Abstract
Oncogenesis associated with human T-cell leukemia virus (HTLV) infection is directly linked to the virally encoded transcription factor Tax. To activate HTLV-1 transcription Tax interacts with the cellular protein CREB and the pleiotropic coactivators CBP and p300. While extensively studied, the molecular mechanisms of Tax transcription function and coactivator utilization are not fully understood. Previous studies have focused on Tax binding to the KIX domain of CBP, as this was believed to be the key step in recruiting the coactivator to the HTLV-1 promoter. In this study, we identify a carboxy-terminal region of CBP (and p300) that strongly interacts with Tax and mediates Tax transcription function. Through deletion mutagenesis, we identify amino acids 2003 to 2212 of CBP, which we call carboxy-terminal region 2 (CR2), as the minimal region for Tax interaction. Interestingly, this domain corresponds to the steroid receptor coactivator 1 (SRC-1)-interacting domain of CBP. We show that a double point mutant targeted to one of the putative α-helical motifs in this domain significantly compromises the interaction with Tax. We also characterize the region of Tax responsible for interaction with CR2 and show that the previously identified transactivation domain of Tax (amino acids 312 to 319) participates in CR2 binding. This region of Tax corresponds to a consensus amphipathic helix, and single point mutations targeted to amino acids on the face of this helix abolish interaction with CR2 and dramatically reduce Tax transcription function. Finally, we demonstrate that Tax and SRC-1 bind to CR2 in a mutually exclusive fashion. Together, these studies identify a novel Tax-interacting site on CBP/p300 and extend our understanding of the molecular mechanism of Tax transactivation.
Human T-cell leukemia virus type 1 (HTLV-1) is a complex retrovirus etiologically linked to an aggressive and often fatal malignancy called adult T-cell leukemia (ATL) (54). It is estimated that between 5 and 20 million people are HTLV-1 carriers worldwide; however, only a small percentage of these infected individuals develop ATL (12). ATL develops after a prolonged latency period of up to 30 years postinfection (48) and is characterized by lytic bone lesions, skin abnormalities, and greater than 5% abnormal T cells (reviewed in reference 57). HTLV-1 infection is also associated with the neurodegenerative disorder known as tropical spastic paraparesis or HTLV-1-associated myelopathy (reviewed in references 38, 54, and 60). The disorder is often characterized by demyelination of the nerves of the spinal cord, resulting in weakness or paralysis in muscles of the lower extremities. The molecular basis of HTLV-1-associated diseases is strongly linked to the expression of the virally encoded Tax protein (34).
Tax is a potent transcriptional activator that stimulates HTLV-1 viral gene expression, which leads to high-level virion production in the infected T cell. Three highly conserved 21-bp repeat enhancer elements, located in the HTLV-1 transcriptional control region, are critical to Tax-activated transcription. These elements, which are referred to as viral cyclical response elements (CREs), carry a central binding site for the transcription factor CREB, flanked by highly conserved GC-rich DNA sequences. Tax associates with the HTLV-1 promoter through protein-protein interactions with CREB (13, 61) and protein-DNA interactions with the minor-groove GC-rich sequence (26, 27, 33, 37). The formation of this Tax-containing promoter-bound complex appears to be critical in the recruitment of the multifunctional cellular coactivators CBP and p300. Several previous studies indicate that Tax specifically binds to the KIX and C/H1 domains of CBP/p300 to recruit the coactivators to the HTLV-1 promoter, resulting in strong transcriptional activation of the virus (14, 25, 31, 32).
CBP and p300 are very large (2,441 and 2,414 amino acids, respectively), highly conserved coactivator proteins that serve as mediators in the regulation of gene expression in metazoans (18, 52). Numerous pathways of regulated gene expression converge at CBP and its paralog p300. These include signal-dependent and independent activation, programs of differentiation, embryogenesis, and modulation of apoptotic pathways and pathways involved in transcriptional regulation (reviewed in references 15, 16, 18, and 52). Although CBP was originally named following its identification as a coactivator for Ser133-phosphorylated CREB, the acronym is a misnomer, as CBP is utilized by numerous cellular transcription factors and is likely involved in the transcriptional regulation of nearly all protein-coding genes in the cell. In addition, many viral activator proteins have evolved strategies to take advantage of CBP's coactivator properties (4, 11, 31, 56).
Alterations in CBP expression levels appear to influence the onset of certain types of cancers, as evidenced by Rubinstein-Taybi patients. Rubinstein-Taybi syndrome is caused by mutations or rearrangements in one CBP allele, underscoring the importance of proper intracellular CBP concentrations in normal cellular processes and tumor suppression (40). Translocations into the genes that encode CBP and p300 have also been strongly correlated with the development of treatment-related (chemotherapy) acute myeloid leukemia. Patients with this ailment carry an in-frame translocation of the mixed lineage leukemia (MLL) gene to the CBP gene, suggesting that dysregulation of CBP plays a role in leukemogenesis (42, 47). Furthermore, defects in primitive hematopoiesis were observed in the mice nullizygous for the CBP gene (39, 59). Although about 50% of mice heterozygous for the CBP gene are born alive, a significant percentage exhibit hematopoietic defects and malignancies (30).
CBP recruitment by Tax to the HTLV-1 promoter has been studied extensively in recent years, and the region of Tax interaction with CBP has been mapped to the amino-terminal C/H1 and KIX domains (14, 20, 31, 32, 55). In this report we show that Tax also interacts strongly with another region of CBP. This region, which we call carboxy-terminal region 2 (CR2), is located between amino acids (aa) 2003 and 2212. CR2 is conserved in both CBP and p300, and not surprisingly, we show that Tax binds strongly to the CR2 domain present in both proteins. We identify a carboxy-terminal region of Tax that is predicted to form an amphipathic helix and show that a specific amino acid on the surface of this helix (aa Y312) is responsible for the interaction with CR2. This CR2-interacting carboxy-terminal region of Tax is distinct from the previously identified KIX-interacting domain of Tax, which has been shown to reside between amino acids 81 and 95 (20). Transient-cotransfection assays provide functional evidence supporting the relevance of the Tax-CR2 interaction. Specifically, Tax Y312→E is defective for transactivation in vitro and in vivo, and two additional point mutations in this region of Tax (I315→E and L319→R) are defective for Tax transactivation in vivo. Interestingly, the CR2 region of CBP corresponds to the steroid receptor coactivator 1 (SRC-1)-interacting domain that is critical to coactivator utilization by liganded nuclear hormone receptors (17, 35). Tax has previously been shown to strongly repress transcription mediated through steroid and retinoid receptors (9). We show in this report that Tax competes for SRC-1 binding to CR2 in vitro, suggesting that the mechanisms of Tax repression of nuclear hormone signaling may be mediated through coactivator competition. Together, these data indicate that Tax recruits CBP/p300 through binding to multiple independent sites, illustrating the complex nature of activator-coactivator interactions in transcriptional regulation.
MATERIALS AND METHODS
Cloning, expression, and purification of recombinant proteins.
