Abstract
RNA is more than just a combination of four genetically encoded nucleobases as it carries extra information in the form of epitranscriptomic modifications. Diverse chemical groups attach covalently to RNA to enhance the plasticity of cellular transcriptome. The reversible and dynamic nature of epitranscriptomic modifications allows RNAs to achieve rapid and context-specific gene regulation. Dedicated cellular machinery comprising of writers, erasers, and readers drives the epitranscriptomic signaling. Epitranscriptomic modifications control crucial steps of mRNA metabolism such as splicing, export, localization, stability, degradation, and translation. The majority of the epitranscriptomic modifications are highly abundant in the brain and contribute to activity-dependent gene expression. Thus, they regulate the vital physiological processes of the brain, such as synaptic plasticity, neurogenesis, and stress response. Furthermore, epitranscriptomic alterations influence the progression of several neurologic disorders. This review discussed the molecular mechanisms of epitranscriptomic regulation in neurodevelopmental and neuropathological conditions with the goal to identify novel therapeutic targets.
Keywords: RNA modifications, N6-Methyladenosine, N1-Methyladenosine, Inosine, 5-Methylcytosine, Pseudouridine, Brain, Stroke
Introduction
Analogous to epigenetics representing DNA and histone modifications, the epitranscriptome refers to RNA modifications. To date, 172 types of epitranscriptomic modifications are identified, which are far more diverse than the currently known 51 epigenetic modifications [1, 2]. In an RNA, adenosine, cytidine, guanosine, uridine, and ribose can be modified by the addition of functional groups such as methyl, acyl, thioalkyl, and glycosyl groups [1]. Methyl group modifications are highly pervasive with 72 variants [1]. Apart from the covalent addition of these functional groups, epitranscriptomic modifications also include post-transcriptional nucleobase exchange and isomerization [3]. The well-characterized epitranscriptomic modifications include N6-methyladenosine (m6A), N1-methyladenosine (m1A), inosine (I), 5-methylcytosine (m5C), and pseudouridine (ψ) [4] (Fig. 1). Among various classes of RNAs, the tRNAs show the highest prevalence of epitranscriptomic modifications (~ 25% of tRNAs show modifications), which are thought to stabilize their tertiary structure [5]. However, transcriptome-wide profiling of modified RNAs after selective enrichment showed the widespread distribution of epitranscriptomic modifications in both mRNAs and other classes of noncoding RNAs [3]. Identification of machinery responsible for depositing (writers) and removing (erasers) of m6A in RNAs and deciphering of m6A-modified RNAs (readers) garnered attention to epitranscriptomics. For example, the discovery of m6A demethylase fat mass and obesity-associated protein (FTO) spurred the interest to investigate the reversibility and dynamic existence of other epitranscriptomic modifications [6].
Fig. 1.
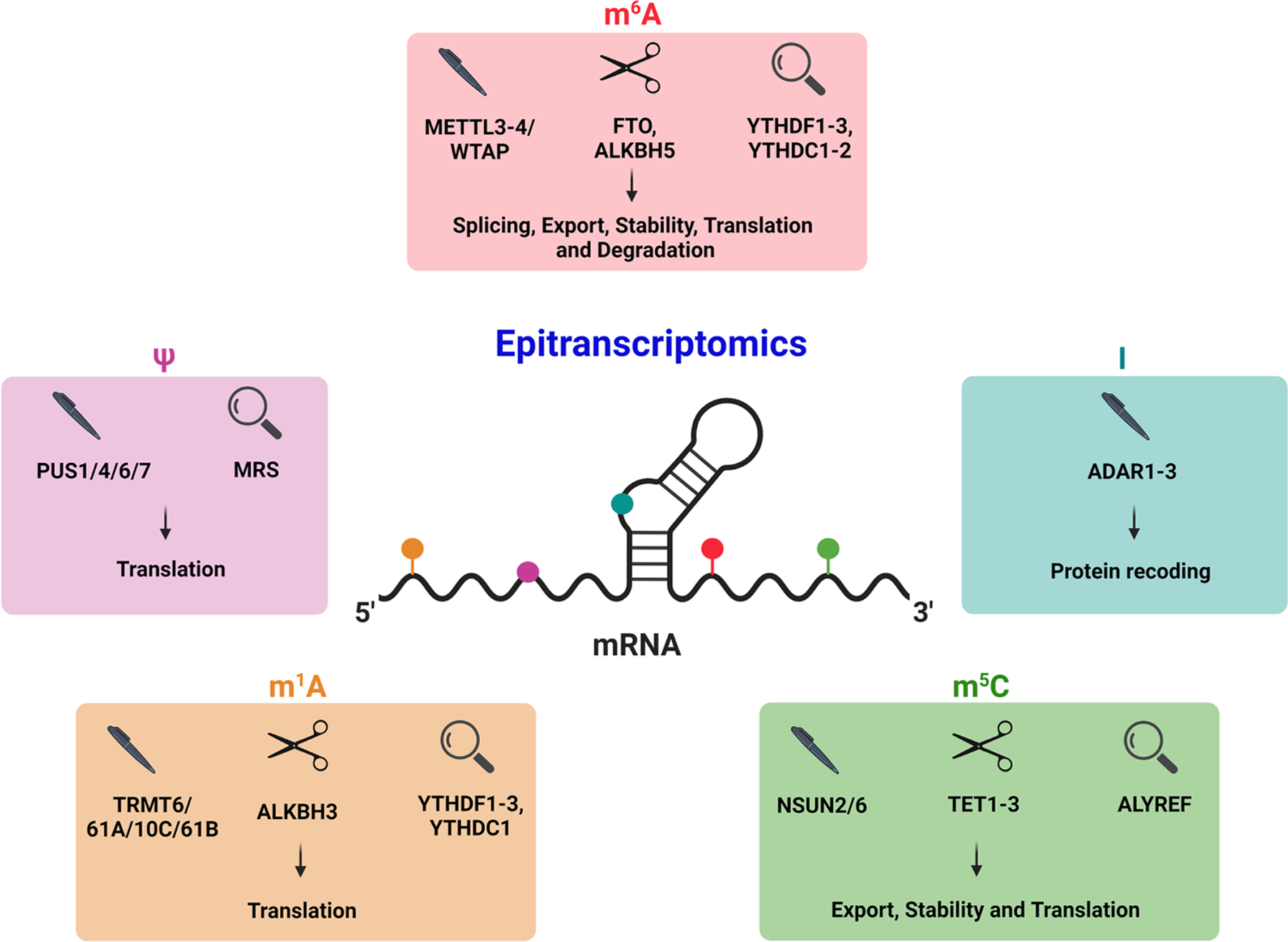
Overview of major epitranscriptomic modifications in the brain. The most abundant and well-studied epitranscriptomic modifications in the brain include N1-methyladenosine (m1A), pseudouridine (ψ), inosine (I), N6-methyladenosine (m6A), and 5-methylcytosine (m5C). The m1A, m6A, and m5C involve the addition of methyl group, whereas I and ψ are base exchange modifications. These modifications are controlled by a set of writers (marker), erasers (scissors), and readers (magnifier) to dictate the fate of modified mRNAs. They regulate various steps of mRNA processing such as splicing, export, localization, stability, translation, degradation, and protein recoding
Epitranscriptomic modifications fine-tune gene expression by regulating multiple steps of mRNA processing such as splicing, export, stability, degradation, and translation [7]. Importantly, several epitranscriptomic modifications, including m6A, m1A, and I, are enriched in the brain and essential for CNS physiological functions such as synaptic transmission and neurogenesis [8–10]. Epitranscriptomic alterations are also associated with neurologic dysfunction, and a better understanding of these molecular mechanisms will provide novel therapeutic targets [4]. This review delineates the role of some of the most abundant and well-studied epitranscriptomic modifications, including m6A, m1A, I, m5C, and ψ, and the key epitranscriptomic enzymes involved in the post-transcriptional gene regulation in brain development and diseases, with an emphasis on acute brain injuries, chronic neurodegeneration, tumorigenesis, and neuropsychiatric disorders.
