Abstract
Background
The TIMELESS–TIPIN complex protects the replication fork from replication stress induced by chemotherapeutic drugs. We hypothesised genetic polymorphisms of the TIMELESS–TIPIN complex may affect the response, progression-free survival (PFS), and overall survival (OS) of cytotoxic drugs in patients with metastatic colorectal cancer (mCRC).
Methods
We analysed data from the MAVERICC trial, which compared FOLFOX/bevacizumab and FOLFIRI/bevacizumab in untreated patients with mCRC. Genomic DNA extracted from blood samples was genotyped using an OncoArray. Eight functional single nucleotide polymorphisms (SNPs) in TIMELESS and TIPIN were tested for associations with clinical outcomes.
Results
In total, 324 patients were included (FOLFOX/bevacizumab arm, n = 161; FOLFIRI/bevacizumab arm, n = 163). In the FOLFOX/bevacizumab arm, no SNPs displayed confirmed associations with survival outcomes. In the FOLFIRI/bevacizumab arm, TIMELESS rs2291739 was significantly associated with OS in multivariate analysis (G/G vs. any A allele, hazard ratio = 3.06, 95% confidence interval = 1.49–6.25, p = 0.004). TIMELESS rs2291739 displayed significant interactions with treatment regarding both PFS and OS.
Conclusions
TIMELESS rs2291739 might have different effects on therapeutic efficacy between oxaliplatin- and irinotecan-based chemotherapies. Upon further validation, our findings may be useful for personalised approaches in the first-line treatment of mCRC.
Subject terms: Colorectal cancer, Predictive markers
Introduction
Replication stress is a hallmark of cancer development [1, 2]. Endogenous and exogenous sources of replication stress promote genomic instability by leading to DNA replication fork collapse and subsequent DNA double-strand breaks (DSBs) [3]. Whereas cells that can activate an adequate DNA damage response (DDR) to DNA damage undergo apoptosis, cells with a faulty DDR system escape from apoptosis and develop mutations and chromosome aberrations that result in tumorigenesis [4, 5]. In this context, cancer cells harbour distinct molecular backgrounds adaptive to oncogene-induced replication stress, thereby shaping an environment that favours survival and encourages tumour growth [6, 7]. Recently, DDR pathway alteration has been targeted in the treatment of cancer using the concept of synthetic lethality [8]. One great success of this strategy is PARP inhibition in ovarian, breast, prostate, and pancreatic cancers with homologous recombination deficiency [9].
TIMELESS and TIMELESS-interacting protein (TIPIN), which form a complex, are components of the replication fork machinery [10]. The TIMELESS–TIPIN complex contributes to full activation of the ATR–Chk1 checkpoint signalling pathway, which plays a central role in preventing fork collapse [11]. In addition to their role in the ATR–Chk1 pathway, TIMELESS and TIPIN also interact with numerous components of the replication machinery, such as the replicative helicase components MCM2-7 and CDC45 and replicative polymerases Polε and Polδ, thereby stabilising the replication fork structure [12]. When DNA lesions transiently stall the leading-strand polymerase without impeding the movement of the rest of the replisome, uncoupling between DNA polymerase and helicase activities can be induced [13]. The TIMELESS–TIPIN complex can sense functional uncoupling of the replisome to transduce a signal to remodel and restart the fork [14]. Otherwise, in the absence of the TIMELESS–TIPIN complex, the fork collapses, and in turn, harmful DSBs are generated [14]. Thus, TIMELESS and TIPIN are necessary for cell survival under replication stress because of their roles in protecting the replication fork in both ATR-dependent and ATR-independent manners. Tumours, such as colorectal cancer (CRC), overexpress these molecules, and this overexpression may serve as a supportive mechanism against replication stress in cancer cells [15–17].
In the standard first-line treatment of metastatic CRC (mCRC), oxaliplatin or irinotecan is used as the cytotoxic agent in combination with 5-fluorouracil. The choice of these agents remains an important clinical question for optimising treatment in individual patients. Oxaliplatin and irinotecan form different structures on the DNA strands which prevent DNA replication in different manners [18, 19]. Thus, although fork protection is a global response to genotoxic treatments, it might be possible that oxaliplatin and irinotecan induce distinct responses that protect the replication fork in tumours [20]. We hypothesised that common and functional single nucleotide polymorphisms (SNPs) within TIMELESS and TIPIN are associated with the different therapeutic effects of oxaliplatin- and irinotecan-based chemotherapies in mCRC. We tested our hypothesis using genetic and clinical data from the MAVERICC trial, a randomised phase II clinical trial of patients with mCRC in the first-line setting [21].
