Purpose of review
Liquid biopsies have emerged as a noninvasive alternative to tissue biopsy with potential applications during all stages of pediatric oncology care. The purpose of this review is to provide a survey of pediatric cell-free DNA (cfDNA) studies, illustrate their potential applications in pediatric oncology, and to discuss technological challenges and approaches to overcome these hurdles.
Recent findings
Recent literature has demonstrated liquid biopsies’ ability to inform treatment selection at diagnosis, monitor clonal evolution during treatment, sensitively detect minimum residual disease following local control, and provide sensitive posttherapy surveillance. Advantages include reduced procedural anesthesia, molecular profiling unbiased by tissue heterogeneity, and ability to track clonal evolution. Challenges to wider implementation in pediatric oncology, however, include blood volume restrictions and relatively low mutational burden in childhood cancers. Multiomic approaches address challenges presented by low-mutational burden, and novel bioinformatic analyses allow a single assay to yield increasing amounts of information, reducing blood volume requirements.
Summary
Liquid biopsies hold tremendous promise in pediatric oncology, enabling noninvasive serial surveillance with adaptive care. Already integrated into adult care, recent advances in technologies and bioinformatics have improved applicability to the pediatric cancer landscape.
Keywords: biomarkers, cell-free DNA, circulating DNA, liquid biopsy
INTRODUCTION
First identified in 1948, cell-free DNA (cfDNA) has become a promising circulating biomarker in oncology [1,2]. Developments in liquid biopsy, the capture of cfDNA, circulating proteins, and circulating tumor cells (CTCs), have led to adoption of technologies across stages of patient management including screening and diagnosis, molecular prognostication, tracking treatment response, detection of minimal residual disease (MRD), monitoring clonal evolution, and posttherapy surveillance [3–7]. The US Food and Drug Administration approved the first digital drop PCR (ddPCR) liquid biopsy tests in 2016, and two next generation sequencing (NGS) liquid biopsy panels in 2020 as companion diagnostics for associated targeted therapies [8–10]. As of the time of publication, no liquid biopsy tests have been Food and Drug Administration (FDA)-approved in pediatrics.
Box 1.
no caption available
Liquid biopsies are of particular interest for pediatric oncology as they are noninvasive, avoiding procedural sedation, and enabling serial sampling. Furthermore, liquid biopsies mitigate diagnostic challenges of tumor heterogeneity and accessibility by capturing genetic material shed throughout the body. This promise, however, comes with unique challenges that have limited wider implementation in pediatrics. Pediatric and adult cancers have differing genomic properties; pediatric cancers have low mutational burden [11–14] with few recurrent hotspots [15▪▪,16–20]. Adult cancers feature more point mutations [single nucleotide variants (SNV)] and insertion/deletion errors (indels), whereas pediatric cancers are characterized by chromosomal structural variations including copy number alterations (CNA), translocations, and fusion genes [12,14,15▪▪,21–23]. This review highlights studies that illustrate the potential applications of cfDNA in pediatric cancers. We discuss technological challenges and emerging approaches to overcome these hurdles.
CELL-FREE DNA OVERVIEW
cfDNA are ∼120–220 bp long fragments [24] of double-stranded DNA found in plasma, cerebral spinal fluid (CSF), saliva, pleural fluid, ascites, stool, aqueous humor, and urine [25–28]. cfDNA molecules are released from healthy and malignant cells through apoptosis, necrosis, and secretion, then cleared from circulation with a half-life of several minutes to 2.5 h [29,30]. cfDNA's rapid clearance and dynamic changes make it an ideal biomarker for ‘real-time’ analyses compared with classic biomarkers like alfa-fetoprotein [31].
cfDNA originating from tumors (circulating tumor DNA; ctDNA) are shorter than normal cfDNA (∼90 to 150 bp) [32,33,34▪]. The portion of overall cfDNA constituted of ctDNA varies with cancer type, tumor location, tumor burden, and metastases. In low-burden and early disease, ctDNA fraction is minute [35,36]. The detection of ctDNA, therefore, requires ultrasensitive methods that detect somatic variations. Thus, a broad understanding of the available technologies and evaluation of these considerations is necessary when selecting an approach for analyzing cfDNA.
TECHNOLOGIES
PCR
PCR specifically amplify targeted cfDNA templates. To improve sensitivity and amplify a minute quantity of ctDNA, ddPCR technology subdivides PCR reactions into numerous nano-liter droplets, and can detect variant allele fractions (VAF) as low as 0.001% [7,37]. Although ddPCR attains high sensitivities, it has limitations. First, it requires a priori knowledge of disease-specific or patient-specific mutations [37], thereby missing any de novo mutations. Similarly, genome-wide surveys of translocations, indels, and CNA are limited, given the focused nature of the assay.
Next generation sequencing
NGS methods do not require prior knowledge of mutations but have worse limits of detection than ddPCR and increased cost. The primary parameters affecting cost are breadth and depth of sequencing. Breadth is the proportion of the genome that is sequenced. NGS breadth varies from a panel of genes of interest (e.g. CAPPseq [38]), to regions of the genome [e.g. whole exome sequencing (WES)], to the entire genome [whole genome sequencing (WGS)]. This gives NGS the capacity to detect both recurrent hotspot mutations and previously unknown or uncommon variants.
