Abstract
Nonsense-mediated mRNA decay (NMD), the loss of mRNAs carrying premature stop codons, is a process by which cells recognize and degrade nonsense mRNAs to prevent possibly toxic effects of truncated peptides. Most mammalian nonsense mRNAs are degraded while associated with the nucleus, but a few are degraded in the cytoplasm; at either site, there is a requirement for translation and for an intron downstream of the early stop codon. We have examined the NMD of a mutant HEXA message in lymphoblasts derived from a Tay-Sachs disease patient homozygous for the common frameshift mutation 1278ins4. The mutant mRNA was nearly undetectable in these cells and increased to approximately 40% of normal in the presence of the translation inhibitor cycloheximide. The stabilized transcript was found in the cytoplasm in association with polysomes. Within 5 h of cycloheximide removal, the polysome-associated nonsense message was completely degraded, while the normal message was stable. The increased lability of the polysome-associated mutant HEXA mRNA shows that NMD of this endogenous mRNA occurred in the cytoplasm. Transfection of Chinese hamster ovary cells showed that expression of an intronless HEXA minigene harboring the frameshift mutation or a closely located nonsense codon resulted in half the normal mRNA level. Inclusion of multiple downstream introns decreased the abundance further, to about 20% of normal. Thus, in contrast to other systems, introns are not absolutely required for NMD of HEXA mRNA, although they enhance the low-HEXA-mRNA phenotype.
Investigators studying genetic disorders have often found that some mutations are associated with very low levels of mRNA. Barring gross DNA alterations, these are usually frameshift or nonsense mutations (15, 21–23, 25, 28, 34, 35, 43, 46, 48, 61, 62). Among them are mutations of the HEXA gene, leading to Tay-Sachs disease (TSD), an autosomally inherited neurodegenerative disorder (26). While numerous mutations have been identified as the cause of TSD, the most common is a 4-bp insertion in exon 11 (1278ins4) of the 14-exon HEXA gene, shortening the reading frame by 100 codons. Patient fibroblasts homozygous for this frameshift mutation (or in compound heterozygosity with a splice site mutation) have undetectable levels of the nonsense-containing mRNA (48), even though transcription is normal (52). It is not the frameshift itself but the premature termination codon 9 nucleotides (nt) downstream that causes the low-mRNA phenotype (12).
The degradation of mRNAs containing premature stop codons, termed nonsense-mediated mRNA decay (NMD), is a process of much broader biological significance than removal of mRNA in genetic disease (4, 10, 33, 40–42, 44, 54, 56, 57, 66; L. E. Maquat, Editorial, Am. J. Hum. Genet. 59:279–286, 1996). Organisms as diverse as yeast (54), worms (51, 56), plants (55), and mammals (41; Maquat, editorial), demonstrate this phenomenon. It is presumed that NMD prevents the synthesis of truncated peptides that could have dominant negative effects on the organism. The process serves as a general surveillance mechanism to abolish aberrant transcripts resulting not only from rare mutations but also from mistakes in RNA processing (56).
Numerous studies in the last decade have addressed the mechanism of NMD. In Saccharomyces cerevisiae, NMD occurs in the cytoplasm while the transcript is associated with polysomes (73). It has been suggested that when an early stop codon is reached, translation stalls and the message is scanned by specific factors. If a certain downstream element is encountered (53, 72), and if specific trans factors are present (1, 2, 20, 36, 53, 60), the message undergoes a type of conformational change that renders it sensitive to a 5′ decapping enzyme independent of the normal poly(A) tail shortening, followed by a 5′-to-3′ exonuclease-mediated degradation (27, 45). Thus, the data show that in yeast, NMD occurs in the cytoplasm (1, 2, 7, 18, 19, 30, 45, 53, 54, 57–60, 73).
However, very few mammalian nonsense messages studied to date show a cytoplasmic mode of decay. These include transcripts for β-globin from bone marrow cells of transgenic mice (38, 39), Rous sarcoma virus gag (4, 5), and selenium-dependent glutathione peroxidase I (44; P. M. Moriarty, C. C. Reddy, and L. E. Maquat, Letter, RNA 3:1369–1373, 1997). Instead, most mammalian nonsense mRNAs display a nuclear or nucleus-associated mechanism of decay. These include transcripts for human triosephosphate isomerase (8, 9, 16, 17, 21, 69, 70), human β-globin in nonerythroid cells (6, 71), hamster dihydrofolate reductase (66), hamster adenine phosphoribosyltransferase (33), mouse major urinary protein (10), and mouse T-cell receptor β (13, 14). With these transcripts, the level of the nonsense mRNA that fractionates with the nucleus is reduced to the same degree as the level in the cytoplasm; furthermore, the nonsense mRNA that escapes into the cytoplasm has normal stability (6, 9, 10, 17, 21, 33, 62, 66). This implies that NMD occurs while the message is still associated with the nucleus. But translation remains essential, as shown by the stabilization of nonsense mRNAs in the presence of translation-inhibiting drugs (13, 14), tRNA suppressors (8), and 5′ stem-loop structures that prevent mRNA translation (8, 65).
Additional suggestions of nuclear involvement in NMD come from observations that exons harboring an early stop codon are often skipped (3, 22, 23), implying recognition of the reading frame in the nucleus. However, this interpretation is not universally accepted, and the suggestion of a reverse transcription-PCR artifact has been offered as an explanation for nonsense-mediated exon skipping (67, 68).