Patients and methods
Patient population and study design
The subjects of this study were patients with mCRC enrolled in the MAVERICC trial (NCT01374425). Patients were randomised to treatment with either FOLFOX (oxaliplatin cohort) or FOLOFIRI (irinotecan cohort), both in combination with bevacizumab. Patients without sufficient peripheral whole blood samples, SNP data, and/or any other relevant data were excluded from this study. All patients provided informed consent for molecular research before study enrollment. The study protocol was approved by the institutional review board of each participating institution and was conducted in accordance with the tenets of the Declaration of Helsinki as well as the Good Clinical Practice and REMARK guidelines.
Genotyping and selecting polymorphisms
Genomic DNA was extracted from peripheral whole blood collected before treatment initiation using a QIAmp Kit (Qiagen, Inc., Valencia, CA, USA) in accordance with the manufacturer’s protocol. The OncoArray of 530 K SNPs was used for genotyping (Illumina, Inc., San Diego, CA, USA). The candidate SNPs within TIMELESS and TIPIN were selected from dbSNP variants (http://www.ncbi.nlm.nih.gov) if the SNPs met both following criteria: (1) minor allele frequency in Caucasians (defined as ‘European’ in 1000 Genomes Project Phase 3 and/or ‘Non-Finnish European’ in gnomAD genomes r3.0) of at least 10% in the Ensemble Genome Browser (https://www.ensembl.org) and (2) missense, 3′-untranslated region (UTR), or intron/5′-UTR variants having potential biological functions based on public databases (https://snpinfo.niehs.nih.gov). SNPs exhibiting linkage disequilibrium with R2 > 0.8 (https://ldlink.nci.nih.gov, among the population of ‘European’) were excluded. In total, eight SNPs (rs2291739, rs3759786, rs8035497, rs11071888, rs28593577, rs11637949, rs6494568, and rs12323975) met the criteria for inclusion in this study (Table S1).
Statistical analysis
The selected SNPs were evaluated for their associations with tumour response, progression-free survival (PFS), and overall survival (OS) based on the dominant and recessive genetic models. The overall response rate (ORR) was calculated as the percentage of patients with a complete or partial response using Response Evaluation Criteria in Solid Tumors version 1.1. PFS was defined as the time from randomisation to disease progression or death from any cause. OS was defined as the time from randomisation to death from any cause. Patients who did not experience any events were censored at the last follow-up date. The correlations of SNPs with ORR were examined using the likelihood ratio test. To test the associations of SNPs with PFS or OS, the Cox proportional hazards regression model and the log-rank test were performed. Multivariate analyses based on the Wald test were performed for tumour response, PFS, and OS. In the multivariate analyses, adjustment was performed for the following covariates: ethnicity, sex, age, Eastern Cooperative Oncology Group performance status, primary tumour site, primary tumour resected, number of metastases, and KRAS status. To formally assess the predictive value of SNPs, the treatment-by-SNP interaction was tested based on the multivariate analysis. All analyses were two-sided at a significance level of 0.05 and were performed using SAS ver. 9.4 software (SAS Institute, Cary, NC, USA).
Results
Patient characteristics
In total, 324 patients were included in this study, including 161 patients in the oxaliplatin cohort and 163 patients in the irinotecan cohort (Fig. S1). Patient characteristics were balanced between the cohorts, excluding the higher proportion of patients aged ≤65 in the oxaliplatin cohort (Table 1).
Table 1.
Patient characteristics.
| Characteristics | Total N = 324 | MAVERICC | P-value | |
|---|---|---|---|---|
| FOLFOX + BEV (Oxaliplatin cohort) N = 161 | FOLFIRI + BEV (Irinotecan cohort) N = 163 | |||
| Sex | 0.93 | |||
| Male | 204 | 101 (62.7%) | 103 (63.2%) | |
| Female | 120 | 60 (37.3%) | 60 (36.8%) | |
| Age | 0.04 | |||
| ≤65 | 218 | 117 (72.7%) | 101 (62.0%) | |
| >65 | 106 | 44 (27.3%) | 62 (38.0%) | |
| Performance status | 0.11 | |||
| ECOG 0 | 178 | 81 (50.3%) | 97 (59.5%) | |
| ECOG 1 | 145 | 79 (49.1%) | 66 (40.5%) | |
| Unknown* | 1 | 1 (0.6%) | 0 (0%) | |
| Primary tumour site | 0.80 | |||
| Right-sided | 131 | 64 (39.8%) | 67 (41.1%) | |
| Left-sided | 193 | 97 (60.2%) | 96 (58.9%) | |
| Number of metastases | 0.67 | |||
| ≤2 | 207 | 101 (62.7%) | 106 (65.0%) | |
| >2 | 117 | 60 (37.3%) | 57 (35.0%) | |
| Primary tumour resected | 0.50 | |||
| No | 301 | 148 (91.9%) | 153 (93.9%) | |
| Yes | 23 | 13 (8.1%) | 10 (6.1%) | |
| Adjuvant chemotherapy | 0.39 | |||
| No | 289 | 146 (90.7%) | 143 (87.7%) | |
| Yes | 35 | 15 (9.3%) | 20 (12.3%) |
P-values was estimated by Chi-square test.
