Abstract
Inflammation is at the forefront of carcinogenesis, tumor progression and resistance to therapy. The Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling axis is a central pathway that mediates the cellular response to inflammation and contributes to carcinogenesis. The JAK/STAT pathway coordinates intercellular communication between tumor cells and their immune microenvironment, and JAK/STAT activation leads to the expression of a variety of proteins involved in cell proliferation, cell survival, stemness, self-renewal, evasion of immunosurveillance mechanisms and overall tumor progression. Activation of JAK/STAT signaling also mediates resistance to radiation therapy or cytotoxic agents and modulates tumor cell responses to molecularly targeted and immune modulating drugs. Despite extensive research focused on understanding its signaling mechanisms and downstream phenotypic and functional consequences in hematological disorders, the importance of JAK/STAT signaling in solid tumor initiation and progression has been underappreciated. We highlight the role of chronic inflammation in cancer, the epidemiological evidence for contribution of JAK/STAT to carcinogenesis, the current cancer prevention measures involving JAK/STAT inhibition and the impact of JAK/STAT signaling activity on cancer development, progression and treatment resistance. We also discuss recent therapeutic advances in targeting key factors within the JAK/STAT pathway with single agents and the use of these agents in combination with other targeted therapies and immune checkpoint inhibitors.
Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling is a central hub bridging inflammation with cancer development, progression and therapy resistance. JAK/STAT inhibitors are being explored as cancer prevention supplements as well as therapeutic agents to sensitize tumors to chemotherapy, radiation, targeted therapy and immunotherapy.
Introduction
Inflammation is a hallmark of cancer, and chronic inflammation is associated with ~15–25% of cancer cases or deaths worldwide (1,2). Inflammation has also been proposed to contribute to carcinogenesis even without obvious signs of inflammatory conditions. Whether clinically evident or occult, inflammation is triggered by a large variety of factors, each of which is individually implicated in increasing the risk of cancer, including infection, injury, environmental exposures, autoimmune disorders and obesity.
Perhaps the best documented epidemiological evidence linking inflammation to cancer is within the digestive system, where inflammation caused by poor diet, certain gut microbiota and infection is prevalent. Unresolved inflammation caused by Helicobacter pylori infection contributes to ~75% of gastric cancers and certain lymphomas (3,4). Chronic inflammation associated with the autoimmune disorders ulcerative colitis and Crohn’s disease increases the risk of developing colon cancer (5). Accessory digestive organs are also highly susceptible to inflammation-related cancers. For instance, inflammation caused by chronic pancreatitis is associated with a higher risk of developing pancreas cancer (6). Hepatitis B or C infection increases the risk of hepatocellular carcinoma (7). Additionally, inflammation instigated by dietary factors such as frequent alcohol consumption and exposure to aflatoxins contributes to esophageal cancer and hepatocellular carcinoma (7). Inflammation also instigates cancer development in tissues outside the gut. The most notable example is the respiratory system, where ~80–90% of lung cancers are attributed to smoking exposures. Carcinogenic toxins in tobacco smoke elicit genetic and epigenetic changes in the cells lining the airways, causing chronic inflammation. Cigarette smoke also contains reactive oxygen species that drive expression of pro-inflammatory genes. Lung cancer risk is also increased by other chronic inflammatory conditions including asbestosis, emphysema, asthma and chronic bronchitis (8).
Intrinsic factors are also in play. With the increased prevalence of obesity worldwide, much attention has been recently given to the pro-inflammatory environment produced by excessive adipose tissue. The relationship between obesity and various cancers, including non–Hodgkin lymphoma and tumors of the breast, colon, esophagus, liver, pancreas, gallbladder, kidney and uterine endometrium, is now firmly established (9). Thus, chronic inflammation is unmistakably an overarching element linked to cancer. Our improved understanding of the mechanistic underpinnings of chronic inflammation in cancer development has led to an increased awareness about cancer prevention, augmented our understanding of tumor biology and expanded research focused on improving cancer therapy.
Mediators of inflammation
Inflammation is marked by elevated levels of cytokines and other secreted factors, including interleukin (IL)-6, IL-8, interferon (IFN)γ, tumor growth factor (TGF)-β, tumor necrosis factor-α, vascular–endothelial growth factor and nitric oxide (NO), among others. These factors are upregulated in response to inflammation and perpetuate the pro-inflammatory environment, leading to increased oxidative stress and subsequent DNA damage. Additionally, they contribute to malignant transformation by altering gene expression, cell proliferation, cellular senescence, cell survival and angiogenesis. Once cancer develops, IL-6 and IL-8, among others, are secreted by tumor infiltrating immune cells, e.g. tumor-associated macrophages, as well as other cells within the tumor microenvironment (TME), such as adipocytes, stromal fibroblasts and the tumor cells themselves (10,11). The secreted cytokine cascades propagate systemic and intratumor feed-forward loops, further amplifying the pro-inflammatory and pro-tumorigenic environment (12), as detailed below.
When a cell recognizes external pro-inflammatory signals, a number of downstream signaling pathways are activated. For example, the nuclear factor-kappaB pathway is activated in numerous immune and epithelial cell types in response to external inflammation or antigens (e.g. IL1, lipopolysaccharides), which then drives cell proliferation and cell survival by upregulating genes involved in inflammation and cancer development (13). Hypoxia inducible factors also play an essential role in this process. Inflamed tissue causes local oxygen concentrations to decline, leading to activation of hypoxia inducible factors in the hypoxic tissues. Hypoxia inducible factors induce upregulation of pro-tumorigenic signals, including tumor growth and metastases (14). One of the best recognized pro-tumorigenic signaling hubs that perpetuates this pro-inflammatory environment is the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway, the topic of this review.
JAK/STAT signaling pathway and its regulation
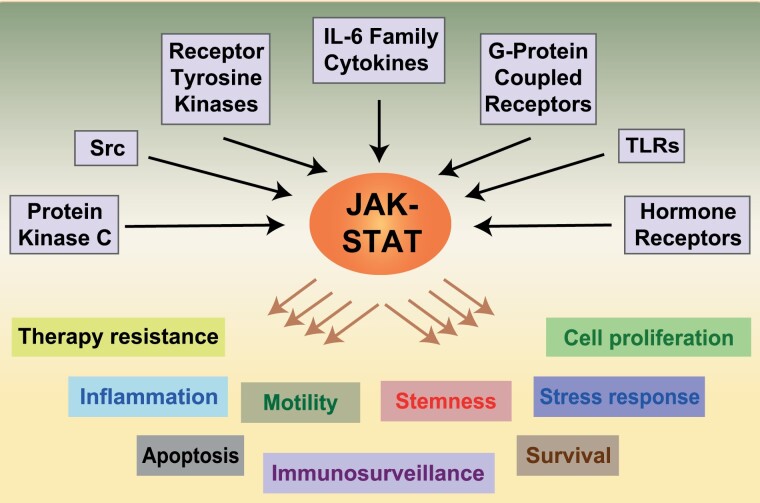
JAK/STAT is an evolutionarily conserved and central pathway required for proper cellular function. Our knowledge of this pathway has been mostly gained from early studies on the immune system. Globally speaking, JAK/STAT links diverse extracellular signals to a wide variety of specific cellular responses, including cell proliferation, motility, survival, apoptosis, inflammation, self-renewal, suppression of antitumor immune response, stress response and other responses depending on the target tissue (Figure 1). The direct external activators include the pro-inflammatory IL-6 family of cytokines such as IL-6, IL-11, IL-31 and Oncostatin M (OSM), as well as the non-IL-6 family cytokines, such as IL-8, IL-10, IL-21, IL-32 and IFNγ. STATs can also be activated by various receptor tyrosine kinases (RTKs), including the human epidermal growth factor receptor (EGFR/HER/ErbB) and vascular–endothelial growth factor R (15), as well as G-protein-coupled receptors, other cytokine receptors such as growth hormone receptors, toll-like receptors, non-RTKs including the Src kinases and other cytoplasmic kinases such as protein kinase C (16). JAK/STAT relays this large variety of signals from the external cellular environment to elicit the intended cellular response.