The strongest evidence that nuclear processes are important for NMD in mammalian cells comes from the failure of intronless minigenes (cDNAs) to reproduce the decay phenotype (50, 66, 70; X. Sun, and L. E. Maquat, Letter, RNA 6:1–8, 2000) and from the requirement for downstream introns (10, 14, 16, 65, 70, 71). To reconcile the nuclear involvement and the requirement for cytoplasmic translation, it has been proposed that exon-exon junctions are tagged upon completion of splicing, producing an mRNP capable of signaling decay when an early stop codon is present at a proper distance from the tag (31). Some transcripts would be translated, recognized as nonsense, and degraded in the cytoplasm, and others would undergo this process while still associated with the nucleus. In this paper, we address the site of decay and the requirement for introns for the NMD of the HEXA transcript.
MATERIALS AND METHODS
Cell culture.
Normal (GM 03299D) and TSD (GM 11852) lymphoblasts (Coriell Institute) were cultured in suspension under 5% CO2 in T75 flasks with RPMI 1640 medium (Life Technologies) containing 10% fetal bovine serum (Irvine Scientific), penicillin and streptomycin, nonessential amino acids, and sodium pyruvate. Cells were passaged 1 to 2 days earlier, so that they would be at a density of about 2 × 106 cells/ml on the day of the experiment. To inhibit translation, 2 × 107 to 3 × 107 lymphoblasts were pelleted, washed once with phosphate-buffered saline, and resuspended in medium containing 28 μg of cycloheximide/ml. To subsequently remove the cycloheximide, cells were pelleted, washed twice with phosphate-buffered saline, and resuspended in medium without the inhibitor. The dose of cycloheximide selected, 28 μg/ml, inhibited translation by 93% after a 4-h incubation, as monitored by [35S]methionine-cysteine incorporation. After removal of cycloheximide, total translation increased to 63% of the initial level within 1 h and remained at that level for an additional 5 h. To inhibit transcription, 5 μg of actinomycin D/ml (29, 33, 66) was added to the cycloheximide-free medium. Chinese hamster ovary cells were cultured under 5% CO2 in 10-cm plates with alpha minimal essential medium (Life Technologies) and 5% fetal bovine serum.
RNA extraction.
Total cellular RNA was extracted with RNA STAT 60 (Tel-Test) as per the supplier's protocol. Nuclear and cytoplasmic RNAs were isolated by methods described by Jakubowski and Roberts (32). Briefly, cells were lysed with 500 μl of lysis buffer (10 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2, 0.3 M sucrose, 0.5% NP-40, 0.25% sodium deoxycholate, and 0.5 U of RNase inhibitor [Boehringer Mannheim]/μl) and loaded onto 500 μl of a cushion buffer (10 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2, and 0.4 M sucrose). The samples were centrifuged for 10 min at 800 × g. The cytoplasmic supernatant was treated with proteinase K, extracted with phenol-chloroform, and precipitated with ethanol. The nuclear pellet was resuspended in 150 μl of high-salt DNase buffer (10 mM Tris-HCl [pH 7.4], 500 mM NaCl, 5 mM MgCl2, and 0.1 mM CaCl2) and incubated for 30 min at 37°C with 100 U of DNase I (Life Technologies). The samples were then treated with proteinase K, extracted with phenol-chloroform, and precipitated with ethanol.
Northern blot analysis.
Fifteen-microgram aliquots of total, nuclear, and cytoplasmic RNA were used for Northern blot analysis. The RNA was subjected to electrophoresis on a 1% agarose–formaldehyde gel and blotted onto a nylon membrane. The blots were stained with methylene blue to visualize the 18S and 28S rRNA and then probed with a 32P-labeled full-length HEXA cDNA, a 360-bp 5′ HEXA cDNA, a 600-bp β-actin cDNA, or an 800-bp neomycin resistance probe. A riboprobe was synthesized from the plasmid pSPU4b, kindly provided by Douglas Black (University of California, Los Angeles), for the 94-nt U4 snRNA probe (11). Radioactivity was visualized and quantitated with a PhosphorImager (Molecular Dynamics).
Polysome profile analysis.
Polysomes were isolated following methods described by Duncan and Burgoyne (24). In brief, 2 × 107 to 3 × 107 lymphoblasts were lysed with 900 μl of lysis buffer (50 mM Tris-HCl [pH 7.4], 100 mM KCl, 5 mM MgCl2, 1% Triton X-100, 1% sodium deoxycholate, 1 mg of heparin/ml, and 20 μg of cycloheximide/ml) and loaded onto a 15-to-50% linear sucrose gradient. The sucrose solutions were made with gradient buffer (50 mM Tris [pH 7.4], 100 mM KCl, and 5 mM MgCl2) and poured into 14- by 89-cm Beckman UltraClear centrifuge tubes using a gradient mixer attached to a peristaltic pump. To disrupt polysomes, 30 mM EDTA was added to the lysis buffer. Samples were centrifuged at 4°C in an SW41 rotor at 40,000 rpm for 2.5 h. Polysome profiles were created with an ISCO model 185 gradient fractionator with a tube piercer, attached to a UA-6 absorbance monitor and fraction collector. Fourteen fractions were collected, and sodium dodecyl sulfate was added to each to a concentration of 0.5%. RNA was isolated by extraction with phenol-chloroform and then chloroform alone and precipitated with 3 M ammonium acetate and ethanol. The entire RNA sample from each fraction was used for Northern analysis.
Construction of minigenes.
Standard molecular biology techniques were used in the construction of the normal and mutant HEXA minigenes and cDNAs. Restriction enzymes were purchased from Pharmacia, and the PCR enzymes TaqPlus Precision PCR system and Pfu DNA polymerase were purchased from Stratagene. All constructs were cloned into the pcDNA3.1(−) vector (Invitrogen) and were thus driven by the cytomegalovirus (CMV) promoter and carried a neomycin resistance (neor) gene. All the mutant constructs except the W392X intronless minigene carried the 1278ins4 mutation in exon 11. The W392X mutation creates a stop codon in exon 11, about 114 nt upstream of the stop codon resulting from 1278ins4.
