Abstract
Purpose
The purpose of this study was to elucidate the effects of interleukin (IL)-38 on experimental autoimmune uveitis (EAU) and its underlying mechanisms.
Methods
Mice with EAU were treated with IL-38, and the retinas and cervical draining lymph nodes (CDLNs) were analyzed by flow cytometry. Single-cell RNA sequencing (scRNA-seq) was conducted to analyze the immune cell profiles of CDLNs from normal, EAU, and IL-38-treated mice.
Results
Administration of IL-38 attenuated EAU symptoms and reduced the proportion of T helper 17 (Th17) and T helper 1 (Th1) cells in the retinas and CDLNs. In scRNA-seq analysis, IL-38 downregulated the IL-17 signaling pathway and reduced the expression of Th17 cell pathogenicity-related genes (Csf2 and Il23r), findings which were also confirmed by flow cytometry. In vitro, IL-38 reduced the granulocyte-macrophage colony-stimulating factor (GM-CSF) stimulation function of IL-23 and inhibited IL-23R expression in Th17 cells. Moreover, when co-cultured with Th17 cells, IL-38 prevented IL-23 production in antigen-presenting cells (APCs).
Conclusions
Our data demonstrate the therapeutic effect of IL-38 on EAU, and suggest that the effect of IL-38 may be caused by dampening of the GM-CSF/IL-23R/IL-23 feedback loop between Th17 cells and APCs.
Keywords: single-cell RNA sequencing, uveitis, GM-CSF/IL-23R/IL-23 loop, interleukin-38
Autoimmune uveitis (AU) is a sight-threatening intraocular inflammatory disorder, which usually has a relapsing-remitting or chronic disease course.1,2 AU belongs to the group of autoimmune diseases of the central nervous system (CNS), which differs from those of the other organs because of the immune-privileged status of the CNS.3,4 Experimental autoimmune uveitis (EAU) is the most widely used animal model of AU and has greatly contributed to our understanding of the underlying mechanisms of the disease as well as treatment development.5 However, the pathogenic mechanisms of AU are still not fully elucidated, and novel treatment regimens with maximal curative effects and minimal side effects need to be developed to bring benefits to patients.
Interleukin (IL)-38, which was discovered in 2001, is the most recent addition to the IL-1 cytokine family.6 IL-1 family cytokines are critical inflammation regulators.7 IL-1 family receptor agonists promote inflammatory reactions, whereas antagonists inhibit them.7 IL-38 is considered an antagonist of IL-1 receptors. It exhibits 43% and 37% to 41% amino acid sequence similarity with IL-36RA and IL-1RA, respectively,6,8 which indicates its anti-inflammatory properties. Exogenous IL-38 is considered an anti-inflammatory drug candidate in various autoimmune diseases, such as psoriasis7 and systemic lupus erythematosus.9 However, its application in AU has not yet been explored.
IL-38 is involved in inflammation and autoimmunity through various immune cells.10,11 In arthritis models, IL-38 reduces the infiltration of macrophages into the synovium and inhibits the production of cytokines by T helper 17 (Th17) cells.12 IL-38 has also been reported to inhibit the secretion of pro-inflammatory cytokines by lipopolysaccharide-activated THP-1 macrophages (a human leukemia monocytic cell line).13 However, an integrated and comprehensive analysis of the regulatory role of IL-38 on various immune cells is still lacking. Single-cell RNA sequencing (scRNA-seq), a cutting-edge and high-throughput tool, is useful for analyzing complex cellular constituents in tissue and blood, and it has helped study the pathogenesis of many diseases.14–16 Thus, this tool provides a chance to unravel the complex immune-regulatory network behind the effects of IL-38.
Lymph nodes (LNs) are critical sites for interaction among various immune cells and initiate autoimmune responses.17 As for eyes, ocular antigens, which may leak from the eyes for any pathological reasons, can be caught by dendritic cells (DCs) and presented to autoreactive T cells in the LNs.18 Then, these T cells are activated, acquire the ability to traverse the blood-eye barrier, and induce a series of pathological changes.19 Cervical draining lymph nodes (CDLNs) are the main draining LNs of the CNS and are involved in the pathogenesis of CNS autoimmune diseases.3 There is efficient drainage of both macromolecules and immune cells from the CNS to CDLNs.3 The scRNA-seq of LNs has been used to demonstrate the mechanisms of diseases, like cancer, infections, and atherosclerosis.20–22 The above findings suggest that CDLNs are ideal for exploring immune changes during EAU. Therefore, in our study, we performed flow cytometry and scRNA-seq on CDLN cells to explore immune cellular changes post-IL-38 treatment.
Methods
Mice
Six- to 8-week-old C57BL/6J female mice were provided by the Guangzhou Animal Experiment Company and were housed under specific pathogen free (SPF) conditions. All animal experiments conformed to ARVO Animal Statement and institutional policies (Sun Yat-Sen University).
EAU Induction and EAU Clinical Score
The mice were subcutaneously injected with a mixture of 2 mg/mL of human interphotoreceptor retinoid-binding protein 1-20 (IRBP1-20, GPTHLFQPSLVLDMAKVLLD; GiL Biochem, Shanghai, China) and complete Freund's adjuvant consisting of 2500 ng of mycobacterium tuberculosis (BD Difco, San Jose, CA, USA) in a 1:1 volume ratio. Additionally, 0.25 µg pertussis toxin (PTX; List Biological Laboratories, Campbell, CA, USA) was injected on the same day and 2 days after immunization.23,24
Funduscopic examination of EAU progress was performed with the fundus camera, and the clinical findings were graded from 0 to 4 based on observable infiltration and vasculitis in the retina.25 The clinical score was assessed in a blinded manner.
Acquirement of Retina Infiltrated Cells
Retinas isolated from the mice were placed in complete RPMI-1640 medium containing collagenase D (Roche, Basel, Switzerland) for 1 hour at 37°C before flow cytometry analysis.
Hematoxylin and Eosin Staining
The eyes of the mice were extracted and placed in 4% paraformaldehyde for 48 hours before being embedded in paraffin for hematoxylin and eosin (H&E) staining. The infiltrating and structural scores for retinal inflammation were graded from 0 to 4, as previously described.25
Treatment of Mice and Cervical Draining Lymph Nodes
Recombinant IL-38 (Adipogen Corporation, San Diego, CA, USA) was dissolved in PBS. The mice received a daily intravenous (IV) injection of IL-38 (10 µg/kg/day) or vehicle control (PBS) for 2 weeks after immunization or for 1 week from the seventh day after immunization.
