Abstract
Cancer is recognized as a preeminent factor of the world’s mortality. Although various modalities have been designed to cure this life-threatening ailment, a significant impediment in the effective output of cancer treatment is heterogeneity. Cancer is characterized as a heterogeneous health disorder that comprises a distinct group of transformed cells to assist anomalous proliferation of affected cells. Cancer stem cells (CSCs) are a leading cause of cancer heterogeneity that is continually transformed by cellular extrinsic and intrinsic factors. They intensify neoplastic cells aggressiveness by strengthening their dissemination, relapse and therapy resistance. Considering this viewpoint, in this review article we have discussed some intrinsic (transcription factors, cell signaling pathways, genetic alterations, epigenetic modifications, non-coding RNAs (ncRNAs) and epitranscriptomics) and extrinsic factors (tumor microenvironment (TME)) that contribute to CSC heterogeneity and plasticity, which may help scientists to meddle these processes and eventually improve cancer research and management. Besides, the potential role of CSCs heterogeneity in establishing metastasis and therapy resistance has been articulated which signifies the importance of developing novel anticancer therapies to target CSCs along with targeting bulk tumor mass to achieve an effective output.
Keywords: Cancer stem cells, tumor heterogeneity, metastasis, therapy resistance
Introduction
With the advancement of cancer genetics, it is evident that cancer is a heterogeneous ailment that involves the acquisition of genetic and epigenetic modifications to assist cancerous cell proliferation, dissemination and therapy resistance [1]. Cancer heterogeneity has been categorized into two distinct types: intertumor heterogeneity and intratumor heterogeneity. Intertumor heterogeneity involves variations in the tumor of the same tissue in different individuals or variations in the tumor of different tissues in the same individual. On the other hand, intratumor heterogeneity encompasses variability within a single tumor. Intertumor heterogeneity lays the foundation of tumor classification in different types and subtypes on the basis of histological appearance, specific expression markers, and divergent genetic profiles. Whereas, intratumor heterogeneity entangles tumor efficacious treatment and complete eradication [2].
CSCs are oncogenic in nature and imputed as key drivers of tumor heterogeneity [3]. Although at first it was presumed that CSCs are a homogenous population of cells, with time it has been proved that CSCs are phenotypically and functionally diverse. There is ample evidence that CSCs hierarchy leads to the intratumor heterogeneity [4-6]. In a contemporary study, Tabuchi and coworkers have unmasked CSCs functional heterogeneity at clonal level. They showed that CSCs in uterine endometrial carcinoma comprised two distinct sub-clones i.e. highly tumorigenic clone and treatment-refractory clone [7]. Moreover, recent literature also represents that CSCs are not static but dynamic populations of cells that are continually transformed by the cells’ extrinsic and intrinsic factors [8].
Bonnet and Dick were the first researchers to disclose CSCs in human acute myeloid leukemia (AML). In accordance with their pioneer work, just like healthy hematopoietic stem cells (HSCs) leukemia stem cells (LSCs) bear CD34+CD38- phenotype [9]. Recent investigations with large sample sizes display that LSCs are not only restricted to CD34+CD38- fraction but CD34+CD38+ fraction also harbors leukemogenic cells [10,11]. Confirming LSCs heterogeneity Taussig and collaborators stated that LSCs in NPM1-mutation harbor CD34- phenotype while CD34+ phenotype is only associated with NPM1-mutated AML that additionally possess higher risk to develop FLT3IDT mutation [12]. Results from some other groups also made explicit that both CD34- and CD34+ phenotypes can exist in NPM1-mutated AML [13].
Like leukemia, solid tumors also harbor several different subpopulations of stem-like cells. In the year 2003, Al-Hajj and his companions first identified breast cancer stem cells (BCSCs) with CD44+CD24-/LLIN- pattern [14]. Later, another group detected an elevated level of ALDH1 in both normal and CSCs. However, cells bearing both CD44+CD24-/L and ALDH+ markers were more malignant than the cells bearing a single marker only [15]. Further describing combination markers for BCSCs enrichment Pece and coworkers revealed that cells with CD24HCD49HDNERH, CD24HCD49HDLL1H and CD49FHDLLHDNERH phenotypes could be utilized as markers for BCSCs identification [16]. On the other hand, to reveal BCSCs heterogeneity, Hwang-Verslues and coworkers reported a highly tumorigenic subpopulation of BCSCs with PROCR+/ESA+ phenotype instead of CD44+CD24-/L and ALDH+ signature [17]. Similarly, in ER-α-negative breast cancer, not only CD44+CD24- and CD44+CD24+ cell populations were found oncogenic but they also found a third more malignant phenotype CD44+CD49fHCD133/2H of BCSCs [18]. Furthermore, heterogeneous markers have been described for enrichment of CSCs in the tumors of other organs [19]. For example in glioblastoma, CD133, CD44, CD15, A2B5, integrin α6 and LICAM markers have been reported for CSCs identification [20,21]. Cell surface markers, in particular CD133, CD44, ALDH, CD166, CD133+ESA+, CD166+CD44+, and CD166+EpCAM+ are exploited for lung cancer stem cells (LCSCs) isolation, whereas for bladder cancer stem cells detection CD44, CD67LR, ALDH1A1, BCMab1 and EMA markers have been described in the literature [22].
In light of these data, it can be stated that CSCs have been found to be highly heterogeneous in different malignancies, and only one type of marker is not enough to isolate or target a particular CSCs from the bulk of the tumor mass. Hence, this review article is intended to summarize recent findings of potential mechanisms of CSCs heterogeneity that how these are equipped with stemness features. At first, we have magnified the intrinsic factors, like transcription factors, signaling pathways, genetic alterations, epigenetic modifications, ncRNAs and epitranscriptomics that we regard as the inherent properties of self-renewal. Secondly, we have pinpointed the role of TME as an extrinsic factor that can significantly alter phenotype of these cells. Additionally, the function of this heterogeneity in cancer dissemination and therapeutic resistance has been documented, along with discussing some already established therapeutic approaches to target CSCs.
Models of CSCs heterogeneity
Historically, two contrasting models were proposed to describe cancer heterogeneity. These models were named: the clonal evolution (CE) model and the cancer stem cell (CSC) model. The CE model was emanated from Darwin’s evolution theory, where a single cell is responsible for tumor initiation. This model states that with time stochastic changes occur within a cell and as a consequence of cellular division these changes can pass to the daughter cells. The daughter cells are prone to acquire more and more perturbations and as a result of natural selection, only the most adopted clone will survive while the clones with less adaptation will extinct out eventually (Figure 1) [23]. On the contrary, the CSC model explicates that cancerous cells harbor a small subgroup of stem cells with cancer-initiating potential. These cells are recognized as “cancer stem cells (CSCs)” and just like healthy stem cells, CSCs hold self-renewable and differentiation capabilities. This model intimates that due to the stem cells differentiation cancerous cells proliferate in a hierarchical fashion and thus, only CSCs are accountable for tumor induction, propagation, infiltration and relapse (Figure 1) [24].
Figure 1.
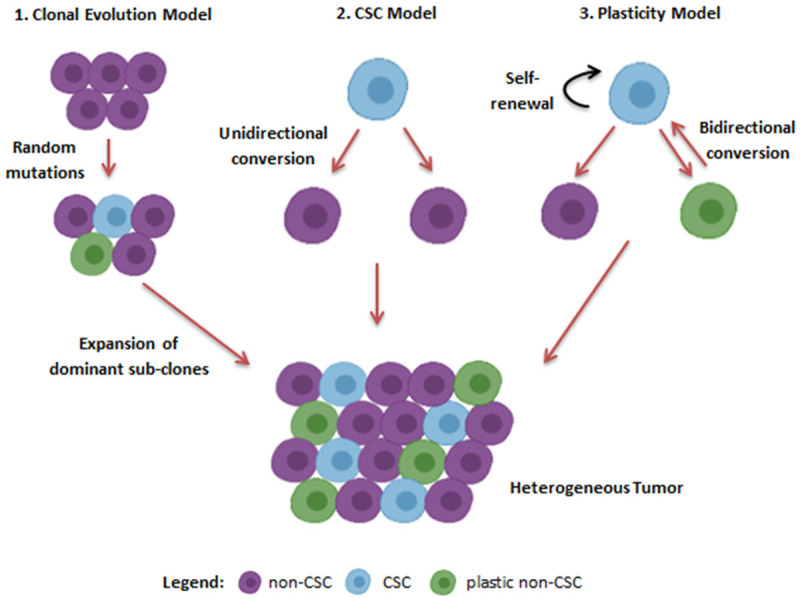
Models of Cancer heterogeneity. According to the clonal evolution model stochastic mutations give raise to the divergent sub-clones which finally make the tumor heterogeneous. The second model or the CSCs model explicates that small population of stem cell like cells within cancer initiates and maintains cancer heterogeneity as they possess the ability to differentiate into any cell type. This model represents that the conversion of CSCs to non-CSCs is irreversible process. On the contrary, according to the plasticity model this conversion is bidirectional. Hence, cancer cells change their phenotype in response to the dynamic microenvironment.
Although in the beginning, it was believed that these models are not mutually exclusive as some cancers follow the CE model, while others follow the CSC model. But this concept was not enough to describe cancer heterogeneity completely. For example, in chronic myeloid leukemia (CML), a cancer that is completely directed by CSCs, imatinib treatment caused clonal evolution [25]. Similarly, phenotypic plasticity has been observed in LSCs upon chemotherapy treatment [26]. These observations led to a third evolving model called the plasticity model, in which CSCs and non-CSCs harbor the ability to interchange their states among each other (Figure 1). Thus, the CE model and CSCs model are not mutually exclusive and the plasticity model further facilitates complicacy to the well-explained paradigm of cancer heterogeneity.
Factors contributing to the CSCs heterogeneity
In order to understand CSCs heterogeneity, it is essential to comprehend how stemness attributes are set up in cancerous cells. Therefore, in this section different intrinsic and extrinsic elements have been discussed which contribute to CSCs stemness initiation and maintenance (Figure 2).
Figure 2.
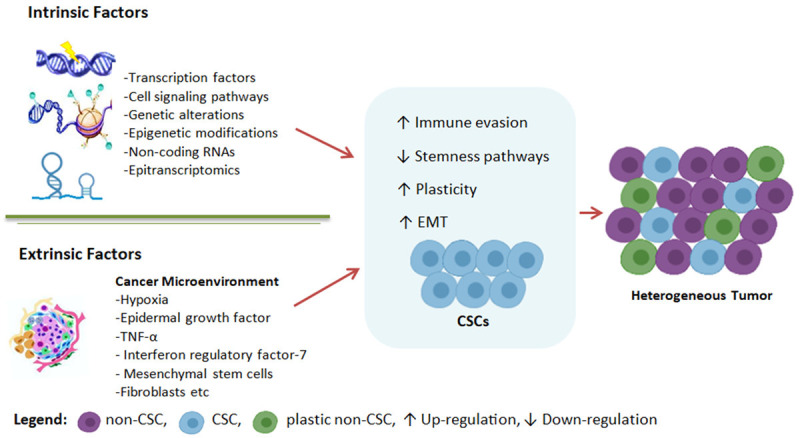
Factors regulating CSCs to promote cancer heterogeneity. Cellular intrinsic and extrinsic factors up-regulate stemness traits in cancerous cells. They also help CSCs to evade immune response and change their phenotype to promote cancer.
Intrinsic factors
Transcription factors
OCT4, SOX2, KLF4, c-MYC and NANOG are recognized as master pluripotency factors as they can reprogram somatic cells into induced pluripotent stem cells by suppressing differentiation-associated genes or upregulating pluripotency and self-renewal related genes. Accumulating evidence shows increased expression of these master regulators in CSCs [27]. For instance, increased expression of OCT4 has been found to be positively linked with chemoresistance and inversely linked with clinical prognosis in the majority of the tumors [28]. Its increased expression also shows an association with glioma grades [29]. Likewise, SOX2 promotes metastasis and cancer stem-cell function in invasive cutaneous squamous cell carcinoma [30]. Gangemi and companions represented that SOX2 is essential for sustaining tumorigenesis and self-renewal of glioblastoma-initiating cells (GICs) as its knockdown halted glioblastoma tumorigenicity and proliferation [31]. KLF4, which is another important transcription factor of pluripotency, has been found to perform a dual role in cancer maintenance. Sometimes, it acts as an oncogene as in the melanoma xenograft model, its depletion causes inhibition of cancer [32]. In the same fashion, its overexpression in osteosarcoma cells caused the formation of osteosarcoma cancer stem cells with chemoresistance and metastasis potential [33]. On the contrary, in some other cancers, its anticancer effect has also been documented as in liver cancer, bladder cancer, non-small-cell lung carcinoma and leukemia its downregulation has been observed [27]. The three different isoforms of c-MYC are also overexpressed in different cancers to maintain CSCs characteristics [27]. Similarly, NR5A2 has been found to increase NANOG expression to maintain stemness of lung cancer [34]. Moreover, these factors also work in a combination of other transcription factors i.e. STAT3, ZIC3 and HESX1 to promote CSCs tumorigenic potential, transformation and metastasis [35].
Besides maintaining stemness properties of CSCs, transcription factors also play a key role in promoting tumor heterogeneity and plasticity. For example, in glioblastoma, a core set of four transcription factors (SOX2, OLIG2, POU3F2 and SALL2) can reprogram differentiated glioblastoma cells into tumor-initiating glioblastoma stem-like cells [36]. A recent study showed that increased c-MYC expression has been found to be linked with intratumor heterogeneity in triple-negative breast cancer cells [37]. Similarly, the transcription factor YB-1 was found to be responsible for the reversion of differentiated tumor cells into CSCs [35]. Hence, it is evident that transcription factors play a dominant role in maintaining tumor heterogeneity by regulating CSCs proliferation and differentiation.
Cell signaling pathways
Many cell signaling pathways that control normal stem cells homeostasis are found to be highly activated or repressed in cancerous cells. Thus, leading to the activation and proliferation of CSCs [27]. These pathways are not independent of each other but form a complicated signaling network to regulate and propagate tumors [38]. Different endogenous or exogenous genes and ncRNAs are implicated in the regulation of these signaling pathways. In addition, these can also stimulate the expression of some downstream genes associated with the apoptosis, proliferation and metastasis of CSCs [38]. In this section, some key signaling pathways are discussed that support CSCs formation and function.
Notch signaling pathway
In mammals, the Notch signaling pathway has four receptors: (Notch1, Notch2, Notch3, and Notch4) and five ligands: (DLL1, DLL3, DLL4, JAG1 and JAG2) [39]. These ligands and receptors are transmembrane proteins that participate in cellular communication [27]. This pathway gets activated when a receptor binds to a ligand on an adjacent cell in a juxtacrine manner, resulting in the translocation of the Notch intracellular domain to the nucleus to activate transcription of targeted genes [40].
The Notch pathway has been found abnormally activated in different malignancies. However, its exact role in the maintenance of CSCs has just been recently understood. It is because different cancers express different Notch receptors and ligands [41]. Hence, it functions as either tumor promoter or suppressor, depending upon the type of cancer involved. As a tumor promoter, Notch signaling is overactivated in pancreatic, gastric, colon and breast cancer cells [42-44]. While this pathway is found suppressed in liver, prostate, lung and skin cancer cells [45-49]. This dual nature of the Notch signaling pathway is controlled by the microenvironment [50]. Studies conducted on the role of Notch pathway in CSCs revealed that its activation promotes the development and differentiation of CSCs. For example, in hepatocellular carcinoma (HCC) and breast cancer, aberrant activation of the Notch1 and Notch4 enhanced metastasis and self-renewal properties of CSCs [51,52]. Likewise, patient-derived pancreatic cancer stem cells (PCSCs) exhibited increased expression of Notch1, Notch3, JAG1, JAG2 and HES1, suggesting the potential role of Notch signaling in CSCs maintenance. Moreover, overexpression of DLL4 in gastric cancer cells resulted in increased self-renewal and metastasis of gastric cancer stem cells (GCSCs) [53]. In glioblastoma, DLL1 requires actin cytoskeleton regulator ARP2/3 complex to activate Notch signaling for maintaining stem cells phenotype [54]. Notch pathway activation is also required for maintaining the undifferentiated state of CSCs, as in adenomas its inhibition caused differentiation of adenoma cells into goblet cells [55]. Furthermore, some endogenous genes also activate the Notch pathway. In cervical cancer, MAP17 sequestrates NUMB to differentiate tumor cells into CSCs [56]. Finally, it has also been observed that different tumorigenic factors also maintain CSCs phenotype by activating Notch. In osteosqamous cell carcinoma cells, TNF-α mediated Notch activation promotes CSCs phenotype [57]. Similarly, hypoxia-induced JAG2 activation enhances the invasiveness properties of lung and BCSCs [58,59]. Additionally, BMP-4 activates the Notch pathway to promote epithelial-to-mesenchymal transition (EMT), a process that is cardinal to maintain cancer cells’ plasticity and stem cell properties in breast cancer cells [60]. Collectively, these studies clearly express that the Notch pathway is essential for the survival and growth of CSCs in different cancers.
