Abstract
The mechanisms by which chemotherapeutic drugs mediate efficacy and toxicity in patients across cancers are not fully understood. A poorly understood aspect of the tumor cell response to chemotherapy is cytokine regulation. Some drug-induced cytokines promote the anti-cancer activity of the drugs, but others may promote proliferation, metastasis, and drug resistance. We evaluated effects of clinical chemotherapeutics oxaliplatin, cisplatin, 5-fluorouracil (5-FU), doxorubicin, paclitaxel, docetaxel, and carboplatin on a panel of 52 cytokines in MCF7 breast cancer (BC) cells. We observed pan-drug effects, such as the upregulation of TRAIL-R2 and Chitinase 3-like 1 and drug-specific effects on interleukin and CXCL cytokines. We compared cytokine regulation in MCF7 BC and HCT116 colorectal cancer (CRC) cells, revealing tissue-specific drug effects such as enhanced upregulation of TRAIL-R2 and downregulation of IFN-β and TRAIL in MCF7 by cisplatin, oxaliplatin, and 5-FU. We found that chemotherapy-inducible transcripts have varying potential for prognostic significance in CRC versus BC. Among the non-prognostic CRC genes that were prognostic in BC were NFKBIA and GADD45A, both of which support anti-cancer drug mechanisms. Thus, we establish a novel 7-drug, 52-cytokine signature in MCF7 BC cells and a 3-drug, 40-cytokine signature in HCT116 CRC cells that suggest drug-specific and tissue-specific cytokine regulation. Distinct differences across prognostic gene signatures in BC and CRC further support tissue specificity in the relative impact of drug-regulated genes on patient survival.
Keywords: Breast cancer, colorectal cancer, cytokines, oxaliplatin, cisplatin, 5-fluorouracil, doxorubicin, paclitaxel, docetaxel, carboplatin
Introduction
Breast cancer (BC) is one of the most common cancers in the U.S. and will account for 14.8% of new cancer cases in 2021 [1]. It is one of the leading causes of death in women. The 5-year survival rate for metastatic BC is 28% [2]. There are five molecular subtypes of BC which include: luminal A (hormone receptor positive, HER2 negative), luminal B (hormone receptor positive, HER2 negative or positive), triple-negative (hormone receptor negative, HER2 negative), HER2-enriched (hormone receptor negative, HER2 positive), and normal-like (similar to luminal A) [3]. Triple-negative breast cancer can be further classified into basal-like 1 (BL1), basal-like 2 (BL2), immunomodulatory (IM), mesenchymal (M), mesenchymal-stem like (MSL), luminal androgen receptor (LAR), and unstable (UNS) [4]. Colorectal cancer (CRC) is similarly deadly with a 5-year survival rate of 13% once the disease reaches distant organs. In recent decades, the incidence rate of colorectal cancer has been rising and it is currently the second leading cause of cancer deaths [5]. A better understanding of the molecular mechanisms of cancer cell responses to current treatment options is needed to improve outcomes for these patients.
Treatment options for BC include surgery, radiation, immunotherapy, hormone therapy, and chemotherapy. Factors such as type of BC and metastasis location determine the treatment options used [6]. Hormone therapy, such as estrogen blockers like tamoxifen, in combination with chemotherapy are used as first-line treatments on hormone receptor positive BC [7]. Tumor grade, estrogen receptor expression, and prognostic genes are all taken into consideration prior to deciding on a combined therapy approach for hormone receptor positive BC [8]. Similar to BC, treatment options for CRC include surgery, radiation, immunotherapy, and chemotherapy. 5-fluorouracil (5-FU) is a frontline therapy for CRC and is often combined with other chemotherapies like oxaliplatin and irinotecan (CPT-11) to improve outcomes in the clinic [9]. Some of the drugs used in CRC treatment overlap with drugs used in BC, yet there is heterogeneity in the response to these drugs across tissue types that is incompletely understood. The mechanisms of action for these therapeutics are detailed in Table 1. Further investigation could direct cancer-specific combination treatments and targeted therapies to limit side effects and inefficacy in patients.
Table 1.
Drugs used in the experiments and their mechanisms of action (drugs chosen are relevant to either, or both, breast and colorectal cancer)
| Drug | Mechanism | References |
|---|---|---|
| oxaliplatin | Platinum-based compound which can arrest DNA synthesis by binding to guanine and cytosine resulting in cross-linking | [34] |
| cisplatin | Platinum-based compound that cross links urine bases to form DNA adducts which induces apoptosis | [35] |
| carboplatin | Platinum-based compound that forms monoadducts by attaching alkyl groups to nucleotides resulting in DNA fragmenting | [36] |
| doxorubicin | Inhibits topoisomerase II resulting in DNA damage and apoptosis; is able to insert itself within DNA base pairs causing DNA strands to break | [37] |
| paclitaxel | Taxane compound that disrupts microtubule growth by preventing depolymerization so that the cell can no longer use its cytoskeleton | [38] |
| docetaxel | Taxane which is a derivative of paclitaxel and inhibits proper microtubule assembly resulting in cell cycle arrest | [39] |
| 5-FU | Inhibits the production of dTMP, which is necessary for DNA replication and repair, resulting in double-stranded breaks | [40] |
Cytokines regulate inflammatory responses that may promote or suppress cancer. The presence of cytokines can result in various effects ranging from apoptosis to promoting metastasis. Interleukins, such as IL-1, IL-6, and IL-15, have been shown to be involved in carcinogenesis [10]. There is much interest in the development of cytokine-based biomarkers in cancer [11]. Biomarkers play a role in diagnosis, prognosis, and determining responses to treatment. For example, HER2 and estrogen receptor expression are key biomarkers in BC that determine response to therapy [12]. Alterations in gene expression, protein expression, or metabolic signatures are considered to be biomarkers. One way to establish biomarkers is by analyzing transcripts that are induced by drugs in vitro [12]. Such work has been completed on the HCT116 CRC cell line, which established a dataset of prognostic drug-regulated gene signatures [13]. Evaluating cytokines and biomarkers is needed to better understand the tumor microenvironment and composition which could improve targeted therapies.
Here, we analyze and compare drug-regulated cytokines in BC and CRC and compare the prognostic value of drug-induced transcripts across BC and CRC. The findings contribute to our understanding of the molecular mechanisms of chemotherapy in cancer cells and in the tumor microenvironment across cancer type, aiding our overall goal of improving outcomes for BC and CRC patients.
Materials and methods
Cell lines and culture conditions
MCF7 human breast cancer cells (obtained from ATCC) were grown in DMEM media supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C, 5% CO2. HCT116 CRC cells were obtained from Bert Vogelstein at Johns Hopkins University.
Establishing IC50 doses
MCF7 cells were treated with doses ranging from 0-50 μM of cisplatin, oxaliplatin, 5-FU, carboplatin, paclitaxel, doxorubicin, or docetaxel for 72 hours in a 96-well plate. Cell viability was measured using an MTT assay and the IC50 dose was determined based on the dose response curve constructed from the collected data. HCT116 cells were treated with doses ranging from 0-80 μM of cisplatin, oxaliplatin, or 5-FU for 72 hours in a 96-well plate. Cell viability was measured using a CellTiterGlo assay and the IC50 dose was determined based on the dose response curve constructed from the collected data.
Cytokine profiling
A total of 5×105 MCF7 cells were plated per well of a 24-well plate and incubated for 12-16 hours prior to treatment with cisplatin, oxaliplatin, 5-FU, carboplatin, paclitaxel, doxorubicin, or docetaxel at the corresponding IC50 concentration. Cell medium was collected 24 hours after treatment and stored at -80°C. Samples were shipped to Brown University to run them in biological duplicate on a Luminex 200 Instrument (R&D LX200-XPON-RUO), which captures cytokines on magnetic antibody-coated beads and measures cytokine levels using a system based on the principles of flow cytometry (see https://www.luminexcorp.com/luminex-100200/#overview for more information). A custom 52-cytokine panel was split into 34-plex and 18-plex assays (R&D LXSAHM) and was run on the Luminex 200 instrument according to the manufacturer’s protocol.
TCGA analysis
Generation of prognostic CRC signatures (Table 3)
Table 3.
