Abstract
At the end of the 35S rRNA gene within ribosomal DNA (rDNA) repeats in Saccharomyces cerevisiae lies an enhancer that has been shown to greatly stimulate rDNA transcription in ectopic reporter systems. We found, however, that the enhancer is not necessary for normal levels of rRNA synthesis from chromosomal rDNA or for cell growth. Yeast strains which have the entire enhancer from rDNA deleted did not show any defects in growth or rRNA synthesis. We found that the stimulatory activity of the enhancer for ectopic reporters is not observed in cells with disrupted nucleolar structures, suggesting that reporter genes are in general poorly accessible to RNA polymerase I (Pol I) machinery in the nucleolus and that the enhancer improves accessibility. We also found that a fob1 mutation abolishes transcription from the enhancer-dependent rDNA promoter integrated at the HIS4 locus without any effect on transcription from chromosomal rDNA. FOB1 is required for recombination hot spot (HOT1) activity, which also requires the enhancer region, and for recombination within rDNA repeats. We suggest that Fob1 protein stimulates interactions between rDNA repeats through the enhancer region, thus helping ectopic rDNA promoters to recruit the Pol I machinery normally present in the nucleolus.
The transcription of large rRNAs in most eukaryotes takes place in a subnuclear structure, the nucleolus, using a distinct RNA polymerase, RNA polymerase I (Pol I). The ribosomal DNA (rDNA) encoding the large precursor rRNA is tandemly repeated in many copies and constitutes the structurally and functionally essential component of the nucleolus. In addition to the transcription of rDNA, subsequent steps, such as rRNA processing and modification and ribosome assembly, all take place within the nucleolus. Furthermore, several important cellular functions distinct from ribosome synthesis appear to take place in the nucleolus, as exemplified by the recent discovery of the presence of proteins in the nucleolus regulating cell cycle progression in mitosis (reviewed in references7 and 21). In addition, there are unique mechanisms regulating the replication and recombination of the tandemly repeated rDNA genes, and proteins involved in these processes are also expected to be present in the nucleolus. An example is the presence of a replication fork barrier (RFB) near the end of every rRNA transcription unit, which might play a role in preventing collision of the Pol I transcription machinery with the DNA replication machinery (2). Thus, studies on rDNA transcription in vivo may have to deal with nucleolar structures relevant to other nucleolar functions.
In the yeast Saccharomyces cerevisiae, there are ∼150 rDNA tandem repeats located on chromosome XII. Each repeat is ∼9.1 kb in size and contains the large 35S rRNA gene transcribed by Pol I and the small 5S rRNA gene transcribed by Pol III (Fig. 1). This study examines the function of the Pol I enhancer element, which was originally identified by Elion and Warner (5, 6). These workers reported that a 190-bp DNA element just distal to the 35S rRNA coding region (shown in Fig. 1A) gave a large stimulation of Pol I transcription in experiments using an rRNA reporter gene carried on a CEN plasmid. Hence, this element is called the enhancer of Pol I activity. Subsequently, many papers on the enhancer were published, generally confirming the Pol I stimulatory activity using various reporter systems, but as to the exact DNA sequence element within this enhancer region, different investigators reported somewhat different conclusions (39). In addition, while the enhancer activity was even reported to be observed in vitro (29), the mechanism of stimulation of Pol I activity by the yeast enhancer and its physiological significance have remained unclear.
FIG. 1.
(A) Structure of rDNA repeats in S. cerevisiae. One repeat unit of rDNA is 9,137 bp and is numbered with respect to the Pol I transcription start site (+1). By convention, one repeat unit is shown as the fragment obtained after digestion of rDNA with SmaI, between positions 8931 and 8932. The locations of the 35S and 5S rRNA genes (black bars; the direction of transcription indicated by arrows), the two nontranscribed spacer regions (NTS1 and NTS2), and enhancer and RFB regions are shown. The orientation with respect to the centromere (CEN) and telomere (TEL) is indicated. Although the enhancer studied by Elion and Warner (5, 6) is the ∼190-bp EcoRI (6743)-HindIII (6931) region, subsequent studies on the HOT1 system showed that the ∼130-bp HindIII (6931)-HpaI (7060) region (called the RFB region [see the text]) is also required for efficient Pol I transcription in this system, and the 320-bp EcoRI-HpaI region was called the E element. Following this nomenclature, we define the enhancer and the E element as shown in the figure, even though the entire E element may represent an enhancer. The three restriction sites used to define these regions are indicated. Other EcoRI, HindIII, and HpaI sites are not shown. The I element required for the HOT1 activity is also indicated. In addition, restriction sites for XbaI (X) and the fragment used as a probe (P) (thin black bar) relevant to experiments shown in Fig. 2B are indicated. (B) Structures of pNOY373 (pPol I), pNOY454 (an E deletion [ΔE] derivative of pNOY373), and pNOY130 (pPol II). (C) PCR analysis of genomic DNA showing that the E element was deleted in rdnΔΔ strain NOY906. DNA was isolated from strains NOY505 (wild type [WT]), NOY903 (ΔΔpPol I), NOY906 (ΔΔpPol IΔE), and NOY892 (ΔΔpPol II) and subjected to PCR analysis using two primers outside the E element as indicated by arrowheads 1 and 2 in panel A. The NOY505 and NOY903 genomic DNA samples (lanes 1 and 2) yielded the 1.1-kb PCR product. The NOY906 sample (lane 3) yielded the 0.8-kb PCR product, confirming the absence of the E element. As references, plasmids pNOY373 (pPol I), pNOY454 (pPol IΔE), and pNOY130 (pPol II) were also subjected to PCR analysis (lanes 6, 7, and 8, respectively). As expected, pPol II plasmid (lane 8) and rdnΔΔ strain NOY892 (lane 4) did not yield any PCR product, because they did not contain DNA sequence corresponding to primer 2. Lane 5 contains size markers (M).
Stimulation of Pol I transcription by the enhancer was also observed in connection with HOT1 activity in yeast. HOT1 was originally identified as a DNA element that stimulates genetic recombination at nearby regions when inserted at a non-rDNA site (11). HOT1 activity requires the I element, which corresponds to the rDNA promoter, and the E element, which includes the enhancer studied by Elion and Warner (5, 6) and the adjacent 130-bp region (the RFB region [Fig. 1A]). Extensive studies have demonstrated that the E element greatly stimulates transcription originated from the I element and that the resultant high level of transcription is correlated with the stimulation of recombination (9, 32). The 130-bp region of the E element was not studied by Elion and Warner (5, 6) for stimulation of Pol I transcription but was shown to be required for both Pol I stimulation and HOT1 activities (nomenclature for DNA elements with the enhancer activity used in this paper discussed in the legend to Fig. 1). This 130-bp region overlaps the region containing the RFB site(s) mentioned above, which allows progression of DNA replication in the direction of 35S rDNA transcription, but not in the opposite direction (3, 13). Because pausing of a replication fork is known, at least in bacterial systems, to stimulate both DNA double-strand breakage and genetic recombination (reviewed recently in reference 26), replication fork pausing at the RFB site could explain stimulation of recombination in the HOT1 system. In fact, based on their discovery of the FOB1 gene, which is required for both RFB activity and HOT1 activity, this model was proposed by Kobayashi and Horiuchi (14) as an alternative to the original explanation of HOT1 activity being solely a consequence of high-level transcription from the Pol I promoter. However, it was recently demonstrated that HOT1 activity can take place in the absence of replication fork blocking (38). Thus, the reason why both HOT1 and RFB activities require FOB1 has become a challenging question.