Wnt signaling pathway
The Wnt pathway is a highly intricate signaling pathway that plays a crucial role in the proliferation and maintenance of CSCs [61]. This pathway has been divided into three subtypes: the canonical Wnt pathway, the non-canonical planar-cell polarity pathway and the non-canonical Wnt-calcium pathway [62]. Among these, the canonical Wnt pathway is the most understood and its inhibition has remained a topic of interest in cancer biology. In general, it incorporates 19 ligands and around 15 receptors [63]. The activation of the Wnt pathway has been found different in different malignancies. In some cases, it gets activated by mutations, like FLT3IDT mutation in AML [64], APC mutation in colorectalcarcinoma [65], AXIN mutation in gastric cancer [66] and β-catenin mutation in liver cancer [67]. While in other cases, growth factors from the TME, like HGF, PDGF, VEGF and PGE2 activate this pathway [68]. Tumor dormancy is a critical phenomenon that ultimately develops cancer metastasis and recurrence. Studies suggest that activated Wnt signaling transforms dormant CSCs into active CSCs to promote tumorigenesis [69]. Moreover, activated Wnt signaling has also been found to be associated with the self-renewal of CSCs. Different proto-oncogenes promote this process by activating the Wnt pathway [70]. For instance, in colorectal cancer, EZH2 enhances colorectal cancer stem cells (CCSCs) expansion by activating Wnt/β-catenin signaling [71]. Whereas in prostate cancer, TERT (a telomerase reverse transcriptase) activates downstream targets of Wnt via making a complex with β-catenin [72]. PKM2 increases the proliferation of BCSCs by activating the Wnt pathway [73]. In the same manner, capillary morphogenesis gene 2 upregulates β-catenin expression to regulate GCSCs phenotype [74].
Aberrant Wnt activation also regulates the dedifferentiation of CSCs. PMP22, which normally causes differentiation of CSCs, was found depleted after Wnt activation [75]. TRAP1 halted the differentiation of CSCs via regulating phosphorylation of the β-catenin [76]. Furthermore, in esophageal squamouscell carcinoma, LGR5 caused Wnt activation to restrain CSCs differentiation [77]. Studies also show the role of Wnt signaling in inhibiting apoptosis of CSCs. DACT1 antagonizes the Wnt/β-catenin pathway to increase apoptosis in BCSCs [78]. Likewise, DKK2-mediated downregulation of β-catenin in BCSCs leads to the Go/G1 phase arrest and apoptosis in breast cancer [79]. Wnt signaling also promotes metastasis of CSCs. In the human colon, pancreatic and lung CSCs upregulation of CD44v6 causes Wnt activation which leads to cancer metastasis and invasion [80-82]. Besides, CDH11 was found to decrease the migration ability of CCSCs by deactivating Wnt [83]. These studies suggest that aberrant Wnt activation plays a significant role in maintaining CSCs’ dedifferentiation, apoptosis inhibition, and invasion.
Hedgehog (HH) signaling pathway
The HH signaling pathway is another complex regulatory network, that promotes tumorigenesis of different organs by regulating CSCs [27]. This pathway is comprised of three extracellular ligands: Sonic hedgehog, Desert hedgehog and Indian hedgehog, PTCH receptor, Smoothened protein (SMO) and three GLI transcription factors: GLI1, GLI2 and GLI3 [27]. Studies show increased expression of HH pathway mediators in different CSCs. In human multiple myeloma-derived progenitor cells, high expression of SMO gene and increased transcriptional activity of GLI1 were observed [84]. In the same manner, human gliomas showed increased expression of HH signaling-associated genes PTCH1, GLI1 and SHH [85]. Upon treatment with HH pathway inhibitor, enhanced expression of master pluripotency factors (SOX2, OCT4 and NANOG) was observed, which caused inhibition of self-renewal and proliferation abilities of CSCs [85]. Furthermore, in the SMO-deficient murine CML model, ectopic expression of the SMO gene enhanced CML progression by increasing the frequency of CSCs [86].
Different oncogenes and tumor suppressor genes also regulate tumorigenesis by modulating HH signaling in CSCs. In medulloblastoma stem cells, BCL6 caused repression of the HH pathway by inhibiting GLI1 and GLI2 [87]. In glioma, SCUBE2 inhibited the HH pathway to halt the proliferation and migration of glioma cancer stem cells [88]. Also, in PCSCs, vasohibin 2 caused the deactivation of SMO, GLI1 and GLI2 [89]. HH signaling also promotes CSCs-mediated tumor metastasis. CSCs isolated from human colon cancer liver metastasis patients exhibited increased expression of GLI1, GLI2 and HIP genes. Besides, these CSCs also expressed a high level of SNAIL 1 (an EMT-associated gene), as compared to nonmetastatic controls [90]. In addition, RUNX3 a tumor suppressor transcription factor inhibited metastasis and stemness properties of CCSCs by causing GLI1 intracellular ubiquitination [91]. These studies indicate that overactivated HH signaling assists CSCs growth, proliferation and metastasis.
JAK/STAT signaling pathway
The JAK/STAT signaling cascade is one of the simplest signaling pathways that gets activated by different ligands, like cytokines, hormones and growth factors [92]. This pathway regulates different important biological processes, such as self-renewal, neurogenesis and hematopoiesis in embryonic stem cells (ESCs) [93]. Several lines of evidence indicate aberrant JAK/STAT activation in different CSCs. For example, overactivation of the JAK/STAT pathway-associated genes, such as STAT1, IFNK, IFNGR, CSF2 and IL-6 has been observed in prostate cancer stem cells (PrCSCs) [94]. Whereas the activated form of STAT3 was found to be upregulated in BCSCs [42]. Overactivation of JAK/STAT signaling also promotes stemness properties in human glioblastoma. As in an analysis of glioblastoma, patient-derived GICs showed TGF-β and LIF-mediated JAK/STAT pathway activation that led to the increased self-renewal and reduced differentiation capabilities in GICs [95]. In endometrial cancer, IL-6 caused activation of the JAK1/STAT3 pathway in ALDHH and CD126+ cells [96]. Furthermore, in ovarian cancer, OCT4 promoted activation of the JAK2/STAT3 pathway in CSCs [97]. The JAK/STAT pathway also plays an important role in the regulation of LSCs [98]. Constitutive JAK/STAT activation has been observed in the LSCs isolated from AML patients. Upon treatment with JAK1/2 inhibitor, LSCs lost their tumorigenic ability as in secondary transplant they failed to develop leukemia in immunocompromised mice [99]. The JAK/STAT signaling also participates in CSCs-mediated therapy resistance. As in colorectal cancer, the JAK2/STAT3/CCND2 axis promoted CSCs growth after radiotherapy by limiting apoptosis and accelerating clonogenicity [100]. Similarly, in myxoid liposarcoma cancer stem cells, JAK/STAT signaling activation led to chemotherapy resistance [101]. Thus, the JAK/STAT pathway serves an important role in promoting stemness properties and CSCs-mediated therapy resistance.
NF-ĸβ signaling pathway
NF-ĸβ is another complex and essential signaling cascade that plays a crucial role in regulating inflammatory and immune responses [102]. Aberrant NF-ĸβ has been observed in hematological malignancies, breast tumors, head and neck squamous cell carcinoma (HNSCC), gastrointestinal, gynecological and genitourinary cancers [27]. Although previously its role was unclear in the regulation of CSCs, recent studies have shed light on its essential role in CSCs functioning, maintenance, proliferation and metastasis. In ovarian cancer, CD44+ cells showed increased expression of different NF-ĸβ pathway-associated genes to promote tumorigenesis [103]. In another study, BCSCs displayed increased level of NF-ĸβ-inducing kinase to support the proliferation and metastasis of BCSCs [104]. IKKβ upregulated LIN28 expression to promote BCSCs lung metastasis [105]. Moreover, in a recent study, Nancy and colleagues have also identified the role of NF-ĸβ signaling cascade in maintaining BCSCs plasticity to promote its aggressiveness and invasion [106]. Similarly, in the colorectal cancer mice model, PGE2-mediated NF-ĸβ activation promoted CSCs expansion and metastasis [107]. Finally, CCR7 in association with its ligand CCL21 caused NF-ĸβ activation in CD133+ CCSCs to enhance their survival, while inhibiting apoptosis [108]. These investigations demonstrate that the activated NF-ĸβ pathway regulates CSCs’ properties to increase tumor metastasis and aggressiveness.
PI3/AKT/mTOR signaling pathway
PI3/AKT/mTOR is an important intracellular signaling pathway that regulates the cell cycle. This pathway has been studied extensively for its role in developing cancer. Moreover, its function in driving therapy resistance in various tumors has also been reported. However, limited data is available regarding its role in stemness maintenance [109]. PI3/AKT/mTOR signaling cascade accelerates metastasis and invasion of PCSCs and PrCSCs [110,111]. Deactivation of PTEN, a tumor suppressor gene, induced PI3K activation in CD133+/CD44+ PrCSCs to stimulate their survival and tumorigenesis [112]. In HNSCC, aberrant PI3K signaling improved the proliferation and metastasis capabilities of ALDH+CD44H CSCs [113]. Overactivation of mTOR has also promoted survival and self-renewal in nasopharyngeal cancer stem cells and BCSCs [114,115]. In HCC, mTORC2 increased the expression of EpCAM [116]. Whereas, in CCSCs, activation of mTORC1 increased ALDH1 activity [117]. In another analysis, activated PI3K/AKT pathway promoted SOX2 expression in CCSCs to induce the production of radiotherapy resistant CSCs [118]. Therefore, it is evident that the PI3K/AKT pathway promotes CSCs-mediated therapy resistance, metastasis and invasion.
The aforementioned CSCs regulatory pathways are not always linear, as in some cases cross-talk among different signaling cascades also occur to regulate the properties of tumor-initiating cells. HH and PI3K pathways work together to regulate biliary tract CSCs and PrCSCs [119,120]. Likewise, the association between NF-ĸβ and Wnt/β-catenin pathway occurs in colorectal cancer to support self-renewal and proliferation of CSCs. In this case, β-catenin increased TNFRSF19 expression to overactivate, NF-ĸβ pathway [121]. However, in some cases, negative association also occurs between these regulatory pathways. For example in colon, breast and liver cancer, NF-ĸβ activation caused deactivation of β-catenin/TCF activity by increasing tumor suppressor LZTS2 [122-124]. Cross-talk of CSCs regulatory pathways also participate in establishing therapy resistance in tumor cells. For instance, PI3K and Notch pathways work in association to drive PI3K therapy resistance in triple-negative breast cancer [125]. In HH activated medulloblastoma a mice model, activation of PI3K was observed to sustain therapy-resistant CSCs in the perivascular niche [126]. Moreover, the association of these signaling pathways also regulates CSCs to promote tumor growth and metastasis. IL-6/JAK/STAT3 and TGF-β/Smad signaling work together to promote LCSCs metastasis [127]. The combination of Notch, IKK/NF-ĸβ and some other pathways enhanced the proliferation and migration of CD133+ skin cancer stem cells [128]. Finally, the binding of IL-17E to IL-17B has been observed in HCC to activate NF-ĸβ and JAK/STAT3 pathways for regulating CSCs growth [129].
Genetic alterations
In the journey of cell division, cells agglomerate various genomic alterations just by chance. In an exquisite study, Tomasetti and Vogelstein revealed a strong correlation between no of stem cell division and tissue-associated cancer risk. This work indicates that stem cells are the main players of arising cancer in humans [130]. Although neoplastic transformation can eventuate in any cell type i.e. stem cells, progenitor cells, or differentiated cells, the stem cells are preferentially dominant targets for malignant transformation [131]. Thus, CSCs utilize normal molecular mechanisms of healthy stem cells, like self-renewal and proliferation to initiate and propagate cancer [24]. This phenomenon can be observed well in hematological malignancies in which the acquisition of different forms of chromosomal translocations in HSCs produce different types of leukemia. For example, BCR-ABL translocation in these progenitor cells originates CML [132], MLL-ENL translocation in CD34+ hematopitic stem cells and progenitor cells leads to B-lineage and monocytic leukemia [133] and AML1-ETO in HSCs leads to AML [134]. It is well known that TET2 deletion in the hematopoietic region is linked with CMML/MPN, while FLT3IDT mutation in TET2 deprived hematopoietic stem cells transforms them to AML [135]. Furthermore, these two germline perturbations synergistically exert a pathogenic impact on the bone marrow niche by promoting leukemogenesis [135]. These types of genetic mutations have also been observed in the initiation of solid tumors as PTEN loss in human neural stem cells results in the formation of glioblastoma stem cells (GBSCs) [136]. Lee and coworkers have provided direct molecular genetic proof that neural stem cells that reside in the brain’s subventricular zone carry driver mutations to originate glioblastoma [137]. Similarly, SMAD4 and PTEN mutated LRG5+ stem cells originate invasive intestinal-type gastric cancer in mice [138].
Oncogenic transformations have also been reported to be responsible for promoting the aggressiveness of cancer [139]. In a recent study, Weng and colleagues showed that KrasG12D/p53 loss caused upregulation of cancer stem cell-like traits (CD24, CD133 and EpCAM) in the prostate cancer mice model, which enhanced tumorigenic potential and bone metastasis of prostate cancer [140]. Moreover, oncogenic transformation in stem cells can also produce immune surveillance response in tumor cells. For instance, neoplastic alterations in human mesenchymal stem cells reduced their immunogenicity and increased their ability to inhibit mitogen-driven T-cells proliferation [141]. In the same manner, mutated stem cells are found to express an elevated level of immune modulators such as CTLA4 and CD274 (PD-L1) [142]. A current study confirms this phenomenon by revealing that PARP upregulation in LSCs inhibited NKG2D-L expression which led to evade immune response [143].
Just like a healthy stem cell, normal differentiated cells also depict a permissive pool for CSCs generation [144]. The two most prevalent brain tumors i.e. neuroblastoma and glioblastoma are presumed to be initiated from the dedifferentiation of mature neurons and glial cells [145]. Köhler and colleagues have provided in vivo evidence that differentiated, mature pigment-producing melanocytes cells are cells of origin of melanoma. Using the lineage tracing approach they showed that pigment-producing melanocytes dedifferentiate and expand to originate BRAFV600E induced melanoma [146]. Similarly, the oncogenic isoform of C/EBPα dedifferentiates hepatocytes into CSCs to initiate HCC and aggressive hepatoblastoma [147].
Epigenetic modifications
Even though genetic perturbations play a major role in tumorigenesis, these changes alone are not enough to establish all hallmarks of cancer. Therefore, genetic and epigenetic alterations go hand in hand to maintain and propagate tumors. For instance, loss of APC in LRG5+ intestinal CSCs led to DNA methylation to further increase CSCs signature [148]. Additionally, Arg-882-mutated DNMT3a in combination with RAS-mutation induced LSCs to originate AML [149]. Likewise, mutated DNMT3a induced CpG hypermethylation and histone acetylation to deregulate stemness pathways [149]. Furthermore, irregular DNA methylation has been observed frequently in different malignancies [150,151]. In Aldeflour+ BCSCs, DNA hypermethylation of TAP gene helps to evade T cells [152].
Epigenetic alterations also regulate the plasticity of CSCs. As the existence of functionally distinctive cellular subtypes i.e. CSCs and non-CSCs in tumor leads to heterogeneity, epigenetic modifications can effectively switch CSCs into non-CSCs in established cancers. An example of this mechanism can be observed in liver cancer, in which modulation of Nanog promoter methylation results in the switch between CSCs and non-CSCs [153]. Emerging evidence suggests that epigenetic modifiers contribute significantly to sustain and support oncogenic aptitude of CSCs. As ZSCAN4 mediated NANOG and OCT3/4 promoter hyperacetylation leads to enhanced CSCs features in HNSCC [154]. Collectively, in light of the mentioned literature, it’s evident that genetic and epigenetic modifications are mandatory factors for installing and maintaining stemness traits.
Non-coding RNAs
Although, in the beginning, there was consensus among the scientific society that only a minority of the human genome (less than 2%) encodes for protein while the remaining major portion (98%) was considered junk. With the advancement of scientific research, RNA-sequencing technology has revealed the importance of this junk portion in the regulation of gene expression [155]. Non-coding RNAs in particular, long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) perform a crucial role in carcinogenesis as they can regulate cellular growth and differentiation by reprogramming the transcriptome [156,157]. In recent years, their pivotal role in CSCs progression and stemness maintenance has been studied extensively and it is well-mentioned that they are key regulators of stemness properties and stem cell plasticity [158,159]. For example, the lncRNA MALAT1 and THOR maintain gastric cancer stemness by regulating SOX2 expression post-transcriptionally. This lncRNA binds to the SOX2 mRNA and strengthens its stability and expression [160,161]. Similarly, lncRNA-HAND2-AS1 has been found to increase stemness of HCC and non-small cell lung cancer (NSCLC) through activating BPM signaling and suppressing TGF-β1 respectively [162,163]. Correspondingly, lncGATA6 perpetuates intestinal cancer stemness and stimulates tumorigenesis [164]. In AML, the enhanced expression of HOTAIR in LSCs led to the inhibition of p15, which is a tumor suppressor gene. Thus, lncRNA could serve as a potential therapeutic marker as its knockdown could increase the survival time of AML patients [165]. Furthermore, some oncogenic alterations could also affect the transcription of lncRNAs which are necessary for CSCs stemness maintenance and chemoresistance [166].
Correspondingly, elevated level of miRNAs has been found in different malignancies to promote stemness features. In HCC, the epigenetically altered miR-494 has been found to be linked with stem cell phenotype and sorafenib resistance [167]. Another study shows that miR-217 enhanced hepatocellular carcinoma stem cells phenotype via targeting DkK1 gene expression [157]. While in cutaneous squamous cell carcinoma, miR-142-5p produced CSCs by inhibiting PTEN [168]. Besides regulating stemness-associated factors, these non-coding RNAs also participate in the regulation of the process of EMT [159]. BCSCs, the most studied CSC in solid tumors, prevail in two dynamic states i.e. mesenchymal-like (CD24-CD44+) and epithelial-like (ALDH+). This interconversion aids in the invasion and dissemination of breast tumors at distinct sites. Studies also illustrate that different miRNAs, in particular, miR-9, miR-100, miR-221 and miR-155 give raise to EMT state, whereas, miR-93, miR-200 and miR-205 are associated with the induction of mesenchymal-to-epithelial state [159,169].
