Abstract
Unfolded protein response (UPR) is a stress response that is specific to the endoplasmic reticulum (ER). UPR is activated upon accumulation of unfolded (or misfolded) proteins in the ER's lumen to restore protein folding capacity by increasing the synthesis of chaperones. In addition, UPR also enhances degradation of unfolded proteins and reduces global protein synthesis to alleviate additional accumulation of unfolded proteins in the ER. Herein, we describe a cell-based ultra-high throughput screening (uHTS) campaign that identifies a small molecule that can modulate UPR and ER stress in cellular and in vivo disease models. Using asialoglycoprotein receptor 1 (ASGR) fused with Cypridina luciferase (CLuc) as reporter assay for folding capacity, we have screened a million small molecule library and identified APC655 as a potent activator of protein folding, that appears to act by promoting chaperone expression. Furthermore, APC655 improved pancreatic β cell viability and insulin secretion under ER stress conditions induced by thapsigargin or cytokines. APC655 was also effective in preserving β cell function and decreasing lipid accumulation in the liver of the leptin-deficient (ob/ob) mouse model. These results demonstrate a successful uHTS campaign that identified a modulator of UPR, which can provide a novel candidate for potential therapeutic development for a host of metabolic diseases.
KEY WORDS: β cells, Unfolded protein response, Small molecules, Protein folding, Endoplasmic reticulum, Chaperones, Cell signaling, Diabetes, ER stress, Liver, Pancreas, Metabolic diseases
Abbreviations: ASGR, asialoglycoprotein receptor 1; ATF4, activating transcription factor 4; ATF6, activating transcription factor 6α/β; BID, twice a day; CLuc, Cypridina luciferase; EGFP-VSVG, enhanced green fluorescence protein-vesicular stomatitis virus ts045 G protein; ER, endoplasmic reticulum; ERP72, endoplasmic reticulum proteins 72; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GLuc, Gaussia luciferase; GRP78, 78-kDa glucose-regulated protein; GRPRP94, glucose-regulated protein 94; GSIS, glucose stimulated insulin secretion; IKKβ, inhibitor of nuclear factor kappa-B kinase subunit beta; IL1β, interleukin 1β; INFγ, interferon gamma; i.p., intraperitoneal; IRE1, inositol requiring enzyme 1α/β; NASH, nonalcoholic steatohepatitis; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; Nod, non-obese diabetic; OGTT, oral glucose tolerance test; PERK, PKR-like ER kinase; SP1/2, serine protease1/2; T1/2D, type1/2 diabetes; TG, thapsigargin; Tm, tunicamycin; TNFα, tumor necrosis factor alpha; uHTS, ultra-high throughput screening; UPR, unfolded protein response; XBP1, X-box-binding protein 1
Graphical abstract
A cell-based high throughput screening identified a small molecule that improves pancreatic beta cell viability and function through increased UPR chaperones and ER folding capacity.

1. Introduction
The endoplasmic reticulum (ER) is an important organelle to regulate the folding capacity of the cell and maintain protein homeostasis. Perturbations of the ER due to accumulation of unfolded proteins, abnormal calcium regulation or chemical stress are associated with several diseases including metabolic and neurological syndromes1. This perturbation of the ER homeostasis is known as ER stress, which leads to the activation of the unfolded protein response (UPR)2. Indeed, the ER can sense the unfolded proteins accumulating in the lumen and can regulate protein expression and degradation accordingly. Underpinning this process is the 78-kDa glucose-regulated protein (GRP78, also known as Bip)3. GRP78, an ER chaperone that is located in the ER lumen, activates UPR by binding to the three transmembrane UPR sensors: inositol requiring enzyme 1α/β (IRE1)4, PKR-like ER kinase (PERK)5, and activating transcription factor 6α/β (ATF6)6. During protein homeostasis conditions, GRP78 is bound to the three UPR sensors, keeping them in an inactive state. However, in the case of accumulated unfolded proteins in the lumen of the ER, GRP78 as a chaperone binds to the unfolded proteins and releases and activates the three UPR sensors on the membrane of the ER4,7. When activated, IRE1 undergoes dimerization and autophosphorylation through the kinase domain present in the cytosolic portion5,8, 9, 10. After phosphorylation, the endonuclease activity of IRE1 induces the splicing of the main downstream target, X-box-binding protein 1 (XBP1) mRNA, producing an active transcription factor that regulates the expression of genes necessary for improving the protein folding capacity, protein degradation and protein export from the cells11,12. Upon dimerization and phosphorylation, PERK's main function is to block the translation of new proteins, by phosphorylating and inhibiting the eukaryotic translation initiation factor-2α and activating the transcription of the activating transcription factor 4 (ATF4), which, in turn, can direct an antioxidant response and induce the expression of DNA damage-inducible transcript 3, also known as C/EBP homologous protein (CHOP), a protein implicated in the UPR-induced apoptosis13,14. The last UPR sensor, ATF6, once released from the binding of GRP78, is free to translocate on the Golgi membrane where it is cleaved by two proteases, S1P and S2P, releasing a cytosolic portion that acts as transcription factor and induces the transcription of genes coding for chaperones, such as glucose-regulated protein 94 (GRP94), GRP78 and catalase15. The induction of the ATF6 branch of the UPR is considered a protective response by increasing the ER folding capacity. Indeed, the activation of the ATF6 pathway is shown to be protective for several diseases including renal and cerebral ischemia/reperfusion15 and type 2 diabetes (T2D)16, 17, 18. The pancreatic β cells have a highly developed ER, required for the biosynthesis and folding of pre-insulin, with subsequent trafficking to the Golgi, packaging into granules, conversion to mature insulin and secretion in response to high glucose. For this reason, supporting protein homeostasis is particularly important for the function and survival of these cells.
