Abstract
Effective endosomal escape after cellular uptake represents a major challenge in the field of nanodelivery, as the majority of drug payloads must localize to subcellular compartments other than the endosomes in order to exert activity. In nature, viruses can readily deliver their genetic material to the cytosol of host cells by triggering membrane fusion after endocytosis. For the influenza A virus, the hemagglutinin (HA) protein found on its surface fuses the viral envelope with surrounding membrane at endosomal pH values. Here, biomimetic nanoparticles capable of endosomal escape were fabricated using a membrane coating derived from cells engineered to express HA on their surface. When evaluated in vitro, these virus-mimicking nanoparticles were able to deliver an mRNA payload to the cytosolic compartment of target cells, resulting in the successful expression of the encoded protein. When the mRNA-loaded nanoparticles were administered in vivo, protein expression levels were significantly increased in both local and systemic delivery scenarios. We therefore conclude that utilizing genetic engineering approaches to express viral fusion proteins on the surface of cell membrane-coated nanoparticles is a viable strategy for modulating the intracellular localization of encapsulated cargoes.
Keywords: cell membrane coating, genetic engineering, endosomal escape, hemagglutinin, mRNA
Graphical Abstract
Cell membrane-coated nanoparticles are engineered to express a viral fusion protein, thus enabling them to exhibit improved endosomal escape properties. It is demonstrated that these virus-mimicking nanocarriers are able to deliver mRNA payloads to the cytosolic compartment after cellular uptake, enhancing expression of the encoded proteins both in vitro and in vivo.
Achieving the proper subcellular localization of drug payloads is essential for maximizing their therapeutic potential. In the case of nanotherapeutics, delivery to the cytosol, which houses cellular machinery that is the target for a wide range of therapeutics,[1–3] has long presented a major challenge.[4, 5] Notably, nanodelivery vehicles must overcome the barrier posed by the endolysosomal pathway, which cells oftentimes use to sequester and degrade foreign objects.[6, 7] Endosomes transition from weakly acidic early endosomes to more acidic late endosomes, ultimately fusing with lysosomes, where nanoparticles can be destroyed by acids and enzymes.[6, 8, 9] One method to escape from this pathway is by the proton sponge approach, whereby nanoparticles are fabricated with buffering capabilities that enable them to rupture endosomes by osmotic swelling.[10–12] An alternate method is to destabilize the cell’s exterior plasma membrane during endocytosis and prior to endosome formation, thus creating leaky endosomes.[13, 14] The endosomal pathway can also be avoided altogether by using nanoparticles whose shape and charge allow them to transport directly across the plasma membrane and into the cytosol.[15, 16] Unfortunately, many of these conventional strategies for cytosolic delivery are known to cause cytotoxicity,[17, 18] making them difficult for clinical translation.
Most viruses require the delivery of their genetic material into the cytosol in order to replicate.[19] As such, many viruses have evolved methods that enable them to escape the endosomal compartment to avoid destruction.[20] In the case of the influenza virus, the hemagglutinin (HA) protein present on its surface helps to serve this purpose.[21, 22] After being expressed, HA is converted to its mature form through proteolytic cleavage, resulting in the generation of two subunits.[23] The HA1 subunit allows the virus to attach to the plasma membrane of target cells in order to initiate endocytosis.[24, 25] After endocytic uptake, the HA2 subunit undergoes a conformational change triggered by lowered pH that facilitates the fusion of the viral envelope with endosomal membrane.[26–28] For the influenza A virus, different HA subtypes bind to different sialic acid receptors, which can significantly impact infectivity towards different host species.[29, 30] For example, the H1 subtype is found on strains such as H1N1, which is known to infect humans and was the cause of the 2009 flu pandemic.[31] Influenza A virus carrying the H5 or H7 subtype, commonly known as the bird flu, is known to mostly infect avian hosts, although some human infections have been reported.[29]
Cell membrane coating is an emerging top-down approach for bestowing nanocarriers with enhanced biointerfacing capabilities.[32, 33] For example, erythrocyte membranes have been used to prolong nanoparticle circulation time,[34] whereas cancer cell membranes[35] and platelet membranes[36] have been leveraged for targeted drug delivery. More recently, genetic engineering approaches have been employed to generate cell membrane enriched with a specific surface marker, thus enabling researchers to purposefully manipulate the functionality of cell membrane-coated nanoformulations.[37, 38] These engineered nanoparticles can be equipped with complex surface proteins that would otherwise be infeasible to incorporate using conventional synthetic approaches. In this work, we engineered a cell membrane-coated nanoparticle to display HA, thus enabling the resulting nanocarrier to exhibit virus-mimicking endosomal escape properties and enhanced cytosolic delivery (Figure 1). Given the recent interest in mRNA-based vaccines,[39] we elected to evaluate the ability of our nanoformulation to deliver model mRNA payloads both in vitro and in vivo. Overall, the reported approach represents a compelling strategy for further improving the utility of cell membrane-coated nanocarriers, particularly for the delivery drugs that require cytosolic localization.