In this paper, we first describe our experiments examining the proposed function of the Pol I enhancer directly by measuring synthesis of rRNA and its function to support cell growth, rather than using reporters fused to the Pol I promoter as was done previously. We found that, contrary to the general belief, the enhancer is not directly involved in rDNA transcription by Pol I, and that deletion of the entire enhancer element from the yeast genome does not produce any significant negative effects on rDNA transcription or cell growth relative to those of control strains with the enhancer. We then describe experiments designed to explain why earlier experiments using various reporter systems led to the conclusion that the enhancer element stimulates transcription. Our results demonstrate that the enhancer element is required in ectopic reporter systems to recruit Pol I and Pol I-specific factors, which are localized mostly in the nucleolus, and that FOB1 plays an important role in this process. Some relevant earlier observations such as the requirement of FOB1 for HOT1 activity are discussed in light of these new findings.
MATERIALS AND METHODS
Media, strains, plasmids, and genetic methods.
YEPD medium contains 1% yeast extract, 2% peptone, and 2% glucose. YEPGal medium is the same as YEPD medium except 2% galactose was substituted for glucose. Synthetic glucose (SD) medium (2% glucose, 0.67% yeast nitrogen base) was supplemented with tryptophan and required bases as described by Sherman et al. (30) and is called SD complete medium. Synthetic galactose (SGal) medium is the same as SD medium except 2% galactose was substituted for glucose. Cells were grown at 30°C.
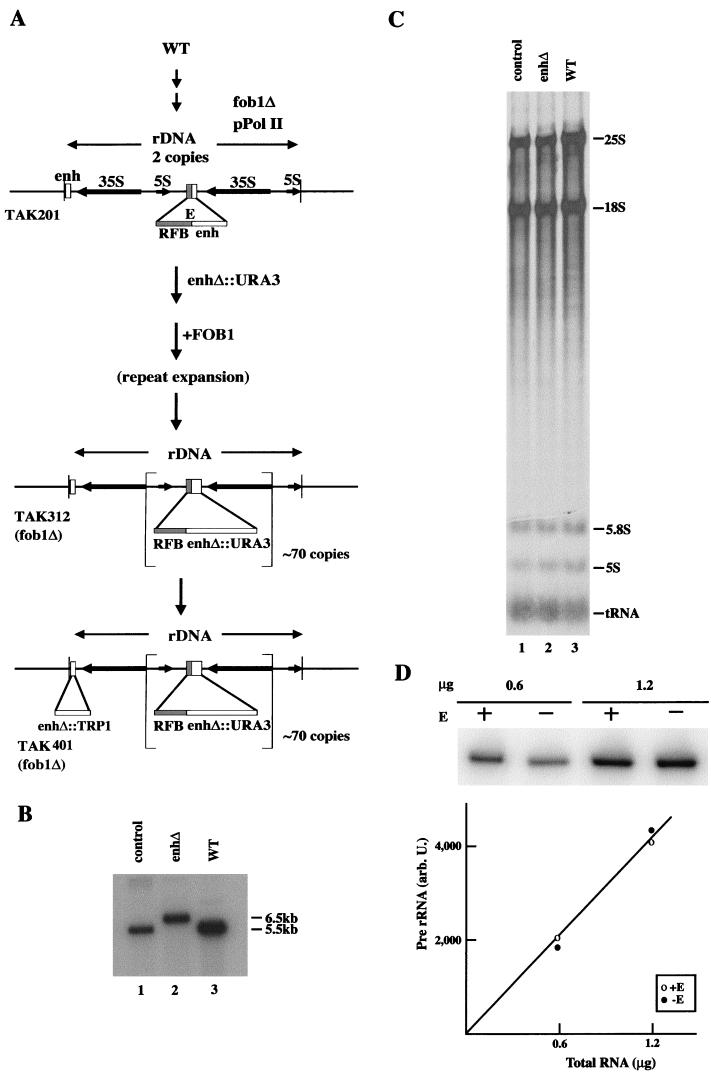
The yeast strains and plasmids used in this study are described in Table 1. Plasmid pNOY454 carrying an E-element deletion was constructed from pNOY373. This plasmid carries the XhoI site (CTCGAG; positions 6738 to 6743) and NotI site (CGGCCG; positions 7068 to 7073) flanking the E element (37). The plasmid was digested with XhoI and NotI, blunt ended with T4 DNA polymerase, and ligated with T4 DNA ligase. In the resulting plasmid, pNOY454, the entire E element has been deleted and the XhoI site has been regenerated. Deletion of the E element in pNOY454 and in strain NOY906 was confirmed by PCR analysis with a primer set, primers 1 and 2. Primer 1, GGGTACTGGCAGGAG, hybridizes 76 bp upstream of the E-element EcoRI site at position +6744. Primer 2, TTTGGATCCGAGTAGTGTAGTGGGTGAC, hybridizes 762 bp downstream of the E-element HpaI site at position +7062 (Fig. 1A). Construction of strain TAK201, which has all the rDNA repeats deleted except for two copies of the 35S rRNA gene (Fig. 2), was described previously (15) together with construction of strains TAK312 and TAK314. An outline of the method is included in Fig. 2A. TAK312 carries approximately 70 rDNA repeats, in which the enhancer is deleted from all copies except for one at the centromere-proximal boundary (Fig. 2A). This last copy was replaced with TRP1 by transformation using a DNA fragment which carries the TRP1 gene flanked by two DNA sequences, one ∼500 bp downstream and the other ∼300 bp upstream of this last enhancer copy (190-bp EcoRI-HindIII). TAK401 is one of Trp+ transformants obtained in this way, whose structure is shown in Fig. 2A. Deletion of the last enhancer was confirmed by PCR using two primers flanking this enhancer. Like TAK312, TAK401 also carries approximately 70 rDNA repeats as judged by contour-clamped homogeneous electric field electrophoresis. Strains TAK320, TAK321, TAK322, and TAK323 were constructed as described previously (14). The HOT1 DNA, which consists of the ∼250-bp SmaI-EcoRI fragment (I element) and the ∼320-bp EcoRI-HpaI fragment (E element) (32), is inserted within the BIK1 gene, which is adjacent to HIS4, in TAK320 and TAK321. The HOT1 recombination system constructed in this work was similar to that used by Voelkel-Meiman et al. (35). Stimulation of recombination by HOT1 was confirmed by measuring the frequency of Ura− cells using SD complete medium with and without 5-fluoroorotic acid (5-FOA) as described previously (14). Strains NOY1003 and NOY1004 were constructed from TAK320 and TAK322, respectively, by replacing the RAD52 coding region by a DNA containing LEU2.
TABLE 1.
