Abstract
Dietary exposure to aflatoxins is a significant risk factor in the development of hepatocellular carcinomas (HCCs). Following bioactivation by microsomal P450s, the reaction of aflatoxin B1 (AFB1) with guanine (Gua) in DNA leads to the formation of stable AFB1-FapyGua adducts. In contrast to most base modifications that result in destabilization of the DNA duplex, the AFB1-FapyGua adduct increases the thermal stability of DNA via 5′-interface intercalation and base-stacking interactions. Although it was anticipated that this stabilization might make these lesions difficult to repair relative to helix distorting modifications, prior studies have shown that both the nucleotide and base excision repair pathways participate in the removal of the AFB1-FapyGua adduct. Specifically for base excision repair, we previously showed that the DNA glycosylase NEIL1 excises AFB1-FapyGua and catalyzes strand scission in both synthetic oligodeoxynucleotides and liver DNA of exposed mice. Since it is anticipated that error-prone replication bypass of unrepaired AFB1-FapyGua adducts contributes to cellular transformation and carcinogenesis, the structural and thermodynamic parameters that modulate the efficiencies of these repair pathways are of considerable interest. We hypothesized that the DNA sequence context in which the AFB1-FapyGua adduct is formed might modulate duplex stability and consequently alter the efficiencies of NEIL1-initiated repair. To address this hypothesis, site-specific AFB1-FapyGua adducts were synthesized in three sequence contexts, with the 5′ neighbor nucleotide being varied. DNA structural stability analyses were conducted using UV absorbance- and NMR-based melting experiments. These data revealed differentials in thermal stabilities associated with the 5′-neighbor base pair. Single turnover kinetic analyses using the NEIL1 glycosylase demonstrated corresponding sequence-dependent differences in the repair of this adduct, such that there was an inverse correlation between the stabilization of the duplex and the efficiency of NEIL1-mediated catalysis.
Keywords: Base Excision Repair, hepatocellular carcinoma, formamidopyrimidines, DNA sequence context effects, DNA structure, base-stacking, DNA thermal stability
Graphical Abstract

Created with BioRender.com
Introduction
The base excision repair (BER) and nucleotide excision repair (NER) pathways are integral to the maintenance of genomic stability following DNA modification by environmental toxicants as well as the induction of base damage by various endogenously produced species, including reactive oxygen and nitrogen species [reviewed in (1–3)]. Although most forms of DNA damage are generally presumed to be exclusively repaired by one pathway or another, there is growing evidence that some DNA modifications can be repaired by both and that proteins which were initially assigned to either NER or BER, can play critical roles in both pathways (2, 3). One example of the overlap in substrate specificities includes a subset of oxidatively-induced base damage (2, 3).
The BER pathway is initiated by DNA glycosylases that search for damaged sites by probing the DNA structure via non-specific interactions, bind their respective substrates, and hydrolyze the N-glycosidic bond, thus producing apurinic/apyrimidinic (AP) sites [reviewed in (4)]. A subset of DNA glycosylases possess a lyase activity that catalyzes a single-stranded break in the DNA backbone through the β- or β,δ-elimination of the AP site [reviewed in (5)]. NEIL1 is a key human DNA glycosylase/AP lyase responsible for the removal of oxidatively-induced lesions [reviewed in (6)]. Its substrates include 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) (7–11), 4,6-diamino-5-formamidopyrimidine (FapyAde) (7–11), thymine glycol (ThyGly) (8–16), 5-hydroxycytosine (5-OH-Cyt) (10, 11, 13, 17), and the secondary oxidation products of 8-hydroxyguanine (8-OH-Gua), such as guanidinohydantoin (Gh) and diastereomers of spiroiminodihydantoin (Sp1 and Sp2) (9, 13–16, 18, 19). Consistent with this broad substrate specificity, NEIL1 also excises methyl- and nitrogen mustard-FapyGua adducts (16, 20), psoralen-induced DNA crosslinks (21, 22), and other lesions (19, 23). There are two different forms of intracellular nuclear NEIL1 that arise from the processing of pre-mRNA by adenosine deaminase ADAR1, in which a single adenosine is converted to inosine. This deamination results in an amino acid change in NEIL1 of Lys242 to Arg242 (K242R) and is referred to as the edited form (10, 11, 13, 17). Unless otherwise noted, throughout this investigation, we will use the edited form of the enzyme and refer to it as NEIL1.
In addition to the base damage described above, we reported that the substrate range of NEIL1 extends to the alkylated FapyGua adduct induced by aflatoxin B1 (AFB1) exposures (10, 11, 24). This adduct arises from dietary consumption of foods, such as corn and peanuts that are contaminated with AFB1-producing molds, including Aspergillus flavis [reviewed in (25)]. Once ingested, liver microsomal P450s activate AFB1 to an epoxide that subsequently reacts at N7-Gua in duplex DNA (Fig. 1). The resulting N7 cationic species undergoes either hydrolytic depurination, resulting in an AP site, or imidazole ring-opening to form the 8,9-dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxyaflatoxin B1 (AFB1-FapyGua) adduct (Fig. 1). AFB1-FapyGua is mutagenic, primarily producing G to T transversions (26, 27). This mutation is the dominant base substitution found in genomes of AFB1-exposed cells or animals (28–30) and also in human hepatocellular carcinomas (HCC) that are associated with AFB1 exposure (28, 29, 31, 32). To mitigate the effects of AFB1 exposure, NEIL1 initiates AFB1-FapyGua removal and catalyzes β,δ-eliminations, generating both 5′ and 3′ phosphates (10, 11, 24). Under physiological temperature conditions, the rates of excision by the edited and unedited enzyme forms were 0.18 ± 0.01 min−1 and 0.26 ± 0.01 min−1, respectively (10, 11, 24). The removal of AFB1-FapyGua by NEIL1 was unanticipated given that prior work suggested that the NER pathway was exclusively responsible for the repair of this sterically bulky and intercalated lesion (33–35). However, comparative analyses of the effects of deficiencies in NER (XPA−/−) versus BER (Neil1−/−) on AFB1-induced carcinogenesis in murine models revealed a greater susceptibility to HCC formation with Neil1 deficiency (24).
Figure 1.