Isolated CDLN cells from the EAU group were cultured with IRBP1-20 alone or IRBP1-20 plus IL-23 (20 ng/mL; PeproTech, Rocky Hill, NJ, USA) with or without IL-38 (10 ng/mL) at 37°C for 72 hours and analyzed by flow cytometry.
Flow Cytometric Analysis
After staining with live/dead dye for 15 minutes at 4°C, surface markers (Biolegend, San Diego, CA, USA) were used to label the harvested cells: CD4 (catalog 100434), CD25 (catalog 102016), IL-23R (catalog 150907), CXCR3 (catalog 126506), and CCR6 (catalog 129819). For intracellular labeling, cells were cultured for 4 hours with PMA (5 ng/mL), Brefeldin A (1000 ng/mL), and ionomycin (500 ng/mL), followed by fixation and permeabilization before labeling with IL-22 (catalog 516409), IL-17A (catalog 506930), IFN-γ (catalog 505808), Foxp3 (catalog 11-5773-82), granulocyte-macrophage colony-stimulating factor (GM-CSF; catalog 2131415), and IL-10 antibodies (catalog 505007). Finally, the cells were measured by flow cytometry and further analyzed using FlowJo software (version 10.0.7).
ELISAs
Serum levels of IL-23 and GM-CSF in mice from different groups were tested using mouse GM-CSF ELISA kits and mouse IL-23 ELISA kits (Thermo Fisher Scientific, Waltham, MA, USA), respectively, according to the manufacturer's instructions.
Co-Cultivation of CD11C+ APCs With CD4+CCR6+CXCR3-T Cells
CD4+CCR6+CXCR3- T cells were separated by cell sorting; CD11C+ APCs were separated using a CD11C positive selection kit (STEMCELL). Sorted CD4+CCR6+ CXCR3- T cells and CD11C+ APCs were co-cultured at a ratio of 2:1 with or without anti-GM-CSF (10 µg/mL; R&D Systems) or IL-38(10 ng/mL) for 24 hours. IL-23 levels in the supernatants were measured using mouse IL-23 ELISA kit (Thermo Fisher Scientific).
Adoptive Transfer Experiment
The CD4+ lymphocytes of the EAU mice were pulsed for 3 days with IRBP1-20 (20 µg/mL) followed by resuspension with PBS before injection into normal mice (2 × 107 cells per mouse).
scRNA-Seq
A reagent kit (10x Genomics) was used to generate the scRNA libraries. The initial processing of the sequenced data was performed using CellRanger (version 3.1.0). Data were integrated and clustered using the Seurat R package (version 2.3.4).26 Quality control steps: cells with < 300 genes or mitochondrial gene ratio > 15% were removed. Batch effects were removed, and the data were integrated using the harmony R package (version 1.0). Differentially expressed genes (DEGs) among different mouse groups (EAU/Normal, IL-38/EAU) on distinct cell types were generated using “FindMarkers” functions of the Seurat package with Wilcoxon Rank Sum test. Genes with | Log2 (fold change) | > 0.25 and P values < 0.05 were used for further analysis.
Gene Functional Annotation
Gene ontology (GO) analysis was performed using the Metascape webtool27 according to the DEG between different groups. The q-value was calculated using the Banjamini–Hochberg procedure and used as the metric for GO terms in order to account for multiple testing. Among the top 50 enriched GO terms across different cell types, 5 to 10 GO terms or pathways that were associated with EAU were graphed with the heatmap (version 1.0.12) and ggplot2 package.28
Data Availability Statement
The single-cell sequencing data have been deposited at Genome Sequence Archive (GSA) with the project number PRJCA006109 and GSA accession number CRA005276.
Results
IL-38-Mediated Amelioration of EAU and Inhibition of Teff Cell Infiltration into the Retina
To examine the function of IL-38 in EAU, we developed an EAU model using IRBP1-20 and treated it with IL-38 (10 µg/kg/d). Obvious chorioretinal lesions, cell infiltration, and extensive retinal folding emerged in the EAU mice. Such symptoms were ameliorated in IL-38-treated mice and they exhibited significantly lower clinical and pathological scores than IL-38-untreated EAU mice (Figs. 1A–D; Supplementary Fig. S1A). To test the therapeutic effect of IL-38 on established EAU, we injected IL-38 into mice with established EAU (those on the seventh day after the initial immunization) for 1 week. We found that the therapeutic effect of IL-38 was still present, but was not better than that of IL-38 preventive treatment (see Supplementary Fig. S1B).
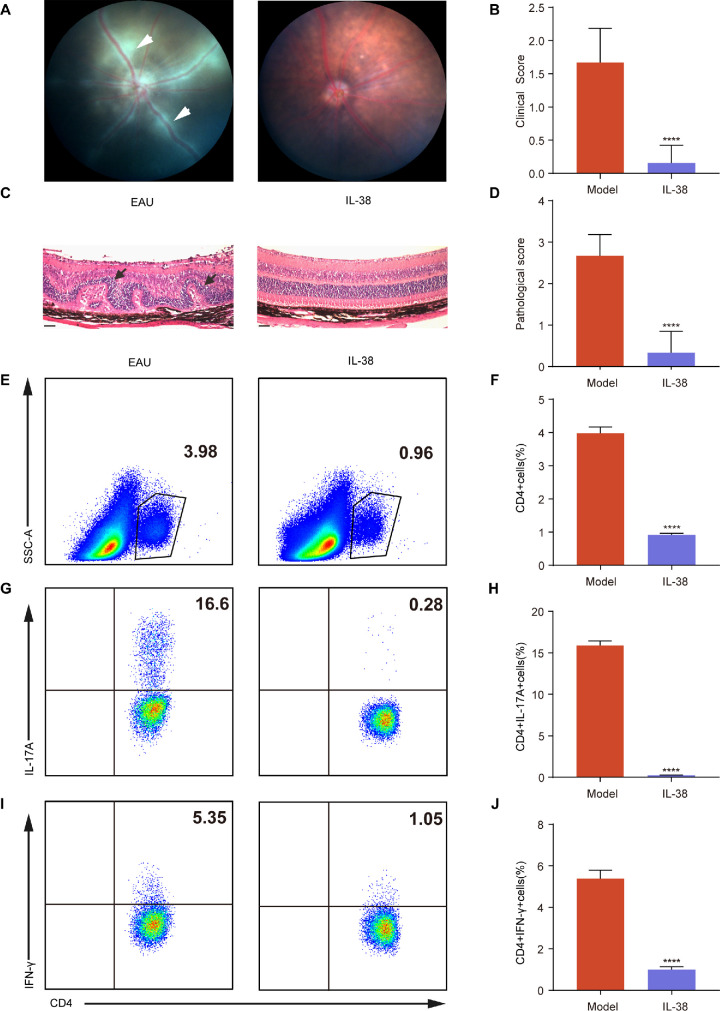
Figure 1.