Figure 1.
Pathways activating JAK/STAT signaling. The JAK–STAT pathway is activated by diverse receptors, including those for IL-6 and IL-6 family cytokines, RTKs, G-protein-coupled receptors, toll-like receptors (TLRs), hormone receptors and the intracellular kinases, Src and protein kinase C. Activated STAT induces a genetic program that promotes various cellular processes that are depicted in the lower part of the diagram, including cancer therapy resistance, inflammation, apoptosis, cell motility, immunosurveillance, stemness, survival, stress response and cell proliferation.
In contrast to RTKs, the cell surface receptors that directly activate the JAK/STAT pathway generally lack catalytic activity. However, once the ligand engages with its receptor, additional proteins are recruited, and in effect, the activated receptor complex acts as an RTK. The proteins recruited share some common themes but there are distinct differences depending on the receptor. The receptor then undergoes a conformational change and induces the recruitment of subunits. For IL-6R, this subunit is the β-receptor glycoprotein 130 (gp130) homodimer. Engagement of the β-receptor subunit then leads to activation of one or more members of the receptor-associated JAK family, of which there are four (JAK1, JAK2, JAK3 and Tyk2). The engaged JAK proteins then trans-phosphorylate each other as well as phosphorylate certain tyrosine residues on the cytoplasmic tail of the receptor complex. These tyrosine-phosphorylated subunits provide a high-affinity docking site for a domain on STAT proteins, called SRC homology 2 (SH2), which also serves as a docking site for proteins in other downstream pathways, including PI3K/AKT, RAS/RAF/MEK/MAPK, and more recently identified, Hippo/YAP (17).
STAT proteins themselves are among the most potent and conserved transcription factors. The family comprises seven members (STAT1, 2, 3, 4, 5a, 5b and 6), all of which are typically located in the cytoplasm when inactive. Because the STAT proteins share similar structural arrangement of their functional motifs, knowledge from one member may generally be applied to the others, although differences in their structure, expression levels, subcellular localization and others contribute to distinct and sometimes divergent cellular responses. For the purposes of this review, we will not only focus on the common themes across the family members but also provide a few protein-specific examples. STATs are activated by phosphorylation on a C-terminal tyrosine residue. Once activated, STAT dissociates from JAK, undergoes stable dimerization, translocates to the nucleus, binds to specific palindromic DNA elements and regulates the expression of hundreds of target genes (Figure 2). The best known palindromic DNA elements recognized by many of the STAT proteins are often called the IFNγ-stimulated response element sites.
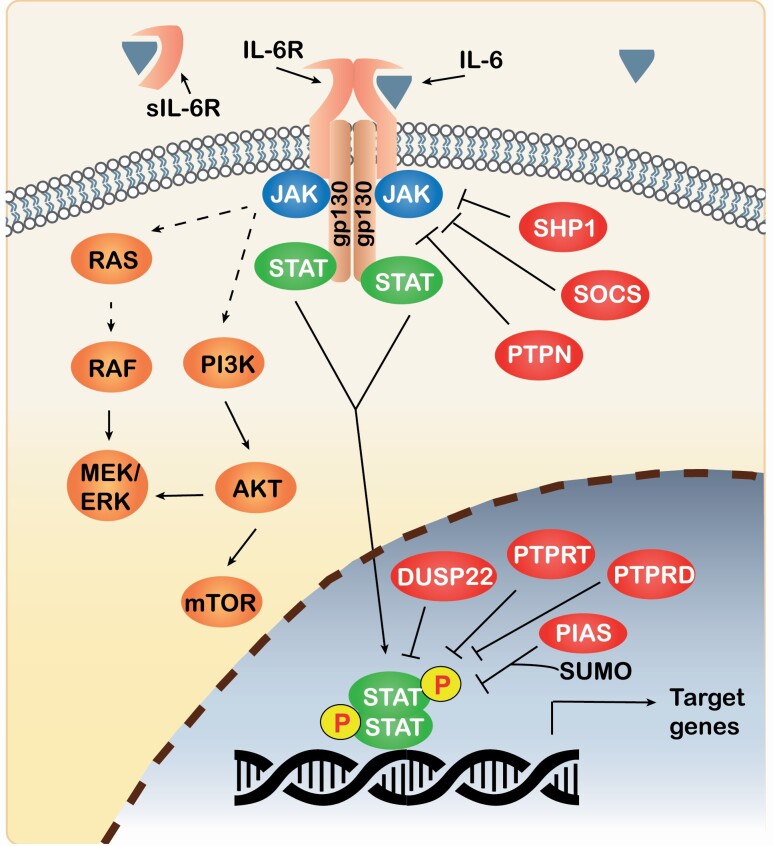
Figure 2.
Schematic representation of STAT3 activation and its pathways. STAT3 is activated when IL-6 binds the IL-6 receptor, which triggers the recruitment of a gp130 homodimer and JAK, followed by a series of phosphorylations, leading to activation of STAT. Activated STAT dimers translocate to the nucleus, bind to specific DNA elements and induce the transcription of STAT target genes. Phosphorylated JAK also leads to activation of the RAS/RAF/MEK/ERK and PI3K/AKT pathways. The negative regulators of the pathway include SHP1, SOCS, PTPNs, DUSP22, PTPRT, PTPRD and PIAS. JAK/STAT signaling can also be attenuated by secretion of soluble IL-6R (sIL-6R), which binds extracellular IL-6.
JAK/STAT signaling from the receptor to the nucleus is a simple design, but as with other pathways that appear simple (18), the wide diversity of cellular effectors and outcomes are compounded by 60 or more cytokines and growth factors channeling into this pathway, and the large set of non-overlapping downstream gene signatures. This complexity is further broadened because both JAK and STAT proteins have additional functions. For example, STAT3 can localize to the mitochondria to promote oxidative phosphorylation and membrane permeability, whereas JAKs phosphorylate histones (16). The intriguing question becomes how are cell-type and context-dependent specificities achieved when so few pathway members are responsible for coordinating the intended response?
One answer lies with evidence that subtle changes in expression of specific JAK/STAT pathway proteins cause qualitative differences in the efficiency, intensity or duration of STAT signaling, which can be calibrated with the receptor activated (16). For example, oscillating levels of STAT1 and STAT4 dictate responsiveness to cytokines in CD8+ T cells and NK cells (19,20), whereas changes in STAT5 availability profoundly influence T-cell functions (21). In addition, the ligands or intracellular proteins activating JAK/STAT can engage one or more JAK and/or STAT proteins, which guides the magnitude and direction of responses. For example, IL-12 plays a distinct role in cell-mediated immune responses by inducing Th1 differentiation of CD4+ T cells through STAT4 activation (22). However, IFNα also activates STAT4 in the same cells, so pathway specificity cannot be entirely explained by STAT4 activation. Type I and II IFNs underpin both innate and adaptive antitumor immune responses in which the availability of cellular STAT1, along with the activation of STAT3, STAT4 and STAT5, facilitates crucial pathway crosstalk (23). The differential cellular responses are also dictated by (i) nuances in optimal DNA binding elements for the various STAT proteins, (ii) a complex combination of post-translational modifications, (iii) subcellular localization of STATs, (iv) recruitment of cell-type or context-dependent co-activators and (v) epigenetic modulation of target genes (24,25).