Depth refers to the average number of times a base pair is sequenced. Shallow WGS [e.g. 0.1–2x ultra-low pass WGS (ULP-WGS)] accurately detects CNAs [39] at low cost but has poor sensitivity for specific somatic variants. As depth increases, SNVs, indels, and translocations may be identified. However, as assay cost increases, deep-sequencing panels typically restrict breadth, focusing on smaller genomic regions and missing abnormalities outside of those regions.
Methylation, fragmentomics, and transcriptomics
Although previous methods identify genetic variations to characterize ctDNA, recent techniques leverage other ctDNA signatures. Pediatric cancer's low mutational burden may be better suited to alternative markers or combinatorial approaches. Methylation, for example, is a promising pediatric marker as epigenetic dysregulation is a recurrent characteristic of childhood cancers [40]. The circulating methylome can be measured in targeted cfMeDIP-seq [41] or ddMethyLight assays [42] as well as genome-wide approaches [43]. Methylation fingerprints may also define cfDNA tissues of origin [44,45]. Emerging bioinformatic techniques, such as fragmentomics and transcriptomics, leverage nonrandom differences in cfDNA fragments and sequence coverage for in silico enrichment of ctDNA [33,34▪,46], and infer transcriptome profiles based on chromatin availability and nucleosome footprints [47–50,51▪▪].
CLINICAL APPLICATIONS OF LIQUID BIOPSIES
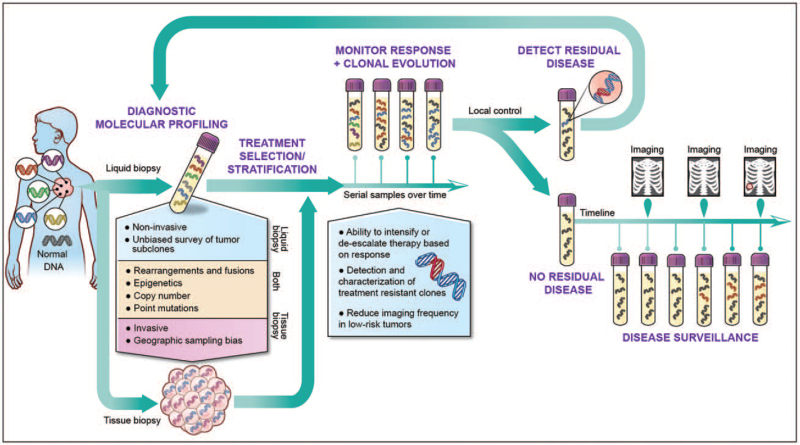
The diversity of assays available to profile cfDNA is matched by the range of potential applications for these tools in the clinic (Fig. 1).
FIGURE 1.
Clinical applications of liquid biopsy in pediatric oncology. Liquid biopsies capture genetic material shed throughout the body, enabling noninvasive molecular profiling without the confounding variable of tumor heterogeneity. Liquid biopsies can, therefore, inform care and treatment selection at diagnosis, monitor clonal evolution during treatment, sensitively detect minimum residual disease following local control and provide sensitive posttherapy surveillance. Multiple studies have demonstrated that liquid biopsies have the potential to detect relapsed disease before radiographically evident.
Molecular profiling and treatment selection
Sequencing from multiple regions of the same tumor reveals significant intratumor genetic heterogeneity [52,53]. Up to 69% of detected somatic mutations are absent from other regions of the same tumor [52], leading to potential sampling biases during biopsy. Liquid biopsy of cfDNA captures genetic materials released from multiple tissue regions and separate disease foci, mitigating sampling biases. Current adult clinical trials employ cfDNA for risk stratification and treatment selection [7,54].
Liquid biopsy could similarly be used to inform management of pediatric cancers, guiding treatment intensification and de-escalation. Intralesional and interlesional heterogeneity of MYCN and ALK, for example, is well described in neuroblastoma [55,56]. Accurate assessment of MYCN, ALK, and segmental chromosome alterations (SCA), has prognostic implications [57–59] and is incorporated into treatment selection. Intermediate-risk neuroblastoma patients without MYCN amplification or specific SCA maintain excellent outcomes with treatment reduction [60]; therefore, genomic misclassification would lead to inappropriate de-escalation of treatment. Multiple studies demonstrate sensitive and tissue concordant molecular characterization of these genes and SCA in neuroblastoma using liquid biopsies [61–65]. The promise of liquid biopsy for risk stratification is further illustrated in pediatric sarcomas. In Ewing sarcoma (EWS) and osteosarcoma, the presence of detectable ctDNA alone predicts inferior outcomes. Furthermore, 8q gain in osteosarcoma cfDNA portended poorer 3-year event-free survival (EFS) (60 vs. 80.9%) [66], and EWS high-risk co-mutations of STAG and TP53[67] are detectable in cfDNA [68].
Implementation of liquid biopsy for genomic characterization of Wilms tumor could reconcile international treatment paradigms. European consortium paradigms recommend neoadjuvant chemotherapy without tissue diagnosis because of the prevalence of favorable disease and risk of metastatic seeding with biopsy or surgery. In contrast, North American algorithms recommend upfront nephrectomy for biology-guided therapy. In small studies, TP53 mutations were detectable in 100% of prenephrectomy urine cfDNA [69] and tumor-confirmed mutations were identified in plasma [70]. cfDNA profiles of poor prognosis TP53 mutations [71,72] and 1p/16q alterations [70], therefore, could guide therapy without risking seeding tumor.
