Abstract
Infectious disease outbreaks pose a significant threat to the conservation of chimpanzees (Pan troglodytes) and all threatened nonhuman primates. Characterizing and mitigating these threats to support the sustainability and welfare of wild populations is of the highest priority. In an attempt to understand and mitigate the risk of disease for the chimpanzees of Gombe National Park, Tanzania, we initiated a long-term health-monitoring program in 2004. While the initial focus was to expand the ongoing behavioral research on chimpanzees to include standardized data on clinical signs of health, it soon became evident that the scope of the project would ideally include diagnostic surveillance of pathogens for all primates (including people) and domestic animals, both within and surrounding the National Park. Integration of these data, along with in-depth post-mortem examinations, have allowed us to establish baseline health indicators to inform outbreak response. Here we describe the development and expansion of the Gombe Ecosystem Health project, review major findings from the research and summarize the challenges and lessons learned over the past 16 years. We also highlight future directions and present the opportunities and challenges that remain when implementing studies of ecosystem health in a complex, multi-species environment.
Keywords: chimpanzees, human-primate interactions, ecosystem health, disease transmission
Introduction
Infectious disease outbreaks pose a significant threat to the conservation of chimpanzees (Pan troglodytes) and all threatened nonhuman primates. Characterizing and mitigating these threats to support the sustainability and welfare of wild populations is of the highest priority. In an attempt to understand and mitigate the risk of disease for the chimpanzees of Gombe National Park, Tanzania, we initiated a long-term health-monitoring program in 2004. Concurrently, there has been a growing appreciation of the extent to which human and nonhuman animal health are affected by their interaction, or “interface”. Many diseases with profound historical impacts, such as smallpox, HIV, plague, and most recently COVID-19 (Morens et al., 2020), originated in nonhuman animal populations (Wolfe et al., 2007). A review of 1415 microbial spp. known to be pathogenic to humans determined that 868 (61%) are zoonoses (Taylor et al, 2001). Many pathogens have a broad host range, such that humans and populations of wild and domestic animals serve as reservoirs of infection for one another. Close contact, such as keeping livestock and pets, butchering animals, or simply sharing the same environment, can provide opportunities for pathogens to move between humans and other animals. For example, research at Gombe Stream Research Centre (GSRC; Gombe National Park, Tanzania) has played a key role in determining that the AIDS pandemic originated from human contact with chimpanzees, likely through hunting and butchering apes for the bushmeat trade (Sharp & Hahn, 2011). Recognition of the interface between wildlife, domestic animal and human health has led our project and others to expand from a single-species focus to a more holistic ecosystem health approach.
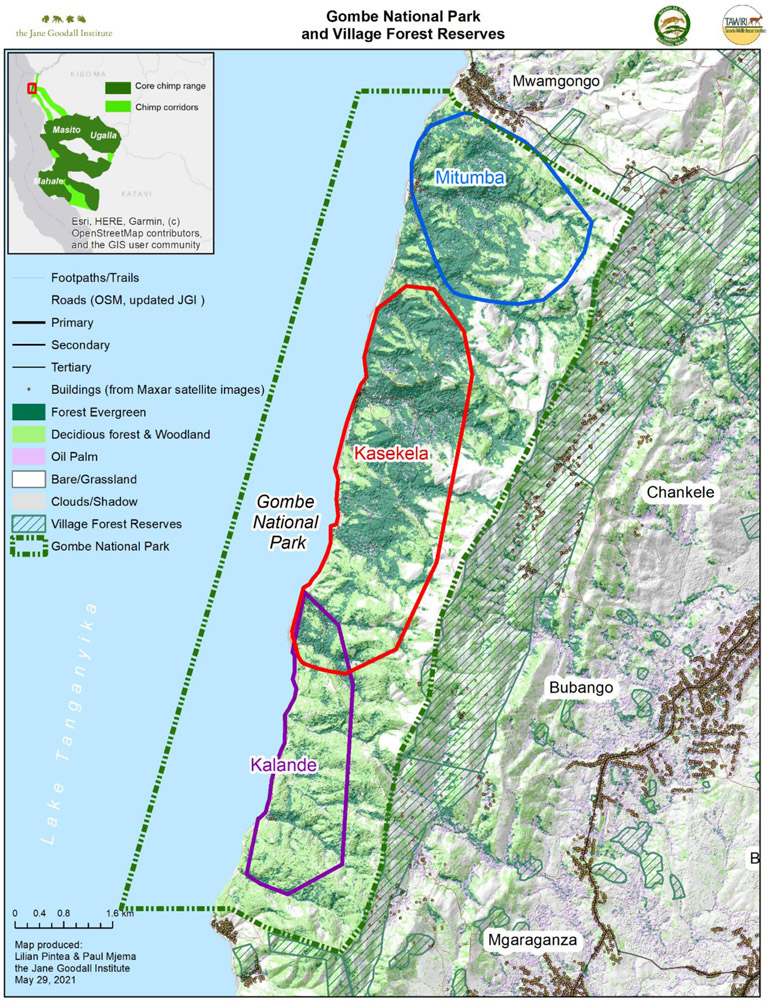
Gombe National Park is a small (land area=35.69 km2) protected area located on the western border of Tanzania and is currently home to three chimpanzee communities (see Figure 1) and several other primate species, including olive baboons (Papio anubis), red colobus monkeys (Piliocolobus tephrosceles), red-tailed monkeys (Cercopithecus ascanius schmidti), blue monkeys (C. mitis doggetti), and vervets (Chlorocebus pygerythrus). The central Kasekela community of chimpanzees was habituated in the early 1960s by J. Goodall and standardized behavioral data collection on individuals commenced in 1969. This community has contained a median of 52 individuals (range: 39-62 individuals) (Wilson et al. 2020). Habituation of the northern Mitumba community of chimpanzees began in the mid-1980s and behavioral data collection commenced in the mid-1990s (Pusey et al., 2008). The Mitumba community is smaller, containing a median of 25 individuals (range: 20-29 individuals) (Wilson et al. 2020). Members of the Kalande community in the south of the park have been monitored regularly since 1999 (Rudicell et al., 2010), but remain largely unhabituated. This community was estimated to have 19 individuals in 2002, but only 5 individuals by 2020 (Wilson et al. 2020). A long-term study on baboons, currently led by D.A. Collins, began in 1967 (Ransom, 1981) and has documented the lives of over 1260 individual baboons since it began (Bakuza, 2020). In addition, the Gombe Hybrid Monkey project, led by K. Detwiler has documented the behavior and population genetics of the Cercopithecus monkeys since 2004 (Detwiler et al., 2005).
Figure 1.
Map of the Greater Gombe Ecosystem illustrating the human-wildlife-domestic animal interface. Within Gombe National Park, chimpanzee community ranges are outlined in blue (Mitumba community), red (Kasekela community) and purple (Kalande community).
Since the inception of standardized observational data collection on the Gombe chimpanzees, the major cause of death appears to have been infectious disease (as determined by observable clinical signs of illness), although the specific pathogen responsible has rarely been confirmed (Williams et al., 2008). While the genesis of these infections is often unknown, there has been a presumed link to humans since proximity to humans has been linked to disease transmission across the human-ape interface. For example, studies have implicated humans as the source of respiratory outbreaks at both Taï National Park, Côte d'Ivoire (Köngden et al., 2008) and Kibale National Park, Uganda (Scully et al., 2018; Negrey et al., 2019). In fact, chimpanzees at Taï suffered from a human coronavirus that was genetically identical at the loci tested to a virus found during screening of a researcher (Patrono et al., 2018), highlighting the threat of the current SARS-CoV-2 pandemic to wild chimpanzee populations (Gillespie et al. 2020). In addition to these human-sourced infections, naturally occurring pathogens such as Ebola (Formenty et al., 1999) and anthrax (Leendertz et al., 2004) have also been implicated as sources of mortality in wild chimpanzee populations. At Gombe, there is evidence of periodic outbreaks of disease of various presumed causes (Williams et al., 2009), including sarcoptic mange (see Figure 2; Walton et al., 2004), polio (Goodall, 1986), and a myriad of other gastrointestinal and respiratory infections.
Figure 2.
Chimpanzee Fifi during the outbreak of Sarcoptic mange (left) and her juvenile son, Faustino, who was not severely affected. Fifi fully recovered from the outbreak, but her infant son (not pictured) was one of three infants to die during the outbreak. Photo by Anne Pusey.
As the chimpanzee population in the park decreased over time from an estimated 120-150 individuals in the 1960s to approximately 90 individuals in 2020 (Wilson et al., 2020), fear arose that an infectious disease outbreak could cause irreparable harm to the population. In hopes of better understanding the potential causes and consequences of disease, researchers and park authorities collaborated in the development of a long-term program aimed at monitoring the health status of chimpanzees. These initial efforts then expanded to other primates and to areas outside the park. Our contribution here is a summary of our efforts to:
build a (minimally invasive) syndromic health-monitoring program parallel to the long-term behavioral data collection program to establish baseline parameters of health;
integrate monitoring with ongoing noninvasive research on Simian Immunodeficiency Virus (SIVcpz), and support expansion/validation of non-invasive diagnostic investigation;
build upon chimpanzees as a focal point to examine potential pathogen exchange among and between humans, domestic animals and wildlife in the Greater Gombe Ecosystem;
illustrate the lessons we learned throughout the evolution of the project, including the importance of developing local expertise and partnerships
briefly describe the future directions of the program based upon these findings.
Our overarching goal is to provide a detailed example of the development of the Gombe Ecosystem Health Program. We recognize that all study areas have their own goals, complexities, and constraints, and that there is no “one size fits all” approach. Nevertheless, we hope that by sharing our program’s evolution, other projects can apply our lessons and findings as appropriate to their area.
1). Development of the chimpanzee health-monitoring system
Historic health monitoring
Throughout the history of the Gombe study, health-related information was collected periodically and in a variety of formats (see Lonsdorf et al., 2006 for an overview of data types). In the early decades of the study, researchers collected health data in response to outbreaks or more acute/obvious health events (e.g. severe wounds). Observers would record lists of individuals accompanied by descriptions of observable signs of ill health (hereafter: clinical signs) and/or written descriptive reports. Research staff also made efforts to collect samples from deceased chimpanzees for pathological investigation, and visiting researchers conducted analyses of gastrointestinal parasites (detailed below). These early efforts provided proof of concept and set the stage for the integrated monitoring program that is in place today.