Yeast strains and plasmids used in this study
| Strain or plasmid | Description |
|---|---|
| Strains | |
| NOY505 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 |
| NOY891 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY353 |
| NOY892 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY130 |
| NOY903 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY130 and pNOY373 |
| NOY906 | MATaade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rdnΔΔ::HIS3 carrying pNOY130 and pNOY454 |
| NOY907 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY130 and YEp351 |
| NOY908 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY373 |
| NOY995 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 carrying pNOY130 |
| NOY999 | MAT? ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 fob1Δ::LEU2 |
| NOY1003 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rad52Δ::LEU2 HIS4 his4Δ::URA3 bik1Δ::HOT1 (Fig. 5A) |
| NOY1004 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rad52Δ::LEU2 HIS4 his4Δ::URA3 (Fig. 5A) |
| TAK201 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔ-2 fob1Δ::HIS3 carrying pNOY353 (Fig. 2A) |
| TAK312 | Same as TAK201, but all of the expanded rDNA repeats carry the enhΔ::URA3 mutation and do not carry any plasmid (Fig. 2A) |
| TAK314 | Same as TAK312, but the expanded rDNA repeats do not carry the enhΔ::URA3 mutation |
| TAK320 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 HIS4 his4Δ::URA3 bik1Δ::HOT1 (see Fig. 5A) |
| TAK321 | Same as TAK320 but fob1Δ::HIS3 |
| TAK322 | Same as TAK320 but BIK1 (no HOT1 insertion) |
| TAK323 | Same as TAK322 but fob1Δ::HIS3 |
| TAK401 | Same as TAK312 but one enhancer remained at the centromere-proximal boundary is replaced by TRP1 (see Fig. 2A) |
| Plasmids | |
| YEp351 | High-copy-number plasmid carrying LEU2, 2μm, amp (8) |
| rR10 | A derivative of YCp50 carrying URA3, CEN, amp, and a rDNA repeat in which the majority of the 35S coding region is replaced by T7 DNA (5) (Fig. 4A) |
| rR8 | Same as rR10, but the enhancer is absent (5) (Fig. 4A) |
| pNOY130 | A derivative of high-copy-number plasmid pNOY102 (18) carrying GAL7-35S rDNA, 5S rDNA, URA3, 2μm, amp (37) (Fig. 1B) |
| pNOY353 | A derivative of high-copy-number plasmid pTV3 (25) carrying GAL7-35S rDNA, 5S rDNA, TRP1 2μm, amp (19) |
| pNOY373 | A derivative of high-copy-number plasmid YEp351 carrying rDNA with promoter starting from −206 and with the XhoI-NotI flanked E element, LEU2, 2μm, amp (37) (Fig. 1B) |
| pNOY454 | A derivative of pNOY373 with the E element deleted (Fig. 1B) |
FIG. 2.
(A) Construction of strain TAK401, in which the enhancer was deleted from all rDNA repeats. Strain TAK201 was constructed from the wild-type (WT) strain. Strain TAK201 has two copies of rDNA covering the 5S-NTS2–35S region and a single copy of the intact NTS1 between the two copies of rDNA. The E-element region is shown as expanded. The enhancer element was replaced by a URA3 DNA segment, and repeat expansion was initiated by introduction of FOB1. The structure of rDNA repeats in strain TAK312 constructed in this way is shown. TAK401 was then constructed by replacing one remaining enhancer copy with TRP1 as shown in the figure. (B) Southern analysis of DNA from strains TAK314, TAK401, and NOY999. DNA was isolated from TAK314, TAK401, and NOY999 (lanes 1, 2, and 3, respectively), digested with XbaI, subjected to agarose gel electrophoresis, and probed with the radioactive DNA shown in Fig. 1A (thin black bar labeled P). An autoradiogram is shown. TAK314 (control) and NOY999 (WT) carry intact 9.1-kb rDNA repeats which yielded the expected 5.5-kb XbaI fragment detected by the probe. TAK401 (enhΔ) carries ∼70 copies of 10.1-kb repeats which yielded the expected 6.5-kb XbaI band. No band corresponding to 5.5 kb was observed, indicating that all the expanded repeat unit contained the enhΔ::URA3 mutation. (C) Accumulation of [14C]uracil-labeled RNA in the three strains analyzed in panel B, strains TAK314, TAK401, and NOY999 (lanes 1, 2, and 3, respectively). Cultures growing exponentially in SD complete medium containing 5 μg of uracil per ml were diluted to a cell density giving an A600 of 0.07 using the same medium containing [14C]uracil (0.1 μCi/ml) and incubated for 5 h (to an A600 of approximately 0.7). The amounts of total [14C]uracil-labeled RNA accumulated per cell density were the same for the three strains within experimental errors (data not shown). RNA was then isolated, and equal amounts of radioactive RNA were analyzed by acrylamide-agarose gel electrophoresis. An autoradiogram is shown. (D) Primer extension analysis of rDNA transcription. RNA was prepared from TAK314 (+E) and TAK401 (−E), and 0.6 and 1.2 μg were used to determine the amounts of the 5′ end of 35S rRNA as indicated. Radioactive bands visualized by a PhosphorImager are shown in the gel, and the results of quantification (in arbitrary units) are shown in the graph.
Other procedures.
Analysis of RNA synthesis by [3H]uridine or [14C]uracil incorporation was performed as described previously (18). Northern analysis of RNA and Southern analysis of DNA were done by standard procedures (27). T7 reporter transcripts from plasmids YCprR8 (rR8) and YCprR10 (rR10) were detected with a 32P-labeled antisense riboprobe prepared with pSP-T7(+) by the method of Johnson and Warner (10). Analysis of the 5′ end of 35S rRNA by primer extension was performed by using a 32P-labeled primer which hybridizes to the 35S rRNA external transcribed spacer as described previously (36). Analysis of transcript from the Pol I promoter in the HOT1 system was done with the primer described by Stewart and Roeder (32) which is complementary to chromosomal sequences downstream from the site of HOT1 insertion. Autoradiograms were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
Enhancer deletion in rdnΔΔ strains.
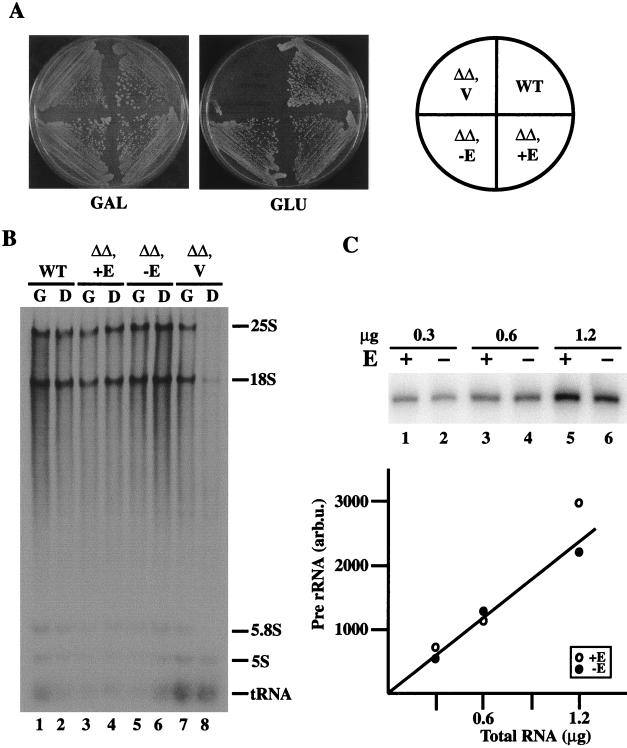
We have previously constructed yeast strains in which all the chromosomal rDNA repeats had been deleted (which we will call rdnΔΔ here). Growth is supported by a helper plasmid carrying, in addition to the 5S rRNA gene, either the 35S rRNA gene transcribed from the native promoter by Pol I (Pol I plasmid or pPol I) or the 35S rRNA gene fused to the GAL7 promoter for transcription by Pol II (Pol II plasmid or pPol II). This system has made it possible to assess the expression of rDNA by measuring the actual rRNA synthesized and its ability to support cell growth, rather than by using reporter mini-rDNA genes (19, 37). Two Pol I plasmids, one with the 320-bp E element (pNOY373) and one without the 320-bp E element (pNOY454), were constructed (Fig. 1B) and introduced into rdnΔΔ strain NOY892, which carries a Pol II plasmid (pNOY130). The growth of NOY892 without the Pol I plasmid (or with only the control vector plasmid) can take place only on galactose, but not on glucose, because cell growth depends on the synthesis of 35S rRNA from the GAL7-35S rDNA fusion gene on the Pol II plasmid (Fig. 3A). As expected, introduction of the control Pol I plasmid, pNOY373, allowed the strain to grow on glucose. To our initial surprise, the Pol I plasmid with the E-element deletion (ΔE) also allowed growth on glucose (Fig. 3A). No differences in colony size (Fig. 3A) or growth rate measured in liquid culture (in YEPD or YEPGal medium) for the two strains were observed (Table 2). We measured plasmid copy numbers in these strains and found no significant difference between the two strains (70 to 100 copies per genome in both strains). We have also confirmed the complete absence of the E element in DNA from the strain carrying the ΔE Pol I plasmid by PCR using two primers flanking the E element (Fig. 1A and C).