Treatment response and clonal evolution
Although mortality from primary pediatric cancers has decreased, intensification of treatments has led to increased treatment-related morbidity [73]. Clinical trials, therefore, have examined response-adapted protocols that reduce long-term morbidities in hematologic malignancies [74–76]. In pediatric solid tumors, response-adapted therapy is hindered by lack of serial biopsies; response markers are limited to imaging [77], and necrosis estimates on postneoadjuvant resections [78]. Liquid biopsies offer noninvasive, serial monitoring to assess tumor burden, clonal evolution, and epigenetic changes.
In a small osteosarcoma cohort, patients with more than 80% necrosis in resections had undetectable ctDNA following initiation of neoadjuvant therapy. Patients with less than 70% necrosis; however, had persistence of ctDNA throughout therapy [64]. Although larger validation studies are needed, these data suggest that ctDNA may predict prognostic postneoadjuvent percentage necrosis in osteosarcoma [78]. Similarly, dynamic changes in ctDNA have been shown to correlate with tumor burden in Wilms tumor [79], hepatoblastoma [80], retinoblastoma [81–83], EWS [51▪▪,77,84], and neuroblastoma [61,62]. Additionally, circulating epigenetic signatures approximate tumor burden and response. Data suggest that regional differences in coverage over DNase I hypersensitive sites (DHSs), a surrogate for chromatin status and epigenetic signatures [49,51▪▪], infer tissue type and can estimate tumor burden [51▪▪]. Applebaum et al. measured epigenetic signatures using 5-hydroxymethylcytosine (5-hmC) to predict disease burden and response in metastatic neuroblastoma. 5hmC deposition on MYCN predicted relapse and end of induction total 5-hmC levels are prognostic. In two patients with no clinical evidence of disease, 5hmC profiling predicted subsequent relapse [85▪].
Finally, serial cfDNA can monitor clonal evolution and identify therapy-resistant subclones. A landmark study in lung cancer reported that cfDNA is more sensitive than tissue biopsy for detection of acquired resistance to erlotinib through EGFRT790M mutations [86]. In pediatrics, cfDNA temporally resolved heterogeneity in progressive neuroblastoma with a mean of 22 new SNV between diagnostic and subsequent samples, including 17 commonly acquired relapse-specific mutations [61]. Similarly, Barris et al.[87] identified TP53 mutations in relapsed osteosarcoma plasma not identified in initial tumor or germline. In medulloblastoma, dynamic changes in CSF cfDNA methylation mirror tissue changes during treatment and progression [43].
Detection of minimum residual disease and posttherapy surveillance
cfDNA's short half-life [29,30] and representation of spatial heterogeneity [88] make it an ideal biomarker to detect postoperative minimum residual disease (MRD) and early recurrence. In adults, postoperative persistence of ctDNA portends a more than 80% risk of relapse in colorectal cancers [89] and urothelial carcinomas [90]. These findings suggest that ctDNA positivity postneoadjuvent therapy or surgery could inform clinical decisions regarding follow-up frequency and need for adjuvant or radiation therapy. Indeed, the addition of immunotherapy to urothelial carcinoma patients with postoperative ctDNA improved disease-free survival and overall survival [90]. Furthermore, application of cfDNA for MRD may enable earlier relapse detection. In lung cancer, recurrence of detectable ctDNA identified relapse a median of 5.2 months earlier than imaging [91]; in colon and breast cancer, cfDNA outperforms biochemical biomarkers CEA and CA 15-3 for early, sensitive detection of recurrence [92,93].
To date, there have been no large MRD pediatric studies using cfDNA; however, case series demonstrate feasibility. Hayashi et al. used tumor-informed EWS-ETS ddPCR to detect fusion genes in plasma from three EWS patients. Two patients had persistent postoperative ctDNA and clinically relapsed. The only ctDNA-negative patient remained in remission [84]. In osteosarcoma, three of seven patients followed with targeted NGS liquid biopsy relapsed, all of whom had recurrence of detectable ctDNA prior to radiographic relapse [87]. Circulating mutant Rb1 becomes undetectable following enucleation of intraocular retinoblastoma [94,95] but was again detectable in regionally recurrent or metastatic relapsed disease [83]. Finally, previously detectable circulating CTNNB1 in hepatoblastoma becomes undetectable following total resection with no histological or radiographic evidence of residual disease [80].
CHALLENGES AND FUTURE DIRECTIONS
Rapid technological advances coupled with exciting preliminary studies in pediatric histologies portend an important future for cfDNA in pediatric oncology; however, several important considerations must be considered in this population.
Blood draw limitations
An assay's limit of detection is constrained by the number of unique sequencing reads generated. Each milliliter of blood has approximately 1000 genome equivalents [96] and sequencing to a depth greater than unique genome equivalents results in duplicated reads with no improvement in limits of detection [97▪▪]. Theoretically, this presents a barrier in pediatrics because of weight-based limits in blood draw volume. Kahana-Edwin et al., however, highlight that the percentage of total blood volume collected during weight-based draws in pediatrics is consistent with adults. Furthermore, even early-stage pediatric cancers often represent a larger tumor burden relative to patient size than adult counterparts. Taken together, pediatric assays should capture a proportional or higher ratio of ctDNA:cfDNA because of relative tumor burden in similarly staged diseases [98]. Indeed, pediatric studies in neuroblastoma [65,99], EWS [66], and hepatoblastoma [80] have demonstrated the feasibility of sensitive detection from less than 1 ml of plasma, and studies of unilateral intraocular retinoblastoma, characterized by exceptionally small tumor volumes, detect ctDNA in plasma [83,100].