BEV bevacizumab, ECOG Eastern Cooperative Oncology Group.
*Unknown group was not included in the analysis.
Associations of SNPs in TIMELESS and TIPIN with clinical outcomes in the oxaliplatin cohort
In the oxaliplatin cohort, univariate analysis did not show significant associations between the tested SNPs and tumour response, whereas TIPIN rs11637949 was significantly associated with tumour response in multivariate analysis (Table 2, Tables S2, S3). Conversely, two SNPs exhibited significant associations with survival outcomes in univariate analysis (Tables 3, 4, Tables S2, S3). Specifically, the G/G genotype of TIMELESS rs2291739 was associated with better PFS than any A allele (median PFS, 13.9 months vs. 9.5 months, hazard ratio [HR] = 0.51, 95% confidence interval [CI] = 0.32–0.83, p = 0.006), and the A/A genotype of TIPIN rs8035497 was linked to worse OS than any G allele (median OS, 19.2 months vs. 25.5 months, HR = 1.96, 95% CI = 1.02–3.75, p = 0.04). However, these associated were not confirmed in multivariate analysis (Tables 3, 4, Table S3).
Table 2.
Univariate and multivariate analyses for the association between SNPs and tumour response.
| SNP/genetic model | Oxaliplatin cohort | Irinotecan cohort | ||
|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |
| Pa | Pb | Pa | Pb | |
| TIMELESS rs2291739 | ||||
| Dominant | 0.64 | 0.85 | 0.90 | 0.89 |
| Recessive | 0.39 | 0.84 | 0.79 | 0.29 |
| TIPIN rs3759786 | ||||
| Dominant | 0.97 | 0.90 | 0.80 | 0.28 |
| Recessive | NA | NA | NA | NA |
| TIPIN rs8035497 | ||||
| Dominant | 0.63 | 0.63 | 0.87 | 0.61 |
| Recessive | 0.52 | 0.55 | 0.43 | 0.21 |
| TIPIN rs11071888 | ||||
| Dominant | 0.47 | 0.77 | 0.22 | 0.56 |
| Recessive | 0.63 | 0.92 | 0.56 | 0.96 |
| TIPIN rs28593577 | ||||
| Dominant | 0.71 | 0.91 | 0.96 | 0.83 |
| Recessive | 1.00 | 0.65 | 0.70 | 0.30 |
| TIPIN rs11637949 | ||||
| Dominant | 0.61 | 0.23 | 0.73 | 0.68 |
| Recessive | 0.06 | 0.04 | 0.83 | 0.56 |
| TIPIN rs6494568 | ||||
| Dominant | 0.72 | 0.25 | 0.74 | 0.50 |
| Recessive | NA | NA | NA | NA |
| TIPIN rs12323975 | ||||
| Dominant | 0.79 | 0.79 | 0.56 | 0.58 |
| Recessive | NA | NA | NA | NA |
Significant values are indicated in bold characters. TIPIN rs3759786, TIPIN rs6494568, and TIPIN rs12323975 were not assessed with recessive genetic model because there were no patients having homozygous genotype of the recessive allele in these SNPs.
NA not assessed, SNP single nucleotide polymorphism.
aP-values were based on likelihood ratio test.
bP-values were based on Wald test in the multivariate model.
Table 3.
Univariate and multivariate analyses for the association between SNPs and PFS.