IL-38-mediated amelioration of EAU and Inhibition of Teff Cell infiltration into the Retina. (A) The representative fundus images of eyes in EAU mice and IL-38-treated mice after immunization at day 14 (n = 6). The white arrows indicate inflammatory exudation and vascular deformation. (B) Clinical scores of EAU mice and IL-38-treated mice (n = 6). (C) The H&E staining of EAU mice and IL-38-treated mice after immunization at day 14 (n = 6). Scale bars, 20 mm. The black arrows indicate retinal folding. (D) Histological scores of EAU model mice and IL-38-treated mice (n = 6). (E–J) The proportion of CD4+ (gated on living cells) E to F, CD4+IL-17A+(Th17) cells (gated on CD4+ cells) G and H, and CD4+IFN-γ+ (Th1) cells (gated on CD4+ cells) (I–J) infiltrated into retina after immunization at day 14 in EAU mice and IL-38-treated mice (n = 6). Data shown as mean ± SD from three independent experiments. Data were analyzed using unpaired two-tailed Student t-tests, ****P < 0.0001.
Ocular-infiltrating T effector (Teff) cells, particularly T helper 1 (Th1) and Th17 cells, play a decisive role in the induction of AU.19 We performed flow cytometry and observed that the proportion of total infiltrating CD4+ T cells, IL-17A-producing CD4+ T cells, and IFN-γ-producing CD4+ T cells were much lower in IL-38-treated mice than in EAU mice (see Figs. 1E–J). Thus, IL-38 could prevent CD4+ T cells, especially Th17 and Th1 cells, from infiltrating into the retina.
Decreased Teff Cells and Increased Regulatory T Cells in CDLNs After IL-38 Treatment
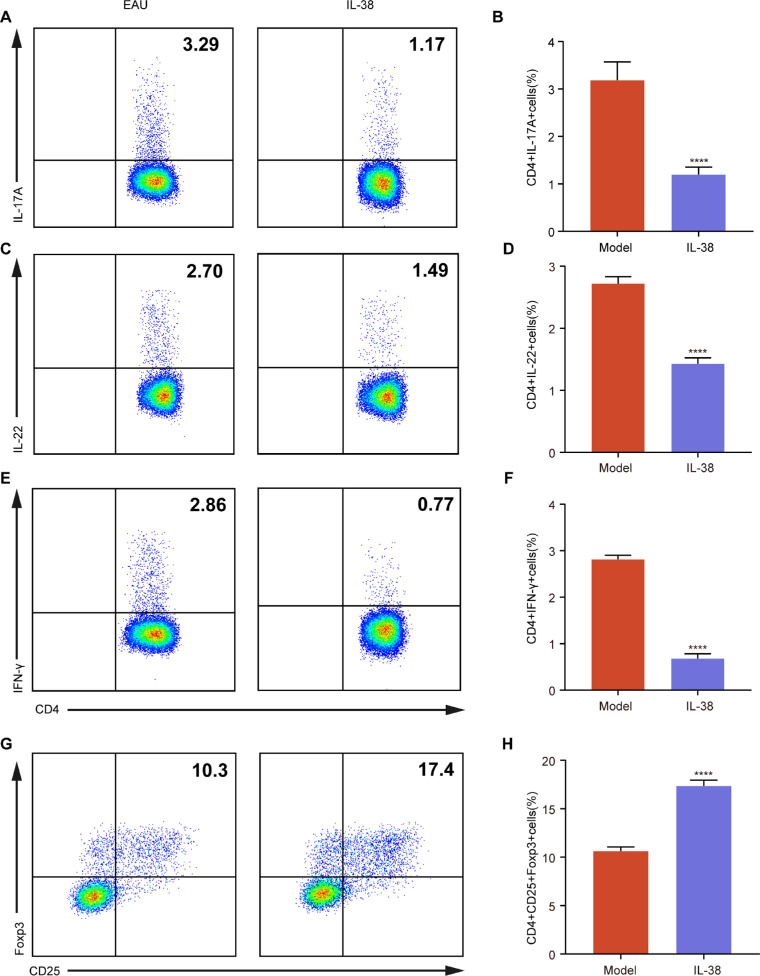
Th1 and Th17 cells can promote autoimmunity,29,30 whereas regulatory T cells (Tregs) can attenuate it.31 In our study, IL-38 reduced the proportion of IL-17A- and IL-22-producing Th17 cells and IFN-γ-producing Th1 cells in CDLNs (Figs. 2A–F), but increased the proportion of CD25+ Foxp3+ Tregs (Figs. 2G, 2H), suggesting that IL-38 could restore the balance between Teff cells and Tregs in CDLNs in EAU.
Figure 2.
Decreased Teff Cells and Increased Regulatory T cells (Tregs) in CDLNs after IL-38 treatment. (A–H) The proportions of CD4+IL-17A+ A and B, CD4+IL-22+ C and D, CD4+IFN-γ+ (E, F), and CD4+CD25+Foxp3+ (G, H) cells on the gate of CD4+ cells in CDLNs after immunization at day 14 in EAU mice and IL-38-treated mice (n = 6). Data shown as mean ± SD from three independent experiments. Data were analyzed using unpaired two-tailed Student t-tests, ****P < 0.0001.