N6-Methyladenosine (m6A)
The m6A over-represents the mammalian epitranscriptome with an abundance ranging 0.1–0.4% of total adenines [11]. It is prevalent at the RRACH sequence motif (R can be adenosine or cytosine or uracil, A is adenosine that can be modified to m6A, C is cytosine, and H can be guanosine or adenosine) present in the 3′-UTR of mRNAs and deposited by the multi-subunit writer complex consisting of methyltransferase like (METTL) 3 and 4 and Wilms tumor-associated protein (WTAP) [8, 12]. Two demethylases, FTO and alkB homology (ALKBH) 5, erase m6A methylation [6, 13]. The m6A readers bind and recruit diverse cellular machinery to the methylated mRNAs to mediate the downstream signaling. The well-characterized m6A readers include YT521-B homology (YTH) domain family/containing proteins (YTHDF1, 2, and 3, and YTHDC1 and 2), each with distinct functions [14]. The m6A methylation is known to regulate multiple steps of mRNA processing such as splicing, export, localization, stability, translation, and degradation, thereby fine-tuning the gene expression [4]. Notably, CNS shows the highest m6A abundance in the mammalian body [8]. As m6A and its machinery are highly enriched in axons and dendrites, it is thought to mediate the activity-dependent gene regulation [15]. The m6A levels gradually increase in the brain during development and are indispensable for neurogenesis, axonogenesis, and gliogenesis [4]. The FTO-deficient mice show severe developmental brain deficits, including microcephaly, postnatal growth retardation, and aberrant dopaminergic neurotransmission [16]. Interestingly, m6A crosstalk with epigenetic modifications such as H3K27me3 at the proliferation-related gene loci during neurogenesis [17].
The m6A also controls axon guidance and elongation by promoting the local translation of key axonal transcripts such as Gap43 and Robo3.1 [18, 19]. In the adult brain, m6A acts as a circadian pacesetter by decreasing the levels of crucial clock transcripts such as CK1δ, which otherwise prolongs the oscillation [20]. More importantly, m6A shapes the local proteome at the synapse by regulating the translation of over 1,000 synaptic plasticity-related transcripts in a YTHDF1-dependent manner [15]. Therefore, attenuating m6A-reliant neural plasticity impairs adaptive behaviors such as anxiety, depression, addiction, and cognition [21]. Moreover, neuronal m6A is sensitive to stressful stimuli such as heat shock and hypoxia, suggesting its dynamic regulatory potential [22, 23]. Because of these striking abilities, m6A signaling is implicated in the progression of acute CNS injuries, chronic neurodegeneration, tumorigenesis, and neuropsychiatric disorders [4]. For example, we recently reported that cerebral ischemia downregulates FTO, thereby inducing m6A hypermethylation in inflammatory transcripts such as Tnf-α and IL-6, and apoptotic transcripts such as Fas and Bcl2a1c [24]. Curtailing m6A hypermethylation by FTO overexpression was shown to decrease oxygen–glucose deprivation-induced apoptosis in the cortical neurons [25]. In contrast, traumatic brain injury decreases METTL3 expression in the hippocampus, leading to hypomethylation of neuronal metabolism-related transcripts such as Pde12 and Qsox2 [26]. Furthermore, spinal cord injury (SCI) was shown to induce hypomethylation of neural regeneration-related transcripts such as Hsp90ab1 and Igf2bp1, preventing their degradation [27]. Therefore, m6A drives neurotoxic or neuroprotective signaling after acute injury to the CNS in a context-specific manner. Interestingly, FTO protein levels were shown to be significantly elevated in both Alzheimer’s disease (AD) and Parkinson’s disease (PD) rodent brains [28, 29]. Additionally, FTO knockout ameliorated the AD-associated cognitive decline, and its inhibition reduced PD-associated neuronal loss in rodents [28, 29]. Thus, targeting FTO appears to be a viable therapeutic approach to prevent chronic neurodegeneration.
The m6A dysregulation due to increased expression of writers (METTL3 and WTAP) and erasers (FTO and ALKBH5) was shown to predict poor prognosis and survival in glioblastoma multiforme (GBM) patients [30]. Of note, METT3 is proposed to act as both an oncogene and a tumor suppressor [31, 32], whereas FTO and ALKBH5 display oncogenic potential as their inhibition reduced glioma growth and tumorigenicity in mice [30]. Mechanistically, altered m6A methylation of several master regulators, including RNA editing enzymes such as ADAR1 and transcription factors such as Sox2 and FOXM1, drives GBM pathogenesis [33, 34]. The SNPs in m6A effectors such as FTO, ALKBH5, and YTHDC2 are also associated with various neuropsychiatric disorders such as major depressive disorder, attention-deficit/hyperactivity disorder, and autism spectrum disorder [35–37]. Overall, m6A warrants further research to evaluate its therapeutic potential in various neurological diseases due to its high abundance and pervasive functional repertoire.
N1-Methyladenosine (m1A)
Although the m1A in the mammalian RNA was first identified in 1961, it remains a poorly characterized epitranscriptomic mark [38]. The m1A was initially discovered in tRNAs and rRNAs and recently shown in nuclear and mitochondrial mRNAs [9, 39–41]. Approximately 0.02% of adenines in the human transcriptome are m1A modified, and therefore, m1A is ten times lower in abundance than m6A [9, 41]. Under alkaline conditions, m1A undergoes Dimroth rearrangement to form m6A, making it challenging to measure accurately [42]. In contrast to m6A, the m1A is predominantly localized in the 5′-UTR region near the translation initiation sites [40]. The positive charge conferred by m1A is thought to influence RNA–protein interactions, thereby correlating with the translation efficiency [9]. Interestingly, m1A methylases are organelle-specific with cytosolic m1A deposited by tRNA methyltransferase (TRMT) 6/61A complex, and mitochondrial m1A deposited by TRMT10C/61B complex [40]. Like m6A, m1A is also a reversible epitranscriptomic mark and erased by ALKBH3 in the mRNA and ALKBH1 in the tRNA [39, 43]. Based on the proteomic profiling of the m1A interactome, recent studies proposed YTH domain-containing proteins as the putative m1A readers [44, 45]. Despite its low abundance, m1A is dynamically regulated by cellular stress. Specifically, ischemia reduces and heat shock increases the global m1A levels [9, 23]. Moreover, m1A safeguards mRNAs in the stress granules during heat shock stress and promotes their translation during recovery, indicating its protective function [46]. Notably, the brain has the highest abundance of m1A, followed by kidney, muscle, heart, and liver [9]. However, as the transcriptome-wide profile of m1A in the brain is not yet evaluated, its function in brain physiology and diseases is not yet understood. Piecemeal evidence indicates the association of m1A machinery with GBM progression. The expression levels of TRMT6 and TRMT61A were found to be higher in the aggressive form of GBM compared to the low-grade tumors [47]. Furthermore, the mRNA expression of TRMT61A was observed to be decreased in GBM cells treated with an anticancer agent [48]. The m1A is embedded within the tertiary fold configuration in the tRNA and is thought to be crucial for stabilizing its three-dimensional structure [49]. The tRNAs are cleaved to produce tRNA halves (tiRNAs) by ribonuclease angiogenin following cellular stress, exposing the m1A tag [50, 51]. Interestingly, the m1A-tagged tiRNA levels were shown to be significantly elevated after stroke [52]. Furthermore, treatment of anti-inflammatory, neuroprotective agent minocycline reduced the m1A-tagged tiRNA levels in PC12 cells subjected to oxygen–glucose deprivation [53]. More importantly, the plasma concentration of the m1A, an indirect measure of tiRNA levels, is significantly increased in both ischemic and hemorrhagic stroke patients compared to healthy controls and correlated with the infarct size and hematoma volume [54]. All these studies show the possibility of m1A as a stroke biomarker. Furthermore, m1A levels were reported to be significantly increased in the urine of AD patients compared to control subjects [55]. However, the role of m1A in mRNA metabolism after stroke and other neurological diseases remains to be investigated in detail.