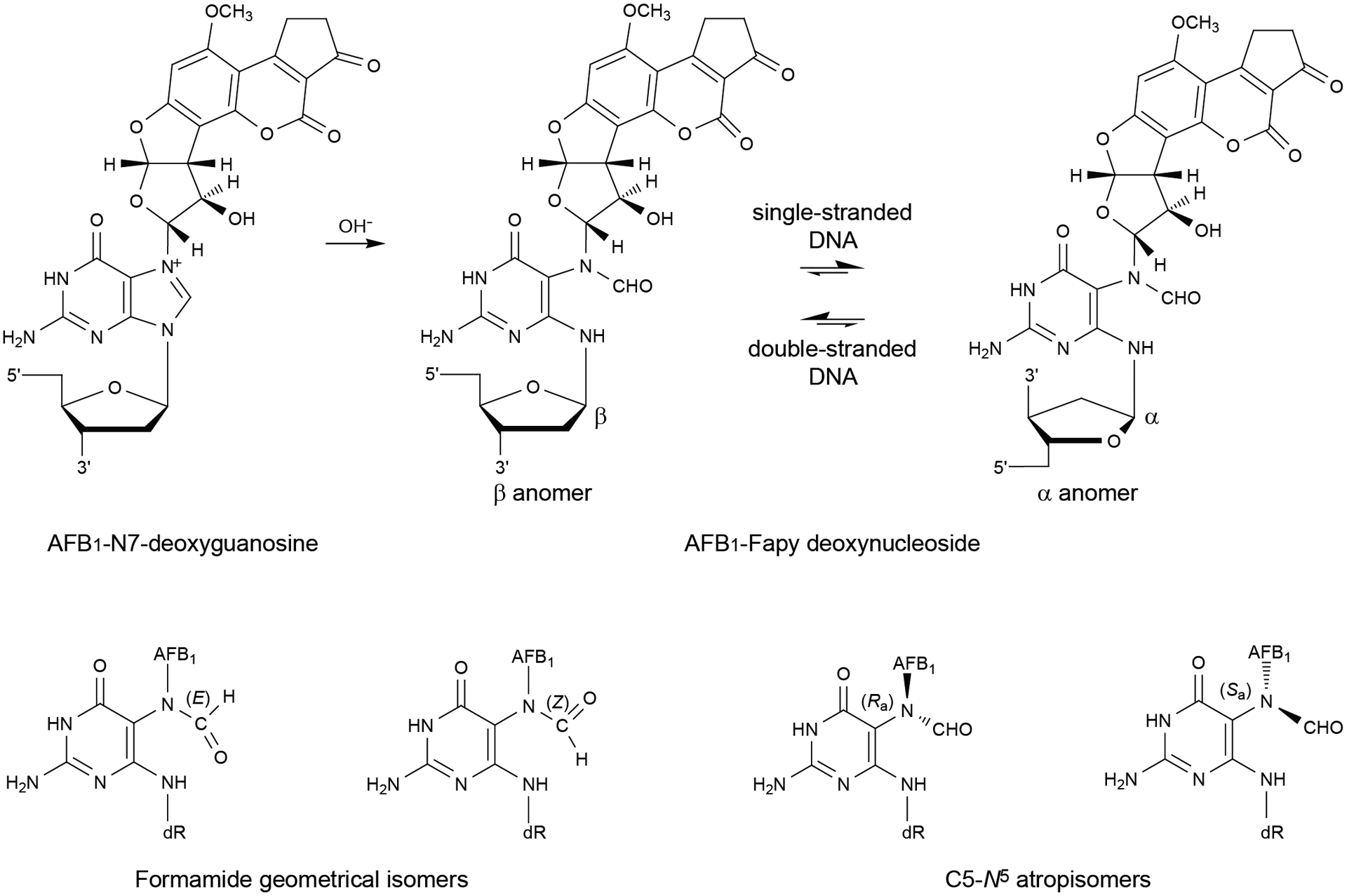
Equilibrium chemistry of the AFB1-FapyGua adduct in DNA.
Damage recognition by DNA glycosylases depends in part, on deviations of DNA structure from its canonical geometry (4, 36, 37), but such structural alterations imposed by the lesion can be influenced by local sequence context. These interactions between the nearby nucleotides and the adduct may not only dictate its coordinates in space, but also modulate the equilibrium between various conformational and configurational isomers of the adduct (38). This may be especially important for AFB1-FapyGua adducts. AFB1-FapyGua adducts exist in DNA as a complex mixture of configurational and conformational isomers (Fig. 1) (27, 39). Among these are interconvertible α- and β-anomers (39). Both are observed in single-stranded DNA, whereas the β-anomer is favored in duplex DNA (39). The β-anomer intercalates in duplex DNA with the AFB1 moiety on the 5′ face of the modified base, forms strong stacking interactions, and stabilizes DNA to thermal denaturation (40–42). In contrast, the α-anomer, while also intercalated, disrupts the DNA phosphodiester backbone and exhibits less favorable stacking interactions, thereby reducing the thermal stability of DNA (40–42). The thermal stability of lesion-containing DNA can be a factor in the efficiency of BER machinery, since DNA glycosylases achieve specific substrate recognition, in part, by extrahelical extrusion of the target nucleotide and fitting the base in an active site pocket [reviewed in (4, 43)].
Additionally, the AFB1-FapyGua adduct exhibits geometrical isomerism involving the formamide moiety (39). If the nucleotide located 3′ with respect to the AFB1-FapyGua adduct is dA, a hydrogen bond forms between the formyl oxygen and N6-dA, and if the nucleotide located 3′ with respect to the AFB1-FapyGua adduct is dC, a hydrogen bond forms between the formyl oxygen and N4-dC. This explains the presence of the E geometrical isomer in these sequences; otherwise the Z isomer is favored (38–40). At the nucleoside level, the AFB1-FapyGua adduct exhibits atropisomers about the C5-N5 bond (39). This has not been observed in duplex DNA, probably because 5′-intercalation of the AFB1 moiety enforces the Ra axial conformation for the C5-N5 bond (40–42).
Thus, the purpose of this study was to determine whether there is a correlation between the efficiency of NEIL1-mediated excision of AFB1-FapyGua and sequence-dependent features of modified DNA. Since intercalation of the AFB1-FapyGua occurs on the 5′ side of the adducted nucleotide, to investigate the local sequence context effects, we chose to vary the 5′ nucleotide to be either dA, dT, or dC, while holding constant the 3′ nucleotide as dC. These alterations were predicted to differentially stabilize the duplex, and thus, modulate the kinetic efficiency of NEIL1-mediated incision.
Materials and Methods
Chemicals.
Unmodified oligodeoxynucleotides were purchased from the Midland Certified Reagent Co. (Midland, TX) and Integrated DNA Technologies, Inc. (Coralville, Iowa). AFB1 was purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). Caution: AFB1 is a potent liver toxin and is a human carcinogen. Crystalline AFB1 is particularly hazardous due to its electrostatic nature and therefore, should be handled using appropriate containment procedures in a well-ventilated hood and wearing a respiratory mask to prevent inhalation. AFB1 can be destroyed by oxidation with NaOCl. It should be presumed that AFB1-exo-8,9-epoxide is also toxic and a human carcinogen; hence, manipulations should be carried out with suitable containment procedures.
Adduct Synthesis.
The 11-mer oligodeoxynucleotides containing a site-specific AFB1-FapyGua were prepared according to published procedures (38). Each of the oligodeoxynucleotides was purified through reverse-phase HPLC using a C18 column (Gemini C18 250 mm × 10 mm, Phenomenix, Inc., Torrance, CA) and subsequently, concentrations were determined by UV absorbance at 260 nm. Dimethyldioxirane (DMDO), which was used for the oxidation of AFB1, was synthesized as described (44). DMDO in acetone was dried over heat-activated micro-sieves (5 Å), stored at −20 °C, and used within 1 week of preparation. AFB1-exo-8,9-epoxide was prepared by oxidizing AFB1 in the presence of DMDO, as described (45). The 11-mer single-stranded oligodeoxynucleotides, 5′-d(CCATAGCTACC)-3′, 5′-d(CCATTGCTACC)-3′ and 5′-d(CCATCGCTACC)-3′ (70 nmole each), each containing a unique N7-Gua alkylation site, were annealed at an equimolar concentration with short complementary scaffold oligodeoxynucleotides, 5′-d(TAGCTA)-3′, 5′-d(TAGCAA)-3′ and 5′-d(TAGCGA)-3′, respectively, to form a double-stranded DNA structure around the target site. The reactions were conducted in 10 mM sodium phosphate buffer (pH 7.5), containing 100 mM NaCl and 50 μM Na2EDTA at 4 °C. For each sequence context, a 12-fold molar excess of AFB1-exo-8,9-epoxide (in anhydrous CH2Cl2) was added to the annealed DNA, and the two-phase mixtures were stirred for 3–4 h at 4 °C to yield the AFB1-N7-Gua cationic adducts. Subsequently, the aqueous phase of each reaction mixture was isolated and AFB1-diol, a byproduct of the reaction, was extracted by washing 2–3 times with anhydrous CH2Cl2. The reaction mixtures were passed over a reverse-phase C18 column at 2 mL/min flow rate, with a linear, 45 min gradient of 1–15% acetonitrile (CH3CN) in 100 mM sodium phosphate buffer (pH 7.5) to separate the unreacted DNA from those containing the N7-dG cationic adducts. Following lyophilization, each of the 11-mer oligodeoxynucleotides containing the cationic adduct was dissolved in 500 μL of 100 mM sodium carbonate buffer (pH 10.5) and stirred at 4 °C overnight to subject the cationic adducts to base hydrolysis and form the imidazole-ring opened AFB1-FapyGua adducts. The 11-mer oligodeoxynucleotides containing the AFB1-FapyGua adducts were isolated by passing the reaction mixtures over a reverse-phase C18 column at 2 mL/min flow rate, with a linear, 45 min gradient of 1–15% acetonitrile (CH3CN) in 100 mM ammonium formate buffer (pH 6.8). The AFB1-FapyGua adducted oligodeoxynucleotides were lyophilized and the concentrations measured by UV 260 nm absorbance. The AFB1-FapyGua adducts in 11-mer oligodeoxynucleotides were analyzed by MALDI-TOF mass spectrometry: for 5′-AXC-3′, calcd. 3607.3, found 3607.3; for 5′-TXC-3′, calcd. 3598.3, found. 3598.8; for 5′-CXC-3′, calcd. 3583.3, found. 3584.4 (X denotes the modified site).