Accumulating evidence link ER stress with impaired β cell function in type 1 and 2 diabetes, as well as peripheral insulin resistance associated with T2D. Elevated levels of UPR markers like CHOP were observed in islets from individuals with type 1 diabetes (T1D)19. Elevated ER stress markers were observed in non-obese diabetic (NOD) mice and leptin-deficient (ob/ob) mice. In addition, administration of chemical chaperones rescues the harmful ER stress response and improves pathophysiological signs of diabetes in both diabetic models, validating UPR modulators as potential therapeutic agents for diabetes20. In ob/ob mice, the overexpression of endogenous chaperones in the liver, such as GRP78, promotes the activation of a protective UPR and the expression of more chaperones, including GRP94 (also known as heat shock protein 90 kDa beta member 1), leading to a clearance of the lipids accumulated in the liver21. Overexpression of ATF6 in the liver has a similar beneficial effect of increasing fatty acid oxidation and protecting against hepatic steatosis with increased expression of endogenous chaperones like GRP7822. Similarly, overexpression of ATF6 in diet-induced obese mice have beneficial effects on insulin sensitivity16. UPR levels are elevated in the pre-diabetic stage of T1D. Previous studies showed that modulating the UPR at this stage, by enhancing the ATF6 pathway and promoting a pro-survival UPR, can prevent the disease onset in a diabetes mouse model18,23. In obese human, tauroursodeoxycholic acid, a bile acid derivative that acts as a chemical chaperone to enhance protein folding and ameliorate ER stress improved hepatic and muscle insulin sensitivity, although target cells and mechanisms require additional studies20,24,25. Thus, the identification of drugs that target the folding capacity of the ER could be beneficial for several disorders, including autoimmune and metabolic diseases. Previously, Fu et al.26 designed a high-throughput functional screening system to measure the protein folding capacity of the ER by tagging asialoglycoprotein receptor 1 with Cypridina luciferase reporter (ASGR-CLuc) and identified azoramide that promotes chaperones expression, protects hepatocytes against chemically induced ER stress, and β cells survival and function and improves glucose handling in a T2D mouse model. We have optimized this system for ultra-high throughput screening (uHTS), screened Calibr's one million compound library and identified APC655 that improves the ER folding capacity, activates the ATF6 pathway, induces chaperones expression and preserves β cell viability and function during stress conditions.
2. Materials and methods
2.1. In vitro cell-based assays
2.1.1. ASGR-CLuc and ATF6-CLuc assay
ASGR-CLuc and AFT6-CLuc cells that were maintained in growth medium (Dulbecco's modified Eagle's medium containing antibiotics and 10% fetal bovine serum) were detached using trypsin. After removing excess trypsin by gentle centrifugation (600×g for 5 min), the cells were resuspended in growth medium containing 2% FBS at a density of 625 cells/μL. Using an automated liquid dispenser, 4 μL of this solution was then dispensed to each well of 1536-well plates (Greiner) that were pre-spotted with 20 nL of various concentrations compound. The plates were incubated for 24 h in the incubator at 37 °C with constant supply of 5% CO2, and 95% humidity. After the incubation period, plates were taken out and each well was supplemented with one μL of growth medium (10% FBS) or one μL of growth medium (10% FBS) containing 1 μmol/L tunicamycin (200 nmol/L final concentration) using an automated liquid dispenser. Plates were put back into the incubator for an additional 24 h. Quantification of secreted ASGR-CLuc or ATF6-CLuc was done by addition of 2 μL of CLuc reagent (Pierce™ Cypridina Luciferase Glow Assay Kit (# 16,171) and the evolved luminescence signal was read using Envision (0.1 s/well, Perkin–Elmer).
2.1.2. Cell culture and in vitro UPR assays
INS-1e β cells that were maintained in growth medium were detached using trypsin. After removing excess trypsin by gentle centrifugation (600×g for 5 min), the cells were resuspended in growth medium at a density of 250 cells/μL. 40 μL of this solution was then dispensed to each well of 384-well plates (Greiner) that were pre-spotted with 100 nL of various concentrations compound. The plates were incubated for 24 h in the incubator at 37 °C with constant supply of 5% CO2, and 95% humidity. After the incubation period, plates were taken out and each well was supplemented with 10 μL of growth medium or 10 μL of growth medium containing 150 nmol/L thapsigargin (TG, 30 nmol/L final concentration). Plates were put back into the incubator for an additional 24 h before further analysis.
2.1.3. Cell viability assay
Cell viability of INS1e cells and primary rat islands was detected by adding 10 μL of CellTiter-Glo® luminescent cell viability assay (#G7473) to each well in 384-well plate. Luminescence signal was read using Envision (0.1 s/well, Perkin–Elmer) to determine cell viabilities.
2.1.4. Caspase 3/7 activity
Caspase 3/7 activity of cells was detected by adding 10 μL of Caspase-Glo® 3/7y assay (#G8092) to each well in 384-well plate. Luminescence signal was read using Envision (0.1 s/well, Perkin–Elmer) to determine the luciferase signal.
2.1.5. pEGFP VSVG trafficking assay
A temperature-sensitive variant (ts045) of vesicular stomatitis virus G protein tagged with GFP (VSVG-GFP)-based assay was used to follow the protein trafficking. ts045-VSVG protein reversibly misfolds and is retained in the ER at 40 °C, but upon temperature shift to 32 °C it correctly folds and is transported out of the ER into the secretory pathway27. HEK293 cells were transfected with the pEGFP VSVG vector and cultured for 16 h at 40 °C and treated with DMSO or APC655 3 mmol/L. After 16 h the temperature was shifted to 32 °C for 15 min. A cellomics technique was used to quantify the P-EGFP-VSVG protein trafficking assay.
2.1.6. ATF6 promoter reporter assay
For the ATF6 promoter reporter assay, HEK293 cells were transfected with the pGL4.39 [luc2P/ATF6 RE/Hygro] vector from Promega (# 9PIE366). Cells were resuspended in growth medium and plated at a density of 250 cells/μL in 384 wells pre-spotted with 100 nL of various concentrations compound. The plates were incubated for 24 h in the incubator at 37 °C with constant supply of 5% CO2, and 95% humidity. After the incubation period, plates were taken out and each well was supplemented with 10 μL of growth medium (10% FBS) or 10 μL of growth medium (10% FBS) containing thapsigargin for a final concentration of 500 nmol/L. Plates were put back into the incubator for an additional 8 h. Quantification of ATF promoter activity was done by addition of 10 μL of Bright-Glo™ luciferase assay system from Promega (#E2650) and the evolved luminescence signal was read using Envision (0.1 s/well, Perkin–Elmer).
