all 64 chimeras are functional binders, validating the repeat unit as a building block and allowing comprehensive functional data to be harvested. The biophysical analysis of the 64 chimeras provides an improved quantitative understanding of the modular contributions to binding made by FxxA and LFDE, so that structure–activity relationships can be drawn up. These efforts are partially motivated by the therapeutic potential of blocking the BRCA2–RAD51 interaction. Unlike typical computational approaches, the affinity enhancement achieved by our shuffling approach requires no a priori knowledge of the binding mechanism. Guided by the fast and reagent-saving evaluation of Kd values in microfluidic droplets, empirical models can be developed that yield novel insights into binding mechanism, and chimeras with improved affinity for use in various diagnostic and therapeutic applications can be obtained. Our crystal structure of a shuffled repeat in complex with monomeric RAD51 allowed detailed investigation of its binding mechanism, and the juxtaposition of functional and structural data will help future efforts at developing BRC peptides with enhanced RAD51 affinity. This work combines rational and combinatorial engineering productively and illustrates a general strategy: knowledge of functional units of proteins that are autonomously folded and functional, bypasses the need to design proteins from scratch, while their shuffling reduces the library complexity vastly, compared to the total sequence randomization typical in directed evolution approaches. Defining such functional modules in chimeragenesis experiments will provide the basis for more sophisticated libraries created by module shuffling, to ultimately reach the goal of eliciting functional proteins more quickly. The approach exemplified for BRC repeats may be taken up in the future for enhancing interactions between combinations of linear motifs (i.e., modules equivalent to the BRC repeats) (64) and corresponding binding domains (e.g., SH3). At a binary level these interactions are typically of low affinity, but multivalent interactions can target longer segments in a protein-binding partner and therefore bind with higher affinity (65). Multivalent interactions will be entropically favored and lead to increased avidity, thus enhancing affinities and specificities of molecular recognition. Examples where multivalent interactions between a polypeptide with several short motifs bind with high affinity to several binding modules in another protein include tandem SH2 domains in ZAP-70 (66), tandem SH3 domains of CAP (67), and dimeric 14-3-3 proteins (68). This framework for de- and reconstruction of binding interfaces, by generating libraries of shuffled binding modules to give chimeras, may become a route to identifying optimal combinations of binding epitopes for protein–protein interactions, to generate biochemical tools in a mix-and-match approach starting with proteins composed of modular units.
Methods
Reagents.
Preparation of BRC4 repeat peptide, N-terminally labeled with fluorescein (BRC4fl, sequence CKEPTLLGFHTASGKKVKIAKESLDKVKNLFDEKEQ) was described previously (49). CHES was from Sigma, Pico-Surf 1 was from Dolomite, HFE-7500 was from 3M.
Plasmid Constructs and Cloning.
For the construction of 8 parental and 56 shuffled BRC peptides, as well as several more mutant peptides, please see SI Appendix, Figs. S1 and S2 and Table S1. The Escherichia coli expression construct for monomeric RAD51 (pBAT4-HumRadA22), has been described previously (46). Cloning of plasmids GFP-NLS, GFP-NLS-BRC8-2, and GFP-NLS-BRC4 for mammalian cell transfection is described in SI Appendix, Fig. S3 and Table S2. All plasmids used in this study may be requested from the corresponding authors.
Protein Expression and Purification.