Prognostic Gene Signatures in CRC
| Prognostic Gene Signatures in CRC | ||
|---|---|---|
| Pan-drug | HIST2H2BA (H2BP1) | PLGLB2 |
| CDKN1A | HIST2H2BE (H2BC21) | PRR11 |
| FAS | HIST2H2BF (H2BC18) | RIT1 |
| GDF15 | HIST2H3A (H3C15); HIST2H3C (H3C14) | STAG1 |
| PPM1D | HIST2H3D (H3C13) | TMEM40 |
| SERPINB5 | HIST2H4A (H4C14); HIST2H4B (H4C15) | Oxaliplatin-specific |
| SESN2 | HIST2H4B (H4C15); HIST2H4A (H4C14) | ACTA2 |
| 5-FU-specific | HIST3H2BB (H2BU1) | LYPD3 |
| HIST1H1C (H1-2) | NEK2 | PHYHIP |
| HIST1H2AC (H2AC6) | SMAD6 | USP53 |
| HIST1H2AE (H2AC8) | CPT-11-specific | Cisplatin-specific |
| HIST1H2AL (H2AC16); HIST1H2BN (H2BC15) | ABHD4 | ATXN7L1 |
| HIST1H2BC (H2BC4) | ANKRA2 | CUX1 |
| HIST1H2BD (H2BC5) | BUB1 | OSGIN1 |
| HIST1H2BG (H2BC8) | CCNB1 | VAV2 |
| HIST1H4E (H4C5) | HPCAL1 | ZFAT |
| HIST1H4H (H4C8) | MXD4; MIR4800 | Cis/Ox-specific |
| HIST2H2AA3 (H2AC18); HIST2H2AA4 (H2AC19) | PI4K2A | SNAI1 |
| HIST2H2AA4 (H2AC19); HIST2H2AA3 (H2AC18) | PIP4K2A | |
Chemotherapy-inducible transcripts have varying potential for prognostic significance in CRC vs. BC. We evaluated previously established prognostic CRC gene signatures [13] for prognostic value in BC. Only one transcript, H2BC4, also had prognostic value (P-value <0.05) in BC (red, bolded text) according to analysis in cBioPortal.
Our lab previously generated prognostic drug-induced gene signatures in HCT116 colorectal cancer cells treated with cisplatin, oxaliplatin, irinotecan (CPT-11), or 5-FU. Briefly, this involved using cBioPortal and TCGA Colorectal Adenocarcinoma PanCancer Atlas database (containing 592 samples with RNA-seq data) to determine the difference in overall survival between groups of patients with low or high mRNA expression (<-1 or >1 standard deviation from the mean of all samples, respectively) of genes within the drug-induced gene signatures [13]. Here, we selected 45 genes from these previously established signatures which had high prognostic value (logrank P-value <0.05), along with an additional 5 genes that were included in the prognostic CRC signature after evaluation of existing literature, to generate a 50-gene prognostic CRC signature. We evaluated this 50-gene signature for prognostic value (logrank P-value <0.05) in breast cancer using cBioPortal. The TCGA Breast Invasive Carcinoma PanCancer Atlas database (containing 1082 samples with RNA-seq data) was used to determine the difference in overall survival between groups of patients with low or high mRNA expression (<-1 or >1 standard deviation from the mean of all samples, respectively) of genes within this 50-gene signature.
Generation of non-prognostic CRC signatures (Table 4)
Table 4.
Non-prognostic Gene Signatures in CRC
| Non-prognostic Gene Signatures in CRC | ||
|---|---|---|
| Pan-drug | FDXR | EGR1 |
| ATF3 | FHIT | ETS1 |
| BTG2 | FMNL2 | FAM214A |
| CD274 | FTO | FGD1 |
| DCP1B | G2E3 | FGD4 |
| E2F7 | GRHL3 | FOS |
| IER5 | HMGA2 | GLIPR1 |
| MDM2 | HS6ST2 | HOXB8 |
| PLK2 | IFIT3 | HS6ST1 |
| POLH | IFNAR2 | KLF6 |
| PRDM1 | IKBIP | KLK6 |
| RNF19B | IPO11; LRRC70 | KLK7 |
| SERTAD1 | IRS2 | KRTAP2-2 |
| SESN1 | ISG15 | LAMB3 |
| TOB1 | KIAA0040 | MAP1S |
| TP53INP1 | KITLG | MBD5 |
| TUBGCP3 | KSR2 | NKTR |
| 5-FU-specific | LAMP3 | NR4A3 |
| ARHGEF39 | LIN37 | NUAK1 |
| CCNG2 | LRBA | RBM24 |
| CENPA | LYNX1 | RIOK3 |
| DTL | MED13L | RNU11 |
| FAM83D | MGAT5 | SAT1 |
| HIST1H2AH (H2AC12) | MSI2 | SHFM1 (SEM1) |
| HIST1H2AJ (H2AC14) | NBAS | SRSF5 |
| HIST1H2BB (H2BC3) | NTPCR | SUSD2 |
| HIST1H2BF (H2BC7) | OR51B5 | TGM2 |
| HIST1H2BH (H2BC9) | OR51I1 | THEM6 |
| HIST1H2BK (H2BC12) | PARD6G | TIGAR |
| HIST1H2BO (H2BC17) | PDLIM3 | TUFT1 |
| HIST1H3A (H3C1) | PIBF1 | ULBP1 |
| HIST1H3B (H3C2) | PPP3CA | VCAN |
| HIST1H3F (H3C7) | PRRG4 | Cisplatin-specific |
| HIST1H3G (H3C8) | PTCH1 | ADAM22 |
| HIST1H4A (H4C1) | PTK2 | ARHGEF3 |
| HIST1H4B (H4C2) | PTPRO | ARID5B |
| HIST1H4D (H4C4) | RAB27B | ATXN1 |
| HIST1H4K (H4C12) | RAB33B | BAZ2B |
| HIST2H2AB (H2AC21) | RABGAP1L | BTBD9 |
| HIST2H2AC (H2AC20) | RAD51B | CDKL5 |
| HIST3H2A (H2AW) | RAET1G | CMIP |
| INCENP | RASA1 | DOCK4 |
| KIRREL | REEP1 | EXT1 |
| MYBL1 | RGS5 | FAM217B |
| PLAU | RHOBTB3 | FUT8 |
| PSMC3IP | RINL | GBX2 |
| RASSF6 | RPS27L | GTF2IRD1 |
| SOX4 | RRM2B | IGF2BP3 |
| TRAF4 | RSRC1 | JARID2 |
| TRIM59 | SDC1 | KAT6B |
| TTK | SFN | KDM4C |
| ZNF658 | SLC29A3 | KLF12 |
| CPT-11-specific | SMAD3 | KLHL29 |
| ABCA12 | SPAG5 | MAML2 |
| AKR1B15 | STAT4 | MYO1E |
| APAF1 | STK17A | NFKBIA |
| APOBEC3C | STK39 | NR6A1 |
| APOBEC3H | TBL1X | OPHN1 |
| ARHGAP10 | TCP11L1 | OR2M3 |
| BAG1 | THUMPD3-AS1 | OR8D1 |
| BCKDHB | TLR3 | PHLPP1 |
| BIRC3 | TMEM63B | PITPNC1 |
| CAMK2D | TNFRSF10C | PRKCE |
| CCDC91 | TP53I3 | PRPF39 |
| CCNB2 | TP73 | PTPRM |
| CDKAL1 | TRIO | SIPA1L3 |
| CDKL2 | TRPM6 | SSH2 |
| CITED2 | VPS13B | TANC1 |
| CMBL | VTI1A | TANC2 |
| CNNM4 | WDR63 (DNAI3) | TBC1D22A |
| COG5 | XPC | TBX3 |
| COL17A1 | XRCC4 | TESK1 |
| CTBP2 | ZNF672 | TIAM1 |
| DEPDC7 | ZNF804A | TMEM133 (ARHGAP42) |
| DIAPH2 | ZNF823 | TNS3 |
| DOCK3 | Oxaliplatin-specific | TSPAN18 |
| DRAM1 | AEN | UVRAG |
| DST | AHNAK2 | ZNF407 |
| DYM | C3orf14 | ZNF426 |
| EFNB1 | CD22; MIR5196 | CisOx-specific |
| EHBP1 | CSRNP1 | ARRDC4 |
| EPN3 | CTSS | GADD45A |
| EXOC6B | DUSP1 | LPAR6 |
| FAF1 | DUSP19 | SLC10A5 |
| ZRSR2 | ||
Non-prognostic CRC signatures have prognostic value in BC. Using drug-induced gene signatures previously established in the lab [13], we established non-prognostic (P-value >0.2) CRC gene signatures using cBioPortal. While these signatures were non-prognostic in CRC, several transcripts were prognostic (P-value <0.05) in BC (red, bolded text).