2.1.7. Glucose stimulated insulin secretion
Rat primary islets (5000 cells in one 10 cm dish) were dispersed following a dispersal protocol; cells were counted and diluted to a final concentration of 5000 cells/100 μL in 96-well “V” bottom non-treated Nunc plates (Nunc#249935) with 5000 cells/well (100 μL/well). Plates were spin at 1000 RPM (RT6000B Sorvall tabletop = 208 RCF) for 5 min room temp, then placed in 37 °C 5% CO2 incubator overnight. INS1 cells are plated at the density of 100,000 cells/100 μL/well in 96 well plate in 11 mmol/L glucose medium overnight. The day after, the cells are treated with APC655 for 16 h. After 16 h, the cells are switched in 5.5 mmol/L RPMI medium and co-treated with APC655 and stressor (palmitate acid 0.2 mmol/L or thapsigargin 25 nmol/L) for 24 h. After 24 h, the cells are incubated in Krebs–Ringer bicarbonate HEPES buffer/2.8 mmol/L glucose for 2 h for serum deprivation. After serum deprivation, the cells are stimulated with different glucose levels (2.8 mmol/L/20 mmol/L) in Krebs–Ringer bicarbonate HEPES buffer/fatty-acid free 0.1% BSA buffer for 2 h. At this point the medium is collected to determine the insulin level using HTRF Insulin Assay Kit (Cisbio Assay, 62INSPEC). Briefly, 10 μL antibodies solution containing two monoclonal antibodies that recognize the insulin was added for each 10 μL of the medium. These antibodies were labeled with fluorophores that are fluorescence resonance energy transfer pair in proximity. Fluorescence resonance energy transfer signal is measured using Envision plate reader with excitation at 320 nm and emission at 665 nm and 615 nm. The cell layer is lysate with RIPA buffer and used to determine protein quantification using the BCA assay. Protein quantification is used to normalize the insulin level.
2.1.8. Western blot
For total protein extract preparation from cell preparation, the cell pellet was lysate with RIPA buffer, sonicated and incubate on the shaker at 4 °C for 30 min and centrifuged at 12,000×g for 15 min at 4 °C. The pellet was discarded. An aliquot of the supernatants was used for protein determination using the BCA protein assay reagent, according to the manufacturer's instruction, while the remainder was used for Western blot analysis. An equal amount of protein from each sample was loaded onto 4%–12% SDS-polyacrylamide gel and transferred to a PVDF membrane. The membranes were incubated with the appropriate primary and secondary antibodies, and the signal was detected using the Odyssey Li-Cor platform and the densitometric analysis was performed using the Image J software (NIH, Bethesda, MA, USA). For total protein extract from livers, a fragment of the liver from each mouse was homogenized in RIPA buffer using the Precellys Lysing Kit and the Precellys 24 homogenizer. Then sonicated and processed the same way of the cell lysate.
2.1.9. Gene expression analysis
For total RNA extract from cell preparation, the total RNA extraction was performed using the RNeasy® Mini Kit (Quiagen), following the manufacture's protocols. Total RNA from liver tissues was extracted using the TRIZOL. Total RNA was converted into cDNA using the high-capacity cDNA-RT kit from Applied Biosystem, according to supplier's instructions. PCR amplification was performed using the PowerUp™ SYBR™ Green Master Mix from Applied Biosystem, following the manufacturer's protocols. For data analysis, the 7500 Software v2.0.5, provided by Applied Biosystem, was used. All PCR reactions were carried out in triplicate (duplicate for the standard curves). All qPCR results are expressed as relative ratio of the target cDNA transcripts to GGAPDH and normalized to that of the reference condition.
2.2. Animal model and in vivo treatment
2.2.1. Animal studies
All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee of Calibr at the Scripps Research Institute and strictly followed the National Institutes of Health guidelines for humane treatment of animals.
2.2.2. Animal
C57BL/6J (JAX stock# 000664) and ob/ob (JAX stock #000632) male mice 12/13 weeks of age were purchased from Jackson Laboratory in Bar Harbor, ME. The mice were ear notched for identification and housed in individually and positively ventilated polysulfonate cages with HEPA filtered air at a density of 3–4 mice per cage. The animal room was lit entirely with artificial fluorescent lighting, with a controlled 12 h light/dark cycle (6 am–6 pm light). The normal temperature and relative humidity ranges in the animal rooms were 22 ± 4 °C and 50 ± 15%, respectively. The animal rooms were set up to have 15 air exchanges per hour. Filtered tap water, acidified to a pH of 2.5–3.0, and normal rodent chow were provided ad libitum.
2.2.3. PK plasma collection
Approximately 75 μL whole blood were collected at 0, 1, 3, 5 and 24 h post dose on study Day 7. Whole blood was collected into lithium heparin plasma separator tubes. Plasma samples were extracted after centrifugation (18,000×g; 4 °C). Plasma was stored at −80 °C for future analyses.
2.2.4. Dosing and non-fasted blood glucose
All animals enrolled on study were dosed twice a day (BID) by intraperitoneal injections at volume of 5 mL/kg. Body weights were recorded each day of dosing in the morning. Non-fasted blood glucose values were measured three times weekly and were assessed at the same time each day pre dose (e.g., between 8 and 10 am).
2.2.5. Oral glucose tolerance tests and measurement of insulin release
Oral glucose tolerance (OGTT) tests were conducted on Day 7 after an overnight fast. For OGTT, initial blood glucose values were determined prior to administration (1.5 g/kg BW) of a 30% glucose solution (300 mg/mL) d-glucose in sterile PBS) by oral gavage. Blood glucose was measured using the AlphaTrak2 at 0,15, 30, 60–90 and 120-min post glucose administration. Any values exceeding the limitation of the glucometer (>750 mg/dL) were diluted 1:1 with sterile PBS and re-measured. Sera insulin was determined using the Mercodia Mouse Ultrasensitive Insulin ELISA Kit (Mercodia Cat. No. 10-1249-01).
2.2.6. Histological analysis and staining quantification
The livers were collected from the mice, fixed in formalin for 24 h and subsequentially frozen. Thereafter, cryosections were prepared for histological and immunohistochemical analyses. Sections stained with oil red O (from Abcam) were photographed and the images were analyzed using Image J.
2.3. Reagents
Thapsigargin (#T9033) and PF429242 (SML0667) were purchased from Millipore Sigma; tunicamycin was purchased from Tocris (#5316). Antibodies and primers used in this study are provided in Supporting Information Tables S1 and S2.