The 64 different GB1-BRC peptide fusion constructs were separately transformed to chemically competent E. coli BL21(DE3). Overnight Luria-Bertani (LB) broth precultures were used to inoculate 20 mL LB broth, which were grown up to midlog phase (OD600 of 0.5). Expression was induced using 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) and cultures were incubated for a further 3 h at 37 °C. Cells were then harvested through centrifugation and lysed by the addition of BugBuster/Benzonase lysis reagent (Novagen, with 5 mM imidazole, 20 mM Tris⋅HCl, 100 mM NaCl, pH 8). The resuspended mixture was incubated for 20 min at room temperature, then loaded directly onto a Ni-NTA protein miniprep column (His Spin Protein Miniprep, Zymo Research). Protein was washed following the manufacturer’s instructions. Proteins were eluted in 500 mM imidazole, 20 mM Tris⋅HCl, 100 mM NaCl, pH 8, 150 µL. Protein concentrations were quantified by ultraviolet (UV) absorption at 280 nm (using a Nanodrop spectrophotometer) and corrected for the presence of truncated side products by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (SI Appendix, Fig. S4). Monomeric RAD51 expression and purification for fluorescence anisotropy measurements was carried out as described previously (46), where monomeric RAD51 was called “HumRadA22.” Full-length HsRAD51 (used for the EMSA assay in SI Appendix, Fig. S11) was prepared based on a protocol described previously (46, 69). Briefly, RAD51 was coexpressed with a BRC4 sequence fused to an N-terminal His-MBP tag in BL21(DE3) Rosetta2 E. coli strain. After initial purification of the His-MBP-BRC4:RAD51 complex on Ni-NTA resin, RAD51 was separated from the His-MBP-BRC4 fusion protein using heparin Sepharose resin. It was then further purified by size exclusion chromatography, concentrated, and flash frozen for storage at −80 °C.
Monomeric RAD51:BRC8-2 Complex Purification for Crystallography.
E. coli BL21(DE3) cells carrying pUBS520 plasmid for rare AGA/AGG encoding tRNA were transformed with GB1-BRC8-2 or monomeric RAD51 constructs and grown at 37 °C in 1 L of 2× YT medium in shaker flasks in the presence of 100 μg/mL ampicillin and 25 μg/mL kanamycin until OD600 of 0.8. Expression was induced with 0.4 mM IPTG for 3 h. Cells were resuspended in 25 mL of 50 mM Tris⋅HCl (pH = 8.0), 100 mM NaCl, 20 mM imidazole and lysed on an Emulsiflex C5 homogenizer (Avestin). Cell lysate was centrifuged at 15,000 × g for 30 min and supernatant collected. GB1-BRC8-2 lysate was loaded on a 3-mL Ni-NTA agarose matrix (Cube Biotech), followed by the application of monomeric RAD51 lysate. Column matrix was washed with five column volumes 50 mM Tris⋅HCl pH 8.0, 100 mM NaCl, 20 mM imidazole. Complex was eluted with 50 mM Tris⋅HCl pH 8.0, 100 mM NaCl, 200 mM imidazole into 2-mL fractions. Fractions containing the proteins of interest were pooled and incubated with 100 μL of 2 mg/mL tobacco etch virus protease overnight at 4 °C. Cleaved GB1 fusion partner was removed from the solution by a second Ni-NTA affinity step, collecting the flow through which contains the monomeric RAD51:BRC8-2 complex. Flow through was concentrated on a centrifugal filter (Amicon, 3000 Da molecular weight cut-off) to 2 mL volume and loaded into a Superdex 75 16/60 prep grade size exclusion column (GE Lifesciences), previously equilibrated with 20 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES) pH 9.5, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA). The complex was eluted at 75 mL, the fractions containing the complex were pooled, and the complex was concentrated to 0.45 mM.
Fluorescence Polarization Competition Assay and Microfluidic-Based Measurements.
FP was measured in nanoliter droplets produced and analyzed in microfluidic devices to quantify bound vs. unbound RAD51:BRC complexes and derive Kd values, essentially as described previously (49). Briefly, nanoliter droplets were generated from a well in which an increasing amount of peptide is allowed to compete with BRC4fl. A four-channel parallelized device was used for all measurements to increase throughput. A 10× objective was used and the power of the 488-nm diode laser was 50 mW. BRC peptides were preloaded into polyethylene (PTE) tubing (internal Ø, 0.38 mm) to avoid cleaning syringes between runs. To this end, 40 µL of each peptide was aspirated in tubing, followed a plug of 10 µL of HFE-7500 and 0.5% Pico-Surf 1 (Sphere Fluidics). Typically, four peptides were preloaded in each tubing, so that 16 samples could be screened at a go. All measurements were performed in CHES buffer pH 9.5, 1% bovine serum albumin (BSA) with HFE-7500 oil and 0.5% Pico-Surf 1 surfactant as carrier phase. Flow rates were 3 μL/min for withdrawal and 40 μL/min for 30 s for peptide injection. Data were fit to a competitive binding model using 12 nM for the Kd of the monomeric RAD51:BRC4fl interaction (49). The concentrations for each droplet in a given titration were calculated from fluidic parameters and stock concentrations of injected peptides as described previously (49).