IL-38 Reprogrammed the Immune Cell Profile During EAU
To explore the mechanisms underlying the therapeutic effects of IL-38, we performed scRNA-seq on CDLN cells from normal, EAU, and IL-38-treated mice (Fig. 3A). After quality control, we obtained a total of 30,658 high-quality cells (normal: 12,781 cells, EAU: 12,626 cells, and IL-38-treated mice: 5,251 cells) for further analysis (see Fig. 3A). Eight immune cell types were identified, including T cells (TCs), B cells (BCs), monocytes (Monos), conventional dendritic cells (cDCs), macrophages (Macros), plasmacytoid dendritic cells (pDCs), natural killer cells (NK), and neutrophils (Neu; see Fig. 3B, Supplementary Figs. S1C, S1D). T and B cells dominated in the CDLN cells, and the cellular composition of CDLNs changed during EAU and after IL-38 treatment (see Figs. 3C, 3D).
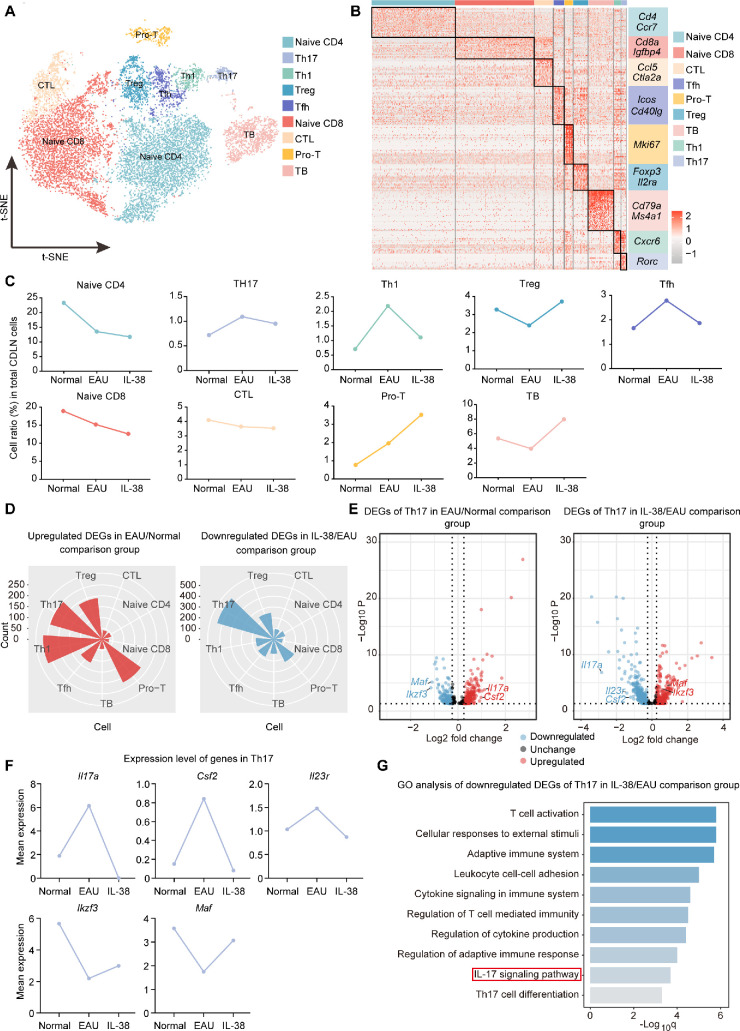
Figure 3.
Study design and scRNA-seq analysis of immune cells. (A) Experimental design for sc-RNA seq. Samples of CDLNs were collected from normal mice, EAU mice, and IL38-treated EAU mice. Each sample included three mice. The gene expression data of samples were obtained via scRNA sequencing for subsequent single-cell analysis. (B) The t-SNE clustering of CDLN cells. (C, D) Pie charts and Bar charts show proportions of each cell type across three groups derived from scRNA-seq data. (E) Volcano plot showing up- and down-regulated DEGs of CDLN cells in the EAU/Normal and IL-38/EAU comparison groups. Red and blue dots indicate up- and down-regulated DEGs, respectively. (F) GO analysis of downregulated DEGs of CDLN cells in the IL-38/EAU comparison groups. The q value was calculated using the Benjamini-Hochberg procedure. (G) Rose diagrams showing the numbers of up- and down-regulated DEGs of CDLN cells in the EAU/Normal and IL-38/EAU comparison groups. (H) GO analysis of downregulated DEGs of T cells in the IL-38/EAU comparison groups. q value was calculated using the Benjamini-Hochberg procedure.
Next, we conducted DEG analysis between EAU and normal mice, and between IL-38-treated and EAU mice. In CDLN cells, naïve phenotype-associated genes (Lef1 and Ccr7) were downregulated, whereas inflammatory genes (S100a8 and S100a11)32,33 were upregulated during EAU (see Fig. 3E). After IL-38 treatment, cell activation-related genes (CD69 and Dusp1) were downregulated, whereas naïve phenotype-associated gene, Igfbp4, and chemokine, Ccr6, were upregulated after IL-38 treatment (see Fig. 3E). Subsequent GO analysis showed that the IL-17 signaling pathway and pathways related to T cell activation were upregulated during EAU but downregulated by IL-38 (see Fig. 3F, Supplementary Fig. S1E). Additionally, Th17 cell differentiation was upregulated during EAU (see Supplementary Fig. S1E). Interestingly, when analyzing each immune cell type separately, myeloid cells showed more DEGs than other cell types during EAU and IL-38 treatment (see Fig. 3G). To further explore the effect of IL-38 on different immune cells, GO analysis was performed. In T cells, IL-38 downregulated the IL-17 signaling pathway, which was prominent among the upregulated pathways during EAU (see Fig. 3H, Supplementary Fig. S2A). Moreover, pathways related to T cell activation and TNF signaling were also downregulated by IL-38 (see Fig. 3H, Supplementary Fig. S2A). The other immune cell types also showed some pathways that changed conversely during EAU and after IL-38 treatment, such as the antigen processing and presentation and positive regulation of T cell activation pathways in cDCs, which were partly driven by altered expression of T cell co-stimulators (CD86 and CD80)34 and MHCII gene (H2-Ab1).35 Similar changes were presented in the antigen processing and presentation pathway in B cells, inflammatory response in monocytes, and IL-6 signaling pathway in macrophages (see Supplementary Figs. S2B, S2D). Overall, these findings indicate that IL-38 inhibits some upregulated genes and their related pathways in EAU.