Using drug-induced gene signatures previously established in the lab [13], we established non-prognostic (P-value >0.2) drug-induced gene signatures in CRC using cBioPortal (analysis as described above). We evaluated this non-prognostic gene signature for prognostic significance (P-value <0.05) in BC.
Statistical analysis
MTT assay
Mean and standard deviation were calculated for each drug treatment concentration.
TCGA analysis
Samples selected in cBioPortal had mRNA expression z-scores relative to all samples. Logrank test P-values were considered to be significant if they were below 0.05.
Cytokine profiling
The mean of biological duplicates was used as a final fold-change value.
Results
Differential sensitivity of MCF7 BC cells to cytotoxic chemotherapeutic drugs
IC50 concentrations were established in the MCF7 cell line for doxorubicin (1.42 μM), paclitaxel (1.59 μM), oxaliplatin (2.7 μM), docetaxel (9.93 μM), 5-fluorouracil (5-FU) (10 μM), cisplatin (29.5 μM), and carboplatin (50 μM) using an MTT assay (Figure 1A-C). The established IC50 concentrations were used for subsequent cytokine profiling experiments. In the HCT116 cell line, IC50 concentrations were established for 5-FU (9.8 μM), oxaliplatin (2 μM), and cisplatin (14.8 μM) using a CellTiterGlo assay (Figure 1D).
Figure 1.
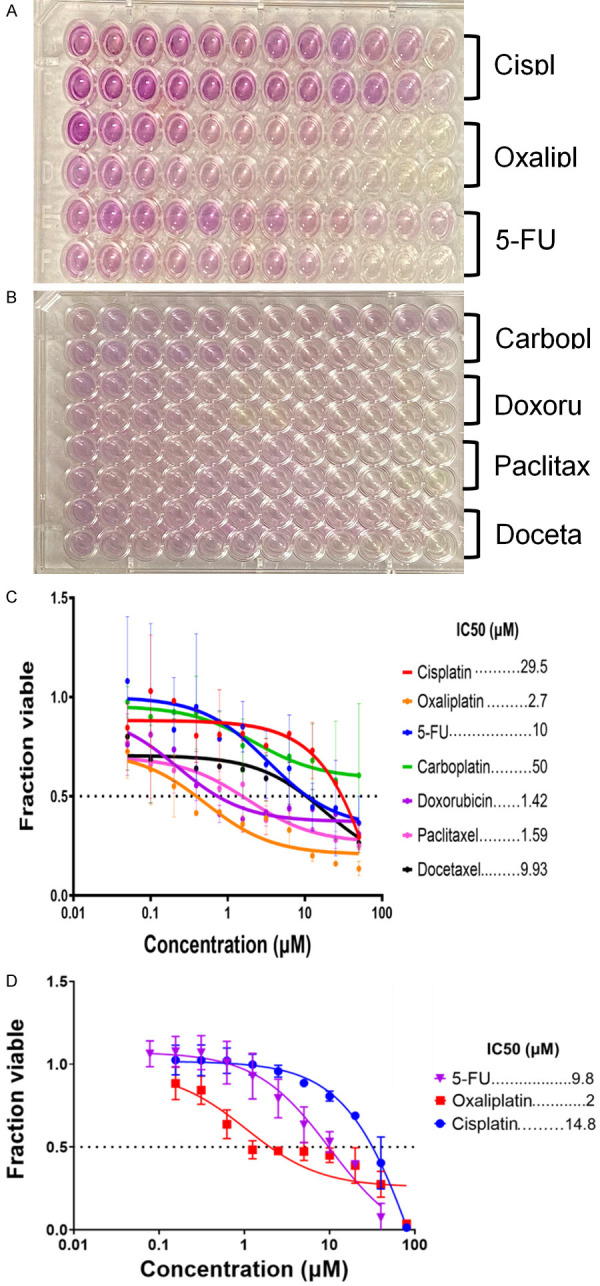
The concentration of each drug needed to kill half of the cells. Images of MCF7 MTT assay plates (A, B) and corresponding IC50 curves generated in GraphPad Prism (C). MCF7 cells were least sensitive to carboplatin and cisplatin and most sensitive to oxaliplatin, doxorubicin, and paclitaxel. The IC50 concentrations were used for treatment of cells for cytokine profiling. IC50 concentrations for HCT116 cells were measured using a CellTiterGlo assay (D).
TRAIL-R2, IFN-β, Chitinase 3-like 1, and TRAIL are the only cytokines out of a panel of 52 that were regulated by a majority of clinically used drugs in BC
Evaluating cytokine production post-drug treatment can reveal information about drug mechanisms and how they mediate pro- and anti-cancer responses in cancer cells. To investigate this, we treated MCF7 BC cells with oxaliplatin, cisplatin, 5-FU, doxorubicin, paclitaxel, docetaxel, or carboplatin at their respective IC50 concentrations, harvested the cell supernatant after 24 hours, and analyzed levels of 52 cytokines using Luminex 200 Instrumentation. The function and impact on survival of these cytokines in BC patients based on existing literature is outlined in Table 2.
Table 2.
Cytokine function
| Cytokine | Function | Predicted Impact on Patient Survival | Reference |
|---|---|---|---|
| 4-1BB/TNFRSF9/CD137 | promotes the movement of monocytes/macrophages to the tumor microenvironment and promotes metastasis of breast cancer to bone | negative | [41] |
| BAFF/BLyS/TNFSF13B | increases stemness in epithelial breast cancer cells which promotes metastasis | negative | [23] |
| C-Reactive Protein/CRP | inflammatory protein that is produced in response to IL-6, elevated levels are associated with more aggressive tumors | negative | [42] |
| CCL2/JE/MCP-1 | recruits and activates tumor-related macrophages that promote tumor growth and angiogenesis | negative | [32] |
| CCL20/MIP-3 alpha | attracts CCR6 which allows breast cancer cells to break through the extracellular matrix resulting in angiogenesis and metastasis | negative | [43] |
| CCL22/MDC | associated with accumulation of tumor-associated macrophages | negative | [32,44] |
| CCL3/MIP-1 alpha | promotes breast cancer cell migration | negative | [32] |
| CCL4/MIP-1 beta | high expression promotes tumor development | negative | [32] |
| CCL5/RANTES | promotes growth, angiogenesis, and metastasis | negative | [20] |
| CCL7/MCP-3/MARC | promotes breast cancer cell proliferation and is involved in metastasis to bone | negative | [45] |
| CCL8/MCP-2 | recruits and activates tumor-related macrophages that promote tumor growth and angiogenesis | negative | [46] |
| Chitinase 3-like 1 | induces pro-angiogenic/pro-tumorigenic factors | negative | [14] |
| general role in cancer | |||
| CXCL10/IP-10/CRG-2 | inhibits tumor growth by inducing Granzyme B, mediates the NF-kB pathway | not determined | [20] |
| CXCL11/I-TAC | induces cancer cell migration | negative | [22] |
| general role in cancer | |||
| CXCL13/BLC/BCA-1 | increases cell proliferation, levels of IL-1, TNF, and TGF; decreases apoptosis | negative | [20] |
| CXCL14/BRAK | inhibits tumor angiogenesis, proliferation, and invasion of cancer cells | positive | [20] |
| CXCL5/ENA-78 | promotes tumor angiogenesis | negative | [20] |
| CXCL9/MIG | inhibits chemotaxis to endothelial cells | positive | [20] |
| Fas Ligand/TNFSF6 | induces apoptosis; cancer cells that are able to escape the effects of the immune system can use this cytokine to mediate their survival | not determined | [47] |
| general role in cancer | |||
| Fas/TNFRSF6/CD95 | involved in eliminating tumor-infiltrating immune cells and tissue destruction | negative | [48] |
| Ferritin | downregulation is associated with increased apoptosis | positive | [49] |
| general role in cancer | |||
| G-CSF | high levels can induce the formation of neutrophilic extracellular traps which promote breast cancer cell migration | negative | [50] |
| GM-CSF | inactivates VEGF and blocks angiogenesis | positive | [51] |
| Granzyme B | used by cytotoxic T lymphocytes and natural killer cells to induce cell death | positive | [52] |
| general role in cancer | |||
| IFN-alpha | induces the activation of the STAT1 pathway | negative | [53,54] |
| general role in cancer | |||
| IFN-β | can induce autophagy in response to IFN-β treatment | not determined | [15] |
| IFN-gamma | inhibits cell proliferation | positive | [55] |
| IFN-gamma R1/CD119 | inhibits cell proliferation | positive | [55] |
| IL-1 beta/IL-1F2 | makes the tumor more aggressive and invasive | not determined | [21] |
| IL-10 | high levels are associated with metastasis | not determined | [21] |
| IL-12/IL-23 p40 | promotes inflammation and angiogenesis | negative | [56] |
| IL-15 | increases and activates natural killer cells | not determined | [57] |
| IL-17/IL-17A | inhibits apoptosis through NF-kB activation, promotes angiogenesis, makes cells more aggressive | negative | [29,58] |
| IL-2 | high levels associated with more aggressive tumors | not determined | [21] |
| IL-21 | promotes proliferation and invasiveness of breast cancer cells | negative | [21] |
| IL-4 | induces apoptosis in breast cancer cells by producing macrophages and eosinophils | positive | [59] |