| SNP/genetic model | Oxaliplatin cohort | Irinotecan cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
| HR (95% CI) | Pa | HR (95% CI) | Pb | HR (95% CI) | Pa | HR (95% CI) | Pb | |
| TIMELESS rs2291739 | ||||||||
| Dominant | 1.03 (0.69–1.55) | 0.87 | 1.25 (0.73–2.15) | 0.41 | 1.44 (0.90–2.30) | 0.13 | 1.26 (0.73–2.16) | 0.40 |
| Recessive | 0.51 (0.32–0.83) | 0.006 | 0.60 (0.31–1.17) | 0.12 | 1.48 (0.97–2.26) | 0.07 | 1.49 (0.77–2.87) | 0.25 |
| TIPIN rs3759786 | ||||||||
| Dominant | 0.78 (0.47–1.29) | 0.33 | 0.88 (0.48–1.64) | 0.69 | 0.90 (0.57–1.42) | 0.65 | 1.06 (0.59–1.90) | 0.85 |
| Recessive | NA | NA | NA | NA | NA | NA | NA | NA |
| TIPIN rs8035497 | ||||||||
| Dominant | 1.22 (0.86–1.75) | 0.27 | 1.27 (0.81–1.99) | 0.30 | 0.87 (0.59–1.27) | 0.46 | 0.80 (0.47–1.35) | 0.40 |
| Recessive | 1.20 (0.68–2.15) | 0.53 | 1.31 (0.66–2.60) | 0.44 | 1.20 (0.58–2.49) | 0.62 | 0.93 (0.37–2.38) | 0.89 |
| TIPIN rs11071888 | ||||||||
| Dominant | 0.84 (0.57–1.24) | 0.37 | 0.90 (0.52–1.56) | 0.70 | 1.12 (0.77–1.65) | 0.55 | 1.04 (0.61–1.76) | 0.90 |
| Recessive | 0.18 (0.03–1.29) | 0.05 | 0.23 (0.03–1.86) | 0.09 | 1.59 (0.65–3.93) | 0.31 | 0.84 (0.24–2.91) | 0.79 |
| TIPIN rs28593577 | ||||||||
| Dominant | 1.02 (0.71–1.46) | 0.92 | 1.06 (0.67–1.68) | 0.80 | 0.92 (0.62–1.38) | 0.70 | 0.85 (0.50–1.45) | 0.55 |
| Recessive | 1.24 (0.63–2.45) | 0.54 | 1.62 (0.76–3.46) | 0.24 | 1.22 (0.56–2.64) | 0.61 | 0.99 (0.39–2.52) | 0.99 |
| TIPIN rs11637949 | ||||||||
| Dominant | 1.31 (0.91–1.89) | 0.15 | 1.27 (0.80–1.99) | 0.31 | 1.23 (0.84–1.80) | 0.28 | 1.17 (0.71–1.91) | 0.54 |
| Recessive | 1.41 (0.68–2.92) | 0.35 | 1.61 (0.69–3.78) | 0.29 | 1.04 (0.53–2.08) | 0.90 | 0.91 (0.41–2.02) | 0.81 |
| TIPIN rs6494568 | ||||||||
| Dominant | 1.26 (0.81–1.95) | 0.31 | 1.11 (0.60–2.06) | 0.74 | 0.84 (0.51–1.37) | 0.48 | 0.88 (0.42–1.81) | 0.72 |
| Recessive | NA | NA | NA | NA | NA | NA | NA | NA |
| TIPIN rs12323975 | ||||||||
| Dominant | 0.86 (0.53–1.40) | 0.55 | 0.83 (0.44–1.56) | 0.56 | 1.16 (0.69–1.95) | 0.58 | 0.74 (0.37–1.51) | 0.40 |
| Recessive | NA | NA | NA | NA | NA | NA | NA | NA |
Significant values are indicated in bold characters. TIPIN rs3759786, TIPIN rs6494568, and TIPIN rs12323975 were not assessed with recessive genetic model because there were no patients having homozygous genotype of the recessive allele in these SNPs.
CI confidence interval, HR hazard ratio, NA not assessed, PFS progression-free survival, SNP single nucleotide polymorphism.
aP-values were based on log-rank test.
bP-values were based on Wald test in the multivariate Cox proportional hazards regression model.
Table 4.
Univariate and multivariate analyses for the association between SNPs and OS.
| SNP/genetic model | Oxaliplatin cohort | Irinotecan cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
| HR (95% CI) | Pa | HR (95% CI) | Pb | HR (95% CI) | Pa | HR (95% CI) | Pb | |
| TIMELESS rs2291739 | ||||||||
| Dominant | 1.03 (0.64–1.66) | 0.90 | 1.41 (0.76–2.60) | 0.27 | 1.32 (0.70–2.48) | 0.39 | 1.47 (0.73–2.96) | 0.27 |
| Recessive | 0.63 (0.34–1.14) | 0.12 | 0.83 (0.36–1.94) | 0.66 | 2.01 (1.19–3.41) | 0.008 | 3.06 (1.49–6.25) | 0.004 |
| TIPIN rs3759786 | ||||||||
| Dominant | 0.78 (0.42–1.45) | 0.44 | 0.68 (0.32–1.42) | 0.28 | 0.80 (0.42–1.50) | 0.48 | 1.03 (0.47–2.27) | 0.94 |