(i) Construct 1.
Two HEXA minigenes (12) with four introns upstream and three downstream of the mutation were a kind gift from Richard L. Proia (National Institutes of Health). These minigenes, normal (TKα) and mutant (Mut TKα), were digested with SalI, and the 8.8-kb fragment was cloned into the XhoI linearized pcDNA3.1(−) in order to transfer them from a herpes simplex thymidine kinase promoter to a CMV promoter.
(ii) Construct 2.
The two minigenes were digested with KpnI and partially digested with SalI to purify a 6.5-kb fragment, which was ligated to a 1.45-kb SalI/KpnI fragment PCR amplified from the normal HEXA cDNA. This intermediate plasmid was linearized with KpnI and ligated to a 200-bp KpnI fragment from the original normal and mutant minigenes, digested with SalI, and finally cloned into XhoI-linearized pcDNA3.1(−).
(iii) Construct 3.
Two bands of about 3 and 7.9 kb were gel purified after digestion of the normal and mutant construct 2 with HpaI and HindIII. The 3-kb fragment was further cut with Psp1406I to yield a 550-bp fragment, which was then ligated to the 7.9-kb HpaI/HindIII fragment and a 420-bp HindIII/Psp1406I fragment PCR amplified from the normal HEXA cDNA.
(iv) Construct 4.
Normal 2-kb HEXA cDNA was gel purified after digestion with XbaI and HindIII and then cloned into pcDNA3.1(−). The intermediate construct was cut with KpnI to release a 72-bp fragment, which was then replaced with a 273-bp genomic KpnI fragment with or without the 4-bp mutation.
(v) Construct 5.
A 5.4-kb PvuII/EcoRI fragment of construct 1 was cloned into pGEMEX-1 (Promega) and then digested with PvuII and BamHI to release a 4.6-kb vector. This vector was ligated to a 820-bp PvuII/BamHI fragment obtained either from the normal or mutant HEXA cDNA. This intermediate construct was digested with EcoRI yielding a 1.4-kb band that was ligated to an 8-kb EcoRI fragment of construct 1.
(vi) Construct 6.
The mutant cDNAs (harboring the 1278ins4 or the W392X mutation) were both created with the PCR-based QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. Briefly, the normal HEXA cDNA in the pGEM-3Zf(+) (Promega) vector was PCR amplified by Pfu DNA polymerase (Stratagene), known for its high fidelity, with primers harboring the desired mutation. The nicked DNA was used to transform XL1-Blue ultracompetent cells after the parental DNA had been digested with DpnI. The new mutant cDNAs were sequenced to confirm the presence of the mutation. They were then digested with XbaI and PstI to release a 2-kb fragment, which was cloned into pBluescript II KS(+) (Stratagene). This intermediate construct was cut with XbaI and EcoRI to yield a 2.1-kb band that was finally cloned into pcDNA3.1(−).
Transfections.
CHO cells were transiently transfected with the various constructs using the Lipofectamine Plus system (Life Technologies) according to the manufacturer's directions. Transfection was carried out in 10-cm plates with 4 μg of DNA in serum-free medium for 3 h. Serum was added to 5%, and about 40 h later, RNA was extracted for analysis as described above.
RESULTS
Translation is required for decay of nonsense HEXA mRNA.
The effect of inhibiting translation on nonsense mRNA abundance was examined by treating normal and TSD lymphoblasts with cycloheximide. Total RNA was analyzed by Northern blotting, which in these and subsequent experiments showed three distinguishable HEXA species—a major species of 2.1 kb and two minor species of 2.6 and 2.4 kb (Fig. 1 and 2). The 2.1-kb mRNA migrated in the region of the 18S rRNA, which occasionally disrupted the hybridization signal. The minor 2.6-kb transcript hybridized with several nonoverlapping HEXA probes and disappeared within 2 h of actinomycin D treatment (data not shown); it is therefore thought to be a pre-mRNA. It was present in normal and mutant cells, appeared to be insensitive to NMD, and was not studied further. The other minor species, of 2.4 kb, hybridized to an alternative 3′ untranslated region probe (data not shown), showing that it was the previously described HEXA mRNA with a longer 3′ untranslated region (47). Though the 2.4-kb message was subject to NMD, only the abundant full-length 2.1-kb mRNA was used for all analyses.
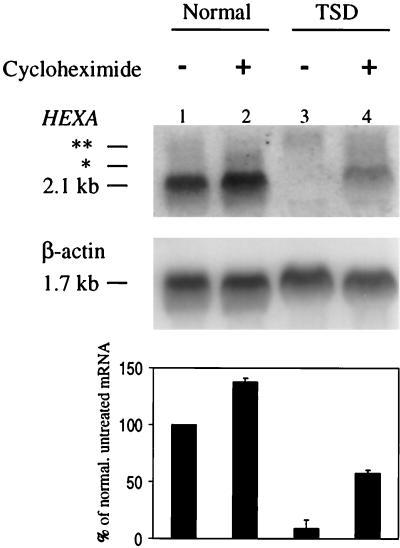
FIG. 1.
Stabilization of the nonsense TSD mRNA with cycloheximide. (Top) Northern blot analysis of total RNA isolated from lymphoblasts, either normal (lanes 1 and 2) or TSD (lanes 3 and 4), with and without a 4-h cycloheximide treatment. (Bottom) PhosphorImager quantitation of the average amount of HEXA mRNA in the above lanes and in two other experiments, normalized to β-actin. Error bars indicate standard deviations. The mRNA measured is the major species of 2.1 kb. The two minor species (2.4 [∗] and 2.6 [∗∗] kb, respectively) are described in the text.