Inosine (I)
In addition to the covalent modifications of RNA like m6A and m1A, epitranscriptomic regulation also comprises RNA editing that involves alteration of the mRNA nucleotides relative to the genomic sequence. The conversion of adenosine to inosine (A-to-I) by adenosine deaminase that acts on RNA (ADAR) family of enzymes is an example of this type of modification that is highly prevalent with over 2 million sites in the human transcriptome [56]. To date, 3 mammalian ADARs have been identified, with ADAR1 and 2 being enzymatically active and ADAR3 being a brain-specific negative modulator of editing [57]. A-to-I editing occurs in the double-stranded RNA structures found within the Alu repetitive elements of pre-mRNAs and noncoding RNAs [58]. Although the sequence motif preferred by ADARs remains elusive, several trans regulators such as Pin1, WWP2, and AIMP2 were shown to control the editing activity [10, 59]. Greater than 75% of these sites are present in the intronic regions, whereas only 0.17% occur in the exonic regions of mRNAs, and the remaining are intergenic [60]. Interestingly, inosine is recognized by the translation machinery as guanosine, and therefore, A-to-I editing directly modulates the function of ion channels and neurotransmitter receptors, including GluA2, 5-HT2CR, and Kv1.1 [61]. A-to-I editing was also shown in the noncoding RNAs, where it influenced splicing, translation, and miRNA targeting [62]. Brain shows the highest expression of ADAR1 and 2 and A-to-I editing activity in the mammalian body [10]. Furthermore, neurons exhibit a higher editing activity relative to other cell types within the brain [63]. A-to-I editing is known to be elevated spatiotemporally during brain development and its dysregulation was extensively reported in several neurological disorders [64–66]. A-to-I editing was shown to be reduced in the transcripts such as 5-HT2CR and Kv1.1 following SCI due to the downregulation of ADAR2 [67]. Similarly, the editing levels in GluA2 mRNA were reported to be reduced after cerebral ischemia due to the loss of ADAR2 and its overexpression protected the hippocampal neurons from ischemic injury [68]. Recent studies also reported a significant decrease in the total RNA editing during GBM progression [69] along with downregulation of ADAR2 and upregulation of ADAR3 in GBM patients [70, 71]. Furthermore, several transcripts related to neuronal signaling such as GluA2, GluK1, GluK2, and GABRA3 and glioma growth such as CDC14B were shown to undergo hypo-editing in GBM [69, 72, 73]. In medulloblastomas, A-to-I editing was observed to be diminished in a key Hedgehog signaling-related transcript—GLI1, subsequently inhibiting its transcriptional activity [74]. The A-to-I editing of several glutamate receptors such as GRIA2, 3, and 4 and ion channels such as UNC80 was significantly downregulated in the hippocampus and frontal and temporal lobes of AD patients compared to healthy controls [75]. Another recent study reported differential RNA editing of the transcripts related to endocytic and inflammatory pathways such as TREM2 and BIN1 in the blood of the multi-ethnic AD disease cohort [76]. ADeditome database cataloged 108,010 RNA editing events from 1,524 AD patient samples that were associated with disease progression [77]. The hypo-editing of GluA2 due to the loss of ADAR2 was shown to be related to the death of motor neurons in amyotrophic lateral sclerosis (ALS) patients [78]. Furthermore, AAV9-mediated overexpression of ADAR2 in the ADAR2-deficient mice prevented motor neuron death and improved motor function [79]. Taken together, ADAR2-mediated epitranscriptomic editing of neural receptors is impaired in brain tumors and in acute and chronic neurodegenerative conditions.
5-Methylcytosine (m5C)
5-Methylcytosine (5mC) is a well-characterized epigenetic modification in DNA. The rRNAs, tRNAs, and mRNAs are also shown to undergo methylation of cytosine at the 5th position (m5C) [80]. The abundance of m5C in RNAs is variable across species, ranging from 0.03 to 0.1% of total cytosines [81]. Similar to m1A, the m5C is also enriched near the translation initiation sites in the 5′ UTR region of mRNA [82]. Two isoforms of NOP2/Sun domain family (NSUN) methyltransferases were shown to deposit m5C in the mRNA based on the sequence context. While NSUN2 methylates the CNGGG motif in the 5′ UTR, NSUN6 methylates the CTCCA motif in the 3′ UTR [83, 84]. Furthermore, the ten-eleven translocation (Tet) family of enzymes Tet1, Tet2, and Tet3 that oxidize 5mC to 5-hydroxymethylcytosine (5hmC) in DNA also catalyze m5C to hm5C in the RNA, but the prevalence of hm5C in comparison to m5C is very low (1:5,000) [85]. The mRNA export adaptor Aly/REF export factor (ALYREF) directly binds to m5C and promotes the nuclear export of the methylated mRNAs [82]. In addition, Y-box binding protein 1 (YBX1) binds and stabilizes the m5C-modified mRNAs during maternal to zygotic transition in zebrafish [86]. Furthermore, m5C in the coding regions of mRNAs positively correlates with their translation efficiency, particularly m5C coordinates with m6A to promote the translation of p21 mRNA [83, 87]. NSUN6-dependant m5C methylation in the 3′ UTR was shown to promote translation termination [84]. Intriguingly, m5C is induced at the sites of DNA double-stranded breaks and serves as a signal to recruit the DNA repair proteins such as RAD51 and RAD52 to promote homologous recombination [88]. This indicates that the regulatory potential of m5C and m6A might be similar despite a 3–10 times lower abundance of m5C [89]. Despite their overlapping functions, the transcriptome-wide crosstalk between m6A and m5C is not yet evaluated. The m5C seems to be important for normal brain function. In humans, a missense mutation in the NSUN2 gene was linked to intellectual disability [90]. Furthermore, NSUN2 is enriched in the Purkinje cells of the cerebellum, hinting that mutant NSUN2 interferes with the GABAergic cerebellar circuitry in humans [90]. A causal link was identified between homozygous splice mutation in NSUN2 and Dubowitz syndrome, clinically characterized by neurological abnormalities such as microcephaly and behavioral deficits like speech delay [91]. More importantly, NSUN2 knockout mice show severe neurodevelopmental defects, such as decreased neuronal cell size and impaired synaptogenesis [92]. Mechanistically, loss of NSUN2 in the neuroepithelial stem cells of the developing human brain causes hypomethylation and subsequent cleavage of tRNAs, ultimately inhibiting the migration and differentiation of neural progenitors [93]. Moreover, the loss of m5C in noncoding vault RNAs due to NSUN2 deficiency impairs the generation of microRNA-like molecules that regulate the intellectual disability-associated ion channel proteins [94]. A recent study found that the mRNA expression of NSUN6 was significantly decreased in human GBM samples compared to healthy controls [84]. These studies indicate that altered m5C might promote neurological dysfunction in various conditions. It was also reported that hm5C is widespread in RNAs in the brainstem, hippocampus, amygdala, cortex, and cerebellum of mice [95]. Furthermore, the abundance of RNA hm5C was shown to be decreased significantly in the hippocampus, substantia nigra, and striatum of the mice subjected to MPTP-induced PD [95]. This indicates the potential interplay between m5C and hm5C in RNAs during neuropathological conditions.
