Abstract
Ankyrin repeats are well-known structural modules that mediate interactions between a wide spectrum of proteins. The regulatory factor X with ankyrin repeats (RFXANK) is a subunit of a tripartite RFX complex that assembles on promoters of major histocompatibility complex class II (MHC II) genes. Although it is known that RFXANK plays a central role in the nucleation of RFX, it was not clear how its ankyrin repeats mediate the interactions within the complex and with other proteins. To answer this question, we modeled the RFXANK protein and determined the variable residues of the ankyrin repeats that should contact other proteins. Site-directed alanine mutagenesis of these residues together with in vitro and in vivo binding studies elucidated how RFXAP and CIITA, which simultaneously interact with RFXANK in vivo, bind to two opposite faces of its ankyrin repeats. Moreover, the binding of RFXAP requires two separate surfaces on RFXANK. One of them, which is located in the ankyrin groove, is severely affected in the FZA patient with the bare lymphocyte syndrome. This genetic disease blocks the expression of MHC II molecules on the surface of B cells. By pinpointing the interacting residues of the ankyrin repeats of RFXANK, the mechanism of this subtype of severe combined immunodeficiency was revealed.
Major histocompatibility complex class II (MHC II) molecules are crucial players in the immune response. These cell surface glycoproteins are constitutively expressed on antigen-presenting cells and can be induced on other cell types by gamma interferon (4, 5, 9, 20). They present processed antigens to helper T cells and initiate immune responses (10). Different subtypes of human MHC II molecules are transcribed from TATA-less promoters that contain conserved S, X, and Y boxes (4, 5, 9, 14, 16, 20). Protein complexes that bind to these proximal promoter elements finally attract the class II transactivator (CIITA) by an as-yet-unknown mechanism (4, 5, 9, 14, 16, 20, 33). S and X boxes bind a tripartite regulatory factor X (RFX) complex, while the Y box binds the nuclear factor Y (NFY) complex (14). Congenital absence of MHC II molecules on B cells is known as the bare lymphocyte syndrome (BLS) (20). Its unique phenotypic outcome is the result of diverse genetic backgrounds. While the genes for MHC II determinants remain intact, different mutations have been found in four trans-acting factors, namely, RFX5, RFXAP, RFXANK(B), and CIITA, defining four complementation groups of BLS (12, 22, 24, 31).
The complex architecture of proteins that are directly or indirectly bound to MHC II promoters is achieved by multiple protein-protein interactions within the RFX and NFY complexes, between these complexes, and with CIITA, the master switch that triggers the transcription of MHC II genes (8, 11, 14, 33). The RFX complex is composed of three subunits, namely, RFX5, RFXAP, and RFXANK(B). We and others have shown how the RFX complex assembles (11, 26). Whereas RFXAP interacts with the two other subunits via its C-terminal, glutamine-rich domain (11, 26), RFX5 contacts the RFX complex via two separate regions that surround its DNA-binding domain (11). RFXANK or RFX(B) (henceforth called RFXANK) is a 33-kDa protein with three distinct domains (11, 22, 24). The potential role of its N-terminal imperfect PEST sequence is still unknown. The C-terminal portion of RFXANK contains at least three ankyrin repeats. RFXANK is therefore the only protein within the RFX complex that contains well-established modules for protein-protein interactions. The domain between the PEST-like sequence and the first ankyrin repeat has been suggested to make contacts with DNA, although it lacks any recognizable DNA-binding consensus sequence.
Ankyrin repeats are one of the most common protein sequence motifs, with each of them consisting of 33 residues (30). They have been found in proteins as different as Cdk inhibitors, signal transduction and transcriptional regulators, cytoskeletal organizers, developmental regulators, and toxins. Their presence in such a colorful palette of functionally diverse proteins suggests that their role is of more of a structural than a functional nature. Indeed, these protein scaffolding modules mediate protein-protein interactions in a number of different biological systems, from microbes to humans (30). The number of ankyrin repeats varies from only 2 in plutonium, a small protein from Drosophila (2), to more than 20 in ankyrin, a well-studied ubiquitous adapter protein that links membrane proteins with the spectrin-based cytoskeleton (23). Elucidation of the three-dimensional structure of the ankyrin repeats by X-ray crystallography and nuclear magnetic resonance techniques offered an insight into their conserved, stable backbone. Certain amino acid residues of the backbone play only an architectural role by making multiple intramolecular interactions, mainly hydrogen and hydrophobic ones. However, ankyrin repeats can easily handle a broad diversity of their binding partners by containing variable residues, insertions, and deletions between single repeats and by stacking in different numbers. Thus, it is not surprising that there are no specific ankyrin-binding motifs in their target proteins, which can also vary considerably in their shape and size (30). Potential binding surfaces on the ankyrin repeats are all solvent-exposed parts that contain variable residues.
Although it has been suggested that the ankyrin repeats of RFXANK mediate protein-protein interactions within the RFX complex (11, 26), no informative mapping on RFXANK has been done. Furthermore, the involvement of ankyrin repeats in protein-protein interactions that go beyond the RFX complex has not been addressed. Our preliminary experiments showed that RFXANK binds multiple protein partners. We wanted to investigate how the smallest subunit of the RFX complex successfully mediates these protein-protein interactions and what is the role of ankyrin repeats in this process. However, deletion mapping was not informative, and we found the structure-function analysis, based on a three-dimensional structure prediction for RFXANK, more useful. By combining a well-studied ankyrin fold with site-directed alanine mutagenesis, we showed how its multiple binding sites recruit the interacting proteins, and in this way we mapped precisely the ankyrin-centered interactions on MHC II promoters.