FIG. 3.
(A) Growth of rdnΔΔ strains in the presence of helper plasmids with and without the E element. Strains NOY995 (wild type [WT]), NOY903 (ΔΔ, +E), NOY906 (ΔΔ, −E), and NOY907 (vector) (ΔΔ, V) were grown on YEPGal medium (GAL) at 30°C for 5 days and on YEPD medium (GLU) at 30°C for 2 days. (B) Polyacrylamide-agarose gel electrophoresis of RNA synthesized in the strains shown in panel A. Cells were grown in SGal complete medium. At a cell density (A600) of 0.2, the cultures were divided: one part was shifted to SD complete medium (D) and the other was kept in the same medium (G). One hour after the shift, cells were labeled with [3H]uridine (162 mCi/mg; 50 μCi/ml) for 1 h. RNA was isolated from each culture, and samples containing approximately equal radioactivities were subjected to electrophoresis. An autoradiogram of the gel is shown. (C) Primer extension analysis of rDNA transcription. RNA was prepared from strains NOY903 (rdnΔΔ) carrying pNOY373 (+E) and NOY906 (rdnΔΔ) carrying pNOY454 (−E), and 0.3, 0.6, and 1.2 μg were used to determine the amounts of the 5′ end of 35S rRNA as indicated. Radioactive bands visualized by a PhosphorImager are shown in the gel, and the results of quantification (in arbitrary units) are shown in the graph.
TABLE 2.
Growth rates and RNA accumulation rates of rdnΔΔ strains carrying helper Pol I plasmids with or without the E element and a control RDN1 straina
| Strain | Pol I plasmidb | Galactose
|
Glucose
|
||
|---|---|---|---|---|---|
| RNA (%) | DT (h) | RNA (%) | DT (h) | ||
| NOY995 (RDN1) | − | 100 | 2.2 | 100 | 1.7 |
| NOY903 (rdnΔΔ) | +E | 54 ± 10 | 2.7 | 60 ± 7 | 2.0 |
| NOY906 (rdnΔΔ) | −E | 68 ± 6 | 2.6 | 77 ± 2 | 2.0 |
| NOY907 (rdnΔΔ) | V | 26 ± 4 | 6.5 | 12 ± 2 | |
In the [3H]uridine labeling experiment shown in Fig. 3, aliquots were taken 1 h after labeling and RNA was precipitated with trichloroacetic acid. The amounts of 3H-labeled RNA were measured and are expressed as percentages of the values obtained with the control strain, NOY995. The values obtained from this experiment and two additional experiments are averaged and shown in the table. Growth rates of the four strains were measured in media containing galactose (YEPGal) and glucose (YEPD) and are shown as doubling time (DT) (in hours).
−, no Pol I plasmid; +E and −E, Pol I plasmid with and without the E element, respectively; V, vector.
We next compared the rate of accumulation of rRNA by [3H]uridine incorporation in synthetic glucose medium. Although total [3H]uridine incorporation in these rdnΔΔ strains was reduced somewhat relative to that in the wild-type strain which does not have the rdn deletion (consistent with a small reduction in their growth rates), no further decrease due to the E-element deletion was observed (Table 2). (In fact, total [3H]uridine incorporation was slightly higher [∼28%] in the E-element deletion strain. The significance of this slight increase was not studied.)
Analysis of radioactive RNAs by gel electrophoresis demonstrated that there was no significant difference in the synthesis of rRNAs relative to that of total RNA in the two rdnΔΔ strains, one with and one without the E element (Fig. 3B, lanes 4, 6, and 8). The amounts of the unstable 5′ end of 35S precursor rRNA, which reflect rDNA transcription rates by Pol I, in the cells were also measured by primer extension. Again, no significant difference was observed between the two rdnΔΔ strains (Fig. 3C). We conclude that the absence of the E element on helper Pol I plasmid in rdnΔΔ strains causes no significant decrease in rDNA transcription rate and does not lead to a decrease in the growth rate of the cells.
Growth and rDNA transcription in a strain in which the enhancer is removed from all the chromosomal rDNA repeats.
In connection with analyses of cis elements required for chromosomal rDNA repeat expansion, we constructed a yeast strain in which most of the rDNA repeats are deleted, leaving two copies of rDNA covering the 5S-NTS2–35S region and a single copy of the intact NTS1 between the two copies. Growth of this strain is supported by the presence of the helper Pol II plasmid pNOY353 (15) (Fig. 2A). This strain is made fob1 by deletion to prevent FOB1-dependent repeat expansion. Thus, it was possible to perform mutagenesis of the intact NTS1 region first and then expand the rDNA containing the desired mutation by introduction of FOB1 on a plasmid by transformation. It was shown that the RFB region is essential for repeat expansion whereas the enhancer is not essential but has a stimulatory role. Substitution of the 190-bp EcoRI-HindIII corresponding to the enhancer with a URA3 fragment allowed repeat expansion, although the rate of expansion was reduced. Strain TAK312 was obtained in this way and was shown to have approximately 70 copies of the expanded rDNA repeats with the coamplified URA3 fragment replacing the enhancer (15) (Fig. 2A). Because there is a single enhancer copy present at the centromere-proximal boundary of the rDNA and this copy was not deleted in strain TAK312, strain TAK401 was constructed from TAK312, replacing this last copy of the enhancer with TRP1 (Fig. 2A). Control strain TAK314 was constructed following the same steps but without the mutational substitution of the enhancer and had approximately the same number (∼70) of rDNA repeats. Following the rDNA repeat expansion, loss of the FOB1-containing plasmid was selected in order to stabilize rDNA repeat numbers. The helper Pol II plasmid was spontaneously lost because the chromosomal rDNA repeat expansion allowed these strains to grow in its absence. The substitution of the enhancer by URA3 in all the expanded rDNA repeats in TAK312 was demonstrated previously (15) and was confirmed by a different Southern analysis of genomic DNA prepared from TAK401 in this study (Fig. 2B).
Strain TAK401 essentially had the same growth rate and rRNA accumulation rate as the control strain (TAK314) (Fig. 2C and other data not shown). The rates of rDNA transcription in TAK401 and TAK314 were also compared by measuring the amounts of the unstable 5′ end of 35S rRNA by primer extension, as was done in the experiment shown in Fig. 3C. No significant difference was observed between the two strains, TAK401 and TAK314 (Fig. 2D). These results are consistent with the conclusion obtained using the rdnΔΔ strains described above, and it appears that the enhancer element is not required for rDNA transcription even in the context of the normal tandemly repeated rDNA structure on chromosome XII.
Plasmid Pol I reporter genes with and without the enhancer.
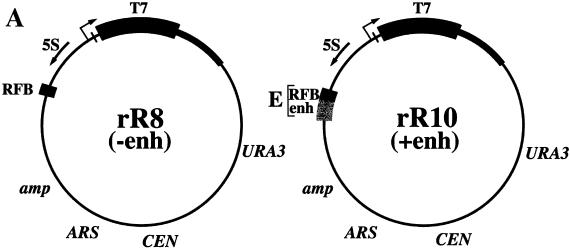
We obtained two CEN/ARS plasmids carrying the Pol I reporter genes used in the original Pol I enhancer experiments (5, 6), rR8 and rR10, from J. R. Warner. These plasmids carry a fragment of Escherichia coli phage T7 DNA fused to the rDNA promoter together with the upstream NTS1–5S-NTS2 region. Plasmid rR10 retains the intact NTS1, but plasmid rR8 has the enhancer element deleted (Fig. 4A). The two plasmids were individually introduced into our standard wild-type strain NOY505 by transformation. Several independent Ura+ transformants obtained after introduction of rR8 and those obtained after introduction of rR10 were examined. The expression of the T7 reporter was analyzed by Northern blotting (see the legend to Fig. 4 for details).