Effects of IL-38 on T and B Cell Subsets During EAU
T cells are known to play the main role in EAU development.19 Thus, we focused on the effect of IL-38 on T cell subsets, including naïve CD4+ T cells (naïve CD4), Th17 cells, Th1 cells, Tregs that suppress autoimmunity, T follicular helper cells (Tfh), naïve CD8+ T cells (naïve CD8), cytotoxic CD8+ T lymphocytes (CTL), proliferative T cells (Pro-T), and TB cells expressing both T and B cell markers (Figs. 4A, 4B, Supplementary Fig. S3A). In scRNA-seq analysis, the proportion of Th1 cells was enhanced during EAU, and reduced after IL-38 treatment, whereas Tregs showed an opposing tendency (Fig. 4C). The change of Th17 cell proportion was not obvious, but its trend was consistent with the above flow cytometry results (see Figs. 2A, 4C). Interestingly, DEG analysis revealed that Th17 cells exhibited the most downregulated genes after IL-38 treatment, indicating the IL-38-induced transcriptional changes in Th17 cells was more significant than the proportional change (see Figs. 4C, 4D). In contrast, Th1 cells only exhibited plentiful DEGs during EAU but not after IL-38 treatment (see Figs. 4C, 4D). Naïve CD4+ T cells give rise to effector T cells,36 so we conducted GO analysis on them and observed that IL-17 signaling and leukocyte differentiation pathways were inhibited by IL-38 treatment (see Supplementary Figs. S3B, S3C). These results suggest that Th17 cells and their related pathways may be critical mediators of the effect of IL-38. Thus, we conducted DEG analysis on Th17 cells and found that the genes encoding cytokines IL-17A (Il17a), GM-CSF (Csf2), and IL-23R (Il23r), which are related to Th17 cell pathogenicity37,38 were upregulated during EAU and inhibited by IL-38 treatment (see Figs. 4E, 4F). In contrast, the IL-10-inducing transcription factors, Maf39 and Ikzf3,40 showed a converse expression changes in Th17 cells (see Figs. 4E, 4F). Furthermore, GO analysis of the DEGs in Th17 cells showed that the IL-17 signaling pathway, which was upregulated during EAU, was inhibited by IL-38 treatment (see Fig. 4G, Supplementary Fig. S3D).
Figure 4.
Effects of IL-38 on T and B Cell Subsets During EAU. (A) The t-SNE clustering of T cell subsets. (B) Heatmap of discriminative gene sets for T cell subsets. (C) Line charts show proportions of each T cell subsets in three groups derived from scRNA-seq data. (D) Rose diagrams showing the numbers of up- and down-regulated DEGs of T cell subsets in the EAU/Normal and IL-38/EAU comparison groups. (E) Volcano plot showing up- and down-regulated DEGs of Th17 cells in the EAU/Normal and IL-38/EAU comparison groups. Red and blue dots indicate up- and down-regulated DEGs, respectively. (F) Line chart of mean expression of Il17a, Csf2, Il23r, Maf, and Ikzf3 from Th17 cells across the three groups. (G) GO analysis of downregulated DEGs of Th17 cells in the IL-38/EAU comparison groups. The q value was calculated using the Benjamini-Hochberg procedure.
We also explored the effects of IL-38 on B cell subsets: naïve (NBCs), germinal (GBCs), and plasma B cells (PBCs; Supplementary Figs. S4A–C). IL-38 inhibited the change in the proportion of NBCs and GBCs during EAU (Supplementary Fig. S4D). GO analysis of the downregulated DEGs revealed that the MAPK signaling and IL-17 signaling pathways of NBCs, and the regulation of RNA splicing pathway of GBCs were downregulated by IL-38 (Supplementary Figs. S4E, S4F).
IL-38 Mediated Attenuation of Th17 Cell Pathogenicity During EAU
GM-CSF production, mainly by Th17 cells, can be induced by IL-23 from APCs through the IL-23R in Th17 cells.41 Expression of GM-CSF marks the pathogenicity of Th17 cells, whereas GM-CSF deficiency causes a loss of pathogenicity.37,42 IL-10, on the other hand, is mainly expressed by non-pathogenic Th17 cells.38 Thus, we explored the effect of IL-38 on Th17 cell pathogenicity at the protein level. IL-38 treatment significantly reduced the proportion of IL-17A+GM-CSF+ T cells among the total CD4+ T cells (Figs. 5A, 5B). It also reduced the proportion of GM-CSF+ and IL-23R+ cells and increased the proportion of IL-10+ cells in Th17 cells (Figs. 5C–H). These results support the impaired Th17 cell pathogenicity after IL-38 treatment and suggest that IL-23 signaling may be involved in this process.
Figure 5.
IL-38 mediated attenuation of Th17 Cell Pathogenicity during EAU. (A, B) The proportion of GM-CSF+ IL-17A+ cells (gated on CD4+ cells) in EAU mice and IL-38-treated mice (n = 6). (C-H) The proportion of GM-CSF+ C to D, IL-23R+ E and F, and IL-10+ G and H cells on the gate of Th17 cells (CD4+IL-17A+ cells) in EAU mice and IL-38-treated mice (n = 6). Data shown as mean ± SD from three independent experiments. Data were analyzed using unpaired two-tailed Student t-tests, ****P < 0.0001.
IL-38 Mediated Inhibition of Th17 Cell Response to IL-23 and Reduction of IL-23 Secretion by APCs Co-Cultured With Th17 Cells
IL-23 can bind to IL-23R in Th17 cells, thus facilitating GM-CSF secretion.41 Thus, we tested the effect of IL-38 on the response of Th17 cells to IL-23 signaling. We cultured CDLN cells isolated from EAU mice with IRBP1-20 alone or IRBP1-20 plus IL-23 with or without IL-38. IL-23 treatment increased the proportion of IL-23R+ and GM-CSF+ Th17 cells, and this effect was inhibited by IL-38 (Figs. 6A–D), whereas no changes were observed in IL-10+ Th17 cells (see Figs. 6E, 6F). Thus, the GM-CSF stimulation function of IL-23 was attenuated by IL-38 treatment, an effect that might result from the decreased IL-23R expression in Th17 cells.
Figure 6.