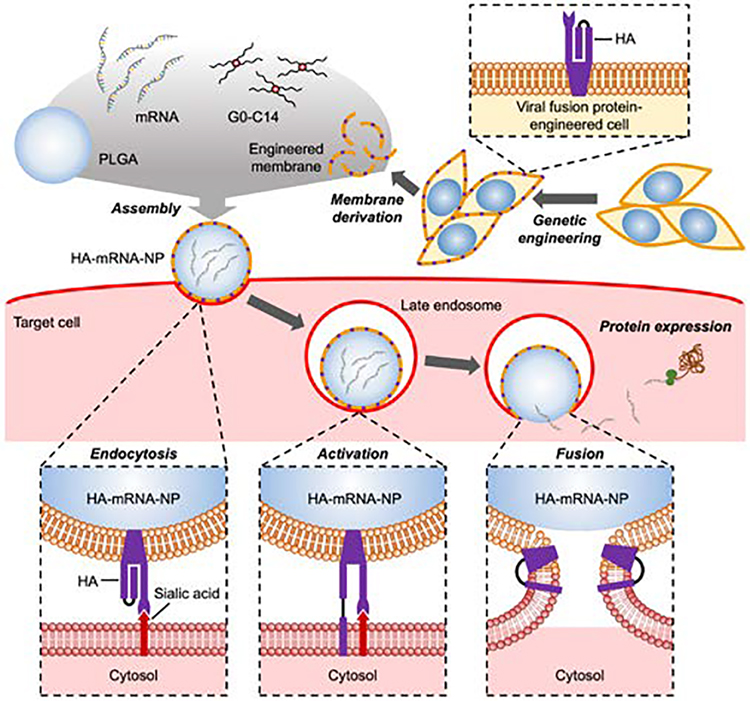
Figure 1.
Schematic illustration of genetically engineered cell membrane-coated nanoparticles for the cytosolic delivery of mRNA. Cells are genetically engineered to express the influenza virus fusion protein hemagglutinin (HA). Then, the membrane from the engineered cells is isolated and coated onto poly(lactic-co-glycolic acid) (PLGA) nanoparticle cores that are loaded with mRNA with the help of the cationic lipid-like molecule G0-C14. After the final HA-mRNA-NP formulation is endocytosed by a target cell, the lowered pH in the late endosomes causes a conformation change in HA. The activated HA then triggers membrane fusion, enabling escape of the mRNA payload into the cytosol, where it can be translated into a protein product.