The expression and purification of GST-C/H1-KIXaa302–683 and GST-CR2aa1894–2221 have previously been described (51). The six glutathione S-transferase (GST)–CR2 deletion mutants were prepared by PCR amplification of the mouse CBP cDNA sequence (pRC/RSV-CBP) (36) corresponding to aa 1894 to 2150, 1894 to 2100, 1894 to 2003, 2003 to 2212, 2003 to 2150, 2100 to 2212, and 2055 to 2150. These PCR fragments were inserted into the BamHI and EcoRI sites of pGEX2T (Amersham Pharmacia Biotech) and transformed into Escherichia coli BL21(DE3)pLysS cells, and the GST fusion proteins were purified by glutathione-agarose chromatography. The four double point mutants of GST- CR2aa2003–2212 were made by PCR amplification of the CBP cDNA sequence using the QuickChange site-directed mutagenesis kit (Stratagene). The PCR products were cloned, and the proteins expressed and purified as described above. Histidine-tagged CR2 (His6-CR2) was made by PCR amplification of the CBP cDNA sequence corresponding to amino acids 2003 to 2212, with attB and attR sites at their respective 5′ and 3′ ends for cloning into the Gateway system (Life Technologies, Inc.). This PCR fragment was inserted into the attB and attR sites of pDonr206 (Gateway) and subsequently cloned into pDest17 (Gateway), a His6 fusion vector for expression and purification by nickel chelate chromatography as previously described (14). The CR2 region from human p300, encompassing aa 1970 to 2193 (corresponding to mouse CBP CR2 aa 2003 to 2212) was cloned by PCR amplification of the relevant sequence with attB and attR sites at their respective 5′ and 3′ ends and insertion into pDonr206 (Gateway) and then into pDest15 (Gateway), a GST fusion vector. The final clone was transformed in BL21(DE3)pLysS cells, expressed, and purified by glutathione-agarose chromatography. All purified proteins were dialyzed against TM buffer (50 mM Tris-HCl [pH 7.9], 100 mM KCl, 12.5 mM MgCl2, 1 mM EDTA [pH 8.0], 1 mM dithiothreitol, 0.1% [vol/vol] Tween 20, 20% [vol/vol] glycerol) and stored at −70°C.
CREB (13) and Tax-His6 (62) were expressed and purified as previously described (14). CREB was serine-133 phosphorylated by protein kinase A for use in the in vitro transcription assays as previously described (14). Tax Y312→E was made using the QuickChange site-directed mutagenesis kit (Stratagene) in the pTaxHis6 background (62) and purified by nickel chelate chromatography. Tax K88→A was expressed and purified as previously described (20). GST-Tax (full length) and GST-Taxaa286–337 (8) were made by appropriate PCR amplification of the Tax cDNA sequence and inserted into the BamHI site of pGEX2T (Amersham Pharmacia Biotech). The GST-Tax proteins were purified using glutathione-agarose chromatography as described above. Purified proteins were stored at −70°C in TM buffer. Full-length SRC-1 (45) was transcribed and translated using the TNT Quick-Coupled in vitro transcription/translation system (Promega). SRC-1 was labeled with [35S]methionine during the in vitro transcription-translation reaction.
GST pulldown assays.
All GST pulldown experiments were performed using 12.5 μl of glutathione-agarose beads equilibrated in pulldown buffer (20 mM HEPES [pH 7.9], 12.5 mM MgCl2, 10 μM ZnSO4, 25 mM KCl, 0.5 mM EDTA [pH 8.0], 10% [vol/vol] glycerol, 0.05% [vol/vol] Nonidet P-40). The purified GST proteins were incubated with the equilibrated beads for 2 h at 4°C, washed, and incubated with the second protein overnight at 4°C. The reactions were washed three times with pulldown buffer, and bound proteins were separated by electrophoresis sodium dodecyl sulfate–12% polyacrylamide gel (SDS–12% PAGE), transferred to nitrocellulose, and analyzed by Western blot. The following antibodies were used in the GST pulldown experiments: anti-His antibody (H-15; Santa Cruz Biotechnology), anti-GST antibody (Sigma), and anti-Tax antibody (epitope corresponding to the 13 carboxy-terminal aa). The GST pulldown competition assays were carried out by incubating 5 pmol of GST or GST-CR2aa2003–2212 with 12.5 μl of glutathione-agarose beads equilibrated in pulldown buffer for 2 h at 4°C, washed, and incubated with radiolabeled SRC-1 (0.1 μl) and increasing amounts of wild-type Tax or Tax Y312→E overnight at 4°C. The reactions were washed four times with pulldown buffer, and bound proteins were separated by electrophoresis on SDS–12% PAGE. Bound, labeled SRC-1 was detected by PhosphorImager analysis. Tax binding was detected by Western blot analysis using an anti-His6 antibody (H-15; Santa Cruz Biotechnology).
Yeast two-hybrid assay.
Full-length Tax (pTaxHis6) (62) was cloned into a TRP1-marked Gal4 activation domain construct, pDest22 (Gateway). The KIX domain (aa 588 to 683) and the CR2 domain (aa 2003 to 2212) were generated by PCR amplification of their respective domains from the CBP cDNA sequence (pRC/RSV-CBP) and subsequently cloned into a LEU2-marked Gal4 DNA-binding domain construct, pDest21 (Gateway). Different fusion constructs and a promoter containing the HIS3 gene with four upstream Gal4 binding sites were transformed into Saccharomyces cerevisiae strain MaV103 (relevant genotype: MATa leu2-3,112 trp1-901 his3Δ200 gal4Δ gal80Δ GAL1::LACZ GAL1::HIS3@lys2 SPA-L10::URA3) for two-hybrid analysis. Protein-protein interactions were detected by cell growth phenotypes in the presence of aminotriazole on plates lacking histidine (21).
Mammalian expression plasmids, cell culture, and transient-cotransfection assays.
The Tax expression plasmid pHTLV-I Tax (6), the c-fos expression plasmid RSV-c-fos (3), and the luciferase reporter plasmids viral CRE-Luc (14) and AP-1-Luc (53) have been described previously. The cytomegalovirus (CMV) expression plasmids for CR2aa2003–2212 and CR1aa1515–1895 were prepared by PCR amplification of the CBP cDNA sequence corresponding to their respective amino acids, incorporating the attB and attR sites on the ends of the PCR fragment. The ΔCR2aa2003–2212 L2068→A/L2071→A CMV expression plasmid was prepared by PCR amplification of the GST-CR2aa2003–2212 double point mutant plasmid, incorporating the attB and attR sites on the ends of the PCR fragment. These fragments were cloned into pDonr206 (Gateway) and then into the CMV promoter-driven expression vector pDest26 (Gateway). The Gal4-Tax point mutants Y312→E, I315→E, and L319→R were made in the Tax cDNA sequence in the pGal4-Tax-S expression plasmid (8) using the QuickChange site-directed mutagenesis kit (Stratagene). Transient-cotransfection assays were performed in the HTLV-I-negative human T-lymphocyte cell line Jurkat. Cells were cultured in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and penicillin-streptomycin. Transient cotransfections were performed with 106 cells in unsupplemented medium, a constant amount of DNA, and Lipofectamine (Life Technologies, Inc.) for 5 h. Supplemented medium was then added to each transfection and incubated for an additional 24 h. Cells were lysed and assayed for luciferase activity using a Turner Designs model TD 20-e luminometer. The Renilla luciferase plasmid was used as an internal control.
Recombinant plasmids and in vitro transcription assays.
The 4TxRE G-less cassette carries four copies of the third 21-bp Tax-responsive elements (TxRE) located upstream of the HTLV-1 core promoter (2). Preinitiation complexes were formed on 100 ng of DNA template in TM buffer supplemented with 10 μM acetyl-coenzyme A (Sigma) by the addition of the indicated amounts of CREB and/or Tax and 70 μg of CEM nuclear extract (10) in a final reaction volume of 30 μl. The reactions were incubated for 60 min at 30°C. RNA synthesis was initiated by the addition of 250 μM each ATP, CTP, and UTP and 12 μM GTP plus 0.07 μM [α-32P]UTP (3,000 Ci/mmol) and incubated for an additional 35 min at 30°C. RNase T1 was then added to the reaction for an additional 20 min at 37°C. The reactions were terminated by the addition of stop solution (133 mM NaCl, 0.5% [vol/vol] SDS, 3.3 mM Tris-HCl [pH7.9]) with recovery standard. RNA was purified with phenol-chloroform, precipitated with ethanol, and analyzed by 6.5% urea denaturing PAGE. Gels were dried and visualized by PhosphorImager analysis.
RESULTS
Identification of carboxy-terminal region of CBP that interacts with Tax.