Crystallography of Monomeric RAD51:BRC8-2 Complex.
Monomeric RAD51:BRC8-2 complex was crystallized using sitting-drop vapor diffusion in a 96-well MRC plate format. A total of 40 mM adenosine diphosphate (ADP)/Mg2+ water solution was added to 0.45 mM complex in a 1:9 ratio. A total of 200 nL of the complex was then mixed with 200 nL of the crystallization condition using a Mosquito liquid handling robot (TTP Labtech). Crystals were observed in 0.2 M NH4Cl, 20% (wt/vol) polyethylene glycol 3350 and used directly for data collection without the need for further optimization of the crystallization conditions.
A crystal was cryocooled in liquid nitrogen without the application of a cryoprotectant, and diffraction data were collected at Diamond Light Source (Harwell, UK) synchrotron radiation source. Images were processed with autoPROC (70). Molecular replacement phasing method was used with unliganded monomeric RAD51 structure (HumRadA22, PDB: 5KDD) as a search model. The structure was refined without peptide first and the peptide was built into the clearly visible electron density manually (SI Appendix, Fig. S10). Manual real-space refinement was done in Coot (71) and automated refinement with phenix.refine (72) and autoBUSTER (73). Crystallographic data and refinement statistics are shown in SI Appendix, Table S5. The final model contains seven complexes of monomeric RAD51 complexed with BRC8-2 peptide and one monomeric RAD51 chain with no peptide (chain H). Chain H has poorly defined electron density, which is likely caused by the lower number of crystal contacts it makes compared to other monomeric RAD51 molecules in the asymmetric unit. Individual atomic B factors were not refined for chain H. The protein structure is fully defined in all of the complexes, but the peptide density had more variable quality. Chains B and J represent the best-defined monomeric RAD51:BRC8-2 complex and were used in the analysis. The coordinates and corresponding structure factors have been deposited to the PDB under accession no. 6HQU. Contact area within the complex of both monomeric RAD51:BRC8-2 (6HQU) and RAD51:BRC4 (1n0w) was calculated (for atoms within 3.9-Å distance of atoms of the other binding partner) using a Pymol script written by Martin Christen (contact_surface v.3.0, available at https://pymolwiki.org/index.php/Contact_Surface). The script was adapted for Python3 using the 2to3 program (https://docs.python.org/2/library/2to3.html).
Cell Line.
U2OS cell line (ATCC, HTB-96) was grown in Dulbecco's modified Eagle medium (DMEM) supplement with 10% fetal bovine serum (FBS) (Gibco, 11573397) and 100 U/mL penicillin/streptomycin (Gibco, 15140122) at 37 °C and 5% CO2.
Transfection and Cell Treatment.
Cells were transfected using Lipofectamine 3000 Transfection Reagent (Invitrogen) following manufacture’s protocol. Plasmid DNA and transfection reagent amounts were scaled for a 10-cm dish: 4 μg DNA and 7.75 μL Lipofectamine 3000.
Approximately 18 h after transfection, cells were either exposed to 3 Gy caesium-137 g-irradiation (GammaCell 1000, Atomic Energy of Canada, Ltd) or unirradiated and allowed to recover for 3 h before being collected for analysis.
Immunostaining.
Coverslips were washed twice in phosphate-buffered saline (PBS) before fixation with 4% paraformaldehyde/PBS for 15 min, then washed three times in PBS and permeabilized in 0.5% Triton-X/PBS for 7 min. Coverslips were washed three times in PBS, blocked for at least 30 min in 1% BSA-Fraction V (A3059-50G, Sigma-Aldrich)/PBS, and followed by a 1-h incubation at room temperature with RAD51 (RAD51 H-92, sc-8349, Santa Cruz) primary antibody diluted 1:100 in 1% BSA-Fraction V/PBS. The coverslips were washed three times with PBS, then incubated with anti-rabbit Alexa Fluor 647 secondary antibody (A21244, Invitrogen) diluted 1:500 in 1% BSA-Fraction V/PBS for 45 min in the dark at room temperature. Coverslips were wash three times in PBS, mounted onto slides using ProLong Gold Antifade Mountant with DAPI (P36941, Invitrogen), and stored at 4 °C for further analysis.