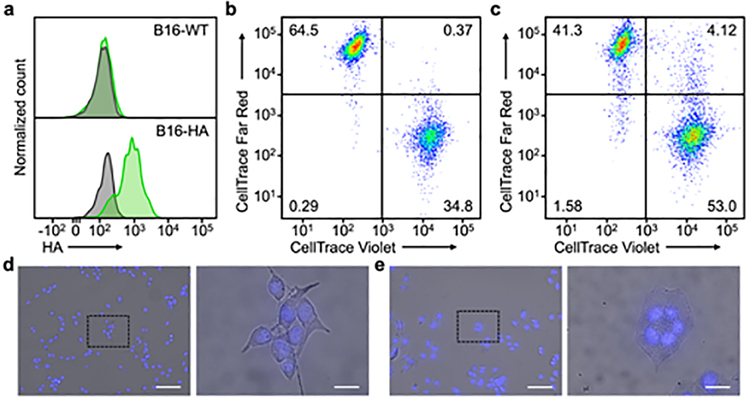
HA subtype H7 was chosen as a model viral protein for expression due to its strong ability to promote fusion.[40–42] Additionally, the fact that H7 targets α2,3-linked sialic acid enabled us to evaluate our platform in vivo using murine models.[30] Wild-type B16F10 cells (denoted ‘B16-WT’) were transfected with an expression plasmid encoding for H7, yielding engineered cells (denoted ‘B16-HA’) with high levels of the viral fusion protein on their surface (Figure 2a). As B16-WT is known to express α2,3-linked sialic acid,[43] a cell–cell fusion study was used to evaluate the functionality of the HA transgene in vitro (Figure 2b,c). B16-HA cells were divided into two aliquots, which were then stained with either CellTrace Violet or CellTrace Far Red. After combining the two dye-labeled aliquots together, the cell mixture was incubated with L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-treated trypsin for HA maturation before being subjected to endosomal pH to promote fusion activity. Following 2 h of incubation, the cells were analyzed by flow cytometry, which revealed a significant population of cells positive for both CellTrace Violet and CellTrace Far Red. This indicated that the HA on the surface of the engineered cells was active and could promote cell–cell fusion. In contrast, flow cytometric analysis of B16-WT cells subjected to an identical experimental protocol showed a negligible population of double-positive cells. It should be noted that our analysis could not identify fusion events between cells labeled with the same dye or distinguish events between only two cells versus those involving three or more cells. This suggests that the rate of fusion could be significantly higher than the double-positive percentage reported in our data, especially given that daughter cells resulting from the mitotic division of the same parent cell are more likely to fuse with each other as a result of their close proximity. Next, in order to visualize the cell fusion, either B16-WT or B16-HA cells were incubated with TPCK-treated trypsin and incubated under endosomal pH values (Figure 2d,e). Upon inspection under a fluorescence microscope, syncytia with multiple nuclei were observed among the B16-HA cells, providing a clear indication of cell–cell fusion. No signs of fusion were observed for the B16-WT cells.
Figure 2.
Fusion activity of HA on engineered cells. a) Flow cytometric analysis of HA expression on B16-WT and B16-HA cells (gray: isotype, green: anti-HA). b,c) Flow cytometric analysis of B16-WT (b) and B16-HA (c) cells co-incubated with themselves after half of the cell population was labeled with CellTrace Violet and the other half with CellTrace Far Red. d,e) Visualization of syncytia formation among B16-WT (d) and B16-HA (e) cells by optical microscopy (blue: nuclei). Scale bar = 100 μm (left) and 20 μm (right).
After confirming the successful expression of HA, the engineered B16-HA cells were harvested, and their membrane was derived as previously described.[44] The purified cell membrane was then coated onto preformed poly(lactic-co-glycolic acid) (PLGA) nanoparticle cores using a sonication process.[45] The PLGA nanoparticle cores were loaded with mRNA using a double emulsion method with the assistance of the cationic lipid-like molecule G0-C14.[46, 47] The resulting mRNA-loaded nanoparticles coated with B16-WT membrane (denoted ‘WT-mRNA-NP’) and the engineered B16-HA membrane (denoted ‘HA-mRNA-NP’) both had an average diameter of approximately 185 nm and a zeta potential of approximately −20 mV (Figure 3a,b). Transmission electron microscopy of negatively stained HA-mRNA-NP verified that the membrane was properly coated onto the polymeric cores (Figure 3c). In order to probe for the presence of HA on the purified cell membrane and on the nanoformulations, western blotting analysis was performed (Figure 3d). HA was clearly present on the membrane derived from B16-HA, as well as the final HA-mRNA-NP formulation. As expected, no signal was detected from the membrane of B16-WT or from WT-mRNA-NP. The long-term stability of WT-mRNA-NP and HA-mRNA-NP was evaluated by monitoring their size for 8 weeks when suspended in 10% sucrose solution at 4 °C (Figure 3e). No significant changes in size were detected during this period. Finally, mRNA loading was studied by measuring the fluorescent signal from a Cy5-labeled mRNA payload. It was determined that the encapsulation efficiency and drug loading yield were approximately 54% and 1 μg/mg of PLGA, respectively (Figure 3f).