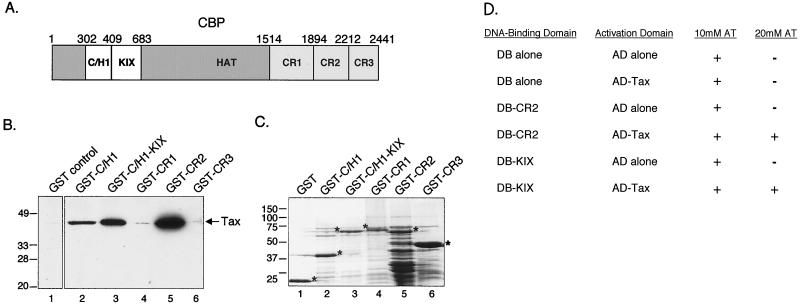
Several previous studies have shown that Tax physically interacts with the C/H1 and KIX domains of CBP to activate transcription (14, 20, 31, 32). We were interested in testing whether additional regions of CBP may participate in Tax transactivation. To test this hypothesis, several GST-CBP fusion proteins spanning the carboxy-terminal half of the coactivator were bound to glutathione-agarose beads and used in a GST pulldown assay with full-length purified, recombinant Tax. All of these GST-CBP fusion constructs are illustrated in Fig. 1A. Tax bound strongly to one of the three carboxy-terminal regions of CBP (Fig. 1B, lane 5). This region, which we call carboxy-terminal region 2 (CR2), encompasses CBP aa 1894 to 2212. As positive controls, we also tested the binding of Tax to the amino-terminal C/H1 domain (aa 302 to 409) and to a region that encompasses both C/H1 and KIX (aa 302 to 683) (Fig. 1B, lanes 2 and 3). We observed markedly stronger Tax binding to CR2 than to these other CBP regions. A Coomassie-stained gel showing the GST proteins used in the pulldown assay is shown in Fig. 1C. The interaction between Tax and CR2 was also confirmed using the yeast two-hybrid assay (Fig. 1D).
FIG. 1.
Tax binds to both the CR2 and C/H1-KIX domains of CBP. (A) Schematic representation of the 2,441-aa cellular coactivator CBP. The regions tested for Tax binding are indicated. HAT, histone acetyltransferase domain. (B) Tax binds to the CR2 domain in vitro. Purified recombinant Tax (50 pmol) was incubated with GST alone or the indicated GST-CBP fusion protein (50 pmol each). As controls, Tax binding to GST-C/H1aa302–409 and GST-C/H1-KIXaa302–683 was also assayed. The bound proteins were separated by SDS–12% PAGE, transferred to nitrocellulose, and detected using an anti-His6 antibody. The positions of bound Tax and protein molecular size standards are indicated (in kilodaltons). (C) Coomassie-stained SDS–12% PAGE showing the GST fusion proteins used in panel B. Asterisks denote GST fusion proteins, and protein molecular size standards are indicated in kilodaltons. (D) Tax binds to the CR2 domain in vivo. Growth phenotypes of the designated DNA-binding (DB) and activation domain (AD) constructs were assayed by streaking cells on plates lacking histidine and containing 10 or 20 mM aminotriazole (AT). Plates were analyzed following 5 days of incubation at 30°C.
Critical CR2 amino acids required for Tax binding.
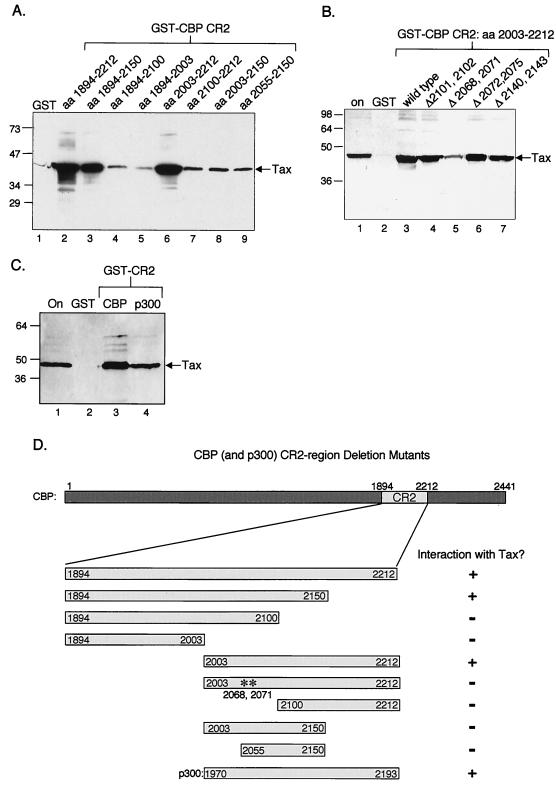
We next determined the minimal region of CR2 competent for Tax binding. Progressive deletions of CR2 revealed that aa 2150 (Fig. 2A, lane 3) represents the carboxy-terminal border, and aa 2003 (Fig. 2A, lane 6) represents the amino-terminal border competent for wild-type interaction with Tax. Further deletions from either terminus significantly reduced the CR2 interaction with Tax (Fig. 2A). These data indicate that the minimal region of CR2 competent for interaction with Tax should reside between aa 2003 and 2150. Unexpectedly, this region in isolation (GST-CR2aa2003–2150) was not competent for Tax binding (Fig. 2A, lane 8). Rather, a larger region, encompassing aa 2003 to 2212, was required for Tax binding (Fig. 2A, lane 6). This observation suggests that amino acids near either end of this domain participate in Tax binding and that while removal of one has no detectable effect on the interaction, removal of both termini abolishes Tax binding. This hypothesis fits with secondary structural analysis that predicts that this region of CBP has the potential to form multiple α-helices (45). It is possible that a critical number of interacting motifs are required for Tax binding and that the strength of the Tax-CR2 interaction is a function of the absolute number of helical motifs present in the fragment.
FIG. 2.
Identification and characterization of minimal CR2 domain. (A) Tax interacts with aa 2003 to 2212 of CBP in vitro. Purified recombinant Tax (50 pmol) was incubated with GST alone or the indicated GST-CR2 deletion mutants (50 pmol). As a positive control, Tax binding to the full-length region of GST-CR2aa1894–2212 was also tested. The bound proteins were separated by SDS–12% PAGE, transferred to nitrocellulose, and detected using an anti-His6 antibody. Positions of bound Tax and protein molecular size standards are indicated (in kilodaltons). The Western blot was stripped and reprobed with anti-GST to ensure that equal amounts of GST fusion protein were used in the assay. (B) Tax is defective for interaction with the double point mutant ΔCR2 L2068→A/L2071→A. Purified recombinant Tax (50 pmol) was assayed for its ability to bind to GST alone or the GST-CR2aa2003–2212 double point mutants F2101→A/I2102→A, L2068→A/L2071→A, L2072→A/L2075→A, and L2140→A/L2143→A (50 pmol each). Tax binding to wild-type GST-CR2aa2003–2212 was tested as a positive control. The bound proteins were separated by SDS–12% PAGE, transferred to nitrocellulose, and detected using an anti-His6 antibody. Bound Tax and protein molecular size standards are indicated (in kilodaltons). Five percent of the Tax onput is shown in lane 1. (C) Tax binds equally well to the CR2 domains derived from CBP and p300. Purified recombinant Tax (50 pmol) was incubated with GST alone or the GST-CR2 region from CBP (aa 2003 to 2212) or p300 (aa 1970 to 2193) (50 pmol each). The bound proteins were electrophoresed on SDS–12% PAGE, transferred to nitrocellulose, and detected using an anti-His6 antibody. Bound Tax and protein molecular size standards are indicated. Asterisks indicate the double point mutant (in kilodaltons) in GST-ΔCR2 L2068→A/L2071→A. Bound Tax was quantified using ImageQuant, and the results indicated that the intensities were nearly equal. (D) Summary of CR2-Tax interactions.