Cells were visualized using a Nikon Eclipse e-400 microscope with 60× objective. Images were processed and analyzed for GFP signal and RAD51 foci using CellProfiler 4.0.6, and statistical analysis was performed using GraphPad Prism 9.1.0.
Flow Cytometry.
Cells were trypsinized, washed twice in PBS, and fixed by gently vortexing while adding 1 mL ice-cold 70% ethanol dropwise. Samples were stored for a minimum of 12 h at −20 °C. Prior to flow cytometry analysis, cells were spun down, washed twice in PBS, and resuspended in around 0.5 mL staining solution (5 μg/mL propidium iodide [P3566, Thermo Fisher Scientific], 100 μg/mL RNase A [R5503-100MG, Sigma-Aldrich] in PBS) and incubated for at least 30 min in the dark at room temperature. Cells were analyzed on a BD FACSymphony A3 Flow Cytometer (BD Biosciences) and cell cycle profiles were generated after gating GFP-positive cells using FlowJo v10.7.1 software. A detailed description of the gating strategy is found in SI Appendix, Fig. S13. Around 10,000 GFP-positive cells were analyzed per condition and experiment.
Supplementary Material
Acknowledgments
We thank Diamond Light Source for access to macromolecular crystallography beam line i03 (proposal mx18548) and for the data that contributed to these results. We are also grateful for access to and support by the X-Ray Crystallographic and Biophysical Research Facilities at the Department of Biochemistry, University of Cambridge. This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) (BB/K013629/1) and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement 695669). L.H.L. received a Marie Curie individual fellowship from the European Commission (grant 659029), M.B. received a fellowship from the Schweizerischer Nationalfonds, T.P. received a studentship from the Medical Research Council Doctoral Training Partnership, and F.H. is an ERC Advanced Investigator (grant 695669). P.Z.-V. and J.A.D. were supported by Cancer Research UK (C7905/A25715). C.F.K. acknowledges funding from the UK Engineering and Physical Sciences Research Council (grants EP/L015889/1 and EP/H018301/1), the Wellcome Trust (grants 3-3249/Z/16/Z and 089703/Z/09/Z), and the UK MRC (grants MR/K015850/1 and MR/K02292X/1).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017708118/-/DCSupplemental.
Data Availability
SI Appendix contains detailed descriptions of the cloning of bacterial expression constructs for the 64 shuffled BRC peptide variants, cloning of mammalian expression constructs, and notes on the soluble expression of the shuffled BRC peptide variants. Also included is a description of ITC used to cross-validate the microfluidic measurements, single concentration point measurements carried out with microfluidics, and exemplary titrations carried out by microfluidics. The fluorescence anisotropy data obtained for the 64 separate titrations as well as the Matlab script used in the analysis have been uploaded as separate files. The supplementary data also contain an analysis on the effect of shuffling of BRC peptides and in particular on the effect of the exact shuffle cutoff point placement. X-ray crystallography electron density map images, data collection, and refinement statistics are also to be found in SI Appendix. Additional cell images highlighting the pan-nuclear signal of RAD51 are also included in SI Appendix. The coordinates and corresponding structure factors for the monomeric RAD51:BRC8-2 complex have been deposited to the PDB under accession code 6HQU. As described previously (49), the transformation from intensity maps into anisotropy values from image data was carried out with a custom Matlab code available on GitHub (https://github.com/quantitativeimaging/icetropy). A custom Matlab script used to fit Kd values for the unlabeled competitive GB1-BRC peptides can be found in SI Appendix, Datasets S1–S4. All other study data are included in the article and/or supporting information.