To prepare double-stranded DNA for melting experiments, each of the AFB1-modified 11-mer oligodeoxynucleotides was annealed with its full-length complementary oligodeoxynucleotide in 10 mM sodium phosphate buffer (pH 7.5) containing 100 mM NaCl and 50 μM Na2EDTA. The mixture was heated to 85 °C for 5 min and cooled slowly to room temperature. The annealed oligodeoxynucleotide duplexes were purified from residual single-stranded oligodeoxynucleotides using hydroxyapatite chromatography with a gradient of 10–200 mM sodium phosphate (pH 6.8). The modified duplexes were desalted using Sephadex G25 gel filtration chromatography (Bio-Rad Labs), followed by lyophilization.
UV Melting Experiments.
Thermal melting experiments were carried out in a 10 mm pathlength-quartz cuvette (1 mL volume) on a Varian Cary 4E spectrophotometer (Agilent Technologies, Santa Clara, California). The samples contained 5 μM unmodified or modified duplexes in a 10 mM sodium phosphate buffer (pH 7.0), 1 M NaCl and 50 μM Na2EDTA. The temperature was increased from 20 °C to 90 °C at a rate of 0.5 °C/min with a hold time of 2 min, and subsequently, the temperature was decreased to the starting temperature at the same rate of temperature change. Absorbance was monitored at 260 nm. The melting points (Tm) were determined from the maxima of the absorbance versus temperature curves obtained from the first derivatives in each case. All melting point determinations were carried out a minimum of three independent runs.
NMR Spectroscopy.
Samples for the unmodified and modified duplexes in the sequence contexts of 5′-AXC-3′ and 5′-CXC-3′, were prepared in 10 mM sodium phosphate buffer (pH 7.0), containing 100 mM NaCl and 50 μM Na2EDTA and lyophilized. The oligodeoxynucleotide duplex concentration was 500 μM. To examine exchangeable protons, samples were dissolved in 95:5 H2O:D2O. The volume was maintained at 600 μL and data were collected with cryogenic probes (Bruker Biospin, Inc., Billerica, MA) in 5 mm NMR tubes. Water suppression was accomplished by excitation sculpting (46). 1H experiments were recorded as a function of temperature with 16,384 data points. The chemical shifts were referenced to the H2O resonance at the corresponding temperatures. To assign water exchangeable protons, 2D- NOESY spectra were recorded with 512 real data points in the t1 dimension and 2,048 real data points in the t2 dimension with States-TPPI acquisition mode at 1H frequencies of 800 MHz and 900 MHz. These spectra were obtained at 5 °C with mixing times of 200 ms and relaxation delay of 1.5 s, and were zero filled during processing to obtain matrices of 2,048 × 2,048 points. The program TOPSPIN (Bruker Biospin, Inc., Billerica, MA) was used for the data collection and processing. The proton resonance assignments and peak integration were performed using the program SPARKY (47).
DNA Substrates for NEIL1-catalyzed Reactions.
To construct DNA substrates of a size sufficiently long for binding and incision by NEIL1, each of the AFB1-FapyGua-containing 11-mer oligodeoxynucleotides was ligated with a 9-mer oligodeoxynucleotide (Integrated DNA Technologies, Inc.). The 11-mer fragment had the tetramethylrhodamine (TAMRA) moiety at the 3′ terminus [5′-d(ATCGCTGGA)-TAMRA-3′]. Prior to ligation, the 9-mer oligodeoxynucleotide was phosphorylated at the 5′ terminus by T4 polynucleotide kinase (New England BioLabs) using 10 units of enzyme per 500 pmols DNA substrate. The ligation reaction contained 40 units of T4 DNA ligase (New England BioLabs), 1 mM ATP, and 500 pmol each of the 9-mer, the 11-mer and a scaffold oligodeoxynucleotide. Following overnight incubation at 12 °C, the reaction mixtures were mixed with 2 volumes of DNA denaturing solution (95% formamide and 20 mM Na2EDTA), and the products were resolved by electrophoresis through the 8 M urea - 10% polyacrylamide gel in Tris-borate-Na2EDTA buffer. The 20-mer product bands were excised and eluted with a solution composed of 500 mM ammonium acetate and 10 mM magnesium acetate. The oligodeoxynucleotides were concentrated using Amicon filter units (3K CO), followed by three washes with 10 mM Tris-HCl buffer (pH 7.3) containing 1 mM Na2EDTA. Similar procedures were used to prepare 30-mer modified oligodeoxynucleotides that were constructed by ligation of the 5′-terminal 10-mer fragment [5′-d(GACTACGTAC)-3′], 5′-phosphorylated AFB1-FapyGua-containing 11-mer with either 5′-AXC-3′ or 5′-CXC-3′ sequence, and 5′-phosphorylated TAMRA-labeled 9-mer. DNA concentrations were measured using the Infinite M200 plate reader (TECAN Group Ltd.) with a 525/9 nm excitation filter and a 598/20 nm emission filter using dilutions of the initial 9-mer TAMRA-labeled oligodeoxynucleotide as a reference. The control ThyGly-containing oligodeoxynucleotide (5′-TAMRA-d(TCACCXTCGTACGACTC)-3′, where X denotes the ThyGly site was synthesized by Integrated DNA Technologies, Inc. Double-stranded DNA substrates were prepared by combining the adducted oligodeoxynucleotides with the complementary strand at a molar ratio of 1:1.2 in 20 mM Tris⋅HCl buffer (pH 7.4), 100 mM KCl, and 0.01% (v/v) Tween-20. The reactions were incubated at 90 °C for 2 min, gradually cooled to room temperature, and stored at 4 °C for at least 48 h to establish structural equilibrium.
NEIL1-catalyzed Reactions.
Recombinant His-tagged human NEIL1 glycosylase was expressed and purified as described in our earlier studies (8, 10). To evaluate the relative efficiencies of NEIL1-catalyzed excision of AFB1-FapyGua from duplex DNAs, we modified our previously published methodologies that utilized 32P-labeled oligodeoxynucleotides (10, 11, 24) in favor of fluorescently-labeled oligodeoxynucleotides as described below. Specifically, 100 nM DNA substrate and 2 μM NEIL1 were separately incubated at 37 °C for 3 min, combined at equal volumes, and incubated at 37 °C in 20 mM Tris⋅HCl (pH 7.4), 100 mM KCl, 100 μg/mL BSA, and 0.01% (v/v) Tween-20. The ratio of substrate to active enzyme molecules under these conditions was estimated as ~ 1:6. At selected time points, aliquots were removed and transferred to tubes containing a 5-fold excess volume of DNA denaturing solution (95% formamide and 20 mM Na2EDTA), and heated at 90 °C for 2 min. Although product accumulation in this case is a result of the combined glycosylase and lyase NEIL1 activities, the rate of lyase reaction is known to be significantly faster than rates of excision of AFB1-FapyGua, ThyGly, and many other base lesions (8, 9, 16), and thus, its contribution to the overall reaction rate was anticipated to be negligible. The reaction products were resolved by electrophoresis in a 20% denaturing polyacrylamide gel containing 8 M urea in Tris-borate-Na2EDTA buffer and visualized with the FluorChem M system (ProteinSimple) using a 534 nm LED light source and 593 nm emission filter. The intensities of DNA bands were measured using the FluorChem M built-in software. The product (P) was plotted as a function of time (t) using KaleidaGraph software. The first-order rate constant (kobs), non-specific product (Pns), and extrapolated maximal substrate utilization (S) were obtained from the best fit of the data to equation P= Pns +S(1 − exp−kobs*t). Control reactions with 32P-labeled DNA substrates were performed and analyzed as previously reported (10, 11, 24).