| IL-6 | decreases responsiveness to endocrine and chemotherapy | negative | [18] |
| IL-7 | promotes survival and growth of breast cancer cells | negative | [21] |
| IL-8/CXCL8 | low levels are related with lack of hormone receptors and metastasis | negative | [19] |
| M-CSF | promotes metastasis | negative | [51] |
| PD-L1/B7-H1 | associated with higher tumor grade and increased infiltration of T regulatory cells | negative | [60] |
| Prolactin | plays a role in tumorigenesis | negative | [61] |
| TNF-alpha | pro-tumorigenic and contributes to drug resistance development | negative | [62] |
| TRAIL R2/TNFRSF10B | low levels of membrane-bound associated with metastasis, migration, and invasion of cancer cells | negative | [63] |
| general role in cancer | |||
| TRAIL R3/TNFRSF10C | high levels of membrane-bound associated with metastasis | negative | [64] |
| TRAIL/TNFSF10 | selectively triggers cancer cell death | positive | [17] |
| general role in cancer | |||
| TRANCE/TNFSF11/RANKL | induced by progesterone resulting in the proliferation of mammary progenitor cells | not determined | [65] |
| TREM-1 | triggers amplification of inflammatory responses in the tumor | not determined | [66] |
| role in lung cancer | |||
| VEGF | high levels are associated with tumors that have large sizes, high histological grade | negative | [67] |
| VEGFR3/Flt-4 | promotes breast cancer cell proliferation and survival | negative | [68] |
A panel of 52 cytokines that mediate different pro- or anti- cancer effects were evaluated in the MCF7 and HCT116 cell lines. The function and impact on survival of these cytokines in BC patients based on existing literature is outlined here.
We observed that all of the drugs upregulated Chitinase 3-like 1 (CHI3L1) and TRAIL-R2 (Figure 2A). CHI3L1 induces pro-angiogenic/pro-tumorigenic factors and is associated with a poor prognosis in multiple cancer types including breast and colorectal cancer [14]. Soluble TRAIL-R2 cytokine can serve as a decoy receptor for death receptor 5 (DR5) to inhibit apoptosis and may indicate poor survival when expressed at high levels. Most of the drugs downregulated IFN-β, which is likely beneficial for survival of the patient due to its role in inducing autophagy in response to IFN-β treatment [15,16]. Most of the drugs also downregulated TRAIL, a pro-apoptotic cytokine [17]. In summary, each of the chemotherapeutic drugs regulated cytokines in a way that may exert both positive and negative effects on cancer progression.
Figure 2.
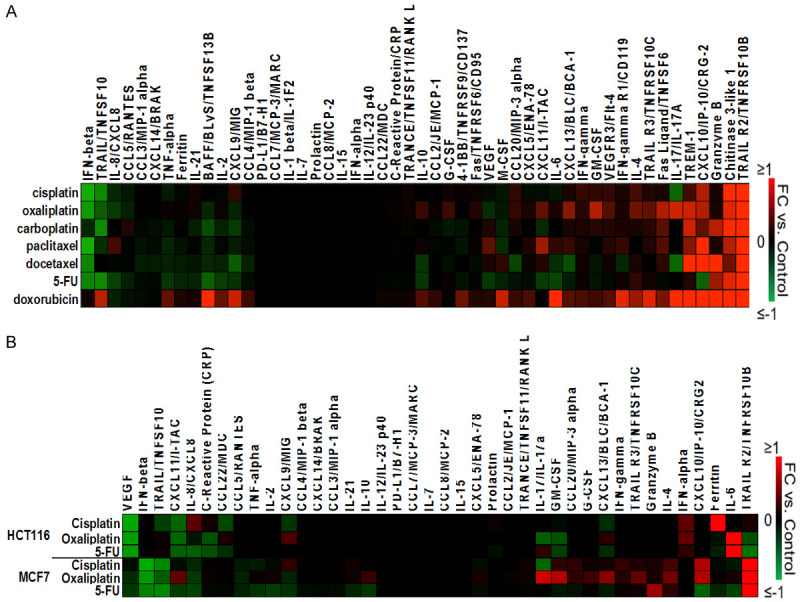
Regulation of cytokines by chemotherapy in the MCF7 breast cancer cell line and HCT116 colorectal cancer cell line. A. IC50 drug concentrations were used to treat MCF7 cells and conditioned media was collected for the profiling of 52 cytokines. B. Comparison of regulation of cytokines by chemotherapy between the MCF7 breast cancer cell line and the HCT116 colorectal cancer line. Respective IC50 drug concentrations were used to treat both cell lines and conditioned media was collected for the profile at 24 hours (MCF7) and at 48 hours (HCT116).
Identification of pan-drug mechanisms of efficacy point toward the development of potential targeted therapies, whereas identification of pan-drug mechanisms of acquired resistance point toward potential combination therapies to improve outcomes.
Drug-specific cytokine induction reveals possible mechanisms of efficacy, toxicity, and/or acquired resistance
In addition to the observed pan-drug cytokine regulation, there were also drug-specific effects, including across some drugs with identical primary targets or similar mechanisms of action (Figure 2A). These drug effects were mostly on cytokines belonging to the IL or CXCL family. Interleukins (IL) and CXCLs often exert a positive effect on cancer progression by mediating responses which aid proliferation.
Taxane-induced cytokine signatures
Docetaxel and Paclitaxel are both taxanes which share similar structure and mechanism of action. Not surprisingly, some cytokines were similarly regulated by these drugs including IFN-β (decreased), BAFF (decreased), CXCL10 (increased), and TRAIL-R2 (increased), although these cytokines were also similarly regulated by other drugs without similar mechanisms of action such as cisplatin and oxaliplatin.
We also noted some striking differences in regulation of cytokines by docetaxel vs. paclitaxel such as TRAIL, IL-8, CXCL11, IL-17, and Granzyme B. Interestingly, the downregulation of IL-17 was observed after treatment with docetaxel and cisplatin, but this effect did not extend to paclitaxel although this drug shares a similar mechanism to docetaxel (Figure 2A). IL-17 is needed for the tumor-promoting activity of myeloid-derived suppressor cells, thus cisplatin and docetaxel may have a unique ability to inhibit the pro-cancer effects of these cells [18].
Also surprising was the observation that paclitaxel was the only drug to upregulate IL-8/CXCL8, a cytokine associated with metastasis and poor prognosis in breast cancer [19,20] (Figure 2A). Although docetaxel and paclitaxel belong to the same class of drugs, their cytokine regulation showed that there may be distinctions in how they mediate their effects.
Platinum-based drug-induced cytokine signatures
Carboplatin, cisplatin, and oxaliplatin share similar platinum-based structures and similar mechanisms of action. Not surprisingly, these drugs had similar effects on some cytokines such as TRAIL (decreased), TRAIL-R2 (increased), Chitinase 3-like 1 (increased), and CXCL10 (increased). However, these drugs had divergent effects on several cytokines in the panel. For example, IL-17 was downregulated by cisplatin, upregulated by oxaliplatin, and not changed by carboplatin. As IL-17 is needed for the tumor-promoting activity of myeloid-derived suppressor cells [18], this observation may indicate an opposite effect on this cell population by cisplatin and oxaliplatin. While carboplatin and cisplatin had no effect on IL-10 and CXCL13 levels, oxaliplatin upregulated these cytokines (Figure 2A). High levels of IL-10 correlate with metastasis and CXCL13 increases BC cell proliferation, indicating a possible pro-cancer effect of oxaliplatin that does not extend to carboplatin or cisplatin [19-21]. CXCL11/I-TAC, responsible for the migration of cancer [22], was upregulated by oxaliplatin but downregulated by carboplatin. Oxaliplatin may pose more harm than carboplatin since it is able to exert more tumor-enhancing effects through several cytokines. Again, despite having similar structures and mechanisms, these drugs were able to have individual effects on some cytokines.