| Recessive | NA | NA | NA | NA | NA | NA | NA | NA |
| TIPIN rs8035497 | ||||||||
| Dominant | 1.13 (0.73–1.75) | 0.59 | 1.41 (0.81–2.43) | 0.22 | 0.59 (0.35–1.01) | 0.05 | 0.53 (0.26–1.10) | 0.08 |
| Recessive | 1.96 (1.02–3.75) | 0.04 | 2.02 (0.93–4.42) | 0.09 | 0.21 (0.03–1.52) | 0.09 | 0.20 (0.02–1.57) | 0.06 |
| TIPIN rs11071888 | ||||||||
| Dominant | 0.64 (0.39–1.06) | 0.08 | 0.63 (0.32–1.24) | 0.17 | 1.32 (0.80–2.20) | 0.28 | 0.85 (0.41–1.76) | 0.66 |
| Recessive | 0.46 (0.06–3.30) | 0.43 | 0.42 (0.05–3.57) | 0.38 | 2.01 (0.73–5.58) | 0.17 | 0.62 (0.15–2.54) | 0.50 |
| TIPIN rs28593577 | ||||||||
| Dominant | 0.87 (0.56–1.36) | 0.54 | 1.00 (0.58–1.75) | 0.99 | 0.78 (0.45–1.35) | 0.37 | 0.64 (0.30–1.35) | 0.23 |
| Recessive | 1.52 (0.69–3.33) | 0.29 | 1.42 (0.59–3.40) | 0.45 | 0.24 (0.03–1.74) | 0.13 | 0.21 (0.03–1.67) | 0.07 |
| TIPIN rs11637949 | ||||||||
| Dominant | 1.04 (0.66–1.64) | 0.86 | 1.01 (0.58–1.76) | 0.96 | 1.95 (1.17–3.24) | 0.009 | 1.81 (0.93–3.51) | 0.08 |
| Recessive | 1.87 (0.81–4.32) | 0.14 | 2.16 (0.84–5.55) | 0.14 | 1.38 (0.59–3.21) | 0.45 | 1.10 (0.43–2.82) | 0.84 |
| TIPIN rs6494568 | ||||||||
| Dominant | 1.39 (0.83–2.33) | 0.21 | 0.99 (0.47–2.07) | 0.98 | 0.92 (0.48–1.76) | 0.79 | 2.18 (0.91–5.19) | 0.09 |
| Recessive | NA | NA | NA | NA | NA | NA | NA | NA |
| TIPIN rs12323975 | ||||||||
| Dominant | 0.70 (0.38–1.29) | 0.25 | 0.67 (0.30–1.46) | 0.29 | 1.21 (0.61–2.39) | 0.58 | 0.54 (0.19–1.49) | 0.21 |
| Recessive | NA | NA | NA | NA | NA | NA | NA | NA |
Significant values are indicated in bold characters. TIPIN rs3759786, TIPIN rs6494568, and TIPIN rs12323975 were not assessed with recessive genetic model because there were no patients having homozygous genotype of the recessive allele in these SNPs.
CI confidence interval, HR hazard ratio, NA not assessed, OS overall survival, SNP single nucleotide polymorphism.
aP-values were based on log-rank test.
bP-values were based on Wald test in the multivariate Cox proportional hazards regression model.
Associations of SNPs in TIMELESS and TIPIN with clinical outcomes in the irinotecan cohort
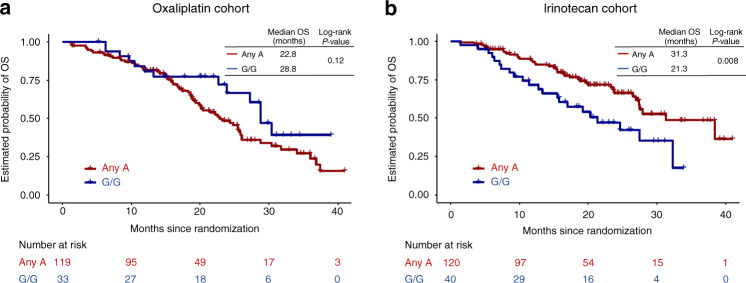
In the irinotecan cohort, univariate and multivariate analyses did not show significant associations between the tested SNPs and tumour response or PFS (Tables 2, 3, Table S4, S5). Conversely, two SNPs exhibited significant associations with OS in univariate analysis univariate (Table 4, Table S4, S5). Specifically, any G allele of TIPIN rs11637949 was linked to worse OS than the A/A genotype using the dominant genetic model (median OS, 23.8 months vs. 32.3 months, HR = 1.95, 95% CI = 1.17–3.24, p = 0.009), and the G/G genotype of TIMELESS rs2291739 was linked to worse OS than any A allele using the recessive genetic model (median OS, 21.3 months vs. 31.3 months, HR = 2.01, 95% CI = 1.19–3.41, p = 0.008, Fig. 1). Multivariate analysis confirmed the significant association between TIMELESS rs2291739 and OS (G/G vs. any A allele, HR = 3.06, 95% CI = 1.49–6.25, p = 0.004, Table 4, Table S5).
Fig. 1. Overall survival of patients TIMELESS rs2291739 variants.
a Oxaliplatin cohort (FOLFOX plus bevacizumab arm). b Irinotecan cohort (FOLFIRI plus bevacizumab arm). Abbreviations: OS, overall survival.