Clonal hematopoiesis of indeterminate potential
An emerging challenge in liquid biopsies is false positives because of somatic mutations present in peripheral blood but not in tumor, termed clonal hematopoiesis of indeterminate potential (CHIP) [101]. A common phenomenon associated with aging, CHIP is the presence of a mutant clone in the blood without evidence of dysplasia, neoplasm, or cytopenia [102]. CHIP are attributed to survival advantages in certain mutations acquired during normal hematopoietic stem cell (HSC) divisions [103]. Studies of serial samples banked 10 years apart suggest that CHIP rarely undergo continued expansion [104] and have minimal risk of transforming into hematologic malignancy [105,106].
CHIP are classically defined as having a VAF of at least 2% [102–105,107]; however, this threshold is based on previous limits of detection using WES [105]. Recent studies using error suppression algorithms suggest that CHIP accumulation likely begins in fetal development. 18.2% of sequenced cord blood samples harbor low frequency (VAF 0.002–0.006) somatic mutations [108] and, during normal hematopoiesis, individuals gain one mutation in HSCs per decade [103,109]. It is hypothesized however, that these mutations do not confer survival advantages in youth [110] and, consequently, VAF of CHIP typically remains less than 0.5% until age 50 years [107,111]. The VAF of CHIP in pediatrics, therefore, remains undetectable by many liquid biopsy assays [7,39]. As technologies continue to push limits of detection in an effort to identify early-stage cancer, low-VAF CHIP may become a relevant source of false-positives in pediatric oncology. Multiomic or combinatorial imaging-cfDNA assays may mitigate this.
CHIP frequency increases in adults who received chemotherapy [105,112,113] and radiation therapy [105,113]. These CHIP are enriched for TP53 mutations [113], the most common somatically mutated gene in pediatric cancer [13], presenting a potential source of false positives in posttherapy surveillance and MRD detection. The only study examining CHIP in pediatric cancer survivors, however, showed no increases using a 14-gene NGS panel [110]. Larger, longitudinal studies including broader panels or WGS with error suppression are necessary for validation.
Low mutational burden
With early-stage disease, increasing the number of targeted mutations improves probability of detection [5]. Pediatric cancers, however, have approximately one-fourteenth of the mutation burden of adult cancers [13], with as few as one SNV per exome [12], limiting targeted panels’ utility in early-stage detection.
One strategy to improve probability of detection is to expand assayed targets by integrating ‘multiomic’ features including CNA [32,34▪,51▪▪], fragmentation profiles [32–34▪,46,51▪▪], protein [114–116], and epigenetics [51▪▪]. CNA is the most common alteration in pediatric solid tumors [13,15▪▪,22] and may be inferred from off-target reads of targeted panels [117] or detected using ULP-WGS [34▪,39,81]. The utility of cfDNA CNA as a biomarker has been shown in neuroblastoma [61,62,65], retinoblastoma [81,82], and EWS [64], and the sensitivity of these assays can be boosted with fragmentomics [32,33,34▪,46]. Our group reported that CNA combined with fragment size analysis not only detects malignancy but can accurately distinguish benign from malignant tumors in neurofibromatosis type 1 (NF1) [34▪]. For small tumors, combinatorial assays with cfDNA and protein have doubled sensitivity in stage I/II pancreatic cancers and lesions measuring less than 1.5 cm [115]. Furthermore, pediatric cancers have a high incidence of mutations in epigenetic regulators [13,15▪▪,118]. Integrating epigenetic and genetic signatures enabled accurate classification of sarcoma types [51▪▪]. Finally, combinatorial approaches may not be multiomic but multimodality. Using a pan-cancer liquid biopsy for surveillance, Lennon et al.[119] increased their positive-predictive value by 45% after integrating imaging features.
CONCLUSION
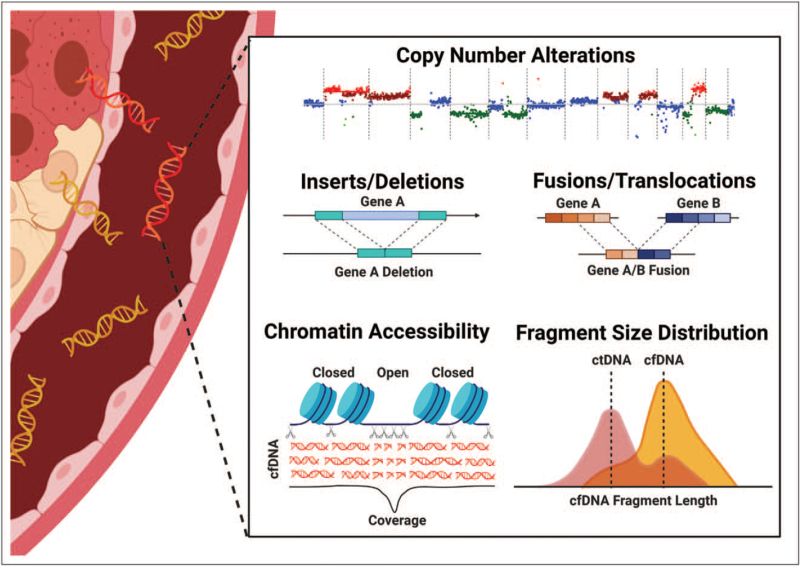
Liquid biopsies have emerged as a noninvasive alternative to tissue biopsy. Despite the potential to allow serial molecular profiling while reducing invasive procedures and anesthesia events, implementation in pediatrics has been limited and liquid biopsy remains only a research tool. This is partly because of challenges presented by relatively low mutational burden. Multiomic strategies show promise for sensitive detection of low mutational burden disease but are expensive and require more blood for parallel assays. Exciting recent studies demonstrate that bioinformatic processing of 12x–35x WGS enables characterization of copy number analysis, small indels, fusions, chromatin accessibility, and detailed fragmentomics from a single assay (Fig. 2) [51▪▪,97▪▪]. These novel approaches offer tremendous promise for childhood cancer by detecting the most common pediatric alterations in childhood cancers, incorporating multiomic input, and reducing required blood and cost.