IL-38 mediated inhibition of Th17 cell Response to IL-23 and reduction of IL-23 secretion by APCs co-cultured with Th17 cells. (A–F) CDLN cells of EAU mice were collected after immunization at day 14 and cultured in three different conditions: IRBP1-20, IRBP1-20 plus IL-23, IRBP1-20 plus IL-23, and IL-38, for 72 hours. The proportion of IL-23R+ A and B, GM-CSF+ C and D, and IL-10+ E and F cells was measured with flow cytometry on the gate of Th17 cells (CD4+IL-17A+ cells). Data shown as mean ± SD of six independent experiments. Data were analyzed using 2-way ANOVA, ****P < 0.0001. ns, not significant. (G) Serum levels of GM-CSF of the normal mice, EAU mice, and IL-38-treated-mice were measured by ELISA (n = 6). Data shown as mean ± SD. Data were analyzed using 1-way ANOVA, ****P < 0.0001. (H) CD11C+ APCs and CD4+CCR6+CXCR3- T cells (Th17) from CDLN of EAU mice were cultured with or without IL-38 or GM-CSF neutralizing antibody. IL-23 level of supernatant was tested by ELISA. Data shown as mean ± SD of six independent experiments. Data were analyzed using 2-way ANOVA, ****P < 0.0001. (I–J) The representative fundus images and clinical scores on day14 after injection of IRBP1-20-specific CD4+ T cells treated with or without IL-38 (n = 6). The white arrows indicate inflammatory exudation and vascular deformation. (K, L) The H&E staining and pathological scores of the two groups (n = 6). Scale bars, 20 mm. The black arrows indicate retinal folding. Data shown as mean ± SD from three independent experiments. Data were analyzed using unpaired 2-tailed Student t-tests, *P < 0.05, **P < 0.01.
APC-secreted IL-23 stimulates Th17 cells to produce GM-CSF, which in turn induces IL-23 secretion by APCs, thus creating a positive feedback loop.43 In our study, serum levels of GM-CSF in IL-38-treated mice were significantly lower than in EAU mice (see Fig. 6G), thus suggesting that IL-23 secretion by APCs may also be affected. We co-cultured CD11C+ APCs and CD4+ CCR6+ CXCR3- T cells (Th17 cells)44 from the CDLNs of EAU mice with or without IL-38 or GM-CSF neutralizing antibody. Th17 cells from EAU mice strongly stimulated IL-23 secretion by APCs, which was inhibited by GM-CSF neutralizing antibodies and IL-38 (see Fig. 6H). This result suggests that the inhibition of GM-CSF secretion in Th17 cells by IL-38 resulted in reduced IL-23 secretion by APCs, thus interrupting the positive feedback loop between Th17 cells and APCs and further decreasing Th17 cell pathogenicity.
IL-38 Mediated Impairment of the EAU Induction Function of IRBP1-20-Specific T Cells
To confirm that IL-38 treatment alleviated EAU through functional regulation of autoreactive CD4+ T cells, an adoptive transfer (AT) test was conducted. From the CDLNs of EAU mice, we isolated CD4+ T cells, cultured them with IRBP1-20 or IRBP1-20 plus IL-38, and then transferred them into normal mice. IL-38 treatment significantly attenuated the EAU induction function of IRBP1-20-specific CD4+ T cells (see Figs. 6I–L, Supplementary Fig. S4G), which confirms that IL-38 exerts a therapeutic effect on EAU by reducing CD4+ T cell pathogenicity.
Discussion
Here, we evaluated the therapeutic potential of IL-38 in EAU and performed a comprehensive analysis of the post-IL-38 treatment immune cell transcriptional landscape of CDLNs using scRNA-seq.
The scRNA-seq allowed a comprehensive analysis of IL-38 effects on various immune cell types, which provided novel insights into the function of IL-38 in EAU. Our GO analysis revealed that the IL-17 signaling pathway was inhibited by IL-38 treatment, which has also been reported in several studies.10,45,46 In cDCs, the upregulated antigen processing and presentation and positive regulation of T cell activation pathway during EAU were inhibited by IL-38 treatment. Combined with the decreased Th17 cell proportion validated by flow cytometry and the reduced Th17 cell differentiation pathway in naïve CD4 in IL-38 treated group, these findings suggested that the effect of IL-38 on APC might impaired Th17 differentiation. Previous studies have also shown the inhibition of pro-inflammatory cytokine secretion in macrophages by IL-38,12 which were also reflected in our data. Therefore, IL-38 treatment changed the gene signatures of various immune cells during EAU.
Further, in our study, investigation of T cell subsets revealed increased Teff and decreased Tregs in EAU mice, which are common in CNS autoimmune diseases.47 This change was inhibited by IL-38 treatment. Th17 cells exert essential pathogenic effects in many autoimmune diseases, including multiple sclerosis and autoimmune uveitis.29 IL-38 is reported to inhibit IL-17 signaling in both patients and animal models.45,48,49 In our study, we found that Th17 cells might be the critical mediators influenced by IL-38, as evidenced by the large number of DEGs, IL-17 signaling pathway, and Th17 differentiation pathway that were downregulated after IL-38 treatment.
Although previous studies have revealed that IL-38 represses Th17 cell function, the underlying mechanisms remain unclear. Among Th17 cells, both non-pathogenic and pathogenic populations have been recognized.42 Non-pathogenic Th17 cells secrete IL-17 and IL-10 and exert protective functions, whereas pathogenic Th17 cells secrete IL-17 and GM-CSF and promote inflammation.50,51 The differentiation of pathogenic Th17 cells requires IL-23 stimulation to induce GM-CSF expression.37 Our study revealed that IL-38 treatment resulted in the suppression of Th17 cell pathogenicity, as evidenced by decreased expression of GM-CSF and upregulation of IL-10. The concomitant downregulation of IL-23R also indicates that compromised Th17 cell pathogenicity is associated with an impaired response to IL-23 signaling. Indeed, our in vitro study suggests that IL-38 treatment inhibits IL-23-induced upregulation of GM-CSF production in Th17 cells. Pan et al. have reported that IL-23R expression is downregulated by IL-38 in PBMCs from patients with thyroid-associated ophthalmopathy.52 However, the specific immune cell type in which IL-23R downregulation occurs is unclear. Our data indicates that the downregulation of IL-23R expression in Th17 cells after IL-38 treatment may account for the poor response of these cells to IL-23 signaling and the reduction of Th17 cell pathogenicity.