5-FU-specific cytokine regulation
5-FU was the only drug to downregulate CXCL10 (Figure 2A), a cytokine that is involved in the NF-kB pathway and which promotes angiogenesis, suggesting a 5-FU-specific mechanism of cancer inhibition. However, CXCL10 can also exert an anti-tumorigenic role by inducing Granzyme B production [20], this upregulation or downregulation may pose a beneficial effect that is likely context dependent.
Doxorubicin-specific cytokine induction
Interestingly, doxorubicin did not downregulate any cytokines in the panel but did upregulate several (Figure 2A). Cytokines uniquely upregulated by doxorubicin include BAFF, CXCL9, and IL-6. BAFF has been shown to increase stemness in epithelial breast cancer cells which is associated with metastasis [23]. IL-6 is responsible for stimulating the growth and invasiveness of MCF7 cells and is involved in the activation of the NF-kB pathway which promotes proliferation and survival of malignant cells as well as anti-estrogen resistance [18].
The promotion of anti-estrogen resistance by doxorubicin through the IL-6 cytokine may suggest that this drug would not be suitable in combination with hormone therapy. Unlike BAFF and IL-6, CXCL9 inhibits chemotaxis to endothelial cells which has a positive prognosis in cancer [20]. Still, the positive role of CXCL9 may not outweigh the harms of BAFF and IL-6.
These observations suggest that patients treated with doxorubicin may benefit further from combination therapies to mitigate these potentially harmful effects. Together, we identify drug-specific effects which enhance our understanding of how these drugs function, directing the development of novel treatment combinations to enhance efficacy or avoid toxicity.
Comparison of drug-induced cytokine signatures across CRC and BC reveal tissue-specific drug effects
It is recognized that widely used cytotoxic chemotherapies such as 5-FU, oxaliplatin, and cisplatin have tissue-specific effects however it remains less clear if these effects extend to cytokine regulation and/or secretion. To investigate this, we measured cytokine levels in cisplatin, oxaliplatin, or 5-FU-treated HCT116 colorectal cancer cells (48-hour treatment) and compared them to drug-induced cytokine levels in MCF7 cells (24-hour treatment) [13].
We observed many differences across cell line, including enhanced downregulation of VEGF and CXCL11 in the HCT116 cells, enhanced TRAIL-R2 upregulation in the MCF7 cells, and enhanced downregulation of IFN-β and TRAIL in the MCF7 cells (Figure 2B).
A limitation of this experiment is that cytokines were measured at different time points post-treatment and it is possible that some cytokines are differentially regulated at 24 hours compared to 48 hours. However, it is likely that striking differences such as downregulation IFN-β and TRAIL, and upregulation of TRAIL-R2, in the MCF7 cell line at 24 hours may extend to 48 hours.
Overall, these findings indicate that MCF7 cells are more prone to chemotherapy-mediated downregulation of TRAIL and upregulation of TRAIL-R2 by cisplatin, oxaliplatin, and 5-FU, both of which are likely to have pro-tumor effects. Future directions could include investigation across other cell types and with other clinically relevant drugs used for treatment of BC.
Chemotherapy-inducible transcripts have varying potential for prognostic significance in CRC versus BC
Our analysis of CRC and BC cytokine regulation after drug treatment revealed some similar drug effects across cancer type, but also revealed some tissue-specific effects of cisplatin, oxaliplatin, and 5-FU. Based on this, we hypothesized that a database previously established in the lab containing prognostic drug-induced gene signatures in HCT116 colorectal cancer cells treated with cisplatin, oxaliplatin, irinotecan (CPT-11), or 5-FU would contain some genes that were similarly prognostic in breast cancer, and some genes that were strikingly different across tissue type [13] (Tables 3 and 4). To investigate this, we used the computational tool cBioPortal and the TCGA Breast Invasive Carcinoma PanCancer Atlas database (containing 1082 samples with RNA-seq data) to determine the difference in overall survival between groups of patients with low or high mRNA expression (<-1 or >1 standard deviation from the mean of all samples, respectively) of genes within this CRC database. Genes were considered prognostic if the log rank P-value was below 0.05.
Only 1 gene (H2BC4) from a list of 50 that were prognostic in CRC (Table 3) was also prognostic in BC (Figure 3), indicating that though some chemotherapy-induced genes play a role in patient survival across tissue type, most are tissue-specific. High levels of H2BC4 correlate with better survival in BC while low levels correlate with better survival in CRC (Figure 3), further demonstrating this tissue specificity.
Figure 3.
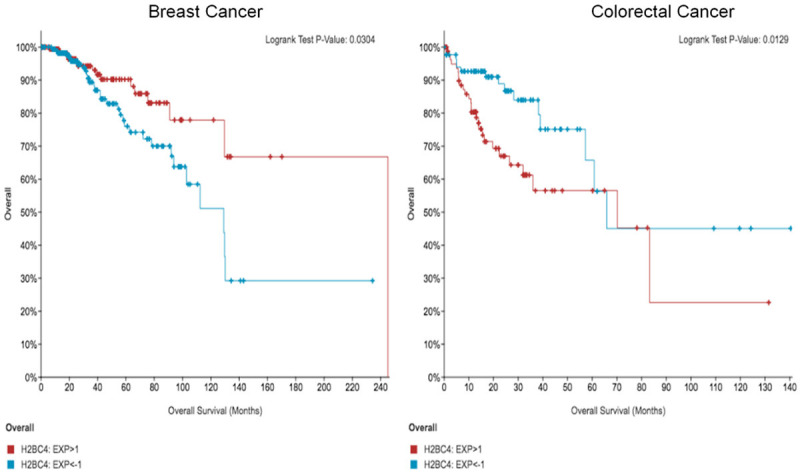
One gene (H2BC4) from a prognostic gene signature in CRC is also prognostic in BC. Kaplan-Meier curves correlate with the bolded gene found in Table 3. High levels of H2BC4 correlate with better survival in BC while low levels correlate with better survival in CRC, suggesting chemotherapy-induced genes play a tissue-specific role in patient survival.
We also established non-prognostic drug-induced CRC gene signatures and evaluated them for prognostic value in BC (Table 4). We found that among the non-prognostic CRC genes, 25 genes were prognostic in BC (Figure 4). This suggests, perhaps unsurprisingly, that there is variation in the contribution of drug-regulated genes to patient survival across organ type.
Figure 4.
A subset of a non-prognostic gene signature in CRC is prognostic in BC. Kaplan-Meier curves correlate with the genes found in Table 4.
Two of the genes that were not prognostic in CRC but were prognostic in BC are NFKBIA and GADD45A (Figure 5). In line with our finding that higher expression levels correlate with better survival, NFKBIA is responsible for inhibiting the NFKB pathway which promotes proliferation of malignant cells [24] and higher levels of GADD45A are associated with increased sensitivity to docetaxel and paclitaxel. Furthermore, NFKB activity inhibits the GADD45 promoter, thus suppressing this pathway’s activity also aids with overcoming drug resistance [25].
Figure 5.