The lncRNA/miRNA regulatory network is highly complex as lncRNA also harbors the ability to silence miRNA. In colorectal cancer, lnc34a epigenetically masks miR-34a to induce asymmetric division of CSCs [170]. On the other hand, LINC00337 in cervical cancer acts like a competing endogenous RNA (ceRNA) of miR-145, where it positively regulates KLF5 translation by inhibiting miR-145 [171]. These non-coding RNAs are not always oncogenic as their tumor suppressor role has also been reported. In recent years, Chen and companions have discovered a novel lncRNA (lnc-LBCS) that inhibits bladder cancer tumorigenesis and chemoresistance by inhibiting SOX2 protein expression [172]. Whereas, miR-15b suppresses TRIM14 to drive cisplatin sensitivity and inhibit CSCs in oral tongue squamous cell carcinoma [173]. Furthermore, depletion of HOTAIRM1 upgraded self-renewal of CSCs and tumor aggressiveness in colorectal cancer and uveal melanoma [174]. In the same fashion, miRNA, let-7g-5p inhibited GBSCs phenotypes by reducing EMT via downregulating VSIG4 [175].
Epitranscriptomics
With the advancement of sequence databases and transcriptome-wide mapping, various post-transcriptional modifications have been identified in both coding and non-coding RNA molecules. At first, these modifications were presumed to be limited to only tRNA and rRNA molecules. However, further investigations revealed that these changes also exist in mRNA molecules. Thus, an emerging group of RNA variations led to the foundation of epitranscriptomics in 2012 [176]. Although previously (epi)genetic alterations, transcription factors and cell signaling pathways were thought to be responsible for stem cells regulation and fate decision, increasing data suggest that RNA-modifications also cause strong dramatic and dynamic changes during cellular reprogramming or lineage decision [177,178]. Hence, epitranscriptomics has emerged as an additional regulatory layer in CSCs stemness maintenance and lineage commitment. Therefore, recent studies are focusing on the role of these RNA modifications in CSCs’ self-renewal differentiation and proliferation. To date, more than 170 different RNA alterations have been reported [179]. Increasing evidence suggests that among these distinct modifications, the two RNA-based modifications: N6-methyladenosine (m6A) and adenosine-to-inosine (A-to-I) play a significant role in CSCs regulation and maintenance [176,180].
N6-methyladenosine modifications (m6A)
N6-methyladenosine is one of the most frequent and prevalent RNA modifications that has been observed in almost all types of RNAs (mRNA, rRNA, tRNA, lncRNA, miRNA and cirRNA) [176,181]. This process of RNA editing involves methylation in the N6-position of adenosine that is mediated by methyltransferases (METTL3/14/16, CBLL1, ZC3H3, WTAP, RBM15/15B, VIRMA and KIAA1429), erased by demethylases (ALKBH5 and FTO) and identified by the reader proteins (IGF2BP1/2/3, YTHDF1/2/3, YTHDC1/2 and hnRNPA2B1) [182]. Accumulating evidence suggests that m6A acts as a molecular switch to control embryonic development [183,184]. In a transcriptome-wide analysis, m6A was found to induce abundant alterations in the core pluripotency factors i.e. SOX2, OCT4, NANOG, and KLF4 [183]. Moreover, METTL3 depleted mice exhibited increased half-life of these key pluripotency genes, which resulted in embryonic lethality [183]. The m6A modifications also control the dedifferentiation of cancer cells. For instance, depletion of m6A methylation led to NANOG translation that caused the reversion of cancer cells to their primitive pluripotent state [185]. Similarly, another group observed increased expression of FTO in leukemia that reduced m6A methylation of RARA and ABS2, thus promoting AML growth and proliferation [186]. FTO-mediated demethylation has also been observed to regulate self-renewal of GBSCs as its inhibition by MA2 impaired GBSCs proliferation and self-renewal [187]. In the same manner, ALKBH5 was found to be highly expressed in GBSCs and its depletion disturbed the proliferation of patient-derived GBSCs by inhibiting FOXM1 expression [188]. Surprisingly, a lncRNA antisense to FOXM1 promoted GBSCs tumorigenesis by accelerating FOXM1 and ALKBH5 transcripts interaction [188]. On the contrary, aberrant m6A alterations promote BCSCs generation [189]. Also, in AML, increased METTL3 activity has been observed that increased translation of c-MYC and BCL2 [190]. Similarly, elevated level of METTL14 has been found in AML cells that lead to the upregulation of MYB and MYC mRNA to regulate LSCs [191]. Visvanathan and coauthors reported that MELTT3-mediated methylation also promotes glioma cancer stem cells dedifferentiation [192]. They also showed that MELTT3 targets SOX2 to sensitize glioma cancer stem cells to γ-radiation [192]. Finally, in colorectal cancer, increased expression level of m6A reader YTHDF1 has been found to assist self-renewal and colonosphere formation via increasing Wnt/β-catenin pathway activity [193]. Overall, depending upon the type of cancer both m6A-mediated methylation and demethylation regulate tumorigenesis by modulating CSCs stemness.
Adenosine to Inosine (A-to-I) editing
A-to-I modification is another important post-transcriptional modification observed in eukaryotes. In this process, hydrolytic deamination of adenosine takes place, which is catalyzed by ADAR-RNA editases: ADAR1, ADAR2 and ADAR3 [194]. During mRNA translation this newly generated inosine base is read as guanosine by the ribosome, thus causing A-to-G transcriptome diversity if occurred in the protein-coding region [195]. A-to-I editing plays an important role in embryonic development and growth. Hartner and his group showed that ADAR1 depletion in mice caused hematopoietic deficits that led to embryonic lethality [196]. Similarly, ADAR2 deficient mice were born and developed normally but died after three weeks of birth [197]. Keeping in view the importance of A-to-I modification in growth and development, it is not surprising that it also participates in CSCs regulation. Notably, A-to-I modifications have been found to regulate stemness features in hematopoietic malignancies. According to Lazzari and coworkers, around 30-50% of multiple myeloma patients show ADAR1 amplification that favores poor prognosis [198]. In another study, ectopic expression of ADAR1 in LSCs led to myeloid progenitor cells expansion and enhanced expression of GSK-3β that promotes LSCs self-renewal by activating β-catenin [199]. Furthermore, ADAR1 knockdown inhibited in vivo engraftment capability of multiple myeloma via attenuating GLI1 activity [198]. ALDAR1 also caused mutation in GLI1 by editing its exon 12, which caused inhibition of HH pathway negative regulation [198]. In a tremendous study, Jiang and collaborators have identified that ADAR1 downregulation may serve as an efficient way to eradicate dormant imatinib-resistant LSCs. According to this study, ADAR1 controls the cell cycle of progenitor cells by inducing A-to-I editing, which led to increase MDM2 expression and p53 inactivation [200].
Besides mRNA, ADAR1-mediated A-to-I editing has also been observed to regulate the cell cycle by deregulating the formation of tumor suppressor miRNAs, leading to accelerate the self-renewal of CSCs. For example, wild-type ALDAR1 was found to inhibit tumor suppressor miR-277 expression, which targets STAT5 to restore imatinib sensitivity in CML [200]. Similarly, A-to-I editing caused inhibition of cell cycle proliferation-associated miR-411 [200]. Lastly, the expression of miR-26a-5p (target of master pluripotency factor MYC) was also repressed by ADAR1 mediated inosine editing [200]. These findings suggest that ADAR1 mediated RNA editing is pivotal for LSCs self-renewal and stemness maintenance. Hence, further studies should be conducted on the elucidation of the role of RNA-based alterations in the regulation of CSCs, which may serve as a novel biomarker for CSCs’ elimination.
Extrinsic factors
The cancer microenvironment
Cancer cells remain ingrained in a complex microenvironment that is usually comprised of various stromal cells, immune cells, cancer secretory factors, hypoxic regions and extracellular matrix [201]. The cancer microenvironment contributes significantly to the modulation of CSCs phenotype. In light of a recent study, it can be stated that external cues from the TME induced phenotypic drift in CSCs. In this study, temozolomide treatment under hypoxic conditions transformed glioma stem cells (GSCs) and their progeny into various cell types, by modifying their surface marks [202]. A contemporaneous study ratifies this view by stating that stimulation of luminal-A breast cancer cells with a combination of tumor microenvironmental factors including TNF-α, epidermal growth factor and estrogen, promoted intra-tumor heterogeneity [203]. Such a microenvironmental network enriched breast cancer cells with chemotherapy-resistant and highly metastatic CSCs with CD44/CD24L phenotype that exhibited high plasticity [203]. It is well-known that an ample number of mesenchymal stem cells are recruited to the stem cell niche. Exposure of MSCs to cancer cell-secreted soluble factors transformed them to chemotherapy-resistant cancer stem-like cells. Further stimulation of these cells with TGF-β enhanced their mesenchymal phenotype and invasion potency [204]. Cancer-associated fibroblasts, a major constituent of tumor stroma, were found to regulate liver cancer-initiating cells via releasing HGF [201]. Accordingly, interferon regulatory factor-7 produced an inflammatory microenvironment to induce angiogenesis and heterogeneity in glioma. It capacitated glioma cells to acquire GSCs state by releasing interleukin-6 [205]. Altogether, CSCs’ fundamental traits and heterogeneity are greatly influenced by different extrinsic factors elicited from the cancer microenvironment.
Cancer stem cell’s heterogeneity and cancer metastasis
Cancer dissemination is considered a major obstacle in oncology. Albeit, various attempts have been made to mitigate and eradicate primary tumors completely, the successful progression of metastatic lesions makes cancer a leading cause of mortality. Accumulative evidence represents that cancer stem cells, the key drivers of cancer metastasis, are genetically and phenotypically diverse in metastatic sites as compared to primary tumors. While studying breast cancer metastasis initiating cells, Lawson and his group unveiled a hierarchical model for metastasis. They found that metastasis cells obtained from low and high burden tissues were heterogeneous. As low-burden metastatic tissues expressed elevated level of stemness, dormancy, EMT and anti-apoptosis genes, while metastatic cells within high burden metastasis tissue were more heterogeneous with increased proliferation and differentiation capacity [206]. The expression level of stemness-associated genes was also found to be diverse in primary and metastatic sites. For instance, LGR5, a well-known marker for CCSCs, expression was remarkably greater at tumor-infiltrating front in contrast to tumor expanding front [207]. Similarly, more CSCs were found in the highest metastatic breast cancer model than in the lowest metastatic model [206].
Circulating tumor cells (CTCs) are associated with enhanced metastasis risk as in the presence of a favorable microenvironment CSCs in CTCs enable them to initiate metastasis [208,209]. A recent study shows that breast cancer CTCs express multiple CSCs marks [208]. Lyberopoulou and coworkers found genetic discordance between CTCs and primary tumors in 5-10% of cases [210]. The existence of a variant isoform of a particular subset of CSCs is also plausible that participates in dissemination. Identifying BCSCs heterogeneity Hu and colleagues reported that CD44v, a variant isoform of CD24-/CD44+/CSCs, was responsible for lung cancer metastasis [211]. Moreover, the expression of CD44v6 was higher in metastatic lesions compared to the initial tumor [80]. Recent studies present a dynamic picture of CSCs in metastasis settings [212,213]. In the journey of establishing metastasis, colorectal CSCs first lose the LGR5 expression mark hence, LGR5- cells migrate to the metastasis site and then convert back to the LGR5+ state for effective metastatic growth [213]. These observations illustrate that CSCs plasticity and heterogeneity play a significant role in cancer dissemination as it enables metastatic cells to adopt the new microenvironment in the secondary site (Figure 3).
Figure 3.
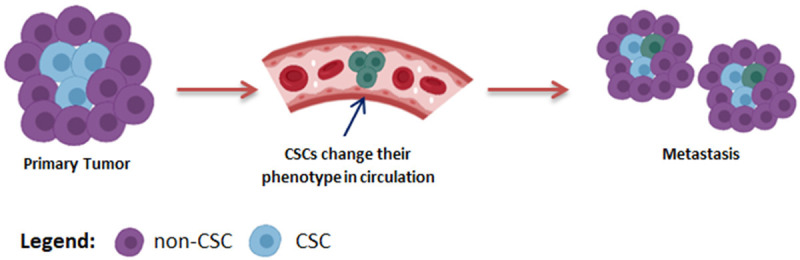
CSCs heterogeneity and plasticity favour metastasis. Cancer stem cells change their phenotype to assist metastasis at secondary site, where they either regain primary tumor characteristics or remain changed.
Cancer stem cell’s heterogeneity and therapy resistance
Although various modalities have been proposed for the effective treatment of cancer, chemotherapy has remained one of the most extensively used methods for the treatment of many types of cancers [214]. Lately, remarkable chemotherapeutic agents have been designed to eliminate primary tumor lesions and their residues which remain left after surgery or radiotherapy. However, heterogeneity in cancer cells renders them resistant to the effects of chemotherapeutic agents [215]. Hence, chemotherapy resistance is a pivotal obstacle in the meaningful outcome of cancer treatment. It is increasingly appreciated that CSCs are more refractory to anticancer therapies as compared to their differentiated counterparts [216]. Therefore, enrichment of CSCs [215] or elevated level of stemness-associated factors like SOX2, OCT4 and NESTIN have been observed in cancer cells upon chemotherapies [217]. Besides, another group of researchers represented that rather than enrichment of CSCs, phenotypic plasticity drives drug resistance [218]. From the CSCs viewpoint, different mechanisms exist that allow therapy to establish tumor heterogeneity. At first, chemotherapy favors the selection of CSCs to change intra-tumor heterogeneity as CSCs harbor the ability to instigate cancerous cells in diverse heterogeneous lineages upon exposure to chemotherapy [215]. Similarly, some cytotoxic drugs like temozolomide can also induce heterogeneity in CSCs which ultimately aid in therapy resistance [219].
Phenotypic plasticity is another mechanism by which therapy can induce heterogeneity in tumor mass. For instance, by utilizing the EMT phenomenon, CSCs expeditiously shifts from the chemotherapy-sensitive (epithelial) phenotype to the chemotherapy-resistant (mesenchymal) phenotype [220]. Recent literature shows that cancerous cells also exploit epigenetic modifications as a protective mechanism to shift phenotypes and escape chemotherapy. As KDM5B epigenetically converts melanoma propagating cells from CD34+ state to CD34- state to escape vemurafenib [221]. In an elegant study, sherma and coworkers carried out a detailed analysis of patient-derived oral squamous cell carcinomas primary cells evolutionary mechanism to cisplatin resistance. They revealed that pre-existing heterogeneity favors the selection of drug-resistant CSCs clones, while within homogenous cancer population epigenetic reprogramming caused transdifferentiation of CSCs into the drug-resistant state [222]. Therefore, it can be stated that CSCs heterogeneity and plasticity is a major hurdle in the effective eradication of the tumor (Figure 4). Hence, new anticancer therapies should be designed to kill CSCs in parallel of targeting bulk tumor cells.
Figure 4.
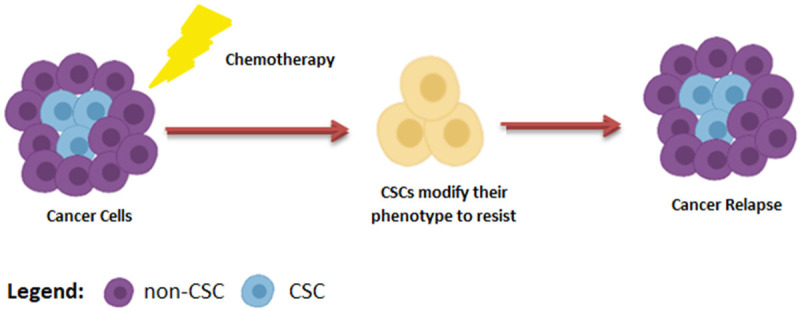
CSCs Heterogeneity favour therapy resistance. Upon chemotherapy CSCs modify their phenotype to resist therapy.
Targeting cancer stem cells
Considering the importance of CSCs in developing tumor heterogeneity, metastasis and therapy resistance, it is important to design novel strategies against them. Undoubtedly, the eradication of CSCs will effectively improve the prognosis and survival rate of cancer patients. In recent decades, several new modalities have been proposed to eliminate CSCs present in the bulk tumor mass. These include disrupting regulatory signaling pathways, targeting CSCs surface markers, disturbing TME and immunotherapy.
Targeting CSCs associated signaling pathways
As mentioned in the previous section, cell signaling pathways play a vital role in the formation and regulation of CSCs. Mainly Wnt, Notch and HH pathways are found to be altered in CSCs as these are the key regulators for the maintenance and survival of these cancer propagating cells. Therefore, these pathways have gained increased attention from researchers to cure cancer.
Targeting notch pathway
The Notch pathway has been found abnormally expressed in many cancers, like leukemia, glioblastoma and lung, breast, colon and pancreatic cancer [223]. Its abnormal functioning supports different aspects of cancer biology i.e. CSCs differentiation, immune evasion, metastasis and angiogenesis [224]. Currently, the three main approaches used to suppress Notch signaling include gamma-secretase inhibitors (GSI), Notch receptors inhibitors, and their combination with other therapies. GSI, the first Notch inhibitor to enter clinical trials, showed potent antitumor activity in different malignancies. For instance, MK-0752 exhibited targeted inhibition of pediatric brain tumors in phase I clinical trials [225]. PF-03084014, an oral GSI, showed anti-neoplastic activity in desmoid tumors in both phase I and II clinical studies [226]. Similarly, MRK-003 expressed anti-CSCs activity in breast cancer [227]. GSI also acts as an adjuvant to chemotherapeutic drugs to eliminate cancer. For example, the combination of PF-03084014 with docetaxel enhanced the chemosensitivity of PrCSCs to docetaxe [228]. Likewise, the combination of DAPT with cisplatin repressed growth and metastasis of lung-resistant osteosarcoma cells [229]. Similar to GSI, DLL4 inhibition also showed promising results to suppress Notch pathway activation. Demcizumab, a humanized monoclonal antibody against DLL4, remarkably decreased tumor growth and CSCs markers in non-small cell lung, breast, pancreatic, ovarian and colon cancers, when used in combination with chemotherapy [230]. It also suppressed CSCs formation and tumor growth in advanced malignancies [230]. ABL001 is another therapeutic antibody, that was found to inhibit GCSCs population, metastasis and invasion either alone or in the combination with irinotecan [231]. Xie and coauthors have identified that increased expression of DLL4 is linked with sunitinib resistance in metastatic renal cell carcinoma. Hence, inhibiting DLL4 and VEGF may serve as an efficient strategy to cure tumors of interest [232].