FIGURE 2.
Moderate depth whole genome sequencing improve detection by enabling multiomic integration from a single assay. Recent studies have demonstrated that, with advanced bioinformatic analysis, WGS to depths of 12x–35x is sufficient for detection of copy number alterations, indels, translocations and fusions, assessment of chromatin accessibility and fragmentomics [51▪▪,97▪▪]. In addition to detecting the most common genomic alterations in childhood cancer, therefore, integration of these output also infers epigenetic and transcriptomic signatures. Combinatorial approaches, previously requiring multiple assays, enhance detection of early stage cancers.
Acknowledgements
NIH Medical Arts created Fig. 1. Images from BioRender were used to create Fig. 2.
Financial support and sponsorship
None.
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1.Mandel P, Metais P. Nuclear acids in human blood plasma. C R Seances Soc Biol Fil 1948; 142:241–243. [PubMed] [Google Scholar]
- 2.Stroun M, Anker P, Lyautey J, et al. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol 1987; 23:707–712. [DOI] [PubMed] [Google Scholar]
- 3.Abbou SD, Shulman DS, DuBois SG, Crompton BD. Assessment of circulating tumor DNA in pediatric solid tumors: the promise of liquid biopsies. Pediatr Blood Cancer 2019; 66:e27595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson D, Fagman H, Dalin MG, Ståhlberg A. Circulating cell-free tumor DNA analysis in pediatric cancers. Mol Aspects Med 2020; 72:100819. [DOI] [PubMed] [Google Scholar]
- 5.Wan J. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017; 17:223–238. [DOI] [PubMed] [Google Scholar]
- 6.Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018; 36:1631–1641. [DOI] [PubMed] [Google Scholar]
- 7.Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med 2018; 379:1754–1765. [DOI] [PubMed] [Google Scholar]
- 8. U.S. Food and Drug Administration. cobas EGFR Mutation Test v2 [Internet]. fda.gov. 2016. Available at: https://www.fda.gov/drugs/resources-information-approved-drugs/cobas-egfr-mutation-test-v2. [Accessed 8 August 2021]. [Google Scholar]
- 9. U.S. Food and Drug Administration. FDA approves first liquid biopsy next-generation sequencing companion diagnostic test [Internet]. fda.gov. 2020 [cited 28 August 2021]. Available at: https://www.fda.gov/news-events/press-announcements/fda-approves-first-liquid-biopsy-next-generation-sequencing-companion-diagnostic-test. [Accessed 8 August 2021]. [Google Scholar]
- 10. U.S. Food and Drug Administration. FDA approves liquid biopsy NGS companion diagnostic test for multiple cancers and biomarkers [Internet]. fda.gov. 2020 [cited 2021 Aug 28]. Available at: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-liquid-biopsy-ngs-companion-diagnostic-test-multiple-cancers-and-biomarkers. [Accessed 8 August 2021]. [Google Scholar]
- 11.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013; 339:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gröbner SN, Worst BC, Weischenfeldt J, et al. The landscape of genomic alterations across childhood cancers. Nature 2018; 555:321–327. [DOI] [PubMed] [Google Scholar]
- 14.Campbell BB, Light N, Fabrizio D, et al. Comprehensive analysis of hypermutation in human cancer. Cell 2017; 171:1042.e10–1056.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15▪▪.Newman S, Nakitandwe J, Kesserwan CA, et al. Genomes for kids: the scope of pathogenic mutations in pediatric cancer revealed by comprehensive DNA and RNA sequencing. Cancer Discov 2021; [Online ahead of print]. [Accessed 1 September 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses a multiomic approach (WGS, WES, RNAseq) to examine tumor and germline from pediatric cancers. The authors demonstrated that, in tissue, combinatorial approaches are necessary to identify the full range of genomic variants in childhood cancer.