In an effort to identify critical amino acids within the CR2 region responsible for the interaction with Tax, we prepared and characterized double point mutations. The amino acids selected for point mutagenesis were chosen based on homology between mouse and human CBP as well as homology between CBP and p300. We also used sequence gazing to select leucine residues that had the potential to form α-helices (and thus the potential to form protein-protein contacts). The selected residues were changed to alanines, as they are the least disruptive to secondary and tertiary structure. Four CR2 constructs were prepared, each carrying two point mutations. These double point mutations were F2101→A/I2102→A, L2068→A/L2071→A, L2072→A/L2075→A, and L2140→A/L2143→A. Figure 2B shows that only the double point mutation L2068→A/L2071→A had a significant effect on Tax binding (lane 5). Interestingly, this mutation disrupts one of the four α-helices that have been predicted to reside within this region (45). Although we cannot rule out the possibility that this CR2 double point mutation disrupts the structure of the full CR2 domain, the data provide further evidence for a specific interaction between Tax and CR2.
Although CBP and p300 are highly homologous, it is unclear whether these two proteins are functionally redundant. CBP and p300 have domains of high amino acid sequence homology (e.g., KIX, >90%), whereas other regions, such as CR2, are more divergent (∼50%). We were therefore interested in testing whether Tax interacts with the corresponding CR2 region present in p300. To address this question, we cloned the corresponding region of p300 (aa 1970 to 2193), fused it to GST, and tested Tax binding in a GST pulldown assay. Figure 2C shows that Tax binds comparably to the CR2 regions from both CBP and p300 (lanes 3 and 4). A summary of the Tax interaction with the various CR2 constructs from CBP/p300 is shown in Fig. 2D.
CR2 domain represses Tax transactivation in vivo.
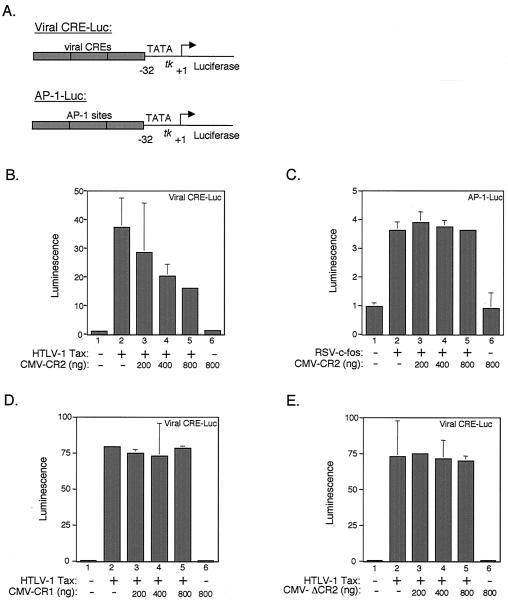
The observation that Tax efficiently binds to the CR2 domain of CBP in vitro led to the hypothesis that expression of CR2 in vivo might compete with active CBP/p300 in the cell, thus inhibiting Tax transactivation. To test this idea, we performed transient-cotransfection assays in HTLV-1-negative Jurkat T cells using a reporter plasmid carrying three copies of the Tax-responsive viral CRE (viral CRE-Luc) driving expression of the luciferase gene (Fig. 3A) (14). We measured Tax transactivation in the presence of increasing amounts of an expression plasmid carrying the CR2 region (aa 2003 to 2212) under the control of the CMV promoter. As expected, expression of Tax strongly activated transcription from the Tax-responsive promoter (Fig. 3B, lanes 1 and 2). Cotransfection of increasing amounts of the CR2 expression plasmid repressed Tax transactivation in a dose-dependent fashion (Fig. 3B, lanes 3 to 5). While the highest amount of CR2 (800 ng) strongly repressed Tax transcription function, it had no effect on reporter plasmid expression in the absence of Tax, suggesting that the effect of CR2 was specific to Tax and that CR2 expression was not toxic to the cells (Fig. 3B, lane 6). It is noteworthy that in these experiments, CR2 may be directly affecting Tax protein levels, as the expression plasmid uses the Tax-responsive HTLV-1 promoter to drive Tax synthesis. Therefore, the repressive effect of CR2 may occur through both reductions in Tax levels and repression of the viral CRE-luciferase reporter plasmid. As a control, we tested CR2 expression on c-fos-dependent transcription from an AP-1-luc reporter plasmid (Fig. 3A). Figure 3C shows that increasing concentrations of CR2 had no effect on c-fos-dependent transcription (lanes 3 to 5), suggesting that CR2 disruption of transcriptional activity is specific to proteins that bind to the CR2 domain. As additional controls, we also tested expression of two CBP molecules that are negative for an interaction with Tax. We have shown that Tax does not interact with CR1 (Fig. 1B) or a form of CR2 carrying the L2068→A/L2071→A double point mutation (ΔCR2) (Fig. 2B). As expected, cotransfection of plasmids expressing these molecules had no effect on Tax transactivation (Fig. 3D and E).
FIG. 3.
CR2aa2003–2212 inhibits Tax-activated transcription in vivo. (A) Schematic illustration of the viral CRE and AP-1-luciferase reporter constructs used in the transient-cotransfection assays. Transient-cotransfection assays were performed in HTLV-1-negative human Jurkat T cells. (B) CMV-CR2aa2003–2212 expression inhibits Tax transactivation. The Tax-responsive viral CRE-Luc reporter plasmid (100 ng) (14) was cotransfected with a constant amount of the HTLV-1 Tax expression plasmid (6) (200 ng) (lanes 2 to 5) and an increasing amount of an expression plasmid for CMV-CR2aa2003–2212 (lanes 3 to 6), as indicated. (C) CMV-CR2aa2003–2212 expression does not affect c-fos-activated transcription in vivo. Transient-cotransfection assays were again performed in Jurkat T cells; however, the AP-1-Luc reporter plasmid (400 ng) (14, 53) was cotransfected with a constant amount of the RSV-c-fos (3) (400 ng) (lanes 2 to 5) and an increasing amount of the CR2 expression plasmid CMV-CR2aa2003–2212 (lanes 3 to 6), as indicated. (D and E) CMV-CR1aa1515–1895 expression and CMV-ΔCR2aa2003–2212 expression of the L2068→A/L2071→A double point mutation does not affect Tax-activated transcription in vivo. The viral CRE-Luc reporter plasmid (100 ng) was cotransfected with a constant amount of the HTLV-1 Tax expression plasmid (6) (200 ng) (lanes 2 to 5) and an increasing amount of the expression plasmid for CMV-CR1aa1515–1895 (panel D, lanes 3 to 6), as indicated, or CMV-ΔCR2aa2003–2212 carrying the L2068→A/L2071→A double point mutation (panel E, lanes 3 to 6), as indicated. In all experiments, a constant amount of the Renilla luciferase reporter plasmid (10 ng) was added to each reaction as an internal control. Luminescence was quantitated with a luminometer, and activation was quantitated relative to expression from the viral CRE-Luc reporter plasmid in the absence of Tax or from the AP-1-Luc reporter plasmid in the absence of c-fos. The values shown are the mean fold activation (in duplicate) ± the standard deviation. The experiments are representative of at least three independent experiments. Western blot analysis using anti-His6 antibody confirmed expression of CMV-CR2 and CMV-ΔCR2 following transfection (data not shown).
Critical Tax amino acids required for CR2 binding.