JAK/STAT signaling can also execute epigenetic changes that modulate gene expression, such as association of STAT2 with HDAC1, acetylation of STAT3 by the histone acetyltransferase p300/CBP, and recruiting DNA methyltransferase 1 (DNMT1) to gene promoters. STAT5 recruits histone methyltransferase EZH2, which is linked to tumor-associated immune suppression (25). Notably, STAT5 represses BMI1, a key component of the polycomb repressor complex 1 (PRC1) that controls cellular self-renewal (26). BMI1 regulates tissue stem cells and is a key target in tumor stem-like cells as it mediates the clonal progression of multiple solid tumors (27), connecting stem cell signaling to JAK/STAT-mediated activities.
Negative regulators of JAK/STAT signaling
As with any cellular pathway, the mechanisms attenuating or inhibiting JAK/STAT signaling are just as important as those that activate it. The specific negative regulators can vary depending on which JAK and STAT proteins are involved. The four main classes of negative regulators of IL6–JAK–STAT3 pathway are (i) certain phosphatases, (ii) suppressors of cytokine signaling (SOCS) proteins, (iii) protein inhibitor of activated STAT (PIAS) proteins and (iv) soluble IL6 receptors (12). Since phosphorylation and dephosphorylation processes play opposing roles, protein tyrosine phosphatases (PTPs) are critical negative regulators. Protein phosphatases participate at multiple levels of control, from the point of surface receptors to the STAT proteins themselves (Figure 2). For example, PTP non-receptor type 2 (PTPN2) is a tumor suppressor that dephosphorylates RTKs and SRC kinases, thus blocking upstream STAT3 activation (28). Other PTPs more directly regulate STAT3 activation, including PTPN1, PTPN6, PTPN9, PTPN11 (encodes SRC homology-2 domain containing phosphatase, SHP2) and dual specificity protein phosphatase 22 (DUSP22). The tumor suppressors PTPRD and PTPRT were demonstrated to dephosphorylate STAT3 itself (29,30). Under basal conditions, JAK1 and JAK2, among other proteins, can phosphorylate SHP2, limiting the activation of STAT in the absence of cytokine signaling. SOCS and SHP2 proteins suppress STAT activation by directly interacting with the kinase domain of JAK or by binding to the tyrosine phosphorylated gp130 subunit (31). As for PIAS proteins, they control the degree and duration of STAT3 activation by inhibiting STAT3 transcriptional activity and regulating its degradation (15). An additional layer of negative control may involve the trans-signaling JAK/STAT pathway (12). There is evidence that a single-nucleotide polymorphism (SNP) alternative splicing, or protein cleavage of IL-6R can produce a secreted soluble form (sIL-6R). Although the precise consequences are debated, sIL-6R can bind extracellular IL-6 and, in some circumstances, reduce the cis-signaling of JAK/STAT in cells expressing IL6R on the cell surface. Cells can also secrete sgp130, allowing it to bind the IL-6/sIL-6R complex. The newly formed IL6/sIL6R/sgp130 complex can then activate the trans-signaling JAK/STAT pathway, even in cells not expressing IL-6R (17). However, the increased levels of sIL-6R/sgp130 might buffer the system by blocking systemic low-grade IL-6 effects (32). Clearly, the built-in checks and balances controlling JAK/STAT signaling suggests that tight control is needed for normal physiology, and dysregulation of the pathway can lead to unwanted cellular consequences.
JAK/STAT pathway in cancer
JAK/STAT is among the top 12 signaling pathways aberrantly regulated in cancer. In solid tumors, persistent phosphorylation of STAT1, STAT3 and STAT5 has been shown in breast, lung, liver and head and neck cancers, generally mediated by upregulated cytokine levels in both autocrine and paracrine manners, and by an enhanced expression of cytokine receptors (33). Whether overexpressed cytokines and their receptors contribute as primary driver(s) of malignant transformation has been contentious. However, it is largely accepted that upregulated JAK/STAT signaling via these mechanisms contributes to cancer aggressiveness by increasing ‘stemness’ or enhancing tumor progression through the induction of an epithelial–mesenchymal transition, as recently reviewed (34).
For many cancer types, including ovarian, cervical, lung, prostate, skin, breast, brain, head and neck and colorectal cancers (CRCs), heightened JAK/STAT signaling is also associated with a worse prognosis, including increased risk for recurrence, and reduced overall survival (35,36). An IFNγ signature, associated with JAK/STAT signaling, contributes to prostate cancer health disparities in the African American (AA) population, with increased risk of aggressive cancer and higher incidence, worse pathological subtypes and higher mortality rates when compared with European Americans (37). Persistent activation of JAK/STAT generates an IFN response (38) and therefore induces an IFN-related DNA damage resistance signature (IRDS) of biomarkers, which is detected in glioblastoma (39), and in breast or prostate cancers more prevalently in AA than European American patients (40,41). Ongoing studies from our groups are evaluating the effects of modulating JAK/STAT signaling on IRDS expression in various tumor and TME cell types, and the potential clinical use of IRDS targets for identifying patients with increased sensitivity to immune checkpoint blockade (ICB) therapy.
Somatic mutations in JAK/STAT pathway genes have been recognized as cancer drivers. Activating JAK mutations are found primarily in hematological cancers but also in some non-hematological cancers (17,42). The most highly studied mutation is JAK2V617F, which leads to constitutive JAK2 activation via destabilization of the pseudokinase domain, preventing its autoinhibitory function on the kinase domain. We recently reported non-recurrent JAK2 mutations in non-small cell lung cancer (NSCLC), which were found throughout the gene. Intriguingly, JAK2 mutations were four times more common in NSCLCs from AAs than European Americans (43,44). These mutations were not the result of clonal hematopoiesis, suggesting they were tumor-specific (43). Their functional consequences are areas of intense ongoing studies.
STAT3 is the most commonly mutated STAT in cancer (17). As with JAK2, STAT3 mutations are prevalent in hematological tumors but also found in various solid tumors (17). Activating STAT3 mutations cluster in the SH2 domain, crucial for STAT3 dimerization. These mutations increase the hydrophobicity of the homodimerization region, causing STAT3 to dimerize more readily, and promoting its nuclear localization and DNA binding (45). Mutations in the STAT3 phosphatases, PTPRD and PTPRT, were reported by Jennifer Grandis’ and Timothy Chan’s groups and were implicated in aberrant activation of STAT3 in head and neck cancers and glioblastoma (29,30). We recently reported the presence of PTPRT mutations in ~25% of lung adenocarcinomas from AAs as compared with only 7% of tumors from European Americans (43), further supporting a population-associated difference in JAK/STAT aberrations in NSCLC (46,47). It is important to note that, as with kinases, phosphatases also have multiple targets. PTPRT dephosphorylates paxillin, resulting in activation of the PI3K/AKT pathway, and PTPRT mutations inhibit T-cell adhesion in CRC (48). The JAK/STAT pathway can also be constitutively activated by RTK mutations such as activating EGFR mutations or by constitutive activation of non-RTKs such as SRC family kinases. Activating gp130 mutations have been observed in over half of inflammatory hepatocellular adenomas developed in cirrhotic livers (49), whereas in hepatocellular carcinoma, rare gp130 mutations were found to be accompanied by beta-catenin-activating mutations (50), suggesting a cooperative mechanism of these signaling pathways in hepatocellular carcinoma. Regardless of the mechanism, our increased knowledge about dysregulated JAK/STAT signaling has led to expanded studies aimed to better understand the impact of the pathway on cancer development.