- 16.Kaseb AO, Sánchez NS, Sen S, et al. Molecular profiling of hepatocellular carcinoma using circulating cell-free DNA. Clin Cancer Res 2019; 25:6107L–6118L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol 2016; 2:1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins S, Yang JC-H, Ramalingam SS, et al. Plasma ctDNA analysis for detection of the EGFR T790M mutation in patients with advanced non-small cell lung cancer. J Thorac Oncol 2017; 12:1061–1070. [DOI] [PubMed] [Google Scholar]
- 19.Sanmamed MF, Fernández-Landázuri S, Rodríguez C, et al. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin Chem 2015; 61:297–304. [DOI] [PubMed] [Google Scholar]
- 20.Schmiegel W, Scott RJ, Dooley S, et al. Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol 2017; 11:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X, Liu Y, Liu Y, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018; 555:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sweet-Cordero EA, Biegel JA. The genomic landscape of pediatric cancers: Implications for diagnosis and treatment. Science 2019; 363:1170–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alcaide M, Cheung M, Hillman J, et al. Evaluating the quantity, quality and size distribution of cell-free DNA by multiplex droplet digital PCR. Sci Rep 2020; 10:12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017; 14:531–548. [DOI] [PubMed] [Google Scholar]
- 26.Husain H, Melnikova VO, Kosco K, et al. Monitoring daily dynamics of early tumor response to targeted therapy by detecting circulating tumor DNA in urine. Clin Cancer Res 2017; 23:4716–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Springer S, Mulvey CL, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med 2015; 7:293ra104-293ra104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Mattos-Arruda L, Mayor R, Ng CKY, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 2015; 6:8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao W, Mei C, Nan X, Hui L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: a qualitative study. Gene 2016; 590:142–148. [DOI] [PubMed] [Google Scholar]
- 30.Lo YMD, Zhang J, Leung TN, et al. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet 1999; 64:218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shim JH, Han S, Lee YJ, et al. Half-life of serum alpha-fetoprotein: an early prognostic index of recurrence and survival after hepatic resection for hepatocellular carcinoma. Ann Surg 2013; 257:708–717. [DOI] [PubMed] [Google Scholar]
- 32.Mouliere F, Mair R, Chandrananda D, et al. Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol Med 2018; 10:e9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouliere F, Chandrananda D, Piskorz AMA, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 2018; 10:eaat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34▪.Szymanski JJ, Sundby RT, Jones PA, et al. Cell-free DNA ultra-low-pass whole genome sequencing to distinguish malignant peripheral nerve sheath tumor (MPNST) from its benign precursor lesion: a cross-sectional study. PLoS Med 2021; 18:e1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first study use cfDNA to identify malignancy in a cancer predisposition syndrome. Copy number analysis with fragmentomics distinguished benign from malignant tumors and cfDNA profiles recapitulated characteristic tissue genomic alterations.
- 35.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14:985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kockerols CCB, Valk PJM, Levin M-D, et al. Digital PCR for BCR-ABL1 quantification in CML: current applications in clinical practice. HemaSphere 2020; 4:e496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014; 20:548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adalsteinsson VAV, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017; 8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filbin M, Monje M. Developmental origins and emerging therapeutic opportunities for childhood cancer. Nat Med 2019; 25:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018; 563:579–583. [DOI] [PubMed] [Google Scholar]
- 42.Cho N-Y, Park J-W, Wen X, et al. Blood-based detection of colorectal cancer using cancer-specific DNA methylation markers. Diagnostics 2020; 11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Zhao S, Lee M, et al. Reliable tumor detection by whole-genome methylation sequencing of cell-free DNA in cerebrospinal fluid of pediatric medulloblastoma. Sci Adv 2020; 6:5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun K, Jiang P, Chan KCA, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci 2015; 112:E5503–E5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gai W, Zhou Z, Agbor-Enoh S, et al. Applications of genetic-epigenetic tissue mapping for plasma dna in prenatal testing, transplantation and oncology. Elife 2021; 10:e64356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cristiano S, Leal A, Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019; 570:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lo YMD, Han DSC, Jiang P, Chiu RWK. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 2021; 372:eaaw3616. [DOI] [PubMed] [Google Scholar]
- 48.Jiang P, Sun K, Tong Y, et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc Natl Acad Sci U S A 2018; 115:E10925–E10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun K, Jiang P, Cheng S, et al. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res 2019; 29:418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivanov M, Baranova A, Butler T, et al. Nonrandom fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics 2015; 16:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51▪▪.Peneder P, Stütz AM, Surdez D, et al. Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden. Nat Commun 2021; 12:3230. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors demonstrate that modest-depth WGS is sufficient to describe cancer-specific chromatin signatures using the novel LIQUORICE alogirthm. Advanced fragmentomic analyses are introduced for classification of low mutation burden cancers. Finally, the authors use integrated genetic/epigenetic analysis to sensitively detect and distinguish between various pediatric sarcomas.