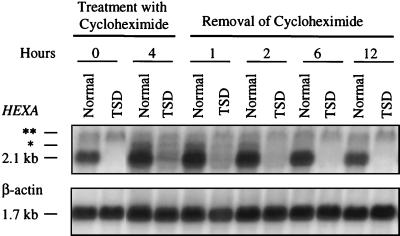
FIG. 2.
Decay of the cycloheximide-stabilized pool of HEXA mRNA. Northern blot analysis of total RNA isolated from lymphoblasts before treatment (0 h), after a 4-h treatment with cycloheximide, and 1, 2, 6, and 12 h following cycloheximide removal. Blots were rehybridized with a β-actin cDNA probe for normalization. Asterisks indicate minor mRNA species as in Fig. 1.
As seen in Fig. 1, the HEXA mRNA was nearly undetectable in TSD lymphoblasts, less than 10% of nonsense-free mRNA, prior to treatment with cycloheximide. After 4 h of treatment, the level of TSD mRNA had increased to 56% of untreated normal mRNA or 40% of cycloheximide-treated normal mRNA. Cycloheximide withdrawal, which allowed translation to resume within an hour, caused the level of the accumulated mutant mRNA to decrease progressively until it reached the low steady-state level in 6 h (Fig. 2). Quantitation of the data in Fig. 2 revealed that half the mutant message had disappeared in about 3 h (data not shown). On the other hand, the half-life of the normal mRNA was estimated using actinomycin D to be more than 30 h (data not shown), far longer than the time the lymphoblasts could be maintained with the transcriptional inhibitor.
Cycloheximide-stabilized nonsense HEXA mRNA is found only in the cytoplasmic fraction.
Cellular fractionation and Northern blot analysis were used to determine the cellular distribution of the normal and nonsense HEXA mRNA. As shown in Fig. 3, in the absence of cycloheximide, the HEXA mRNA was lacking in both the nuclear and cytoplasmic fractions of TSD cells, while it was present almost exclusively in the cytoplasmic fraction of normal cells. After treatment with cycloheximide, the stabilized TSD mRNA was elevated exclusively in the cytoplasmic fraction, with no noticeable changes in the nuclear fraction. (The NMD-insensitive 2.6-kb transcript was seen in all fractions and is not included in the analysis.) Although some leakage from nucleus to cytoplasm may have occurred, the nuclear fraction appeared to be generally intact, as shown by enrichment of the snRNAs U4 (Fig. 3) and U6 (data not shown).
FIG. 3.
Northern blot analysis of nuclear and cytoplasmic HEXA mRNA distribution. Lymphoblasts, before or after a 4-h treatment with cycloheximide, were lysed with buffer containing 0.5% NP-40 and loaded onto a 0.4 M sucrose pad to pellet nuclei. RNA was extracted from the supernatant (cytoplasmic [C]) and the pelleted (nuclear [N]) fractions. Approximately 15 μg of nuclear and cytoplasmic RNA, or about a tenth of the cytoplasmic and about half of the nuclear yield, was used for Northern analysis. Blots were reprobed for the nuclear fraction-enriched U4 snRNA. ∗∗, 2.6-kb NMD-insensitive transcript.
Nonsense HEXA mRNA accumulates on polysomes in the presence of cycloheximide and disappears from polysomes upon removal of the inhibitor.
The cytoplasmic HEXA mRNA distribution was determined by Northern blot analysis of ribosome fractions from normal and TSD lymphoblasts. Fractionation of cell lysates through a sucrose gradient separated ribosomal subunits (40S and 60S), monosomes (80S), and polysomes into 14 fractions. The absorbance profile of each sample at 254 nm showed polysomes to be abundant in fractions 9 to 14 (Fig. 4, left panels). The presence of polysomes in these fractions was verified by the addition of 30 mM EDTA, which disrupts polysomes, to a sample lysate prior to centrifugation. In the presence of EDTA, absorbance shifted from the polysome peaks (fractions 9 to 14) to the upper monosome or soluble phase (fractions 2 to 7), and most of the polysome-associated HEXA message shifted to the lighter fractions (data not shown).
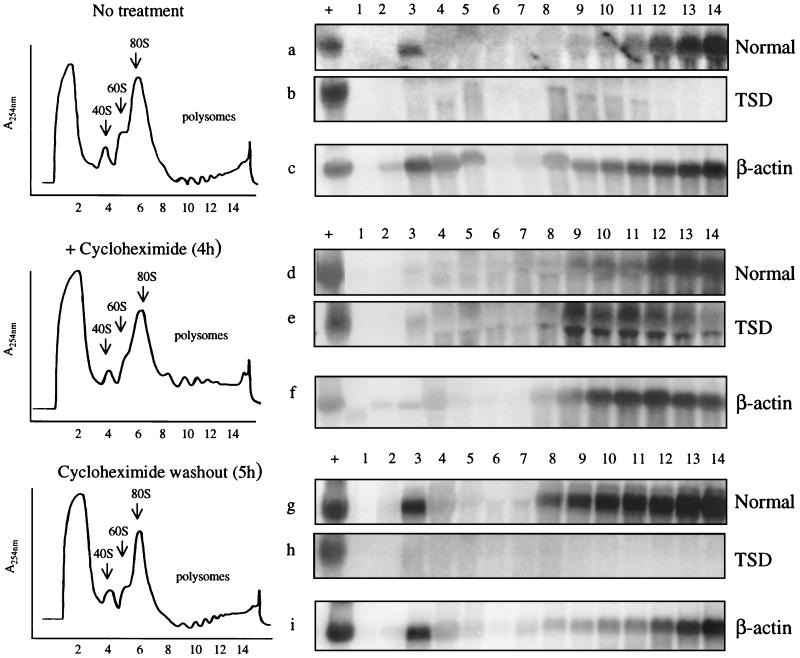
FIG. 4.