The biological role of STATs in the carcinogenic process has been delineated by the use of genetic mouse models and immortalized cell lines. Notably, since STAT3, STAT5a and STAT5b are expressed in most cell types and are activated by various ligands, and other STATs play specific roles in host defenses, deletion of STAT3 leads to embryonic lethality, whereas deletion of STAT5a and STAT5b leads to developmental and immune defects, respectively, in mice (51). STAT3 was first proposed to be an oncogene based on evidence that a constitutively dimerized/activated form can transform immortalized mouse and rat fibroblasts, measured by their ability to grow into tumors in nude mice (52). In the classical mouse skin model of multistage carcinogenesis, where chemical initiators and promoters are directly applied to the skin, John DiGiovanni’s group determined that STAT3 is upregulated and constitutively activated. The primary mechanism causing STAT3 activation in this model was through EGFR, and STAT3 was required during both the initiation and promotion stages of skin carcinogenesis in vivo (53,54).
Although hyper-activated STAT3 is generally associated with tumor progression (12), there is also context-related evidence that STAT3 can act as a tumor suppressor. For example, STAT3 is a negative regulator of intestinal tumor progression in Apc(Min) mice (55). Moreover, inactivation of STAT3 increased tumor formation and progression in the KrasG12D mouse model of lung adenocarcinoma. In this model, STAT3 inhibited IL-8-mediated myeloid-derived tumor infiltration, tumor vascularization and hence tumor progression (56). An additional layer of complexity that awaits further investigation is that STAT1 and STAT3, though sharing common activating stimuli, displaying high sequence homology, and interacting frequently as heterodimers, can still play opposing roles. Although STAT3 generally promotes a pro-tumorigenic role, STAT1 enhances immunosurveillance, triggering an antiproliferative and pro-apoptotic response in tumor cells. The relative abundance of STAT1 or STAT3 may determine their relative activation levels and biological effects in response to common activating stimuli (57).
Epidemiological evidence for JAK/STAT signaling in carcinogenesis
In addition to characterized somatic mutations, certain germline variants in JAK/STAT pathway genes are associated with cancer. In a large case–control study of CRC, SNPs in JAK2, SOCS2, STAT1, STAT3, STAT5A, STAT5B, STAT6 and TYK2 were significantly associated with CRC, and many of these SNPs were associated with CRC survival (58). Of note, the association between several SNPs and CRC was modified by aspirin/NSAID use or smoking status (58), suggesting an interaction between inflammation and the SNPs. Likewise, in a large case–control study of breast cancer, 11 SNPs in JAK1, JAK2 and STAT3 were significantly associated with breast cancer, modified by menopausal status or body-mass index. In this study, SNPs in the JAK/STAT pathway were also significantly associated with breast cancer-specific survival, and these associations were modified by NSAIDs/aspirin use and smoking status, further suggesting a connection between JAK/STAT and lifestyle factors in contributing to cancer (59). These gene–environmental interactions highlight the potential of using genetics to guide behaviors in cancer prevention strategies and further highlight the impact of inflammation-related pathways on cancer development. It is conceivable that similar to how the rs6983267 SNP in chromosome 8q24 was proposed as a biomarker for aspirin use to reduce risk of developing CRC (60), chemoprevention recommendations could be tailored by certain JAK/STAT pathway SNPs in CRC and breast cancer.
Inhibition of the JAK/STAT3 pathway for cancer prevention
As detailed throughout this review, JAK/STAT signaling integrates signals from a wide variety of inputs to upregulate hallmarks of cancer development, including cell growth, differentiation and survival. Thus, inhibition of JAK/STAT has been postulated as a cancer prevention strategy, mostly through the exploration of natural agents, such as phytochemicals. Certain phytochemicals from diverse plant origins have shown promise in exerting chemopreventive effects, at least partly, through attenuating JAK/STAT activation (61). For example, Resveratrol, a polyphenol found in berries, grapes and peanuts, is a potent inhibitor of SRC, STAT1 and STAT3 (62). Chalcones, flavonoids found in certain fruits and vegetables, have promising chemopreventive activity due to their antioxidant and anti-inflammatory effects (63). Chalcones prevent STAT3 phosphorylation in endothelial cells exposed to IL6 (64). Polyphenols extracted from green tea have been the subject of cancer prevention studies since before the turn of the century (65). They are also found in certain fruits and vegetables, and they protect intestinal epithelial cells from inflammation by blocking JAK1 and STAT1 activation (66). It is important to note that although phytochemical natural compounds are well known for their non-toxic effects under physiological conditions and are good candidates for dietary supplements, they are highly unselective. Thus, the mechanisms by which they exert their chemopreventive effects are probably to be multifactorial. For example, Resveratrol also inhibits IGF2, induces AKT1 activation, causes cell cycle arrest by activating p38 MAPK and promotes apoptosis by inducing cleavage of caspase-3 (67). Despite their wide range of targets, it would be worthwhile to systematically test JAK-STAT pathway-targeting phytochemicals in carefully controlled human trials to assess their cancer prevention capabilities.
Targeting JAK/STAT for cancer therapy
Given the central roles of JAK/STAT in cancer, great effort has been put into exploring JAK/STAT inhibitors for cancer therapy. Targeting JAK or STAT proteins with RNA interference (RNAi) or molecularly targeted compounds has been extensively studied, and the different types of JAK and STAT inhibitors were recently reviewed elsewhere (68). JAK/STAT inhibition is clinically effective in certain hematological conditions such as myeloproliferative disorders and diffuse large B-cell lymphoma (12,69), where activating JAK/STAT mutations are prevalent. Initial findings in epithelial cancer cell lines or mouse tumor xenografts have been promising, where JAK/STAT3 abrogation induced marked apoptosis and inhibited cell proliferation (69). In view of the encouraging pre-clinical data, many JAK/STAT inhibitors are being tested in phase I and II clinical trials in unselected patients with solid tumors such as brain tumors and melanomas. However, the outcome of trials on JAK or STAT inhibitors as single agents has been unsatisfactory, primarily due to toxicities and/or a lack of efficacy (12,69). As the field develops more specific inhibitors of the pathway, these new compounds might perform better, with fewer off-target or other-on-target effects. Notably, the trials on JAK/STAT pathway inhibitors in solid tumors were performed on unselected patients, and they did not report the oncogenic driver mutations; thus, it is unclear if the compounds are effective in the subset of tumors with activating JAK/STAT pathway mutations or additional genetic subsets of tumors. The need to select patients is particularly relevant for AAs with NSCLC, who have a higher prevalence of JAK2 and PTPRT mutations (43). Given the speed by which JAK/STAT inhibitors are being developed and optimized for hematological disorders, it is worthwhile not only to examine them more carefully in selected solid tumors across mutational backgrounds but also to examine how they impact the steady states between different STATs and STAT-mediated actions, which could provide unique therapeutic opportunities.
Role of JAK/STAT3 in cancer resistance and combination therapies
JAK/STAT pathway activation is a known mechanism of therapy resistance. An unanswered question is whether JAK/STAT inhibition in combination with other therapies would be effective in certain patients. DNA damage and genotoxic stress caused by standard chemotherapy or radiotherapy evokes an inflammatory response, leading to STAT3 activation. The subsequent cytokine and chemokine upregulation promotes therapy resistance and increases the risk of tumor recurrence (70,71). Moreover, inflammation caused by necrotic cells from surgery or chemoradiotherapy promotes the acquisition of stem-like features in tumor cells through STAT3 activation (34). There has been increasing evidence from in vitro and animal studies that JAK or STAT inhibitors sensitize tumor cells to cytotoxic chemoradiotherapy, a topic reviewed in detail elsewhere (71,72).