Pseudouridine (ψ)
The ψ formed by uridine isomerization is the first epitranscriptomic modification discovered [96]. The ψ is the second most highly abundant epitranscriptomic modification after m6A in mammalian mRNA, observed in ~ 0.3% of the total uridines [97]. The ψ is known to stabilize the secondary structure of tRNAs and rRNAs by providing an extra hydrogen bond, but its prominence in mRNAs is not yet studied in detail [98]. Transcriptome-wide mapping of ψ conservatively identified 260 sites in 238 mRNAs of yeast and HeLa cells [99]. More importantly, ψ is regulated in mRNAs in response to environmental cues such as nutrient deprivation, oxidative stress, and heat shock [97, 99, 100]. Among the 13 pseudouridine synthases (PUSs) that catalyze the pseudouridylation in various types of RNA, PUS 1, 4, 6, and 7 can generate ψ in mRNAs [101–103]. There are no known erasers for this epitranscriptomic mark, suggesting that ψ might be irreversible. A recent study demonstrated that methionine aminoacyl tRNAMet synthetase (MRS) functions as ψ reader and reduces the translation initiation of YEF3 mRNA in the yeast [103]. Another study showed that mRNA pseudouridylation impedes translation elongation [104]. Although these studies show that ψ affects translation fidelity, the mechanism of how it influences the ribosome function remains elusive. Multiple studies linked the aberrant pseudouridylation with neurological disorders. Humans with PUS3 mutation display intellectual disability and PUS1 mutation develop mild cognitive impairment, probably due to perturbed tRNA pseudouridylation [105, 106]. A recent study demonstrated the beneficial role of pseudouridylation in myotonic dystrophy type 2 (DM2) patients. Mechanistically, pseudouridylation within the toxic CCUG repeats moderately prevents the sequestration of splicing regulator Muscleblind-like 1 protein, which otherwise drives the DM2 pathology [107]. Overall, ψ is a widespread epitranscriptomic mark, but its function during brain development and diseases is still unexplored.
Clinical Significance
Epitranscriptomic imbalance is increasingly recognized as a molecular hallmark of various CNS diseases (Fig. 2). Reversing the epitranscriptomic alterations by targeting its machinery has proven to be a novel therapeutic strategy for acute and chronic neurological disorders [4]. However, the evidence for designing drugs to modulate epitranscriptome is still emerging. High-throughput screens identified several small molecules that target m6A effectors. Of note, seven small molecule inhibitors of m6A demethylase FTO are identified [108]. Among them, R-2-hydroxyglutarate displayed anti-glioma activity by inhibiting FTO and thereby modulating the transcription factors c-MYC and CEBPA [109]. Another FTO inhibitor, entacapone, delayed PD-associated motor dysfunction in humans [110, 111]. Furthermore, the redox cofactor nicotinamide adenine dinucleotide phosphate (NADP) was found to be a highly potent small molecule activator of FTO [112]. In addition, STM2457 was found to selectively inhibit m6A methylase subunit—METTL3, whereas the piperazine derivative compound 4 was found to activate the m6A methylase complex [113, 114]. Future studies are needed to explore the therapeutic benefits of these epitranscriptomic drugs in various CNS diseases. For example, NADP and STM2457 may be tested for ischemic stroke as they nullify the m6A hypermethylation (Chokkalla AK, Stroke, 2019). In addition to m6A, certain small molecules target RNA hm5C effectors Tet 1, 2, and 3. Particularly, ascorbic acid activates, whereas dimethyloxallyl glycine inhibits, Tet proteins [115, 116]. However, if these compounds modulate RNA hm5C in CNS diseases remains unknown. Instead of global attenuation of epitranscriptomic modifications by small molecules that target epitranscriptomic enzymes, the development of mRNA substrate-selective inhibitors/activators is highly desired. For example, a short helix-threading peptide binds near the RNA A-to-I editing site in the 5-HT2CR mRNA and selectively inhibits its editing by ADAR2 [117]. Future studies should exploit this concept for selectively modulating the epitranscriptomic modification within a single transcript.
Fig. 2.
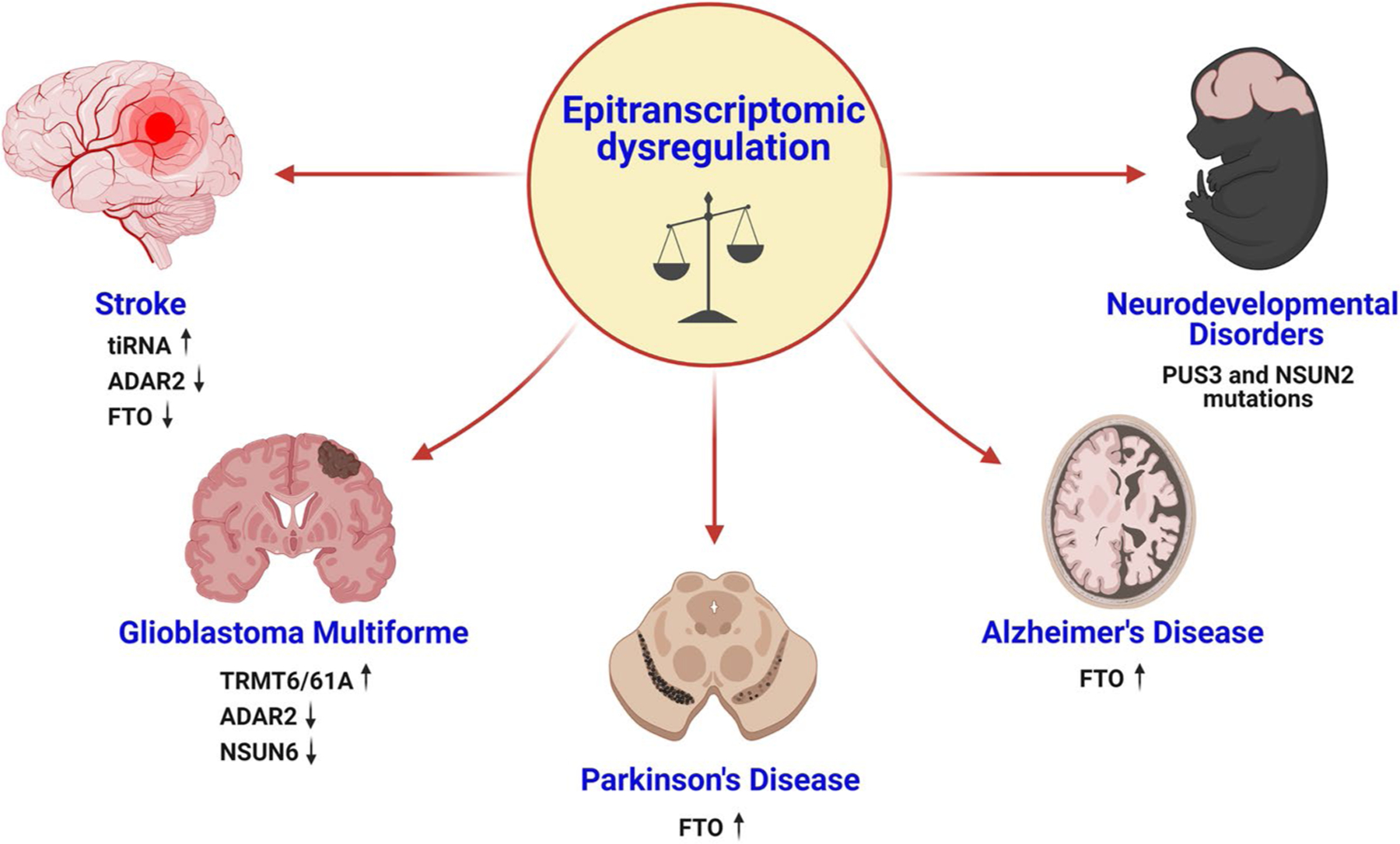
Epitranscriptomic imbalance in major CNS diseases. The altered expression of writers and erasers, including ADAR2, FTO, TRMT6/61A, and NSUN6, mediates several brain disorders such as stroke, glioblastoma multiforme, Parkinson’s disease, and Alzheimer’s disease. Additionally, the mutations in certain epitranscriptomic enzymes such as PUS3 and NSUN2 drive neurodevelopmental disorders
The epitranscriptomic modifications display enormous potential to serve as diagnostic and prognostic biomarkers in addition to their therapeutic utility. During RNA turnover in the cells, unmodified nucleosides such as adenosine are typically recycled via the salvage pathway [118]. In contrast, the modified nucleosides such as inosine are released into the extracellular space and subsequently detected in blood and urine [119, 120]. Mass spectrometry-based profiling identified 20 types of modified nucleosides, including m6A, m1A, I, and m5C, in human plasma [121]. More importantly, these modified nucleosides are highly abundant and accounted for 49% of total nucleosides [121]. Furthermore, pulse-chase labeling revealed that extracellular m6A nucleosides are major byproducts of mRNA and rRNA catabolism [121]. Inosine levels were reported to be markedly elevated in the blood of stroke, multiple sclerosis, and epileptic patients, whereas decreased in the blood of major depressive disorder and saliva of AD subjects [122–126]. Moreover, the urinary concentration of ψ was observed to be significantly increased in patients with AD and post-stroke depression [55, 127]. Additionally, stratification of glioblastoma patients based on RNA A-to-I editing profiles identified a novel sex-dependent high-risk patient subgroup [128]. These studies suggest the possibility of developing point-of-care diagnostic and therapeutic tools for CNS diseases based on epitranscriptomic modifications.
Future Perspectives