Polysome profiles and Northern blot analysis of density gradient fractions of normal and TSD lymphoblasts. Cells were untreated, treated with cycloheximide for 4 h, or treated and then released from cycloheximide for 5 h. Samples were then lysed and centrifuged through a 15-to-50% linear sucrose gradient. A UV absorbance monitor continuously monitored the gradient before separation into 14 fractions. The locations of the 40S and 60S ribosomal subunits, the 80S monosomes, and the polysomes are indicated. RNA was extracted from each fraction for Northern blot analysis. Fifteen micrograms of total RNA from normal GM 03299D cells was loaded on all gels as a positive control for hybridization (lanes +). The entire RNA yield from the gradient fractions was used for the blots. Blots were initially probed with the full-length 2-kb HEXA cDNA probe (a, b, d, e, g, and h) and then reprobed with a β-actin probe (c, f, and i). Polysome profiles and β-actin Northern blots are shown only for preparations from TSD cells, but equivalent preparations from normal cells gave similar results.
The effect of cycloheximide treatment and removal on the nonsense mRNA associated with polysomes is shown in the Northern blots depicted in Fig. 4. Cycloheximide inhibits peptide elongation and freezes polysomes on mRNA. Prior to cycloheximide treatment, there was negligible mutant mRNA associated with polysomes (Fig. 4b), in contrast to a substantial amount of normal mRNA (Fig. 4a). After 4 h of cycloheximide treatment, TSD mRNA appeared in polysomal fractions 8 to 14 (Fig. 4e). Upon removal of the inhibitor, the TSD mRNA disappeared from all polysomal fractions (Fig. 4h) while the normal mRNA continued to associate with polysomes either in the presence of actinomycin D (Fig. 4g) or in its absence (data not shown). Northern blots of the β-actin mRNA from TSD cells showed approximately equal loading (Fig. 4c, f, and i), as did blots of β-actin mRNA from normal cells (data not shown). These experiments show that polysome-associated mutant HEXA mRNA is subject to degradation when translation is resumed.
Introns are not absolutely required but enhance NMD of HEXA mRNA.
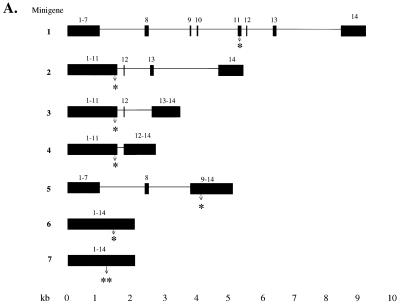
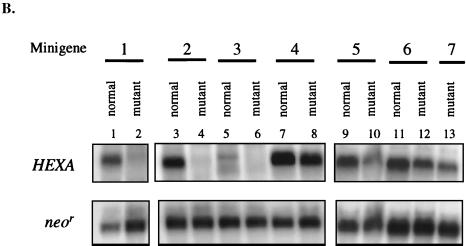
Intron-containing and intronless HEXA minigenes (Fig. 5A), driven by the CMV promoter, were transiently expressed in CHO cells. All but one of the mutant minigenes had the common 1278ins4 mutation, which shortens the reading frame by 100 codons to four-fifths of the normal frame. An additional nonsense cDNA, W392X, was tested for comparison, as it also had been shown to result in low mRNA in cells of TSD patients (61). The nonsense codon W392X is located 117 nt upstream of the nonsense codon of 1278ins4 (Fig. 5A, constructs 6 and 7). For each construct, the HEXA mRNA was subject to Northern blot analysis (Fig. 5B), quantitated by PhosphorImager, and normalized to the neor mRNA transcribed from the same plasmid in order to account for differences in transfection and RNA recovery (Fig. 5C).
FIG. 5.
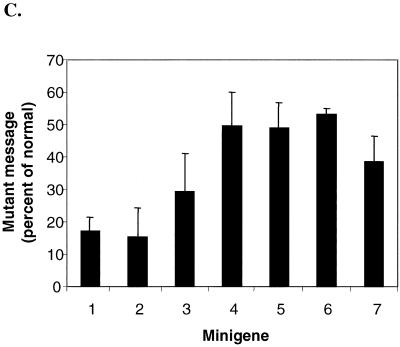
Transient transfection of CHO cells with normal and mutant HEXA minigenes. (A) Seven constructs containing the indicated exons (black box) and introns (black horizontal line) were made. All intron-containing mutant minigenes carried the 1278ins4 mutation (∗) (constructs 1 to 5). This placed the early stop codon 41 nt upstream of intron 11, as in the naturally occurring mutation. Of the two intronless minigenes (6 and 7), the first has the 1278ins4 mutation (∗) and the other has the W392X mutation (∗∗). The sites of these mutations are shown with an arrow. (B) Representative Northern blot analysis of RNA extracted 40 h after transient transfection of CHO cells with the minigenes in panel A. Fifteen micrograms of total RNA was used, and the blots were first probed with the 360-bp HEXA 5′ probe and then reprobed for the neomycin resistance (neor) mRNA. (C) HEXA mRNA levels were quantitated with a PhosphorImager, normalized to neor mRNA levels, and then expressed as a percentage of the normal counterpart. The average of three experiments is shown, with standard deviations.
The mutant minigenes with four introns upstream and/or only three introns downstream of the frameshift mutation, minigenes 1 and 2, expressed nonsense message at a fifth of the normal mRNA, showing that upstream introns are not essential for NMD. Further deletion of the downstream intron 13 (minigene 3) resulted in nonsense mRNA at about a third of normal mRNA level. Minigene 4, with only intron 11, or minigene 5, with the upstream introns 7 and 8 resulted in nonsense mRNA at about half the normal mRNA level. Even minigenes without any introns (cDNAs) produced nonsense mRNAs that were present at lower levels than normal; the 1278ins4 nonsense message was present on average at 53% of normal, while the W392X nonsense message was found to be at 39% of normal (minigenes 6 and 7, respectively). These data show that some NMD of the HEXA mRNA is seen when the nonsense codon is in the terminal or penultimate exon or in an intronless minigene. Maximal NMD is seen if there are two or more downstream introns.