A previous study reported a positive feedback loop between Th17 cells and APCs.43 Our results showed that IL-38 reduced IL-23 production by APCs when co-cultured with Th17 cells, which was similar to the effect of GM-CSF neutralizing antibody. This result suggests that IL-38 may dampen the positive feedback loop of GM-CSF/IL-23R/IL-23 between Th17 and APCs, and this molecular change may underlie the regulatory function of IL-38 in Th17 cells. Our adoptive transfer experiment showed that the EAU induction ability was attenuated in IL-38-treated CD4+ T cells. Since Th17 cells are the key pathogenic CD4+ T cells implicated in EAU, this result suggests that IL-38 can weaken Th17 cell pathogenicity, which accounts for its therapeutic effect.
To the best of our knowledge, our study is the first to reveal the therapeutic effect of exogenous IL-38 on EAU. In this study, we provide a comprehensive atlas of immune cells post-IL-38 treatment. We also identify that IL-38 can weaken Th17 cell pathogenicity and propose a potential underlying molecular regulation. Thus, this study expands our understanding of the immune regulatory mechanisms behind IL-38 and suggests that Th17 cells are central mediators of the effects of IL-38 in EAU.
Supplementary Material
Acknowledgments
Supported by the National Key Research and Development Program of China (2017YFA0105804).
Disclosure: H. Li, None; L. Zhu, None; R. Wang, None; L. Xie, None; Y. Chen, None; R. Duan, None; X. Liu, None; Z. Huang, None; B. Chen, None; Z. Li, None; X. Wang, None; W. Su, None
References
- 1. Bertrand P, Jamilloux Y, Ecochard R, et al.. Uveitis: Autoimmunity… and beyond. Autoimmun Rev . 2019; 18(9): 102351. [DOI] [PubMed] [Google Scholar]
- 2. Diedrichs-Möhring M, Kaufmann U, Wildner G.. The immunopathogenesis of chronic and relapsing autoimmune uveitis - Lessons from experimental rat models. Prog Retin Eye Res . 2018; 65: 107–126. [DOI] [PubMed] [Google Scholar]
- 3. Louveau A, Herz J, Alme MN, et al.. CNS lymphatic drainage and neuroinflammation are regulated by meningeal lymphatic vasculature. Nat Neurosci . 2018; 21(10): 1380–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ransohoff RM, Engelhardt B.. The anatomical and cellular basis of immune surveillance in the central nervous system. Nature Reviews Immunology. 2012; 12(9): 623–635. [DOI] [PubMed] [Google Scholar]
- 5. Pennesi G, Mattapallil MJ, Sun S, et al.. A humanized model of experimental autoimmune uveitis in HLA class II transgenic mice. J Clin Invest . 2003; 111(8): 1171–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin H, Ho AS, Haley-Vicente D, et al.. Cloning and characterization of IL-1HY2, a novel interleukin-1 family member. J Biological Chemistry . 2001; 276(23): 20597–20602. [DOI] [PubMed] [Google Scholar]
- 7. Han Y, Mora J, Huard A, et al.. IL-38 Ameliorates Skin Inflammation and Limits IL-17 Production from γδ T Cells. Cell Rep . 2019; 27(3): 835–846. [DOI] [PubMed] [Google Scholar]
- 8. Bensen JT, Dawson PA, Mychaleckyj JC, Bowden DW.. Identification of a novel human cytokine gene in the interleukin gene cluster on chromosome 2q12-14. J Interferon Cytokine Res: The Official J Intl Soc Interferon Cytokine Res . 2001; 21(11): 899–904. [DOI] [PubMed] [Google Scholar]
- 9. Xu W, Su L, Liu X, et al.. IL-38: A novel cytokine in systemic lupus erythematosus pathogenesis. J Cell Mol Med . 2020; 24(21): 12379–12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garraud T, Harel M, Boutet M, Le Goff B, Blanchard F. The enigmatic role of IL-38 in inflammatory diseases. Cytokine Growth Factor Rev . 2018; 39: 26–35. [DOI] [PubMed] [Google Scholar]
- 11. Xie L, Huang Z, Li H, Liu X, Zheng S, Su W.. IL-38: A New Player in Inflammatory Autoimmune Disorders. Biomolecules . 2019; 9(8): 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mora J, Schlemmer A, Wittig I, et al.. Interleukin-38 is released from apoptotic cells to limit inflammatory macrophage responses. J Mol Cell Biol . 2016; 8(5): 426–438. [DOI] [PubMed] [Google Scholar]
- 13. Yuan XL, Li Y, Pan XH, Zhou M, Gao QY, Li MC.. [Production of recombinant human interleukin-38 and its inhibitory effect on the expression of proinflammatory cytokines in THP-1 cells]. Molekuliarnaia Biologiia . 2016; 50(3): 466–473. [DOI] [PubMed] [Google Scholar]
- 14. Grün D, van Oudenaarden A.. Design and Analysis of Single-Cell Sequencing Experiments. Cell . 2015; 163(4): 799–810. [DOI] [PubMed] [Google Scholar]
- 15. Luo G, Gao Q, Zhang S, Yan B.. Probing infectious disease by single-cell RNA sequencing: Progresses and. Comput Struct Biotec . 2020; 18: 2962–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rantalainen M. Application of single-cell sequencing in human cancer. Brief Funct Genomics . 2018; 17(4): 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gasteiger G, Ataide M, Kastenmüller W.. Lymph node - an organ for T-cell activation and pathogen defense. Immunol Rev . 2016; 271(1): 200–220. [DOI] [PubMed] [Google Scholar]
- 18. Mochizuki M, Sugita S, Kamoi K.. Immunological homeostasis of the eye. Prog Retin Eye Res . 2013; 33: 10–27. [DOI] [PubMed] [Google Scholar]
- 19. Bose T, Diedrichs-Möhring M, Wildner G.. Dry eye disease and uveitis: A closer look at immune mechanisms in animal models of two ocular autoimmune diseases. Autoimmun Rev . 2016; 15(12): 1181–1192. [DOI] [PubMed] [Google Scholar]