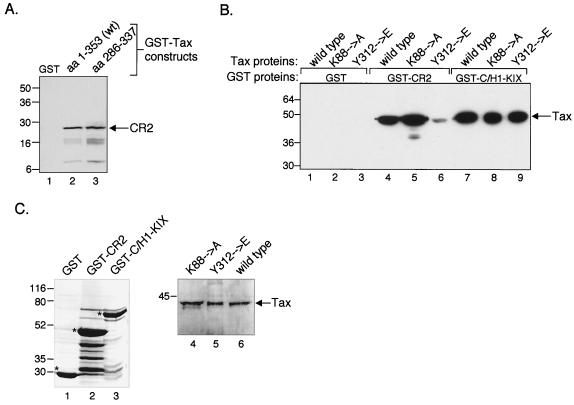
To further characterize the Tax-CR2 interaction, we were interested in determining the region of Tax that is involved in CR2 binding. We prepared deletion mutants of Tax, fused them to GST, and tested their interaction with purified His6-CR2aa2003–2212. Figure 4A shows that CR2 interacted comparably with both full-length Tax and a carboxy-terminal fragment of Tax encompassing aa 286 to 337, suggesting that the CR2-interacting region of Tax resides near the carboxy terminus of the protein. Interestingly, several previous studies have identified several point mutations in this region that significantly reduce or abolish Tax transactivation of HTLV-1 transcription in vivo, suggesting that this region of Tax may serve as an activation domain (43, 44, 46). Since one of the prominent functions of an activation domain is coactivator recruitment, we reasoned that specific amino acids in this region might contact CR2. To address this possibility, we created a point mutation specifically within a subdomain of this region (Tax aa 312 to 319) that we predicted would participate in amphipathic helix formation. This prediction is based on the sequence aro-X-X-φ-φ-X-X-φ (aro represents F or Y, and φ represents bulky hydrophobic groups), where the hydrophobic and aromatic amino acids are predicted to form the face of an amphipathic helix and mediate protein-protein contacts (41). This sequence has been identified in the activation domains of several transcription factors, including CREB and p53 (41). The Tax sequence 312-Y-T-N-I-P-I-S-L-319 matches this conserved sequence, suggesting that it might participate in coactivator binding. To determine whether this sequence in Tax is involved in CR2 binding, we targeted the critical tyrosine at aa 312 and replaced it with a glutamic acid. Purified full-length Tax protein carrying the Y312→E mutation was tested in a GST pulldown assay with GST-CR2aa2003–2212. As a control, we compared the binding of Tax Y312→E with Tax K88→A, a mutant form of Tax that has been reported to be defective for KIX binding (20). Figure 4B shows that Tax protein carrying the Y312→E point mutation was significantly compromised for an interaction with CR2 (lane 6), whereas Tax K88→A was not (lane 5). Tax Y312→E bound GST-C/H1-KIX with apparent wild-type affinity, indicating that the mutation is not globally disruptive to the structure of Tax (Fig. 4B, lane 9). Interestingly, we observed near wild-type binding of Tax K88→A to our GST-C/H1-KIX construct. Although this binding was unexpected, it is likely due to additional contacts between Tax and the C/H1 portion of GST-C/H1-KIX, as we have previously observed strong binding of Tax to this region (32). Coomassie-stained gels of Tax and the GST fusion proteins used in the GST pulldown assay are shown in Fig. 4C.
FIG. 4.
Carboxy-terminal activation domain of Tax interacts with CR2. (A) CR2 binds to the putative transactivation domain of Tax. Purified, recombinant His6-CR2aa2003–2212 (50 pmol) was incubated with GST alone or the indicated GST-Tax fusion constructs (50 pmol). The bound proteins were separated by SDS–10% PAGE, transferred to nitrocellulose, and detected using an anti-His6 antibody. The positions of bound His6-CR2aa2003–2212 and protein molecular size standards are indicated (in kilodaltons). (B) The Tax Y312→E point mutant is defective for interaction with CR2. Purified recombinant wild-type Tax (lanes 1, 4, and 7), Tax K88→A (lanes 2, 5, and 8), or Tax Y312→E (lanes 3, 6, and 9) (50 pmol each) were incubated with GST alone or the indicated GST-CR2aa2003–2212 or GST-C/H1-KIXaa302–683 fusion proteins. The bound proteins were separated by SDS–12% PAGE, transferred to nitrocellulose, and detected using an anti-His6 antibody. Bound Tax and protein molecular size standards are indicated (in kilodaltons). (C) Coomassie-stained gels of the purified proteins used in the experiment shown in panel B. The left panel shows the GST fusion proteins; asterisks denote the relevant protein band. The right panel shows both the wild-type and mutant forms of Tax used in the experiment. Protein molecular size standards are indicated (in kilodaltons).
Functional studies of Tax-CR2 interaction.
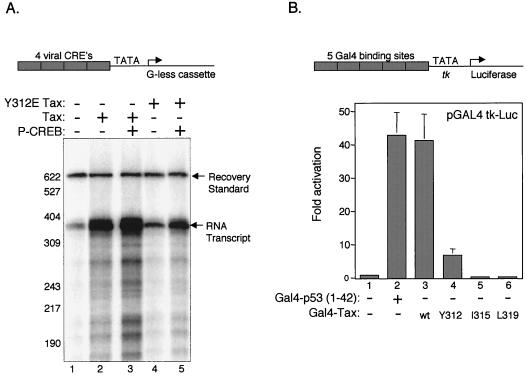
We were then interested in testing whether point mutations in Tax that disrupted the CR2 interaction also disrupted Tax transactivation. We first examined Tax Y312→E activity in an in vitro transcription assay using a plasmid carrying a Tax-responsive promoter driving synthesis of two tandem G-less cassettes. The G-less cassette allows synthesis of a defined 380-nucleotide (nt) guanineless transcript from the closed circular template. The Tax-responsive promoter, which drives the G-less cassette, carries four viral CRE elements cloned immediately upstream of the core HTLV-1 promoter (2). A schematic representation of the promoter construct is shown in Fig. 5A. We performed in vitro transcription assays using nuclear extracts prepared from the HTLV-1-negative human T-cell line CEM. Figure 5A shows the results of the in vitro transcription experiment. As expected, the addition of wild-type Tax alone and in combination with the Ser133-phosphorylated form of CREB produced an increase in transcription from the 4TxRE/G-less template (8- and 15-fold, respectively) (Fig. 5A, lanes 1 to 3). However, the addition of the same amount of Tax protein carrying the Y312→E point mutation produced only a very modest increase in RNA synthesis (twofold) (Fig. 5A, lane 4). The addition of Ser133-phosphorylated CREB to the Tax Y312→E reactions only partially rescued transcriptional activation (fivefold) (Fig. 5A, lane 5). In an attempt to directly evaluate an effect of the Tax Y312→E on coactivator utilization, we tested the effect of full-length p300 in these experiments. We did not, however, observe enhanced Tax transactivation upon p300 addition (data not shown). This observation is consistent with previous reports showing that p300 does not activate transcription in vitro in the absence of chromatin (28, 29).
FIG. 5.
Tax Y312→E is defective for transactivation both in vitro and in vivo. (A) Tax Y312→E is defective for transcriptional activation from a Tax-responsive element promoter. The in vitro transcription assay was performed on a 4T×RE G-less cassette (2) template that carries four tandem copies of the third (promoter-proximal) viral CRE driving expression of a 380-nt RNA (shown schematically above panel 5A). Transcription reaction mixtures contained the 4TxRE G-less template (100 ng), nuclear extract (70 μg of CEM, a human T-cell line) (lanes 1 to 5), purified recombinant wild-type Tax (100 ng) (lanes 2 and 3), purified recombinant Tax Y312→E (100 ng) (lanes 4 and 5), and pCREB (50 ng) (lanes 3 and 5). The positions of the full-length 380-nt RNA transcript, labeled DNA recovery standard, and molecular size markers are indicated (in nucleotides). This experiment is representative of three independent experiments. (B) Tax Y312→E is defective for transactivation in vivo. Transient-cotransfection assays were performed in HTLV-1-negative human T-lymphocyte Jurkat cells. The Gal4-luciferase reporter plasmid (400 ng) (8) (shown schematically above panel B) was cotransfected with Gal4-p53 (aa 1 to 42) (200 ng) (lane 2) or the indicated wild-type or mutant Gal4-Tax expression plasmid (200 ng) (lanes 3 to 6) and a constant amount of Renilla luciferase reporter plasmid (10 ng). The Gal4-Tax point mutations used in this study are Y312→E, I315→E, and L319→R. Luminescence was quantitated with a luminometer, and activation was calculated relative to the activation from the Gal4 reporter plasmid alone in the absence of Tax or p53. The values shown are the mean fold activation of duplicates ± the standard deviation. The experiment shown is representative of three independent trials.