References
- 1.Söding J., Lupas A. N., More than the sum of their parts: On the evolution of proteins from peptides. BioEssays 25, 837–846 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Moore A. D., Björklund Å. K., Ekman D., Bornberg-Bauer E., Elofsson A., Arrangements in the modular evolution of proteins. Trends Biochem. Sci. 33, 444–451 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Bornberg-Bauer E., Albà M. M., Dynamics and adaptive benefits of modular protein evolution. Curr. Opin. Struct. Biol. 23, 459–466 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Maervoet V. E. T., Briers Y., Synthetic biology of modular proteins. Bioengineered 8, 196–202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scaiewicz A., Levitt M., Unique function words characterize genomic proteins. Proc. Natl. Acad. Sci. U.S.A. 115, 6703–6708 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alva V., Lupas A. N., From ancestral peptides to designed proteins. Curr. Opin. Struct. Biol. 48, 103–109 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Kolodny R., Nepomnyachiy S., Tawfik D. S., Ben-Tal N., Bridging themes: Short protein segments found in different architectures. Mol. Biol. Evol. 38, 2191–2208 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolodny R., Searching protein space for ancient sub-domain segments. Curr. Opin. Struct. Biol. 68, 105–112 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Schüler A., Bornberg-Bauer E., Evolution of protein domain repeats in metazoa. Mol. Biol. Evol. 33, 3170–3182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel C., Berzuini C., Bashton M., Gough J., Teichmann S. A., Supra-domains: Evolutionary units larger than single protein domains. J. Mol. Biol. 336, 809–823 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Lapenta F., Jerala R., Design of novel protein building modules and modular architectures. Curr. Opin. Struct. Biol. 63, 90–96 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Jacobs T. M., et al. , Design of structurally distinct proteins using strategies inspired by evolution. Science 352, 687–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunette T. J., et al. , Exploring the repeat protein universe through computational protein design. Nature 528, 580–584 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunette T. J., et al. , Modular repeat protein sculpting using rigid helical junctions. Proc. Natl. Acad. Sci. U.S.A. 117, 8870–8875 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gidley F., Parmeggiani F., Repeat proteins: Designing new shapes and functions for solenoid folds. Curr. Opin. Struct. Biol. 68, 208–214 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Plückthun A., Designed ankyrin repeat proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 55, 489–511 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Pellegrini M., Tandem repeats in proteins: Prediction algorithms and biological role. Front. Bioeng. Biotechnol. 3, 143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Söding J., Remmert M., Biegert A., HHrep: De novo protein repeat detection and the origin of TIM barrels. Nucleic Acids Res. 34, W137–W142 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero-Romero S., Kordes S., Michel F., Höcker B., Evolution, folding, and design of TIM barrels and related proteins. Curr. Opin. Struct. Biol. 68, 94–104 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stemmer W. P. C., Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Crameri A., Raillard S. A., Bermudez E., Stemmer W. P., DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391, 288–291 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Christians F. C., Scapozza L., Crameri A., Folkers G., Stemmer W. P., Directed evolution of thymidine kinase for AZT phosphorylation using DNA family shuffling. Nat. Biotechnol. 17, 259–264 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Voigt C. A., Martinez C., Wang Z.-G., Mayo S. L., Arnold F. H., Protein building blocks preserved by recombination. Nat. Struct. Biol. 9, 553–558 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Bedbrook C. N., et al. , Structure-guided SCHEMA recombination generates diverse chimeric channelrhodopsins. Proc. Natl. Acad. Sci. U.S.A. 114, E2624–E2633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolkman J. A., Stemmer W. P. C., Directed evolution of proteins by exon shuffling. Nat. Biotechnol. 19, 423–428 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Yang H., Long M., Li L., Dean A. M., Evolution of enzymatic activities of testis-specific short-chain dehydrogenase/reductase in Drosophila. J. Mol. Evol. 71, 241–249 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Gao B., Zhu S., Exon shuffling and origin of scorpion venom biodiversity. Toxins (Basel) 9, 10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smithers B., Oates M., Gough J., ‘Why genes in pieces?’