FIG. 4.
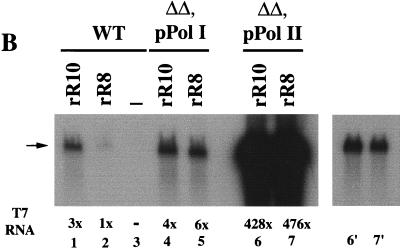
(A) Structures of plasmids rR8 and rR10. The Pol I start site of the 35S rRNA gene (+1) is shown by a bent arrow, and the 5S rRNA gene is shown by an arrow. T7 reporter DNA is shown as a black bar which is followed by the distal part of the 25S rRNA coding region (thick line). The RFB, enhancer (enh), and E element are indicated. The sizes of these DNA elements are not to scale. (B) Expression of the T7 Pol I reporter RNA in strains NOY505 (wild type [WT]), NOY908 (ΔΔ, pPol I), and NOY891 (ΔΔ, pPol II) carrying plasmid rR8 or rR10. RNA was prepared from cells growing exponentially in uracil-deficient SGal medium, and equal amounts of RNA were subjected to Northern analysis first using a 32P-labeled actin probe (not shown), followed by stripping of the probe and reprobing using a 32P-labeled T7 RNA probe. DNA was isolated from the same cultures, and plasmid copy numbers were estimated by Southern analysis using a 32P-labeled URA3 probe and by comparing plasmid URA3 signals to chromosomal URA3 signals. The amounts of T7 reporter RNA (indicated by the arrow) were quantified by PhosphorImager analysis, and relative amounts obtained after normalization to the amounts of actin mRNA and correction for copy number differences are given in the figure. Lane 3 is a sample obtained from NOY505 (WT) without any plasmid (−). The autoradiogram shown for lanes 1 to 7 was exposed for 18 h. Lanes 6′ and 7′ correspond to lanes 6 and 7, respectively, and are from the autoradiogram after 8 min of exposure.
Although a significant degree of variation was observed among independent transformants for both the rR8 and rR10 transformation sets (discussed further below), it was clear that plasmid rR10, which carries the enhancer, showed greater expression of the T7 reporter than rR8 (Fig. 4B, lanes 1 and 2). Using averages of the values for 10 independent transformants for each plasmid, it was calculated that the reporter expression from rR10 was approximately fivefold higher than that from rR8. The results confirm the original results obtained by Elion and Warner (5, 6) but are in apparent conflict with the conclusion described above, namely, the absence of a requirement of the enhancer for the transcription of the intact 35S rRNA. To resolve this discrepancy, we introduced plasmids rR8 and rR10 individually into rdnΔΔ strains, one carrying a helper Pol I plasmid (NOY908) and the other carrying helper Pol II plasmid (NOY891).
Although there was some variability in the expression of the T7 reporter gene among individual transformants derived from the rdnΔΔ strain carrying pPol I, there was no significant difference between the rR8 and rR10 transformant groups, and the expression in those transformants (both rR8 and rR10) was in general higher than in rR10 transformants derived from the wild-type strain (Fig. 4B, lanes 4 and 5). When rR8 or rR10 was introduced into the rdnΔΔ strain carrying pPol II , the reporter T7 expression was strikingly high in both rR8 and rR10 transformants (Fig. 4B, lanes 6 and 7), and there was essentially no significant variability among independent transformants. Reporter T7 expression was nearly 100-fold higher than that in rR8 or rR10 transformants derived from a rdnΔΔ strain carrying pPol I. There was no significant difference in the reporter expression for rR8 and rR10.
FOB1 is required for Pol I transcription in a HOT1 system at the HIS4 region.
Another reporter system where stimulation of Pol I transcription by the enhancer has been well studied is the HOT1 system constructed at the HIS4 region on chromosome III (9, 32). A requirement for the E element in both Pol I transcription and stimulation of recombination has been clearly demonstrated. In addition, it is known that HOT1 activity, i.e., stimulation of recombination, requires FOB1. Thus, we considered the possibility that stimulation of Pol I activities in such a system, which utilizes a reporter promoter present outside the nucleolus, may require FOB1.
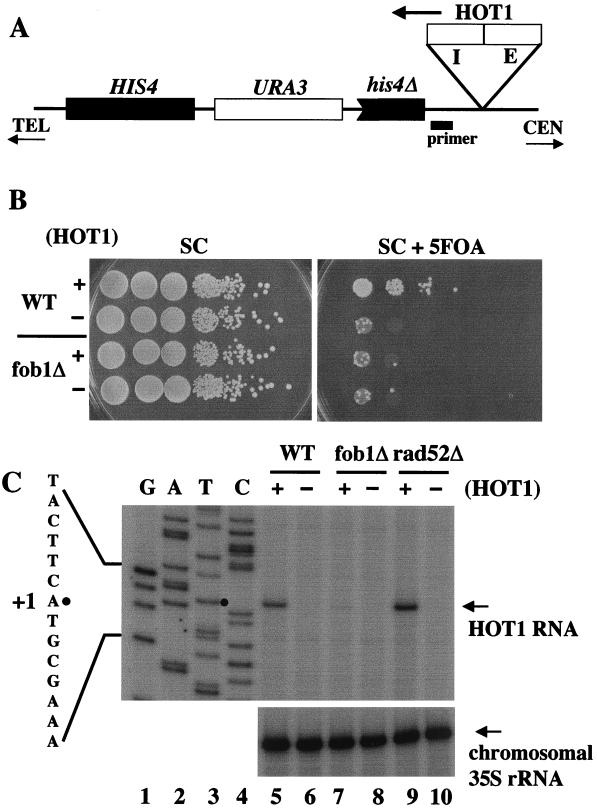
We constructed a HOT1 system at the HIS4 locus of chromosome III which is similar to the system used by Roeder and coworkers in their studies on HOT1 stimulation of recombination (32, 35). This system consists of a direct repeat of (truncated) HIS4 DNA which was inserted downstream of the HOT1-initiated transcription site as shown in Fig. 5A. In this system, recombination between the HIS4 repeats is measured by loss of the intervening URA3 marker, resulting in resistance to 5-FOA. As shown in Fig. 5B (and other results not shown), the frequency of recombination between direct repeats was increased by the presence of the HOT1 DNA by 20- to 100-fold. This HOT1-dependent stimulation of recombination was abolished by a deletion of FOB1, confirming the previous observations (14).
FIG. 5.