Direct veterinary interventions for treatment of health concerns have also occurred over the course of the study. Launching interventions when wild animals fall sick raises challenging questions for both managers and researchers - particularly for those seeking to understand how animals respond to natural threats. At Gombe, efforts have been made to treat sick chimpanzees depending on the severity and/or specifics of the circumstance. For example, during a 1966 outbreak of a polio-like disease, Goodall provided chimpanzees with polio vaccine via bananas (Goodall, 1986). Researchers have attempted to provide medication in bananas on several subsequent occasions, but with varying success since they commonly detect medication and avoid consumption. Researchers and park staff have rarely undertaken interventions requiring immobilization, given the safety concerns associated with anesthetizing arboreal primates.. Additionally, there are concerns for human safety in the presence of potentially aggressive animals, and negative impacts on habituation for behavioral research and tourism (Lukasik, 2002; Travis et al., 2008). Gombe chimpanzees have been immobilized for veterinary treatment in just four instances, to: 1) obtain a biopsy from a bulbous facial growth, 2) examine and treat a testicular wound, 3) amputate a hand after a severe snare injury and 4) examine and treat an extremely ill individual exhibiting severe hind limb weakness (Lonsdorf et al., 2014). In all four cases, the chimpanzee that was immobilized survived the intervention and recovered. In the final case, we were able to collect blood samples in the hopes of obtaining a definitive diagnosis of the cause of illness. Though samples were shipped to the U.S. and Germany and the individual was seroreactive to several viral antigens - including influenza and parainfluenza - none were associated with clinical signs exhibited at the time (Lonsdorf et al. 2014). Thus, even when blood samples can be collected from a sick individual, a diagnosis can remain elusive.
Prospective monitoring of clinical signs
Formal efforts to monitor health at Gombe began in 2000, when veterinarian M. Lukasik implemented a modified health-monitoring data collection template originally created by The Gorilla Doctors (formerly the Mountain Gorilla Veterinary Project) and worked with local diagnostic labs to test samples collected noninvasively during outbreaks (Lukasik-Braum & Spelman, 2008). Because of the complexity of procuring an adequate clinical diagnosis (e.g. collection of invasive samples and need for sample preservation and diagnostic infrastructure) conservationists, researchers and managers typically employ syndromic surveillance, which entails the collection of health data on disease “syndromes” (i.e., respiratory, dermatologic, gastrointestinal) (Henning, 2004). Such surveillance can be combined with noninvasive sampling of biomaterials, such as collection of feces and urine. Following Lukasik’s initial efforts, a comprehensive non-invasive health-monitoring program was initiated by E. Lonsdorf and D. Travis in 2004 with the aim of understanding the impacts of health insults both individually and at the population level (Lonsdorf et al., 2006, 2018; Travis et al., 2008). Working with GSRC staff, they implemented a detailed standardized health datasheet that was collected on every individual who was the target of a focal follow, as well as on any individual observed to be ill. In addition, a weekly health census was conducted to record each individual in the community observed during a particular week and whether he/she had any clinical signs (Lonsdorf et al., 2006, 2018). This system built off of previous data collection efforts and remains in place today, allowing us to create long-term baselines of some indicators and investigate how health insults variably affect individuals in the population. For example, in an analysis of socioecological correlates of clinical signs in the first eight years of the study, Lonsdorf et al. (2018) found that adults are more often observed with diarrhea, loss of body condition, and wounds or lameness compared to immature individuals, while males have a higher probability of being observed with wounds or lameness than females. In contrast, signs of respiratory illness appear unrelated to chimpanzee social or demographic factors.
2). Integration of non-invasive diagnostics
An ongoing challenge for understanding the health status of wild animals is lack of access to physiological and pathological measures. In the case of chimpanzees, an endangered species, non-invasive diagnostic testing has largely been the standard (as mentioned above). However, it is often the case that non-invasive diagnostic tests for pathogen identification and/or other physiological measures simply have not been developed, although this has improved greatly over the past decade (see Calvignac-Spencer et al., 2013). In addition, most diagnostic tests are ordered based upon an “index of suspicion”, but the spectrum of possible pathogens in wild populations are often unknown. There are also many financial and logistical constraints, such as access to appropriate laboratory equipment, reagents and expertise. Despite these challenges, Gombe has served as a demonstration site for many non-invasive diagnostic approaches for wild primates (see Table 1). Using these approaches, we have identified over 30 different pathogens in the chimpanzees (see Table 2).
Table 1.
Description of data types collected as part of the Gombe Ecosystem Health Project, including years and frequency of collection and approximate sample sizes.
| Data type | Description (including species) | Years collected |
Target collection frequency (chimpanzee community of focus*) |
Average # individuals sampled/year |
Approximate # of samples collected |
Key references |
|---|---|---|---|---|---|---|
| Virology | Fresh fecal samples collected from individually identified chimpanzees placed in RNAlater | 2000-present | Annually/biannually from 2000-2009; quarterly thereafter (KK, MT, KL) | 70 | 4836 | (1, 2) |
| Observable clinical signs | Focal health observations and weekly health census | 2004-present | Monthly for focal observations, weekly for census (KK, MT) | 80 (focal) | 13,220 (focal) | (3,4) |
| Parasitology | Fresh fecal samples collected from individually identified chimpanzees placed in 10% neutral buffered formalin | 2006-present | Monthly from 2006-2009; quarterly thereafter (KK, MT) | 73 | 5827 | (5) |
| Tissues | Tissues collected from recovered corpses of any primate found in the park | 2004-present | As needed (all primates) | 3 | 40 | (6) |
| Endocrine | Fresh fecal samples collected from individually identified chimpanzees stored without preservative and frozen until processing | 2009-present | Has varied over time due to project-specific goals (primarily KK) | 54 | 6489 | (7) |
| Spatial data on species overlap | GPS data on movement of domestic dogs, goats and sheep at the park interface | 2010 | 1 dry season, 1 wet season (NA) | NA | Dogs: 8 Goats: 27 Sheep: 2 |
(8) |
| Zoonotic pathogen and antimicrobial resistance screening | Fresh fecal samples collected from targeted individuals and species (chimpanzees, baboons, humans, dogs, goats and sheep | 2010-2011 | Chimpanzees (KK, MT), baboons, humans and domestic animals: 1 dry season, 1 wet season | NA | Chimpanzees: 251 Baboons: 80 Humans: 254 Dogs: 9 Goats: 76 Sheep: 14 |
(9, 10, 11) |
(1) Santiago et al. 2002; (2) Keele et al., 2009; (3) Lonsdorf et al. 2006; (4) Lonsdorf et al. 2018; (5) Gillespie et al. 2010; (6) Terio et al. 2001; (7) Murray et al. 2013; (8) Parsons et al. 2014; (9) Parsons et al. 2015; (10) Deere et al. 2019; (11) Parsons et al. 2021
KK=Kasekela, MT=Mitumba, KL=Kalande.
Table 2.
Pathogens found over the past 15 years in the chimpanzee population of Gombe National Park, Tanzania and their spillover potential.
| Pathogen name | Pathogen type |
Potential spillover: humans |
Potential spillover: livestock/domestic animals |
Reference |
|---|---|---|---|---|
| Rotavirus A | Virus | Yes | Yes | (1) |
| Norovirus GII | Virus | Yes | Yes | (1) |
| Enterovirus | Virus | Yes | Yes | (1) |
| Adenovirus | Virus | Yes | Yes | (1) |
| SIVcpz | Virus | Yes | No | (2), (3) |
| Oesophagostumum sp. | Helminth | Yes | Yes | (3), (4), (5) |
| Strongyloides sp. | Helminth | Yes | Yes | (4) |
| Necator sp. | Helminth | Yes | Yes | (4) |
| Ascaris sp. | Helminth | Yes | Yes | (4) |
| Trichuris sp. | Helminth | Yes | Yes | (4) |
| Probstymayria sp. | Helminth | Unknown | Unknown | (4) |
| Dicrocoeliidae sp. | Helminth | Yes | Yes | (4) |
| Bertiella sp. | Helminth | Yes | No | (4) |
| L1 Strongyle | Helminth | Yes | Yes | (1) |
| Trichostrongylus sp. | Helminth | Yes | Yes | (1) |
| Physaloptera sp. | Helminth | Yes | No | (1) |
| Mammonogamous | Helminth | Unknown | Unknown | (1) |
| Cryptosporidium sp. | Protozoan | Yes | Yes | (6), (7) |
| Giardia spp. | Protozoan | Yes | Yes | (1) |
| Entamoeba histolytica | Protozoan | Yes | Yes | (8) |
| Balantidium coli | Protozoan | Yes | Yes | (4) |
| Iodamoeba sp. | Protozoan | Yes | Yes | (4) |
| Troglodytella sp. | Protozoan | No | No | (4) |
| Troglocorys cava | Protozoan | No | No | (4) |
| Chilomastix sp. | Protozoan | Yes | Yes | (1) |
| Streptococcus pneumoniae pleuropneumonia | Bacteria | Yes | Yes | (9) |
| Enterococcus faecalis | Bacteria | Yes | Yes | (1) |
| Aeromonas spp. | Bacteria | Yes | Yes | (1) |
| Salmonella sp. | Bacteria | Yes | Yes | (1) |
| Shigella sp. | Bacteria | Yes | Yes | (1) |
| Enterotoxigenic E. coli ST and LT | Bacteria | Yes | Yes | (1) |
(1) Gillespie, unpublished data; (2) Keele et al., 2009; (3) Terio et al., 2011; (4) Gillespie et al., 2010 (5) Terio et al., 2018; (6) Parsons et al., 2014; (7) Parsons et al., 2015; (8) Deere et al., 2019; (9) Wolf, Singer, et al., 2019
SIVcpz
The earliest novel non-invasive diagnostic test implemented at Gombe was for SIVcpz (Santiago et al., 2002); work on this project began in 1999, led by B. H. Hahn. While it was originally assumed that the virus would not cause illness in naturally-infected chimpanzees, integration of SIVcpz infection data with demographic and pathology data (see pathology section below) revealed that SIVcpz-infected chimpanzees have a 10-16 fold increased risk of death and can develop CD4+ T cell depletion and AIDS-like immunopathology (Keele et al., 2009; Terio et al., 2011). Moreover, SIVcpz infected females were found to have lower birth rates and higher infant mortality compared to non-infected females (Keele et al., 2009), and one community with a high SIVcpz prevalence suffered a catastrophic population decline (Rudicell et al., 2010). Those individuals that die with AIDS-like pathology exhibit destabilization of their gut microbiome in the months immediately preceding death (Barbian et al., 2015). These results indicate that SIVcpz is indeed pathogenic in its natural chimpanzee host and may contribute to population declines. In addition, samples collected in RNAlater for the SIVcpz project have proven valuable for several other research areas, including non-invasive paternity assignment (Wroblewski et al., 2009), the evolution of the gut microbiome (Moeller et al., 2014, 2015), and studies of great ape genetic diversity and population history (Prado-Martinez et al., 2013).