Collectively, epitranscriptomic modifications form an additional layer to control gene expression during the physiological and pathological processes of the brain. Despite being sparse, their dynamic regulation by writers and erasers drives activity-dependent gene expression in the brain. In addition to the above-discussed modifications, the functions of several less abundant modifications such as N4-acetylcytosine (ac4C), N6,2′-O-dimethyladenosine (m6Am), 2′-O-methylation (Nm), and N7-methylguanosine (m7G) are currently being unfolded [89]. A major challenge in studying these modifications is the lack of specific antibodies and chemical reagents. Several controversial findings reported antibody cross-reactivity among epitranscriptomic modifications. For example, the m1A antibody cross-reacts with m7G, skewing the previously reported transcriptome-wide m1A prevalence [129]. Likewise, m6A antibody cross-reacts with m6Am [130]. Hence, the development of antibody-independent techniques is necessary to map these modifications more accurately. Interestingly, certain epitranscriptomic modifications such as m5C and m6A exist in proximity with each other and co-regulate the fate of the transcript [87]. Hence, future studies should implement holistic approaches to understand their interplay resulting in synergy or competition. For example, techniques such as modified RNA bisulfite sequencing can simultaneously map the co-occurrence of multiple modifications, such as m5C, Ψ, and m1A, at single-nucleotide resolution [131]. Although the writers and erasers for various epitranscriptomic modifications are well-characterized, our understanding of the readers and their mechanisms of action, especially for modifications such as ψ, is limited. Unbiased methods, such as mass spectrometry-based proteome profiling after pulldown with modified bait RNA probes, may be applied to comprehensively elucidate the readers and their interactome [132]. Surprisingly, certain readers like YTHDF proteins bind both m6A and m1A, further convoluting the epitranscriptomic crosstalk [14, 44, 45]. Although several studies exposed the prevalence of epitranscriptomic alterations in CNS diseases, their mechanistic link to disease pathology remains obscure. Importantly, whether these changes serve as disease drivers or merely disease manifestations (cause or effect) has to be examined. This requires evaluating the temporal landscape of epitranscriptomic modifications during disease pathogenesis. For example, the dynamic changes in epitranscriptomic signaling can be investigated by employing auxin-inducible degron systems to achieve transient and sharp degradation of epitranscriptomic machinery at a particular stage of the disease [133]. Moreover, most of the current studies are restricted to just profiling these modifications in CNS disease models. Overall, the application of various knockout/overexpression mouse models and small molecule modulators of epitranscriptomic enzymes will lead to a better understanding of the RNA modifications in neurological disorders.
Funding
This study was partly supported by the NIH grant RO1 NS109459 and AHA grant 20PRE35120233.
Footnotes
Ethics Approval This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of Interest The authors declare no competing interests.
References
- 1.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303–d7. 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sood AJ, Viner C, Hoffman MM. DNAmod: the DNA modification database. J Cheminform. 2019;11(1):30. 10.1186/s13321-019-0349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helm M, Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet. 2017;18(5):275–91. 10.1038/nrg.2016.169. [DOI] [PubMed] [Google Scholar]
- 4.Chokkalla AK, Mehta SL, Vemuganti R. Epitranscriptomic regulation by m(6)A RNA methylation in brain development and diseases. J Cereb Blood Flow Metab. 2020;40(12):2331–49. 10.1177/0271678x20960033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helm M, Alfonzo JD. Posttranscriptional RNA Modifications: playing metabolic games in a cell’s chemical Legoland. Chem Biol. 2014;21(2):174–85. 10.1016/j.chembiol.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–7. 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livneh I, Moshitch-Moshkovitz S, Amariglio N, Rechavi G, Dominissini D. The m(6)A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci. 2020;21(1):36–51. 10.1038/s41583-019-0244-z. [DOI] [PubMed] [Google Scholar]
- 8.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–46. 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530(7591):441–6. 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, et al. Dynamic landscape and regulation of RNA editing in mammals. Nature. 2017;550(7675):249–54. 10.1038/nature24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. 1975;4(4):379–86. 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 12.Schöller E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. Rna. 2018;24(4):499–512. 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the transcriptome: m(6)A-binding proteins. Trends Cell Biol. 2018;28(2):113–27. 10.1016/j.tcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merkurjev D, Hong WT, Iida K, Oomoto I, Goldie BJ, Yamaguti H, et al. Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci. 2018;21(7):1004–14. 10.1038/s41593-018-0173-6. [DOI] [PubMed] [Google Scholar]
- 16.Hess ME, Hess S, Meyer KD, Verhagen LA, Koch L, Brönneke HS, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16(8):1042–8. 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, et al. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21(2):195–206. 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Chen M, Huang H, Zhu J, Song H, Zhu J, et al. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2018;46(3):1412–23. 10.1093/nar/gkx1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuang M, Li X, Zhu J, Zhang J, Niu F, Liang F, et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019;47(9):4765–77. 10.1093/nar/gkz157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fustin JM, Kojima R, Itoh K, Chang HY, Ye S, Zhuang B, et al. Two Ck1δ transcripts regulated by m6A methylation code for two antagonistic kinases in the control of the circadian clock. Proc Natl Acad Sci U S A. 2018;115(23):5980–5. 10.1073/pnas.1721371115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung Y, Goldman D. Role of RNA modifications in brain and behavior. Genes Brain Behav. 2018;17(3):e12444. 10.1111/gbb.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kisliouk T, Rosenberg T, Ben-Nun O, Ruzal M, Meiri N. Early-Life m(6)A RNA Demethylation by fat mass and obesity-associated protein (FTO) influences resilience or vulnerability to heat stress later in life. eNeuro. 2020;7(3):ENE URO.0549–19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fry NJ, Law BA, Ilkayeva OR, Holley CL, Mansfield KD. N(6)-methyladenosine is required for the hypoxic stabilization of specific mRNAs. Rna. 2017;23(9):1444–55. 10.1261/rna.061044.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chokkalla AK, Mehta SL, Kim T, Chelluboina B, Kim J, Vemuganti R. Transient focal ischemia significantly alters the m(6) A epitranscriptomic tagging of RNAs in the brain. Stroke. 2019;50(10):2912–21. 10.1161/strokeaha.119.026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu K, Mo Y, Li D, Yu Q, Wang L, Lin F, et al. N(6)-methyladenosine demethylases Alkbh5/Fto regulate cerebral ischemia-reperfusion injury. Ther Adv Chronic Dis. 2020;11:2040622320916024. 10.1177/2040622320916024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Mao J, Wang X, Lin Y, Hou G, Zhu J, et al. Genome-wide screening of altered m6A-tagged transcript profiles in the hippocampus after traumatic brain injury in mice. Epigenomics. 2019;11(7):805–19. 10.2217/epi-2019-0002. [DOI] [PubMed] [Google Scholar]