DISCUSSION
NMD of mammalian messages occurs most often while the transcript is still associated with the nucleus (6, 8–10, 13, 14, 16, 17, 21, 33, 66, 69–71), though in a few cases it occurs when the mRNA is in the cytoplasm (4, 5, 38, 39, 44; Moriarty et al., letter). Our studies show that NMD of the HEXA mRNA is similar to all other mammalian mRNAs in that ongoing translation is required for decay (Fig. 1 to 4). However, it differs from nucleus-associated NMD (33, 63) in that the cytoplasmic, polysome-associated nonsense HEXA message remains sensitive to decay (Fig. 4); thus, the HEXA transcript may be added to the short list of mammalian messages that follow a cytoplasmic mode of decay. In addition, NMD of the HEXA message differs from that of all mammalian transcripts studied to date (49, 64) in that downstream introns are not absolutely required, although they enhance the process.
Although we have shown degradation of cytoplasmic nonsense HEXA mRNA, we cannot exclude the possibility that some TSD transcripts are degraded while associated with the nucleus. First, we could demonstrate stabilization of the nonsense mRNA pool to only 40% of normal. Second, any increase in nucleus-associated nonsense mRNA in the presence of cycloheximide would not have been observed, because the HEXA mRNA was not detected even in the nuclear fraction of normal cells, implying rapid processing and export of this transcript.
Of the few mammalian mRNAs that follow a cytoplasmic mechanism of decay (5, 38, 39), the glutathione peroxidase I transcript, the one best studied, requires an intron at least 59 nt downstream of the early stop codon (44, 64Moriary et al., letter), as do transcripts that decay while associated with the nucleus (10, 49, 65, 70, 71). To explain the necessity of introns and the requirement of cytoplasmic translation, a model has been proposed in which transcripts, upon completion of splicing, are marked at each exon-exon junction (31, 65, 70, 71). This mRNP would be exported out of the nucleus, translated, and degraded if an early stop codon was reached and there was an exon-exon junction marker at a proper distance downstream. If the proper mRNP structure was not created (i.e., there was no splicing to create a downstream exon-exon junction marker [14], or if the marker was too close to the early stop codon [49, 64]), then that message could escape decay.
Our data showing that intronless HEXA minigenes that harbor nonsense codons give rise to low mRNA add an interesting complexity to the mechanism of NMD; how can the same mechanism operate on intron-containing and intronless genes containing early stop codons with the same effect? As mentioned above, it has been proposed that certain factors bind to exon-exon junctions, and spliceosomal proteins (i.e., SRm160 and hPrp8p) have been found to associate with spliced mRNA at such junctions (37), creating a certain mRNP structure. It could be envisioned that some nonsense transcripts would rely on the process of splicing to create the proper mRNP complex to facilitate NMD, but others, resembling nonsense mRNAs in yeast, would rely partially or exclusively on sequences in cis to create the functionally equivalent mRNP structure. In addition, the mRNP structures derived either with or without splicing could facilitate NMD with different efficiencies. Such may be the case for the HEXA message, for which the decrease in abundance in the absence of introns was not as severe as when introns were present.
To our knowledge, this is the first time intronless nonsense HEXA minigenes have been shown to reproduce, if incompletely, the low-mRNA phenotype. Previously, the level of nonsense HEXA mRNA was found to be normal in transfected COS-1 cells (50). But COS-1 cells may not be suitable for the study of NMD, at least of this transcript. They failed to show NMD when transfected with an intron-containing HEXA minigene, even though the same minigene was subject to NMD in mouse L cells (12).
These results, in the context of the published literature, confirm that NMD in mammalian cells is a multipathway phenomenon, with some transcripts following a nuclear or nucleus-associated pathway and others following a cytoplasmic pathway. It is also possible that the pathways are hierarchical and that nonsense codons can be recognized and degraded, with various efficiencies, at different stages following transcription.
ACKNOWLEDGMENTS
We thank Douglas Black (UCLA) for providing the U4 and U6 snRNA probes and Dohn Glitz (UCLA) and David Greenberg (UCLA) for helpful discussions.
This work was supported in part by National Institutes of Health research grant NS22376.