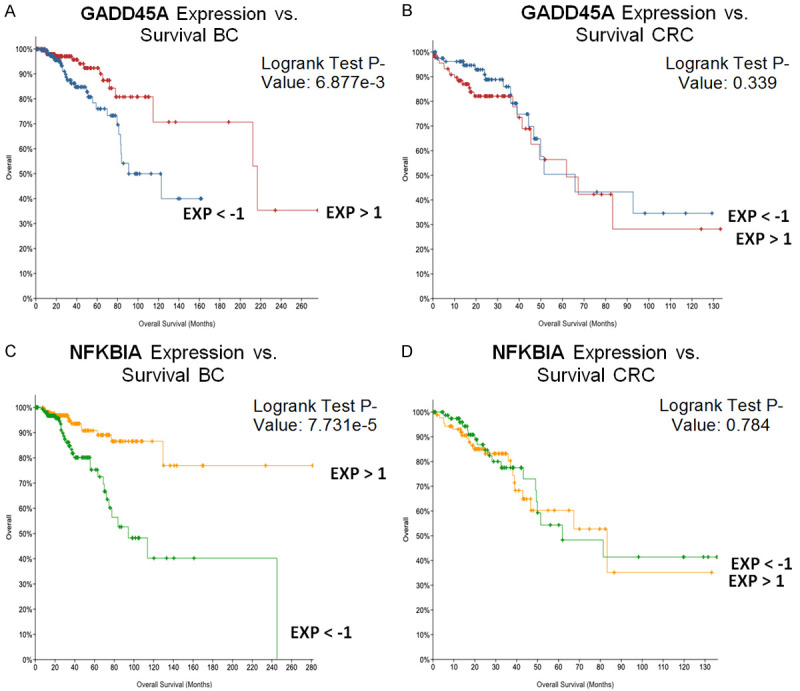
Upregulation of GADD45A and NFKBIA is associated with better overall survival in breast cancer but not colorectal cancer. High expression of GADD45A is associated with better survival in BC (A) but not CRC (B). High expression of NFKBIA correlates with better survival in BC (C) but not CRC (D).
The drug-specific prognostic genes between the two cancers vary due to the heterogeneity in the drug response. These genes should be further evaluated in drug-treated MCF7 cells, to confirm tissue-specific prognostic gene signatures, because we only previously tested their regulation in CRC.
Discussion
Immune responses can play a key role in both cancer development and tumor suppression, and these responses are in part regulated by the identity and magnitude of cytokines present and secreted by cancer cells. We analyzed cytokine responses of MCF7 BC cells to multiple clinically used chemotherapeutic agents in order to gain a better understanding how these drugs mediate anti- or pro- cancer responses.
When treating luminal breast cancer, chemotherapy is used in combination with hormone therapy to improve outcomes [6]. It is possible that additional combinations to combat the upregulation of harmful cytokines, like CHI3L and TRAIL-R2 (upregulated by all seven chemotherapeutic agents investigated here) could even further improve outcomes.
The mechanism by which TRAIL-R2 is increased is unclear. While we previously discovered that TRAIL-R2 (DR5) is a p53 target gene and MCF7 cells have wild-type p53, the soluble TRAIL-R2 may be increased due to the activity of proteases [26]. Proteases are increased in the tumor microenvironment and may oppose the innate immune system in killing BC cells through increase in soluble TRAIL-R2 [27]. It is interesting that in HCT116 colorectal cancer cells, TRAIL-R2 was decreased in the presence of oxaliplatin and 5-FU. While it is well-documented that TRAIL-R2 on the cell surface is upregulated after treatment with DNA-damaging agents in a p53-dependent manner, the effects of these compounds on soluble TRAIL-R2 are much less studied. It is possible that HCT116 and MCF7 have differential protease activity which impacts on the drug effects on soluble TRAIL-R2.
Paclitaxel and docetaxel have similar mechanisms of action, yet they had different effects on cytokine levels. Previous clinical studies have shown that early luminal A breast cancer survival does not benefit from taxane (paclitaxel, docetaxel) addition to hormone therapy but taxane addition does benefit metastatic luminal BC survival [28]. Paclitaxel may pose more harm than docetaxel since it upregulated multiple harmful cytokines (IL-6, CXCL11, IL-8, etc.) that docetaxel either downregulated or had no effect on. Elevated levels of IL-17A have been associated with protection from docetaxel-induced death in MCF7 cells [29], however in the profile docetaxel was able to downregulate this cytokine, suggesting that docetaxel may be effective at overcoming the barrier IL-17 poses to cell death.
Tamoxifen-resistant MCF7 cells have shown increased sensitivity to 5-FU compared to wild-type MCF7 cells [30]. Our investigation revealed that 5-FU downregulated many cytokines that aid cancer progression, further supporting the use of 5-FU after patients have become resistant to tamoxifen.
Protein expression of erbB-2 (HER2) is a predictor of cells’ responsiveness to doxorubicin. Overexpression of this protein is correlated with increased topoisomerase-IIa expression, a target for doxorubicin action [31]. While MFC7 is a HER2 negative cancer, doxorubicin still induced robust cell death and cytokine induction. Doxorubicin may not be the most effective agent for treating luminal breast cancer. The status of erbB-2 may not be the reason for doxorubicin’s lack of effectiveness since the cells were sensitive to the drug. Doxorubicin could contribute to mechanisms of acquired resistance through the upregulation of many harmful cytokines. The apparent unique inability of doxorubicin to down-regulate any cytokines in the panel could indicate a doxorubicin-specific effect in which harmful cytokines are more likely to be upregulated compared to the other drugs.
While cisplatin, oxaliplatin, and carboplatin are all platinum-based drugs that function similarly, they had distinct effects on the cytokine panel. Cisplatin had a unique effect on the IL-17 cytokine that only docetaxel was also able to have. Carboplatin and oxaliplatin had many opposing effects on the panel. The mechanism of each drug may ultimately distinguish their pathway of action to yield varying results in the cell, and the results suggest there may be differences in structurally related drugs.
Genes from the prognostic and non-prognostic CRC signatures that had prognostic significance in BC survival could be further evaluated in drug-treated BC cells. Our cBioPortal analysis found that differential regulation of these genes impacted patient survival differently across cancer type. Most of the genes that were prognostic in CRC were not prognostic in BC. On the other hand, several non-prognostic CRC genes were prognostic in BC. Validation of the regulation of these genes by drugs in BC cells could aid in their development as predictive or prognostic biomarkers. Furthermore, these genes should be assessed in other relevant cancers, for instance, endometrial cancer, to further build upon their prognostic value.
The inflammatory results of the NFKB pathway, mediated by cytokines which include IL-1, IL-6, and IL-12, progress the development of breast cancer thus the NFKBIA gene has a positive effect on survival by inhibiting this [24,32]. GADD45A can serve as a potential target to further study the effects of taxane therapy on luminal breast cancer. Understanding taxane effects may create better guidelines on when it should be implemented in the treatment of advanced or metastatic luminal BC.
The lack of treatment information in the TCGA database is a limitation of this study. Without treatment information, we cannot fully understand the prognostic significance in terms of the drugs that are used for patient treatment. Exploring the effects of these drugs in other BC cell lines and subtypes could create a better understanding of the tumor environment and which chemotherapies are more effective for each subtype.
Other future directions include using a hormone therapy in the study as well as evaluating drug combinations. Tamoxifen is a hormone therapy that is commonly used in luminal breast cancer treatment [33]. Having a better understanding of the tamoxifen immune profile in MCF7 cells may reveal more about what is responsible for the efficacy and toxicity of this treatment. In the clinic, these chemotherapies are not used as single agents, which is why it is important to evaluate what immune response combinations of drugs yield in the cancer cells. Understanding the signals involved in drug responses can lead to the development of novel drug combinations.
Lastly, it will also be vital to analyze the gene signatures and cytokines in other preclinical models such as patient-derived or transgenic organoids as well as patient tissue and blood samples. These models could address the complexity of intra- and inter-tumor heterogeneity. This work should involve evaluation across BC and CRC subtypes to further address tumor heterogeneity.
In summary, we establish a 7-drug, 52-cytokine signature in BC cells and a 3-drug, 40-cytokine signature in CRC cells that suggest drug-specific and tissue-specific cytokine regulation. Differences across prognostic gene signatures in BC and CRC further support tissue specificity in impact of gene expression on patient survival.
Acknowledgements
W.S.E-D. is an American Cancer Society Research Professor and is supported by the Mencoff Family University Professorship at Brown University. This work was supported by the Teymour Alireza P’98, P’00 Family Cancer Research Fund established by the Alireza Family. L.G. contributed to the research as part of the Leadership Alliance National Consortium at Brown University during the summer of 2021. Laboratory facilities at Brown University (W.S.E-D.) and at Cornell University (W.H.S.) were used to complete the experiments. Due to the COVID pandemic that continued during the summer of 2021, students participating in the Leadership Alliance Program were not able to be physically present at the laboratory facilities at Brown University. The work was conducted under the guidance of W.S.E-D. at Brown University. The co-authors are grateful to W.H.S. for allowing L.G. to perform the MCF7 cell culture, cytotoxicity assays and harvesting of samples in connection with this work in her laboratory at Cornell University.
Disclosure of conflict of interest
None.
References
- 1.Cancer stat facts: female breast cancer. NIH NCI surveillance, epidemiology, and end results program. https://seer.cancer.gov/statfacts/html/breast.html.