3. Results
3.1. Phenotypic screening to measure the protein folding capacity of the ER identified APC655 as a UPR modulator
We utilized a reporter system in which ASGR1, a transmembrane receptor whose folding is affected by chemically induced ER stress, is tagged with Cypridina luciferase reporter (ASGR-CLuc fusion protein) and outfitted with a secretion signal to monitor and quantify the folding capacity of the ER26. As an internal control, this cell line also constitutively expressed a Gaussia luciferase, which is not modulated by ER stress. The reporter assay was optimized and miniaturized to 1536 well format suitable for uHTS. Two molecules were used as positive controls: azoramide, which is an UPRmodulator with beneficial effects in obese mice26 and forskolin, a known activator of the enzyme adenylyl cyclase which was identified as a strong activator of the reporter system from a small pilot screening28 (Supporting Information Fig. S1). The treatment of HEK293 cells expressing ASGR-CLuc with thapsigargin and tunicamycin inhibits the folding of ASGR1, thus decreasing the luciferase signal in a dose responsive manner (Fig. 1A and B). We screened compounds at 4 μmol/L concentration in the presence of 100 nmol/L tunicamycin. Compounds that inhibited 60% of the reduction of ASGR-CLuc signal were identified as hits for follow-up studies. The hit compounds were subsequentially confirmed in triplicate at 4 μmol/L (Fig. 1C) and counter-screened for luciferase stabilizers using a non-UPR related CLuc control construct and for cytotoxicity using the cell titer-glo assay in INS1e cells (<30% inhibition). Some of the known small molecules were identified from this screening, validating our screening assay (Fig. S1). Among the confirmed hits (Fig. 1C), APC655 presents as a novel and potent hit in preserving protein folding capacity in dose-responsive manner, both when treated simultaneously or following pre-treatment with tunicamycin (Fig. 1D and E and Supporting Information Fig. S2).
Figure 1.
Ultra-high throughput screening assay and APC655 in ASGR-CLuc reporter assay. (A) Dose-dependent response of the ASGR-CLuc assay to thapsigargin. (B) Dose-dependent response of the ASGR-CLuc assay to tunicamycin. (C) Primary screening output; forskolin and azoramide were used as positive controls in the screening. 0% represents DMSO treated cells, the orange dots represent the positive controls, the grey dots are the HITS and in yellow are the confirmed HITS. The red circled dot is APC655. (D) Chemical structure of APC655. (E) Dose response of APC655-induced increase of ASGR-CLuc secretion with and without tunicamycin 1 μmol/L. Data are shown as percentage to DMSO. Error bars are represented as mean ± SD; n = 3.
3.2. APC655 activates ATF6 pathway and chaperone expression
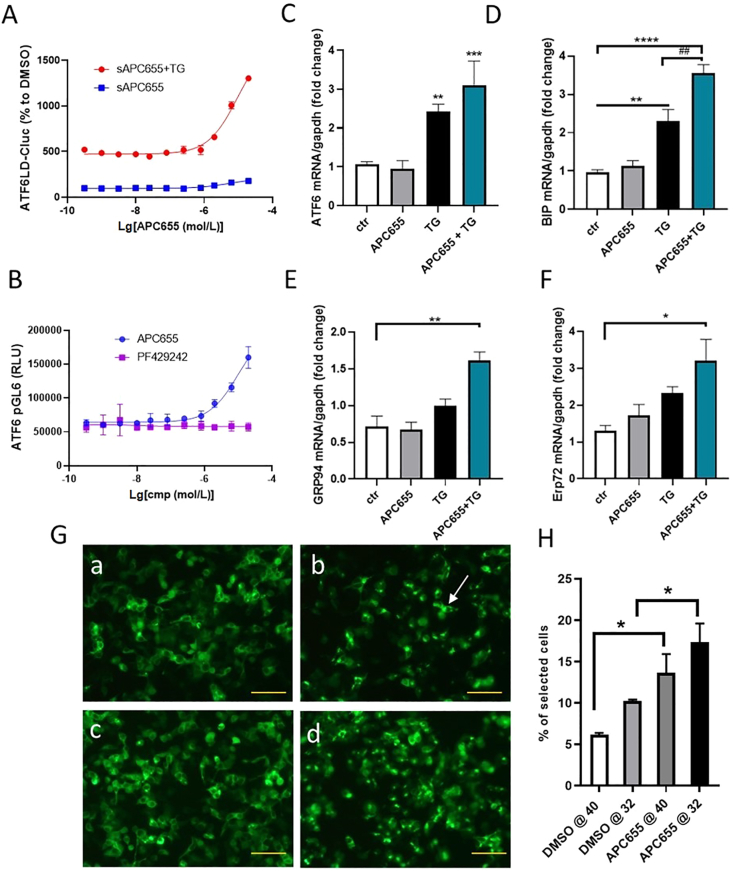
To investigate whether the increased protein folding capacity of the ER induced by APC655 is due to the modulation of the UPR, we analyzed different signaling markers of the three UPR branches. Western blot analysis of proteins involved in the IRE1 and PERK branches of the UPR pathways, such as BIP, IRE1, XBP1 and ATF4, showed that APC655 does not affect these signaling components in the presence of stress (Supporting Information Fig. S3). To investigate the involvement of the ATF6 branch, we looked at ATF6LD-CLuc reporter assay to monitor ATF6 release by GRP7826. APC655 induced the release of ATF6LD in the medium, in a dose-dependent manner and enhanced the effect of 0.5 μmol/L thapsigargin on ATF6 activation (Fig. 2A).
Figure 2.