In order to verify that the protocol modified for TAMRA-labeled DNA substrates was suitable for measuring the kinetic parameters under single turnover conditions, NEIL1-catalyzed reactions were conducted using a double-stranded 17-mer oligodeoxynucleotide that contained a unique ThyGly and the TAMRA moiety on the 5′ terminus of the adducted strand (9, 10). The observed rate constant was ~1.3 min−1, which was in excellent agreement with our published data generated using 32P-labeled DNA (~1.35 min−1 (24)) as well as with the result of another study (~1.3 min−1 (13)).
Results
Experimental Rationale and Design.
Given that the AFB1 moiety of the AFB1-FapyGua adduct intercalates into the DNA helix between the damaged nucleotide and its 5′ neighboring base-pair (38, 40), we hypothesized that the relative extent of DNA stabilization would influence the overall efficiency of NEIL1-catalyzed base release and strand breakage, such that the kinetics of adduct removal would be diminished in a more thermally stable duplex environment. To test this, oligodeoxynucleotides containing site-specific AFB1-FapyGua adducts in three different sequence conexts, with the 5′ neighboring nucleotide being dA, dT, or dC were synthesized, and evaluated using biophysical and biochemical techniques.
Melting Temperature Analyses.
To evaluate the effect of sequence context on thermal stability of AFB1-FapyGua-containing DNAs, the 11-mer oligodeoxynucleotides with a common 5′-d(CCATNXCTACC)-3′ sequence were prepared and annealed with their respective complementary strands. Tm values were calculated based on DNA absorbance at 260 nm as a function of temperature. Each of the AFB1-FapyGua-modified duplexes showed biphasic transitions during heating (Fig. 2, A–C), in contrast to unmodified duplexes that showed a single transition (Fig. 2, D–F). The first transition, attributed to the α-anomer of AFB1-FapyGua, consistently occurred at lower temperature (Tm value below < 47 °C) relative to the corresponding unmodified duplexes. The Tm values of the 5′-AXC-3′, 5′-TXC-3′, and 5′-CXC-3′ modified duplexes attributed to the β-anomer of AFB1-FapyGua, were 61, 64, and 71 °C, respectively (Fig. 2, A–C). In comparison, the Tm values of the corresponding unmodified duplexes were 53, 55, and 59 °C, respectively (Fig. 2, D–F). The observed 8–12 °C increase in Tm values as compared to the corresponding unadducted duplexes was consistent with earlier reports (38, 40, 42). The effect of sequence context on thermal stability of modified duplexes was significant, with the Tm values of the least stable 5′-AXC-3′ sequence versus the most stable 5′-CXC-3′ sequence differing by 10 °C. Thus, our hypothesis concerning the kinetics of NEIL1-mediated incision being modulated by the relative thermal stability could be tested using this set of sequences.
Figure 2.

Thermal melting curves as recorded by UV 260 absorbance. The data for the AFB1-FapyGua modified , , and duplexes are shown in panels A-C, respectively; the data for the corresponding unmodified duplexes, 5′-AGC-3′, 5′-TGC-3′, 5′-CGC-3′ are shown in panels D-F, respectively. The dotted lines represent heating curves; solid lines represent corresponding first derivative curves.
Sequence-dependent NEIL1-catalayzed Excisions of AFB1-FapyGua.
To facilitate investigations of the effect of changes in sequence context on the ability of NEIL1 to remove the AFB1-FapyGua adduct, TAMRA-labeled 20-mers were constructed using site-specifically modified oligodeoxynucleotides. The set of AFB1-FapyGua-containing DNA substrates (Fig. 3 A) was created by ligation of a 9-mer oligodeoxynucleotide to the 3′-termini of 11-mer oligodeoxynucleotides that contained AFB1-FapyGua in either 5′-AXC-3′, 5′-TXC-3′, or 5′-CXC-3′ context. The 9-mer had the TAMRA fluorophore on its 3′ terminus, thus allowing visualization of the resulting 20-mer DNA substrates and the 3′ cleavage products. The modified 20-mer oligodeoxynucleotides were annealed with the corresponding complementary strands (Fig. 3 A), stored at 4 °C for at least 48 h to allow different adduct isomeric species to reach an equilibrium, and incubated with NEIL1 under conditions described in the Materials and Methods. Representative gel images and a plot demonstrating time-dependent product accumulation in different sequence contexts are shown in Fig. 3 B and C, respectively. In all three sequence contexts, product formation followed single exponential kinetics, with a high degree of correlation between the experimental points and the curve (P > 0.99). For the 5′-AXC-3′, 5′-TXC-3′, and 5′-CXC-3′ sequences, the mean observed rate constants (kobs) with standard deviations were calculated from at least three independent experiments. These data demonstrated that the efficiency of AFB1-FapyGua excision from the 5′-AXC-3′ sequence was slightly higher than from the 5′-TXC-3′ sequence (0.22 ± 0.03 versus 0.16 ± 0.03 min−1, P < 0.05). These rates were comparable with those previously measured for the 5′-TXA-3′ sequence (0.17 ± 0.03 min−1 (24) and 0.18 ± 0.01 min−1 (10)). In contrast, the rate of excision from the 5′-CXC-3′ sequence was ~3-fold lower (0.06 ± 0.01 min−1); the difference between this rate and the rates of excision from either 5′-AXC-3′ or 5′-TXC-3′ sequence was statistically significant (P < 0.001).
Figure 3.

NEIL1-catalyzed excision of AFB1-FapyGua. The reactions were conducted using 20-mer DNA substrates (A), where X denotes the adduct site and N shows the position of the variable nucleotide. Representative gels (B) and kinetic curves (C) of NEIL1-catalyzed excision of AFB1-FapyGua were calculated from at least three independent experiments. In a subset of samples, DNA substrates are represented by two bands, with minor double-stranded (upper band) and major single-stranded (lower band) oligodeoxynucleotides. When observed, both bands were included in quantification of the total DNA. Uncertainties (standard deviations) are shown. The data were fit to a single exponential equation (correlation coefficient R > 0.99 for each sequence).
The extrapolated maximal substrate utilization values obtained from the best fit of the data to a single exponential equation demonstrated that on average, ~ 90% of the DNA could be incised by NEIL1 with the remaining DNA fraction being resistant to cleavage (Fig. 3 B & C). These data suggested that AFB1-FapyGua was present in the NEIL1 reactions as a mixture of species that were differentially processed by NEIL1. Since AFB1-FapyGua exists predominantly in the more stable β-anomeric configuration in double-stranded DNA (27, 39) under the conditions in which the kinetic assays were performed, we ascribe the NEIL1-cleavable species to be the β-anomeric AFB1-FapyGua.
The low efficiency of excision from the 5′-CXC-3′ sequence raised concern that the proximity of the adduct to the 5′ terminus of the oligodeoxynucleotide could affect the rate of NEIL1-catalyzed reaction. To address such a possibility, 30-mer substrates were created containing a centrally located AFB1-FapyGua adduct in either the 5′-AXC-3′ or 5′-CXC-3′ sequence context and the incision rates measured (Supporting Fig. S1). Consistent with results obtained using the 20-mer substrates (Fig. 3), the observed rate constants were 0.20 and 0.07 min−1, respectively. In addition, the inefficient excision of AFB1-FapyGua in the 5′-CXC-3′ sequence was confirmed using 32P-labeled DNA and a 2-fold increased enzyme to substrate ratio (Supporting Fig. S2).