- 20. Speranza E, Williamson BN, Feldmann F, et al.. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. SCI Transl Med . 2021; 13(578): eabe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Demeulemeester J, Kumar P, Møller EK, et al.. Tracing the origin of disseminated tumor cells in breast cancer using single-cell sequencing. Genome Biol . 2016; 17(1): 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wolf D, Gerhardt T, Winkels H, et al.. Pathogenic Autoimmunity in Atherosclerosis Evolves From Initially Protective Apolipoprotein B(100)-Reactive CD4(+) T-Regulatory Cells. Circulation . 2020; 142(13): 1279–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agarwal RK, Silver PB, Caspi RR.. Rodent models of experimental autoimmune uveitis. Methods in Molecular Biology (Clifton, N.J.) . 2012; 900: 443–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen J, Caspi RR.. Clinical and Functional Evaluation of Ocular Inflammatory Disease Using the Model of Experimental Autoimmune Uveitis. Methods in Molecular Biology (Clifton, N.J.) . 2019; 1899: 211–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen J, Caspi RR.. Clinical and Functional Evaluation of Ocular Inflammatory Disease Using the Model of Experimental Autoimmune Uveitis. Vol 1899. New York, NY: Springer New York; 2019: 211–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol . 2018; 36(5): 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Y, Zhou B, Pache L, et al.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun . 2019; 10(1): 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ginestet C. ggplot2: Elegant Graphics for Data Analysis. J R Stat Soc A Stat . 2011; 174(1): 245. [Google Scholar]
- 29. Yang J, Sundrud MS, Skepner J, Yamagata T.. Targeting Th17 cells in autoimmune diseases. Trends Pharmacol Sci . 2014; 35(10): 493–500. [DOI] [PubMed] [Google Scholar]
- 30. Bing SJ, Shemesh I, Chong WP, et al.. AS101 ameliorates experimental autoimmune uveitis by regulating Th1 and Th17. J Autoimmun . 2019; 100: 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Esensten JH, Muller YD, Bluestone JA, Tang Q.. Regulatory T-cell therapy for autoimmune and autoinflammatory diseases: The next frontier. J Allergy Clinical Immunol . 2018; 142(6): 1710–1718. [DOI] [PubMed] [Google Scholar]
- 32. D'Amico F, Granata M, Skarmoutsou E, et al.. Biological therapy downregulates the heterodimer S100A8/A9 (calprotectin) expression in psoriatic patients. Inflammation Research: Official Journal of the European Histamine Research Society . 2018; 67(7): 609–616. [DOI] [PubMed] [Google Scholar]
- 33. Elyahu Y, Hekselman I, Eizenberg-Magar I, et al.. Aging promotes reorganization of the CD4 T cell landscape toward extreme regulatory and effector phenotypes. Sci Adv . 2019; 5(8): w8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ni K, O'Neill HC. The role of dendritic cells in T cell activation. Immunol Cell Biol . 1997; 75(3): 223–230. [DOI] [PubMed] [Google Scholar]
- 35. Blum JS, Wearsch PA, Cresswell P.. Pathways of antigen processing. Annu Rev Immunol . 2013; 31: 443–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou L, Chong MMW, Littman DR.. Plasticity of CD4+ T cell lineage differentiation. Immunity . 2009; 30(5): 646–655. [DOI] [PubMed] [Google Scholar]
- 37. El-Behi M, Ciric B, Dai H, et al.. The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol . 2011; 12(6): 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yasuda K, Takeuchi Y, Hirota K.. The pathogenicity of Th17 cells in autoimmune diseases. Semin Immunopathol . 2019; 41(3): 283–297. [DOI] [PubMed] [Google Scholar]
- 39. Pot C, Jin H, Awasthi A, et al.. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. Journal of immunology (Baltimore, MD : 1950) . 2009; 183(2): 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Evans HG, Roostalu U, Walter GJ, et al.. TNF-α blockade induces IL-10 expression in human CD4+ T cells. Nat Commun . 2014; 5: 3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Komuczki J, Tuzlak S, Friebel E, et al.. Fate-Mapping of GM-CSF Expression Identifies a Discrete Subset of Inflammation-Driving T Helper Cells Regulated by Cytokines IL-23 and IL-1β. Immunity . 2019; 50(5): 1289–1304. [DOI] [PubMed] [Google Scholar]
- 42. Lee Y, Awasthi A, Yosef N, et al.. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol . 2012; 13(10): 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. El-Behi M, Ciric B, Dai H, et al.. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol . 2011; 12(6): 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao Y, Yamasaki R, Yamaguchi H, et al.. Oligodendroglial connexin 47 regulates neuroinflammation upon autoimmune demyelination in a novel mouse model of multiple sclerosis. Proc Natl Acad Sci USA . 2020; 117(4): 2160–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luo D, Chen Y, Zhou N, Li T, Wang H.. Blockade of Th17 response by IL-38 in primary Sjögren's syndrome. Mol Immunol . 2020; 127: 107–111. [DOI] [PubMed] [Google Scholar]
- 46. Chai Y, Lin S, Zhang M, et al.. IL-38 is a biomarker for acute respiratory distress syndrome in humans and down-regulates Th17 differentiation in vivo. Clinical immunology (Orlando, Fla.) . 2020; 210: 108315. [DOI] [PubMed] [Google Scholar]
- 47. Korn T, Kallies A.. T cell responses in the central nervous system. Nature Reviews Immunology. 2017; 17(3): 179–194. [DOI] [PubMed] [Google Scholar]
- 48. van de Veerdonk FL, Stoeckman AK, Wu G, et al.. IL-38 binds to the IL-36 receptor and has biological effects on immune cells similar to IL-36 receptor antagonist. Proc Natl Acad Sci USA . 2012; 109(8): 3001–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boutet M, Najm A, Bart G, et al.. IL-38 overexpression induces anti-inflammatory effects in mice arthritis models and in human macrophages in vitro. Ann Rheum Dis . 2017; 76(7): 1304–1312. [DOI] [PubMed] [Google Scholar]
- 50. Langrish CL, Chen Y, Blumenschein WM, et al.. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. The Journal of Experimental Medicine . 2005; 201(2): 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zheng Y, Danilenko DM, Valdez P, et al.. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature . 2007; 445(7128): 648–651. [DOI] [PubMed] [Google Scholar]
- 52. Pan Y, Wang M, Chen X, et al.. Elevated IL-38 inhibits IL-23R expression and IL-17A production in thyroid-associated ophthalmopathy. Int Immunopharmacol . 2021; 91: 107300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-cell sequencing data have been deposited at Genome Sequence Archive (GSA) with the project number PRJCA006109 and GSA accession number CRA005276.