Comparing treatment efficacy between FOLFOX/bevacizumab and FOLFIRI/bevacizumab by TIMELESS rs2291739 genotype
In the patients having any A allele of TIMELESS rs2291739, FOLFIRI/bevacizumab showed significantly better OS (median OS, 31.3 months vs. 22.8 months, HR = 0.56, 95% CI = 0.38–0.83, p = 0.004) and PFS (median PFS, 14.0 months vs. 9.5 months, HR = 0.57, 95% CI = 0.42–0.78, p = <0.001) than FOLFOX/bevacizumab (Fig. S2A, B). In contrast, in the patients harbouring G/G genotype of TIMELESS rs2291739, FOLFIRI/bevacizumab showed worse OS (median OS, 21.3 months vs. 28.8 months, HR = 1.80, 95% CI = 0.90–3.59, p = 0.09) and PFS (median PFS, 9.5 months vs. 13.9 months, HR = 1.62, 95% CI = 0.92–2.85, p = 0.10) than FOLFOX/bevacizumab (Fig. S2C, D).
Treatment-by SNP interaction
In the dominant genetic model, TIPIN rs8035497 had a significant interaction with treatment in terms of OS. In the recessive genetic model, three SNPs exhibited significant interactions with treatment: TIPIN rs8035497 and TIPIN rs28593577 in terms of OS and TIMELESS rs2291739 in terms of both OS and PFS (Table 5).
Table 5.
Treatment-by-SNP interaction test.
| SNP | Dominant genetic model | Recessive genetic model | ||||
|---|---|---|---|---|---|---|
| TR | PFS | OS | TR | PFS | OS | |
| Interaction P | Interaction P | Interaction P | Interaction P | Interaction P | Interaction P | |
| TIMELESS rs2291739 | 0.73 | 0.86 | 0.90 | 0.44 | 0.04 | 0.007 |
| TIPIN rs3759786 | 0.54 | 0.73 | 0.53 | NA | NA | NA |
| TIPIN rs8035497 | 0.34 | 0.07 | 0.02 | 0.87 | 0.55 | 0.01 |
| TIPIN rs11071888 | 0.90 | 0.28 | 0.49 | 0.91 | 0.08 | 0.67 |
| TIPIN rs28593577 | 0.66 | 0.28 | 0.24 | 0.55 | 0.45 | 0.04 |
| TIPIN rs11637949 | 0.61 | 0.99 | 0.15 | 0.12 | 0.73 | 0.49 |
| TIPIN rs6494568 | 0.65 | 0.42 | 0.27 | NA | NA | NA |
| TIPIN rs12323975 | 0.65 | 0.40 | 1.00 | NA | NA | NA |
Significant values are indicated in bold characters. TIPIN rs3759786, TIPIN rs6494568, and TIPIN rs12323975 were not assessed with recessive genetic model because there were no patients having homozygous genotype of the recessive allele in these SNPs.
NA not assessed, OS overall survival, PFS progression-free survival, SNP single nucleotide polymorphism, TR tumour response.
Discussion
Our findings revealed for the first time that genetic variations in the TIMELESS–TIPIN complex are associated with survival outcomes in patients with mCRC treated with irinotecan-based first-line chemotherapy. Our findings highlight the significance of this pathway and its impact on therapeutic efficacy based on the difference in outcomes between irinotecan- and oxaliplatin-based chemotherapies. These data suggest that genes regulating the replication fork in tumour cells are related to the distinct response to cytotoxic agents with different DNA-damaging activities.
We identified a significant association of TIMELESS rs2291739 with OS in patients treated with FOLFIRI plus bevacizumab using the recessive genetic model, and this finding was confirmed in multivariate analyses. The association with PFS was consistent with the OS data, but statistical significance was not reached. Meanwhile, no confirmed associations with clinical outcomes were observed in patients treated with FOLFOX plus bevacizumab. Of note, TIMELESS rs2291739 displayed significant interactions with treatment in terms of both PFS and OS.