- 2.Breast cancer-metastatic: statistics. Cancer.Net, 2021. https://www.cancer.net/cancer-types/breast-cancer-metastatic/statistics.
- 3.Molecular subtypes of breast cancer. Breast Cancer Org: https://www.breastcancer.org/symptoms/types/molecular-subtypes.
- 4.Marra A, Trapani D, Viale G, Criscitiello C, Curigliano G. Practical classification of triple-negative breast cancer: intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. NPJ Breast Cancer. 2020;6:54. doi: 10.1038/s41523-020-00197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89–103. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gradishar WJ, Anderson BO, Abraham J, Aft R, Agnese D, Allison KH, Blair SL, Burstein HJ, Dang C, Elias AD, Giordano SH, Goetz MP, Goldstein LJ, Isakoff SJ, Krishnamurthy J, Lyons J, Marcom PK, Matro J, Mayer IA, Moran MS, Mortimer J, O’Regan RM, Patel SA, Pierce LJ, Rugo HS, Sitapati A, Smith KL, Smith ML, Soliman H, Stringer-Reasor EM, Telli ML, Ward JH, Young JS, Burns JL, Kumar R. Breast cancer, version 3.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2020;18:452–478. doi: 10.6004/jnccn.2020.0016. [DOI] [PubMed] [Google Scholar]
- 7.Hormone therapy for breast cancer. American Cancer Society: https://www.cancer.org/cancer/breast-cancer/treatment/hormone-therapy-for-breast-cancer.html.
- 8.Gao JJ, Swain SM. Luminal A breast cancer and molecular assays: a review. Oncologist. 2018;23:556–565. doi: 10.1634/theoncologist.2017-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020;5:22. doi: 10.1038/s41392-020-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121:2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 11.Kartikasari AER, Huertas CS, Mitchell A, Plebanski M. Tumor-induced inflammatory cytokines and the emerging diagnostic devices for cancer detection and prognosis. Front Oncol. 2021;11:692142. doi: 10.3389/fonc.2021.692142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry NL, Hayes DF. Cancer biomarkers. Mol Oncol. 2012;6:140–146. doi: 10.1016/j.molonc.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlsen L, Schorl C, Huntington K, Hernandez-Borrero L, Jhaveri A, Zhang S, Zhou L, El-Deiry WS. Pan-drug and drug-specific mechanisms of 5-FU, irinotecan (CPT-11), oxaliplatin, and cisplatin identified by comparison of transcriptomic and cytokine responses of colorectal cancer cells. Oncotarget. 2021;12:2006–2021. doi: 10.18632/oncotarget.28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Libreros S, Garcia-Areas R, Iragavarapu-Charyulu V. CHI3L1 plays a role in cancer through enhanced production of pro-inflammatory/pro-tumorigenic and angiogenic factors. Immunol Res. 2013;57:99–105. doi: 10.1007/s12026-013-8459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambjørn M, Ejlerskov P, Liu Y, Lees M, Jäättelä M, Issazadeh-Navikas S. IFNB1/interferon-β-induced autophagy in MCF-7 breast cancer cells counteracts its proapoptotic function. Autophagy. 2013;9:287–302. doi: 10.4161/auto.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He W, Wang Q, Xu J, Xu X, Padilla MT, Ren G, Gou X, Lin Y. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2- and RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy. 2012;8:1811–1821. doi: 10.4161/auto.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mérino D, Lalaoui N, Morizot A, Solary E, Micheau O. TRAIL in cancer therapy: present and future challenges. Expert Opin Ther Targets. 2007;11:1299–1314. doi: 10.1517/14728222.11.10.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esquivel-Velázquez M, Ostoa-Saloma P, Palacios-Arreola MI, Nava-Castro KE, Castro JI, Morales-Montor J. The role of cytokines in breast cancer development and progression. J Interferon Cytokine Res. 2015;35:1–16. doi: 10.1089/jir.2014.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin AR, Wang SE. Cytokines driving breast cancer stemness. Mol Cell Endocrinol. 2014;382:598–602. doi: 10.1016/j.mce.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Yang Z, Lu W, Chen Z, Chen L, Han S, Wu X, Cai T, Cai Y. Chemokines and chemokine receptors: a new strategy for breast cancer therapy. Cancer Med. 2020;9:3786–3799. doi: 10.1002/cam4.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fasoulakis Z, Kolios G, Papamanolis V, Kontomanolis EN. Interleukins associated with breast cancer. Cureus. 2018;10:e3549. doi: 10.7759/cureus.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puchert M, Obst J, Koch C, Zieger K, Engele J. CXCL11 promotes tumor progression by the biased use of the chemokine receptors CXCR3 and CXCR7. Cytokine. 2020;125:154809. doi: 10.1016/j.cyto.2019.154809. [DOI] [PubMed] [Google Scholar]
- 23.Pelekanou V, Notas G, Athanasouli P, Alexakis K, Kiagiadaki F, Peroulis N, Kalyvianaki K, Kampouri E, Polioudaki H, Theodoropoulos P, Tsapis A, Castanas E, Kampa M. BCMA (TNFRSF17) induces APRIL and BAFF mediated breast cancer cell stemness. Front Oncol. 2018;8:301. doi: 10.3389/fonc.2018.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura RE, de Vasconcellos JF, Sarkar D, Libermann TA, Fisher PB, Zerbini LF. GADD45 proteins: central players in tumorigenesis. Curr Mol Med. 2012;12:634–651. doi: 10.2174/156652412800619978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu GS, Burns TF, McDonald ER 3rd, Jiang W, Meng R, Krantz ID, Kao G, Gan DD, Zhou JY, Muschel R, Hamilton SR, Spinner NB, Markowitz S, Wu G, el-Deiry WS. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 27.Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukada I, Araki K, Kobayashi K, Kobayashi T, Horii R, Akiyama F, Takahashi S, Iwase T, Ito Y. Therapeutic effect of taxanes on metastatic breast cancer of various immunohistochemical subtypes. Oncol Lett. 2016;12:663–669. doi: 10.3892/ol.2016.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cochaud S, Giustiniani J, Thomas C, Laprevotte E, Garbar C, Savoye AM, Curé H, Mascaux C, Alberici G, Bonnefoy N, Eliaou JF, Bensussan A, Bastid J. IL-17A is produced by breast cancer TILs and promotes chemoresistance and proliferation through ERK1/2. Sci Rep. 2013;3:3456. doi: 10.1038/srep03456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe T, Oba T, Tanimoto K, Shibata T, Kamijo S, Ito KI. Tamoxifen resistance alters sensitivity to 5-fluorouracil in a subset of estrogen receptor-positive breast cancer. PLoS One. 2021;16:e0252822. doi: 10.1371/journal.pone.0252822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paik S, Bryant J, Park C, Fisher B, Tan-Chiu E, Hyams D, Fisher ER, Lippman ME, Wickerham DL, Wolmark N. erbB-2 and response to doxorubicin in patients with axillary lymph node-positive, hormone receptor-negative breast cancer. J Nat Cancer Inst. 1998;90:1361–1370. doi: 10.1093/jnci/90.18.1361. [DOI] [PubMed] [Google Scholar]
- 32.Ali S, Lazennec G. Chemokines: novel targets for breast cancer metastasis. Cancer Metastasis Rev. 2007;26:401–420. doi: 10.1007/s10555-007-9073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang KT, Kim EK, Jung SH, Lee ES, Kim SI, Lee S, Park HK, Kim J, Oh S, Kim YA Korean Breast Cancer Society. Tamoxifen therapy improves overall survival in luminal A subtype of ductal carcinoma in situ: a study based on nationwide Korean Breast Cancer Registry database. Breast Cancer Res Treat. 2018;169:311–322. doi: 10.1007/s10549-018-4681-6. [DOI] [PubMed] [Google Scholar]
- 34.Alcindor T, Beauger N. Oxaliplatin: a review in the era of molecularly targeted therapy. Curr Oncol. 2011;18:18–25. doi: 10.3747/co.v18i1.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aldossary AS. Review on pharmacology of cisplatin: clinical use, toxicity and mechanism of resistance of cisplatin. Biomed Pharmacol J. 2019:12. [Google Scholar]
- 36.Carboplatin. DrugBank Online, https://go.drugbank.com/drugs/DB00958.