To further examine the role of the CR2-interacting domain in Tax transactivation, we performed transient-cotransfection assays. For these experiments, we used an expression plasmid carrying the full-length Tax protein fused to the DNA-binding domain of Gal4 (8). In this background, we introduced the Y312→E point mutation and two additional point mutations in the activation domain region of Tax. These two new mutations, I315→E and L319→R, were also targeted to the putative amphipathic helix in Tax that likely plays a role in HTLV-1 transcription (see above). Figure 5B shows, as expected, that cotransfection of Gal4-wtTax strongly activated transcription from a reporter plasmid carrying five copies of the Gal4 DNA-binding site (Gal4-Luc; Fig. 5B, compare lanes 1 and 3). Each of the Gal4-Tax point mutants, Y312→E, I315→E, and L319→R, were significantly compromised for Tax transactivation (Fig. 5B, lanes 4 to 6). As a control for promoter activity, we also tested the activation domain of p53 (aa 1 to 42) fused to the Gal4 DNA-binding domain (GaL4-p53aa1–42) in the transient-cotransfection assay (Fig. 5B, compare lanes 1 and 2). These data further support a functional role for this region of Tax in the activation of transcription.
Tax and SRC-1 compete for CR2 binding in vitro.
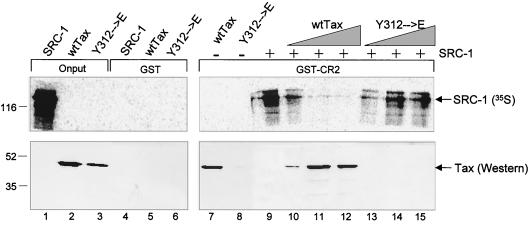
In addition to binding Tax, the CR2 region also corresponds to the SRC-1-interacting domain of CBP that is critical to coactivator utilization by liganded nuclear hormone receptors (17, 35). Previously, Tax has been shown to strongly repress transcription mediated through steroid and retinoid receptors (9). This observation, together with the observation that both Tax and SRC-1 physically interact with CR2, led to the hypothesis that their binding may be mutually exclusive. To directly test this idea, we examined whether increasing concentrations of Tax can displace SRC-1 from CR2 in vitro. As a control, we also tested Y312→E Tax, which is defective for CR2 binding. Glutathione beads were bound with GST-CR2aa2003–2212 and then incubated with in vitro-transcribed and translated full-length SRC-1. Increasing amounts of wild-type Tax or Tax Y312→E were included in the binding reactions containing SRC-1, and the resulting protein-protein interactions were detected by PhosphorImager and Western blot analysis. Figure 6 shows that increasing amounts of Tax dramatically reduced SRC-1 binding to CR2, with a concomitant increase in Tax binding (lanes 10 to 12). As expected, increasing amounts of Tax Y312→E had no effect on SRC-1 binding to CR2 (lanes 13 to 15). Together, these data indicate that the binding of SRC-1 and Tax to CR2 is mutually exclusive in vitro.
FIG. 6.
(A) Tax and SRC-1 binding to CR2aa2003–2212 is mutually exclusive. SRC-1 [35S]-labeled transcription-translation product (0.1 μl) was incubated with GST alone or GST-CR2aa2003–2212 (5 pmol) (lanes 4 and 9 to 15) in the presence of increasing amounts of wild-type Tax protein (5, 50, and 100 pmol) (lanes 10 to 12) or Tax Y312→E (5, 50, and 100 pmol) (lanes 13 to 16). Wild-type (wt) and Y312→E Tax (50 pmol) were also incubated with GST alone (5 pmol) (lanes 5 and 6) or GST-CR2aa2003–2212 (5 pmol) (lanes 7 and 8). Bound proteins were separated by SDS–12% PAGE. Top, PhosphorImager analysis of bound SRC-1. Bottom, Western blot analysis using anti-His6 antibody. The positions of SRC-1 onput (100%) (lane 1), wild-type Tax (5 pmol) (lane 2), and Tax Y312→E (5 pmol) (lane 3) and molecular size markers are indicated (in kilodaltons).
DISCUSSION
The results presented in this report indicate that the HTLV-1 Tax protein interacts with multiple domains of the pleiotropic coactivators CBP and p300. In addition to the interactions with the C/H1 and KIX domains, we show here that Tax strongly interacts with a carboxy-terminal region of CBP, encompassing CBP aa 2003 to 2212. Tax also binds strongly to the corresponding CR2 region in p300. Expression of the CBP-CR2 region alone in vivo represses Tax transactivation function, suggesting that Tax binding to the isolated CR2 region prohibits recruitment of the full-length coactivators. We identified a carboxy-terminal region of Tax, encompassing aa 312 to 319, that participates in the interaction with CR2. A point mutation in this region, Y312→E, causes defects in CR2 binding and Tax transactivation. We show that other point mutations (I315→E and L319→R) in this region also strongly compromise Tax transactivation function. The data in this report suggest that the aa 312 to 319 region of Tax forms a potent activation domain and that this region functions to contact CR2 and thus participate in the recruitment of the coactivator to the HTLV-1 promoter. Further evidence supporting a role for the Tax activation domain in coactivator interaction comes from a recent report showing that M47 Tax (L319→R, L320→S) is partially defective for interaction with full-length p300 in an electrophoretic mobility shift assay (20). Together, these data strengthen previously published reports showing that a variety of point mutations in this region strongly repress Tax transcription function. These include double point mutations at aa 310 to 311, 315 to 316, and 319 to 320 and single point mutations at aa 316, 317, 318, and 320 (44, 46).
We have begun to delineate the molecular basis of the Tax-CR2 interaction. A short sequence in the Tax activation domain conforms precisely to a consensus sequence found in the activation domains of several other transcription factors (41). This consensus sequence, aro-X-X-φ-φ-X-X-φ- is predicted to form an amphipathic helix and therefore participate in protein-protein interactions. The Tax sequence 312-Y-T-N-I-P-I-S-L-319 matches the reported consensus sequence precisely. Furthermore, mutation of the critical tyrosine residue (Y312→E), which is predicted to reside on the face of the helix, dramatically reduced CR2 binding in vitro and Tax transactivation in vitro and in vivo. Mutation of additional residues (I315→E and L319→R) also predicted to reside on the face of the helix also dramatically reduced Tax transactivation in vivo. These data strongly suggest that the activation domain of Tax forms an amphipathic helix-binding surface which mediates CR2 interaction. Previous research has also shown that the M47 Tax mutant is defective for transcriptional activation from the HTLV-1 long terminal repeat (LTR) while completely functional for transactivation from the human immunodeficiency virus type 1 LTR (1, 43). The other half of the activator-coactivator interaction is contributed by CR2 aa 2003 to 2212. Interestingly, this minimal CR2 region carries four predicted α-helical domains (45), which may be critical for interaction with the amphipathic helix of Tax. Consistent with this idea, disruption of a single α-helix by point mutagenesis (L2068→A/L2071→A) significantly reduced Tax binding to CR2. These data define the Tax-CR2 interface and provide a framework for further studies on the molecular structure of the Tax-CR2 interaction.