Enteric Pathogens and Gastrointestinal Parasites
Shortly after implementing systematic prospective health-monitoring, we began a standardized gastrointestinal parasite monitoring program. Historic surveys of the gastrointestinal parasites of Gombe’s chimpanzees span almost five decades (File et al. 1976; Nutter, 1993; Murray et al. 2000), but this was the first longitudinal effort to measure parasite infection in concert with data on behavior and clinical signs. Since the initiation of the project (led by T. Gillespie), more than 30 species of enteric pathogens and gastrointestinal parasites have been detected in the chimpanzee populations (Bakuza & Nkwengulila, 2009; Deere et al., 2019; Gillespie et al., 2010; Parsons et al., 2014, 2015), some of which cause significant illness and can circulate more broadly through the ecosystem (see Table 2).
Other Gombe primates, particularly baboons and vervet monkeys, are also susceptible to a variety of infections. Both species are eaten by chimpanzees as opportunistic prey, and both species sometimes leave the park and raid nearby villages in search of food (Bakuza 2012), creating potential for zoonotic pathogen exchange. A study by Bakuza (2012) identified Physaloptera spp., Trichuris spp., hookworms, and unidentified nematodes in Gombe vervets, with higher infection rates in monkeys ranging near Mwamgongo village where humans were infected with the same parasites. Similarly, parasites associated with diarrheal disease (Cryptosporidium hominis and Entamoeba histolytica) are prevalent in Gombe’s baboons (Parsons et al., 2015; Deere et al., 2019). Further studies are needed to explore the extent of real or potential zoonotic transmission associated with Gombe’s monkey species.
Pathology
To complement non-invasive viral and parasite screening, the veterinary pathology portion of the project was conceptualized in 2004 by K. Terio and M. Kinsel. Pathology, through complete post-mortem examinations, provides important information not just on causes of death for an individual animal but, through compilation of data from multiple individuals, provides the basis for a greater understanding of disease at the species and ecosystem level. Incorporation of pathologic evaluation of tissues has enabled us to identify the impacts of specific pathogens. As noted above, pathologic findings of depleted CD4+ T cells and AIDS-like immunopathology helped refute the assumption that SIVcpz was not pathogenic in its natural host (Keele et al., 2009). Identification of Streptococcus pneumoniae-associated pleuropneumonia and meningitis (Wolf, Singer, et al., 2019) has prompted further research into the potential role of this pathogen in Gombe respiratory disease outbreaks. Post-mortem evaluations include detailed documentation of co-morbidities, contributing to our understanding of ape health generally (Lowenstine et al., 2016; Terio et al., 2011) and providing opportunities to better understand socioecology, behavior and health. Skeletons have also been preserved where possible, providing valuable additional information on the linkages between chimpanzee life history and health (Kirchhoff, 2019).
From its inception, the pathology surveillance program has examined all retrieved nonhuman primate carcasses enabling investigation and comparison of pathogens across the ecosystem. While Oesophagostomum sp. were known to be important pathogens of chimpanzees, comparison across primates identified similar severe disease in baboons, but not in blue monkeys or red colobus (Terio et al., 2018). Parasitological studies have identified Oesophagostomum eggs in cercopithecid monkeys and colobus at other sites (e.g. Gillespie et al., 2004, 2005); thus, these findings emphasize the need to better understand these infection patterns at Gombe.
Respiratory pathogens
Outbreaks of respiratory illness have occurred repeatedly among the Gombe chimpanzees and have occasionally been associated with high mortality (Williams et al., 2008). For example, in 1987, nine individuals died during a respiratory outbreak (17% of the 53 chimpanzees in the Kasekela community; Pusey et al., 2008). To date, the only respiratory pathogens detected in association with mortality and/or respiratory outbreaks have been Streptococcus pneumoniae and S. pyogenes (Mlengeya, 2000; Wolf, Singer, et al., 2019). Fortunately, most recent outbreaks have not resulted in mortality, although that has limited our ability to confirm a diagnosis (Mlengeya, 2000; Wolf, Singer, et al., 2019). However, we are currently validating a non-invasive molecular based panel for respiratory pathogens to identify the likely causes of these outbreaks.
Given the increasing evidence regarding the potential impact of respiratory diseases spread from humans to habituated great ape populations (Gillespie et al., 2008; Köndgen et al., 2008, 2010; Leendertz et al., 2006; Palacios et al., 2011; Williams et al., 2008), an evaluation of our syndromic health monitoring system was undertaken to determine its sensitivity in detecting respiratory outbreaks (Wolf, Singer, et al., 2019). Comparison of syndromic respiratory data collected via daily and weekly monitoring revealed some discrepancies, indicating the possibility of recall bias with the latter (Wolf, Wang, et al., 2019). Although the weekly health census enhances population coverage and provides additional contextual information that is particularly relevant during outbreaks, the lower accuracy of these data required us to limit our evaluation to the data collected by daily health follows. These analyses revealed no strong seasonal or secular (over periods of years) patterns of respiratory disease in habituated communities, indicating that a simple threshold for outbreak detection could be used to signal a potentially developing outbreak. The sensitivity of the outbreak detection threshold was then evaluated using an agent-based model that simulated seeding respiratory outbreaks into the dynamic social network of the Kasekela community during surveillance (Wolf, Wang, et al., 2019), providing insights into patterns of respiratory outbreaks and limitations in surveillance. For example, simulated outbreaks were less frequent and smaller-scale at the end of the long dry season (July - September), when food was less abundant (Pusey et al. 2005) and chimpanzees were more scattered. Model output also revealed that, after controlling for outbreak size, this was also when surveillance sensitivity was lowest, suggesting that outbreaks would be much less likely to be detected by researchers at this time. Collectively, these findings established a threshold of ≥ 2 chimpanzees observed with clinical signs within a week to signal a developing respiratory outbreak. Reaching this threshold triggers an outbreak response protocol that includes increased monitoring and biological sample collection on individuals exhibiting clinical signs and their social contacts. This has also led to the ongoing optimization of protocols for the molecular screening of these non-invasively collected samples (feces and food wadges) for respiratory pathogens.
In addition to efforts to understand and detect respiratory outbreaks in chimpanzees, Gombe served as a feasibility site for the application of noninvasive screening of primates for tuberculosis in locations where spillover risks are of conservation concern (Wolf et al., 2014, 2015). This effort began shortly after a new pathogenic mycobacterial organism was discovered in wild chimpanzees in Taï National Park, Côte d’Ivoire (Coscolla et al., 2013), and marked the first large-scale, noninvasive screening of free-living primate populations for tuberculosis. Researchers conducted repeated fecal screening of habituated chimpanzees (including two ill chimpanzees displaying clinical signs consistent with tuberculosis) and baboons, but found no evidence of tuberculosis (Wolf et al., 2015). Nevertheless, this effort demonstrated the feasibility of screening in locations where spillover risks are a concern.
Physiological monitoring
Health and welfare are not just defined by the presence or absence of infectious disease. Veterinary practice includes the assessment of physiological indicators of well-being and reproductive potential (population viability and health). Over the last few decades, field studies have increasingly incorporated non-invasive monitoring of physiological markers in wild primates (reviewed in Behringer & Deschner, 2017; Higham, 2016). For example, previous efforts to examine non-invasive physiological markers at Gombe focused on reproductive endocrinology (Emery Thompson, 2005). In 2006, C. Murray led an initiative to integrate long-term monitoring program of steroidal hormones (via reproductive and glucocorticoid metabolite concentrations), using noninvasive fecal and urine sampling. Since hormones are metabolized extensively prior to excretion, quantification of hormone concentrations is usually based upon detection of their metabolites. However, metabolite excretion pathways vary by species, so physiological and biological validation is essential (e.g. Bahr et al., 2000; Palme, 2005). Additional challenges in field endocrinology include the processing and storage of samples in situ (since fecal hormone concentrations can be affected by the storage method: Khan et al., 2002; Terio et al., 2002) and the risk of bacterial degradation. While storage in ethanol can prevent hormone degradation (Wasser et al., 1998), shipping ethanol is problematic. Thus, techniques to process and store fecal steroid hormones in the field were developed, validated (Murray et al., 2013) and applied to the investigation of physiological stress in mothers and their offspring (Markham et al., 2014; Murray et al., 2018; Stanton et al., 2015). Specifically, we found that low-ranking females have higher fecal glucocorticoid metabolite (FGM) concentrations in social groups (Markham et al., 2014), and that male offspring born to low-ranking mothers were exposed to higher FGM in utero and showed decreased FGM concentrations as they aged (Murray et al., 2018).