- 52.Gerlinger M, Rowan A, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res 2012; 72:4875–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tie J, Chapman M. Circulating tumour DNA (ctDNA) analysis informing adjuvant chemotherapy in Stage II Colon Cancer [Internet]. Australian New Zealand Clinical Trials Registry [cited 8 September 2021]. Available at: https://www.anzctr.org.au/Trial/Registration/TrialReview.aspx?ACTRN=12615000381583. [Google Scholar]
- 55.Bellini A, Bernard V, Leroy Q, et al. Deep sequencing reveals occurrence of subclonal alk mutations in neuroblastoma at diagnosis. Clin Cancer Res 2015; 21:4913–4921. [DOI] [PubMed] [Google Scholar]
- 56.Theissen J, Boensch M, Spitz R, et al. Heterogeneity of the MYCN oncogene in neuroblastoma. Clin Cancer Res 2009; 15:2085–2090. [DOI] [PubMed] [Google Scholar]
- 57.Janoueix-Lerosey I, Schleiermacher G, Michels E, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol 2009; 27:1026–1033. [DOI] [PubMed] [Google Scholar]
- 58.Schleiermacher G, Mosseri V, London WB, et al. Segmental chromosomal alterations have prognostic impact in neuroblastoma: a report from the INRG project. Br J Cancer 2012; 107:1418–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ambros PF, Ambros IM, Brodeur GM, et al. International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br J Cancer 2009; 100:1471–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Twist C, London WB, Naranjo A, et al. Maintaining outstanding outcomes using response- and biology-based therapy for intermediate-risk neuroblastoma: a report from the Children's Oncology Group study ANBL0531. J Clin Oncol 2014; 32: (15 Suppl): 10006–110006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chicard M, Colmet-Daage L, Clement N, et al. Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin Cancer Res 2018; 24:939–949. [DOI] [PubMed] [Google Scholar]
- 62.Chicard M, Boyault S, Colmet Daage L, et al. Genomic copy number profiling using circulating free tumor DNA highlights heterogeneity in neuroblastoma. Clin Cancer Res 2016; 22:5564–5573. [DOI] [PubMed] [Google Scholar]
- 63.Lodrini M, Sprüssel A, Astrahantseff K, et al. Using droplet digital PCR to analyze MYCN and ALK copy number in plasma from patients with neuroblastoma. Oncotarget 2017; 8:85234–85251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klega K, Imamovic-Tuco A, Ha G, et al. Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children with solid tumors. JCO Precis Oncol 2018; 2018:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kahana-Edwin S, Cain LE, McCowage G, et al. Neuroblastoma molecular risk-stratification of DNA copy number and ALK genotyping via cell-free circulating tumor DNA profiling. Cancers (Basel) 2021; 13:3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shulman DS, Klega K, Imamovic-Tuco A, et al. Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: a report from the Children's Oncology Group. Br J Cancer 2018; 119:615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tirode F, Surdez D, Ma X, et al. St. Jude Children's Research Hospital–Washington University Pediatric Cancer Genome Project and the International Cancer Genome Consortium. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 2014; 4:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shukla NN, Patel JA, Magnan H, et al. Plasma DNA-based molecular diagnosis, prognostication, and monitoring of patients with EWSR1 fusion-positive sarcomas. JCO Precis Oncol 2017; 1:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Treger TD, Chagtai T, Butcher R, et al. Somatic TP53 mutations are detectable in circulating tumor DNA from children with anaplastic Wilms tumors. Transl Oncol 2018; 11:1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dix DB, Seibel NL, Chi Y-Y, et al. Treatment of stage IV favorable histology Wilms tumor with lung metastases: a report from the Children's Oncology Group AREN0533 Study. J Clin Oncol 2018; 36:1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ooms AHAG, Gadd S, Gerhard DS, et al. Significance of TP53 mutation in Wilms tumors with diffuse anaplasia: a report from the Children's Oncology Group. Clin Cancer Res 2016; 22:5582–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maschietto M, Williams RD, Chagtai T, et al. TP53 mutational status is a potential marker for risk stratification in Wilms tumour with diffuse anaplasia. PLoS One 2014; 9:e109924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Armstrong GT, Liu Q, Yasui Y, et al. Late mortality among 5-year survivors of childhood cancer: a summary from the Childhood Cancer Survivor Study. J Clin Oncol 2009; 27:2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Dongen JJ, Seriu T, Panzer-Grümayer ER, et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet (London, England) 1998; 352:1731–1738. [DOI] [PubMed] [Google Scholar]
- 75.Kelly KM, Cole PD, Pei Q, et al. Response-adapted therapy for the treatment of children with newly diagnosed high risk Hodgkin lymphoma (AHOD0831): a report from the Children's Oncology Group. Br J Haematol 2019; 187:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oeffinger KC, Stratton KL, Hudson MM, et al. Impact of risk-adapted therapy for pediatric Hodgkin lymphoma on risk of long-term morbidity: a report from the childhood cancer survivor study. J Clin Oncol 2021; 39:2266–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krumbholz M, Hellberg J, Steif B, et al. Genomic EWSR1 fusion sequence as highly sensitive and dynamic plasma tumor marker in Ewing sarcoma. Clin Cancer Res 2016; 22:4356–4365. [DOI] [PubMed] [Google Scholar]
- 78.Meyers PA, Schwartz CL, Krailo M, et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J Clin Oncol 2005; 23:2004–2011. [DOI] [PubMed] [Google Scholar]
- 79.Miguez ACK, Barros BDF, de Souza JES, et al. Assessment of somatic mutations in urine and plasma of Wilms tumor patients. Cancer Med 2020; 9:5948–5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kahana-Edwin S, McCowage G, Cain L, et al. Exploration of CTNNB1 ctDNA as a putative biomarker for hepatoblastoma. Pediatr Blood Cancer 2020; 67:e28594. [DOI] [PubMed] [Google Scholar]
- 81.Polski A, Xu L, Prabakar RK, et al. Cell-free DNA tumor fraction in the aqueous humor is associated with therapeutic response in retinoblastoma patients. Transl Vis Sci Technol 2020; 9:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berry JL, Xu L, Murphree AL, et al. Potential of aqueous humor as a surrogate tumor biopsy for retinoblastoma. JAMA Ophthalmol 2017; 135:1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abramson DH, Mandelker D, Francis JH, et al. Retrospective evaluation of somatic alterations in cell-free DNA from blood in retinoblastoma. Ophthalmol Sci 2021; 1:100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hayashi M, Chu D, Meyer CF, et al. Highly personalized detection of minimal ewing sarcoma disease burden from plasma tumor DNA. Cancer 2016; 122:3015–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85▪.Applebaum MA, Barr EK, Karpus J, et al. 5-Hydroxymethylcytosine profiles in circulating cell-free DNA associate with disease burden in children with neuroblastoma. Clin Cancer Res 2020; 26:1309–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that epigenetic signatures in cfDNA are a viable marker for risk stratification and prognostication in pediatric cancers. The authors approximate tumor burden and predict risk of relapse in neuroblastoma.