These data revealed that the rate of NEIL1-catalyzed excision of AFB1-FapyGua was modulated by the 5′ neighboring base-pair and that the adduct was removed more efficiently from less thermally stable sequences. Thus, we utilized an NMR-based approach to reveal sequence-dependent differences in intramolecular interactions within modified DNA duplexes that are manifested as increased thermal stability.
NMR-based Characterization of DNA Thermal Stability.
To show that the differences in DNA thermal stability, as monitored by UV absorbance, correlated with specific AFB1-FapyGua-induced structural changes to the duplexes, the thermal melting of 5′-AXC-3′ and 5′-CXC-3′ DNAs was further examined by NMR. The DNA 1H resonances were assigned using established strategies (38, 40, 42, 48–50). To confirm the intercalation of the AFB1 moiety above the 5′-interface of the modified base, sequential NOE connectivities were monitored between adjacent Watson-Crick hydrogen bonded imino protons (50). For each modified duplex, there was an interruption in sequential NOE connectivity between the X6 N1H resonance and the 5′-neighbor base-pair imino proton resonance, T18 N3H [5′-(A5X6C7)-3′:5′-(G16C17T18)-3′ duplex] or G18 N1H [5′-(C5X6C7)-3′:5′-(G16C17G18)-3′ duplex]. This confirmed the 5′-interface intercalation of the AFB1 moiety in both duplexes. At 5 °C, the modified AFB1-FapyGua imino proton resonance shifted upfield in comparison to the unadducted duplexes, X6 N1H, ~0.6 and 0.55 ppm, for 5′-(A5X6C7)-3′:5′-(G16C17T18)-3′ and 5′-(C5X6C7)-3′:5′-(G16C17G18)-3′, respectively; and the 5′-neighbor base pair imino resonance also shifted upfield, T18 N3H by ~0.15 ppm and G18 N1H by ~0.39 ppm (Fig. 4). These upfield shifts further confirmed the 5′-interface intercalation of the AFB1 moiety in each instance. The imino proton resonances of the 5′-(A5X6C7)-3′:5′-(G16C17T18)-3′ and 5′-(C5X6C7)-3′:5′-(G16C17G18)-3′ modified sequences, as compared to the corresponding unmodified sequences, were monitored at higher temperatures. For the modified 5′-(A5X6C7)-3′:5′-(G16C17T18)-3′ duplex, the AFB1-FapyGua and 3′-neighbor base-pair imino-proton resonances, X6 N1H and G16 N1H, respectively, were observed at 50 °C, but broadened due to exchange with water at 55 °C (Fig. 4A). The 5′-neighbor base-pair imino-proton resonance, T18 N3H completely broadened at 50 °C (Fig. 4A). In constrast, for the modified 5′-(C5X6C7)-3′:5′-(G16C17G18)-3′ duplex, the modified AFB1-FapyGua, 5′- and 3′-neighbor base-pair imino proton resonances (X6 N1H, G18 N1H and G16 N1H, respectively) were observed at both 50 and 55 °C (Fig. 4B). For the unadducted 5′-(A5G6C7)-3′:5′-(G16C17T18)-3′ duplex, the G6 N1H 5′- and 3′-neighbor base-pair imino-proton resonances, T18 N1H and G16 N1H, were not detected at 50 and 55 °C (Fig. 4C). Likewise, for the unadducted 5′-(C5G6C7)-3′:5′-(G16C17G18)-3′ duplex, each of the G6 N1H, G18 N1H, and G16 N1H imino proton resonances were observed at 50 °C, but were almost completely broadened at 55 °C (Fig. 4D). Overall, the NMR studies confirmed that the differences in the stabilization of the AFB1-FapyGua-modified duplexes correlated with the 5′-neighbor base-pairs in these sequences, as well as the observed modulation of NEIL1-catalyzed base excision.
Figure 4.

1H NMR spectra collected at 900 MHz as a function of temperature. The AFB1-FapyGua, 5′ and 3′-neighbor base pair imino-proton resonances for the and modified duplexes are shown in panels A and B; the corresponding base pair imino-proton resonance for the unmodified duplexes are shown in panels C and D, respectively.
DISCUSSION
HCCs that arise from the combination of life-long dietary exposures to AFB1 and high frequencies of hepatitis B infection represent the second highest number of environmentally-induced cancers, world-wide, ranking only behind tobacco-associated lung cancers [reviewed in (25)]. To combat the high frequencies of AFB1-associated cancers, it is necessary not only to limit dietary exposures to foods contaminated with Aspergillus flavus, and immunize large populations against hepatitis B infection, but also to understand the chemical biology underlying the bioactivation of AFB1, its propensity to damage DNA, and removal of AFB1-induced DNA damage by DNA repair mechanisms. Such knowledge is critical for discovering biomarkers of disease susceptibility and designing potential intervention strategies.
The traditional view of the division of labor between the two major mechanisms for repair of DNA adducts, BER and NER, suggested that whereas the former is responsible for removal of small, non-distorting base modifications, the latter is involved in removal of bulky and helix-distorting DNA lesions. Consistent with this view, several decades of research, including studies in both bacterial and mammalian experimental systems (33–34), and the protective function of NER against AFB1-induced carcinogenesis in mice (35), supported the conclusion that the AFB1-FapyGua adduct was exclusively a substrate for NER. Thus, our findings that the BER enzyme, NEIL1, excises AFB1-FapyGua in vitro and decreases accumulation of this adduct in the livers of AFB1-exposed mice was unanticipated and revealed another example of shared substrates between BER and NER pathways (10, 11, 24). Beyond this specific example, additional investigations of the substrate specificity of NEIL1 have revealed that it initiates the repair of a variety of psoralen-induced DNA crosslinks (21, 22), which are also DNA modificatons that were generally considered the substrate domain of NER-initiated repair. However, specific, well-documented examples of bulky lesions being BER substrates are rare, with AFB1-FapyGua representing the best described case.
In addition to these examples of traditional NER substrates also being repaired via BER, the ability of NER to recognize subsets of oxidatively-induced base lesions that are classical BER substrates, is well documented [reviewed in (2, 3)]. Thus, the emerging field of cooperation between BER and NER will benefit from future studies on interactions of these mechanisms in repair of AFB1-FapyGua and on their participation in maintaining genomic stability following AFB1 exposure. It is worth emphasizing that relative to deficiency in NER (XPA−/−), lack of NEIL1 conferred a greater susceptibility to AFB1-induced HCC formation in murine model (24).
AFB1-driven cancers are characterized by a specific mutagenic signature dominated by G to T transversions, with a limited percentage of A to G transitions (28, 29, 31, 32). Significantly, the G to T transversions that constitute COSMIC signature 24 attributed to AFB1 exposure (32), are not distributed randomly across all trinucleotide sequence contexts, with a second position G. Instead they are predominantly clustered within 3–4 sequence context hot-spots, with other trinucleotide sequences being nearly devoid of mutations. These observations apply to both investigations utilizing human samples, as well as studies in animal models and cell culture systems (28–30). These data suggest that there are potentially multiple mechanisms through which this signature is manifested. Among these possibilities are sequence-dependent preferential DNA adduct formation, differential rates of conversion of the initially formed AFB1-N7-dG cationic species to AP sites or the ring-opening to the AFB1-FapyGua adduct, differential repair rates, and differential fidelity of translesion synthesis.