REFERENCES
- 1.Atkin A L, Altamura N, Leeds P, Culbertson M R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkin A L, Schenkman L R, Eastham M, Dahlseid J N, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 3.Bach G, Moskowitz S M, Tieu P T, Matynia A, Neufeld E F. Molecular analysis of Hurler syndrome in Druze and Muslim Arab patients in Israel: multiple allelic mutations of the IDUA gene in a small geographic area. Am J Hum Genet. 1993;53:330–338. [PMC free article] [PubMed] [Google Scholar]
- 4.Barker G F, Beemon K. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol. 1991;11:2760–2768. doi: 10.1128/mcb.11.5.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker G F, Beemon K. Rous sarcoma virus RNA stability requires an open reading frame in the gag gene and sequences downstream of the gag-pol junction. Mol Cell Biol. 1994;14:1986–1996. doi: 10.1128/mcb.14.3.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baserga S J, Benz E J., Jr Beta-globin nonsense mutation: deficient accumulation of mRNA occurs despite normal cytoplasmic stability. Proc Natl Acad Sci USA. 1992;89:2935–2939. doi: 10.1073/pnas.89.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 8.Belgrader P, Cheng J, Maquat L E. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci USA. 1993;90:482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belgrader P, Cheng J, Zhou X, Stephenson L S, Maquat L E. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol Cell Biol. 1994;14:8219–8228. doi: 10.1128/mcb.14.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belgrader P, Maquat L E. Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations can facilitate exon skipping. Mol Cell Biol. 1994;14:6326–6336. doi: 10.1128/mcb.14.9.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Black D L, Pinto A L. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol Cell Biol. 1989;9:3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boles D J, Proia R L. The molecular basis of HEXA mRNA deficiency caused by the most common Tay-Sachs disease mutation. Am J Hum Genet. 1995;56:716–724. [PMC free article] [PubMed] [Google Scholar]
- 13.Carter M S, Doskow J, Morris P, Li S, Nhim R P, Sandstedt S, Wilkinson M F. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 14.Carter M S, Li S, Wilkinson M F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 15.Chang J C, Kan Y W. β0 thalassemia, a nonsense mutation in man. Proc Natl Acad Sci USA. 1979;76:2886–2889. doi: 10.1073/pnas.76.6.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng J, Belgrader P, Zhou X, Maquat L E. Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol Cell Biol. 1994;14:6317–6325. doi: 10.1128/mcb.14.9.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng J, Maquat L E. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol. 1993;13:1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czaplinski K, Majlesi N, Banerjee T, Peltz S W. Mtt1 is a Upf1-like helicase that interacts with the translation termination factors and whose overexpression can modulate termination efficiency. RNA. 2000;6:730–743. doi: 10.1017/s1355838200992392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Han X, Weng Y, Perlick H A, Dietz H C, Ter-Avanesyan M D, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czaplinski K, Weng Y, Hagan K W, Peltz S W. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 21.Daar I O, Maquat L E. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol Cell Biol. 1988;8:802–813. doi: 10.1128/mcb.8.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietz H C, Kendzior R J., Jr Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet. 1994;8:183–188. doi: 10.1038/ng1094-183. [DOI] [PubMed] [Google Scholar]
- 23.Dietz H C, Valle D, Francomano C A, Kendzior R J, Jr, Pyeritz R E, Cutting G R. The skipping of constitutive exons in vivo induced by nonsense mutations. Science. 1993;259:680–683. doi: 10.1126/science.8430317. [DOI] [PubMed] [Google Scholar]
- 24.Duncan J S, Burgoyne R D. Characterization of the effects of Ca2+ depletion on the synthesis, phosphorylation and secretion of caseins in lactating mammary epithelial cells. Biochem J. 1996;317:487–493. doi: 10.1042/bj3170487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gossage D L, Norby-Slycord C J, Hershfield M S, Markert M L. A homozygous 5 base-pair deletion in exon 10 of the adenosine deaminase (ADA) gene in a child with severe combined immunodeficiency and very low levels of ADA mRNA and protein. Hum Mol Genet. 1993;2:1493–1494. doi: 10.1093/hmg/2.9.1493. [DOI] [PubMed] [Google Scholar]
- 26.Gravel R A, Clarke J T R, Kaback M M, Mahuran D, Sandhoff K, Suzuki K. The GM2 gangliosidoses. In: Scriver C R, Beaudet A L, Sly W S, Valle D, editors. The metabolic and molecular bases of inherited disease. Vol. 2. New York, N.Y: McGraw-Hill, Inc.; 1995. pp. 2839–2879. [Google Scholar]
- 27.Hagan K W, Ruiz-Echevarria M J, Quan Y, Peltz S W. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamosh A, Rosenstein B J, Cutting G R. CFTR nonsense mutations G542X and W1282X associated with severe reduction of CFTR mRNA in nasal epithelial cells. Hum Mol Genet. 1992;1:542–544. doi: 10.1093/hmg/1.7.542. [DOI] [PubMed] [Google Scholar]
- 29.Hattori Y, Vera J C, Rivas C I, Bersch N, Bailey R C, Geffner M E, Golde D W. Decreased insulin-like growth factor I receptor expression and function in immortalized African Pygmy T cells. J Clin Endocrinol Metab. 1996;81:2257–2263. doi: 10.1210/jcem.81.6.8964861. [DOI] [PubMed] [Google Scholar]
- 30.He F, Peltz S W, Donahue J L, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc Natl Acad Sci USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hentze M W, Kulozik A E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 32.Jakubowski M, Roberts J L. Processing of gonadotropin-releasing hormone gene transcripts in the rat brain. J Biol Chem. 1994;269:4078–4083. [PubMed] [Google Scholar]
- 33.Kessler O, Chasin L A. Effects of nonsense mutations on nuclear and cytoplasmic adenine phosphoribosyltransferase RNA. Mol Cell Biol. 1996;16:4426–4435. doi: 10.1128/mcb.16.8.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kugler W, Enssle J, Hentze M W, Kulozik A E. Nuclear degradation of nonsense mutated beta-globin mRNA: a post-transcriptional mechanism to protect heterozygotes from severe clinical manifestations of beta-thalassemia? Nucleic Acids Res. 1995;23:413–418. doi: 10.1093/nar/23.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J H, Novoradovskaya N, Rundquist B, Redwine J, Saltini C, Brantly M. Alpha 1-antitrypsin nonsense mutation associated with a retained truncated protein and reduced mRNA. Mol Genet Metab. 1998;63:270–280. doi: 10.1006/mgme.1998.2680. [DOI] [PubMed] [Google Scholar]