Targeting hedgehog pathway
Targeting the HH pathway is another strategy to inhibit CSCs proliferation. It also participates in embryonic development and its aberrant activation has been observed in different malignancies [233]. The mechanism of HH pathway-mediated transcription regulation mainly depends upon SMO proteins that cause activation and nuclear transfer of respective transcription factors. At present, FDA has approved three different SMO antagonists to cure basal cell carcinoma (BCC) and AML. These include vismodegib, sonidegib and glasdegib [27].
Vismodegib was the first cyclopamine competitive antagonist, approved in 2021 for metastatic and advanced BCC. In the overall survival analysis of BCC patients, it was found that it can increase the median survival duration to 2.8 years, which was computed as only 2 years with standard treatments. Afterward, exploring the efficacy of vismodegib in medulloblastoma, it was found that it can improve progression-free survival (PFS) of SHH-subtype medulloblastoma patients than the non-SHH subtype cases. Furthermore, vismodegib also showed anti-neoplastic activity in the colon and oral squamous cell carcinoma [234,235]. After vismodegib, FDA approved sonidegib for the treatment of surgery and radiation therapy-resistant BCC patients. In a phase I study of extensive grade small cell lung cancer patients, sonidegib was found effective to sustain PFS when used in combination with etoposide and cisplatinin SOX2 amplified patients [236]. Subsequently, in a phase II trial, it was found to cure 50% of recurrent medulloblastoma patients [237]. Sonidegib has also been found to affect CSCs properties and decreases carcinogenesis in glioblastoma, prostate cancer and renal cancer [238-240].
In 2018, FDA approved glasdegib for the treatment of newly diagnosed AML patients. In phase I trial, it displayed significant results for the treatment of partial hematologic malignancies in Japanese patients [241]. Moreover, in the phase II study, glasdegib in combination with cytarabine and daunorubicin showed promising results to cure AML and high-risk myelodysplastic syndromes [242]. Similarly, the combination of glasdegib and low dose cytarabine was found effective for AML patients which were not appropriate for intensive chemotherapy [243]. Besides these, some other SMO antagonists like taladegib and saridegib have been tested to treat advanced solid tumors and medulloblastoma respectively [244,245].
Targeting Wnt pathway
Activation of the Wnt signaling pathway is also implicated in a variety of cancers [27]. At present, various inhibitors of this pathway are in clinical testing, while most of the anti-Want drugs are in preclinical trials. Ipafricept is a high-quality recombinant protein, that inhibits the Want pathway by targeting FZD8 [246]. Correspondingly, vantictumab inhibits the Wnt signaling by binding with the FZD family receptors. It also reduced the number of CSCs and the size of tumor growth in patient-derived xenograft models of different tumors. Besides, it also exhibited synergy with various chemotherapeutic drugs [247]. Cirmtuzumabis another antibody that antagonized Wnt signaling to suppress stemness signatures in CML patients by targeting the ROR1 receptor [247]. PRI-724 and CWP-2322 are β-catenin inhibitors effective for advanced myeloid malignancies and AML respectively [248]. Recent literature also highlights the effectiveness of using Wnt inhibitors prior to standard-of-care chemotherapeutic medications [249]. Moreover, the combination of C4 and nilotinib has been found efficacious to inhibit Wnt pathways in CML stem cells [250]. Similarly, the triple combinations of ipafricept, NCT02069145 and sorafenib for metastatic pancreatic cancer, Wnt-974, cetuximab and encorafenib for metastatic colorectal cancer and vantictumab, gemcitabine and nab-paclitaxel for HCC are also investigated.
Targeting CSCs microenvironment
The TME serves as an ideal place for maintaining differentiation and tumorigenicity of CSCs. in recent decades, different therapeutic agents have been designed to inhibit signals from the cancer microenvironment, that aid generation and propagation of CSCs. VEGF is an important immunological regulator in the TME [251]. In hypoxic conditions, it causes induction of immunosuppressive cells and vascular endothelial cells to support tumorigenesis [251]. Pristimerin, a natural compound isolated from Hippocrateaceae and Celastraceae, was found to reduce prostate cancer bone metastasis by inhibiting VEGF-mediated vasculogenesis in endothelial progenitor cells [252]. CXCL12 is another important constituent of the TME. It works in association with the CXCR4 receptor to assist cancerous cells to communicate with their microenvironment. Plerixafor is a potent therapeutic drug that targets CXCL12/CXCR4 axis in hematological malignancies. LY2510924 also antagonizes CXCR4 in patients with advanced cancer. In addition, the combination of VEGF and CXCR4 antagonists (mcr84 and POL5551 respectively) has been found effective to increase the survival time of glioblastoma patients by targeting the perivascular niche [253]. Similarly, the combination of plerixafor and high-dose etoposide and cytarabine has shown successful results to cure pediatric resistant or relapsed acute leukemia and myelodysplastic syndrome [254]. Cancer-associated fibroblasts are a heterogeneous group of activated fibroblasts that regulate tumorigenesis in a variety of ways [255]. Some of their subsets have been recognized to regulate CSCs by promoting their generation and propagation [255]. Therefore, directly disrupting the results in improved clinical outcomes. To date, different antagonists have been designed to target surface markers present on their surface. For example, targeting FAP by sibrotuzumab, S1004A by 5C3 and ADC by TEM8 cured different tumors [256,257]. TGF-β is one of the major secretions of CAFs, that regulate cancer stemness and metastasis. Inhibition of TGF-β signaling by LY364947 based nanotherapy was found to be effective to clear BCSCs [258]. In the cancer microenvironment, M2 featured tumor-associated macrophages are found abundantly. Overexpression of the SRC gene in M2-like tumor-associated macrophages caused cisplatin resistance [259]. A recent study revealed that dasatinib treatment caused the elimination of stemness markers and cisplatin-resistant NSCLC [259].
Targeting CSCs-associated surface markers
Targeting CSCs surface markers has emerged as a novel strategy to eliminate cancer. Several monoclonal antibodies have been developed against CSCs biomarkers. Bivatuzumab is an effective monoclonal antibody against CD44v6, which is expressed in many cancers such as melanoma, breast, ovarian, lung and colon cancer [260,261]. Talacotuzumab has been found efficacious to eliminate CD16 and CD123 [262]. Accordingly, adecatumumab can cure hormone-resistant prostate cancer by targeting EpCAM. Recently, a group of researchers has developed a humanized anti-CD271 antibody for targeting CSCs in melanoma and hypopharyngeal cancer [263]. On the contrary, another group has discovered ROR1 specific antibody to inhibit chemotherapy-refractory BSCs [38]. Although significant progress has been made to design antibodies against CSCs surface markers, their plastic nature enables them to evolve in distinct subtypes upon treatment. Therefore, it is important to determine CSCs’ phenotype prior to treatment. Moreover, antibodies in combination with appropriate chemotherapeutic drugs should be used to obtain an optimal therapeutic outcome.
Immunotherapy
Paul Ehrlich, in his pioneer study, convinced the idea that a healthy immune system can fight cancer [27]. Based on this idea, cancer immunotherapy has been emerged as a novel approach to suppress cancer. Recent advances in cancer immunotherapies showed that this approach eradicates CSCs effectively. Different immunotherapies to target CSCs include dendritic cell-based vaccine, adoptive T-cells therapy, oncolytic virotherapy, immune checkpoint inhibitors and combined therapies [264]. Among these, anti-CTLA-4, PD-1 and PD-L1 antibodies have provided significant clinical outcomes in patients with advanced malignancies [265,266]. However, in the majority of cases, the use of single antibodies has remained limited due to the poor treatment response. Thus, combinatorial approaches provide a more remarkable response and increased recovery rate. For example, in the mouse melanoma model, the simultaneous use of anti-PD-L1 and anti-CTLA-4 antibodies along with anti-CSCs dendritic vaccine promoted tumor eradication by eliminating ALDHH [267]. Likewise, oncolytic herpes virus expressing IL-4 along with anti-PD-1 and anti-CTL-4 antibodies cured glioblastoma in mouse models [268]. In addition, PD-1 blockade has been reported to increase the anticancer activity of bladder cancer stem cells vaccine [269].
Conclusion
Cancer is a complex illness that is hard to cure due to its heterogeneity. Recent studies regard CSCs as the main cause of cancer heterogeneity. They make cancerous cells resistant to the therapies and escalate the processes of cancer dissemination, recurrence and therapy resistance. In light of the discussed data, it is evident that both cell intrinsic and extrinsic factors promote cancer aggressiveness by inducing CSCs heterogeneity. Hence, CSCs along with the bulk cancer cells must be targeted to achieve a fruitful outcome of cancer therapy. However, the targeted removal of preexisting CSCs is inadequate as cellular reprogramming induces the generation of de novo CSCs from the differentiated non-cancer cells with divergent characteristics. Therefore, it is needed to design new strategies to target stem cell heterogeneity and plasticity and associated factors that establish and maintain this diversification.
Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (buctrc201910), Beijing-Tianjin-Hebei Basic Research Cooperation Special Project (19JCZDJC65800(Z)), National Key Research and Development Program (2017YFA0105900), National Natural Science Foundation of China (82174531) and Shenzhen Science and Technology Project (JCYJ20180507183842516).
Disclosure of conflict of interest
None.
References
- 1.Seth S, Li CY, Ho IL, Corti D, Loponte S, Sapio L, Del Poggetto E, Yen EY, Robinson FS, Peoples M, Karpinets T, Deem AK, Kumar T, Song X, Jiang S, Kang Y, Fleming J, Kim M, Zhang J, Maitra A, Heffernan TP, Giuliani V, Genovese G, Futreal A, Draetta GF, Carugo A, Viale A. Pre-existing functional heterogeneity of tumorigenic compartment as the origin of chemoresistance in pancreatic tumors. Cell Rep. 2019;26:1518–1532. e1519. doi: 10.1016/j.celrep.2019.01.048. [DOI] [PubMed] [Google Scholar]
- 2.Rich JN. Cancer stem cells: understanding tumor hierarchy and heterogeneity. Medicine (Baltimore) 2016;95(Suppl 1):S2–S7. doi: 10.1097/MD.0000000000004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibata M, Hoque MO. Targeting cancer stem cells: a strategy for effective eradication of cancer. Cancers (Basel) 2019;11:732. doi: 10.3390/cancers11050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan X, Jörg DJ, Cavalli FMG, Richards LM, Nguyen LV, Vanner RJ, Guilhamon P, Lee L, Kushida MM, Pellacani D, Park NI, Coutinho FJ, Whetstone H, Selvadurai HJ, Che C, Luu B, Carles A, Moksa M, Rastegar N, Head R, Dolma S, Prinos P, Cusimano MD, Das S, Bernstein M, Arrowsmith CH, Mungall AJ, Moore RA, Ma Y, Gallo M, Lupien M, Pugh TJ, Taylor MD, Hirst M, Eaves CJ, Simons BD, Dirks PB. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549:227–232. doi: 10.1038/nature23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng H, Pomyen Y, Hernandez MO, Li C, Livak F, Tang W, Dang H, Greten TF, Davis JL, Zhao Y, Mehta M, Levin Y, Shetty J, Tran B, Budhu A, Wang XW. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology. 2018;68:127–140. doi: 10.1002/hep.29778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman M, Gillmor AH, Kunz DJ, Johnston MJ, Nikolic A, Narta K, Zarrei M, King J, Ellestad K, Dang NH, Cavalli FMG, Kushida MM, Coutinho FJ, Zhu Y, Luu B, Ma Y, Mungall AJ, Moore R, Marra MA, Taylor MD, Pugh TJ, Dirks PB, Strother D, Lafay-Cousin L, Resnick AC, Scherer S, Senger DL, Simons BD, Chan JA, Morrissy AS, Gallo M. Intratumoral genetic and functional heterogeneity in pediatric glioblastoma. Cancer Res. 2019;79:2111–2123. doi: 10.1158/0008-5472.CAN-18-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabuchi Y, Hirohashi Y, Hashimoto S, Mariya T, Asano T, Ikeo K, Kuroda T, Mizuuchi M, Murai A, Uno S, Kawai N, Kubo T, Nakatsugawa M, Kanaseki T, Tsukahara T, Saito T, Torigoe T. Clonal analysis revealed functional heterogeneity in cancer stem-like cell phenotypes in uterine endometrioid adenocarcinoma. Exp Mol Pathol. 2019;106:78–88. doi: 10.1016/j.yexmp.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Hirata A, Hatano Y, Niwa M, Hara A, Tomita H. Heterogeneity of colon cancer stem cells. Adv Exp Med Biol. 2019;1139:115–126. doi: 10.1007/978-3-030-14366-4_7. [DOI] [PubMed] [Google Scholar]
- 9.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 10.Taussig DC, Miraki-Moud F, Anjos-Afonso F, Pearce DJ, Allen K, Ridler C, Lillington D, Oakervee H, Cavenagh J, Agrawal SG, Lister TA, Gribben JG, Bonnet D. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood. 2008;112:568–575. doi: 10.1182/blood-2007-10-118331. [DOI] [PubMed] [Google Scholar]
- 11.Sarry JE, Murphy K, Perry R, Sanchez PV, Secreto A, Keefer C, Swider CR, Strzelecki AC, Cavelier C, Récher C, Mansat-De Mas V, Delabesse E, Danet-Desnoyers G, Carroll M. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2Rγc-deficient mice. J Clin Invest. 2011;121:384–395. doi: 10.1172/JCI41495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taussig DC, Vargaftig J, Miraki-Moud F, Griessinger E, Sharrock K, Luke T, Lillington D, Oakervee H, Cavenagh J, Agrawal SG, Lister TA, Gribben JG, Bonnet D. Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34(-) fraction. Blood. 2010;115:1976–1984. doi: 10.1182/blood-2009-02-206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerber JM, Zeidner JF, Morse S, Blackford AL, Perkins B, Yanagisawa B, Zhang H, Morsberger L, Karp J, Ning Y, Gocke CD, Rosner GL, Smith BD, Jones RJ. Association of acute myeloid leukemia’s most immature phenotype with risk groups and outcomes. Haematologica. 2016;101:607–616. doi: 10.3324/haematol.2015.135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell stem cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Hwang-Verslues WW, Kuo WH, Chang PH, Pan CC, Wang HH, Tsai ST, Jeng YM, Shew JY, Kung JT, Chen CH, Lee EY, Chang KJ, Lee WH. Multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem cell markers. PLoS One. 2009;4:e8377. doi: 10.1371/journal.pone.0008377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer MJ, Fleming JM, Lin AF, Hussnain SA, Ginsburg E, Vonderhaar BK. CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor-negative breast cancer. Cancer Res. 2010;70:4624–4633. doi: 10.1158/0008-5472.CAN-09-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22:457–472. doi: 10.1038/cr.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dirkse A, Golebiewska A, Buder T, Nazarov PV, Muller A, Poovathingal S, Brons NHC, Leite S, Sauvageot N, Sarkisjan D, Seyfrid M, Fritah S, Stieber D, Michelucci A, Hertel F, Herold-Mende C, Azuaje F, Skupin A, Bjerkvig R, Deutsch A, Voss-Böhme A, Niclou SP. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun. 2019;10:1787. doi: 10.1038/s41467-019-09853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Lin K, Yang Z, Han N, Quan X, Guo X, Li C. Bladder cancer stem cells: clonal origin and therapeutic perspectives. Oncotarget. 2017;8:66668–66679. doi: 10.18632/oncotarget.19112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iseghohi SO. Cancer stem cells may contribute to the difficulty in treating cancer. Genes Dis. 2016;3:7–10. doi: 10.1016/j.gendis.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16:41. doi: 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah NP, Skaggs BJ, Branford S, Hughes TP, Nicoll JM, Paquette RL, Sawyers CL. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J Clin Invest. 2007;117:2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baykal-Köse S, Acikgoz E, Yavuz AS, Gönül Geyik Ö, Ateş H, Sezerman OU, Özsan GH, Yüce Z. Adaptive phenotypic modulations lead to therapy resistance in chronic myeloid leukemia cells. PLoS One. 2020;15:e0229104. doi: 10.1371/journal.pone.0229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, Zhang G, Wang X, Dong Z, Chen F, Cui H. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5:8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohiuddin IS, Wei SJ, Kang MH. Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165432. doi: 10.1016/j.bbadis.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y, Cao X, Ling EA, Hao A. Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia. 2009;57:724–733. doi: 10.1002/glia.20800. [DOI] [PubMed] [Google Scholar]