Examination of HOT1 recombination activity and HOT1-promoted transcription at the HIS4 region. (A) HOT1-dependent recombination system used. The location of the HOT1 segment consisting of the Pol I promoter (I) and the E element is indicated, as is the location of the primer used for primer extension to measure HOT1 RNA. The orientation with respect to the centromere (CEN) and telomere (TEL) is shown by arrows. (B) HOT1-dependent stimulation of recombination between two tandemly repeated H1S4 DNA sequences. Strains TAK320 (HOT1, +), TAK322 (no HOT1 insertion, −), TAK321 (fob1Δ; HOT1, +), and TAK323 (fob1Δ; no HOT1 insertion, −) were grown in SD complete medium (SC) lacking uracil. The frequencies of Ura− recombinants were then determined by spotting aliquots of 10-fold serial dilutions of the cultures on SC with and without 5-FOA. (C) The four strains analyzed in panel B, together with two rad52 strains with the HOT1 insertion (NOY1003) (+) and without the HOT1 insertion (NOY1004) (−) were grown in SC medium lacking uracil, and RNA was isolated. Equal amounts of RNA (20 μg for HOT1 RNA and 2 μg for chromosomal 35S rRNA) were used for primer extension analyses using the primer indicated in panel A to measure the amount of RNA (HOT1 RNA) initiated at the Pol I promoter inserted at the HIS4 region (top gel) and using another primer to measure the amounts of the 5′ end of 35S rRNA transcribed from the chromosomal rDNA repeats (bottom gel). The primer extension products were analyzed by gel electrophoresis adjacent to products (lanes 1 to 4) obtained by dideoxy-chain termination sequencing reactions with the same primer using DNA obtained from TAK320. The sequence shown is complementary to the sequence read from the sequencing reaction and corresponds to positions −6 to +8, indicating that the initiation site of HOT1 RNA is identical to the 35S rRNA start site (the A at position +1 indicated by a small solid circle). We note that the amount of HOT1 RNA in the rad52 mutant (lane 9) was approximately twofold higher than that in the control FOB1 strain (lane 5). The significance of this observation is not known.
We then performed primer extension experiments to measure the amounts of transcripts specifically initiated from the Pol I promoter in the HOT1 system. The transcript initiating at position +1 of the Pol I initiation site was detected in the FOB1 genetic background (Fig. 5C, top gel, lane 5). In contrast, only a very small amount (∼3% relative to FOB1) of the corresponding transcript was observed for the HOT1 system in the fob1 mutant (Fig. 5C, lane 7). Unstable 5′-end transcripts from the Pol I promoters in the chromosomal rDNA were also analyzed by primer extension using the same RNA samples. As expected from the growth rates of these strains, which were approximately the same, no significant effect of the fob1 mutation was observed (Fig. 5C, bottom gel, lanes 5 to 8). We conclude that FOB1 is specifically required for increased transcription initiation from the Pol I promoter associated with the E element located at the HIS4 region.
FOB1-dependent Pol I transcription in the HOT1 system does not require RAD52.
FOB1 is known to be required for efficient recombination within rDNA repeats (4, 12). Therefore, there was the possibility that the HOT1 element, the Pol I promoter plus the E element, on chromosome III was integrated into the rDNA repeats on chromosome XII using homologous recombination, which could be the basis for the requirement of FOB1 for efficient Pol I transcription in the HOT1 system. Such a scenario seems unlikely, since a homologous recombination between the two chromosomes would yield a dicentric chromosome and an acentric chromosome due to the orientation of the HOT1 element used in the present experiments (Fig. 1A and 5A). In addition, examination of the HIS4 gene by fluorescence in situ hybridization using a specific HIS4 probe showed no significant difference in the location of the HIS4 gene with respect to the nucleolus in strains with and without the HOT1 element at the HIS4 region; in a majority of cells, the HIS4 gene was localized outside the nucleolus in both types of strains (M. Oakes and M. Nomura, unpublished data). Nevertheless, the possibility of recombinational events between the two chromosomes in a small fraction of cells could not be excluded. Therefore, we examined whether FOB1-dependent Pol I transcription in the HOT1 system required RAD52. The RAD52 gene product is required for most homologous recombinational events (reviewed in reference 31) including recombination within the rDNA repeats that leads to production of extrachromosomal rDNA circles (20) and rDNA repeat expansion (T. Kobayashi, unpublished data). As shown in Fig. 5C (compare lane 9 to lane 5; for quantitative comparison, see the figure legend), deletion of RAD52 did not decrease the transcription from the Pol I promoter in the HOT1 system. Thus, we conclude that the E-element (enhancer)-dependent Pol I transcription in the HOT1 system does not involve RAD52-mediated homologous recombination; that is, the FOB1-dependent Pol I transcription in this system appears to take place almost certainly without integration of the HOT1-HIS4 region into the chromosomal rDNA repeats.
DISCUSSION
The yeast Pol I enhancer is dispensable for Pol I transcription.
The experiments described in this paper demonstrate that the yeast Pol I enhancer, which has been assumed to be important for rDNA transcription, is completely dispensable for rDNA transcription and cell growth. First, rdnΔΔ strains with all chromosomal rDNA deleted and carrying a helper Pol I plasmid did not show any significant differences in rRNA synthesis rate or cell growth rate, regardless of whether the single rDNA repeat on the plasmid had a complete deletion of the enhancer (together with the adjacent RFB region). Second, a yeast strain was constructed in which approximately 70 copies of an rDNA unit were tandemly repeated at the natural RDN1 locus of chromosome XII, but all the enhancer elements were deleted, a terminal copy replaced by a TRP1 fragment and all others replaced by a URA3 DNA fragment. No significant difference in rRNA synthesis rate or cell growth rate was observed between this strain and a control strain without the enhancer deletion.
Previous experiments which led to the conclusion of a large stimulation of Pol I transcription by the enhancer were mostly performed with artificial reporter gene systems set outside the nucleolus. Our interpretation is that the stimulation of transcription by the enhancer in these systems is probably due to inefficient accessibility of the Pol I promoter to the Pol I transcription machinery, which is localized to the nucleolus. First, we confirmed stimulation by the enhancer of Pol I transcription of the ectopic reporter in standard (i.e., RDN1) strains. However, the expression of the reporter gene without the enhancer was greatly increased in rdnΔΔ strains, especially in the rdnΔΔ strain carrying pPol II. In these strains, no significant difference was observed between reporter plasmids with and without the enhancer. It was previously shown that in rdnΔΔ strains, the crescent nucleolar structure does not exist; in the case of the helper Pol I plasmid, many mininucleoli with Pol I and Pol I factors are formed and preferentially localized to the nuclear periphery. In the case of the helper Pol II plasmid, helper plasmid molecules apparently coalesce, forming one (or two) round nucleolus at a more internal location(s) (19). In the latter case, both Pol I and Pol I factors are spread throughout most of the nuclear region without colocalization with the Pol II-type nucleolus (19). Thus, in the rdnΔΔ strains carrying pPol II, both rR8 and rR10 plasmids may be equally accessible to Pol I and Pol I factors, and without any competition with other DNA elements for the use of Pol I and Pol I factors, expression of the T7 reporter gene may be maximal. The results suggest a correlation between stimulation of Pol I transcription by the enhancer for the plasmid reporter system and the presence of an intact nucleolar structure. It should be noted that plasmids carrying the rDNA reporter gene are introduced into the wild-type cells by transformation using the URA3 gene (or other Pol II genes) for selection. Therefore, it appears likely that these plasmids take a subnuclear localization outside the nucleolus (as was found by fluorescence in situ hybridization; Oakes and Nomura, unpublished), that the efficiency of recruitment of Pol I and Pol I factors is low, and that the enhancer somehow increases this efficiency. The variability of the reporter gene expression among transformants in the wild-type background, but not in rdnΔΔ strains carrying pPol II, might also be explained on the basis of heterogeneity in subnuclear localization of the reporter plasmids.