- 27.Xing L, Cai Y, Yang T, Yu W, Gao M, Chai R, et al. Epitranscriptomic m6A regulation following spinal cord injury. J Neurosci Res. 2020. 10.1002/jnr.24763. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Ren Y, Mao K, Hua F, Yang Y, Wei N, et al. FTO is involved in Alzheimer’s disease by targeting TSC1-mTOR-Tau signaling. Biochem Biophys Res Commun. 2018;498(1):234–9. 10.1016/j.bbrc.2018.02.201. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Yu C, Guo M, Zheng X, Ali S, Huang H, et al. Downregulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem Neurosci. 2019;10(5):2355–63. 10.1021/acschemneuro.8b00657. [DOI] [PubMed] [Google Scholar]
- 30.Dong Z, Cui H. The emerging roles of rna modifications in glioblastoma. Cancers (Basel). 2020;12(3):736. 10.3390/cancers12030736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–34. 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visvanathan A, Patil V, Abdulla S, Hoheisel JD, Somasundaram K. N6-Methyladenosine landscape of glioma stem-like cells: METTL3 is essential for the expression of actively transcribed genes and sustenance of the oncogenic signaling. Genes (Basel). 2019;10(2):141. 10.3390/genes10020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591–606.e6. 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, et al. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37(4):522–33. 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 35.Choudhry Z, Sengupta SM, Grizenko N, Thakur GA, Fortier ME, Schmitz N, et al. Association between obesity-related gene FTO and ADHD. Obesity (Silver Spring). 2013;21(12):E738–44. 10.1002/oby.20444. [DOI] [PubMed] [Google Scholar]
- 36.Du T, Rao S, Wu L, Ye N, Liu Z, Hu H, et al. An association study of the m6A genes with major depressive disorder in Chinese Han population. J Affect Disord. 2015;183:279–86. 10.1016/j.jad.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 37.Liu X, Shimada T, Otowa T, Wu YY, Kawamura Y, Tochigi M, et al. Genome-wide association study of autism spectrum disorder in the East Asian populations. Autism Res. 2016;9(3):340–9. 10.1002/aur.1536. [DOI] [PubMed] [Google Scholar]
- 38.Dunn DB. The occurrence of 1-methyladenine in ribonucleic acid. Biochim Biophys Acta. 1961;46:198–200. 10.1016/0006-3002(61)90668-0. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016;12(5):311–6. 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, et al. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell. 2017;68(5):993–1005.e9. 10.1016/j.molcel.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017;551(7679):251–5. 10.1038/nature24456. [DOI] [PubMed] [Google Scholar]
- 42.Macon JB, Wolfenden R. 1-Methyladenosine. Dimroth rearrangement and reversible reduction Biochemistry. 1968;7(10):3453–8. 10.1021/bi00850a021. [DOI] [PubMed] [Google Scholar]
- 43.Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell. 2016;167(7):1897. 10.1016/j.cell.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 44.Dai X, Wang T, Gonzalez G, Wang Y. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal Chem. 2018;90(11):6380–4. 10.1021/acs.analchem.8b01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seo KW, Kleiner RE. YTHDF2 recognition of N(1)-methyladenosine (m(1)A)-modified RNA is associated with transcript destabilization. ACS Chem Biol. 2020;15(1):132–9. 10.1021/acschembio.9b00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alriquet M, Calloni G, Martínez-Limón A, Delli Ponti R, Hanspach G, Hengesbach M, et al. The protective role of m1A during stress-induced granulation. J Mol Cell Biol. 2021;12(11):870–80. 10.1093/jmcb/mjaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macari F, El-Houfi Y, Boldina G, Xu H, Khoury-Hanna S, Ollier J, et al. TRM6/61 connects PKCα with translational control through tRNAi(Met) stabilization: impact on tumorigenesis. Oncogene. 2016;35(14):1785–96. 10.1038/onc.2015.244. [DOI] [PubMed] [Google Scholar]
- 48.Mongiardi MP, Savino M, Falchetti ML, Illi B, Bozzo F, Valle C, et al. c-MYC inhibition impairs hypoxia response in glioblastoma multiforme. Oncotarget. 2016;7(22):33257–71. 10.18632/oncotarget.8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson JT, Droogmans L. Biosynthesis and function of 1-methyladenosine in transfer RNA. In: Grosjean H, editor. Fine-tuning of RNA functions by modification and editing. Heidelberg: Berlin Heidelberg: Springer Berlin; 2005. p. 121–39. [Google Scholar]
- 50.Mishima E, Inoue C, Saigusa D, Inoue R, Ito K, Suzuki Y, et al. Conformational change in transfer RNA is an early indicator of acute cellular damage. J Am Soc Nephrol. 2014;25(10):2316–26. 10.1681/asn.2013091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mishima E, Jinno D, Akiyama Y, Itoh K, Nankumo S, Shima H, et al. Immuno-northern blotting: detection of RNA modifications by using antibodies against modified nucleosides. PLoS One. 2015;10(11):e0143756. 10.1371/journal.pone.0143756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato K, Rashad S, Niizuma K, Tominaga T. Stress induced tRNA halves (tiRNAs) as biomarkers for stroke and stroke therapy; pre-clinical study. Neuroscience. 2020;434:44–54. 10.1016/j.neuroscience.2020.03.018. [DOI] [PubMed] [Google Scholar]
- 53.Elkordy A, Rashad S, Shehabeldeen H, Mishima E, Niizuma K, Abe T, et al. tiRNAs as a novel biomarker for cell damage assessment in in vitro ischemia-reperfusion model in rat neuronal PC12 cells. Brain Res. 2019;1714:8–17. 10.1016/j.brainres.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 54.Ishida T, Inoue T, Niizuma K, Konno N, Suzuki C, Inoue T, et al. Prediction of functional outcome in patients with acute stroke by measuring tRNA derivatives. Cerebrovasc Dis. 2020;49(6):639–46. 10.1159/000511627. [DOI] [PubMed] [Google Scholar]
- 55.Lee SH, Kim I, Chung BC. Increased urinary level of oxidized nucleosides in patients with mild-to-moderate Alzheimer’s disease. Clin Biochem. 2007;40(13–14):936–8. 10.1016/j.clinbiochem.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 56.Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O’Connell MA, et al. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods. 2013;10(2):128–32. 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. Rna. 2000;6(5):755–67. 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42((Database issue)):D109–13. 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marcucci R, Brindle J, Paro S, Casadio A, Hempel S, Morrice N, et al. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. Embo j. 2011;30(20):4211–22. 10.1038/emboj.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramaswami G, Li JB. Identification of human RNA editing sites: a historical perspective. Methods. 2016;107:42–7. 10.1016/j.ymeth.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Behm M, Öhman M. RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet. 2016;32(3):165–75. 10.1016/j.tig.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 62.Nishikura K Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–49. 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gal-Mark N, Shallev L, Sweetat S, Barak M, Billy Li J, Levanon EY, et al. Abnormalities in A-to-I RNA editing patterns in CNS injuries correlate with dynamic changes in cell type composition. Sci Rep. 2017;7:43421. 10.1038/srep43421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaidan H, Ramaswami G, Golumbic YN, Sher N, Malik A, Barak M, et al. A-to-I RNA editing in the rat brain is age-dependent, region-specific and sensitive to environmental stress across generations. BMC Genomics. 2018;19(1):28. 10.1186/s12864-017-4409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hwang T, Park CK, Leung AK, Gao Y, Hyde TM, Kleinman JE, et al. Dynamic regulation of RNA editing in human brain development and disease. Nat Neurosci. 2016;19(8):1093–9. 10.1038/nn.4337. [DOI] [PubMed] [Google Scholar]