Figure 3.
Nanoparticle characterization. a,b) Size (a) and surface zeta potential (b) of WT-mRNA-NP and HA-mRNA-NP as measured by dynamic light scattering (n = 3; mean + SD). c) Representative transmission electron microscopy image of HA-mRNA-NP negatively stained with uranyl acetate. Scale bar = 200 nm. d) Western blot probing for HA on B16-WT membrane (WT mem.), B16-HA membrane (HA mem.), WT-mRNA-NP, and HA-mRNA-NP. e) Size of WT-mRNA-NP and HA-mRNA-NP when stored in 10% sucrose over 8 weeks (n = 3; mean ± SD). f) Encapsulation efficiency (EE) and drug loading (DL) of mRNA into HA-mRNA-NP (n = 3; mean + SD).
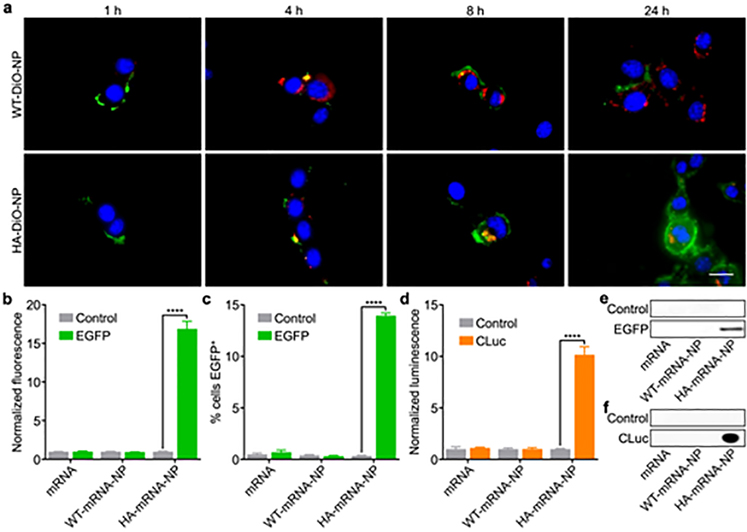
In order to qualitatively visualize endosomal escape, PLGA cores were loaded with the fluorescent dye benzoxazolium, 3-octadecyl-2-[3-(3-octadecyl-2(3H)-benzoxazolylidene)-1-propenyl]-, perchlorate (DiO) and coated with the membrane from either B16-WT or B16-HA (denoted ‘WT-DiO-NP’ or ‘HA-DiO-NP’, respectively). After 1, 4, 8, and 24 h of incubation with WT-DiO-NP or HA-DiO-NP, B16-WT cells were stained with Hoechst 33342 and LysoTracker Red DND-99 prior to the imaging (Figure 4a and Figure S1). At the 1 h timepoint, nanoparticles could only be seen bound to the surface of the cells, while colocalization of the nanoparticles and the endosomes was observed at 4 h, indicating endocytosis for both formulations. After another 4 h, some nanoparticle signal was visualized outside of the endosomes for HA-DiO-NP, indicating endosomal escape, while intracellular WT-DiO-NP signal was still colocalized with the endosomes. At 24 h after starting the incubation, signal from HA-DiO-NP permeated the cytosol, and there was little to no evidence of cytosolic delivery for WT-DiO-NP. It was also observed that cells treated with HA-DiO-NP had attenuated LysoTracker signal, which may be explained by their reduced endolysosomal load resulting from endosomal escape.[48, 49]
Figure 4.