Previously, sequence-dependent repair of DNA lesions by BER has been hypothesized to provide a mechanism for the non-random distribution of mutations resulting in the occurrence of hot spots in genomes (51). Relative to aflatoxin-induced mutagenesis and subsequent carcinogenesis, the efficiency of the BER pathway in limiting carcinogenesis has been previously shown in our study demonstrating that knockout of Neil1 rendered these mice ~3–4 times more susceptible to AFB1-induced HCCs than wild-type mice (24). This effect was greater than that observed in NER-deficient mice, which showed only ~1.3-fold enhancement in carcinogenesis (33). These observations led us to speculate that the sequence context of AFB1-FapyGua adducts may modulate the efficiency by which they are repaired and thus, be a contributor to the final mutagenic signature.
The current investigation was designed to assess whether the catalytic efficiency of NEIL1 on these lesions was dependent on a local sequence context in which only the 5′ nucleotide was varied, while keeping the 3′ nucleotide constant. The present data demonstrate that the catalytic efficiency of NEIL1 is correlated with the thermal stability of the adducted DNA, such that AFB1-FapyGua is removed more efficiently from sequences with lower Tm. Our results predict that certain sequence contexts will potentially be more readily repaired by NEIL1. Ultimately, this may contribute to the characteristic mutagenic signature observed in AFB1-associated tumors, as more easily repaired sequence contexts may manifest as sites of lower mutation rates, while less easily repaired sequences may be enriched in the ultimate mutation signature. The differential thermal stability of AFB1-damaged DNA could be a contributing factor in AFB1-induced carcinogenesis, since in addition to BER, it might affect additional aspects of DNA processing. To establish these relationships, it will be necessary to examine mutation signatures generated in mice that are deficient in either Neil1 or key components of the global or transcription-coupled NER pathways.
Prior studies demonstrated that the effect of sequence context on glycosylase activity may be both DNA damage- and glycosylase-specific. For example, human methylpurine DNA glycosylase excised 7-methyl-Gua and hypoxanthine in a sequence-dependent manner, but excision of 3-methyl-Ade was comparable in all sequences (52–54). The efficiency of repair of methyl-FapyGua in different sequence contexts showed variations of ~7-fold for Escherichia coli Fpg and ~17-fold for human OGG1, with preferred and unpreferred sequences being glycosylase-specific (55). In contrast to methyl-FapyGua, excision of 8-OH-Gua by human OGG1 was not influenced by the 5′-flanking nucleotide but was modulated by the 3′ neighbor (56, 57). Whereas mouse Neil3 preferred ThyGly in the telomere sequence context in double-stranded DNA, human NEIL1, OGG1, and NTH1 did not show such a preference (15). When similar comparative analyses were performed with Gh-containing DNA, NEIL1, but not Neil3, showed enhanced activity on the telomere sequence (15). Here, we not only identified an additional lesion that is excised by NEIL1 in a sequence-dependent manner, but also found an inverse correlation between the thermal DNA stability and the efficiency of NEIL1-mediated catalysis.
The inter-relationships between sequence-dependent physical features of damaged DNA, the efficiencies of BER enzymes, and damage-induced mutagenesis have been the focus of several studies, with recent progress in assessing the mutational landscape in genomes further facilitating advancements in this field [reviewed in (51) and (15, 54, 56–59)]. Although these investigations revealed that both thermodynamic and structural factors can be involved in sequence-dependent modulation of the catalytic efficiency, the attempts to directly correlate the thermal stability of modified duplex DNA with the excision rates are limited. Our previous analyses demonstrated that the efficiency of excision of an A opposite to a G by the E. coli MutY glycosylase could not be predicted from the melting temperature of mismatch-containing oligodeoxynucleotides with varying local sequence contexts (60). These data suggest that although the presence of the mismatch decreases duplex stability relative to corresponding control DNA, such sequence-dependent differences do not significantly contribute to lowering the activation energy. In the case of AFB1-FapyGua, the stabilization of DNA structure is hypothesized to raise the energy for chemistry to occur, with the effect of sequence context on excision rates being apparent. Thus, we speculate that other DNA stabilizing lesions will follow a similar trend as observed for AFB1-FapyGua.
The 5′-CXC-3′ modified sequence exhibited the greatest thermal stability, as compared to the 5′-AXC-3′ and 5′-TXC-3′ modified sequences. The higher Tm value for the 5′-CXC-3′ duplex can be attributed to the presence of the 5′-neighbor C5:G18 base pair, which presumably stabilizes AFB1-moiety intercalation above the 5′-face of the modified base. The sequence-dependent thermal stability differences in the AFB1 modified 5′-AXC-3′ and 5′-CXC-3′ duplexes were also observed in the imino-proton resonances of the NMR-based melting experiments. Analyses of these data revealed that in the 5′-AXC-3′ modified duplex (as compared to the 5′-CXC-3′ modified duplex), the resolution of the 5′-neighbor base imino proton resonance showed a broadening and solvent exchange at lower temperatures relative to loss of resolution in the 3′-neighbor base imino proton resonance.These data confirmed the importance of the 5′-neighbor base pair as opposed to the 3′-neighbor base pair with respect to stabilizing AFB1 intercalation. Presumably, stacking interactions become stronger in AFB1-FapyGua-modified duplexes because the AFB1 moiety intercalates parallel to the DNA base pairs with minimal helical bending (40). Overall, these observations are consistent with the notion that for AFB1-FapyGua adducts, the AFB1 moiety intercalates and imparts stability to the modified and neighbor base pairs (40, 41).
The analyses of kinetics of NEIL1-catalyzed excision demonstrated that in all three sequence contexts, product formation followed single exponential functions, with ~90% of the AFB1-FapyGua adducts being good substrates for NEIL1 (Fig. 3 B & C). Based on prior structural analyses showing that AFB1-FapyGua predominatly exists in the more stable β-anomeric configuration in double-stranded DNA (27, 39), we ascribe the NEIL1-cleavable species to be the β-anomeric AFB1-FapyGua. The remaining adducts must be the α anomers. Consistent with this interpretation, the presence of both α and β anomers of the AFB1-FapyGua adducts was apparent from the analyses of melting curves at low temperatures (Fig. 2 A–C). Germane to these conclusions, we have previously shown that the nitrogen mustard-FapyGua adduct was excised by NEIL1 following a bi-component, not a single exponential function (16, 20). The interpretation of this bi-component kinetic curve was that the major preferentially excised species was the β-anomeric adduct and that the second route of product generation was through a slow, rate-limiting conversion of the α anomer into the β anomer, followed by fast NEIL1-catalyzed incision of DNA containing the β anomer. The active involvement of NEIL1 in the α to β anomerization was also previously proposed (16, 20). Since in the case of AFB1-FapyGua, our investigations have revealed no evidence for a second, slower rate of product formation on the time-scale of the incision reaction, we infer that the α to β anomerization, whether NEIL1-induced or independent of NEIL1, was extremely slow.
In conclusion, we demonstrated that the efficiency of the DNA glycosylase NEIL1 in removing the AFB1-FapyGua adduct was significantly affected by the identity of the 5′ neighbor nucleotide, with an inverse correlation observed between the degree of stabilization of DNA structure in the presence of lesion and the rate of NEIL1-catalyzed excision. This differential, sequence-dependent initiation of repair of AFB1-FapyGua may contribute to mechanisms that define the mutagenic signature of AFB1 exposures.
Supplementary Material
FUNDING SOURCES
This work was supported by National Institutes of Health (NIH) grants R01 CA-55678, R01 ES-029357, and P01 CA-160032 (M.P.S. and R.S.L.). The Vanderbilt-Ingram Cancer Center is funded by NIH grant P30 CA-068485. Funding for the NMR spectrometers was provided by in part by instrumentation grants S10 RR-05805, S10 RR-025677, and National Science Foundation Instrumentation Grant DBI 0922862, the latter funded by the American Recovery and Reinvestment Act of 2009 (Public Law 111-5). Vanderbilt University assisted with the purchase of NMR instrumentation. R.S.L. acknowledges support from the National Institute of Environmental Health Sciences (R01 ES-031086) and from the Oregon Institute of Occupational Health Sciences at Oregon Health & Science University via funds from the Division of Consumer and Business Services of the State of Oregon (ORS 656.630). Funding for open access charge: National Institutes of Health.