Our findings are supported by the consistent mechanisms of action and tumour biology. First, TIMELESS rs2291739 is a missense variant that causes a protein function-altering amino acid change. Second, the molecular mechanisms protecting the replication fork are differently affected by irinotecan and oxaliplatin even though both drugs are DNA-damaging agents. Irinotecan is a topoisomerase I (TOP1) inhibitor that inhibits the dissociation of the TOP1 cleavage complex (TOP1-cc), a transient structure formed in front of the replication fork that permits TOP1 to resolve topological stress [22]. In the presence of irinotecan, stabilised TOP1-cc collides with the replication fork during DNA replication and transcription, resulting in DSBs [19]. TIMELESS was identified as a TOP1-binding factor [23]. Interestingly, a previous study revealed that the TIMELESS–TIPIN complex destabilises TOP1-cc by interacting with TOP1, which prevents the generation of irinotecan-induced DSBs [24]. These data support our findings of an association between a functional SNP of TIMELESS and OS of irinotecan-treated patients. Specifically, patients with the TIMELESS rs2291739 G/G genotype had worse OS than those with any A allele, suggesting that the G/G genotype is associated with increased function of the TIMELESS–TIPIN complex, thereby protecting cancer cells against irinotecan-induced cytotoxicity. Meanwhile, oxaliplatin is a platinum compound forming two types of DNA crosslinks: intra-strand crosslinks on the same strand of DNA and inter-strand crosslinks between the two complementary strands of DNA [18, 25]. Platinum drugs produce a high proportion of intra-strand crosslinks, which contribute to their cytotoxic activity [26]. This type of crosslinks can avoid the development of DSBs if uncoupling activities between DNA polymerase and helicase are properly sensed by the TIMELESS–TIPIN complex in associated with checkpoint mechanisms [13]. Conversely, inter-strand crosslinks are more toxic because they physically block DNA replication and induce DSBs by covalently linking both DNA strands [27]. The repair of inter-strand crosslinks is highly dependent on homologous recombination and Fanconi anaemia pathways [28, 29]. We speculate that these mechanisms of inter-strand crosslinks, which are independent of TIMELESS–TIPIN complex function, attenuated the impact of TIMELESS rs2291739 on clinical outcomes in oxaliplatin-treated patients. Based on these molecular bases, we supposed that the function of the TIMELESS–TIPIN complex is more relevant to the efficacy of irinotecan than to that of oxaliplatin.
Our findings suggest a novel approach of clinical decision making based on the TIMELESS rs2291739 genotype. Specifically, FOLFIRI plus bevacizumab may be a favourable treatment option for the patients with any A allele because our results showed it led to better survival outcomes compared to FOLFOX plus bevacizumab in this patient subset. In contrast, FOLFOX plus bevacizumab may be favourable for the patients with G/G genotype because this patient subset was more likely to benefit from FOLFOX plus bevacizumab than from FOLFIRI plus bevacizumab. This personalised strategy in the choice of backbone chemotherapy should be assessed in further clinical studies.
This study had several limitations. Because of the retrospective design, the results require validation in prospective clinical trials. Furthermore, we tested the association between SNPs and the efficacy of oxaliplatin and irinotecan in one study cohort, however, these data are preliminary and the predictive value of SNPs need to be confirmed. Thus, further validation studies are needed. However, the significant results demonstrated by the formal interaction test in this study support the predictive potential of TIMELESS rs2291739 concerning the selection of cytotoxic agents in the first-line setting.
In conclusion, our study provided the first evidence that germline polymorphisms in replication fork-protecting genes were associated with the efficacy of FOLFIRI plus bevacizumab, but not with that of FOLFOX plus bevacizumab, in patients with mCRC. Our findings may be useful for personalised approaches in selecting cytotoxic drugs in the first-line treatment of mCRC, and validation in prospective studies is warranted.
Supplementary information
Acknowledgements
We thank all patients who contributed to this study.
Author contributions
HA primarily planned, designed, and drafted the manuscript. H-JL supervised and administered the project, and acquired funding. YX and JM did statistical analyses. All authors made substantial contributions to data collection and drafting the manuscript. All authors read and approved the final manuscript.
Funding information
This work was supported by the National Cancer Institute [P30CA 014089 to HJL], Gloria Borges WunderGlo Foundation, Dhont Family Foundation, Daniel Butler Memorial Fund, Victoria and Philip Wilson Research Fund, and San Pedro Peninsula Cancer Guild.
Data availability
The data sets used and analysed during this study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All patients provided informed consent for molecular research prior to study enrollment. The study protocol was approved by the Institutional Review Boards of each participating institution and was conducted in accordance with the tenets of the Declaration of Helsinki, as well as the Good Clinical Practice and REMARK guidelines.
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01592-7.