Following those efforts, we validated a technique to characterize the immune system antibody immunoglobulin A (IgA), from chimpanzee fecal samples (Lantz et al., 2018). IgA is proposed to indicate general mucosal health and long-term stress in humans and other species. Similar to previous studies (Hau et al., 2001; Hucklebridge et al., 1998; Pihl & Hau, 2003), we found that an acute stress response induced a concomitant immune response as indicated by similar profiles in response to a stress challenge (Lantz et al., 2018). Researchers at other chimpanzee sites have developed methods that further integrate measures of health and physiological stress. For example, Emery Thompson et al. (2009) showed that urinary C-peptide levels (a measure of energy balance) of male chimpanzees at Kanyawara were significantly lower during a respiratory outbreak despite high food availability. More recently, Behringer et al., (2020) reported that urinary cortisol increased during a respiratory outbreak in the Taï chimpanzees. The development of these non-invasive methods will allow researchers to investigate if and when the immune and physiological stress responses are decoupled, which may help differentiate short-term from chronic stress.
Our capacity for sample collection, in situ processing and storage has vastly expanded over the course of the study. We have continued to collect both fecal and urine samples throughout, building a biological sample bank that can be used for diverse research projects as new physiological methods emerge. For example, banked fecal samples are now being used to characterize dietary stable isotope profiles and dietary transitions during development, and frozen urine samples are being used to determine water balance and hydration risk in our study population. We have also expanded our capacity to store and ship urine samples that can be processed for protein hormones, including vasopressin and oxytocin, which are proximate drivers of social bonding in primates and other species (e.g. chimpanzees: Carter et al., 2008; Crockford et al., 2013; Wittig et al., 2014).
3). Expansion to the ecosystem
While Gombe’s chimpanzees are relatively isolated from other chimpanzees, they exist within a complex interconnected community of species. There are numerous opportunities for zoonotic disease transmission including between people and wildlife within the park, or people, domestic animals, and pests within the anthropogenic mosaic where chimpanzees range outside the park. Understanding and responding to such community pathogen dynamics requires a One Health framework with input and collaboration from diverse stakeholders across the ecosystem in question. In 2009, T. Gillespie, E. Lonsdorf, and D. Travis began building collaborations with the Tanzanian Ministry of Health, the Kigoma District Veterinary Office, TANAPA, and communities around the national park to develop a One Health framework with support from the Morris Animal Foundation. This effort integrated diagnostic surveillance for Gombe’s chimpanzees and baboons, as well as the people and domesticated animals within the Greater Gombe Ecosystem with animal movement data, survey data, and chimpanzee observational health data.
This work has uncovered a complex system of pathogen exchange among humans, chimpanzees, baboons, and the domestic animals surrounding the park. For example, although it was known that domestic animals entered the park, Parsons et al. (2014) quantified the frequency and nature of these incursions as well as chimpanzee overlap with domestic animals outside the park. This work highlighted areas where chimpanzees raided crops as a potential hotspot for chimpanzee pathogen exposure due to high visitation rates by domestic animals (especially goats) and the presence of potentially infected animal feces (Parsons et al., 2014). Parsons et al. (2015) then used a One Health framework to screen human and nonhuman primates (NHPs) for a set of zoonotic pathogens and found complex dynamics for Cryptosporidium. For example, humans, baboons and a subset of chimpanzees were infected with Cryptosporidium hominis subtype IfA12G2 (a human subtype), while another subset of chimpanzees was infected with C. suis, a pig subtype (Parsons et al., 2015). In contrast, all positive domestic animals (goats and sheep) were infected with C. xiaoi (a bovis-like sheep subtype). The dominance of C. hominis subtype IfA12G2 among humans and NHPs suggests regular cross-species transmission, while the finding of C. suis in chimpanzees is novel. Additionally, this simultaneous screening of Gombe’s chimpanzees, baboons, and human populations demonstrated high prevalence of Entamoeba histolytica (Deere et al. 2019), the pathogen responsible for amoebic dysentery, as well as high prevalence of genes encoding antimicrobial resistance (Parsons et al. 2021). These findings suggest that zoonotic exchange of pathogenic protozoa, as well as bacteria harboring resistance genes are additional conservation and public health concerns that deserve further study (Parsons et al. 2021).
4). Summary and lessons learned
Summary of scientific findings and activities
We began our project with the assumption that disease was an important conservation threat for Gombe’s chimpanzees. However, we knew very little about the population impacts of disease, what pathogens were important, and how pathogens flowed through the system. Sixteen years later, we have implemented a multipronged non-invasive approach to identifying health concerns (see Table 1), documented the population-level impacts of disease (Rudicell et al., 2010; Williams et al., 2008; Wilson et al., 2020) and identified numerous pathogens of concern (see Table 2). We also have a better understanding of the pathogenic effects of SIVcpz (Keele et al., 2009) and the importance of other causes of death in the population, such as conspecific aggression and renal failure (Terio et al., 2011). We have assessed our surveillance systems and updated our respiratory outbreak response protocols as a result (Wolf, Singer, et al., 2019; Wolf, Wang, et al., 2019). In addition, we have integrated physiological monitoring (e.g. Murray et al., 2018) and validated a non-invasive test for IgA (Lantz et al., 2018). Our large biobank of samples has proven useful for a variety of other projects as novel laboratory approaches are validated (e.g. Bibollet-Ruche et al., 2019; Moeller et al., 2015; Ozga et al., 2020; Wroblewski et al., 2009). Finally, we have begun to broaden our approach outside of our original target species and area, to incorporate other NHPs, domestic animals, and humans both inside and outside the park (Deere et al., 2019; Parsons et al., 2014, 2015).
Training initiatives
Implementation of an integrated field health-monitoring program requires not only transdisciplinary expertise and a network of international collaborating laboratories, but is dependent upon highly trained personnel and successful partnerships on the ground. This requires that all partners have some underlying knowledge of conservation biology, the principles of shared leadership and One Health approaches, and a dedication to capacity building and transnational organizational development. While funding for such efforts can be difficult to obtain, we structured our original proposals such that an onsite Tanzanian program manager and/or veterinarian had primary oversight for implementation of the project, which justified requests for training support. Training opportunities varied throughout the project but have included participation in Pan African Sanctuary Alliance (PASA) veterinary workshops, a yearlong leadership development program, augmented by a month-long multidisciplinary training course hosted by the Lincoln Park Zoo in Chicago, support for travel to other field sites and sanctuaries to participate in veterinary procedures, and remote mentoring of graduate degree work. To date, the project has supported three doctoral students, one Fogarty Fellow, one Tanzanian MS student, and three Tanzanian veterinarians specializing in great ape health. These professionals are now employed in the region by the University of Dar es Salaam, Tanzanian National Parks (TANAPA), Tanzanian Wildlife Research Institute (TAWIRI), and the Jane Goodall Institute (JGI). The project has also provided advanced training in laboratory and research techniques to several other staff. Additionally, the American Association of Zoo Veterinarians (AAZV) supported the development of a formalized necropsy training program, including an onsite necropsy and biosafety training workshop for all veterinary staff from JGI, TANAPA and TAWIRI. This program was designed and implemented by Drs. K. Terio and M. Kinsel of the University of Illinois Zoological Pathology Program in conjunction with veterinarians from Lincoln Park Zoo and has provided didactic training support to great ape field sites and sanctuaries globally for the past fifteen years.
Risk management - implementing health and safety rules
When Gombe was first established as a national park, access was limited to research and park staff. However, by the mid-1970s, tourists began to visit the park as well. During the first decades of research at Gombe, researchers did not yet appreciate the risk of transmitting diseases between humans and chimpanzees and interacted closely with them, including provisioning them with bananas (Pusey et al., 2007). By the 1990s, however, researchers had become more sensitive to concerns about disease transmission. After a respiratory outbreak in 1996, researchers provisioning the Mitumba chimpanzees began sterilizing the bananas before giving them to chimpanzees (Pusey et al., 2008). Following a respiratory outbreak in the Kasekela community in 2000 that led to two chimpanzee deaths, researchers stopped all provisioning of chimpanzees, and worked with park staff to codify and implement regulations to reduce risk of transmitting disease to chimpanzees and other wildlife (Collins, 2003; Pusey et al., 2007). Researchers had long sought to keep at least 5 m from chimpanzees, to ensure that the chimpanzees were not disturbed by being followed. Following recommendations of Homsy (1999), protocols adopted in 2000 stipulated a minimum distance of 7.5 m for researchers, and 10 m for tourists, who are more likely to carry unfamiliar pathogens. Tourist visits were also restricted to no longer than one hour, and in groups no larger than six people. Visiting researchers were also asked to provide proof of vaccinations and complete a 7-day quarantine prior to following chimpanzees (Collins, 2003). In addition, because chimpanzees and baboons often move through areas of the park where staff live, GSRC researchers moved staff families out of the park, built wire mesh cages around the front of staff houses to prevent access to household items, and introduced a shift system to reduce the numbers of staff present full time. Improvements to infrastructure included the covering of garbage pits and pit latrines, and (starting in 2012) plumbing and flush toilets. Since 2017, observers have been required to wear face masks when in the presence of chimpanzees.
Lessons learned through project development
Our ecohealth project began as a complement to an ongoing, long-term behavioral research project. In addition, we began with decades of archival data about health concerns and information from previous studies on potential pathogens, especially respiratory viruses and gastrointestinal parasites. Our project was chimpanzee-focused at the start, given the depth of existing data and concerns about the impacts of disease on the population. While implementing observational health data collection was relatively straightforward, we also developed aspects that included parasitology and pathology, which proved more challenging (see below). Eventually, we expanded to other non-invasive diagnostics both within and outside the park. Given that our ecohealth project capitalized on an existing decades-long behavioral study, our program development may look quite different from efforts starting in a new area. Other programs may, for example, target multiple species and/or prioritize the protected area interface from the beginning. Nevertheless, there are several relevant lessons learned regardless of the project focus. First, it goes without saying that developing strong local partnerships with the governing bodies of the geographical area and the relevant wildlife and health authorities is paramount. We were fortunate that our original collaborating institution, the JGI, had many of these partnerships already through its long-term research collaborations at Gombe and Tacare, JGI’s community-driven conservation approach in the region. JGI’s Tacare program integrates conservation science into local-decision making processes (Pintea et al., 2012; Pintea 2016; Pintea et al., 2016) and lessons learned from those efforts reinforce suggestions that knowledge should not be seen as something to be transferred from researchers to stakeholders, but rather as a “process of relating that involves negotiation …among partners” (Roux et al., 2006). This requires additional considerations as to how One Health studies are designed and implemented with local communities and how the processes and results are perceived and filtered through existing beliefs, traditions, values, experiences, and concerns (Toomey et al., 2017). It has been suggested that collaborative research approaches such as participation action research (PAR) could be used to create more “research-implementation spaces” where scientists, local communities, decision-makers and other partners could engage, collaborate, identify and understand how data, information and knowledge are produced and used, by whom and for whom (Toomey et al. 2017). Thus, newer projects need to take care to not only understand the governing landscape, especially in the case of studying potentially pathogenic and zoonotic disease, but to develop a community-centered approach where site-specific values and involvement guide all aspects of the research process (Broesch et al., 2020).