STAT3 activation contributes to intrinsic and acquired resistance against an increasing number of targeted therapies in oncogene-addicted solid tumors. Targeted therapies against oncogenic drivers are generally effective initially, but the treated tumors eventually develop adaptive or acquired resistance, resulting in recurrence. A key example is in NSCLC, where patients with a RTK inhibitor (TKI)-sensitive, activating EGFR mutation are eligible for TKI-directed therapy. EGFR-mutant lung tumors respond well initially to these TKIs, but acquire resistance, after a median of 9–19 months, depending on the drug used (73–75). A potential strategy to delay or prevent the development of acquired resistance is to combine the initial TKI treatment with a compound that inhibits intrinsic or adaptive resistance, to eliminate surviving cells that would later develop acquired resistance. Jeff Settleman’s group determined that many TKI treatments can induce STAT3 activation through IL-6/JAK1 and FGFR/PI3K (70). Activated STAT3 was a key factor contributing to the resistance to TKIs, because knock-down or inhibition of STAT3 restored sensitivity to TKIs (70). The same group observed a similar trend in tumors with other oncogenic drivers, such as HER2, ALK and MET. When tumor cells were treated with a TKI or MEK inhibitor, it engaged a positive feedback loop to activate STAT3, which limited the overall drug response. Their seminal report suggested that the co-inhibition of the oncogenic driver and STAT3 could prevent or delay the development of acquired resistance and potentially prolong progression-free survival. These data have been supported by other groups (76). Moreover, STAT3 can be activated by signaling through RTKs and PI3K, independently of JAK1/2, further supporting the combination of TKIs with STAT3 inhibitors.
SHP2 (encoded by PTPN11) is a negative regulator of JAK/STAT and is required for RAS-mediated RTK activation (77). SHP2 inhibitors were presented as a viable therapeutic option for RTK-driven cancers but were ineffective in KRAS-mutant tumors (78). PTPN11 knockout profoundly inhibited tumor development in KRAS-mutant models of pancreatic ductal adenocarcinoma and NSCLC. Deletion or inhibition of SHP2 in established tumors only delayed tumor progression but synergy was observed when both SHP2 and MEK were targeted in patient-derived organoids and xenografts of pancreas and lung cancer, therefore presenting dual SHP2/MEK inhibition as a potentially targeted therapy for KRAS-mutant cancers (79). In unselected patients, clinical trials have unsurprisingly not been as encouraging. Combination of EGFR inhibitors with the JAK1/2 inhibitor ruxolitinib in two clinical trials was ineffective in NSCLC patients. In this setting, ruxolitinib was administered to recurrent tumors, after acquired resistance to the TKI had already developed (80,81). A clinical trial in previously untreated EGFR-mutant lung cancer would directly test if JAK/STAT3 inhibition prevents or delays tumor progression.
Role of JAK/STAT3 in cancer immunotherapy
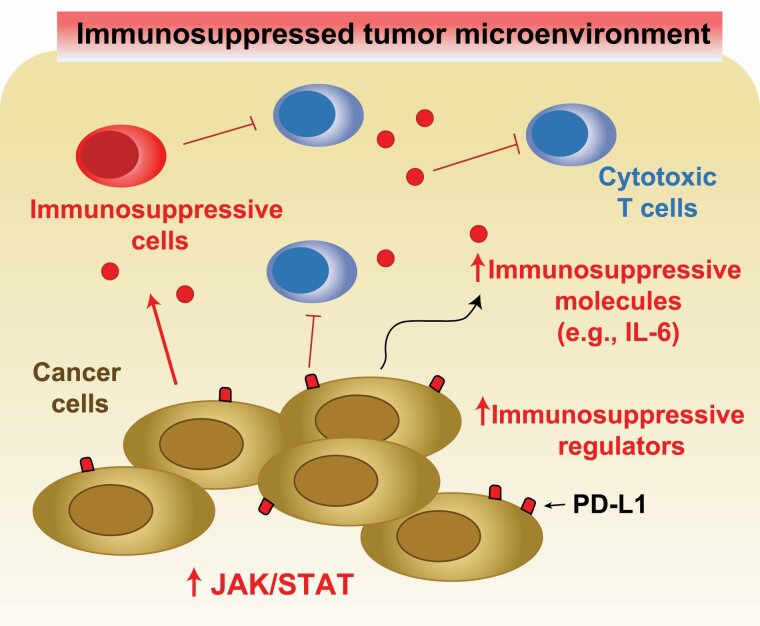
A timely, yet unanswered question is whether immunotherapy combined with JAK/STAT3 pathway inhibitors would be an effective therapeutic option for select cancer patients. Programmed cell death-1 (PD-L1) is a member of the family of immune checkpoint proteins that inhibits the T-cell response. When PD-L1 is expressed on the surface of tumor cells, it binds with PD-1 receptors on nearby activated T cells, leading to the inhibition of cytotoxic T cells and reduction of the antitumor response within the TME (Figure 3). Immune checkpoint inhibitors (ICIs), such as those targeting PD-1, act by re-engaging the antitumor T-cell response (82). PD-L1 is a STAT3 target gene, and STAT3 activation is associated with high PD-L1 expression in numerous cancer types (83,84). In addition, increased IL-6 secretion caused by activated STAT3 results in the recruitment of myeloid-derived suppressor cells and shifts the Th1/Th2 balance toward a Th2 phenotype, further suppressing the actions of antitumor cytotoxic T cells (Figure 3). Frameshift JAK1 mutations observed in prostate, urinary and endometrial cancers have been linked to altered immune cells in the TME, and the JAK2V617F mutation observed in NSCLC has been linked to alterations in tumor cell PD-L1. Alternatively, in melanoma, JAK1 and JAK2 inactivating mutations correlated with PD-L1 loss in the TME, resulting from dampened IFN signaling, which might contribute to these patients’ poor response to ICIs (85).
Figure 3.
JAK/STAT3 activation induces a tumor immunosuppressive microenvironment. STAT3 increases expression of PD-L1, which suppresses the action of cytotoxic T cells. Upregulated secretion of IL-6 caused by activated STAT3 recruits myeloid-derived suppressor cells to the TME and suppresses the antitumor immune response. IL-6 shifts the Th1/Th2 balance toward a Th2 phenotype, further supporting a tumor immunosuppressive environment.
An obvious question is, what impact would JAK/STAT3 inhibitors have on ICB therapy efficacy? Targeting JAK or STAT3 could have a synergistic antitumor effect with ICIs (83), partly because the same immune checkpoint pathway is hit twice by two different mechanisms. The JAK/STAT3 pathway also regulates the immune TME independently of PD-L1. For example, activated STAT3 attenuates dendritic cell differentiation, which decreases CD8 T-cell activation (86). Consistent with this notion, activation of JAK/STAT by prolonged IFNγ signaling might contribute to adaptive resistance to ICIs, and blocking IFNγ signaling can restore responses to ICIs, at least in part by expanding distinct T-cell populations (87). A few trials addressing this important point are ongoing, including a trial in metastatic CRC combining pembrolizumab with BBI-608, a compound that attenuates STAT3 activation (88).
JAK/STAT pathway inhibition has the potential to improve responses to other types of immunotherapy. For example, chimeric antigen receptor (CAR) T-cell therapy is a promising cancer immunotherapy that has been successful in leukemia and lymphoma and is being explored in solid tumors. An unfortunate side effect of CAR T-cell therapy is cytokine release syndrome, with an abundant secretion of cytokines including IFNγ and IL-6. The JAK1 inhibitor, itacitinib, reduced or even prevented cytokine release caused by CAR T cells, in both in vitro and in vivo models (89), and a phase II clinical trial combining itacitinib with CAR T-cell therapy is ongoing (NCT04071366).