APC655 enhances ER protein folding capacity through the activation of the ATF6 pathway and chaperones expression, during stress. (A) APC655 promotes the release of ATF6LD in a dose-dependent way, with or without treatment with 0.5 μmol/L TG. (B) APC655 induces the ATF6 promoter; the SP1 inhibitor PF429242 was used as control in the assay. (C) ATF6 mRNA levels are increased by TG treatment in INS1e cells and boosted by the co-treatment with 5 μmol/L APC655. (D) BIP mRNA level is increased by 6 h TG treatment in INS1e cells and boosted by the co-treatment with 5 μmol/L APC655. (E) ERP72 (PDI) mRNA level is increased by 5 μmol/L APC655 in co-treatment with TG for 6 h in INS1e cells. (F) The chaperone GRP94 expression is increased by 5 μmol/L APC655 co-treatment with TG. (G, H) P-EGFP-VSVG protein trafficking assay after 16 h at 40 °C with DMSO (a) or 3 mmol/L APC655 (c) and after 15 min at 32 °C with DMSO (b) or 3 mmol/L APC655 (d). The arrow indicates the folded P-EGFP-VSVG protein H. Cellomics quantification of P-EGFP-VSVG protein trafficking assay, normalized on number of nuclei and expressed as % of total cells analyzed, Scale bar = 50 μm. Error bars are represented as mean ± SD; n = 4–6. Significance was determined by one-way ANOVA test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
To further delineate whether APC655 affects the expression level of ATF6, we generated an ATF6 reporter assay to study the promoter activity of ATF6. For the promoter reporter assay we used PF429242, an inhibitor of S1P, as control in the experiment29, which provides a contrast to the activating effect of thapsigargin on the ATF6 promoter (Supporting Information Fig. S4). While APC655 has the capacity to induce the ATF6 reporter, the S1P inhibitor did not (Fig. 2B). ATF6 is also upregulated by APC655 in the presence of stress at the mRNA level in INS1e cells pretreated overnight with APC655 5 μmol/L and co-treated with thapsigargin 100 nmol/L for 6 h (Fig. 2C). To confirm the activation of ATF6 by APC655 in vitro, we overexpressed HEK293 cells with EGFP-ATF6 construct and observed higher EGFP signal in cells treated with thapsigargin (100 nmol/L) and APC655 (5 mmol/L) compared to control (Supporting Information Fig. S5). These data suggest that APC655 enhances the upregulation of the ATF6 branch of UPR during stress conditions. As a measure of ATF6 activation, we also analyzed the ATF6 downstream targets by qPCR in INS1e cells treated with thapsigargin, showing the upregulation of the chaperones GRP78, GRP94 and ERP72 (ER protein 72; also known as protein disulfide-isomerase A4), all known targets of ATF6 (Fig. 2D–F). Interestingly, chaperones expression was also associated with the reestablishment of the protein trafficking in the cells as shown in Fig. 2G and H by the pEGFP-VSVG trafficking assay. Taken together these data show that APC655 is a modulator of the ATF6 pathway and induces the expression of endogenous chaperones, thus incrementing the protein folding capacity of the ER in INS1e during stress.
3.3. APC655 protect β cells from chemically induced ER-stress dependent cell death
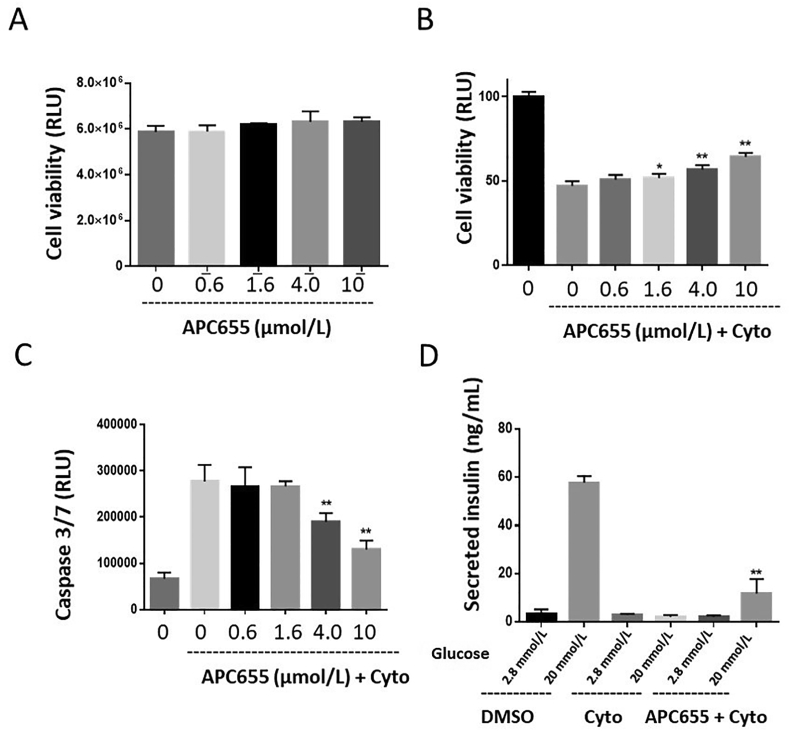
Since APC655 promotes the expression of chaperones and increases the protein folding capacity of the ER, we asked whether this is translated into the protection or improved β cell survival in the various ER stress-induced cell death. We used β cells carrying the Akita mutation (C96Y) in the insulin gene, which leads to incorrect folding of the insulin protein30,31. Blocking protein degradation with the proteasome inhibitor MG132 in these cells leads to the accumulation of the unfolded insulin in the ER and the subsequent UPR activation and cell death. As shown in Fig. 3A, Akita cells treated with MG132 showed 90% reduction of cell viability, and cotreatment with APC655 protected the Akita cells from UPR-induced cell death. In the glucolipotoxicity settings, treatment of INS1e cells with 25 mmol/L glucose and 500 μmol/L palmitate for 48 h leads to significant reduction of cell viability and APC655 protected cell survival in dose–response (Fig. 3B).
Figure 3.
APC655 protects cells survival and function in INS1e cells. (A) Cell viability assay performed with Cell Titer Glo Kit in INS1 Akita cells treated in dose–response with APC655, with or without the proteasome inhibitor MG132. (B) Cell Titer Glo assay performed in INS1e cells treated with APC655 (2, 6 and 20 μmol/L) with or without 25 mmol/L glucose and 500 μmol/L palmitate; Control (ctr) is the cells without any treatment, DMSO are the cells only treated with the stressor. (C) Caspase 3/7 activation performed in INS1e cells treated with APC655 (2, 6 and 20 μmol/L) with or without IFNγ (500 ng/mL) and IL1β (50 ng/mL); ctr is the cells without any treatment, DMSO are the cells only treated with the stressor. (D) glucose stimulated insulin secretion (GSIS) in INS1e cells treated with APC655 (0.2, one and 5 μmol/L) and stressed with 20 nmol/L thapsigargin. Error bars are represented as mean ± SD; n = 4–6. Significance was determined by one-way ANOVA test. ∗∗P < 0.01, ∗∗∗∗P < 0.0001.
Cytokines have been directly implicated in the pathogenesis of type 1 diabetes (T1D) as the major drivers of inflammation and play crucial roles in controlling ongoing β cell destruction32. The in vitro treatment of INS1e cells with interferon g (IFNγ, 500 ng/mL) and interleukin-1b (IL1β, 50 ng/mL) leads to the activation of caspase 3/7, and co-treatment with APC655 reduced the caspase 3/7 activation in dose response (Fig. 3C). As APC655 protects pancreatic β cells from the UPR-induced apoptosis, next we tested whether this improved β cell survival is associated with the protection of β cells function. β cells release insulin in response to high glucose (glucose stimulated insulin secretion, GSIS) in INS1e cells is greatly reduced by the treatment with the ER stressor thapsigargin. To our satisfaction, APC655 treatment totally recovered the attenuated β cell function. At 5 μmol/L concentration, the total insulin secreted at 20 mmol/L glucose stimulation surpassed the normal GSIS response without thapsigargin stressor, suggesting an enhancement of β cell viability and function (Fig. 3D).