ABBREVIATIONS
- BER
base excision repair
- NER
nucleotide excision repair
- AP site
apurinic/apyrimidinic site
- FapyGua
2,6-diamino-4-hydroxy-5-formamidopyrimidine
- FapyAde
4,6-diamino-5-formamidopyrimidine
- ThyGly
thymine glycol
- 5-OH-Cyt
5-hydroxy-cytosine
- 8-OH-Gua
8-hydroxyguanine
- Gh
guanidinohydantoin
- Sp
spiroiminohydantoin
- 7-methyl-Gua
7-methyl-guanine, 3-methyl-Ade, 3-methyl-adenine
- AFB1
aflatoxin B1
- AFB1-FapyGua
8,9-dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxyaflatoxin B1
- HCC
hepatocellular carcinoma
- TAMRA
tetramethylrhodamine
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Supplemental Figures S1–S2 (PDF)
REFERENCES
- (1).Demple B (2020) Special problems for Base Excision Repair in coping with oxidatively-induced DNA damage, In DNA Damage, DNA Repair and Disease (Dizdaroglu M and Lloyd RS, Eds.) pp 202–217, Royal Society of Chemistry, London. [Google Scholar]
- (2).Kumar N, Moreno NC, Feltes BC, Menck CF and Houten BV (2020) Cooperation and interplay between base and nucleotide excision repair pathways: From DNA lesions to proteins. Genet Mol Biol 43, e20190104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kumar N, Raja S and Van Houten B (2020) The involvement of nucleotide excision repair proteins in the removal of oxidative DNA damage. Nucleic Acids Res 48, 11227–11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Endutkin AV and Zharkov DO (2020) Substrate specificities of DNA glycosylases, In DNA Damage, DNA Repair and Disease (Dizdaroglu M and Lloyd RS, Eds.) pp 173–201, Royal Society of Chemistry, London. [Google Scholar]
- (5).McCullough AK, Minko IG, Nilsen A, Nagarajan S and Lloyd RS (2020) Modulation of DNA glycosylase activities via small molecules, In DNA Damage, DNA Repair and Disease (Dizdaroglu M. and Lloyd RS, Eds.) pp 321–345, Royal Society of Chemistry, London. [Google Scholar]
- (6).Dizdaroglu M, Coskun E and Jaruga P (2017) Repair of oxidatively induced DNA damage by DNA glycosylases: Mechanisms of action, substrate specificities and excision kinetics. Mutat Res 771, 99–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M and Mitra S (2002) Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A 99, 3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Roy LM, Jaruga P, Wood TG, McCullough AK, Dizdaroglu M and Lloyd RS (2007) Human polymorphic variants of the NEIL1 DNA glycosylase. J Biol Chem 282, 15790–15798. [DOI] [PubMed] [Google Scholar]
- (9).Jacobs AC, Calkins MJ, Jadhav A, Dorjsuren D, Maloney D, Simeonov A, Jaruga P, Dizdaroglu M, McCullough AK and Lloyd RS (2013) Inhibition of DNA glycosylases via small molecule purine analogs. PLoS One 8, e81667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Minko IG, Vartanian VL, Tozaki NN, Linde OK, Jaruga P, Coskun SH, Coskun E, Qu C, He H, Xu C, Chen T, Song Q, Jiao Y, Stone MP, Egli M, Dizdaroglu M, McCullough AK and Lloyd RS (2019) Characterization of rare NEIL1 variants found in East Asian populations. DNA Repair (Amst) 79, 32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Minko IG, Vartanian VL, Tozaki NN, Coskun E, Coskun SH, Jaruga P, Yeo J, David SS, Stone MP, Egli M, Dizdaroglu M, McCullough AK and Lloyd RS (2020) Recognition of DNA adducts by edited and unedited forms of DNA glycosylase NEIL1. DNA Repair (Amst) 85, 102741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bandaru V, Sunkara S, Wallace SS and Bond JP (2002) A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 1, 517–529. [DOI] [PubMed] [Google Scholar]
- (13).Krishnamurthy N, Zhao X, Burrows CJ and David SS (2008) Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry 47, 7137–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yeo J, Goodman RA, Schirle NT, David SS and Beal PA (2010) RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc Natl Acad Sci U S A 107, 20715–20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhou J, Liu M, Fleming AM, Burrows CJ and Wallace SS (2013) Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem 288, 27263–27272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Prakash A, Carroll BL, Sweasy JB, Wallace SS and Doublie S (2014) Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: activity, structure, and the effect of editing. DNA Repair (Amst) 14, 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Morland I, Rolseth V, Luna L, Rognes T, Bjoras M and Seeberg E (2002) Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res 30, 4926–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hailer MK, Slade PG, Martin BD, Rosenquist TA and Sugden KD (2005) Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 4, 41–50. [DOI] [PubMed] [Google Scholar]
- (19).McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ and David SS (2013) Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J Am Chem Soc 135, 13851–13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Minko IG, Christov PP, Li L, Stone MP, McCullough AK and Lloyd RS (2019) Processing of N5-substituted formamidopyrimidine DNA adducts by DNA glycosylases NEIL1 and NEIL3. DNA Repair (Amst) 73, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Couve S, Mace-Aime G, Rosselli F and Saparbaev MK (2009) The human oxidative DNA glycosylase NEIL1 excises psoralen-induced interstrand DNA cross-links in a three-stranded DNA structure. J Biol Chem 284, 11963–11970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Martin PR, Couve S, Zutterling C, Albelazi MS, Groisman R, Matkarimov BT, Parsons JL, Elder RH and Saparbaev MK (2017) The human DNA glycosylases NEIL1 and NEIL3 excise psoralen-induced DNA-DNA cross-links in a four-stranded DNA structure. Sci Rep 7, 17438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Slyvka A, Mierzejewska K and Bochtler M (2017) Nei-like 1 (NEIL1) excises 5-carboxylcytosine directly and stimulates TDG-mediated 5-formyl and 5-carboxylcytosine excision. Sci Rep 7, 9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Vartanian V, Minko IG, Chawanthayatham S, Egner PA, Lin YC, Earley LF, Makar R, Eng JR, Camp MT, Li L, Stone MP, Lasarev MR, Groopman JD, Croy RG, Essigmann JM, McCullough AK and Lloyd RS (2017) NEIL1 protects against aflatoxin-induced hepatocellular carcinoma in mice. Proc Natl Acad Sci U S A 114, 4207–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).McCullough AK and Lloyd RS (2019) Mechanisms underlying aflatoxin-associated mutagenesis - Implications in carcinogenesis. DNA Repair (Amst) 77, 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lin YC, Li L, Makarova AV, Burgers PM, Stone MP and Lloyd RS (2014) Molecular basis of aflatoxin-induced mutagenesis-role of the aflatoxin B1-formamidopyrimidine adduct. Carcinogenesis 35, 1461–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Smela ME, Hamm ML, Henderson PT, Harris CM, Harris TM and Essigmann JM (2002) The aflatoxin B1 formamidopyrimidine adduct plays a major role in causing the types of mutations observed in human hepatocellular carcinoma. Proc Natl Acad Sci U S A 99, 6655–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chawanthayatham S, Valentine CC 3rd, Fedeles BI, Fox EJ, Loeb LA, Levine SS, Slocum SL, Wogan GN, Croy RG and Essigmann JM (2017) Mutational spectra of aflatoxin B1 in vivo establish biomarkers of exposure for human hepatocellular carcinoma. Proc Natl Acad Sci U S A 114, E3101–E3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Huang MN, Yu W, Teoh WW, Ardin M, Jusakul A, Ng AWT, Boot A, Abedi-Ardekani B, Villar S, Myint SS, Othman R, Poon SL, Heguy A, Olivier M, Hollstein M, Tan P, Teh BT, Sabapathy K, Zavadil J and Rozen SG (2017) Genome-scale mutational signatures of aflatoxin in cells, mice, and human tumors. Genome Res 27, 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Volkova NV, Meier B, Gonzalez-Huici V, Bertolini S, Gonzalez S, Vohringer H, Abascal F, Martincorena I, Campbell PJ, Gartner A and Gerstung M (2020) Mutational signatures are jointly shaped by DNA damage and repair. Nat Commun 11, 2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhang W, He H, Zang M, Wu Q, Zhao H, Lu LL, Ma P, Zheng H, Wang N, Zhang Y, He S, Chen X, Wu Z, Wang X, Cai J, Liu Z, Sun Z, Zeng YX, Qu C and Jiao Y (2017) Genetic features of aflatoxin-associated hepatocellular carcinoma. Gastroenterology 153, 249–262 e242. [DOI] [PubMed] [Google Scholar]