Paradoxically, loss of PD-L1 expression caused by JAK/STAT inhibition could reduce the efficacy of PD-1 inhibitors. When the JAK/STAT pathway is intact, IFNγ provides signals to cancer cells to inactivate antitumor T cells by the adaptive expression of PD-L1 (82), thereby specifically escaping their cytotoxic effects. Antoni Ribas’ group recently reported that loss-of-function alterations in JAK1/2 occurred in cases of acquired resistance to anti-PD1 therapy (pembrolizumab) in melanoma (90). This may be explained by a lack of adaptive PD-L1 expression from the loss of response to IFNγ signaling (91). JAK/STAT pathway alterations might also impact other ICB therapies. Loss of JAK2 by genomic deletion in melanoma was identified in non-responders to the anti-CTLA-4 therapy, ipilimumab, suggesting JAK2 loss and consequential loss of IFNγ response is associated with primary resistance to anti-CTLA-4 therapy (92). Thus, combining STAT3 inhibitors with ICB and/or sequential treatment strategies merit additional and extensive pre-clinical investigations.
The crosstalk between different STATs in the context of cancer therapy also requires further consideration. In the presence of specific TME, different stimuli can modulate the relative intensity and duration of activation of STAT1 and STAT3 levels, thus providing a favorable microenvironment for neoplastic transformation and growth. STAT1 blocks tumor cell cycle progression and inhibits angiogenesis, therefore triggering, in most cases, antiproliferative and pro-apoptotic responses in tumor cells. STAT3’s role in promoting tumor metastasis and therapy resistance continues to emerge, as does the impact of STAT3 as a suppressor of immune cell function in the TME. Many of the inflammatory mediators produced by tumor cells upon STAT3 inactivation are typically STAT1 targets (e.g. CXCL10, CCL5 and ICAM1), suggesting that reciprocal regulation between STAT3 and STAT1 may occur in tumors. Additionally, IFNβ activates STAT1 and STAT2, which form a complex with IRF9 to create the transcription factor complex ISGF3. IFNβ/P-ISGF3 signaling was shown to induce mesenchymal-to-epithelial transition, differentiation into a less aggressive epithelial state, reduced migratory potential and reduced tumor sphere forming capabilities (93). Importantly, IFNβ and OSM/STAT3 signaling pathways strongly oppose one another. OSM represses the transcription of IFNβ, thereby eliminating autocrine and paracrine IFNβ-mediated activation of ISGF3 in both tumor and immune cells (93). STAT5 is also essential for many immune cell functions and activated STAT5 dampens antitumor immune function through CD4+/CD25+T-regs, a subset of T cells that contribute to tumor progression and metastasis (94). Systemic suppression of STAT5 activity could undermine NK cell-mediated tumor immunity and promote tumor progression (95). Therefore, reducing the aberrant activation of STAT5 without complete ablation or specific suppression of STAT5 in immune-suppressive T-reg cells could prove beneficial. Furthermore, activating STAT1 and STAT2 within the TME cells could prevent metastasis and enhance the therapeutic efficacy of ICBs.
Concluding remarks
Chronic inflammation plays a complex role in driving tumorigenesis. The JAK/STAT signaling pathway is a major nexus that bridges inflammation with cancer. The large repertoire of ligands and receptors triggering JAK/STAT signaling, the numerous pathways that JAK/STAT signaling interacts with, and the hundreds of related STAT target genes lay a strong testament to the importance of coordinating JAK/STAT signaling. Numerous pre-clinical and molecular characterization studies revealed that the JAK/STAT pathway is a central contributor to tumor initiation and progression. Epidemiological evidence also supports the role of the JAK/STAT pathway in carcinogenesis. Since JAK or STAT inhibitors are routinely used to treat a variety of inflammatory conditions (96), including to possibly mitigate COVID-19 symptoms (97), an unresolved question in the field is whether inhibition of JAK/STAT decreases cancer risk. Despite the very promising pre-clinical data in cancer cells or xenografts, the outcome of clinical trials on JAK/STAT inhibitors as single agents has been disappointing because of poor efficacy or toxicity. The intricacies of the JAK/STAT signaling could be better deciphered by utilizing (i) advanced single-cell analyses to resolve heterogeneity and integrated signaling redundancies in tumor, immune and other stromal cell subsets at the resolution of individual cells (98); (ii) modeling of clonal and TME interactions using patient-derived models (99); and (iii) rigorously devised patient selection criteria in precision medicine clinical trials to identify more effective and synergistic therapies. Perhaps a feasible approach to target JAK/STAT signaling will include a combinatorial targeted therapy, particularly now that we are empowered by a more in depth understanding of tumor heterogeneity and knowledge of the intrinsic signaling redundancy mechanisms. With the recent advances in immunotherapy, combining ICB with JAK/STAT inhibition may be pursued with caution, by carefully considering the pleotropic impact of JAK/STAT signaling on the tumor and its microenvironment.
Funding
NIH NCI (R01 CA239093) to S.R.P.; NIH NCI (P30 CA072720) to H.E.S., H.K. and S.R.P.; Rutgers Cancer Institute of New Jersey Cancer Health Equity Pilot Award (to S.R.P.); National Cancer Institute (R01 CA226746) to H.E.S.; National Institutes of Health-Leidos (HHSN 2612008000001E) to H.E.S.; New Jersey Health Foundation (ISFP 7) to H.E.S.; New Jersey Commission on Spinal Cord Research (CSR 19IRG014) to H.E.S.; Intramural Research Programs of the National Institute for Minority Health and Health Disparities (to B.M.R.); Center for Cancer Research, National Cancer Institute (to B.M.R.); National Cancer Institute (R01CA233662) to H.K.; V Foundation (T2019-012) to H.K.; New Jersey Commission for Cancer Research (to S.R.P.); American Lung Association (to S.R.P.).