- 30.Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, Brohée S, Salmon I, Dubois C, del Marmol V, Fuks F, Beck B, Blanpain C. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 31.Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 32.Riverso M, Montagnani V, Stecca B. KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth. Oncogene. 2017;36:3322–3333. doi: 10.1038/onc.2016.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi XT, Li YL, Zhang YQ, Xu T, Lu B, Fang L, Gao JQ, Yu LS, Zhu DF, Yang B, He QJ, Ying MD. KLF4 functions as an oncogene in promoting cancer stem cell-like characteristics in osteosarcoma cells. Acta Pharmacol Sin. 2019;40:546–555. doi: 10.1038/s41401-018-0050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye T, Li J, Sun Z, Liu Y, Kong L, Zhou S, Tang J, Wang J, Xing HR. Nr5a2 promotes cancer stem cell properties and tumorigenesis in nonsmall cell lung cancer by regulating Nanog. Cancer Med. 2019;8:1232–1245. doi: 10.1002/cam4.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang F, Cui P, Lu Y, Zhang X. Requirement of the transcription factor YB-1 for maintaining the stemness of cancer stem cells and reverting differentiated cancer cells into cancer stem cells. Stem Cell Res Ther. 2019;10:233. doi: 10.1186/s13287-019-1360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, Curry WT, Martuza RL, Rivera MN, Rossetti N, Kasif S, Beik S, Kadri S, Tirosh I, Wortman I, Shalek AK, Rozenblatt-Rosen O, Regev A, Louis DN, Bernstein BE. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta N, Jung K, Wu C, Alshareef A, Alqahtani H, Damaraju S, Mackey JR, Ghosh S, Sabri S, Abdulkarim BS, Bigras G, Lai R. High Myc expression and transcription activity underlies intra-tumoral heterogeneity in triple-negative breast cancer. Oncotarget. 2017;8:28101–28115. doi: 10.18632/oncotarget.15891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clara JA, Monge C, Yang Y, Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells-a clinical update. Nat Rev Clin Oncol. 2020;17:204–232. doi: 10.1038/s41571-019-0293-2. [DOI] [PubMed] [Google Scholar]
- 39.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 40.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 41.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 42.Zhou W, Fu XQ, Zhang LL, Zhang J, Huang X, Lu XH, Shen L, Liu BN, Liu J, Luo HS, Yu JP, Yu HG. The AKT1/NF-kappaB/Notch1/PTEN axis has an important role in chemoresistance of gastric cancer cells. Cell Death Dis. 2013;4:e847. doi: 10.1038/cddis.2013.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu F, Stutzman A, Mo YY. Notch signaling and its role in breast cancer. Front Biosci. 2007;12:4370–4383. doi: 10.2741/2394. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Li B, Ji ZZ, Zheng PS. Notch1 regulates the growth of human colon cancers. Cancer. 2010;116:5207–5218. doi: 10.1002/cncr.25449. [DOI] [PubMed] [Google Scholar]
- 45.Gupta A, Wang Y, Browne C, Kim S, Case T, Paul M, Wills ML, Matusik RJ. Neuroendocrine differentiation in the 12T-10 transgenic prostate mouse model mimics endocrine differentiation of pancreatic beta cells. Prostate. 2008;68:50–60. doi: 10.1002/pros.20650. [DOI] [PubMed] [Google Scholar]
- 46.Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, Devgan V, Lieb J, Raffoul W, Hohl D, Neel V, Garlick J, Chiorino G, Dotto GP. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konishi J, Yi F, Chen X, Vo H, Carbone DP, Dang TP. Notch3 cooperates with the EGFR pathway to modulate apoptosis through the induction of bim. Oncogene. 2010;29:589–596. doi: 10.1038/onc.2009.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viatour P, Ehmer U, Saddic LA, Dorrell C, Andersen JB, Lin C, Zmoos AF, Mazur PK, Schaffer BE, Ostermeier A, Vogel H, Sylvester KG, Thorgeirsson SS, Grompe M, Sage J. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J Exp Med. 2011;208:1963–1976. doi: 10.1084/jem.20110198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mol Med. 2004;14:779–786. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- 50.Li L, Tang P, Li S, Qin X, Yang H, Wu C, Liu Y. Notch signaling pathway networks in cancer metastasis: a new target for cancer therapy. Med Oncol. 2017;34:180. doi: 10.1007/s12032-017-1039-6. [DOI] [PubMed] [Google Scholar]
- 51.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 52.Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, Bundred NJ, Clarke RB. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70:709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miao ZF, Xu H, Xu HM, Wang ZN, Zhao TT, Song YX, Xu YY. DLL4 overexpression increases gastric cancer stem/progenitor cell self-renewal ability and correlates with poor clinical outcome via Notch-1 signaling pathway activation. Cancer Med. 2017;6:245–257. doi: 10.1002/cam4.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang C, Hai L, Zhu M, Yu S, Li T, Lin Y, Liu B, Zhou X, Chen L, Zhao P, Zhou H, Huang Y, Zhang K, Ren B, Yang X. Actin cytoskeleton regulator Arp2/3 complex is required for DLL1 activating Notch1 signaling to maintain the stem cell phenotype of glioma initiating cells. Oncotarget. 2017;8:33353–33364. doi: 10.18632/oncotarget.16495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 56.Garcia-Heredia JM, Lucena-Cacace A, Verdugo-Sivianes EM, Pérez M, Carnero A. The cargo protein MAP17 (PDZK1IP1) regulates the cancer stem cell pool activating the Notch pathway by abducting NUMB. Clin Cancer Res. 2017;23:3871–3883. doi: 10.1158/1078-0432.CCR-16-2358. [DOI] [PubMed] [Google Scholar]
- 57.Lee SH, Hong HS, Liu ZX, Kim RH, Kang MK, Park NH, Shin KH. TNFα enhances cancer stem cell-like phenotype via Notch-Hes1 activation in oral squamous cell carcinoma cells. Biochem Biophys Res Commun. 2012;424:58–64. doi: 10.1016/j.bbrc.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie M, Zhang L, He CS, Xu F, Liu JL, Hu ZH, Zhao LP, Tian Y. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J Cell Biochem. 2012;113:1501–1513. doi: 10.1002/jcb.24019. [DOI] [PubMed] [Google Scholar]
- 59.Xing F, Okuda H, Watabe M, Kobayashi A, Pai SK, Liu W, Pandey PR, Fukuda K, Hirota S, Sugai T, Wakabayshi G, Koeda K, Kashiwaba M, Suzuki K, Chiba T, Endo M, Mo YY, Watabe K. Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene. 2011;30:4075–4086. doi: 10.1038/onc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi S, Yu J, Park A, Dubon MJ, Do J, Kim Y, Nam D, Noh J, Park KS. BMP-4 enhances epithelial mesenchymal transition and cancer stem cell properties of breast cancer cells via Notch signaling. Sci Rep. 2019;9:11724. doi: 10.1038/s41598-019-48190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsui WH. Cancer stem cell signaling pathways. Medicine (Baltimore) 2016;95(Suppl 1):S8–S19. doi: 10.1097/MD.0000000000004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katoh M, Katoh M. Molecular genetics and targeted therapy of WNT-related human diseases (review) Int J Mol Med. 2017;40:587–606. doi: 10.3892/ijmm.2017.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13:513–532. doi: 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhavanasi D, Klein PS. Wnt signaling in normal and malignant stem cells. Curr Stem Cell Rep. 2016;2:379–387. doi: 10.1007/s40778-016-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morin PJ. beta-catenin signaling and cancer. Bioessays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 66.Mazzoni SM, Fearon ER. AXIN1 and AXIN2 variants in gastrointestinal cancers. Cancer Lett. 2014;355:1–8. doi: 10.1016/j.canlet.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clements WM, Wang J, Sarnaik A, Kim OJ, MacDonald J, Fenoglio-Preiser C, Groden J, Lowy AM. beta-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 2002;62:3503–3506. [PubMed] [Google Scholar]
- 68.Patel S, Alam A, Pant R, Chattopadhyay S. Wnt signaling and its significance within the tumor microenvironment: novel therapeutic insights. Front Immunol. 2019;10:2872. doi: 10.3389/fimmu.2019.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pan T, Xu J, Zhu Y. Self-renewal molecular mechanisms of colorectal cancer stem cells. Int J Mol Med. 2017;39:9–20. doi: 10.3892/ijmm.2016.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen JF, Luo X, Xiang LS, Li HT, Zha L, Li N, He JM, Xie GF, Xie X, Liang HJ. EZH2 promotes colorectal cancer stem-like cell expansion by activating p21cip1-Wnt/β-catenin signaling. Oncotarget. 2016;7:41540–41558. doi: 10.18632/oncotarget.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang K, Guo Y, Wang X, Zhao H, Ji Z, Cheng C, Li L, Fang Y, Xu D, Zhu HH, Gao WQ. WNT/β-catenin directs self-renewal symmetric cell division of hTERT(high) prostate cancer stem cells. Cancer Res. 2017;77:2534–2547. doi: 10.1158/0008-5472.CAN-16-1887. [DOI] [PubMed] [Google Scholar]
- 73.Zhao Z, Song Z, Liao Z, Liu Z, Sun H, Lei B, Chen W, Dang C. PKM2 promotes stemness of breast cancer cell by through Wnt/β-catenin pathway. Tumour Biol. 2016;37:4223–4234. doi: 10.1007/s13277-015-4121-8. [DOI] [PubMed] [Google Scholar]
- 74.Ji C, Yang L, Yi W, Xiang D, Wang Y, Zhou Z, Qian F, Ren Y, Cui W, Zhang X, Zhang P, Wang JM, Cui Y, Bian X. Capillary morphogenesis gene 2 maintains gastric cancer stem-like cell phenotype by activating a Wnt/β-catenin pathway. Oncogene. 2018;37:3953–3966. doi: 10.1038/s41388-018-0226-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cai W, Chen G, Luo Q, Liu J, Guo X, Zhang T, Ma F, Yuan L, Li B, Cai J. PMP22 regulates self-renewal and chemoresistance of gastric cancer cells. Mol Cancer Ther. 2017;16:1187–1198. doi: 10.1158/1535-7163.MCT-16-0750. [DOI] [PubMed] [Google Scholar]
- 76.Lettini G, Sisinni L, Condelli V, Matassa DS, Simeon V, Maddalena F, Gemei M, Lopes E, Vita G, Del Vecchio L, Esposito F, Landriscina M. TRAP1 regulates stemness through Wnt/β-catenin pathway in human colorectal carcinoma. Cell Death Differ. 2016;23:1792–1803. doi: 10.1038/cdd.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lv Z, Yu JJ, Zhang WJ, Xiong L, Wang F, Li LF, Zhou XL, Gao XY, Ding XF, Han L, Cai YF, Ma W, Wang LX. Expression and functional regulation of stemness gene Lgr5 in esophageal squamous cell carcinoma. Oncotarget. 2017;8:26492–26504. doi: 10.18632/oncotarget.15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yin X, Xiang T, Li L, Su X, Shu X, Luo X, Huang J, Yuan Y, Peng W, Oberst M, Kelly K, Ren G, Tao Q. DACT1, an antagonist to Wnt/β-catenin signaling, suppresses tumor cell growth and is frequently silenced in breast cancer. Breast Cancer Res. 2013;15:R23. doi: 10.1186/bcr3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mu J, Hui T, Shao B, Li L, Du Z, Lu L, Ye L, Li S, Li Q, Xiao Q, Qiu Z, Zhang Y, Fan J, Ren G, Tao Q, Xiang T. Dickkopf-related protein 2 induces G0/G1 arrest and apoptosis through suppressing Wnt/β-catenin signaling and is frequently methylated in breast cancer. Oncotarget. 2017;8:39443–39459. doi: 10.18632/oncotarget.17055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S, Cocorullo G, Gulotta G, Dieli F, De Maria R, Stassi G. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 81.Matzke-Ogi A, Jannasch K, Shatirishvili M, Fuchs B, Chiblak S, Morton J, Tawk B, Lindner T, Sansom O, Alves F, Warth A, Schwager C, Mier W, Kleeff J, Ponta H, Abdollahi A, Orian-Rousseau V. Inhibition of tumor growth and metastasis in pancreatic cancer models by interference with CD44v6 signaling. Gastroenterology. 2016;150:513–525. e510. doi: 10.1053/j.gastro.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 82.Su J, Wu S, Wu H, Li L, Guo T. CD44 is functionally crucial for driving lung cancer stem cells metastasis through Wnt/β-catenin-FoxM1-Twist signaling. Mol Carcinog. 2016;55:1962–1973. doi: 10.1002/mc.22443. [DOI] [PubMed] [Google Scholar]
- 83.Li L, Ying J, Li H, Zhang Y, Shu X, Fan Y, Tan J, Cao Y, Tsao SW, Srivastava G, Chan AT, Tao Q. The human cadherin 11 is a pro-apoptotic tumor suppressor modulating cell stemness through Wnt/β-catenin signaling and silenced in common carcinomas. Oncogene. 2012;31:3901–3912. doi: 10.1038/onc.2011.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peacock CD, Wang Q, Gesell GS, Corcoran-Schwartz IM, Jones E, Kim J, Devereux WL, Rhodes JT, Huff CA, Beachy PA, Watkins DN, Matsui W. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci U S A. 2007;104:4048–4053. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao C, Chen A, Jamieson CH, Fereshteh M, Abrahamsson A, Blum J, Kwon HY, Kim J, Chute JP, Rizzieri D, Munchhof M, VanArsdale T, Beachy PA, Reya T. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tiberi L, Bonnefont J, van den Ameele J, Le Bon SD, Herpoel A, Bilheu A, Baron BW, Vanderhaeghen P. A BCL6/BCOR/SIRT1 complex triggers neurogenesis and suppresses medulloblastoma by repressing Sonic Hedgehog signaling. Cancer Cell. 2014;26:797–812. doi: 10.1016/j.ccell.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 88.Guo E, Liu H, Liu X. Overexpression of SCUBE2 inhibits proliferation, migration, and invasion in glioma cells. Oncol Res. 2017;25:437–444. doi: 10.3727/096504016X14747335734344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang F, Ma L, Zhang Z, Liu X, Gao H, Zhuang Y, Yang P, Kornmann M, Tian X, Yang Y. Hedgehog signaling regulates epithelial-mesenchymal transition in pancreatic cancer stem-like cells. J Cancer. 2016;7:408–417. doi: 10.7150/jca.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Varnat F, Duquet A, Malerba M, Zbinden M, Mas C, Gervaz P, Ruiz i Altaba A. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol Med. 2009;1:338–351. doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim BR, Na YJ, Kim JL, Jeong YA, Park SH, Jo MJ, Jeong S, Kang S, Oh SC, Lee DH. RUNX3 suppresses metastasis and stemness by inhibiting Hedgehog signaling in colorectal cancer. Cell Death Differ. 2020;27:676–694. doi: 10.1038/s41418-019-0379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rane SG, Reddy EP. Janus kinases: components of multiple signaling pathways. Oncogene. 2000;19:5662–5679. doi: 10.1038/sj.onc.1203925. [DOI] [PubMed] [Google Scholar]
- 93.Chambers I. The molecular basis of pluripotency in mouse embryonic stem cells. Cloning Stem Cells. 2004;6:386–391. doi: 10.1089/clo.2004.6.386. [DOI] [PubMed] [Google Scholar]
- 94.Birnie R, Bryce SD, Roome C, Dussupt V, Droop A, Lang SH, Berry PA, Hyde CF, Lewis JL, Stower MJ, Maitland NJ, Collins AT. Gene expression profiling of human prostate cancer stem cells reveals a pro-inflammatory phenotype and the importance of extracellular matrix interactions. Genome Biol. 2008;9:R83. doi: 10.1186/gb-2008-9-5-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peñuelas S, Anido J, Prieto-Sánchez RM, Folch G, Barba I, Cuartas I, García-Dorado D, Poca MA, Sahuquillo J, Baselga J, Seoane J. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 96.van der Zee M, Sacchetti A, Cansoy M, Joosten R, Teeuwssen M, Heijmans-Antonissen C, Ewing-Graham PC, Burger CW, Blok LJ, Fodde R. IL6/JAK1/STAT3 signaling blockade in endometrial cancer affects the ALDHhi/CD126+ stem-like component and reduces tumor burden. Cancer Res. 2015;75:3608–3622. doi: 10.1158/0008-5472.CAN-14-2498. [DOI] [PubMed] [Google Scholar]