Another system we used to study the Pol I enhancer is a HOT1 system similar to the HOT1 system in which the requirement of the E element for Pol I transcription was established (32). We have discovered that a fob1 mutation abolishes Pol I transcription in this ectopic system without any effect on the transcription of chromosomal rDNA. The HOT1 system analyzed was integrated at the HIS4 region of (heterologous) chromosome III, which is localized outside the nucleolus. Thus, we would expect that the Pol I promoter in this HOT1 system to be poorly accessible to the Pol I transcription machinery. In fact, we have found that without some special mechanism, which apparently requires both the enhancer and the Fob1 protein, transcription from this ectopic Pol I promoter is extremely inefficient. It was previously observed that the effect of the enhancer was higher when the T7 Pol I reporter was integrated at the URA3 locus compared to the effect observed in the rR8-rR10 plasmid system; as much as a nearly 100-fold stimulation of Pol I transcription by the enhancer was reported for the integrated reporter system (10). The Pol I promoter on rR8 without the enhancer appears to have a limited but higher accessibility to the Pol I machinery in the nucleolus than the same reporter integrated at URA3. However, even plasmid DNAs in this Pol I reporter system appear to be primarily extranucleolar. Our preliminary examination of localization of the rR8 and rR10 plasmids by fluorescence in situ hybridization analysis indicated that even the rR10 plasmid, which has the enhancer and the RFB regions, is localized outside the nucleolus in a majority of cells in which the plasmid signal was clearly distinguishable (Oakes and Nomura, unpublished). (In preliminary experiments, we have examined the expression of the reporter T7 RNA carried by rR8 and rR10 plasmids in a fob1 mutant. While the results varied among individual transformants, on average, there was no significant difference between the two reporter plasmids. However, the data in this case were not as clear as those seen for the HOT1 system. This might be due to the presence of the RFB region in plasmid rR8 [Fig. 4], the region which was clearly shown to be required for efficient Pol I transcription in the HOT1 system [32]. In addition, the plasmid reporter system shows high variability in the reporter expression in different transformants, making a reliable analysis rather difficult. The role of FOB1 in Pol I transcription in plasmid reporter systems will require further study.)
Some earlier studies on yeast Pol I enhancers were done using reporters integrated within chromosomal rDNA repeats. Here, the reported effects of deletion of the E element (the enhancer and the RFB regions) were smaller than those observed using reporter systems integrated at heterologous chromosomal loci or plasmid reporter systems; the difference between a reporter missing the E element on one side and a control reporter was approximately twofold, and the difference between a reporter missing the E element on both sides and the control reporter was approximately fourfold (1, 16). Nevertheless, it was argued from the data that the observed effects may have been small because of the presence of the E element in many other rDNA repeats at the RDN1 locus. These rDNA repeats would presumably have exerted a stimulatory effect at a distance on the reporter, and the importance of the E element in overall rRNA synthesis in vivo was emphasized (16). Although these data are more difficult to explain, it should be noted that the E element comprises the RFB region, which is essential for rDNA repeat expansion (and presumably also contraction) (15). In fact, copy numbers of the control reporter were described to vary more widely than those of the reporter missing one or both of the adjacent E elements (16). Thus, without any means to reduce reporter copy number fluctuation, e.g., by the use of a fob1 mutation, it might be difficult to completely eliminate experimental errors due to copy number fluctuation. In any event, the present experiments demonstrate that deletion of the enhancer from all the rDNA repeats did not cause any significant reduction in rRNA synthesis rate or cell growth rate, an observation which is not consistent with the direct function of the enhancer in transcription stimulation proposed in these previous studies.
Possible function(s) of Pol I enhancer and FOB1.
Two alternative models explain our observations that the enhancer (the E element) is required only for stimulation of Pol I transcription in ectopic systems, such as the HOT1 system at the HIS4 region, but not for Pol I transcription of the rRNA genes in the chromosomal rDNA repeat context. The first model assumes that Pol I, Pol I factors, and Fob1p are present mostly in the nucleolus but are also present in lower amounts outside the nucleolus and that the E element and Fob1p play direct roles in transcription by somehow facilitating recruitment of Pol I and Pol I factors to ectopic Pol I promoters. The second model assumes that both the enhancer (the E element) and Fob1p do not play a direct role in transcription and that both are required for an efficient interaction between ectopic Pol I promoters with the nucleolus, perhaps specifically with the chromosomal rDNA, and thus indirectly stimulate transcription of ectopic Pol I promoters. The first model implies a physiological significance of the proposed (hypothetical) role of the enhancer and Fob1p in transcription of chromosomal rDNA repeats, perhaps under conditions of extremely reduced concentrations of the Pol I machinery in the nucleolus. While we cannot exclude this model completely, we have failed to find growth conditions where one finds a significant difference in growth rate or rRNA synthesis rate for a strain with the intact enhancer present versus a strain with deletion of the entire enhancer from rDNA copies. In addition, the first model proposes an additional (new) function for Fob1p, whereas the second model gives a unified explanation for this and other known functions of Fob1p, as discussed below. Therefore, we favor the second model and suggest that the physiological function(s) of the enhancer and Fob1p are not directly related to Pol I transcription but are concerned mainly with facilitating interactions between rDNA repeats that are important for rDNA repeat expansion and contraction.
Fob1p is localized to the nucleolus (4). It was previously demonstrated that FOB1 is required for rDNA repeat expansion and contraction (12) and formation of extrachromosomal rDNA circles (4). These recombinational events require pairing of different rDNA repeat units. The E element consists of the enhancer and the RFB region, and both elements are required for stimulation of Pol I transcription in the HOT1 system (32). It was recently shown that the RFB region is essential for repeat expansion and the enhancer region is not essential but has a stimulatory role in this process (15). As shown in this paper, stimulation of Pol I transcription by FOB1 and the E element in the HOT1 system almost certainly takes place in the absence of recombinational events. Therefore, we speculate that the role of Fob1p in recombination within rDNA repeats is to facilitate initial pairing (or interaction) of different repeats by the use of the RFB and the enhancer regions (i.e., the E element) and that stimulation by Fob1p of a similar pairing (or interaction) between the E element in the HOT1 system and the corresponding regions in the chromosomal rDNA repeats may be the basis of the stimulation of recruitment of the Pol I machinery to the Pol I promoter in the HOT1 system. After this initial common pairing step involving the Fob1p protein and the E element, subsequent steps leading to repeat expansion on one hand and those leading to recruitment of the Pol I machinery on the other are obviously expected to be different. Thus, the requirement of some DNA cis elements in addition to the E element for repeat expansion (15) but not for transcription stimulation in the HOT1 system (35) is not inconsistent with the model suggested here.
There are some unique features of the enhancer activity found in the earlier studies using the plasmid reporter system (5, 6). These features are orientation independence, action from either upstream or downstream of the reporter gene, and action over long distances (more than 2 kb from the promoter), which are quite different from known yeast enhancers for Pol II. In addition, using the Pol I reporter integrated at the URA3 locus, it was observed that transcription of the reporter gene increases roughly in proportion to the number of enhancer elements (10). These observations are highly consistent with the model proposed here that the enhancer element increases the probability of localization of the reporter gene to be near the nucleolus, perhaps through its interaction with rDNA, rather than by stimulating Pol I transcription directly.
In addition to its requirement for the HOT1 activity, the FOB1 gene is known to be required for RFB activities (14). As for cis elements required for RFB activities, the work by Ward and coworkers (38) showed that some DNA elements are shared with those required for HOT1 activity, but others are required for one activity but not for the other. Although RFB activities were demonstrated using plasmids carrying cloned DNA containing the E element or the RFB region (3, 13, 38), the observed activities were reported to be very weak compared to the activities in the chromosomal context (3). This situation resembles that observed for transcription of a reporter gene; transcription of a Pol I reporter on a plasmid was reported to be much weaker relative to transcription of the same reporter integrated into the chromosomal rDNA repeats (16). It may be interesting to examine whether interactions of rDNA repeats through the E element might play a role in forming, at or near this element, a presumptive macromolecular structure that would achieve the RFB function.
Role of FOB1 in HOT1 activity.