- (32).Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, Islam SMA, Lopez-Bigas N, Klimczak LJ, McPherson JR, Morganella S, Sabarinathan R, Wheeler DA, Mustonen V, Getz G, Rozen SG and Stratton MR (2020) The repertoire of mutational signatures in human cancer. Nature 578, 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Alekseyev YO, Hamm ML and Essigmann JM (2004) Aflatoxin B1 formamidopyrimidine adducts are preferentially repaired by the nucleotide excision repair pathway in vivo. Carcinogenesis 25, 1045–1051. [DOI] [PubMed] [Google Scholar]
- (34).Leadon SA, Tyrrell RM and Cerutti PA (1981) Excision repair of aflatoxin B1-DNA adducts in human fibroblasts. Cancer Res 41, 5125–5129. [PubMed] [Google Scholar]
- (35).Takahashi Y, Nakatsuru Y, Zhang S, Shimizu Y, Kume H, Tanaka K, Ide F and Ishikawa T (2002) Enhanced spontaneous and aflatoxin-induced liver tumorigenesis in Xeroderma pigmentosum group A gene-deficient mice. Carcinogenesis 23, 627–633. [DOI] [PubMed] [Google Scholar]
- (36).Lee AJ and Wallace SS (2017) Hide and seek: How do DNA glycosylases locate oxidatively damaged DNA bases amidst a sea of undamaged bases? Free Radic Biol Med 107, 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kladova OA, Grin IR, Fedorova OS, Kuznetsov NA and Zharkov DO (2019) Conformational dynamics of damage processing by human DNA glycosylase NEIL1. J Mol Biol 431, 1098–1112. [DOI] [PubMed] [Google Scholar]
- (38).Li L, Brown KL, Ma R and Stone MP (2015) DNA sequence modulates geometrical isomerism of the trans-8,9-dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)- 9-hydroxy aflatoxin B1 adduct. Chem Res Toxicol 28, 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Brown KL, Deng JZ, Iyer RS, Iyer LG, Voehler MW, Stone MP, Harris CM and Harris TM (2006) Unraveling the aflatoxin-FAPY conundrum: structural basis for differential replicative processing of isomeric forms of the formamidopyrimidine-type DNA adduct of aflatoxin B1. J Am Chem Soc 128, 15188–15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Mao H, Deng Z, Wang F, Harris TM and Stone MP (1998) An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR. Biochemistry 37, 4374–4387. [DOI] [PubMed] [Google Scholar]
- (41).Giri I and Stone MP (2002) Thermal stabilization of the DNA duplex by adducts of aflatoxin B1. Biopolymers 65, 190–201. [DOI] [PubMed] [Google Scholar]
- (42).Brown KL, Voehler MW, Magee SM, Harris CM, Harris TM and Stone MP (2009) Structural perturbations induced by the α-anomer of the aflatoxin B1 formamidopyrimidine adduct in duplex and single-strand DNA. J Am Chem Soc 131, 16096–16107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Mullins EA, Rodriguez AA, Bradley NP and Eichman BF (2019) Emerging roles of DNA glycosylases and the Base Excision Repair pathway. Trends Biochem Sci 44, 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Adam W, Chan YY, Cremer D, Gauss J, Scheutzow D and Schindler M (1987) Spectral and chemical properties of dimethyldioxirane as determined by experiment and ab initio calculations. J Org Chem 52, 2800–2803. [Google Scholar]
- (45).Baertschi SW, Raney KD, Stone MP and Harris TM (1988) Preparation of the 8,9-epoxide of the mycotoxin aflatoxin B1: the ultimate carcinogenic species. J Am Chem Soc 110, 7929–7931. [Google Scholar]
- (46).Hwang TL and Shaka AJ (1995) Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Mag Reson, Series A 112, 275–279. [Google Scholar]
- (47).Lee W, Tonelli M and Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Patel DJ, Shapiro L and Hare D (1987) DNA and RNA: NMR studies of conformations and dynamics in solution. Q Rev Biophys 20, 35–112. [DOI] [PubMed] [Google Scholar]
- (49).Reid BR (1987) Sequence-specific assignments and their use in NMR studies of DNA structure. Q Rev Biophys 20, 2–28. [DOI] [PubMed] [Google Scholar]
- (50).Boelens R, Scheek RM, Dijkstra K and Kaptein R (1985) Sequential assignment of imino- and amino-proton resonances in 1H NMR spectra of oligonucleotides by two-dimensional NMR spectroscopy. Application to a lac operator fragment. J Magn Reson 62, 378–386. [Google Scholar]
- (51).Donigan KA and Sweasy JB (2009) Sequence context-specific mutagenesis and base excision repair. Mol Carcinog 48, 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Xia L, Zheng L, Lee HW, Bates SE, Federico L, Shen B and O’Connor TR (2005) Human 3-methyladenine-DNA glycosylase: effect of sequence context on excision, association with PCNA, and stimulation by AP endonuclease. J Mol Biol 346, 1259–1274. [DOI] [PubMed] [Google Scholar]
- (53).Ye N, Holmquist GP and O’Connor TR (1998) Heterogeneous repair of N-methylpurines at the nucleotide level in normal human cells. J Mol Biol 284, 269–285. [DOI] [PubMed] [Google Scholar]
- (54).Li M, Ko T and Li S (2015) High-resolution digital mapping of N-methylpurines in human cells reveals modulation of their induction and repair by nearest-neighbor nucleotides. J Biol Chem 290, 23148–23161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Asagoshi K, Yamada T, Terato H, Ohyama Y, Monden Y, Arai T, Nishimura S, Aburatani H, Lindahl T and Ide H (2000) Distinct repair activities of human 7,8-dihydro-8-oxoguanine DNA glycosylase and formamidopyrimidine DNA glycosylase for formamidopyrimidine and 7,8-dihydro-8-oxoguanine. J Biol Chem 275, 4956–4964. [DOI] [PubMed] [Google Scholar]
- (56).Sassa A, Beard WA, Prasad R and Wilson SH (2012) DNA sequence context effects on the glycosylase activity of human 8-oxoguanine DNA glycosylase. J Biol Chem 287, 36702–36710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Allgayer J, Kitsera N, von der Lippen C, Epe B and Khobta A (2013) Modulation of base excision repair of 8-oxoguanine by the nucleotide sequence. Nucleic Acids Res 41, 8559–8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Talhaoui I, Couve S, Gros L, Ishchenko AA, Matkarimov B and Saparbaev MK (2014) Aberrant repair initiated by mismatch-specific thymine-DNA glycosylases provides a mechanism for the mutational bias observed in CpG islands. Nucleic Acids Res 42, 6300–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Völker J, Plum GE, Gindikin V and Breslauer KJ (2019) Dynamic DNA energy landscapes and substrate complexity in triplet repeat expansion and DNA repair. Biomolecules 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Sanchez AM, Volk DE, Gorenstein DG and Lloyd RS (2003) Initiation of repair of A/G mismatches is modulated by sequence context. DNA Repair (Amst) 2, 863–878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