Endosomal escape and mRNA transfection in vitro. a) Fluorescent visualization of B16-WT cells incubated with WT-DiO-NP and HA-DiO-NP for 1, 4, 8, and 24 h (blue: nuclei, red: endosomes, green: nanoparticles). Scale bar = 20 μm. b,c) Normalized fluorescence (b) and % EGFP+ (c) of B16-WT cells after incubation with EGFP mRNA in free form, loaded within WT-mRNA-NP, or loaded within HA-mRNA-NP (n = 3; mean + SD). d) Normalized luminescence of B16-WT cells after incubation with CLuc mRNA in free form, loaded within WT-mRNA-NP, or loaded within HA-mRNA-NP (n = 3; mean + SD). ****p < 0.0001; Student’s t-test. e,f) Western blots of cell lysate probing for EGFP (e) or culture supernatant probing for CLuc (f) after treatment of B16-WT with EGFP mRNA (e) or CLuc mRNA (f) in free form, loaded within WT-mRNA-NP, or loaded within HA-mRNA-NP.
Next, we quantitatively evaluated the ability of HA-mRNA-NP to successfully deliver functional mRNA cargoes for protein translation. First, HA-mRNA-NP was formulated with mRNA encoding for enhanced green fluorescent protein (EGFP) as a model payload. Transfecting B16-WT cells with the resulting formulation led to a significant 17-fold increase in mean EGFP fluorescence and an elevated percentage of EGFP+ cells compared to HA-mRNA-NP loaded with an irrelevant control mRNA (Figure 4b,c). As a secondary means of validating our platform in vitro, the experiment was repeated using Cypridina luciferase (CLuc) mRNA as the payload. Similar results were observed, where cells transfected with HA-mRNA-NP loaded with CLuc mRNA showed a significant 10-fold increase in bioluminescent signal compared to cells treated with the same nanocarrier but loaded with irrelevant control mRNA (Figure 4d). The successful expression of both EGFP and CLuc after treatment with the corresponding HA-mRNA-NP formulation was further validated by western blotting analysis (Figure 4e,f).
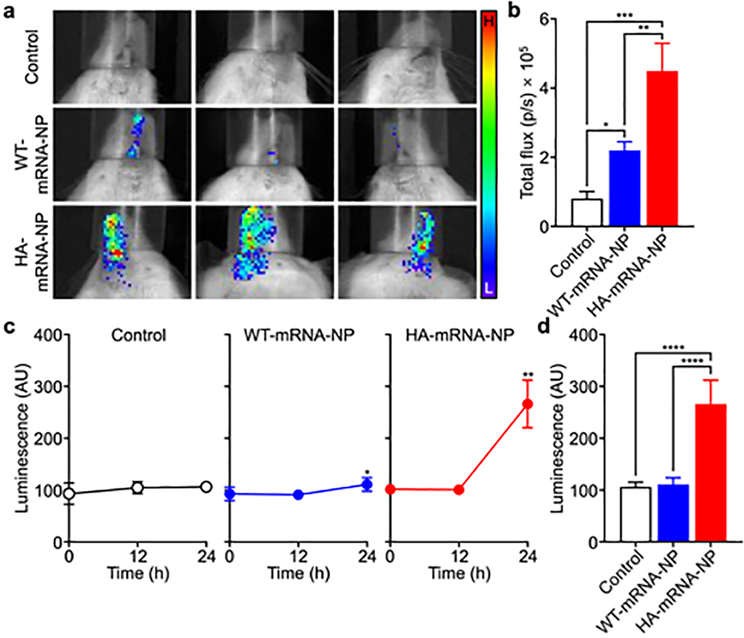
After confirming successful protein translation in vitro, we next assessed the ability of the engineered nanoformulation to achieve transfection in vivo. First, local delivery of CLuc mRNA was evaluated by administering WT-mRNA-NP or HA-mRNA-NP to mice via the intranasal route (Figure 5a,b). At 24 h after administration of the nanoparticles, the mice were injected with Cypridina luciferin, and bioluminescence activity was evaluated using a live animal imaging system. Compared with the untreated controls, a small amount of signal was detected in mice treated with WT-mRNA-NP. Significantly stronger bioluminescence was detected for the mice treated with HA-mRNA-NP, demonstrating the ability of the engineered HA to promote efficient mRNA delivery in vivo. When the signals were quantified, it was determined that the total flux for the HA-mRNA-NP group was more than 2-fold higher than that of the WT-mRNA-NP group. The same nanoformulations were then evaluated for their ability to elevate the serum levels of a secreted payload after systemic delivery (Figure 5c,d). Mice were intravenously administered with each formulation, and their blood was sampled at 12 and 24 h after injection to monitor for CLuc activity. As expected, the untreated control group showed no changes in CLuc signal throughout the study. While there was a slight increase in bioluminescence for the WT-mRNA-NP group at 24 h, the signal for the HA-mRNA-NP group was significantly elevated at the same timepoint. Overall, the results demonstrated that the engineering of cell membrane-coated nanocarriers to express HA can lead to more efficient mRNA delivery in vivo after both local and systemic administration. We also tracked antibody production against HA with repeated weekly dosing and did not observe any significant increase in anti-HA titers over the course of 1 month (Figure S2). This is consistent with the reported low immunogenicity of HA subtype H7[50] and suggests that our platform may be effective across multiple administrations.