Other important lessons learned surround specific logistical considerations. A primary challenge for ecosystem health work is the time and cost associated with legally and safely exporting samples (with the necessary preservation techniques) for analysis in laboratories outside of the country of origin. For example, permitting and shipping regulations change often both in the country of export and import, applying and renewing permits can take months, and maintaining a cold chain is extraordinarily difficult and expensive. If at all possible, partnering with in-country laboratories and experts is ideal. Not only does this reduce the headaches associated with permitting and shipping and specimen integrity during storage and transport, it promotes local investigators, in-country collaborations and local program capacity. This was not possible when our project began due to the specialized nature of some of the laboratory techniques we were implementing, but with the development of new and lower-tech diagnostic techniques, field-friendly infrastructure, and opportunities for remote consultation and training, this is becoming more possible. Newer projects should seek out these collaborations or seek to work collaboratively to develop the necessary infrastructure if it does not exist. For example, members of our team recently completed a project supported by the National Science Foundation to upgrade our laboratory and diagnostic capacities within Gombe. Investing in the human infrastructure is also critically important, so that projects can contribute to the growth of in-country expertise and partner effectively with the communities living in or around wild primates. We recognize that some projects may not have the resources that more mature research sites have. In these cases, we recommend prioritizing collection of fecal samples in RNAlater and, when possible, necropsies on deceased individuals with tissues preserved in both formalin and RNAlater. Such samples can be banked at ambient temperature until the appropriate diagnostic partners and funding support can be identified. Importantly, specific training is required to conduct necropsies with appropriate attention to biosafety, so pathology/biosafety experts should be consulted. The training materials developed by the Zoo Pathology Program can be found here: https://vdl.vetmed.illinois.edu/zoological-pathology-program. All of the above items take time to develop and implement, and require a long-term view of goals and objectives. The length of the project and the stability of the funding for the duration of the project should be considered from the start.
5). Future directions
Even with the luxury of 60 years of behavioral and demographic data at Gombe, and 16+ years of comprehensive health-monitoring data, there is still much more to do. The expansion of the project beyond chimpanzees and beyond the national park to include a more holistic ecosystem- and community-driven approach has illustrated the connections between human and wildlife health. As we continue, our research efforts will be embedded in local decision-making processes and connected to existing national systems in collaboration with TANAPA, TAWIRI and the Tanzanian Ministry of Health. We also plan to implement surveillance of biting insects, which serve as mechanical vectors for pathogens by moving them between individuals and between species and may act as sentinels for emerging outbreaks (Gogarten et al., 2019). This will be implemented in coordination with JGI, Microsoft Research and AI for Earth Program that uses mosquitos, robotic mosquito traps, metagenomic tools and cloud computing to detect pathogens and monitor ecosystem health in Gombe (see Project Premonition https://www.microsoft.com/en-us/research/project/project-premonition/).
We also plan to expand our spatial framework. A new Gombe One Health Hub project is using Esri’s ArcGIS mapping technologies to integrate wildlife and health data with other data layers into a unified geodatabase in the cloud. These data layers include wildlife presence and human land use activities reported by the village forest monitors (Pintea, 2016), biodiversity surveys, and human land use indicators, land cover and habitat change from 50-cm resolution satellite imagery from Maxar and Planet. Project partners will use these layers to create dynamic risk maps by overlaying heat maps of human density, habitat change, chimpanzees and other wildlife presence, and domestic animal movement throughout the Greater Gombe Ecosystem. The Gombe One Health Hub will use dashboards and story maps to communicate and share these maps with government decision-makers and other stakeholders.
Since the initiation of SIVcpz screening, Gombe has served as a demonstration site for new non-invasive diagnostic development and we will continue to focus future efforts in this area. Historically, new pathogen discoveries occurred following investigation of atypical symptoms in an affected population with microbiological detection limited to what could be cultured from a biological specimen. This approach has skewed our view of microbial diversity and appreciation for the complex microbiological interactions that manifest as clinical disease (Hugenholtz & Tyson, 2008). However, recent advances in molecular diagnostics, including metagenomics techniques, make it possible to detect and characterize unculturable pathogens and investigate the presence of genes and genomes from a mixed community of organisms rather than an individual (Bergner et al., 2020; Schloss & Handelsman, 2005). Metagenomics provides a broader view of the community structure and metabolic potential of a community (Hugenholtz & Tyson, 2008). These types of studies are likely to identify novel sequences from clinical specimens and in combination with health monitoring of humans, non-human primates and domesticated animals, correlate clinical illness with imbalances in commensal species and/or the acquisition of specific gene sequences from known pathogens (Finkbeiner et al., 2008; Wolfe et al., 2007; Zhang et al., 2006). As we advance our metagenomic analyses, which will require shipment of samples, we aim to develop in parallel, a system for rapid, on-site diagnostics via standard PCR and sequencing. Improved on-site diagnostics will also provide new avenues to understand, in real-time, disease risks allowing for a more informed approach to any potential future interventions.
Real-time data collection, on-site detection of pathogens, and a better understanding of the human-domesticated animal-wildlife contact structure within and around Gombe will improve our ability to understand the ecology and epidemiology of zoonotic pathogens. The COVID-19 pandemic has re-emphasized the threat of zoonotic disease to the primates in Gombe (Gillespie et al., 2020), and extreme precautions, including the temporary suspension of research, have been employed to safeguard their future. However, it remains difficult to prevent chimpanzees from dying when they become ill or injured. A demographic analysis by Bronikowski et al. (2011) suggested that the Gombe chimpanzee population is failing to maintain replacement-level reproductive rates. Thus, additional extreme measures such as actively intervening to provide veterinary treatment (as is currently done for mountain gorillas: Robbins et al. 2011) may become necessary to ensure the survival of Gombe chimpanzees. While our original focus on chimpanzees remains a priority, the way forward requires an integration of complexity science approaches with participatory action research at the wildlife-human-domestic animal interface.
Acknowledgements
The authors thank the Jane Goodall Institute and Tanzania National Parks (TANAPA) for initiating and supporting the 60 + year research tradition at Gombe, including the current health-monitoring project. In addition, our deepest gratitude goes to the Gombe Stream Research Centre staff, without whom our work would not be possible. Special thanks are due to the Honorable Dr. Titus Mlengeya Kamani and to the Gorilla Doctors for key support of this research at its inception. Permission to carry out research at Gombe was granted by the Government of Tanzania, Tanzania National Parks, Tanzania Commission for Science and Technology, and the Tanzania Wildlife Research Institute. This work was supported by grants from the US Fish and Wildlife Great Ape Conservation Fund, the Arcus Foundation, the Leo S. Guthman Foundation, athe National Institutes of Health (R01 AI58715, R01 AI120810, R00 HD057992), the National Science Foundation (FSML 1624552), the Morris Animal Foundation (MAF D09ZO-041, MAF D09ZO-634), Emory Global Health Institute, and the Emory Synergy II Program. Additionally, monetary support and invaluable time and effort were provided by staff and volunteers at Lincoln Park Zoo’s Davee Center for Epidemiology and Endocrinology, Lester E. Fisher for the Study and Conservation of Apes, and Emory University. We are grateful to Michael Reid, Zarin Machanda and an anonymous reviewer for helpful comments that improved the manuscript. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The authors declare no conflicts of interest.