- 36.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 37.Le Hir H, Moore M J, Maquat L E. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 38.Lim S K, Maquat L E. Human beta-globin mRNAs that harbor a nonsense codon are degraded in murine erythroid tissues to intermediates lacking regions of exon I or exons I and II that have a cap-like structure at the 5′ termini. EMBO J. 1992;11:3271–3278. doi: 10.1002/j.1460-2075.1992.tb05405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim S K, Sigmund C D, Gross K W, Maquat L E. Nonsense codons in human beta-globin mRNA result in the production of mRNA degradation products. Mol Cell Biol. 1992;12:1149–1161. doi: 10.1128/mcb.12.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci USA. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 42.Maquat L E, Kinniburgh A J. A beta zero-thalassemic beta-globin RNA that is labile in bone marrow cells is relatively stable in HeLa cells. Nucleic Acids Res. 1985;13:2855–2867. doi: 10.1093/nar/13.8.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menon K P, Neufeld E F. Evidence for degradation of mRNA encoding alpha-l-iduronidase in Hurler fibroblasts with premature termination alleles. Cell Mol Biol (Noisy-le-Grand) 1994;40:999–1005. [PubMed] [Google Scholar]
- 44.Moriarty P M, Reddy C C, Maquat L E. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 46.Myerowitz R, Costigan F C. The major defect in Ashkenazi Jews with Tay-Sachs disease is an insertion in the gene for the alpha-chain of beta-hexosaminidase. J Biol Chem. 1988;263:18587–18589. [PubMed] [Google Scholar]
- 47.Myerowitz R, Piekarz R, Neufeld E F, Shows T B, Suzuki K. Human beta-hexosaminidase alpha chain: coding sequence and homology with the beta chain. Proc Natl Acad Sci USA. 1985;82:7830–7834. doi: 10.1073/pnas.82.23.7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myerowitz R, Proia R L. cDNA clone for the alpha-chain of human beta-hexosaminidase: deficiency of alpha-chain mRNA in Ashkenazi Tay-Sachs fibroblasts. Proc Natl Acad Sci USA. 1984;81:5394–5398. doi: 10.1073/pnas.81.17.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagy E, Maquat L E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 50.Nishimoto J, Tanaka A, Nanba E, Suzuki K. Expression of the beta-hexosaminidase alpha subunit gene with the four-base insertion of infantile Jewish Tay-Sachs disease. J Biol Chem. 1991;266:14306–14309. [PubMed] [Google Scholar]
- 51.Page M F, Carr B, Anders K R, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paw B H, Neufeld E F. Normal transcription of the beta-hexosaminidase alpha-chain gene in the Ashkenazi Tay-Sachs mutation. J Biol Chem. 1988;263:3012–3015. [PubMed] [Google Scholar]
- 53.Peltz S W, Brown A H, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 54.Peltz S W, He F, Welch E, Jacobson A. Nonsense-mediated mRNA decay in yeast. Prog Nucleic Acid Res Mol Biol. 1994;47:271–298. doi: 10.1016/s0079-6603(08)60254-8. [DOI] [PubMed] [Google Scholar]
- 55.Petracek M E, Nuygen T, Thompson W F, Dickey L F. Premature termination codons destabilize ferredoxin-1 mRNA when ferredoxin-1 is translated. Plant J. 2000;21:563–569. doi: 10.1046/j.1365-313x.2000.00705.x. [DOI] [PubMed] [Google Scholar]
- 56.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 57.Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 58.Ruiz-Echevarría M J, González C I, Peltz S W. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruiz-Echevarria M J, Peltz S W. Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J. 1996;15:2810–2819. [PMC free article] [PubMed] [Google Scholar]
- 60.Ruiz-Echevarría M J, Yasenchak J M, Han X, Dinman J D, Peltz S W. The upf3 protein is a component of the surveillance complex that monitors both translation and mRNA turnover and affects viral propagation. Proc Natl Acad Sci USA. 1998;95:8721–8726. doi: 10.1073/pnas.95.15.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shore S, Tomczak J, Grebner E E, Myerowitz R. An unusual genotype in an Ashkenazi Jewish patient with Tay-Sachs disease. Hum Mutat. 1992;1:486–490. doi: 10.1002/humu.1380010606. [DOI] [PubMed] [Google Scholar]
- 62.Smit L S, Nasr S Z, Iannuzzi M C, Collins F S. An African-American cystic fibrosis patient homozygous for a novel frameshift mutation associated with reduced CFTR mRNA levels. Hum Mutat. 1993;2:148–151. doi: 10.1002/humu.1380020217. [DOI] [PubMed] [Google Scholar]
- 63.Stephenson L S, Maquat L E. Cytoplasmic mRNA for human triosephosphate isomerase is immune to nonsense-mediated decay despite forming polysomes. Biochimie. 1996;78:1043–1047. doi: 10.1016/s0300-9084(97)86728-4. [DOI] [PubMed] [Google Scholar]
- 64.Sun X, Moriarty P M, Maquat L E. Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position. EMBO J. 2000;19:4734–4744. doi: 10.1093/emboj/19.17.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze M W, Kulozik A E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Urlaub G, Mitchell P J, Ciudad C J, Chasin L A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989;9:2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valentine C R. The association of nonsense codons with exon skipping. Mutat Res. 1998;411:87–117. doi: 10.1016/s1383-5742(98)00010-6. [DOI] [PubMed] [Google Scholar]
- 68.Valentine C R, Heflich R H. The association of nonsense mutation with exon-skipping in hprt mRNA of Chinese hamster ovary cells results from an artifact of RT-PCR. RNA. 1997;3:660–676. [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J, Maquat L E. Evidence that the decay of nucleus-associated nonsense mRNA for human triosephosphate isomerase involves nonsense codon recognition after splicing. RNA. 1996;2:235–243. [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang J, Sun X, Qian Y, LaDuca J P, Maquat L E. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J, Sun X, Qian Y, Maquat L E. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang S, Ruiz-Echevarria M J, Quan Y, Peltz S W. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol Cell Biol. 1995;15:2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang S, Welch E M, Hogan K, Brown A H, Peltz S W, Jacobson A. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA. 1997;3:234–244. [PMC free article] [PubMed] [Google Scholar]