Figure 5.
mRNA transfection in vivo. a) Visualization of bioluminescent signal from mice intranasally administered with WT-mRNA-NP and HA-mRNA-NP loaded with CLuc mRNA (H: high signal, L: low signal). b) Quantification of the total flux from the images in (a) (n = 3; mean + SD). *p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA. c) Bioluminescence over time in the serum of mice intravenously administered with WT-mRNA-NP and HA-mRNA-NP loaded with CLuc mRNA (n = 5; mean ± SD). *p < 0.05, **p < 0.01 (compared to 0 h); Student’s t-test. d) Bioluminescence in the serum of mice 24 h after intravenous administration with WT-mRNA-NP and HA-mRNA-NP loaded with CLuc mRNA (n = 5; mean + SD). ****p < 0.0001; one-way ANOVA.
In this work, cell membrane engineered to express a viral fusion protein was used to coat the surface of mRNA-loaded nanoparticle cores, enabling the resulting HA-mRNA-NP formulation to mimic the ability of some viruses to achieve endosomal escape. Influenza A virus HA subtype H7 bound to α2,3-linked sialic acid on the surface of murine cells, thus triggering endocytic uptake. In the late endosomes, the lowered pH caused the HA to induce membrane fusion, thus allowing the nanoparticle contents to be unloaded into the cytosol. To prove our concept, we tested the ability of the engineered cell membrane-coated nanoparticles to escape the endosomal compartment and promote the expression of two model reporter genes in vitro. In both cases, the HA-expressing nanoparticles significantly outperformed a control formulation fabricated using the membrane of wild-type cells lacking the viral transgene. When tested in vivo, HA-mRNA-NP loaded with CLuc mRNA was able to significantly elevate levels of the encoded protein in both local and systemic administration scenarios.
Effective methods for mRNA delivery are highly desirable, particularly given the recent interest in mRNA vaccines driven by the COVID-19 pandemic.[39] Endosomal escape represents one of the key obstacles in mRNA nanodelivery since the payload needs to be present within the cytosol in order to carry out its biological function. As we have demonstrated here, utilizing naturally occurring viral fusion proteins such as influenza virus HA could provide an elegant solution to this challenge. By leveraging cell membrane coating technology in conjunction with genetic engineering, we were able to present HA in its natural context on the surface of nanoparticles, a task that would otherwise be difficult to achieve using conventional functionalization methods. Future studies will be required to validate the utility of this approach for specific mRNA applications such as vaccination and gene therapy. Ultimately, continued research along these lines may yield novel strategies for controlling the subcellular localization of drug payloads, helping to further expand the utility of biomimetic nanomedicine.
Supplementary Material
Acknowledgements
This work is supported by the Defense Threat Reduction Agency Joint Science and Technology Office for Chemical and Biological Defense under Grant Number HDTRA1–18-1–0014 and HDTRA1–21-1–0010. J.H.P. was supported by a National Institutes of Health 5T32CA153915 training grant from the National Cancer Institute.