Footnotes
Ethics statement
All work reviewed herein was non-invasive and complied with all ethical guidelines for the use of animals in research as well as all legal requirements of Tanzania, where the work was conducted.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- Bahr NI, Palme R, Möhle U, Hodges JK, & Heistermann M (2000). Comparative aspects of the metabolism and excretion of cortisol in three individual nonhuman primates. General and Comparative Endocrinology, 117(3), 427–438. 10.1006/gcen.1999.7431 [DOI] [PubMed] [Google Scholar]
- Bakuza JS (2012). Epidemiology of Schistosoma mansoni infection in sympatric humans and non-human primates in the Gombe ecosystem, Tanzania. In PhD Thesis. http://theses.gla.ac.uk/3652/1/2012bakuzaphd.pdf [Google Scholar]
- Bakuza JS (2020). Epidemiological significance of parasitic infections in olive baboons (Papio anubis) at Gombe National Park, Tanzania. Tanzania Journal of Science, 46(1), 137–150. [Google Scholar]
- Bakuza JS, & Nkwengulila G (2009). Variation over time in parasite prevalence among free-ranging chimpanzees at Gombe National Park, Tanzania. International Journal of Primatology, 30(1), 43–53. 10.1007/s10764-008-9329-7 [DOI] [Google Scholar]
- Barbian HJ, Li Y, Ramirez M, Klase Z, Lipende I, Mjungu D, … & Bushman FD (2018). Destabilization of the gut microbiome marks the end-stage of simian immunodeficiency virus infection in wild chimpanzees. American Journal of Primatology, 80(1), e22515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer V, & Deschner T (2017). Non-invasive monitoring of physiological markers in primates. Hormones and Behavior, 91, 3–18. 10.1016/j.yhbeh.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Behringer V, Preis A, Wu DF, Crockford C, Leendertz FH, Wittig RM & Deschner T (2020). Urinary cortisol increases during a respiratory outbreak in wild chimpanzees. Frontiers in Veterinary Science, 7, 485. 10.3389/fvets.2020.00485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergner LM, Orton RJ, Benavides JA, Becker DJ, Tello C, Biek R, & Streicker DG (2020). Demographic and environmental drivers of metagenomic viral diversity in vampire bats. Molecular Ecology, 29, 26–39. 10.1111/mec.15250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibollet-Ruche F, Russell RM, Liu W, Stewart-Jones GBE, Sherrill-Mix S, Li Y, Learn GH, Smith AG, Gondim MVP, Plenderleith LJ, Decker JM, Easlick JL, Wetzel KS, Collman RG, Ding S, Finzi A, Ayouba A, Peeters M, Leendertz FH, … Hahn BH (2019). CD4 receptor diversity in chimpanzees protects against SIV infection. Proceedings of the National Academy of Sciences of the United States of America, 116(8), 3229–3238. 10.1073/pnas.1821197116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broesch T, Crittenden AN, Beheim BA, Blackwell AD, Bunce JA, Colleran H, Hagel K, Kline M, McElreath R, Nelson RG, Pisor AC, Prall S, Pretelli I, Purzycki B, Quinn EA, Ross C, Scelza B, Starkweather K, Stieglitz J, & Mulder MB (2020). Navigating cross-cultural research: methodological and ethical considerations. Proceedings. Biological Sciences, 287(1935), 20201245. 10.1098/rspb.2020.1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronikowski AM, Altmann J, Brockman DK, Cords M, Fedigan LM, Pusey A, Stoinski T, Morris WF, Strier KB, & Alberts SC (2011). Aging in the natural world: comparative data reveal similar mortality patterns across primates. Science, 331, 1325–1328. 10.1126/science.1201571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvignac-Spencer S, Merkel K, Kutzner N, Kühl H, Boesch C, Kappeler PM, Metzger S, Schubert G, & Leendertz FH (2013). Carrion fly-derived DNA as a tool for comprehensive and cost-effective assessment of mammalian biodiversity. Molecular Ecology, 22(4), 915–924. 10.1111/mec.12183 [DOI] [PubMed] [Google Scholar]
- Carter CS, Grippo AJ, Pournajafi-Nazarloo H, Ruscio MG, & Porges SW (2008). Oxytocin, vasopressin and sociality. Progress in Brain Research, 170(08), 331–336. 10.1016/S0079-6123(08)00427-5 [DOI] [PubMed] [Google Scholar]
- Collins A (2003). Health Guidelines for Visiting Researchers in Gombe National Park to Minimize Risk of Disease Transmission among Primates. Pan Africa News, 10(1), 1–3. 10.5134/143425 [DOI] [Google Scholar]
- Coscolla M, Lewin A, Metzger S, Maetz-Rennsing K, Calvignac-Spencer S, Nitsche A, Dabrowski P, Radonic A, Niemann S, Parkhill J, Couacy-Hymann E, Feldman J, Comas I, Boesch C, Gagneux S, & Leendertz F (2013). Novel Mycobacterium tuberculosis complex isolate from a wild chimpanzee. Emerging Infectious Diseases, 19(6), 10.3201/eid1906.121012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockford C, Wittig RM, Langergraber K, Ziegler TE, Zuberbühler K, & Deschner T (2013). Urinary oxytocin and social bonding in related and unrelated wild chimpanzees. Proceedings of the Royal Society B: Biological Sciences, 280(1755). 10.1098/rspb.2012.2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deere JR, Parsons MB, Lonsdorf EV, Lipende I, Kamenya S, Collins DA, Travis DA, & Gillespie TR (2019). Entamoeba histolytica infection in humans, chimpanzees and baboons in the Greater Gombe Ecosystem, Tanzania. Parasitology, 146(9), 1116–1122. 10.1017/S0031182018001397 [DOI] [PubMed] [Google Scholar]
- Detwiler KM, Burrell AS, & Jolly CJ (2005). Conservation implications of hybridization in African cercopithecine monkeys. International Journal of Primatology, 26(3), 661–684. 10.1007/s10764-005-4372-0 [DOI] [Google Scholar]
- Emery Thompson M, Muller MN, Wrangham RW, Lwanga JS & Potts KB (2009). Urinary C-peptide tracks seasonal and individual variation in energy balance in wild chimpanzees. Hormones and Behavior, 55, 299–305. (DOI: 10.1016/j.yhbeh.2008.11.005) [DOI] [PubMed] [Google Scholar]
- File SK, McGrew WC, & Tutin CE (1976). The intestinal parasites of a community offeral chimpanzees, Pan troglodytes schweinfurthii. The Journal of Parasitology, 62(2), 259–261. 10.2307/3279280 [DOI] [PubMed] [Google Scholar]
- Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, & Wang D (2008). Metagenomic analysis of human diarrhea: Viral detection and discovery. PLoS Pathogens, 4(2). 10.1371/journal.ppat.1000011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formenty P, Boesch C, Wyers M, Steiner C, Donati F, Dind F, Walker F, & Le Guenno B (1999). Ebola virus outbreak among wild chimpanzees living in a rain forest of Cote d’Ivoire. Journal of Infectious Diseases, 179(SUPPL. 1), 120–126. 10.1086/514296 [DOI] [PubMed] [Google Scholar]
- Frank DN, & Pace NR (2008). Gastrointestinal microbiology enters the metagenomics era. Current Opinion in Gastroenterology, 24(1), 4–10. 10.1097/MOG.0b013e3282f2b0e8 [DOI] [PubMed] [Google Scholar]
- Gillespie TR, Greiner EC, & Chapman CA (2004). Gastrointestinal parasites of the guenons of western Uganda. Journal of Parasitology, 90(6), 1356–1360. 10.1645/GE-311R [DOI] [PubMed] [Google Scholar]
- Gillespie TR, Greiner EC, & Chapman CA (2005). Gastrointestinal parasites of the colobus monkeys of Uganda. Journal of Parasitology, 91(3), 569–573. 10.1645/GE-434R [DOI] [PubMed] [Google Scholar]
- Gillespie TR, & Leendertz FH (2020). Great-ape health in human pandemics. Nature, 579, 497. 10.3928/0098-9134-19830501-03 [DOI] [PubMed] [Google Scholar]
- Gillespie TR, Lonsdorf EV, Canfield EP, Meyer DJ, Nadler Y, Raphael J, Pusey AE, Pond J, Pauley J, Mlengeya T, & Travis DA (2010). Demographic and ecological effects on patterns of parasitism in eastern chimpanzees (Pan troglodytes schweinfurthii) in Gombe National Park, Tanzania. American Journal of Physical Anthropology, 143(4), 534–544. 10.1002/ajpa.21348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie TR, Nunn CL, & Leendertz FH (2008). Integrative approaches to the study of primate infectious disease: implications for biodiversity conservation and global health. Yearbook of Physical Anthropology, 51, 53–69. 10.1002/ajpa.20949 [DOI] [PubMed] [Google Scholar]
- Gogarten JF, Düx A, Mubemba B, Pléh K, Hoffmann C, Mielke A, Müller-Tiburtius J, Sachse A, Wittig RM, Calvignac-Spencer S, & Leendertz FH (2019). Tropical rainforest flies carrying pathogens form stable associations with social nonhuman primates. Molecular Ecology, 28(18), 4242–4258. 10.1111/mec.15145 [DOI] [PubMed] [Google Scholar]
- Goodall J (1986). The Chimpanzees of Gombe: patterns of behavior. Harvard University Press. [Google Scholar]
- Hau J, Andersson E, & Carlsson HE (2001). Development and validation of a sensitive ELISA for quantification of secretory IgA in rat saliva and faeces. Laboratory Animals, 35(4), 301–306. 10.1258/0023677011911822 [DOI] [PubMed] [Google Scholar]
- Henning KJ (2004). What is syndromic surveillance? MMWR Supplements, 53, 5–11. [PubMed] [Google Scholar]
- Higham JP (2016). Field endocrinology of nonhuman primates: past, present, and future. Hormones and Behavior, 84, 145–155. 10.1016/j.yhbeh.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Homsy J (1999). Ape tourism and human diseases: how close should we get? Tourism, February, 70. http://www.igcp.org/library/ [Google Scholar]
- Hucklebridge F, Clow A, & Evans P (1998). The relationship between salivary secretory immunoglobulin A and cortisol: Neuroendocrine response to awakening and the diurnal cycle. International Journal of Psychophysiology, 31(1), 69–76. 10.1016/S0167-8760(98)00042-7 [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, & Tyson GW (2008). Metagenomics. Nature, 455(25), 481–483. [DOI] [PubMed] [Google Scholar]
- Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, Li Y, Learn GH, Beasley TM, Schumacher-Stankey J, Wroblewski E, Mosser A, Raphael J, Kamenya S, Lonsdorf EV, Travis DA, Mlengeya T, Kinsel MJ, Else JG, … Hahn BH (2009). Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature, 460(7254), 515–519. 10.1038/nature08200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MZ, Altmann J, Isani SS, & Yu J (2002). A matter of time: Evaluating the storage of fecal samples for steroid analysis. General and Comparative Endocrinology, 128(1), 57–64. 10.1016/S0016-6480(02)00063-1 [DOI] [PubMed] [Google Scholar]
- Kirchhoff CA (2019). Life and Death in the Gombe Chimpanzees: Skeletal Analysis as an Insight into Life History (Barrett L (ed.); Developmen). Springer. [Google Scholar]
- Köndgen S, Kuhl H, N’Goran PK, Walsh PD, Schenk S, Ernst N, Biek R, Formenty P, Mtz-Rensing K, Schweiger B, Junglen S, Ellerbrok H, Nitsche A, Briese T, Lipkin WI, Pauli G, Boesch C, & Leendertz FH (2008). Pandemic Human Viruses Cause Decline of Endangered Great Apes. Current Biology, 18(4), 260–264. 10.1016/j.cub.2008.01.012 [DOI] [PubMed] [Google Scholar]
- Köndgen S, Schenk S, Pauli G, Boesch C, & Leendertz FH (2010). Noninvasive monitoring of respiratory viruses in wild chimpanzees. EcoHealth, 7(3), 332–341. 10.1007/s10393-010-0340-z [DOI] [PubMed] [Google Scholar]
- Lantz EL, Lonsdorf EV, Heintz MR, Murray CM, Lipende I, Travis DA, & Santymire RM (2018). Non-invasive quantification of immunoglobulin A in chimpanzees (Pan troglodytes schweinfurthii) at Gombe National Park, Tanzania. American Journal of Primatology, 80(1), 1–9. 10.1002/ajp.22558 [DOI] [PubMed] [Google Scholar]
- Leendertz FH, Ellerbrok H, Boesch C, Couacy-Hymann E, Mätz-Rensing K, Hakenbeck R, Bergmann C, Abaza P, Junglen S, Moebius Y, Vigilant L, Formenty P, & Pauli G (2004). Anthrax kills wild chimpanzees in a tropical rainforest. Nature, 430(6998), 451–452. 10.1038/nature02722 [DOI] [PubMed] [Google Scholar]
- Leendertz FH, Pauli G, Maetz-Rensing K, Boardman W, Nunn C, Ellerbrok H, Jensen SA, Junglen S, & Christophe B (2006). Pathogens as drivers of population declines: The importance of systematic monitoring in great apes and other threatened mammals. Biological Conservation, 131(2), 325–337. 10.1016/j.biocon.2006.05.002 [DOI] [Google Scholar]
- Lonsdorf E, Travis D, Ssuna R, Lantz E, Wilson M, Gamble K, Terio K, Leendertz F, Ehlers B, Keele B, Hahn B, Gillespie T, Pond J, Raphael J, & Collins A (2014). Field immobilization for treatment of an unknown illness in a wild chimpanzee (Pan troglodytes schweinfurthii) at Gombe National Park, Tanzania: Findings, challenges, and lessons learned. Primates, 55(1), 89–99. 10.1007/s10329-013-0372-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdorf EV, Travis D, Pusey AE, & Goodall J (2006). Using retrospective health data from the Gombe chimpanzee study to inform future monitoring efforts. American Journal of Primatology, 68, 897–908. 10.1002/ajp.20296 [DOI] [PubMed] [Google Scholar]
- Lonsdorf EV, Gillespie TR, Wolf TM, Lipende I, Raphael J, Bakuza J, Murray CM, Wilson ML, Kamenya S, Mjungu D, Collins DA, Gilby IC, Stanton MA, Terio KA, Barbian HJ, Li Y, Ramirez M, Krupnick A, Seidl E, … Travis DA (2018). Socioecological correlates of clinical signs in two communities of wild chimpanzees (Pan troglodytes) at Gombe National Park, Tanzania. American Journal of Primatology, 80(1), 1–20. 10.1002/ajp.22562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstine LJ, McManamon R, & Terio KA (2016). Comparative Pathology of Aging Great Apes: Bonobos, Chimpanzees, Gorillas, and Orangutans. Veterinary Pathology, 53(2), 250–276. 10.1177/0300985815612154 [DOI] [PubMed] [Google Scholar]
- Lukasik-Braum M, & Spelman L (2008). Chimpanzee respiratory disease and visitation rules at Mahale and Gombe National Parks in Tanzania. American Journal of Primatology, 70(8), 734–737. 10.1002/ajp.20568 [DOI] [PubMed] [Google Scholar]
- Lukasik M (2002). Establishing a long-term veterinary project for free-ranging chimpanzees in Tanzania. Pan Africa News, 9(2), 13–17. 10.5134/143417 [DOI] [Google Scholar]
- Markham AC, Santymire RM, Lonsdorf EV, Heintz MR, Lipende I, & Murray CM (2014). Rank effects on social stress in lactating chimpanzees. Animal Behaviour, 87(C), 195–202. 10.1016/j.anbehav.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlengeya T (2000). TANAPA Veterinary Department Annual Report 2000/2001. Respiratory disease outbreak in the chimpanzee population of Gombe National Park. [Google Scholar]
- Moeller AH, Li Y, Ngole EM, Ahuka-Mundeke S, Lonsdorf EV, Pusey AE, Peeters M, Hahn BH, & Ochman H (2014). Rapid changes in the gut microbiome during human evolution. Proceedings of the National Academy of Sciences of the United States of America, 111(46), 16431–16435. 10.1073/pnas.1419136111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Peeters M, Ayouba A, Ngole EM, Esteban A, Hahn BH, & Ochman H (2015). Stability of the gorilla microbiome despite simian immunodeficiency virus infection. Molecular Ecology, 24(3), 690–697. 10.1111/mec.13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens DM, Breman JG, Calisher CH, Doherty PC, Hahn BH, Keusch GT, Kramer LD, LeDuc JW, Monath TP, & Taubenberger JK (2020). The origin of COVID-19 and why it matters. American Journal of Tropical Medicine and Hygiene, 103(3), 955–959. 10.4269/ajtmh.20-0849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CM, Heintz MR, Lonsdorf EV, Parr LA, & Santymire RM (2013). Validation of a field technique and characterization of fecal glucocorticoid metabolite analysis in wild chimpanzees (Pan troglodytes). American Journal of Primatology, 75(1), 57–64. 10.1002/ajp.22078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CM, Stanton MA, Wellens KR, Santymire RM, Heintz MR, & Lonsdorf EV (2018). Maternal effects on offspring stress physiology in wild chimpanzees. American Journal of Primatology, 80(1), 1–12. 10.1002/ajp.22525 [DOI] [PubMed] [Google Scholar]
- Murray S, Stem C, Boudreau B, & Goodall J (2000). Intestinal parasites of baboons (Papio cynocephalus anubis) and chimpanzees (Pan troglodytes) in Gombe. Journal of Zoo and Wildlife Medicine, 31(2), 176–178. 10.1638/1042-7260(2000)031 [DOI] [PubMed] [Google Scholar]
- Negrey JD, Reddy RB, Scully EJ, Phillips-Garcia S, Owens LA, Langergraber KE, Mitani JC, Emery Thompson M, Wrangham RW, Muller MN, Otali E, Machanda Z, Hyeroba D, Grindle KA, Pappas TE, Palmenberg AC, Gern JE, & Goldberg TL (2019). Simultaneous outbreaks of respiratory disease in wild chimpanzees caused by distinct viruses of human origin. Emerging Microbes and Infections, 8(1), 139–149. 10.1080/22221751.2018.1563456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutter FB (1993). A Comparison of Gastrointestinal Parasites in Two Communities of Chimpanzees at Gombe National Park, Tanzania. Tufts University School of Veterinary Medicine. [Google Scholar]
- Ozga AT, Webster TH, Gilby IC, Wilson MA, Nockerts RS, Wilson ML, Pusey AE, Li Y, Hahn BH, & Stone AC (2020). Urine as a high-quality source of host genomic DNA from wild populations. Molecular Ecology Resources, March, 1–13. 10.1111/1755-0998.13260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer C, Collins DA, Sindimwo A, & Goodall J (1995). Reproductive constraints on aggressive competition in female baboons. Nature, 373, 60–63. [DOI] [PubMed] [Google Scholar]
- Palacios G, Lowenstine LJ, Cranfield MR, Gilardi KVK, Lukasik-Braum M, Kinani J-F, Mudakikwa A, Nyirakaragire E, Bussetti AV, Savji N, Hutchison S, Egholm M, & Lipkin WI (2011). Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerging Infectious Diseases, 17(4), 711–713. 10.3201/eid1704100883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palme R (2005). Measuring fecal steroids: Guidelines for practical application. Annals of the New York Academy of Sciences, 1046, 75–80. 10.1196/annals.1343.007 [DOI] [PubMed] [Google Scholar]
- Parsons MB, Gillespie TR, Lonsorf EV, Travis D, Lipende I, Gilagiza B, Kamenya S, Pintea L, & Vazquez-Prokopec GM (2014). Global positioning system data-loggers: A tool to quantify fine-scale movement of domestic animals to evaluate potential for zoonotic transmission to an endangered wildlife population. PLoS Neglected Tropical Diseases, 9(11), 1–7. 10.1371/journal.pone.0110984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MB, Travis D, Lonsdorf EV, Lipende I, Roellig DMA, Kamenya S, Zhang H, Xiao L, & Gillespie TR (2015). Epidemiology and molecular characterization of Cryptosporidium spp. in humans, wild primates, and domesticated animals in the Greater Gombe Ecosystem, Tanzania. PLoS Neglected Tropical Diseases, 9(2), 1–14. 10.1371/journal.pntd.0003529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MB, Travis DA, Lonsdorf EV, Lipende I, Elchoufi D, Gilagiza B, Collins DA, Kamenya S, Tauxe R, Gillespie TR (2021). Antimicrobial resistance creates threat to chimpanzee health and conservation in the wild. Pathogens, 10(4), 477. 10.3390/pathogens10040477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrono LV, Samuni L, Corman VM, Nourifar L, Röthemeier C, Wittig RM, Drosten C, Calvignac-Spencer S, & Leendertz FH (2018). Human coronavirus OC43 outbreak in wild chimpanzees, Côte d’Ivoire. Emerging Microbes and Infections, 7(1). 10.1038/s41426-018-0121-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihl L, & Hau J (2003). Faecal corticosterone and immunoglobulin A in young adult rats. Laboratory Animals, 37(2), 166–171. 10.1258/00236770360563822 [DOI] [PubMed] [Google Scholar]
- Pintea L (2016). Geodesign Restores Chimpanzee Habitats in Tanzania. ArcNews. https://www.esri.com/about/newsroom/arcnews/geodesign-restores-chimpanzee-habitats-in-tanzania/ [Google Scholar]