- 97.Ruan Z, Yang X, Cheng W. OCT4 accelerates tumorigenesis through activating JAK/STAT signaling in ovarian cancer side population cells. Cancer Manag Res. 2018;11:389–399. doi: 10.2147/CMAR.S180418. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98.Fasouli ES, Katsantoni E. JAK-STAT in early hematopoiesis and leukemia. Front Cell Dev Biol. 2021;9:669363. doi: 10.3389/fcell.2021.669363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cook AM, Li L, Ho Y, Lin A, Li L, Stein A, Forman S, Perrotti D, Jove R, Bhatia R. Role of altered growth factor receptor-mediated JAK2 signaling in growth and maintenance of human acute myeloid leukemia stem cells. Blood. 2014;123:2826–2837. doi: 10.1182/blood-2013-05-505735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park SY, Lee CJ, Choi JH, Kim JH, Kim JW, Kim JY, Nam JS. The JAK2/STAT3/CCND2 axis promotes colorectal cancer stem cell persistence and radioresistance. J Exp Clin Cancer Res. 2019;38:399. doi: 10.1186/s13046-019-1405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dolatabadi S, Jonasson E, Lindén M, Fereydouni B, Bäcksten K, Nilsson M, Martner A, Forootan A, Fagman H, Landberg G, Åman P, Ståhlberg A. JAK-STAT signalling controls cancer stem cell properties including chemotherapy resistance in myxoid liposarcoma. Int J Cancer. 2019;145:435–449. doi: 10.1002/ijc.32123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 103.Gonzalez-Torres C, Gaytan-Cervantes J, Vazquez-Santillan K, Mandujano-Tinoco EA, Ceballos-Cancino G, Garcia-Venzor A, Zampedri C, Sanchez-Maldonado P, Mojica-Espinosa R, Jimenez-Hernandez LE, Maldonado V. NF-κB participates in the stem cell phenotype of ovarian cancer cells. Arch Med Res. 2017;48:343–351. doi: 10.1016/j.arcmed.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 104.Vazquez-Santillan K, Melendez-Zajgla J, Jimenez-Hernandez LE, Gaytan-Cervantes J, Muñoz-Galindo L, Piña-Sanchez P, Martinez-Ruiz G, Torres J, Garcia-Lopez P, Gonzalez-Torres C, Ruiz V, Avila-Moreno F, Velasco-Velazquez M, Perez-Tapia M, Maldonado V. NF-kappaΒ-inducing kinase regulates stem cell phenotype in breast cancer. Sci Rep. 2016;6:37340. doi: 10.1038/srep37340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen C, Cao F, Bai L, Liu Y, Xie J, Wang W, Si Q, Yang J, Chang A, Liu D, Liu D, Chuang TH, Xiang R, Luo Y. IKKβ enforces a LIN28B/TCF7L2 positive feedback loop that promotes cancer cell stemness and metastasis. Cancer Res. 2015;75:1725–1735. doi: 10.1158/0008-5472.CAN-14-2111. [DOI] [PubMed] [Google Scholar]
- 106.Espinoza-Sánchez NA, Enciso J, Pelayo R, Fuentes-Pananá EM. An NFκB-dependent mechanism of tumor cell plasticity and lateral transmission of aggressive features. Oncotarget. 2018;9:26679–26700. doi: 10.18632/oncotarget.25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang D, Fu L, Sun H, Guo L, DuBois RN. Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology. 2015;149:1884–1895. e1884. doi: 10.1053/j.gastro.2015.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rizeq B, Malki MI. The role of CCL21/CCR7 chemokine axis in breast cancer progression. Cancers. 2020;12:1036. doi: 10.3390/cancers12041036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xia P, Xu XY. PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application. Am J Cancer Res. 2015;5:1602–1609. [PMC free article] [PubMed] [Google Scholar]
- 110.Chang L, Graham PH, Hao J, Ni J, Bucci J, Cozzi PJ, Kearsley JH, Li Y. Acquisition of epithelial-mesenchymal transition and cancer stem cell phenotypes is associated with activation of the PI3K/Akt/mTOR pathway in prostate cancer radioresistance. Cell Death Dis. 2013;4:e875. doi: 10.1038/cddis.2013.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fitzgerald TL, Lertpiriyapong K, Cocco L, Martelli AM, Libra M, Candido S, Montalto G, Cervello M, Steelman L, Abrams SL, McCubrey JA. Roles of EGFR and KRAS and their downstream signaling pathways in pancreatic cancer and pancreatic cancer stem cells. Adv Biol Regul. 2015;59:65–81. doi: 10.1016/j.jbior.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 112.Dubrovska A, Kim S, Salamone RJ, Walker JR, Maira SM, García-Echeverría C, Schultz PG, Reddy VA. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc Natl Acad Sci U S A. 2009;106:268–273. doi: 10.1073/pnas.0810956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Keysar SB, Le PN, Miller B, Jackson BC, Eagles JR, Nieto C, Kim J, Tang B, Glogowska MJ, Morton JJ, Padilla-Just N, Gomez K, Warnock E, Reisinger J, Arcaroli JJ, Messersmith WA, Wakefield LM, Gao D, Tan AC, Serracino H, Vasiliou V, Roop DR, Wang XJ, Jimeno A. Regulation of head and neck squamous cancer stem cells by PI3K and SOX2. J Natl Cancer Inst. 2017;109:djw189. doi: 10.1093/jnci/djw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang C, Zhang Y, Zhang Y, Zhang Z, Peng J, Li Z, Han L, You Q, Chen X, Rao X, Zhu Y, Liao Z. Downregulation of cancer stem cell properties via mTOR signaling pathway inhibition by rapamycin in nasopharyngeal carcinoma. Int J Oncol. 2015;47:909–917. doi: 10.3892/ijo.2015.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, Margolick JB, Liotta LA, Petricoin E 3rd, Zhang Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci U S A. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nishitani S, Horie M, Ishizaki S, Yano H. Branched chain amino acid suppresses hepatocellular cancer stem cells through the activation of mammalian target of rapamycin. PLoS One. 2013;8:e82346. doi: 10.1371/journal.pone.0082346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Park JH, Kim YH, Shim S, Kim A, Jang H, Lee SJ, Park S, Seo S, Jang WI, Lee SB, Kim MJ. Radiation-activated PI3K/AKT pathway promotes the induction of cancer stem-like cells via the upregulation of SOX2 in colorectal cancer. Cells. 2021;10:135. doi: 10.3390/cells10010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sharma N, Nanta R, Sharma J, Gunewardena S, Singh KP, Shankar S, Srivastava RK. PI3K/AKT/mTOR and sonic hedgehog pathways cooperate together to inhibit human pancreatic cancer stem cell characteristics and tumor growth. Oncotarget. 2015;6:32039–32060. doi: 10.18632/oncotarget.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zuo M, Rashid A, Churi C, Vauthey JN, Chang P, Li Y, Hung MC, Li D, Javle M. Novel therapeutic strategy targeting the Hedgehog signalling and mTOR pathways in biliary tract cancer. Br J Cancer. 2015;112:1042–1051. doi: 10.1038/bjc.2014.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schön S, Flierman I, Ofner A, Stahringer A, Holdt LM, Kolligs FT, Herbst A. β-catenin regulates NF-κB activity via TNFRSF19 in colorectal cancer cells. Int J Cancer. 2014;135:1800–1811. doi: 10.1002/ijc.28839. [DOI] [PubMed] [Google Scholar]
- 122.Gwak J, Park S, Cho M, Song T, Cha SH, Kim DE, Jeon YJ, Shin JG, Oh S. Polysiphonia japonica extract suppresses the Wnt/beta-catenin pathway in colon cancer cells by activation of NF-kappaB. Int J Mol Med. 2006;17:1005–1010. [PubMed] [Google Scholar]
- 123.Thyssen G, Li TH, Lehmann L, Zhuo M, Sharma M, Sun Z. LZTS2 is a novel beta-catenin-interacting protein and regulates the nuclear export of beta-catenin. Mol Cell Biol. 2006;26:8857–8867. doi: 10.1128/MCB.01031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Albanese C, Wu K, D’Amico M, Jarrett C, Joyce D, Hughes J, Hulit J, Sakamaki T, Fu M, Ben-Ze’ev A, Bromberg JF, Lamberti C, Verma U, Gaynor RB, Byers SW, Pestell RG. IKKalpha regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol Biol Cell. 2003;14:585–599. doi: 10.1091/mbc.02-06-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bhola NE, Jansen VM, Koch JP, Li H, Formisano L, Williams JA, Grandis JR, Arteaga CL. Correction: treatment of triple-negative breast cancer with TORC1/2 inhibitors sustains a drug-resistant and notch-dependent cancer stem cell population. Cancer Res. 2019;79:875. doi: 10.1158/0008-5472.CAN-18-4087. [DOI] [PubMed] [Google Scholar]
- 126.Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008;22:436–448. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu RY, Zeng Y, Lei Z, Wang L, Yang H, Liu Z, Zhao J, Zhang HT. JAK/STAT3 signaling is required for TGF-β-induced epithelial-mesenchymal transition in lung cancer cells. Int J Oncol. 2014;44:1643–1651. doi: 10.3892/ijo.2014.2310. [DOI] [PubMed] [Google Scholar]
- 128.Quan XX, Hawk NV, Chen W, Coupar J, Lee SK, Petersen DW, Meltzer PS, Montemarano A, Braun M, Chen Z, Van Waes C. Targeting Notch1 and IKKα enhanced NF-κB activation in CD133(+) skin cancer stem cells. Mol Cancer Ther. 2018;17:2034–2048. doi: 10.1158/1535-7163.MCT-17-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Luo Y, Yang Z, Su L, Shan J, Xu H, Xu Y, Liu L, Zhu W, Chen X, Liu C, Chen J, Yao C, Cheng F, Zhang C, Ma Q, Shen J, Qian C. Non-CSCs nourish CSCs through interleukin-17E-mediated activation of NF-κB and JAK/STAT3 signaling in human hepatocellular carcinoma. Cancer Lett. 2016;375:390–399. doi: 10.1016/j.canlet.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 130.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lytle NK, Barber AG, Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat Rev Cancer. 2018;18:669–680. doi: 10.1038/s41568-018-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Holyoake TL, Vetrie D. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood. 2017;129:1595–1606. doi: 10.1182/blood-2016-09-696013. [DOI] [PubMed] [Google Scholar]
- 133.Reimer J, Knöß S, Labuhn M, Charpentier EM, Göhring G, Schlegelberger B, Klusmann JH, Heckl D. CRISPR-Cas9-induced t(11;19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica. 2017;102:1558–1566. doi: 10.3324/haematol.2017.164046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Di Genua C, Norfo R, Rodriguez-Meira A, Wen WX, Drissen R, Booth CAG, Povinelli B, Repapi E, Gray N, Carrelha J, Kettyle LM, Jamieson L, Neo WH, Thongjuea S, Nerlov C, Mead AJ. Cell-intrinsic depletion of Aml1-ETO-expressing pre-leukemic hematopoietic stem cells by K-Ras activating mutation. Haematologica. 2019;104:2215–2224. doi: 10.3324/haematol.2018.205351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ramdas B, Mali RS, Palam LR, Pandey R, Cai Z, Pasupuleti SK, Burns SS, Kapur R. Driver mutations in leukemia promote disease pathogenesis through a combination of cell-autonomous and niche modulation. Stem Cell Reports. 2020;15:95–109. doi: 10.1016/j.stemcr.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Duan S, Yuan G, Liu X, Ren R, Li J, Zhang W, Wu J, Xu X, Fu L, Li Y, Yang J, Zhang W, Bai R, Yi F, Suzuki K, Gao H, Esteban CR, Zhang C, Izpisua Belmonte JC, Chen Z, Wang X, Jiang T, Qu J, Tang F, Liu GH. PTEN deficiency reprogrammes human neural stem cells towards a glioblastoma stem cell-like phenotype. Nat Commun. 2015;6:10068. doi: 10.1038/ncomms10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee JH, Lee JE, Kahng JY, Kim SH, Park JS, Yoon SJ, Um JY, Kim WK, Lee JK, Park J, Kim EH, Lee JH, Lee JH, Chung WS, Ju YS, Park SH, Chang JH, Kang SG, Lee JH. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature. 2018;560:243–247. doi: 10.1038/s41586-018-0389-3. [DOI] [PubMed] [Google Scholar]
- 138.Li XB, Yang G, Zhu L, Tang YL, Zhang C, Ju Z, Yang X, Teng Y. Gastric Lgr5(+) stem cells are the cellular origin of invasive intestinal-type gastric cancer in mice. Cell research. 2016;26:838–49. doi: 10.1038/cr.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao R, Kaakati R, Liu X, Xu L, Lee AK, Bachelder R, Li CY, Hollenbeck ST. CRISPR/Cas9-mediated BRCA1 knockdown adipose stem cells promote breast cancer progression. Plast Reconstr Surg. 2019;143:747–756. doi: 10.1097/PRS.0000000000005316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Weng CC, Ding PY, Liu YH, Hawse JR, Subramaniam M, Wu CC, Lin YC, Chen CY, Hung WC, Cheng KH. Mutant Kras-induced upregulation of CD24 enhances prostate cancer stemness and bone metastasis. Oncogene. 2019;38:2005–2019. doi: 10.1038/s41388-018-0575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miranda A, Funes JM, Sánchez N, Limia CM, Mesa M, Quezada SA, Pérez R, de León J. Oncogenic transformation can orchestrate immune evasion and inflammation in human mesenchymal stem cells independently of extrinsic immune-selective pressure. Cancer Res. 2015;75:3032–42. doi: 10.1158/0008-5472.CAN-14-3276. [DOI] [PubMed] [Google Scholar]
- 142.Zhu L, Finkelstein D, Gao C, Shi L, Wang Y, López-Terrada D, Wang K, Utley S, Pounds S, Neale G, Ellison D, Onar-Thomas A, Gilbertson RJ. Multi-organ mapping of cancer risk. Cell. 2016;166:1132–1146. e1137. doi: 10.1016/j.cell.2016.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Paczulla AM, Rothfelder K, Raffel S, Konantz M, Steinbacher J, Wang H, Tandler C, Mbarga M, Schaefer T, Falcone M, Nievergall E, Dörfel D, Hanns P, Passweg JR, Lutz C, Schwaller J, Zeiser R, Blazar BR, Caligiuri MA, Dirnhofer S, Lundberg P, Kanz L, Quintanilla-Martinez L, Steinle A, Trumpp A, Salih HR, Lengerke C. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature. 2019;572:254–259. doi: 10.1038/s41586-019-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chaffer C, Brueckmann I, Scheel C, Kaestli A, Wiggins P, Rodrigues L, Brooks M, Reinhardt F, Su Y, Polyak K, Arendt L, Kuperwasser C, Bierie B, Weinberg R. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bahmad HF, Mouhieddine TH, Chalhoub RM, Assi S, Araji T, Chamaa F, Itani MM, Nokkari A, Kobeissy F, Daoud G, Abou-Kheir W. The Akt/mTOR pathway in cancer stem/progenitor cells is a potential therapeutic target for glioblastoma and neuroblastoma. Oncotarget. 2018;9:33549–33561. doi: 10.18632/oncotarget.26088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Köhler C, Nittner D, Rambow F, Radaelli E, Stanchi F, Vandamme N, Baggiolini A, Sommer L, Berx G, van den Oord JJ, Gerhardt H, Blanpain C, Marine JC. Mouse cutaneous melanoma induced by mutant BRaf arises from expansion and dedifferentiation of mature pigmented melanocytes. Cell Stem Cell. 2017;21:679–693. e676. doi: 10.1016/j.stem.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 147.Cast A, Valanejad L, Wright M, Nguyen P, Gupta A, Zhu L, Shin S, Timchenko N. C/EBPα-dependent preneoplastic tumor foci are the origin of hepatocellular carcinoma and aggressive pediatric liver cancer. Hepatology. 2018;67:1857–1871. doi: 10.1002/hep.29677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bruschi M, Garnier L, Cleroux E, Giordano A, Dumas M, Bardet A, Kergrohen T, Quesada S, Cesses P, Weber M, Gerbe F, Jay P. Loss of Apc rapidly impairs DNA methylation programs and cell fate decisions in Lgr5+ intestinal stem cells. Cancer Res. 2020;80:2101–2113. doi: 10.1158/0008-5472.CAN-19-2104. [DOI] [PubMed] [Google Scholar]
- 149.Lu R, Wang P, Parton T, Zhou Y, Chrysovergis K, Rockowitz S, Chen WY, Abdel-Wahab O, Wade PA, Zheng D, Wang GG. Epigenetic perturbations by Arg882-mutated DNMT3A potentiate aberrant stem cell gene-expression program and acute leukemia development. Cancer Cell. 2016;30:92–107. doi: 10.1016/j.ccell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y, Jiang G, Lu M, Zhang Z, Yin J, Zeng S, Chen X, Deng M, Jia X, Gu Y, Chen D, Zheng G, He Z. Downregulation of FOXO3a by DNMT1 promotes breast cancer stem cell properties and tumorigenesis. Cell Death Differ. 2020;27:966–983. doi: 10.1038/s41418-019-0389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Norollahi SE, Mansour-Ghanaei F, Joukar F, Ghadarjani S, Mojtahedi K, Gharaei Nejad K, Hemmati H, Gharibpoor F, Khaksar R, Samadani AA. Therapeutic approach of Cancer stem cells (CSCs) in gastric adenocarcinoma; DNA methyltransferases enzymes in cancer targeted therapy. Biomed Pharmacother. 2019;115:108958. doi: 10.1016/j.biopha.2019.108958. [DOI] [PubMed] [Google Scholar]
- 152.Sultan M, Vidovic D, Paine AS, Huynh TT, Coyle KM, Thomas ML, Cruickshank BM, Dean CA, Clements DR, Kim Y, Lee K, Gujar SA, Weaver ICG, Marcato P. Epigenetic silencing of TAP1 in aldefluor(+) Breast cancer stem cells contributes to their enhanced immune evasion. Stem Cells. 2018;36:641–654. doi: 10.1002/stem.2780. [DOI] [PubMed] [Google Scholar]
- 153.Liu S, Cheng K, Zhang H, Kong R, Wang S, Mao C, Liu S. Methylation status of the nanog promoter determines the switch between cancer cells and cancer stem cells. Adv Sci (Weinh) 2020;7:1903035. doi: 10.1002/advs.201903035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Portney B, Arad M, Gupta A, Brown R, Khatri R, Lin P, Hebert A, Angster K, Silipino L, Meltzer W, Taylor R, Zalzman M. ZSCAN4 facilitates chromatin remodeling and promotes the cancer stem cell phenotype. Oncogene. 2020;39:4970–4982. doi: 10.1038/s41388-020-1333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cui M, Chang Y, Fang QG, Du W, Wu JF, Wang JH, Liu ST, Luo SX. Non-coding RNA Pvt1 promotes cancer stem cell-like traits in nasopharyngeal cancer via inhibiting miR-1207. Pathol Oncol Res. 2018;25:1411–1422. doi: 10.1007/s12253-018-0453-1. [DOI] [PubMed] [Google Scholar]