3.4. APC655 improves β cell viability and function from cytokine induced ER-stress in primary islets
Next, we tested whether the improved survival in INS1e cells can be translated into primary β cells from freshly isolated rat islets. APC655 treatment alone does not impact the β cell viability in rat islets (Fig. 4A). We have developed the cytokine stress assay in primary rat islets. β cell viability was greatly reduced with the treatment of a cytokine cocktail composed of IFNγ (100 ng/mL), IL1β (5 ng/mL) and TNFβ (50 ng/mL) and APC655 treatment reduced this loss of viability in dose response (Fig. 4B). Similarly, the caspase activation induced by the cytokine cocktail was greatly inhibited by APC655 treatment (Fig. 4C). In a similar way to INS1e cells, the islet function is totally abolished by the treatment of cytokine cocktail, and APC655 partially recovered its GSIS function, although the effect is mild, potentially due to the harsh assay conditions (Fig. 4D). All these results suggest that APC655 can have beneficial effects in diabetes and potentially protect β cells survival during stress in diabetes models.
Figure 4.
APC655 protects cells survival and function in primary rat islets. (A) Cell viability assay in primary rat islets treated with APC655 for 24 h at different doses (0.6, 1.6, 4 and 10 μmol/L). (B) Cell viability assay in primary rat islets treated with APC655 for 24 h at different doses (0.6, 1.6, 4 and 10 μmol/L), together with DMSO or IFNγ 100 ng/mL and IL1β 5 ng/mL. (C) Caspase 3/7 activation performed in rat islets treated with APC655 (0.6, 1.6, 4 and 10 μmol/L), together with DMSO or IFNγ 100 ng/mL, IL1β 5 ng/mL and TNFα 50 ng/mL. (D) Glucose stimulated insulin secretion (GSIS) in rat islets treated with APC655 (5 μmol/L) and stressed with cytokines (IFNγ 25 ng/mL and IL1β 1.25 ng/mL). Error bars are represented as mean ± SD; n = 4–6. Significance was determined by one-way ANOVA test. ∗P < 0.05, ∗∗P < 0.01.
3.5. APC655 decreased body weight, lowered fast blood glucose and improved liver steatosis in the ob/ob mice
Leptin deficient ob/ob mice are characterized by insulin resistance and liver lipid accumulation. Previous work demonstrated that the ER stress level in this mouse model is high in both pancreatic β cells and in hepatocytes33. Encouraged by the in vitro protection of β cell viability and function in INS1e and primary islets, we tested this compound in the ob/ob mouse model. Intraperitoneal (ip) administration of APC655 at 15 and 50 mg/kg twice a day (BID) for 7 days reduced body weight (Fig. 5A) and fasting glucose (Fig. 5B) in ob/ob mice. In addition, APC655 also shows a trend in improving glucose handling as demonstrated in the OGTT (Fig. 5C and D).
Figure 5.
APC655 decreased body weight, lowered fasted glucose, showed trend improved blood glucose and decreases lipids accumulation in the liver of ob/ob mice. (A) Body weight in grams of naïve and ob/ob mice treated daily with the vehicle, APC655 (15 mg/kg) and APC655 (50 mg/kg) for 6 days. (B) Fast glucose analysis in naïve and ob/ob mice treated daily with the vehicle, APC655 (15 mg/kg) and APC655 (50 mg/kg) for 6 days. (C) OGTT in naïve and ob/ob mice treated daily with the vehicle, APC655 (15 mg/kg) and APC655 (50 mg/kg) for 6 days. (D) Area under the curve (AUC) of OGTT. (E) Oil red O staining in liver sections from naïve and ob/ob mice treated daily with the vehicle, APC655 (15 mg/kg) and APC655 (50 mg/kg) for 6 days. Scale bar = 50 μm. (F) Liver weight in grams. (G) Oil red O staining quantification. Error bars are represented as mean ± SD; n = 4–6. Significance was determined by one-way ANOVA test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Interestingly, a significant reduction of liver size is observed with APC655 treatment (50 mg/kg) in the ob/ob mice during takedown. Indeed, the liver weight normalized by the body weight showed a significant reduction in the mice treated with APC655 at 50 mg/kg and a trend of reduction in the ob/ob mice at 15 mg/kg treatment (Fig. 5F). We also analyzed the lipids accumulation in the livers from the vehicle and APC655 treated ob/ob mice, using the oil red O staining (Fig. 5E). As shown in Fig. 5E and quantified in Fig. 5G, lipids staining is strongly reduced in ob/ob mice treated with APC655 50 mg/kg, pointing to a potential beneficial effect of APC655 in liver steatosis and NASH.
The interesting phenotypic improvement of liver steatosis prompted us to investigate potential mechanisms. The link between obesity, UPR and inflammation is not completely understood but there is much evidence that UPR and inflammation are connected, regulated by each other and involved in obesity34, 35, 36. We analyzed inflammatory response and UPR chaperones in the liver from the ob/ob mice treated with the vehicle or APC655 and compared them with the naïve control mice. The activation of NF-κB by UPR has been reported as a common signaling cascade in the three UPR branches37. Western blot analysis of the total proteins isolated from livers of ob/ob mice showed that the treatment with APC655 modulated NF-κB signaling pathway, as demonstrated by the reduced NF-κB (P65) phosphorylation and increased protein level of the NF-κB inhibitor IKKβ (Fig. 6A–C). Also, protein levels and mRNA levels of GRP94 were upregulated in the livers from ob/ob mice treated with APC655, supporting our in vitro observations (Fig. 6D–F).