The observation that Tax contacts the CR2 region of CBP and p300 indicates that at least three distinct sites on the coactivator may participate in mediating Tax transactivation. Several previous studies have shown that Tax also binds to the KIX and C/H1 domains of CBP and that these interactions may be relevant in Tax transactivation (14, 20, 31, 32, 55). It is interesting that distinct regions of Tax (defined by the K88→A and Y312→E point mutations) make independent contacts on CBP/p300, suggesting that discrete coactivator contacts can be made simultaneously. Functional studies suggest that independent coactivator contacts contribute to Tax function, such as KIX and CR2, when expressed in isolation, each repressing Tax transactivation approximately 50% (14) (Fig. 3B). This evidence suggests that the distinct Tax-CBP interactions occur simultaneously and perhaps cooperate to enhance coactivator-mediated transcriptional activation. It is not currently known whether Tax binding simultaneously at multiple sites in CBP/p300 leads to more efficient coactivator recruitment or whether the individual interactions promote dissimilar effects on Tax-activated transcription. It is interesting that other transcription factors bind to multiple domains of CBP which could be important for reconfiguring the CBP molecule for chromatin remodeling and/or recruitment of the RNA polymerase II transcriptional machinery (22, 51). Most notably, the tumor suppressor p53 also binds to the KIX domain as well as a C-terminal domain of CBP, whereby Tax and p53 exhibit mutually exclusive binding to the KIX domain (19, 51). The fact that Tax binds as a dimer on the HTLV-1 promoter in the nucleoprotein complex also provides further potential for multiple interactions with CBP and p300 as well as with other components of the transcription machinery (23, 50).
It is interesting that the CR2 region of CBP defined in this study precisely overlaps the SRC-1 interaction domain of CBP (24, 28, 45, 58). SRC-1 is a prominent member of a family of coactivators that bind to this specific region of CBP and mediate transcriptional activation of nuclear hormone receptors (17, 35). Given the overlap of the amino acids required for Tax binding and SRC-1 binding (aa 2003 to 2212 and 2058 to 2130, respectively), it not surprising that the binding of these two proteins to CR2 in vitro is mutually exclusive. These data are consistent with a recent study showing that Tax is a potent inhibitor of nuclear receptor-activated transcription mediated through SRC-1 (9). Interestingly, a double point mutation in Tax, at aa 319 and 320 (M47) (46), relieved the repression of nuclear hormone signaling. This observation is not unexpected, as this double point mutation likely disrupts the amphipathic helix in Tax and thus the Tax-CR2 interaction. Reduced Tax binding to CR2 would give SRC-1 greater access to the limiting coactivator and relieve the repression. Although the report did not identify a mechanism of Tax repression of nuclear hormone transcription function, the authors did suggest that CBP may be involved. These data provide strong corroborating evidence for a functionally relevant Tax-CR2 interaction in vivo.
In summary, the studies presented herein define a new domain on CBP/p300 that is involved in Tax transactivation and demonstrate the complexity of activator-coactivator interactions in mediating gene regulation. Several previous studies have provided extensive evidence for Tax binding to the KIX domain, and more recently to the C/H1 domain, resulting in competition with other transcription factors that bind to these regions (5, 7, 32, 49, 51, 53). Our demonstration here that Tax binds to an additional region of CBP/p300 has further implications for Tax derailment of CBP coactivator function in the HTLV-1-infected cell. These studies provide the biochemical foundation for future work on the molecular interactions at the CR2-Tax interface as well as the biological consequences of this novel interaction.
ACKNOWLEDGMENTS
We thank D. Heery (University of Leicester) for the pSG5-hSRC1e expression plasmid and Karen Van Orden, Jeanne Mick, Melissa Gonzales, and Raji Edayathumangalam for their significant contributions to this research. We also thank Steve McBryant and Holli Giebler for critical reading of the manuscript.
This study was supported by Public Health Service grant CA-55035 from the National Cancer Institute.
REFERENCES
- 1.Adya N, Giam C-Z. Distinct regions in human T cell lymphotropic virus type I Tax mediate interactions with activator protein CREB and basal transcription. J Virol. 1995;69:1834–1841. doi: 10.1128/jvi.69.3.1834-1841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson M G, Scoggin K E, Simbulan-Rosenthal C M, Steadman J A. Identification of Poly(ADP-ribose) polymerase as a transcriptional coactivator of the human T-cell leukemia virus type 1 Tax protein. J Virol. 2000;74:2169–2177. doi: 10.1128/jvi.74.5.2169-2177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angel P, Allegretto E A, Okino S T, Hattori K, Boyle W J, Hunter T, Karin M. Oncogene jun encodes a sequence specific trans-activator similar to AP-1. Nature. 1988;332:166–171. doi: 10.1038/332166a0. [DOI] [PubMed] [Google Scholar]
- 4.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 5.Ariumi Y, Kaida A, Lin J Y, Hirota M, Masui O, Yamaoka S, Taya Y, Shimotohno K. HTLV-1 tax oncoprotein represses the p53-mediated trans-activation function through coactivator CBP sequestration. Oncogene. 2000;19:1491–1499. doi: 10.1038/sj.onc.1203450. [DOI] [PubMed] [Google Scholar]
- 6.Brady J, Jeang K-T, Duvall J, Khoury G. Identification of p40x-responsive regulatory sequences within the human T-cell leukemia virus type I long terminal repeat. J Virol. 1987;61:2175–2181. doi: 10.1128/jvi.61.7.2175-2181.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colgin M A, Nyborg J K. The human T-cell leukemia virus type 1 oncoprotein Tax inhibits the transcriptional activity of c-Myb through competition for the CREB binding protein. J Virol. 1998;72:9396–9399. doi: 10.1128/jvi.72.11.9396-9399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connor L M, Oxman M N, Brady J N, Marriott S J. Twenty-one base pair repeat elements influence the ability of a Gal4-Tax fusion protein to transactivate the HTLV-I long terminal repeat. Virology. 1993;195:569–577. doi: 10.1006/viro.1993.1408. [DOI] [PubMed] [Google Scholar]
- 9.Doucas V, Evans R M. Human T-cell leukemia retrovirus-Tax protein is a repressor of nuclear receptor signaling. Proc Natl Acad Sci USA. 1999;96:2633–2638. doi: 10.1073/pnas.96.6.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dynan W S. DNase I footprinting as an assay for mammalian gene regulatory proteins. Genet Eng. 1987;9:75–87. [Google Scholar]
- 11.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franchini G. Molecular mechanisms of human T-cell leukemia/lymphotropic virus type I infection. Blood. 1995;86:3619–3639. [PubMed] [Google Scholar]
- 13.Franklin A A, K. M F, Uittenbogaard M N, Brauweiler A, Utaisincharoen P, Matthews M-A H, Dynan W S, Hoeffler J P, Nyborg J K. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) response and cAMP element-binding protein (CREB) J Biol Chem. 1993;268:21225–21231. [PubMed] [Google Scholar]
- 14.Giebler H A, Loring J E, Van Orden K, Colgin M A, Garrus J E, Escudero K W, Brauweiler A, Nyborg J K. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol Cell Biol. 1997;17:5156–5164. doi: 10.1128/mcb.17.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giles R H, Peters D J, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 16.Giordano A, Avantagglati M L. p300 and CBP: partners for life and death. J Cell Physiol. 1999;181:218–230. doi: 10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 17.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 18.Goodman R H, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 19.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 20.Harrod R, Tang Y, Nicot C, Lu H S, Vassilev A, Nakatani Y, Giam C Z. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol Cell Biol. 1998;18:5052–5061. doi: 10.1128/mcb.18.9.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill D E, Hope I A, Macke J P, Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986;234:451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- 22.Jayaraman G, Srinivas R, Duggan C, Ferreira E, Swaminathan S, Somasundaram K, Williams J, Hauser C, Kurkinen M, Dhar R, Weitzman S, Buttice G, Thimmapaya B. p300/cAMP-responsive element-binding protein interactions with ets-1 and ets-2 in the transcriptional activation of the human stromelysin promoter. J Biol Chem. 1999;274:17342–17352. doi: 10.1074/jbc.274.24.17342. [DOI] [PubMed] [Google Scholar]