References
- [1].Li Y, Wang J, Wientjes MG, Au JL, Adv. Drug Deliv. Rev 2012, 64, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mocan L, Matea C, Tabaran FA, Mosteanu O, Pop T, Mocan T, Iancu C, Int. J. Nanomedicine 2015, 10, 5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dam DH, Lee JH, Sisco PN, Co DT, Zhang M, Wasielewski MR, Odom TW, ACS Nano 2012, 6, 3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fu A, Tang R, Hardie J, Farkas ME, Rotello VM, Bioconjug. Chem 2014, 25, 1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Field LD, Delehanty JB, Chen Y, Medintz IL, Acc. Chem. Res 2015, 48, 1380. [DOI] [PubMed] [Google Scholar]
- [6].He B, Lin P, Jia Z, Du W, Qu W, Yuan L, Dai W, Zhang H, Wang X, Wang J, Zhang X, Zhang Q, Biomaterials 2013, 34, 6082. [DOI] [PubMed] [Google Scholar]
- [7].Biswas S, Torchilin VP, Adv. Drug Deliv. Rev 2014, 66, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huotari J, Helenius A, EMBO J. 2011, 30, 3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Scott CC, Vacca F, Gruenberg J, Semin. Cell Dev. Biol 2014, 31, 2. [DOI] [PubMed] [Google Scholar]
- [10].Zhang C, An T, Wang D, Wan G, Zhang M, Wang H, Zhang S, Li R, Yang X, Wang Y, J. Control. Release 2016, 226, 193. [DOI] [PubMed] [Google Scholar]
- [11].Zhou Z, Badkas A, Stevenson M, Lee JY, Leung YK, Int. J. Pharm 2015, 487, 81. [DOI] [PubMed] [Google Scholar]
- [12].Smith SA, Selby LI, Johnston APR, Such GK, Bioconjug. Chem 2019, 30, 263. [DOI] [PubMed] [Google Scholar]
- [13].Convertine AJ, Benoit DS, Duvall CL, Hoffman AS, Stayton PS, J. Control. Release 2009, 133, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tran KK, Zhan X, Shen H, Adv. Healthc. Mater 2014, 3, 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wu XA, Choi CH, Zhang C, Hao L, Mirkin CA, J. Am. Chem. Soc. 2014, 136, 7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hinde E, Thammasiraphop K, Duong HT, Yeow J, Karagoz B, Boyer C, Gooding JJ, Gaus K, Nat. Nanotechnol 2017, 12, 81. [DOI] [PubMed] [Google Scholar]
- [17].Mukherjee SP, Byrne HJ, Nanomedicine 2013, 9, 202. [DOI] [PubMed] [Google Scholar]
- [18].Lv H, Zhang S, Wang B, Cui S, Yan J, Control J Release 2006, 114, 100. [DOI] [PubMed] [Google Scholar]
- [19].Staring J, Raaben M, Brummelkamp TR, Cell Sci J 2018, 131, jcs216259. [DOI] [PubMed] [Google Scholar]
- [20].Hamilton BS, Whittaker GR, Daniel S, Viruses 2012, 4, 1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Russell CJ, Hu M, Okda FA, Trends Microbiol. 2018, 26, 841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Boonstra S, Blijleven JS, Roos WH, Onck PR, van der Giessen E, van Oijen AM, Annu. Rev. Biophys 2018, 47, 153. [DOI] [PubMed] [Google Scholar]
- [23].Lazarowitz SG, Choppin PW, Virology 1975, 68, 440. [DOI] [PubMed] [Google Scholar]
- [24].Tatulian SA, Tamm LK, Biochemistry 2000, 39, 496. [DOI] [PubMed] [Google Scholar]
- [25].Thoennes S, Li ZN, Lee BJ, Langley WA, Skehel JJ, Russell RJ, Steinhauer DA, Virology 2008, 370, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xu R, Wilson IA, Virol J 2011, 85, 5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kim CS, Epand RF, Leikina E, Epand RM, Chernomordik LV, J. Biol. Chem 2011, 286, 13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Han X, Bushweller JH, Cafiso DS, Tamm LK, Nat. Struct. Biol 2001, 8, 715. [DOI] [PubMed] [Google Scholar]