Figure 6.
sAPC655 decreases NF-κB pathway activation in the liver from ob/ob mice. Western blot analysis (A) and quantification (B and C) of NF-κB pathway in the total proteins extracted from liver of naïve mice or ob/ob mice treated with the vehicle or APC55 (15 and 50 mg/kg). (D) GRP94 protein levels analyzed by Western blot in the total proteins extracted from liver of naïve mice or ob/ob mice treated with the vehicle or APC55 (15 and 50 mg/kg) and quantification (E). (F) GRP94 mRNA level in livers from ob/ob mice treated with the vehicle or APC55 (15 and 50 mg/kg), measured by qPCR. Error bars are represented as mean ± SD; n = 4–6. Significance was determined by the one-way ANOVA test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Next, we analyzed the expression level of genes involved in the UPR pathways. APC655 treatment highlighted a trend but not significant increase of chaperone expression in the livers of ob/ob mice treated with APC655 (Supporting Information Fig. S6). Taken together, these data suggest that the upregulation of chaperones might be consequential to the reduced inflammation and suggests a more systemic mechanism of action of APC655 in vivo.
4. Discussion
ER is important for calcium storage, lipid biosynthesis, protein synthesis and trafficking. Defects in the ER function translate into accumulation of unfolded proteins in the lumen of the ER and result in the activation of the UPR pathways. Abnormal and unresolved UPR is associated with the development of T1D and T2D and is responsible in part for the loss and malfunction of pancreatic β cells23,38. As the main function of the UPR is to restore the ER homeostasis by promoting the expression of chaperones and blocking the translation of new proteins, an unresolved ER-stress can transition into the apoptotic cell death. A “protective” UPR could be beneficial for the cells in order to restore protein homeostasis; however, it is not easy to reach the exact balance between protein synthesis and degradation, preventing the ER-stress induced cell death and promoting proteostasis, thus preserving cell survival. The discovery of a small molecule that can improve the ER folding capacity by stimulating the chaperones expression and, at the same time, preserving cell viability, might be the key to preventing and/or curing many metabolic diseases. Indeed, it has been previously showed that the treatment with chemical chaperones such as tauroursodeoxycholic acid or phenyl butyric acid reduces the diabetes in T1D mouse models when administered in pre-diabetic stage and this protection is lost in B-cell-specific ATF6a-deficient mice25.
Here, we took advantage of a high throughput phenotypic screening assay and identified APC655 as a small molecule UPR modulator that could rescue ER protein folding capacity. We found that in β cells, APC655 can repristinate the protein folding capacity of the ER during stress conditions by boosting the ATF6 pathway and enhancing the expression of chaperons. APC655 showed a protective effect on β cell viability and function in both β cell lines and primary islets. Activation of ATF6 and chaperones expression are known to be associated with a protection of hepatic steatosis in diet-induced insulin resistant mice. The expression of a dominant negative ATF6 in high-fat-high-sucrose diet-fed mice increases the susceptibility to develop hepatic steatosis and insulin resistance. It has been shown that ATF6 can stimulate fatty acid oxidation in hepatic cells through the activation of peroxisome proliferator-activated receptors (PPARα), thus maintaining metabolic homeostasis22. The treatment of ob/ob mice showed a protective effect of APC655 on glucose handling and on the lipid accumulation in the liver. Our in vivo data in the liver of ob/ob mice showed a strong downregulation of the NF-κB inflammatory pathway with the treatment of APC655 and the gene expression analysis of the UPR markers showed significant increase of GRP94 and XBP1, with a trend of increase in ATF6 and BIP and ATF4, indicating an increase in chaperone expression and potential folding capacity, resulting in the improvement of UPR and the ER stress. While the crosstalk between UPR and inflammation is not well defined, there are evidences that they can both regulate each other23. Nevertheless, both pathways are crucial for a complex disease like hepatic steatosis and NASH and the identification of a small molecule like APC655 could provide an interesting tool compound for mechanistic studies and therapeutic application in protein homeostasis and inflammation in complex diseases like diabetes, obesity and NASH.
5. Conclusions
In this study, APC655, a new small molecule enhancer of the ER protein folding capacity, was identified from a uHTS campaign. APC655 promotes the expression of chaperones and activates the ATF6 branch of the UPR during stress conditions in vitro. Moreover, APC655 preserves β cells viability and function when challenged with stress in vitro. In vivo treatment of APC655 in ob/ob mice reduces the NF-κB inflammatory pathway activation and lipid accumulation in the liver while improving glucose handling.
Acknowledgments
The work was supported by the Juvenile Diabetes Research Foundation (JDRF) [3-PAR-2016-241-I-X, US]. We thank Gökhan Hotamisligil for providing the reporter cell lines and Patricia Kilian, Andrew Rakeman, Peter Lomedico, and Frank Martin (JDRF, USA) for helpful discussions.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.05.018.
Author contributions
Valeria Marrocco, Tuan Tran, Siying Zhu, Seung Hyuk Choi, Sijia Li, Mitch Hull, Nikki Rogers, Matthew S. Tremblay, and Weijun Shen conducted and supervised the high throughput screening and biological characterization. Ana M. Gamo, Jason Roland, and Arnab K. Chatterjee conducted and supervised the chemistry and synthesis. Qiangwei Fu, Van Nguyen-Tran, and Sean Joseph conducted and supervised the in vivo pharmacology studies. Nikki Rogers and Matthew S. Tremblay conceived the project idea. Sean Joseph, Arnab K. Chatterjee, Nikki Rogers, Matthew S. Tremblay, and Weijun Shen supervised the work. Valeria Marrocco, Siying Zhu, Nikki Rogers, Matthew S. Tremblay, and Weijun Shen wrote the paper.