- 156.Liu B, Sun L, Song E. Non-coding RNAs regulate tumor cell plasticity. Sci China Life Sci. 2013;56:886–890. doi: 10.1007/s11427-013-4554-5. [DOI] [PubMed] [Google Scholar]
- 157.Jiang C, Yu M, Xie X, Huang G, Peng Y, Ren D, Lin M, Liu B, Liu M, Wang W, Kuang M. miR-217 targeting DKK1 promotes cancer stem cell properties via activation of the Wnt signaling pathway in hepatocellular carcinoma. Oncol Rep. 2017;38:2351–2359. doi: 10.3892/or.2017.5924. [DOI] [PubMed] [Google Scholar]
- 158.Prabhu KS, Raza A, Karedath T, Raza SS, Fathima H, Ahmed EI, Kuttikrishnan S, Therachiyil L, Kulinski M, Dermime S, Junejo K, Steinhoff M, Uddin S. Non-coding RNAs as regulators and markers for targeting of breast cancer and cancer stem cells. Cancers (Basel) 2020;12:351. doi: 10.3390/cancers12020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xu J, Liu S. Noncoding RNAs in cancer cell plasticity. Adv Exp Med Biol. 2016;927:173–189. doi: 10.1007/978-981-10-1498-7_6. [DOI] [PubMed] [Google Scholar]
- 160.Xiao Y, Pan J, Geng Q, Wang G. LncRNA MALAT1 increases the stemness of gastric cancer cells via enhancing SOX2 mRNA stability. FEBS Open Bio. 2019;9:1212–1222. doi: 10.1002/2211-5463.12649. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 161.Song H, Xu Y, Shi L, Xu T, Fan R, Cao M, Xu W, Song J. LncRNA THOR increases the stemness of gastric cancer cells via enhancing SOX9 mRNA stability. Biomed Pharmacother. 2018;108:338–346. doi: 10.1016/j.biopha.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 162.Miao F, Chen J, Shi M, Song Y, Chen Z, Pang L. LncRNA HAND2-AS1 inhibits non-small cell lung cancer migration, invasion and maintains cell stemness through the interactions with TGF-β1. Biosci Rep. 2019;39:BSR20181525. doi: 10.1042/BSR20181525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang Y, Zhu P, Luo J, Wang J, Liu Z, Wu W, Du Y, Ye B, Wang D, He L, Ren W, Wang J, Sun X, Chen R, Tian Y, Fan Z. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 2019;38:e101110. doi: 10.15252/embj.2018101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhu P, Wu J, Wang Y, Zhu X, Lu T, Liu B, He L, Ye B, Wang S, Meng S, Fan D, Wang J, Yang L, Qin X, Du Y, Li C, He L, Ren W, Wu X, Tian Y, Fan Z. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat Cell Biol. 2018;20:1134–1144. doi: 10.1038/s41556-018-0194-0. [DOI] [PubMed] [Google Scholar]
- 165.Gao S, Zhou B, Li H, Huang X, Wu Y, Xing C, Yu X, Ji Y. Long noncoding RNA HOTAIR promotes the self-renewal of leukemia stem cells through epigenetic silencing of p15. Exp Hematol. 2018;67:32–40. e33. doi: 10.1016/j.exphem.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 166.Zhao Y, Li Y, Sheng J, Wu F, Li K, Huang R, Wang X, Jiao T, Guan X, Lu Y, Chen X, Luo Z, Zhou Y, Hu H, Liu W, Du B, Miao S, Cai J, Wang L, Zhao H, Ying J, Bi X, Song W. P53-R273H mutation enhances colorectal cancer stemness through regulating specific lncRNAs. J Exp Clin Cancer Res. 2019;38:379. doi: 10.1186/s13046-019-1375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pollutri D, Patrizi C, Marinelli S, Giovannini C, Trombetta E, Giannone FA, Baldassarre M, Quarta S, Vandewynckel YP, Vandierendonck A, Van Vlierberghe H, Porretti L, Negrini M, Bolondi L, Gramantieri L, Fornari F. The epigenetically regulated miR-494 associates with stem-cell phenotype and induces sorafenib resistance in hepatocellular carcinoma. Cell Death Dis. 2018;9:4. doi: 10.1038/s41419-017-0076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bai X, Zhou Y, Chen P, Yang M, Xu J. MicroRNA-142-5p induces cancer stem cell-like properties of cutaneous squamous cell carcinoma via inhibiting PTEN. J Cell Biochem. 2018;119:2179–2188. doi: 10.1002/jcb.26379. [DOI] [PubMed] [Google Scholar]
- 169.Liu S, Clouthier SG, Wicha MS. Role of microRNAs in the regulation of breast cancer stem cells. J Mammary Gland Biol Neoplasia. 2012;17:15–21. doi: 10.1007/s10911-012-9242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang L, Bu P, Ai Y, Srinivasan T, Chen HJ, Xiang K, Lipkin SM, Shen X. A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. Elife. 2016;5:e14620. doi: 10.7554/eLife.14620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Han Q, Wu W, Cui Y. LINC00337 regulates KLF5 and maintains stem-cell like traits of cervical cancer cells by modulating miR-145. Front Oncol. 2020;10:1433. doi: 10.3389/fonc.2020.01433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 172.Chen X, Xie R, Gu P, Huang M, Han J, Dong W, Xie W, Wang B, He W, Zhong G, Chen Z, Huang J, Lin T. Long noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin Cancer Res. 2019;25:1389–1403. doi: 10.1158/1078-0432.CCR-18-1656. [DOI] [PubMed] [Google Scholar]
- 173.Wang X, Guo H, Yao B, Helms J. miR-15b inhibits cancer-initiating cell phenotypes and chemoresistance of cisplatin by targeting TRIM14 in oral tongue squamous cell cancer. Oncol Rep. 2017;37:2720–2726. doi: 10.3892/or.2017.5532. [DOI] [PubMed] [Google Scholar]
- 174.Li F, Xu Y, Xu X, Ge S, Zhang F, Zhang H, Fan X. lncRNA HotairM1 depletion promotes self-renewal of cancer stem cells through HOXA1-nanog regulation loop. Mol Ther Nucleic Acids. 2020;22:456–470. doi: 10.1016/j.omtn.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang XH, Qian Y, Li Z, Zhang NN, Xie YJ. Let-7g-5p inhibits epithelial-mesenchymal transition consistent with reduction of glioma stem cell phenotypes by targeting VSIG4 in glioblastoma. Oncol Rep. 2016;36:2967–2975. doi: 10.3892/or.2016.5098. [DOI] [PubMed] [Google Scholar]
- 176.Liang W, Lin Z, Du C, Qiu D, Zhang Q. mRNA modification orchestrates cancer stem cell fate decisions. Mol Cancer. 2020;19:38. doi: 10.1186/s12943-020-01166-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Aguilo F, Zhang F, Sancho A, Fidalgo M, Di Cecilia S, Vashisht A, Lee DF, Chen CH, Rengasamy M, Andino B, Jahouh F, Roman A, Krig SR, Wang R, Zhang W, Wohlschlegel JA, Wang J, Walsh MJ. Coordination of m(6)A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell. 2015;17:689–704. doi: 10.1016/j.stem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, Carter AC, Flynn RA, Zhou C, Lim KS, Dedon P, Wernig M, Mullen AC, Xing Y, Giallourakis CC, Chang HY. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, Helm M, Bujnicki JM, Grosjean H. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acid Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jiang Q, Crews LA, Holm F, Jamieson CHM. RNA editing-dependent epitranscriptome diversity in cancer stem cells. Nat Rev Cancer. 2017;17:381–392. doi: 10.1038/nrc.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.McElhinney J, Hasan A, Sajini A. The epitranscriptome landscape of small noncoding RNAs in stem cells. Stem Cells. 2020;38:1216–1228. doi: 10.1002/stem.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, Ben-Haim MS, Eyal E, Yunger S, Pinto Y, Jaitin DA, Viukov S, Rais Y, Krupalnik V, Chomsky E, Zerbib M, Maza I, Rechavi Y, Massarwa R, Hanna S, Amit I, Levanon EY, Amariglio N, Stern-Ginossar N, Novershtern N, Rechavi G, Hanna JH. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 184.Zhang M, Zhai Y, Zhang S, Dai X, Li Z. Roles of N6-methyladenosine (m(6)A) in stem cell fate decisions and early embryonic development in mammals. Front Cell Dev Biol. 2020;8:782. doi: 10.3389/fcell.2020.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113:E2047–2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, Qin X, Tang L, Wang Y, Hong GM, Huang H, Wang X, Chen P, Gurbuxani S, Arnovitz S, Li Y, Li S, Strong J, Neilly MB, Larson RA, Jiang X, Zhang P, Jin J, He C, Chen J. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, Riggs AD, He C, Shi Y. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, Majumder S, He C, Huang S. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. e596. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zheng F, Du F, Zhao J, Wang X, Si Y, Jin P, Qian H, Xu B, Yuan P. The emerging role of RNA N6-methyladenosine methylation in breast cancer. Biomark Res. 2021;9:39. doi: 10.1186/s40364-021-00295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, Schulman J, Famulare C, Patel M, Klimek VM, Garrett-Bakelman FE, Melnick A, Carroll M, Mason CE, Jaffrey SR, Kharas MG. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, Sheng Y, Wang Y, Wunderlich M, Zhang B, Dore LC, Su R, Deng X, Ferchen K, Li C, Sun M, Lu Z, Jiang X, Marcucci G, Mulloy JC, Yang J, Qian Z, Wei M, He C, Chen J. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–205. e199. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 193.Bai Y, Yang C, Wu R, Huang L, Song S, Li W, Yan P, Lin C, Li D, Zhang Y. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zipeto MA, Jiang Q, Melese E, Jamieson CH. RNA rewriting, recoding, and rewiring in human disease. Trends Mol Med. 2015;21:549–559. doi: 10.1016/j.molmed.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 195.Xu LD, Öhman M. ADAR1 editing and its role in cancer. Genes (Basel) 2018;10:12. doi: 10.3390/genes10010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 198.Lazzari E, Mondala PK, Santos ND, Miller AC, Pineda G, Jiang Q, Leu H, Ali SA, Ganesan AP, Wu CN, Costello C, Minden M, Chiaramonte R, Stewart AK, Crews LA, Jamieson CHM. Alu-dependent RNA editing of GLI1 promotes malignant regeneration in multiple myeloma. Nat Commun. 2017;8:1922. doi: 10.1038/s41467-017-01890-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jiang Q, Crews LA, Barrett CL, Chun HJ, Court AC, Isquith JM, Zipeto MA, Goff DJ, Minden M, Sadarangani A, Rusert JM, Dao KH, Morris SR, Goldstein LS, Marra MA, Frazer KA, Jamieson CH. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2013;110:1041–1046. doi: 10.1073/pnas.1213021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jiang Q, Isquith J, Zipeto MA, Diep RH, Pham J, Delos Santos N, Reynoso E, Chau J, Leu H, Lazzari E, Melese E, Ma W, Fang R, Minden M, Morris S, Ren B, Pineda G, Holm F, Jamieson C. Hyper-editing of cell-cycle regulatory and tumor suppressor RNA promotes malignant progenitor propagation. Cancer Cell. 2019;35:81–94. e87. doi: 10.1016/j.ccell.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lau EY, Lo J, Cheng BY, Ma MK, Lee JM, Ng JK, Chai S, Lin CH, Tsang SY, Ma S, Ng IO, Lee TK. Cancer-associated fibroblasts regulate tumor-initiating cell plasticity in hepatocellular carcinoma through c-Met/FRA1/HEY1 signaling. Cell Rep. 2016;15:1175–1189. doi: 10.1016/j.celrep.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 202.Dirkse A, Golebiewska A, Buder T, Nazarov P, Muller A, Poovathingal S, Brons N, Leite S, Nicolas S, Sarkisjan D, Seyfrid M, Fritah S, Stieber D, Michelucci A, Hertel F, Herold-Mende C, Azuaje F, Skupin A, Bjerkvig R, Niclou S. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun. 2019;10:1187. doi: 10.1038/s41467-019-09853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Weitzenfeld P, Meshel T, Ben-Baruch A. Microenvironmental networks promote tumor heterogeneity and enrich for metastatic cancer stem-like cells in luminal-A breast tumor cells. Oncotarget. 2016;7:81123–81143. doi: 10.18632/oncotarget.13213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.El-Badawy A, Ghoneim MA, Gabr MM, Salah RA, Mohamed IK, Amer M, El-Badri N. Cancer cell-soluble factors reprogram mesenchymal stromal cells to slow cycling, chemoresistant cells with a more stem-like state. Stem Cell Res Ther. 2017;8:254. doi: 10.1186/s13287-017-0709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Jin X, Kim SH, Jeon HM, Beck S, Sohn YW, Yin J, Kim JK, Lim YC, Lee JH, Kim SH, Kang SH, Pian X, Song MS, Park JB, Chae YS, Chung YG, Lee SH, Choi YJ, Nam DH, Choi YK, Kim H. Interferon regulatory factor 7 regulates glioma stem cells via interleukin-6 and Notch signalling. Brain. 2012;135:1055–1069. doi: 10.1093/brain/aws028. [DOI] [PubMed] [Google Scholar]
- 206.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY, Yaswen P, Goga A, Werb Z. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zheng Z, Yu H, Huang Q, Wu H, Fu Y, Shi J, Wang T, Fan X. Heterogeneous expression of Lgr5 as a risk factor for focal invasion and distant metastasis of colorectal carcinoma. Oncotarget. 2018;9:30025–30033. doi: 10.18632/oncotarget.23144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Savelieva OE, Tashireva LA, Kaigorodova EV, Buzenkova AV, Mukhamedzhanov RK, Grigoryeva ES, Zavyalova MV, Tarabanovskaya NA, Cherdyntseva NV, Perelmuter VM. Heterogeneity of stemlike circulating tumor cells in invasive breast cancer. Int J Mol Sci. 2020;21:2780. doi: 10.3390/ijms21082780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Park HA, Brown SR, Kim Y. Cellular mechanisms of circulating tumor cells during breast cancer metastasis. Int J Mol Sci. 2020;21:5040. doi: 10.3390/ijms21145040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lyberopoulou A, Aravantinos G, Efstathopoulos EP, Nikiteas N, Bouziotis P, Isaakidou A, Papalois A, Marinos E, Gazouli M. Mutational analysis of circulating tumor cells from colorectal cancer patients and correlation with primary tumor tissue. PLoS One. 2015;10:e0123902. doi: 10.1371/journal.pone.0123902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hu J, Li G, Zhang P, Zhuang X, Hu G. A CD44v(+) subpopulation of breast cancer stem-like cells with enhanced lung metastasis capacity. Cell Death Dis. 2017;8:e2679. doi: 10.1038/cddis.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H, Dijkgraaf GJP, Piskol R, de Sauvage FJ. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 213.Fumagalli A, Oost KC, Kester L, Morgner J, Bornes L, Bruens L, Spaargaren L, Azkanaz M, Schelfhorst T, Beerling E, Heinz MC, Postrach D, Seinstra D, Sieuwerts AM, Martens JWM, van der Elst S, van Baalen M, Bhowmick D, Vrisekoop N, Ellenbroek SIJ, Suijkerbuijk SJE, Snippert HJ, van Rheenen J. Plasticity of Lgr5-negative cancer cells drives metastasis in colorectal cancer. Cell Stem Cell. 2020;26:569–578. e567. doi: 10.1016/j.stem.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Abbas Z, Rehman S. An overview of cancer treatment modalities. 2018 [Google Scholar]
- 215.Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, Lee YK, Kwon HY. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018;2018:5416923. doi: 10.1155/2018/5416923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Safa AR, Saadatzadeh MR, Cohen-Gadol AA, Pollok KE, Bijangi-Vishehsaraei K. Glioblastoma stem cells (GSCs) epigenetic plasticity and interconversion between differentiated non-GSCs and GSCs. Genes Dis. 2015;2:152–163. doi: 10.1016/j.gendis.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Das P, Pillai S, Rakib A, Khanam J, Gopalan V, Lam A, Islam M. Plasticity of cancer stem cell: origin and role in disease progression and therapy resistance. Stem Cell Rev Rep. 2020;16:397–412. doi: 10.1007/s12015-019-09942-y. [DOI] [PubMed] [Google Scholar]