Extensive studies done on the HOT1 system have demonstrated that strong transcription by Pol I stimulates intra- and interchromosomal recombination (9, 32, 35). Stimulation of mitotic recombination by high levels of transcription was also demonstrated for RNA Pol II (28, 33) and may be a general phenomenon related to changes in DNA supercoiling and torsional stress. We have now demonstrated that the requirement for FOB1 in HOT1 activity can be entirely accounted for by its requirement for stimulation of Pol I transcription by the E element (including the enhancer) in ectopic Pol I promoter systems outside the nucleolus. Thus, our finding supports the original conclusion of transcription-associated stimulation of recombination and explains why FOB1 is required for HOT1 activity.
Pol I terminator is dispensable.
Although there were disagreements on the exact sites of yeast Pol I transcription termination in earlier studies (see, e.g., reference 34), extensive in vivo and in vitro studies performed by Reeder and coworkers led to the conclusion that there are two terminators, one at a site 93 nucleotides downstream of the 3′ end of 25S rRNA, which requires binding of the Reb1 protein at a nearby site, and the second at a site 250 bp downstream of the 3′ end of 25S rRNA (23). Although the present work was not designed to study transcription termination, the 190-bp enhancer element we studied contains the Reb1 binding site essential for termination and the second fail-safe terminator. As described in this paper, deletion of the enhancer region, i.e., deletion of the two Pol I terminators, does not cause a significant decrease in the synthesis of rRNA or in growth rate, either in rdnΔΔ strains or in the context of chromosomal rDNA repeats. It appears that transcription termination at these two proposed sites is not required for processing of rRNA to form functional ribosomes. How and where Pol I transcription terminates in these enhancer deletion strains have not been studied.
Pol I enhancers in higher eukaryotes.
The S. cerevisiae Pol I enhancer studied in this work is structurally different from Pol I enhancers in higher eukaryotes, which consist of several repeats of a sequence that is similar to a portion of the gene promoter (22). Thus, results and conclusions obtained for the yeast enhancer in the present study may not apply to those in higher eukaryotes. Nevertheless, we would like to note that most of the published experiments were done using cloned enhancer DNA fragments in ectopic expression systems. In addition, the assays used for enhancer function were mostly by competition assays rather than actual stimulation of transcription in cis. Thus, the possibility that these intergenic elements might have a function(s) only indirectly related to Pol I transcription (rather than direct enhancement of Pol I transcription) cannot be excluded. It is interesting that rDNA enhancer elements in Drosophila melanogaster are known to play an essential role in X-Y meiotic chromosome pairing (17, 24). It may be prudent to await more rigorous future experimentation before accepting the conclusion that these enhancer elements really function directly in stimulating Pol I transcription within the native chromosomal rDNA genes in vivo.
ACKNOWLEDGMENTS
The first two authors, Hobert Wai and Katsuki Johzuka, contributed equally to this work.
We thank Jonathan R. Warner for providing plasmids and for helpful discussions; C. Greer, S. M. Arfin, and S. Jinks-Robertson for critical reading of the manuscript; and M. Oakes and S. VanAmburg for help in preparation of the manuscript.
This work was supported in part by Public Health Service grant GM35949 from the National Institute of Health (to M.N.) and by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.H. and T.K.) and the Ministry of Health, Labour and Welfare, Japan (to T.K.).
REFERENCES
- 1.Banditt M, Koller T, Sogo J M. Transcriptional activity and chromatin structure of enhancer-deleted rRNA genes in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4953–4960. doi: 10.1128/mcb.19.7.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brewer B J, Fangman W L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 3.Brewer B J, Lockshon D, Fangman W L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 4.Defossez P A, Prusty R, Kaeberlein M, Lin S J, Ferrigno P, Silver P A, Keil R L, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 5.Elion E A, Warner J R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984;39:663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- 6.Elion E A, Warner J R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia S N, Pillus L. Net results of nucleolar dynamics. Cell. 1999;97:825–828. doi: 10.1016/s0092-8674(00)80794-1. [DOI] [PubMed] [Google Scholar]
- 8.Hill J E, Meyers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 9.Huang G S, Keil R L. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics. 1995;141:845–855. doi: 10.1093/genetics/141.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson S P, Warner J R. Unusual enhancer function in yeast rRNA transcription. Mol Cell Biol. 1989;9:4986–4993. doi: 10.1128/mcb.9.11.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keil R L, Roeder G S. cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi T, Heck D J, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi T, Hidaka M, Nishizawa M, Horiuchi T. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol Gen Genet. 1992;233:355–362. doi: 10.1007/BF00265431. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi T, Nomura M, Horiuchi T. Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:136–147. doi: 10.1128/MCB.21.1.136-147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkens T, van der Sande C A F M, Dekker A F, van Heerikhuizen H, Planta R J. A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J. 1992;11:4665–4674. doi: 10.1002/j.1460-2075.1992.tb05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKee B D, Karpen G H. Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell. 1990;61:61–72. doi: 10.1016/0092-8674(90)90215-z. [DOI] [PubMed] [Google Scholar]
- 18.Nogi Y, Yano R, Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oakes M, Aris J P, Brockenbrough J S, Wai H, Vu L, Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park P U, Defossez P-A, Guarente L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3848–3856. doi: 10.1128/mcb.19.5.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pederson T. Survey and summary: the plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeder R H. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol. 1999;62:293. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- 23.Reeder R H, Guevara P, Roan J G. Saccharomyces cerevisiae RNA polymerase I terminates transcription at the Reb1 terminator in vivo. Mol Cell Biol. 1999;19:7369–7376. doi: 10.1128/mcb.19.11.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren X, Eisenhour L, Hong C, Lee Y, McKee B D. Roles of rDNA spacer and transcription unit sequences in X-Y meiotic chromosome pairing in Drosophila melanogaster males. Chromosoma. 1997;106:29–36. doi: 10.1007/s004120050221. [DOI] [PubMed] [Google Scholar]
- 25.Rose M D, Broach J R. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- 26.Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break.”. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Saxe D, Datta A, Jinks-Robertson S. Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol Cell Biol. 2000;20:5404–5414. doi: 10.1128/mcb.20.15.5404-5414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz M C, Choe S Y, Reeder R H. In vitro definition of the yeast RNA polymerase I enhancer. Mol Cell Biol. 1993;13:2644–2654. doi: 10.1128/mcb.13.5.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 31.Shinohara A, Ogawa T. Homologous recombination and the roles of double-strand breaks. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 32.Stewart S E, Roeder G S. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 34.van der Sande C A F M, Kulkens T, Kramer A B, de Wijs I J, van Heerikhuizen H, Klootwijk J, Planta R J. Termination of transcription by yeast RNA polymerase I. Nucleic Acids Res. 1989;17:9127–9146. doi: 10.1093/nar/17.22.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voelkel-Meiman K, Keil R L, Roeder G S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 36.Vu L, Siddiqi I, Lee B-S, Josaitis C A, Nomura M. RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc Natl Acad Sci USA. 1999;96:4390–4395. doi: 10.1073/pnas.96.8.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wai H H, Vu L, Oakes M, Nomura M. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 2000;28:3524–3534. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward T R, Hoang M L, Prusty R, Lau C K, Keil R L, Fangman W L, Brewer B J. Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: shared sequences but independent activities. Mol Cell Biol. 2000;20:4948–4957. doi: 10.1128/mcb.20.13.4948-4957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warner J R, Ikonomova R, Johnson S P, Nierras C R, Wang K. The role of the enhancer element in rRNA transcription in Saccharomyces cerevisiae. In: Paule M R, editor. Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. Georgetown, Tex: Springer-Verlag and R. G. Landes Co.; 1998. pp. 243–254. [Google Scholar]