Conflict of Interest Statement: H.E.S. reports honoraria and consulting fees from Janssen outside the submitted work. H.E.S. has patents through Rutgers University and equity in a start-up company, Celvive Inc. outside the submitted work. S.R.P. has patents through New York Medical College, National Cancer Institute, National Institutes of Health and Rutgers University, outside the submitted work. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Glossary
Abbreviations
- AA
African American
- CRC
colorectal cancer
- EGFR
epidermal growth factor receptor
- ICB
immune checkpoint blockade
- ICI
immune checkpoint inhibitor
- IFN
interferon
- IL
interleukin
- JAK
Janus kinase
- NSCLC
non-small cell lung cancer
- PD-L1
programmed cell death-1
- PTP
protein tyrosine phosphatase
- PTPN
protein tyrosine phosphatase non-receptor
- RTK
receptor tyrosine kinase
- SNP
single-nucleotide polymorphism
- STAT
signal transducer and activator of transcription
- TGF
tumor growth factor
- TKI
RTK inhibitor
- TME
tumor microenvironment
References
- 1. Kuper, H. et al. (2000) Infections as a major preventable cause of human cancer. J. Intern. Med., 248, 171–183. [DOI] [PubMed] [Google Scholar]
- 2. Perwez Hussain, S. et al. (2007) Inflammation and cancer: an ancient link with novel potentials. Int. J. Cancer, 121, 2373–2380. [DOI] [PubMed] [Google Scholar]
- 3. Ghimire, P. et al. (2011) Primary gastrointestinal lymphoma. World J. Gastroenterol., 17, 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Herrera, V. et al. (2009) Helicobacter pylori and gastric adenocarcinoma. Clin. Microbiol. Infect., 15, 971–976. [DOI] [PubMed] [Google Scholar]
- 5. Clarke, W.T. et al. (2019) Colorectal cancer surveillance in inflammatory bowel disease: practice guidelines and recent developments. World J. Gastroenterol., 25, 4148–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hussain, S.P. (2016) Pancreatic cancer: current progress and future challenges. Int. J. Biol. Sci., 12, 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang, J.D. et al. (2019) A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol., 16, 589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ballaz, S. et al. (2003) The potential contributions of chronic inflammation to lung carcinogenesis. Clin. Lung Cancer, 5, 46–62. [DOI] [PubMed] [Google Scholar]
- 9. Iyengar, N.M. et al. (2016) Obesity and cancer mechanisms: tumor microenvironment and inflammation. J. Clin. Oncol., 34, 4270–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumari, N. et al. (2016) Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol., 37, 11553–11572. [DOI] [PubMed] [Google Scholar]
- 11. Walter, M. et al. (2009) Interleukin 6 secreted from adipose stromal cells promotes migration and invasion of breast cancer cells. Oncogene, 28, 2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson, D.E. et al. (2018) Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol., 15, 234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taniguchi, K. et al. (2018) NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol., 18, 309–324. [DOI] [PubMed] [Google Scholar]
- 14. Triner, D. et al. (2016) Hypoxia-inducible factors: a central link between inflammation and cancer. J. Clin. Invest., 126, 3689–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levy, D.E. et al. (2002) Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol., 3, 651–662. [DOI] [PubMed] [Google Scholar]
- 16. O’Shea, J.J. et al. (2015) The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu. Rev. Med., 66, 311–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lokau, J. et al. (2019) Activating mutations of the gp130/JAK/STAT pathway in human diseases. Adv. Protein Chem. Struct. Biol., 116, 283–309. [DOI] [PubMed] [Google Scholar]
- 18. Capaccione, K.M. et al. (2013) The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis, 34, 1420–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miyagi, T. et al. (2007) High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J. Exp. Med., 204, 2383–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gil, M.P. et al. (2012) Regulating type 1 IFN effects in CD8 T cells during viral infections: changing STAT4 and STAT1 expression for function. Blood, 120, 3718–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Villarino, A. et al. (2016) Signal transducer and activator of transcription 5 (STAT5) paralog dose governs T cell effector and regulatory functions. Elife, 5, e08384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cho, S.S. et al. (1996) Activation of STAT4 by IL-12 and IFN-alpha: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J. Immunol., 157, 4781–4789. [PubMed] [Google Scholar]
- 23. Villarino, A.V. et al. (2017) Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat. Immunol., 18, 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stark, G.R. et al. (2012) The JAK-STAT pathway at twenty. Immunity, 36, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mandal, M. et al. (2011) Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol., 12, 1212–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gilbert, S. et al. (2015) Activated STAT5 confers resistance to intestinal injury by increasing intestinal stem cell proliferation and regeneration. Stem Cell Rep., 4, 209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bansal, N. et al. (2016) BMI-1 targeting interferes with patient-derived tumor-initiating cell survival and tumor growth in prostate cancer. Clin. Cancer Res., 22, 6176–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shields, B.J. et al. (2013) TCPTP regulates SFK and STAT3 signaling and is lost in triple-negative breast cancers. Mol. Cell. Biol., 33, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peyser, N.D. et al. (2015) Loss-of-function PTPRD mutations lead to increased STAT3 activation and sensitivity to STAT3 inhibition in head and neck cancer. PLoS One, 10, e0135750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Veeriah, S. et al. (2009) The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc. Natl. Acad. Sci. USA, 106, 9435–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rane, S.G. et al. (2000) Janus kinases: components of multiple signaling pathways. Oncogene, 19, 5662–5679. [DOI] [PubMed] [Google Scholar]
- 32. Garbers, C. et al. (2015) The IL-6/gp130/STAT3 signaling axis: recent advances towards specific inhibition. Curr. Opin. Immunol., 34, 75–82. [DOI] [PubMed] [Google Scholar]
- 33. Sansone, P. et al. (2012) Targeting the interleukin-6/Jak/stat pathway in human malignancies. J. Clin. Oncol., 30, 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin, W. (2020) Role of JAK/STAT3 signaling in the regulation of metastasis, the transition of cancer stem cells, and chemoresistance of cancer by epithelial–mesenchymal transition. Cells, 9, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thomas, S.J. et al. (2015) The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer, 113, 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shang, A.Q. et al. (2017) Relationship between HER2 and JAK/STAT-SOCS3 signaling pathway and clinicopathological features and prognosis of ovarian cancer. Cancer Biol. Ther., 18, 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wallace, T.A. et al. (2008) Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res., 68, 927–936. [DOI] [PubMed] [Google Scholar]
- 38. Cheon, H. et al. (2014) Interferons and their stimulated genes in the tumor microenvironment. Semin. Oncol., 41, 156–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duarte, C.W. et al. (2012) Expression signature of IFN/STAT1 signaling genes predicts poor survival outcome in glioblastoma multiforme in a subtype-specific manner. PLoS One, 7, e29653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weichselbaum, R.R. et al. (2008) An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. USA, 105, 18490–18495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang, W. et al. (2018) IFNL4-ΔG allele is associated with an interferon signature in tumors and survival of African-American men with prostate cancer. Clin. Cancer Res., 24, 5471–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Riedlinger, G. et al. (2019) Association of JAK2-V617F mutations detected by solid tumor sequencing with coexistent myeloproliferative neoplasms. JAMA Oncol., 5, 265–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mitchell, K.A. et al. (2019) Recurrent PTPRT/JAK2 mutations in lung adenocarcinoma among African Americans. Nat. Commun., 10, 5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arauz, R.F. et al. (2020) Whole-exome profiling of NSCLC among African Americans. J. Thorac. Oncol., 15, 1880–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koskela, H.L. et al. (2012) Somatic STAT3 mutations in large granular lymphocytic leukemia. N. Engl. J. Med., 366, 1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pine, S.R. et al. (2016) Differential serum cytokine levels and risk of lung cancer between African and European Americans. Cancer Epidemiol. Biomarkers Prev., 25, 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meaney, C.L. et al. (2017) Identification of serum inflammatory markers as classifiers of lung cancer mortality for stage I adenocarcinoma. Oncotarget, 8, 40946–40957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao, Y. et al. (2017) Regulation of paxillin-p130-PI3K-AKT signaling axis by Src and PTPRT impacts colon tumorigenesis. Oncotarget, 8, 48782–48793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Calderaro, J. et al. (2016) Inflammatory hepatocellular adenomas developed in the setting of chronic liver disease and cirrhosis. Mod. Pathol., 29, 43–50. [DOI] [PubMed] [Google Scholar]