- 218.Skowron MA, Niegisch G, Fritz G, Arent T, van Roermund JG, Romano A, Albers P, Schulz WA, Hoffmann MJ. Phenotype plasticity rather than repopulation from CD90/CK14+ cancer stem cells leads to cisplatin resistance of urothelial carcinoma cell lines. J Exp Clin Cancer Res. 2015;34:144. doi: 10.1186/s13046-015-0259-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Welker AM, Jaros BD, An M, Beattie CE. Changes in tumor cell heterogeneity after chemotherapy treatment in a xenograft model of glioblastoma. Neuroscience. 2017;356:35–43. doi: 10.1016/j.neuroscience.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Martin-Castillo B, Lopez-Bonet E, Cuyàs E, Viñas G, Pernas S, Dorca J, Menendez JA. Cancer stem cell-driven efficacy of trastuzumab (Herceptin): towards a reclassification of clinically HER2-positive breast carcinomas. Oncotarget. 2015;6:32317–32338. doi: 10.18632/oncotarget.6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Liu X, Zhang SM, McGeary MK, Krykbaeva I, Lai L, Jansen DJ, Kales SC, Simeonov A, Hall MD, Kelly DP, Bosenberg MW, Yan Q. KDM5B promotes drug resistance by regulating melanoma-propagating cell subpopulations. Mol Cancer Ther. 2019;18:706–717. doi: 10.1158/1535-7163.MCT-18-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sharma A, Cao EY, Kumar V, Zhang X, Leong HS, Wong AML, Ramakrishnan N, Hakimullah M, Teo HMV, Chong FT, Chia S, Thangavelu MT, Kwang XL, Gupta R, Clark JR, Periyasamy G, Iyer NG, DasGupta R. Longitudinal single-cell RNA sequencing of patient-derived primary cells reveals drug-induced infidelity in stem cell hierarchy. Nat Commun. 2018;9:4931. doi: 10.1038/s41467-018-07261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 224.Venkatesh V, Nataraj R, Thangaraj GS, Karthikeyan M, Gnanasekaran A, Kaginelli SB, Kuppanna G, Kallappa CG, Basalingappa KM. Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig. 2018;5:5. doi: 10.21037/sci.2018.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hoffman LM, Fouladi M, Olson J, Daryani VM, Stewart CF, Wetmore C, Kocak M, Onar-Thomas A, Wagner L, Gururangan S, Packer RJ, Blaney SM, Gajjar A, Kun LE, Boyett JM, Gilbertson RJ. Phase I trial of weekly MK-0752 in children with refractory central nervous system malignancies: a pediatric brain tumor consortium study. Childs Nerv Syst. 2015;31:1283–1289. doi: 10.1007/s00381-015-2725-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kummar S, O’Sullivan Coyne G, Do KT, Turkbey B, Meltzer PS, Polley E, Choyke PL, Meehan R, Vilimas R, Horneffer Y, Juwara L, Lih A, Choudhary A, Mitchell SA, Helman LJ, Doroshow JH, Chen AP. Clinical activity of the γ-secretase inhibitor PF-03084014 in adults with desmoid tumors (aggressive fibromatosis) J. Clin. Oncol. 2017;35:1561–1569. doi: 10.1200/JCO.2016.71.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kondratyev M, Kreso A, Hallett RM, Girgis-Gabardo A, Barcelon ME, Ilieva D, Ware C, Majumder PK, Hassell JA. Gamma-secretase inhibitors target tumor-initiating cells in a mouse model of ERBB2 breast cancer. Oncogene. 2012;31:93–103. doi: 10.1038/onc.2011.212. [DOI] [PubMed] [Google Scholar]
- 228.Wang L, Zi H, Luo Y, Liu T, Zheng H, Xie C, Wang X, Huang X. Inhibition of Notch pathway enhances the anti-tumor effect of docetaxel in prostate cancer stem-like cells. Stem Cell Res Ther. 2020;11:258. doi: 10.1186/s13287-020-01773-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Dai G, Deng S, Guo W, Yu L, Yang J, Zhou S, Gao T. Notch pathway inhibition using DAPT, a γ-secretase inhibitor (GSI), enhances the antitumor effect of cisplatin in resistant osteosarcoma. Mol Carcinog. 2019;58:3–18. doi: 10.1002/mc.22873. [DOI] [PubMed] [Google Scholar]
- 230.Smith DC, Eisenberg PD, Manikhas G, Chugh R, Gubens MA, Stagg RJ, Kapoun AM, Xu L, Dupont J, Sikic B. A phase I dose escalation and expansion study of the anticancer stem cell agent demcizumab (anti-DLL4) in patients with previously treated solid tumors. Clin Cancer Res. 2014;20:6295–6303. doi: 10.1158/1078-0432.CCR-14-1373. [DOI] [PubMed] [Google Scholar]
- 231.Kim DH, Lee S, Kang HG, Park HW, Lee HW, Kim D, Yoem DH, Ahn JH, Ha E, You WK, Lee SH, Kim SJ, Chun KH. Synergistic antitumor activity of a DLL4/VEGF bispecific therapeutic antibody in combination with irinotecan in gastric cancer. BMB Rep. 2020;53:533–538. doi: 10.5483/BMBRep.2020.53.10.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Xie M, He CS, He ZH, Lin QZ. Clinical and prognostic implications of delta-like ligand 4 and hypoxia-inducible factors in metastatic renal cell carcinoma (mRCC) patients treated with sunitinib as first-line therapy. J Clinic Oncol. 2013;31:e15567. [Google Scholar]
- 233.Petrirena GJ, Masliah-Planchon J, Sala Q, Pourroy B, Frappaz D, Tabouret E, Graillon T, Gentet JC, Delattre O, Chinot O, Padovani L. Recurrent extraneural sonic hedgehog medulloblastoma exhibiting sustained response to vismodegib and temozolomide monotherapies and inter-metastatic molecular heterogeneity at progression. Oncotarget. 2018;9:10175–10183. doi: 10.18632/oncotarget.23699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Freitas RD, Dias RB, Vidal MTA, Valverde LF, Gomes Alves Costa R, Damasceno AKA, Sales CBS, Siquara da Rocha LO, Dos Reis MG, Soares MBP, Coletta RD, Pereira TA, Bezerra DP, Gurgel Rocha CA. Inhibition of CAL27 oral squamous carcinoma cell by targeting hedgehog pathway with vismodegib or itraconazole. Front Oncol. 2020;10:563838. doi: 10.3389/fonc.2020.563838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wu C, Hu S, Cheng J, Wang G, Tao K. Smoothened antagonist GDC-0449 (Vismodegib) inhibits proliferation and triggers apoptosis in colon cancer cell lines. Exp Ther Med. 2017;13:2529–2536. doi: 10.3892/etm.2017.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Pietanza MC, Litvak AM, Varghese AM, Krug LM, Fleisher M, Teitcher JB, Holodny AI, Sima CS, Woo KM, Ng KK, Won HH, Berger MF, Kris MG, Rudin CM. A phase I trial of the Hedgehog inhibitor, sonidegib (LDE225), in combination with etoposide and cisplatin for the initial treatment of extensive stage small cell lung cancer. Lung Cancer. 2016;99:23–30. doi: 10.1016/j.lungcan.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kieran MW, Chisholm J, Casanova M, Brandes AA, Aerts I, Bouffet E, Bailey S, Leary S, MacDonald TJ, Mechinaud F, Cohen KJ, Riccardi R, Mason W, Hargrave D, Kalambakas S, Deshpande P, Tai F, Hurh E, Geoerger B. Phase I study of oral sonidegib (LDE225) in pediatric brain and solid tumors and a phase II study in children and adults with relapsed medulloblastoma. Neuro Oncol. 2017;19:1542–1552. doi: 10.1093/neuonc/nox109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Fu J, Rodova M, Nanta R, Meeker D, Van Veldhuizen PJ, Srivastava RK, Shankar S. NPV-LDE-225 (Erismodegib) inhibits epithelial mesenchymal transition and self-renewal of glioblastoma initiating cells by regulating miR-21, miR-128, and miR-200. Neuro Oncol. 2013;15:691–706. doi: 10.1093/neuonc/not011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Nanta R, Kumar D, Meeker D, Rodova M, Van Veldhuizen PJ, Shankar S, Srivastava RK. NVP-LDE-225 (Erismodegib) inhibits epithelial-mesenchymal transition and human prostate cancer stem cell growth in NOD/SCID IL2Rγ null mice by regulating Bmi-1 and microRNA-128. Oncogenesis. 2013;2:e42. doi: 10.1038/oncsis.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.D’Amato C, Rosa R, Marciano R, D’Amato V, Formisano L, Nappi L, Raimondo L, Di Mauro C, Servetto A, Fulciniti F, Cipolletta A, Bianco C, Ciardiello F, Veneziani BM, De Placido S, Bianco R. Inhibition of Hedgehog signalling by NVP-LDE225 (Erismodegib) interferes with growth and invasion of human renal cell carcinoma cells. Br J Cancer. 2014;111:1168–1179. doi: 10.1038/bjc.2014.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Minami Y, Minami H, Miyamoto T, Yoshimoto G, Kobayashi Y, Munakata W, Onishi Y, Kobayashi M, Ikuta M, Chan G, Woolfson A, Ono C, Shaik MN, Fujii Y, Zheng X, Naoe T. Phase I study of glasdegib (PF-04449913), an oral smoothened inhibitor, in Japanese patients with select hematologic malignancies. Cancer Sci. 2017;108:1628–1633. doi: 10.1111/cas.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Cortes JE, Douglas Smith B, Wang ES, Merchant A, Oehler VG, Arellano M, DeAngelo DJ, Pollyea DA, Sekeres MA, Robak T, Ma WW, Zeremski M, Naveed Shaik M, Douglas Laird A, O’Connell A, Chan G, Schroeder MA. Glasdegib in combination with cytarabine and daunorubicin in patients with AML or high-risk MDS: phase 2 study results. Am J Hematol. 2018;93:1301–1310. doi: 10.1002/ajh.25238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Cortes JE, Heidel FH, Hellmann A, Fiedler W, Smith BD, Robak T, Montesinos P, Pollyea DA, DesJardins P, Ottmann O, Ma WW, Shaik MN, Laird AD, Zeremski M, O’Connell A, Chan G, Heuser M. Randomized comparison of low dose cytarabine with or without glasdegib in patients with newly diagnosed acute myeloid leukemia or high-risk myelodysplastic syndrome. Leukemia. 2019;33:379–389. doi: 10.1038/s41375-018-0312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ueno H, Kondo S, Yoshikawa S, Inoue K, Andre V, Tajimi M, Murakami H. A phase I and pharmacokinetic study of taladegib, a Smoothened inhibitor, in Japanese patients with advanced solid tumors. Invest New Drugs. 2018;36:647–656. doi: 10.1007/s10637-017-0544-y. [DOI] [PubMed] [Google Scholar]
- 245.Lee MJ, Hatton BA, Villavicencio EH, Khanna PC, Friedman SD, Ditzler S, Pullar B, Robison K, White KF, Tunkey C, LeBlanc M, Randolph-Habecker J, Knoblaugh SE, Hansen S, Richards A, Wainwright BJ, McGovern K, Olson JM. Hedgehog pathway inhibitor saridegib (IPI-926) increases lifespan in a mouse medulloblastoma model. Proc Natl Acad Sci U S A. 2012;109:7859–7864. doi: 10.1073/pnas.1114718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Le PN, McDermott JD, Jimeno A. Targeting the Wnt pathway in human cancers: therapeutic targeting with a focus on OMP-54F28. Pharmacol Ther. 2015;146:1–11. doi: 10.1016/j.pharmthera.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Gurney A, Axelrod F, Bond CJ, Cain J, Chartier C, Donigan L, Fischer M, Chaudhari A, Ji M, Kapoun AM, Lam A, Lazetic S, Ma S, Mitra S, Park IK, Pickell K, Sato A, Satyal S, Stroud M, Tran H, Yen WC, Lewicki J, Hoey T. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A. 2012;109:11717–11722. doi: 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Cortes JE, Faderl S, Pagel J, Jung CW, Yoon SS, Koh Y, Pardanani AD, Hauptschein RS, Lee KJ, Lee JH. Phase 1 study of CWP232291 in relapsed/refractory acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) J. Clin. Oncol. 2015;33:7044–7044. [Google Scholar]
- 249.Fischer MM, Cancilla B, Yeung VP, Cattaruzza F, Chartier C, Murriel CL, Cain J, Tam R, Cheng CY, Evans JW, O’Young G, Song X, Lewicki J, Kapoun AM, Gurney A, Yen WC, Hoey T. WNT antagonists exhibit unique combinatorial antitumor activity with taxanes by potentiating mitotic cell death. Sci Adv. 2017;3:e1700090. doi: 10.1126/sciadv.1700090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Agarwal P, Zhang B, Ho Y, Cook A, Li L, Mikhail FM, Wang Y, McLaughlin ME, Bhatia R. Enhanced targeting of CML stem and progenitor cells by inhibition of porcupine acyltransferase in combination with TKI. Blood. 2017;129:1008–1020. doi: 10.1182/blood-2016-05-714089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tamura R, Tanaka T, Akasaki Y, Murayama Y, Yoshida K, Sasaki H. The role of vascular endothelial growth factor in the hypoxic and immunosuppressive tumor microenvironment: perspectives for therapeutic implications. Med Oncol. 2019;37:2. doi: 10.1007/s12032-019-1329-2. [DOI] [PubMed] [Google Scholar]
- 252.Huang S, He P, Peng X, Li J, Xu D, Tang Y. Pristimerin inhibits prostate cancer bone metastasis by targeting PC-3 stem cell characteristics and VEGF-induced vasculogenesis of BM-EPCs. Cell Physiol Biochem. 2015;37:253–268. doi: 10.1159/000430350. [DOI] [PubMed] [Google Scholar]
- 253.Barone A, Sengupta R, Warrington NM, Smith E, Wen PY, Brekken RA, Romagnoli B, Douglas G, Chevalier E, Bauer MP, Dembowsky K, Piwnica-Worms D, Rubin JB. Combined VEGF and CXCR4 antagonism targets the GBM stem cell population and synergistically improves survival in an intracranial mouse model of glioblastoma. Oncotarget. 2014;5:9811–9822. doi: 10.18632/oncotarget.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Cooper TM, Sison EAR, Baker SD, Li L, Ahmed A, Trippett T, Gore L, Macy ME, Narendran A, August K, Absalon MJ, Boklan J, Pollard J, Magoon D, Brown PA. A phase 1 study of the CXCR4 antagonist plerixafor in combination with high-dose cytarabine and etoposide in children with relapsed or refractory acute leukemias or myelodysplastic syndrome: a pediatric oncology experimental therapeutics investigators’ consortium study (POE 10-03) Pediatr Blood Cancer. 2017;64:10. doi: 10.1002/pbc.26414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Huang TX, Guan XY, Fu L. Therapeutic targeting of the crosstalk between cancer-associated fibroblasts and cancer stem cells. Am J Cancer Res. 2019;9:1889–1904. [PMC free article] [PubMed] [Google Scholar]
- 256.Zhen Z, Tang W, Wang M, Zhou S, Wang H, Wu Z, Hao Z, Li Z, Liu L, Xie J. Protein nanocage mediated fibroblast-activation protein targeted photoimmunotherapy to enhance cytotoxic t cell infiltration and tumor control. Nano Lett. 2017;17:862–869. doi: 10.1021/acs.nanolett.6b04150. [DOI] [PubMed] [Google Scholar]
- 257.Szot C, Saha S, Zhang XM, Zhu Z, Hilton MB, Morris K, Seaman S, Dunleavey JM, Hsu KS, Yu GJ, Morris H, Swing DA, Haines DC, Wang Y, Hwang J, Feng Y, Welsch D, DeCrescenzo G, Chaudhary A, Zudaire E, Dimitrov DS, St Croix B. Tumor stroma-targeted antibody-drug conjugate triggers localized anticancer drug release. J Clin Invest. 2018;128:2927–2943. doi: 10.1172/JCI120481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zuo ZQ, Chen KG, Yu XY, Zhao G, Shen S, Cao ZT, Luo YL, Wang YC, Wang J. Promoting tumor penetration of nanoparticles for cancer stem cell therapy by TGF-β signaling pathway inhibition. Biomaterials. 2016;82:48–59. doi: 10.1016/j.biomaterials.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 259.Huang WC, Kuo KT, Wang CH, Yeh CT, Wang Y. Cisplatin resistant lung cancer cells promoted M2 polarization of tumor-associated macrophages via the Src/CD155/MIF functional pathway. J Exp Clin Cancer Res. 2019;38:180. doi: 10.1186/s13046-019-1166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Rupp U, Schoendorf-Holland E, Eichbaum M, Schuetz F, Lauschner I, Schmidt P, Staab A, Hanft G, Huober J, Sinn HP, Sohn C, Schneeweiss A. Safety and pharmacokinetics of bivatuzumab mertansine in patients with CD44v6-positive metastatic breast cancer: final results of a phase I study. Anticancer Drugs. 2007;18:477–485. doi: 10.1097/CAD.0b013e32801403f4. [DOI] [PubMed] [Google Scholar]
- 261.Porcellini S, Asperti C, Corna S, Cicoria E, Valtolina V, Stornaiuolo A, Valentinis B, Bordignon C, Traversari C. CAR T cells redirected to CD44v6 control tumor growth in lung and ovary adenocarcinoma bearing mice. Front Immunol. 2020;11:99. doi: 10.3389/fimmu.2020.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.He SZ, Busfield S, Ritchie DS, Hertzberg MS, Durrant S, Lewis ID, Marlton P, McLachlan AJ, Kerridge I, Bradstock KF, Kennedy G, Boyd AW, Yeadon TM, Lopez AF, Ramshaw HS, Iland H, Bamford S, Barnden M, DeWitte M, Basser R, Roberts AW. A Phase 1 study of the safety, pharmacokinetics and anti-leukemic activity of the anti-CD123 monoclonal antibody CSL360 in relapsed, refractory or high-risk acute myeloid leukemia. Leuk Lymphoma. 2015;56:1406–1415. doi: 10.3109/10428194.2014.956316. [DOI] [PubMed] [Google Scholar]
- 263.Morita S, Mochizuki M, Wada K, Shibuya R, Nakamura M, Yamaguchi K, Yamazaki T, Imai T, Asada Y, Matsuura K, Sugamura K, Katori Y, Satoh K, Tamai K. Humanized anti-CD271 monoclonal antibody exerts an anti-tumor effect by depleting cancer stem cells. Cancer Lett. 2019;461:144–152. doi: 10.1016/j.canlet.2019.07.011. [DOI] [PubMed] [Google Scholar]
- 264.Badrinath N, Yoo SY. Recent advances in cancer stem cell-targeted immunotherapy. Cancers. 2019;11:310. doi: 10.3390/cancers11030310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Delyon J, Mateus C, Lefeuvre D, Lanoy E, Zitvogel L, Chaput N, Roy S, Eggermont AM, Routier E, Robert C. Experience in daily practice with ipilimumab for the treatment of patients with metastatic melanoma: an early increase in lymphocyte and eosinophil counts is associated with improved survival. Ann Oncol. 2013;24:1697–1703. doi: 10.1093/annonc/mdt027. [DOI] [PubMed] [Google Scholar]
- 266.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zheng F, Dang J, Zhang H, Xu F, Ba D, Zhang B, Cheng F, Chang AE, Wicha MS, Li Q. Cancer stem cell vaccination with PD-L1 and CTLA-4 blockades enhances the eradication of melanoma stem cells in a mouse tumor model. J Immunother. 2018;41:361–368. doi: 10.1097/CJI.0000000000000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Saha D, Martuza RL, Rabkin SD. Macrophage polarization contributes to glioblastoma eradication by combination immunovirotherapy and immune checkpoint blockade. Cancer Cell. 2017;32:253–267. e255. doi: 10.1016/j.ccell.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Shi X, Zhang X, Li J, Mo L, Zhao H, Zhu Y, Hu Z, Gao J, Tan W. PD-1 blockade enhances the antitumor efficacy of GM-CSF surface-modified bladder cancer stem cells vaccine. Int J Cancer. 2018;142:2106–2117. doi: 10.1002/ijc.31219. [DOI] [PubMed] [Google Scholar]