- [29].Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD, Proc. Natl. Acad. Sci. U. S. A 2004, 101, 4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL, J. Virol 2006, 80, 7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Monsalvo AC, Batalle JP, Lopez MF, Krause JC, Klemenc J, Hernandez JZ, Maskin B, Bugna J, Rubinstein C, Aguilar L, Dalurzo L, Libster R, Savy V, Baumeister E, Aguilar L, Cabral G, Font J, Solari L, Weller KP, Johnson J, Echavarria M, Edwards KM, Chappell JD, Crowe JE Jr., Williams JV, Melendi GA, Polack FP, Nat. Med 2011, 17, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fang RH, Kroll AV, Gao W, Zhang L, Adv. Mater 2018, 30, 1706759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fang RH, Jiang Y, Fang JC, Zhang L, Biomaterials 2017, 128, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hu CM, Zhang L, Aryal S, Cheung C, Fang RH, Zhang L, Proc. Natl. Acad. Sci. U. S. A 2011, 108, 10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fang RH, Hu CM, Luk BT, Gao W, Copp JA, Tai Y, O’Connor DE, Zhang L, Nano Lett. 2014, 14, 2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang S, Duan Y, Zhang Q, Komarla A, Gong H, Gao W, Zhang L, Small Struct. 2020, 1, 2000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jiang Y, Krishnan N, Zhou J, Chekuri S, Wei X, Kroll AV, Yu CL, Duan Y, Gao W, Fang RH, Zhang L, Adv. Mater 2020, 32, 2001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Park JH, Jiang Y, Zhou J, Gong H, Mohapatra A, Heo J, Gao W, Fang RH, Zhang L, Sci. Adv 2021, 7, eabf7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Verbeke R, Lentacker I, De Smedt SC, Dewitte H, Control J Release 2021, 333, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Galloway SE, Reed ML, Russell CJ, Steinhauer DA, PLoS Pathog. 2013, 9, e1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Su B, Wurtzer S, Rameix-Welti MA, Dwyer D, van der Werf S, Naffakh N, Clavel F, Labrosse B, PLoS One 2009, 4, e8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ma MJ, Yang Y, Fang LQ, Trends Microbiol. 2019, 27, 93. [DOI] [PubMed] [Google Scholar]
- [43].Chang WW, Yu CY, Lin TW, Wang PH, Tsai YC, Biochem. Biophys. Res. Commun 2006, 341, 614. [DOI] [PubMed] [Google Scholar]
- [44].Kroll AV, Fang RH, Jiang Y, Zhou J, Wei X, Yu CL, Gao J, Luk BT, Dehaini D, Gao W, Zhang L, Adv. Mater 2017, 29, 1703969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Copp JA, Fang RH, Luk BT, Hu CM, Gao W, Zhang K, Zhang L, Proc. Natl. Acad. Sci. U. S. A 2014, 111, 13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Islam MA, Xu Y, Tao W, Ubellacker JM, Lim M, Aum D, Lee GY, Zhou K, Zope H, Yu M, Cao W, Oswald JT, Dinarvand M, Mahmoudi M, Langer R, Kantoff PW, Farokhzad OC, Zetter BR, Shi J, Nat. Biomed. Eng 2018, 2, 850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Xu X, Xie K, Zhang XQ, Pridgen EM, Park GY, Cui DS, Shi J, Wu J, Kantoff PW, Lippard SJ, Langer R, Walker GC, Farokhzad OC, Proc. Natl. Acad. Sci. U. S. A 2013, 110, 18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Martinez-Fabregas J, Prescott A, van Kasteren S, Pedrioli DL, McLean I, Moles A, Reinheckel T, Poli V, Watts C, Nat. Commun 2018, 9, 5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yoon J, Bang SH, Park JS, Chang ST, Kim YH, Min J, Appl. Biochem. Biotechnol 2011, 163, 1002. [DOI] [PubMed] [Google Scholar]
- [50].Blanchfield K, Kamal RP, Tzeng WP, Music N, Wilson JR, Stevens J, Lipatov AS, Katz JM, York IA, Influenza Other Respir. Viruses 2014, 8, 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.