- 37.StatPearls Publishing; 2020. Jul 26, Doxorubicin. https://www.ncbi.nlm.nih.gov/books/NBK459232/ [Google Scholar]
- 38.Paclitaxel. DrugBank Online, https://go.drugbank.com/drugs/DB01229.
- 39.StatPearls Publishing; 2020. Dec 2, Docetaxel. https://www.ncbi.nlm.nih.gov/books/NBK537242/ [Google Scholar]
- 40.StatPearls Publishing; 2021. Jul 18, Fluorouracil. https://www.ncbi.nlm.nih.gov/books/NBK549808/ [Google Scholar]
- 41.Jiang P, Gao W, Ma T, Wang R, Piao Y, Dong X, Wang P, Zhang X, Liu Y, Su W, Xiang R, Zhang J, Li N. CD137 promotes bone metastasis of breast cancer by enhancing the migration and osteoclast differentiation of monocytes/macrophages. Theranostics. 2019;9:2950–2966. doi: 10.7150/thno.29617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asegaonkar SB, Asegaonkar BN, Takalkar UV, Advani S, Thorat AP. C-reactive protein and breast cancer: new insights from old molecule. Int J Breast Cancer. 2015;2015:145647. doi: 10.1155/2015/145647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osuala KO, Sloane BF. Many roles of CCL20: emphasis on breast cancer. Postdoc J. 2014;2:7–16. [PMC free article] [PubMed] [Google Scholar]
- 44.Sun J, Sun J, Song B, Zhang L, Shao Q, Liu Y, Yuan D, Zhang Y, Qu X. Fucoidan inhibits CCL22 production through NF-κB pathway in M2 macrophages: a potential therapeutic strategy for cancer. Sci Rep. 2016;6:35855. doi: 10.1038/srep35855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Cai Y, Liu L, Wu Y, Xiong X. Crucial biological functions of CCL7 in cancer. PeerJ. 2018;6:e4928. doi: 10.7717/peerj.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farmaki E, Kaza V, Chatzistamou I, Kiaris H. CCL8 promotes postpartum breast cancer by recruiting M2 macrophages. iScience. 2020;23:101217. doi: 10.1016/j.isci.2020.101217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossin A, Miloro G, Hueber AO. TRAIL and FasL functions in cancer and autoimmune diseases: towards an increasing complexity. Cancers (Basel) 2019;11:639. doi: 10.3390/cancers11050639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Müllauer L, Mosberger I, Grusch M, Rudas M, Chott A. Fas ligand is expressed in normal breast epithelial cells and is frequently up-regulated in breast cancer. J Pathol. 2000;190:20–30. doi: 10.1002/(SICI)1096-9896(200001)190:1<20::AID-PATH497>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 49.Min PB, Connor JR. Role of ferritin in cancer biology. J Cancer Sci Ther. 2015;7:155–160. [Google Scholar]
- 50.Liu L, Liu Y, Yan X, Zhou C, Xiong X. The role of granulocyte colony-stimulating factor in breast cancer development: a review. Mol Med Rep. 2020;21:2019–2029. doi: 10.3892/mmr.2020.11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eubank TD, Roberts RD, Khan M, Curry JM, Nuovo GJ, Kuppusamy P, Marsh CB. Granulocyte macrophage colony-stimulating factor inhibits breast cancer growth and metastasis by invoking an anti-angiogenic program in tumor-educated macrophages. Cancer Res. 2009;69:2133–2140. doi: 10.1158/0008-5472.CAN-08-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rousalova I, Krepela E. Granzyme B-induced apoptosis in cancer cells and its regulation (review) Int J Oncol. 2010;37:1361–1378. doi: 10.3892/ijo_00000788. [DOI] [PubMed] [Google Scholar]
- 53.Critchley-Thorne RJ, Simons DL, Yan N, Miyahira AK, Dirbas FM, Johnson DL, Swetter SM, Carlson RW, Fisher GA, Koong A, Holmes S, Lee PP. Impaired interferon signaling is a common immune defect in human cancer. Proc Natl Acad Sci U S A. 2009;106:9010–9015. doi: 10.1073/pnas.0901329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Provance OK, Lewis-Wambi J. Deciphering the role of interferon alpha signaling and microenvironment crosstalk in inflammatory breast cancer. Breast Cancer Res. 2019;21:59. doi: 10.1186/s13058-019-1140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.García-Tuñón I, Ricote M, Ruiz AA, Fraile B, Paniagua R, Royuela M. Influence of IFN-gamma and its receptors in human breast cancer. BMC Cancer. 2007;7:158. doi: 10.1186/1471-2407-7-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gangemi S, Minciullo P, Adamo B, Franchina T, Ricciardi GR, Ferraro M, Briguglio R, Toscano G, Saitta S, Adamo V. Clinical significance of circulating interleukin-23 as a prognostic factor in breast cancer patients. J Cell Biochem. 2012;113:2122–2125. doi: 10.1002/jcb.24083. [DOI] [PubMed] [Google Scholar]
- 57.Gillgrass AE, Chew MV, Krneta T, Ashkar AA. Overexpression of IL-15 promotes tumor destruction via NK1.1+ cells in a spontaneous breast cancer model. BMC Cancer. 2015;15:293. doi: 10.1186/s12885-015-1264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song X, Wei C, Li X. The potential role and status of IL-17 family cytokines in breast cancer. Int Immunopharmacol. 2021;95:107544. doi: 10.1016/j.intimp.2021.107544. [DOI] [PubMed] [Google Scholar]
- 59.Nagai S, Toi M. Interleukin-4 and breast cancer. Breast Cancer. 2000;7:181–186. doi: 10.1007/BF02967457. [DOI] [PubMed] [Google Scholar]
- 60.Soliman H, Khalil F, Antonia S. PD-L1 expression is increased in a subset of basal type breast cancer cells. PLoS One. 2014;9:e88557. doi: 10.1371/journal.pone.0088557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zinger M, McFarland M, Ben-Jonathan N. Prolactin expression and secretion by human breast glandular and adipose tissue explants. J Clin Endocrinol Metab. 2003;88:689–696. doi: 10.1210/jc.2002-021255. [DOI] [PubMed] [Google Scholar]
- 62.Cruceriu D, Baldasici O, Balacescu O, Berindan-Neagoe I. The dual role of tumor necrosis factor-alpha (TNF-α) in breast cancer: molecular insights and therapeutic approaches. Cell Oncol (Dordr) 2020;43:1–18. doi: 10.1007/s13402-019-00489-1. [DOI] [PubMed] [Google Scholar]
- 63.Tahir RA, Sehgal SA, Khattak NA, Khan Khattak JZ, Mir A. Tumor necrosis factor receptor superfamily 10B (TNFRSF10B): an insight from structure modeling to virtual screening for designing drug against head and neck cancer. Theor Biol Med Model. 2013;10:38. doi: 10.1186/1742-4682-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Labovsky V, Martinez LM, Davies KM, de Luján Calcagno M, García-Rivello H, Wernicke A, Feldman L, Matas A, Giorello MB, Borzone FR, Choi H, Howard SC, Chasseing NA. Prognostic significance of TRAIL-R3 and CCR-2 expression in tumor epithelial cells of patients with early breast cancer. BMC Cancer. 2017;17:280. doi: 10.1186/s12885-017-3259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sigl V, Jones LP, Penninger JM. RANKL/RANK: from bone loss to the prevention of breast cancer. Open Biol. 2016;6:160230. doi: 10.1098/rsob.160230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho CC, Liao WY, Wang CY, Lu YH, Huang HY, Chen HY, Chan WK, Chen HW, Yang PC. TREM-1 expression in tumor-associated macrophages and clinical outcome in lung cancer. Am J Respir Crit Care Med. 2008;177:763–770. doi: 10.1164/rccm.200704-641OC. [DOI] [PubMed] [Google Scholar]
- 67.Liu Y, Tamimi RM, Collins LC, Schnitt SJ, Gilmore HL, Connolly JL, Colditz GA. The association between vascular endothelial growth factor expression in invasive breast cancer and survival varies with intrinsic subtypes and use of adjuvant systemic therapy: results from the Nurses’ Health Study. Breast Cancer Res Treat. 2011;129:175–184. doi: 10.1007/s10549-011-1432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varney ML, Singh RK. VEGF-C-VEGFR3/Flt4 axis regulates mammary tumor growth and metastasis in an autocrine manner. Am J Cancer Res. 2015;5:616–628. [PMC free article] [PubMed] [Google Scholar]