- 23.Jin D Y, Jeang K T. Transcriptional activation and self-association in yeast: protein-protein dimerization as a pleiotropic mechanism of HTLV-I Tax function. Leukemia. 1997;11:3–6. [PubMed] [Google Scholar]
- 24.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 25.Kashanchi F, Duvall J F, Kwok R P, Lundblad J R, Goodman R H, Brady J N. The coactivator CBP stimulates human T-cell lymphotrophic virus type I Tax transactivation in vitro. J Biol Chem. 1998;273:34646–34652. doi: 10.1074/jbc.273.51.34646. [DOI] [PubMed] [Google Scholar]
- 26.Kimzey A L, Dynan W S. Identification of a human T-cell leukemia virus type I tax peptide in contact with DNA. J Biol Chem. 1999;274:34226–34232. doi: 10.1074/jbc.274.48.34226. [DOI] [PubMed] [Google Scholar]
- 27.Kimzey A L, Dynan W S. Specific regions of contact between human T-cell leukemia virus type I Tax protein and DNA identified by photocross-linking. J Biol Chem. 1998;273:13768–13775. doi: 10.1074/jbc.273.22.13768. [DOI] [PubMed] [Google Scholar]
- 28.Kraus W L, Kadonaga J T. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kundu T K, Palhan V B, Wang Z, An W, Cole P A, Roeder R G. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol Cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- 30.Kung A L, Rebel V I, Bronson R T, Ch'ng L E, Sieff C A, Livingston D M, Yao T P. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok R P, Laurance M E, Lundblad J R, Goldman P S, Shih H, Connor L M, Marriott S J, Goodman R H. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 32.Lemasson I, Nyborg J K. HTLV-I tax repression of p73b is mediated through competition for the C/H1 domain of CBP. J Biol Chem. 2001;276:15720–15727. doi: 10.1074/jbc.M100131200. [DOI] [PubMed] [Google Scholar]
- 33.Lenzmeier B A, Giebler H A, Nyborg J K. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol Cell Biol. 1998;18:721–731. doi: 10.1128/mcb.18.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenzmeier B A, Nyborg J K. Molecular mechanisms of viral transcription and cellular deregulation associated with the HTLV-I Tax protein. Gene Ther Mol Biol. 1999;3:327–345. [Google Scholar]
- 35.Leo C, Chen J D. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 36.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 37.Lundblad J R, Kwok R P, Laurance M E, Huang M S, Richards J P, Brennan R G, Goodman R H. The human T-cell leukemia virus-1 transcriptional activator Tax enhances cAMP-responsive element-binding protein (CREB) binding activity through interactions with the DNA minor groove. J Biol Chem. 1998;273:19251–19259. doi: 10.1074/jbc.273.30.19251. [DOI] [PubMed] [Google Scholar]
- 38.Manns A, Hisada M, La Grenade L. Human T-lymphotropic virus type I infection. Lancet. 1999;353:1951–1958. doi: 10.1016/s0140-6736(98)09460-4. [DOI] [PubMed] [Google Scholar]
- 39.Oike Y, Takakura N, Hata A, Kaname T, Akizuki M, Yamaguchi Y, Yasue H, Araki K, Yamamura K, Suda T. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood. 1999;93:2771–2779. [PubMed] [Google Scholar]
- 40.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C, Masuno M, Tommerup N, van Ommen G J, Goodman R H, Peters D J, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 41.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator: coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 42.Rowley J D, Reshmi S, Sobulo O, Musvee T, Anastasi J, Raimondi S, Schneider N R, Barredo J C, Cantu E S, Schlegelberger B, Behm F, Doggett N A, Borrow J, Zeleznik-Le N. All patients with the T(11;16)(q23;p13.3) that involves MLL and CBP have treatment-related hematologic disorders. Blood. 1997;90:535–541. [PubMed] [Google Scholar]
- 43.Semmes O J, Jeang K T. Definition of a minimal activation domain in human T-cell leukemia virus type I Tax. J Virol. 1995;69:1827–1833. doi: 10.1128/jvi.69.3.1827-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Semmes O J, Jeang K T. Mutational analysis of human T-cell leukemia virus type I Tax: regions necessary for function determined with 47 mutant proteins. J Virol. 1992;66:7183–7192. doi: 10.1128/jvi.66.12.7183-7192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheppard H M, Harries J C, Hussain S, Bevan C, Heery D M. Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1. Mol Cell Biol. 2001;21:39–50. doi: 10.1128/MCB.21.1.39-50.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith M R, Greene W C. Identification of HTLV-I tax trans-activator mutants exhibiting novel transcriptional phenotypes. Genes Dev. 1990;4:1875–1885. doi: 10.1101/gad.4.11.1875. [DOI] [PubMed] [Google Scholar]
- 47.Sobulo O M, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett N A, Rowley J D, Zeleznik-Le N J. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3) Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sodroski J. The human T-cell leukemia virus (HTLV) transactivator (Tax) protein. Biochim Biophys Acta. 1992;1114:19–29. doi: 10.1016/0304-419x(92)90003-h. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki T, Uchida-Toita M, Yoshida M. Tax protein of HTLV-1 inhibits CBP/p300-mediated transcription by interfering with recruitment of CBP/p300 onto DNA element of E-box or p53 binding site. Oncogene. 1999;18:4137–4143. doi: 10.1038/sj.onc.1202766. [DOI] [PubMed] [Google Scholar]
- 50.Tie F, Adya N, Greene W C, Giam C Z. Interaction of the human T-lymphotropic virus type 1 Tax dimer with CREB and the viral 21-base-pair repeat. J Virol. 1996;70:8368–8374. doi: 10.1128/jvi.70.12.8368-8374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Orden K, Giebler H A, Lemasson I, Gonzales M, Nyborg J K. Binding of p53 to the KIX domain of CREB binding protein: a potential link to human T-cell leukemia virus, type I-associated leukemogenesis. J Biol Chem. 1999;274:26321–26328. doi: 10.1074/jbc.274.37.26321. [DOI] [PubMed] [Google Scholar]
- 52.Van Orden K, Nyborg J K. Insight into the tumor suppressor function of CBP through the viral oncoprotein Tax. Gene Expr. 2000;9:29–36. doi: 10.3727/000000001783992678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Orden K, Yan J P, Ulloa A, Nyborg J K. Binding of the human T-cell leukemia virus Tax protein to the coactivator CBP interferes with CBP-mediated transcriptional control. Oncogene. 1999;18:3766–3772. doi: 10.1038/sj.onc.1202703. [DOI] [PubMed] [Google Scholar]
- 54.Watanabe T. HTLV-1-associated diseases. Int J Hematol. 1997;66:257–278. doi: 10.1016/s0925-5710(97)00077-7. [DOI] [PubMed] [Google Scholar]
- 55.Yan J P, Garrus J E, Giebler H A, Stargell L A, Nyborg J K. Molecular interactions between the coactivator CBP and the human T-cell leukemia virus Tax protein. J Mol Biol. 1998;281:395–400. doi: 10.1006/jmbi.1998.1951. [DOI] [PubMed] [Google Scholar]
- 56.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 57.Yao J, Wigdahl B. Human T cell lymphotropic virus type I genomic expression and impact on intracellular signaling pathways during neurodegenerative disease and leukemia. Front Biosci. 2000;5:D138–D168. doi: 10.2741/yao. [DOI] [PubMed] [Google Scholar]
- 58.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 60.Zaninovic V. On the etiology of tropical spastic paraparesis and human T-cell lymphotropic virus-I-associated myelopathy. Int J Infect Dis. 1999;3:168–176. doi: 10.1016/s1201-9712(99)90041-3. [DOI] [PubMed] [Google Scholar]
- 61.Zhao L J, Giam C Z. Human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator, Tax, enhances CREB binding to HTLV-I 21-base-pair repeats by protein-protein interaction. Proc Natl Acad Sci USA. 1992;89:7070–7074. doi: 10.1073/pnas.89.15.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao L J, Giam C Z. Interaction of the human T-cell lymphotrophic virus type I (HTLV-I) transcriptional activator Tax with cellular factors that bind specifically to the 21-base-pair repeats in the HTLV-I enhancer. Proc Natl Acad Sci USA. 1991;88:11445–11449. doi: 10.1073/pnas.88.24.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]