- 86.Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol 2017; 28:784–790. [DOI] [PubMed] [Google Scholar]
- 87.Barris DM, Weiner SB, Dubin RA, et al. Detection of circulating tumor DNA in patients with osteosarcoma. Oncotarget 2018; 9:12695–12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pereira B, Chen CT, Goyal L, et al. Cell-free DNA captures tumor heterogeneity and driver alterations in rapid autopsies with pretreated metastatic cancer. Nat Commun 2021; 12:3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Henriksen TVV, Tarazona N, Frydendahl A, et al. Serial circulating tumor DNA analysis to assess recurrence risk, benefit of adjuvant therapy, growth rate and early relapse detection in stage III colorectal cancer patients. J Clin Oncol 2021; 39: (15 Suppl): 3540-3540. [Google Scholar]
- 90.Powles T, Assaf ZJ, Davarpanah N, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature 2021; 595:432–437. [DOI] [PubMed] [Google Scholar]
- 91.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017; 7:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tie J, Cohen JD, Wang Y, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut 2019; 68:663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dawson S-J, Tsui DWY, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013; 368:1199–1209. [DOI] [PubMed] [Google Scholar]
- 94.Abramson DH. Retinoblastoma: saving life with vision. Annu Rev Med 2014; 65:171–184. [DOI] [PubMed] [Google Scholar]
- 95.Abramson DH, Fabius AWM, Issa R, et al. Advanced unilateral retinoblastoma: the impact of ophthalmic artery chemosurgery on enucleation rate and patient survival at MSKCC. PLoS One 2015; 10:e0145436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Devonshire AS, Whale AS, Gutteridge A, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem 2014; 406:6499–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97▪▪.Zviran A, Schulman RC, Shah M, et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat Med 2020; 26:1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes how the current paradigm of deep sequencing of cfDNA with targeted panels is limited in low-disease burden settings by low tumor fraction with few unqiue genomic equivlanets. They demonstrate that modest depth (35x) WGS improves sensitivity by increasting breadth to integrate the cumulative signals of multiple genomic features and alterations.
- 98.Kahana-Edwin S, Cain LE, Karpelowsky J. Roadmap to liquid biopsy biobanking from pediatric cancers-challenges and opportunities. Biopreserv Biobank 2021; 19:124–129. [DOI] [PubMed] [Google Scholar]
- 99.Trigg RM, Shaw JA, Turner SD. Opportunities and challenges of circulating biomarkers in neuroblastoma. Open Biol 2019; 9:190056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kothari P, Marass F, Yang JL, et al. Cell-free DNA profiling in retinoblastoma patients with advanced intraocular disease: an MSKCC experience. Cancer Med 2020; 9:6093–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hu Y, Ulrich BC, Supplee J, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res 2018; 24:4437–4443. [DOI] [PubMed] [Google Scholar]
- 102.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015; 126:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Silver AJ, Bick AG, Savona MR. Germline risk of clonal haematopoiesis. Nat Rev Genet Nat Res 2021; 22:603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Young AL, Tong RS, Birmann BM, Druley TE. Clonal hematopoiesis and risk of acute myeloid leukemia. Haematologica 2019; 104:2410–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bhattacharya R, Bick AG. Clonal hematopoiesis of indeterminate potential: an expanding genetic cause of cardiovascular disease. Curr Atheroscler Rep 2021; 23:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014; 20:1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bick AG, Weinstock JS, Nandakumar SK, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nat 2020; 586:763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wong WH, Tong S, Druley TE. Error-corrected sequencing of cord bloods identifies pediatric AML-associated clonal hematopoiesis. Blood 2017; 130: (Suppl 1): 2687–12687. [Google Scholar]
- 109.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012; 150:264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Collord G, Park N, Podestà M, et al. Clonal haematopoiesis is not prevalent in survivors of childhood cancer. Br J Haematol 2018; 181:537–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Laurie CC, Laurie CA, Rice K, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet 2012; 44:642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Olszewski AJ, Chorzalska AD, Kim AS, et al. Clonal haematopoiesis of indeterminate potential among cancer survivors exposed to myelotoxic chemotherapy. Br J Haematol 2019; 186:e31–e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bolton KL, Ptashkin RN, Gao T, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet 2020; 52:1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen G, Chen J, Liu H, et al. Comprehensive identification and characterization of human secretome based on integrative proteomic and transcriptomic data. Front Cell Dev Biol 2019; 7:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cohen JD, Javed AA, Thoburn C, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci USA 2017; 114:10202–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multianalyte blood test. Science 2018; 359:926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yao R, Yu T, Qing Y, et al. Evaluation of copy number variant detection from panel-based next-generation sequencing data. Mol Genet Genomic Med 2019; 7:e00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huether R, Dong L, Chen X, et al. The landscape of somatic mutations in epigenetic regulators across 1000 pediatric cancer genomes. Nat Commun 2014; 5:3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lennon AM, Buchanan AH, Kinde I, et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 2020; 369:eabb9601. [DOI] [PMC free article] [PubMed] [Google Scholar]