- 66.Wahlstedt H, Daniel C, Ensterö M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19(6):978–86. 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Narzo AF, Kozlenkov A, Ge Y, Zhang B, Sanelli L, May Z, et al. Decrease of mRNA editing after spinal cord injury is caused by down-regulation of ADAR2 that is triggered by inflammatory response. Sci Rep. 2015;5:12615. 10.1038/srep12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peng PL, Zhong X, Tu W, Soundarapandian MM, Molner P, Zhu D, et al. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006;49(5):719–33. 10.1016/j.neuron.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 69.Patil V, Pal J, Mahalingam K, Somasundaram K. Global RNA editome landscape discovers reduced RNA editing in glioma: loss of editing of gamma-amino butyric acid receptor alpha subunit 3 (GABRA3) favors glioma migration and invasion. PeerJ. 2020;8:e9755. 10.7717/peerj.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17(11):1586–95. 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oakes E, Anderson A, Cohen-Gadol A, Hundley HA. Adenosine deaminase that acts on RNA 3 (ADAR3) binding to glutamate receptor subunit B pre-mRNA inhibits RNA editing in glioblastoma. J Biol Chem. 2017;292(10):4326–35. 10.1074/jbc.M117.779868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galeano F, Rossetti C, Tomaselli S, Cifaldi L, Lezzerini M, Pezzullo M, et al. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene. 2013;32(8):998–1009. 10.1038/onc.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, et al. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem. 2008;283(11):7251–60. 10.1074/jbc.M708316200. [DOI] [PubMed] [Google Scholar]
- 74.Shimokawa T, Rahman MF, Tostar U, Sonkoly E, Ståhle M, Pivarcsi A, et al. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biol. 2013;10(2):321–33. 10.4161/rna.23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khermesh K, D’Erchia AM, Barak M, Annese A, Wachtel C, Levanon EY, et al. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease. Rna. 2016;22(2):290–302. 10.1261/rna.054627.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gardner OK, Wang L, Van Booven D, Whitehead PL, Hamilton-Nelson KL, Adams LD, et al. RNA editing alterations in a multi-ethnic Alzheimer disease cohort converge on immune and endocytic molecular pathways. Hum Mol Genet. 2019;28(18):3053–61. 10.1093/hmg/ddz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu S, Yang M, Kim P, Zhou X. ADeditome provides the genomic landscape of A-to-I RNA editing in Alzheimer’s disease. Brief Bioinform. 2021. 10.1093/bib/bbaa384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aizawa H, Sawada J, Hideyama T, Yamashita T, Katayama T, Hasebe N, et al. TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol. 2010;120(1):75–84. 10.1007/s00401-010-0678-x. [DOI] [PubMed] [Google Scholar]
- 79.Yamashita T, Chai HL, Teramoto S, Tsuji S, Shimazaki K, Muramatsu S, et al. Rescue of amyotrophic lateral sclerosis phenotype in a mouse model by intravenous AAV9-ADAR2 delivery to motor neurons. EMBO Mol Med. 2013;5(11):1710–9. 10.1002/emmm.201302935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023–33. 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huber SM, van Delft P, Mendil L, Bachman M, Smollett K, Werner F, et al. Formation and abundance of 5-hydroxymethylcytosine in RNA. Chembiochem. 2015;16(5):752–5. 10.1002/cbic.201500013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27(5):606–25. 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang T, Chen W, Liu J, Gu N, Zhang R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat Struct Mol Biol. 2019;26(5):380–8. 10.1038/s41594-019-0218-x. [DOI] [PubMed] [Google Scholar]
- 84.Selmi T, Hussain S, Dietmann S, Heiß M, Borland K, Flad S, et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 2021;49(2):1006–22. 10.1093/nar/gkaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136(33):11582–5. 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, et al. RNA 5-Methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell. 2019;75(6):1188–202.e11. 10.1016/j.molcel.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 87.Li Q, Li X, Tang H, Jiang B, Dou Y, Gorospe M, et al. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J Cell Biochem. 2017;118(9):2587–98. 10.1002/jcb.25957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen H, Yang H, Zhu X, Yadav T, Ouyang J, Truesdell SS, et al. m(5)C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat Commun. 2020;11(1):2834. 10.1038/s41467-020-16722-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22(2):119–31. 10.1038/s41576-020-00295-8. [DOI] [PubMed] [Google Scholar]
- 90.Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90(5):856–63. 10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S, et al. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet. 2012;49(6):380–5. 10.1136/jmedgenet-2011-100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33(18):2020–39. 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S, et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Reports. 2017;8(1):112–24. 10.1016/j.stemcr.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4(2):255–61. 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miao Z, Xin N, Wei B, Hua X, Zhang G, Leng C, et al. 5-hydroxymethylcytosine is detected in RNA from mouse brain tissues. Brain Res. 2016;1642:546–52. 10.1016/j.brainres.2016.04.055. [DOI] [PubMed] [Google Scholar]
- 96.Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem. 1957;227(2):907–15. [PubMed] [Google Scholar]