Conflicts of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lin J.H., Walter P., Yen T.S. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karagoz G.E., Acosta-Alvear D., Walter P. The unfolded protein response: detecting and responding to fluctuations in the protein-folding capacity of the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2019;11:a033886. doi: 10.1101/cshperspect.a033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner B.M., Pincus D., Gotthardt K., Gallagher C.M., Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5:a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori K., Ma W., Gething M.J., Sambrook J. A transmembrane protein with a CDC2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 5.Harding H.P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 6.Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox J.S., Shamu C.E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 8.Prischi F., Nowak P.R., Carrara M., Ali M.M. Phosphoregulation of Ire 1 RNase splicing activity. Nat Commun. 2014;5:3554. doi: 10.1038/ncomms4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shamu C.E., Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 10.Tirasophon W., Welihinda A.A., Kaufman R.J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y., Sun S., Sha H., Liu Z., Yang L., Xue Z., et al. Emerging roles for XBP1, a sUPeR transcription factor. Gene Expr. 2010;15:13–25. doi: 10.3727/105221610x12819686555051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerner A.G., Upton J.P., Praveen P.V., Ghosh R., Nakagawa Y., Igbaria A., et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metabol. 2012;16:250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DuRose J.B., Scheuner D., Kaufman R.J., Rothblum L.I., Niwa M. Phosphorylation of eukaryotic translation initiation factor 2 alpha coordinates rRNA transcription and translation inhibition during endoplasmic reticulum stress. Mol Cell Biol. 2009;29:4295–4307. doi: 10.1128/MCB.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fribley A., Zhang K., Kaufman R.J. Regulation of apoptosis by the unfolded protein response. Methods Mol Biol. 2009;559:191–204. doi: 10.1007/978-1-60327-017-5_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackwood E.A., Azizi K., Thuerauf D.J., Paxman R.J., Plate L., Kelly J.W., et al. Pharmacologic ATF6 activation confers global protection in widespread disease models by reprograming cellular proteostasis. Nat Commun. 2019;10:187. doi: 10.1038/s41467-018-08129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozcan L., Ghorpade D.S., Zheng Z., de Souza J.C., Chen K., Bessler M., et al. Hepatocyte DACH1 is increased in obesity via nuclear exclusion of HDAC4 and promotes hepatic insulin resistance. Cell Rep. 2016;15:2214–2225. doi: 10.1016/j.celrep.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Wang X., Yang B., Wang H., Ma Z., Lu Z., et al. 3β-Hydroxysteroid-delta24 reductase (DHCR24) protects pancreatic beta cells from endoplasmic reticulum stress-induced apoptosis by scavenging excessive intracellular reactive oxygen species. J Diabetes Res. 2020;2020:3426902. doi: 10.1155/2020/3426902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engin F., Yermalovich A., Nguyen T., Hummasti S., Fu W., Eizirik D.L., et al. Restoration of the unfolded protein response in pancreatic beta cells protects mice against type 1 diabetes. Sci Transl Med. 2013;5:211ra156. doi: 10.1126/scitranslmed.3006534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marhfour I., Lopez X.M., Lefkaditis D., Salmon I., Allagnat F., Richardson S.J., et al. Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia. 2012;55:2417–2420. doi: 10.1007/s00125-012-2604-3. [DOI] [PubMed] [Google Scholar]
- 20.Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R.O., et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye R., Jung D.Y., Jun J.Y., Li J., Luo S., Ko H.J., et al. GRP78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010;59:6–16. doi: 10.2337/db09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X., Zhang F., Gong Q., Cui A., Zhuo S., Hu Z., et al. Hepatic ATF6 increases fatty acid oxidation to attenuate hepatic steatosis in mice through peroxisome proliferator-activated receptor alpha. Diabetes. 2016;65:1904–1915. doi: 10.2337/db15-1637. [DOI] [PubMed] [Google Scholar]
- 23.Engin F. ER stress and development of type 1 diabetes. J Invest Med. 2016;64:2–6. doi: 10.1097/JIM.0000000000000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kars M., Yang L., Gregor M.F., Mohammed B.S., Pietka T.A., Finck B.N., et al. Tauroursodeoxycholic acid may improve liver and muscle but not adipose tissue insulin sensitivity in obese men and women. Diabetes. 2010;59:1899–1905. doi: 10.2337/db10-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bronczek G.A., Vettorazzi J.F., Soares G.M., Kurauti M.A., Santos C., Bonfim M.F., et al. The bile acid TUDCA improves beta-cell mass and reduces insulin degradation in mice with early-stage of type-1 diabetes. Front Physiol. 2019;10:561. doi: 10.3389/fphys.2019.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu S., Yalcin A., Lee G.Y., Li P., Fan J., Arruda A.P., et al. Phenotypic assays identify azoramide as a small-molecule modulator of the unfolded protein response with antidiabetic activity. Sci Transl Med. 2015;7:292ra98. doi: 10.1126/scitranslmed.aaa9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Presley J.F., Cole N.B., Schroer T.A., Hirschberg K., Zaal K.J., Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 28.Cunha D.A., Ladriere L., Ortis F., Igoillo-Esteve M., Gurzov E.N., Lupi R., et al. Glucagon-like peptide-1 agonists protect pancreatic beta-cells from lipotoxic endoplasmic reticulum stress through upregulation of BiP and JunB. Diabetes. 2009;58:2851–2862. doi: 10.2337/db09-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lebeau P., Byun J.H., Yousof T., Austin R.C. Pharmacologic inhibition of S1P attenuates ATF6 expression, causes ER stress and contributes to apoptotic cell death. Toxicol Appl Pharmacol. 2018;349:1–7. doi: 10.1016/j.taap.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 30.Liu M., Haataja L., Wright J., Wickramasinghe N.P., Hua Q.X., Phillips N.F., et al. Mutant INS-gene induced diabetes of youth: proinsulin cysteine residues impose dominant-negative inhibition on wild-type proinsulin transport. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss M.A. Diabetes mellitus due to the toxic misfolding of proinsulin variants. FEBS Lett. 2013;587:1942–1950. doi: 10.1016/j.febslet.2013.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J., Liu J., Li L., LanY, Liang Y. Cytokines in type 1 diabetes: mechanisms of action and immunotherapeutic targets. Clin Transl Immunology. 2020;9:e1122. doi: 10.1002/cti2.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L., Li P., Fu S., Calay E.S., Hotamisligil G.S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metabol. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amen O.M., Sarker S.D., Ghildyal R., Arya A. Endoplasmic reticulum stress activates unfolded protein response signaling and mediates inflammation, obesity, and cardiac dysfunction: therapeutic and molecular approach. Front Pharmacol. 2019;10:977. doi: 10.3389/fphar.2019.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hotamisligil G.S. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hummasti S., Hotamisligil G.S. Endoplasmic reticulum stress and inflammation in obesity and diabetes. Circ Res. 2010;107:579–591. doi: 10.1161/CIRCRESAHA.110.225698. [DOI] [PubMed] [Google Scholar]
- 37.Garg A.D., Kaczmarek A., Krysko O., Vandenabeele P., Krysko D.V., Agostinis P. ER stress-induced inflammation: does it aid or impede disease progression?. Trends Mol Med. 2012;18:589–598. doi: 10.1016/j.molmed.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 38.Engin F., Nguyen T., Yermalovich A., Hotamisligil G.S. Aberrant islet unfolded protein response in type 2 diabetes. Sci Rep. 2014;4:4054. doi: 10.1038/srep04054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.