-revisited. Nucleic Acids Res. 47, 4970–4973 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W., Zhang J., Alvarez C., Llopart A., Long M., The origin of the Jingwei gene and the complex modular structure of its parental gene, yellow emperor, in Drosophila melanogaster. Mol. Biol. Evol. 17, 1294–1301 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Reichen C., et al. , Computationally designed armadillo repeat proteins for modular peptide recognition. J. Mol. Biol. 428, 4467–4489 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Ernst P., Plückthun A., Advances in the design and engineering of peptide-binding repeat proteins. Biol. Chem. 398, 23–29 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Ernst P., et al. , Structure-guided design of a peptide lock for modular peptide binders. ACS Chem. Biol. 15, 457–468 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Bork P., Blomberg N., Nilges M., Internal repeats in the BRCA2 protein sequence. Nat. Genet. 13, 22–23 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Venkitaraman A. R., Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science 343, 1470–1475 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Wong A. K., Pero R., Ormonde P. A., Tavtigian S. V., Bartel P. L., RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 272, 31941–31944 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Carreira A., Kowalczykowski S. C., Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 108, 10448–10453 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowalczykowski S. C., Molecular mimicry connects BRCA2 to Rad51 and recombinational DNA repair. Nat. Struct. Biol. 9, 897–899 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Pellegrini L., et al. , Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420, 287–293 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Rajendra E., Venkitaraman A. R., Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res. 38, 82–96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo T., Pellegrini L., Venkitaraman A. R., Blundell T. L., Sequence fingerprints in BRCA2 and RAD51: Implications for DNA repair and cancer. DNA Repair (Amst.) 2, 1015–1028 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Nomme J., et al. , Inhibition of filament formation of human Rad51 protein by a small peptide derived from the BRC-motif of the BRCA2 protein. Genes Cells 13, 471–481 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Scott D. E., et al. , Small-molecule inhibitors that target protein-protein interactions in the RAD51 family of recombinases. ChemMedChem 10, 296–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott D. E., et al. , A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death. Cell Chem. Biol. 28, 835–847.e5 10.1016/j.chembiol.2021.02.006. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bignell G., Micklem G., Stratton M. R., Ashworth A., Wooster R., The BRC repeats are conserved in mammalian BRCA2 proteins. Hum. Mol. Genet. 6, 53–58 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Warren M., et al. , Structural analysis of the chicken BRCA2 gene facilitates identification of functional domains and disease causing mutations. Hum. Mol. Genet. 11, 841–851 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Moschetti T., et al. , Engineering archeal surrogate systems for the development of protein-protein interaction inhibitors against human RAD51. J. Mol. Biol. 428, 4589–4607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gielen F., et al. , Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS). Proc. Natl. Acad. Sci. U.S.A. 113, E7383–E7389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomme J., et al. , Design of potent inhibitors of human RAD51 recombinase based on BRC motifs of BRCA2 protein: Modeling and experimental validation of a chimera peptide. J. Med. Chem. 53, 5782–5791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gielen F., et al. , Quantitative affinity determination by fluorescence anisotropy measurements of individual nanoliter droplets. Anal. Chem. 89, 1092–1101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roehrl M. H. A., Wang J. Y., Wagner G., A general framework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein-protein interactions by fluorescence polarization. Biochemistry 43, 16056–16066 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Cole D. J., et al. , Interrogation of the protein-protein interactions between human BRCA2 BRC repeats and RAD51 reveals atomistic determinants of affinity. PLOS Comput. Biol. 7, e1002096 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanyam S., Jones W. T., Spies M., Spies M. A., Contributions of the RAD51 N-terminal domain to BRCA2-RAD51 interaction. Nucleic Acids Res. 41, 9020–9032 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erijman A., Rosenthal E., Shifman J. M., How structure defines affinity in protein-protein interactions. PLoS One 9, e110085 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan S. S. F., et al. , BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59, 3547–3551 (1999). [PubMed] [Google Scholar]