- Pintea L, Pusey AE, Wilson ML, Gilby IC, Collins DA, Kamenya S, & Goodall J (2012). Long-term changes in the ecological factors surrounding the chimpanzees of Gombe National Park: Impacts on Biodiversity and Ecosystems. In Plumptre AJ (Ed.), Long Term Changes in Africa’s Rift Valley: Impacts on Biodiversity and Ecosystems. Nova Science Publishers. [Google Scholar]
- Pintea L, Mtiti ER, Mavanza M, Abdallah F, Kashula A, Mjungu D, Collins DA, Kamenya S, 2016. 20 years and counting: Adaptive management of chimpanzee habitats in the Greater Gombe Ecosystem, Tanzania. Conservation Measures Partnership Open Standards for the Practice of Conservation. https://docs.google.com/document/d/1WXt0tSDKfEIbraoG4jcAL7hVx0MDEap2Lfdtu50748a3FM/edit?usp=sharing [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, Cagan A, Theunert C, Casals F, Laayouni H, Munch K, Hobolth A, Halager AE, Malig M, Hernandez-Rodriguez J, … Marques-Bonet T (2013). Great ape genetic diversity and population history. Nature, 499(7459), 471–475. 10.1038/nature12228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusey AE, Oehlert GW, Williams JM, & Goodall J (2005). Influence of ecological and social factors on body mass of wild chimpanzees. International Journal of Primatology, 26(1), 3–31. 10.1007/s10764-005-0721-2 [DOI] [Google Scholar]
- Pusey AE, Pintea L, Wilson ML, Kamenya S, & Goodall J (2007). The contribution of long-term research at Gombe National Park to chimpanzee conservation. Conservation Biology, 21(3), 623–634. 10.1111/j.1523-1739.2007.00704.x [DOI] [PubMed] [Google Scholar]
- Pusey AE, Wilson ML, & Anthony Collins D (2008). Human impacts, disease risk, and population dynamics in the chimpanzees of Gombe National Park, Tanzania. American Journal of Primatology, 70(8), 738–744. 10.1002/ajp.20567 [DOI] [PubMed] [Google Scholar]
- Ransom TW (1981). Beach troop of the Gombe. Bucknell University Press. [Google Scholar]
- Robbins MM, Gray M, Fawcett KA, Nutter FB, Uwingeli P, Mburanumwe I, Kagoda E, Basabose A, Stoinski TS, Cranfield MR, Byamukama J, Spelman LH, & Robbins AM (2011). Extreme conservation leads to recovery of the virunga mountain gorillas. PLoS ONE, 6(6). 10.1371/journal.pone.0019788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux DJ, Rogers KH, Biggs HC, Ashton PJ & Sergeant A (2006). Bridging the science–management divide: moving from unidirectional knowledge transfer to knowledge interfacing and sharing. Ecology and Society 11(1): 4. http://www.ecologyandsociety.org/vol11/iss1/art4/ [Google Scholar]
- Rudicell RS, Jones JH, Wroblewski EE, Learn GH, Li Y, Robertson JD, Greengrass E, Grossmann F, Kamenya S, Pintea L, Mjungu DC, Lonsdorf EV, Mosser A, Lehman C, Collins DA, Keele BF, Goodall J, Hahn BH, Pusey AE, & Wilson ML (2010). Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathogens, 6(9). 10.1371/journal.ppat.1001116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Rodenburg CM, Kamenya S, Bibollet-Ruche F, Gao F, Bailes E, Meleth S, Soong SJ, Kilby JM, Moldoveanu Z, Fahey B, Muller MN, Ayouba A, Nerrienet E, McClure HM, Heeney JL, Pusey AE, Collins DA, Boesch C, … Hahn BH (2002). SIVcpz in wild chimpanzees. Science, 295(5554), 465. 10.1126/science.295.5554.465 [DOI] [PubMed] [Google Scholar]
- Schloss PD, & Handelsman J (2005). Metagenomics for studying unculturable microorganisms: Cutting the Gordian knot. Genome Biology, 6(8), 6–9. 10.1186/gb-2005-6-8-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully EJ, Basnet S, Wrangham RW, et al. (2018). Lethal respiratory disease associated with human Rhinovirus C in wild chimpanzees, Uganda, 2013. Emerging Infectious Diseases, 24(2), 267–274. doi: 10.3201/eid2402.170778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, & Hahn BH (2011). Origins of HIV and the AIDS Pandemic. Cold Spring Harbor Perspectives in Medicine. 10.1101/cshperspect.a006841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton MA, Heintz MR, Lonsdorf EV, Santymire RM, Lipende I, & Murray CM (2015). Maternal behavior and physiological stress levels in wild chimpanzees (Pan troglodytes schweinfurthii) International Journal of Primatology, 36(3), 473–488. 10.1007/s10764-015-9836-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terio KA, Brown JL, Moreland R, & Munson L (2002). Comparison of different drying and storage methods on quantifiable concentrations of fecal steroids in the cheetah. Zoo Biology, 21(3), 215–222. 10.1002/zoo.10036 [DOI] [Google Scholar]
- Terio KA, Kinsel MJ, Raphael J, Mlengeya T, Lipende I, Kirchhoff CA, Gilagiza B, Wilson ML, Kamenya S, Estes JD, Keele BF, Rudicell RS, Liu W, Patton S, Collins A, Hahn BH, Travis DA, & Lonsdorf EV (2011). Pathologic lesions in chimpanzees (Pan trogylodytes schweinfurthii) from Gombe National Park, Tanzania, 20042010. Journal of Zoo and Wildlife Medicine, 42(4), 597–607. 10.1638/2010-0237.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terio KA, Lonsdorf EV, Kinsel MJ, Raphael J, Lipende I, Collins A, Li Y, Hahn BH, Travis DA, & Gillespie TR (2018). Oesophagostomiasis in non-human primates of Gombe National Park, Tanzania. American Journal of Primatology, 80(1), 1–9. 10.1002/ajp.22572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ME (2005). Reproductive endocrinology of wild female chimpanzees (Pan troglodytes schweinfurthii): Methodological considerations and the role of hormones in sex and conception. American Journal of Primatology, 67(1), 137–158. 10.1002/ajp.20174 [DOI] [PubMed] [Google Scholar]
- Toomey AH, Knight AT, & Barlow J (2017). Navigating the space between research and implementation in conservation. Conservation Letters, 10(5), 619–625. [Google Scholar]
- Travis DA, Lonsdorf EV, Mlengeya T, & Raphael J (2008). A science-based approach to managing disease risks for ape conservation. American Journal of Primatology, 70(8), 745–750. 10.1002/ajp.20566 [DOI] [PubMed] [Google Scholar]
- Vourc’h G, Plantard O, & Morand S (2012). How Does Biodiversity Influence the Ecology of Infectious Disease? In Morand S, Beaudeau F, & Cabaret J (Eds.), New Frontiers of Molecular Epidemiology of Infectious Diseases (pp. 291–309). Springer Netherlands. 10.1007/978-94-007-2114-2_13 [DOI] [Google Scholar]
- Walton SF, Dougall A, Pizzutto S, Holt D, Taplin D, Arlian LG, Morgan M, Currie BJ, & Kemp DJ (2004). Genetic epidemiology of Sarcoptes scabiei (Acari: Sarcoptidae) in northern Australia. International Journal for Parasitology, 34(7), 839–849. 10.1016/j.ijpara.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Wasser SK, Risler L, & Steiner RA (1988). Excreted steroids in primate feces over the menstrual cycle and pregnancy. Biology of Reproduction, 39(4), 862–872. 10.1095/biolreprod39.4.862 [DOI] [PubMed] [Google Scholar]
- Williams JM, Lonsdorf EV, Wilson ML, Schumacher-Stankey J, Goodall J, & Pusey AE (2008). Causes of death in the Kasekela chimpanzees of Gombe National Park, Tanzania. American Journal of Primatology, 70(8), 766–777. 10.1002/ajp.20573 [DOI] [PubMed] [Google Scholar]
- Wilson ML, Lonsdorf EV, Mjungu DC, Kamenya S, Kimaro EW, Collins DA, Gillespie TR, Travis DA, Lipende I, Mwacha D, Ndimuligo SA, Pintea L, Raphael J, Mtiti ER, Hahn BH, Pusey AE, & Goodall J (2020.). Research and conservation in the Greater Gombe Ecosystem: Challenges and opportunities. Biological Conservation. 252, 108853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig RM, Crockford C, Deschner T, Langergraber KE, Ziegler TE, & Zuberbühler K (2014). Food sharing is linked to urinary oxytocin levels and bonding in related and unrelated wild chimpanzees. Proceedings of the Royal Society B: Biological Sciences, 281(1778). 10.1098/rspb.2013.3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf TM, Mugisha L, Shoyama FM, O ’ Malley MJ, Flynn JL, Asiimwe B, Travis DA, Singer RS, & Sreevatsan S (2015). Noninvasive test for tuberculosis detection among primates. Emerging Infectious Diseases, 21(3), 468–470. 10.3201/eid2103.140052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf TM, Singer RS, Lonsdorf EV, Maclehose R, Gillespie TR, Lipende I, Raphael J, Terio K, Murray C, Pusey A, Hahn BH, Kamenya S, Mjungu D, & Travis DA (2019). Syndromic surveillance of respiratory disease in free-living chimpanzees. EcoHealth. 10.1007/s10393-019-01400-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf TM, Sreevatsan S, Travis D, Mugisha L, & Singer RS (2014). The risk of tuberculosis transmission to free-ranging great apes. American Journal of Primatology, 76(1), 2–13. 10.1002/ajp.22197 [DOI] [PubMed] [Google Scholar]
- Wolf TM, Wang W. “Annie,” Lonsdorf EV, Gillespie TR, Pusey A, Gilby IC, Travis DA, & Singer RS (2019). Optimizing syndromic health surveillance in free ranging great apes: The case of Gombe National Park. Journal of Applied Ecology, 56(3), 509–518. 10.1111/1365-2664.13284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Dunavan CP, & Diamond J (2007). Origins of major human infectious diseases. Nature, 447(7142), 279–283. 10.1038/nature05775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski EE, Murray CM, Keele BF, Schumacher-Stankey JC, Hahn BH, & Pusey AE (2009). Male dominance rank and reproductive success in chimpanzees, Pan troglodytes schweinfurthii. Animal Behaviour, 77(4), 873–885. 10.1016/j.anbehav.2008.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SWL, Hibberd ML, Liu ET, Rohwer F, & Ruan Y (2006). RNA viral community in human feces: Prevalence of plant pathogenic viruses. PLoS Biology, 4(1), 0108–0118. 10.1371/journal.pbio.0040003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.