References
- 1.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability-an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8. doi: 10.1038/nrm2858.. [DOI] [PubMed] [Google Scholar]
- 2.Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol. 2015;10:425–48. doi: 10.1146/annurev-pathol-012414-040424.. [DOI] [PubMed] [Google Scholar]
- 3.Berti M, Cortez D, Lopes M. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat Rev Mol Cell Biol. 2020;21:633–51. doi: 10.1038/s41580-020-0257-5.. [DOI] [PubMed] [Google Scholar]
- 4.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–42. doi: 10.1038/nature05327.. [DOI] [PubMed] [Google Scholar]
- 5.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482.. [DOI] [PubMed] [Google Scholar]
- 6.Lecona E, Fernandez-Capetillo O. Replication stress and cancer: it takes two to tango. Exp cell Res. 2014;329:26–34. doi: 10.1016/j.yexcr.2014.09.019.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartek J, Mistrik M, Bartkova J. Thresholds of replication stress signaling in cancer development and treatment. Nat Struct Mol Biol. 2012;19:5–7. doi: 10.1038/nsmb.2220.. [DOI] [PubMed] [Google Scholar]
- 8.Cleary JM, Aguirre AJ, Shapiro GI, D’Andrea AD. Biomarker-guided development of DNA repair inhibitors. Mol Cell. 2020;78:1070–85. doi: 10.1016/j.molcel.2020.04.035.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mateo J, Lord CJ, Serra V, Tutt A, Balmana J, Castroviejo-Bermejo M, et al. A decade of clinical development of PARP inhibitors in perspective. Ann Oncol. 2019;30:1437–47. doi: 10.1093/annonc/mdz192.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leman AR, Noguchi E. Local and global functions of Timeless and Tipin in replication fork protection. Cell cycle. 2012;11:3945–55. doi: 10.4161/cc.21989.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kemp MG, Akan Z, Yilmaz S, Grillo M, Smith-Roe SL, Kang TH, et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J Biol Chem. 2010;285:16562–71. doi: 10.1074/jbc.M110.110304.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho WH, Kang YH, An YY, Tappin I, Hurwitz J, Lee JK. Human Tim-Tipin complex affects the biochemical properties of the replicative DNA helicase and DNA polymerases. Proc Natl Acad Sci USA. 2013;110:2523–7. doi: 10.1073/pnas.1222494110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–52. doi: 10.1101/gad.1301205.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Errico A, Costanzo V. Mechanisms of replication fork protection: a safeguard for genome stability. Crit Rev Biochem Mol Biol. 2012;47:222–35. doi: 10.3109/10409238.2012.655374.. [DOI] [PubMed] [Google Scholar]
- 15.Mazzoccoli G, Panza A, Valvano MR, Palumbo O, Carella M, Pazienza V, et al. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int. 2011;28:841–51. doi: 10.3109/07420528.2011.615182.. [DOI] [PubMed] [Google Scholar]
- 16.Pasero P, Tourriere H. Overexpression of the Fork Protection Complex: a strategy to tolerate oncogene-induced replication stress in cancer cells. Mol Cell Oncol. 2019;6:1607455. doi: 10.1080/23723556.2019.1607455.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bianco JN, Bergoglio V, Lin YL, Pillaire MJ, Schmitz AL, Gilhodes J, et al. Overexpression of Claspin and Timeless protects cancer cells from replication stress in a checkpoint-independent manner. Nat Commun. 2019;10:910. doi: 10.1038/s41467-019-08886-8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perego P, Robert J. Oxaliplatin in the era of personalized medicine: from mechanistic studies to clinical efficacy. Cancer Chemother Pharmacol. 2016;77:5–18. doi: 10.1007/s00280-015-2901-x.. [DOI] [PubMed] [Google Scholar]
- 19.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977.. [DOI] [PubMed] [Google Scholar]
- 20.Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–79. doi: 10.1083/jcb.201406099.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parikh AR, Lee FC, Yau L, Koh H, Knost J, Mitchell EP, et al. MAVERICC, a randomized, biomarker-stratified, phase II study of mFOLFOX6-bevacizumab versus FOLFIRI-bevacizumab as first-line chemotherapy in metastatic colorectal cancer. Clin Cancer Res. 2019;25:2988–95. doi: 10.1158/1078-0432.CCR-18-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, et al. Repair Topoisomerase I-Mediated DNA Damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/s0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park H, Sternglanz R. Identification and characterization of the genes for two topoisomerase I-interacting proteins from Saccharomyces cerevisiae. Yeast. 1999;15:35–41. doi: 10.1002/(SICI)1097-0061(19990115)15:1<35::AID-YEA340>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 24.Hosono Y, Abe T, Higuchi M, Kajii K, Sakuraba S, Tada S, et al. Tipin functions in the protection against topoisomerase I inhibitor. J Biol Chem. 2014;289:11374–84. doi: 10.1074/jbc.M113.531707.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–90. doi: 10.1016/s1470-2045(01)00454-5.. [DOI] [PubMed] [Google Scholar]
- 26.Kartalou M, Essigmann JM. Recognition of cisplatin adducts by cellular proteins. Mutat Res. 2001;478:1–21. doi: 10.1016/s0027-5107(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 27.Hashimoto S, Anai H, Hanada K. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ. 2016;38:9. doi: 10.1186/s41021-016-0037-9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–80. doi: 10.1038/nrc3088.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Yehoyada M, Wang LC, Kozekov ID, Rizzo CJ, Gottesman ME, Gautier J. Checkpoint signaling from a single DNA interstrand crosslink. Mol Cell. 2009;35:704–15. doi: 10.1016/j.molcel.2009.08.014.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used and analysed during this study are available from the corresponding author on reasonable request.