- 55.Chen C. F., Chen P. L., Zhong Q., Sharp Z. D., Lee W. H., Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J. Biol. Chem. 274, 32931–32935 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Trenner A., Godau J., Sartori A. A., A short BRCA2-derived cell-penetrating peptide targets RAD51 function and confers hypersensitivity toward PARP inhibition. Mol. Cancer Ther. 17, 1392–1404 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Martinez J. S., et al. , BRCA2 regulates DMC1-mediated recombination through the BRC repeats. Proc. Natl. Acad. Sci. U.S.A. 113, 3515–3520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chatterjee G., Jimenez-Sainz J., Presti T., Nguyen T., Jensen R. B., Distinct binding of BRCA2 BRC repeats to RAD51 generates differential DNA damage sensitivity. Nucleic Acids Res. 44, 5256–5270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Przetocka S., et al. , CtIP-mediated fork protection synergizes with BRCA1 to suppress genomic instability upon DNA replication stress. Mol. Cell 72, 568–582.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Cochran A. G., Skelton N. J., Starovasnik M. A., Tryptophan zippers: Stable, monomeric beta -hairpins. Proc. Natl. Acad. Sci. U.S.A. 98, 5578–5583 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merritt H. I., Sawyer N., Arora P. S., Bent into shape: Folded peptides to mimic protein structure and modulate protein function. Pept. Sci. (Hoboken) 112, e24145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ochiai K., et al. , Valine 1532 of human BRC repeat 4 plays an important role in the interaction between BRCA2 and RAD51. FEBS Lett. 585, 1771–1777 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Scott D. E., Marsh M., Blundell T. L., Abell C., Hyvönen M., Structure-activity relationship of the peptide binding-motif mediating the BRCA2:RAD51 protein-protein interaction. FEBS Lett. 590, 1094–1102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Roey K., Gibson T. J., Davey N. E., Motif switches: Decision-making in cell regulation. Curr. Opin. Struct. Biol. 22, 378–385 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Mammen M., Choi S.-K., Whitesides G. M., Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Engl. 37, 2754–2794 (1998). [DOI] [PubMed] [Google Scholar]
- 66.Isakov N., et al. , ZAP-70 binding specificity to T cell receptor tyrosine-based activation motifs: The tandem SH2 domains of ZAP-70 bind distinct tyrosine-based activation motifs with varying affinity. J. Exp. Med. 181, 375–380 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao D., et al. , Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin. J. Struct. Biol. 187, 194–205 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Stevers L. M., de Vink P. J., Ottmann C., Huskens J., Brunsveld L., A thermodynamic model for multivalency in 14-3-3 protein-protein interactions. J. Am. Chem. Soc. 140, 14498–14510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brouwer I., et al. , Two distinct conformational states define the interaction of human RAD51-ATP with single-stranded DNA. EMBO J. 37, e98162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vonrhein C., et al. , Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bricogne G., et al. , BUSTER (Version 2.10.3, Global Phasing Ltd., Cambridge, UK, 2017). https://www.globalphasing.com/buster/. Accessed 2 November 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SI Appendix contains detailed descriptions of the cloning of bacterial expression constructs for the 64 shuffled BRC peptide variants, cloning of mammalian expression constructs, and notes on the soluble expression of the shuffled BRC peptide variants. Also included is a description of ITC used to cross-validate the microfluidic measurements, single concentration point measurements carried out with microfluidics, and exemplary titrations carried out by microfluidics. The fluorescence anisotropy data obtained for the 64 separate titrations as well as the Matlab script used in the analysis have been uploaded as separate files. The supplementary data also contain an analysis on the effect of shuffling of BRC peptides and in particular on the effect of the exact shuffle cutoff point placement. X-ray crystallography electron density map images, data collection, and refinement statistics are also to be found in SI Appendix. Additional cell images highlighting the pan-nuclear signal of RAD51 are also included in SI Appendix. The coordinates and corresponding structure factors for the monomeric RAD51:BRC8-2 complex have been deposited to the PDB under accession code 6HQU. As described previously (49), the transformation from intensity maps into anisotropy values from image data was carried out with a custom Matlab code available on GitHub (https://github.com/quantitativeimaging/icetropy). A custom Matlab script used to fit Kd values for the unlabeled competitive GB1-BRC peptides can be found in SI Appendix, Datasets S1–S4. All other study data are included in the article and/or supporting information.