- 97.Li X, Zhu P, Ma S, Song J, Bai J, Sun F, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol. 2015;11(8):592–7. 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 98.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23(24):5020–6. 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–6. 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–62. 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Safra M, Nir R, Farouq D, Vainberg Slutskin I, Schwartz S. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 2017;27(3):393–406. 10.1101/gr.207613.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carlile TM, Martinez NM, Schaening C, Su A, Bell TA, Zinshteyn B, et al. mRNA structure determines modification by pseudouridine synthase 1. Nat Chem Biol. 2019;15(10):966–74. 10.1038/s41589-019-0353-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Levi O, Arava YS. Pseudouridine-mediated translation control of mRNA by methionine aminoacyl tRNA synthetase. Nucleic Acids Res. 2021;49(1):432–43. 10.1093/nar/gkaa1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Eyler DE, Franco MK, Batool Z, Wu MZ, Dubuke ML, Dobosz-Bartoszek M, et al. Pseudouridinylation of mRNA coding sequences alters translation. Proc Natl Acad Sci U S A. 2019;116(46):23068–74. 10.1073/pnas.1821754116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shaheen R, Han L, Faqeih E, Ewida N, Alobeid E, Phizicky EM, et al. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet. 2016;135(7):707–13. 10.1007/s00439-016-1665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cao M, Donà M, Valentino ML, Valentino L, Semplicini C, Maresca A, et al. Clinical and molecular study in a long-surviving patient with MLASA syndrome due to novel PUS1 mutations. Neurogenetics. 2016;17(1):65–70. 10.1007/s10048-015-0465-x. [DOI] [PubMed] [Google Scholar]
- 107.deLorimier E, Hinman MN, Copperman J, Datta K, Guenza M, Berglund JA. Pseudouridine modification inhibits Muscleblind-like 1 (MBNL1) binding to CCUG repeats and minimally structured RNA through reduced RNA flexibility. J Biol Chem. 2017;292(10):4350–7. 10.1074/jbc.M116.770768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou LL, Yang CG. Targeting epitranscriptomic proteins for therapeutic intervention. Biochemistry. 2020;59(2):125–7. 10.1021/acs.biochem.9b00755. [DOI] [PubMed] [Google Scholar]
- 109.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6) A/MYC/CEBPA signaling. Cell. 2018;172(1–2):90–105.e23. 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schrag A Entacapone in the treatment of Parkinson’s disease. Lancet Neurol. 2005;4(6):366–70. 10.1016/s1474-4422(05)70098-3. [DOI] [PubMed] [Google Scholar]
- 111.Peng S, Xiao W, Ju D, Sun B, Hou N, Liu Q, et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci Transl Med. 2019;11:488. 10.1126/scitranslmed.aau7116. [DOI] [PubMed] [Google Scholar]
- 112.Wang L, Song C, Wang N, Li S, Liu Q, Sun Z, et al. NADP modulates RNA m(6)A methylation and adipogenesis via enhancing FTO activity. Nat Chem Biol. 2020;16(12):1394–402. 10.1038/s41589-020-0601-2. [DOI] [PubMed] [Google Scholar]
- 113.Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593(7860):597–601. 10.1038/s41586-021-03536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Selberg S, Blokhina D, Aatonen M, Koivisto P, Siltanen A, Mervaala E, et al. Discovery of small molecules that activate RNA methylation through cooperative binding to the METTL3–14-WTAP complex active site. Cell Rep. 2019;26(13):3762–71.e5. 10.1016/j.celrep.2019.02.100. [DOI] [PubMed] [Google Scholar]
- 115.Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell. 2017;170(6):1079–95.e20. 10.1016/j.cell.2017.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Matuleviciute R, Cunha PP, Johnson RS, Foskolou IP. Oxygen regulation of TET enzymes. FEBS J. 2021. 10.1111/febs.15695. [DOI] [PubMed] [Google Scholar]
- 117.Schirle NT, Goodman RA, Krishnamurthy M, Beal PA. Selective inhibition of ADAR2-catalyzed editing of the serotonin 2c receptor pre-mRNA by a helix-threading peptide. Org Biomol Chem. 2010;8(21):4898–904. 10.1039/c0ob00309c. [DOI] [PubMed] [Google Scholar]
- 118.Lane AN, Fan TW. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015;43(4):2466–85. 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Willmann L, Erbes T, Krieger S, Trafkowski J, Rodamer M, Kammerer B. Metabolome analysis via comprehensive two-dimensional liquid chromatography: identification of modified nucleosides from RNA metabolism. Anal Bioanal Chem. 2015;407(13):3555–66. 10.1007/s00216-015-8516-6. [DOI] [PubMed] [Google Scholar]
- 120.Mitchell EP, Evans L, Schultz P, Madsen R, Yarbro JW, Gehrke CW, et al. Modified nucleosides in human serum. J Chromatogr. 1992;581(1):31–40. 10.1016/0378-4347(92)80444-u. [DOI] [PubMed] [Google Scholar]
- 121.Ogawa A, Nagiri C, Shihoya W, Inoue A, Kawakami K, Hiratsuka S, et al. N(6)-methyladenosine (m(6)A) is an endogenous A3 adenosine receptor ligand. Mol Cell. 2021;81(4):659–74.e7. 10.1016/j.molcel.2020.12.038. [DOI] [PubMed] [Google Scholar]
- 122.Polachini CR, Spanevello RM, Casali EA, Zanini D, Pereira LB, Martins CC, et al. Alterations in the cholinesterase and adenosine deaminase activities and inflammation biomarker levels in patients with multiple sclerosis. Neuroscience. 2014;266:266–74. 10.1016/j.neuroscience.2014.01.048. [DOI] [PubMed] [Google Scholar]
- 123.Dale N, Tian F, Sagoo R, Phillips N, Imray C, Roffe C. Point-of-care measurements reveal release of purines into venous blood of stroke patients. Purinergic Signal. 2019;15(2):237–46. 10.1007/s11302-019-09647-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Beamer E, Lacey A, Alves M, Conte G, Tian F, de Diego-Garcia L, et al. Elevated blood purine levels as a biomarker of seizures and epilepsy. Epilepsia. 2021;62(3):817–28. 10.1111/epi.16839. [DOI] [PubMed] [Google Scholar]
- 125.Zhou X, Liu L, Lan X, Cohen D, Zhang Y, Ravindran AV, et al. Polyunsaturated fatty acids metabolism, purine metabolism and inosine as potential independent diagnostic biomarkers for major depressive disorder in children and adolescents. Mol Psychiatry. 2019;24(10):1478–88. 10.1038/s41380-018-0047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liang Q, Liu H, Zhang T, Jiang Y, Xing H, Zhang A-h. Metabolomics-based screening of salivary biomarkers for early diagnosis of Alzheimer’s disease. RSC Advances. 2015;5(116):96074–9. 10.1039/C5RA19094K. [DOI] [Google Scholar]
- 127.Liang ZH, Jia YB, Li ZR, Li M, Wang ML, Yun YL, et al. Urinary biomarkers for diagnosing poststroke depression in patients with type 2 diabetes mellitus. Diabetes Metab Syndr Obes. 2019;12:1379–86. 10.2147/dmso.S215187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Silvestris DA, Picardi E, Cesarini V, Fosso B, Mangraviti N, Massimi L, et al. Dynamic inosinome profiles reveal novel patient stratification and gender-specific differences in glioblastoma. Genome Biol. 2019;20(1):33. 10.1186/s13059-019-1647-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Grozhik AV, Olarerin-George AO, Sindelar M, Li X, Gross SS, Jaffrey SR. Antibody cross-reactivity accounts for widespread appearance of m(1)A in 5′UTRs. Nat Commun. 2019;10(1):5126. 10.1038/s41467-019-13146-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12(8):767–72. 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Khoddami V, Yerra A, Mosbruger TL, Fleming AM, Burrows CJ, Cairns BR. Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc Natl Acad Sci U S A. 2019;116(14):6784–9. 10.1073/pnas.1817334116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, et al. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 2017;24(10):870–8. 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yesbolatova A, Saito Y, Kitamoto N, Makino-Itou H, Ajima R, Nakano R, et al. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat Commun. 2020;11(1):5701. 10.1038/s41467-020-19532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]