- 50. Rebouissou, S. et al. (2009) Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature, 457, 200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kiu, H. et al. (2012) Biology and significance of the JAK/STAT signalling pathways. Growth Factors, 30, 88–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bromberg, J.F. et al. (1999) Stat3 as an oncogene. Cell, 98, 295–303. [DOI] [PubMed] [Google Scholar]
- 53. Chan, K.S. et al. (2004) Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J. Clin. Invest., 114, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim, D.J. et al. (2009) Constitutive activation and targeted disruption of signal transducer and activator of transcription 3 (Stat3) in mouse epidermis reveal its critical role in UVB-induced skin carcinogenesis. Oncogene, 28, 950–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Musteanu, M. et al. (2010) Stat3 is a negative regulator of intestinal tumor progression in Apc(Min) mice. Gastroenterology, 138, 1003–1011.e5. [DOI] [PubMed] [Google Scholar]
- 56. Grabner, B. et al. (2015) Disruption of STAT3 signalling promotes KRAS-induced lung tumorigenesis. Nat. Commun., 6, 6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lui, V.W. et al. (2007) Antiproliferative mechanisms of a transcription factor decoy targeting signal transducer and activator of transcription (STAT) 3: the role of STAT1. Mol. Pharmacol., 71, 1435–1443. [DOI] [PubMed] [Google Scholar]
- 58. Slattery, M.L. et al. (2013) JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Mol. Carcinog., 52, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Slattery, M.L. et al. (2014) Genetic variation in the JAK/STAT/SOCS signaling pathway influences breast cancer-specific mortality through interaction with cigarette smoking and use of aspirin/NSAIDs: the Breast Cancer Health Disparities Study. Breast Cancer Res. Treat., 147, 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nan, H. et al. (2013) Aspirin use, 8q24 single nucleotide polymorphism rs6983267, and colorectal cancer according to CTNNB1 alterations. J. Natl. Cancer Inst., 105, 1852–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bose, S. et al. (2020) Targeting the JAK/STAT signaling pathway using phytocompounds for cancer prevention and therapy. Cells, 9, 1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kotha, A. et al. (2006) Resveratrol inhibits Src and Stat3 signaling and induces the apoptosis of malignant cells containing activated Stat3 protein. Mol. Cancer Ther., 5, 621–629. [DOI] [PubMed] [Google Scholar]
- 63. Mahapatra, D.K. et al. (2015) Anti-cancer chalcones: structural and molecular target perspectives. Eur. J. Med. Chem., 98, 69–114. [DOI] [PubMed] [Google Scholar]
- 64. Liu, Y.C. et al. (2007) Chalcone inhibits the activation of NF-kappaB and STAT3 in endothelial cells via endogenous electrophile. Life Sci., 80, 1420–1430. [DOI] [PubMed] [Google Scholar]
- 65. Embola, C.W. et al. (2001) Urinary excretion of N-OH-2-amino-3-methylimidazo[4,5-f]quinoline-N-glucuronide in F344 rats is enhanced by green tea. Carcinogenesis, 22, 1095–1098. [DOI] [PubMed] [Google Scholar]
- 66. Nunes, C. et al. (2016) Red wine polyphenol extract efficiently protects intestinal epithelial cells from inflammation via opposite modulation of JAK/STAT and Nrf2 pathways. Toxicol. Res. (Camb)., 5, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Khan, K. et al. (2020) Resveratrol, curcumin, paclitaxel and miRNAs mediated regulation of PI3K/Akt/mTOR pathway: go four better to treat bladder cancer. Cancer Cell Int., 20, 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bousoik, E. et al. (2018) “Do we know jack” about JAK? A closer look at JAK/STAT signaling pathway. Front. Oncol., 8, 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Beebe, J.D. et al. (2018) Two decades of research in discovery of anticancer drugs targeting STAT3, how close are we? Pharmacol. Ther., 191, 74–91. [DOI] [PubMed] [Google Scholar]
- 70. Lee, H.J. et al. (2014) Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell, 26, 207–221. [DOI] [PubMed] [Google Scholar]
- 71. Spitzner, M. et al. (2014) STAT3: a novel molecular mediator of resistance to chemoradiotherapy. Cancers (Basel), 6, 1986–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hall, W.A. et al. (2020) Cytokines, JAK-STAT signaling and radiation-induced DNA repair in solid tumors: novel opportunities for radiation therapy. Int. J. Biochem. Cell Biol., 127, 105827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rosell, R. et al. ; Spanish Lung Cancer Group. (2009) Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med., 361, 958–967. [DOI] [PubMed] [Google Scholar]
- 74. Soria, J.C. et al. ; FLAURA Investigators. (2018) Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med., 378, 113–125. [DOI] [PubMed] [Google Scholar]
- 75. Mok, T.S.K. et al. ; KEYNOTE-042 Investigators. (2019) Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet, 393, 1819–1830. [DOI] [PubMed] [Google Scholar]
- 76. Shien, K. et al. (2017) JAK1/STAT3 activation through a proinflammatory cytokine pathway leads to resistance to molecularly targeted therapy in non-small cell lung cancer. Mol. Cancer Ther., 16, 2234–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chan, G. et al. (2008) The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev., 27, 179–192. [DOI] [PubMed] [Google Scholar]
- 78. Chen, Y.N. et al. (2016) Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature, 535, 148–152. [DOI] [PubMed] [Google Scholar]
- 79. Ruess, D.A. et al. (2018) Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat. Med., 24, 954–960. [DOI] [PubMed] [Google Scholar]
- 80. Yu, H.A. et al. (2017) A phase ½ trial of ruxolitinib and erlotinib in patients with EGFR-mutant lung adenocarcinomas with acquired resistance to erlotinib. J. Thorac. Oncol., 12, 102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Park, J.S. et al. (2019) A phase Ib study of the combination of afatinib and ruxolitinib in EGFR mutant NSCLC with progression on EGFR-TKIs. Lung Cancer, 134, 46–51. [DOI] [PubMed] [Google Scholar]
- 82. Pardoll, D.M. (2012) The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer, 12, 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Song, T.L. et al. (2018) Oncogenic activation of the STAT3 pathway drives PD-L1 expression in natural killer/T-cell lymphoma. Blood, 132, 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fujita, Y. et al. (2015) The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol. Ther., 23, 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shin, D.S. et al. (2017) Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov., 7, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nefedova, Y. et al. (2005) Activation of dendritic cells via inhibition of Jak2/STAT3 signaling. J. Immunol., 175, 4338–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Benci, J.L. et al. (2016) Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell, 167, 1540–1554.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shinozaki, E. et al. (2018) Multicenter phase I/II trial of BBI608 and pembrolizumab combination in patients with metastatic colorectal cancer (SCOOP Study): EPOC1503. J. Clin. Oncol., 36, 3530. [Google Scholar]
- 89. Huarte, E. et al. (2020) Itacitinib (INCB039110), a JAK1 inhibitor, reduces cytokines associated with cytokine release syndrome induced by CAR T-cell therapy. Clin. Cancer Res., 26, 6299–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zaretsky, J.M. et al. (2016) Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med., 375, 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shin, D.S. et al. (2017) Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov., 7, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gao, J. et al. (2016) Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell, 167, 397–404.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Doherty, M.R. et al. (2019) The opposing effects of interferon-beta and oncostatin-M as regulators of cancer stem cell plasticity in triple-negative breast cancer. Breast Cancer Res., 21, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nishikawa, H. et al. (2014) Regulatory T cells in cancer immunotherapy. Curr. Opin. Immunol., 27, 1–7. [DOI] [PubMed] [Google Scholar]
- 95. Gotthardt, D. et al. (2016) STAT5 is a key regulator in NK cells and acts as a molecular switch from tumor surveillance to tumor promotion. Cancer Discov., 6, 414–429. [DOI] [PubMed] [Google Scholar]
- 96. Pencik, J. et al. (2016) JAK-STAT signaling in cancer: from cytokines to non-coding genome. Cytokine, 87, 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Matsuyama, T. et al. (2020) An aberrant STAT pathway is central to COVID-19. Cell Death Differ., 27, 3209–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Marusyk, A. et al. (2020) Intratumor heterogeneity: the Rosetta stone of therapy resistance. Cancer Cell, 37, 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chadwick, M. et al. (2020) Rapid processing and drug evaluation in glioblastoma patient-derived organoid models with 4D bioprinted arrays. iScience, 23, 101365. [DOI] [PMC free article] [PubMed] [Google Scholar]