MATERIALS AND METHODS
Cell culture.
COS cells were grown in Dulbecco's modified Eagle's medium. Media were supplemented with 10% fetal bovine serum, 100 mM l-glutamine, and 50 μg each of penicillin and streptomycin per ml.
Plasmid constructions.
Myc epitope-tagged pEF-RFXANK and hemagglutinin (HA) epitope-tagged pEF-RFXAP plasmid constructs were generated as described before (26). HA epitope-tagged wild-type CIITA protein was generated by PCR and inserted into the EcoRI-SpeI sites of the modified pEF-BOS vector (1). The glutathione S-transferase (GST)-RFXANK plasmid construct was described before (26). Deletion mutants of RFXANK were created by PCR. The primer sequences were as follows: the forward primer F (5′-GCTTCGGGATCCATGGAGCTTACCCAGCCTGCA-3′) and the reverse primers R1 (5′-GCTTCGGAATTCCTACTGGAAGAGCTTGAGGATGTG-3′) for RFXANK(1–251), R2 (5′-GCTTCGGAATTCCTAGCCTCGGGCCAGCAAGGCCTC-3′) for RFXANK(1–213), R3 (5′-GCTTCGGAATTCCTAGTCACGCTCCAGCAGCAGCCC-3′) for RFXANK(1–180), and R4 (5′-GCTTCGGAATTCCTAACCCCACTCCAGCAGGAAGCG-3′) for RFXANK(1–147). Amplified products were ligated into the BamHI-EcoRI sites in frame with the coding region of the GST gene in pGEX-2TK (Amersham Pharmacia Biotech, Piscataway, N.J.). All cDNAs were confirmed by DNA sequencing. The pT7T3-RFXAP and pSV-CIITA plasmid constructs were described before (13, 26).
Site-directed mutagenesis.
Mutagenesis of the ankyrin repeats of RFXANK was performed by using a Transformer Site-Directed Mutagenesis Kit (Clontech Laboratories, Palo Alto, Calif.) according to the manufacturer's instructions. The template for mutagenesis was the GST-RFXANK plasmid construct. The mutagenic primers were designed as follows: 5′-CCTCGTCAACAAGCCAGCGGCCGCGGCCTTCACCCCCCTC-3′ for GST-RFXANK-β1 (contains D121A, E122A, R123A, and G124A substitutions in the cDNA of the wild-type RFXANK protein), 5′-GCCGACCCCCACATCCTGGCGGCCGCGGCCGAGAGCGCCCTGTCG-3′ for GST-RFXANK-β2 (contains K155A, E156A, and R157A substitutions), 5′-GGACATCAACATCTATGCGGCCGCGGCCGGGACGCCACTGC-3′ for GST-RFXANK-β3 (contains D187A, W188A, N189A, and G190A substitutions), 5′-GCTGACCTCACCACCGAAGCCGCGGCCGCGTACACCCCGATGG-3′ for GST-RFXANK-β4 (contains D221A, S222A, and G223A substitutions), 5′-GAGAGATTGAGACCGTTGCGTTCCTGCTGGCGGCCGGTGCCGACCCCCAC-3′ for GST-RFXANK-OH1 (contains R141A, E145A, and W146A substitutions), 5′-GTGGGGCTGCTGCTGGCGGCCGACGTGGACATCAACATCTATGATTGG-3′ for GST-RFXANK-OH2 (contains G174A, E178A, and R179A substitutions), 5′-CACGTGAAATGCGTTGCGGCCTTGCTGGCCGCGGGCGCTGACCTCACCAC-3′ for GST-RFXANK-OH3 (contains E207A and R212A substitutions), 5′-GGAGGGACGCCACTGGCGGCCGCTGCGGCCGGGAACCACGTGAAATGCG-3′ for GST-RFXANK-IH3 (contains L195A, Y196A, V198A, and R199A substitutions), 5′-GCACAGGCGGCTACACAGCCATTGTGGGGCTGCTGCTGG-3′ for GST-RFXANK-turn2 (contains a D171A substitution), 5′-GCGCGGGAACCACGTGGCGTGCGTTGAGGCCTTGCTGGCCCG-3′ for GST-RFXANK-turn3 (contains a K204A substitution), 5′-GGCCCTGGGATACCGGGCGGTGCAACAGGTGATCGAGAACC-3′ for GST-RFXANK-turn4 (contains a K237A substitution), and 5′-GGAATGGAGGGACGCCACTGCCGTACGCTGTGCGCGGGAACCACG-3′ for GST-RFXANK-FZA (contains an L195P substitution). The selection primer was the same for all mutagenesis reactions (5′-CGCGCTGTTAGCGGCGCCATTAAGTTCTGTCTCGGC-3′) and changes a unique ApaI restriction site in the GST-RFXANK plasmid construct. All mutants were confirmed by DNA sequencing.
Immunoprecipitation and Western blotting.
At >48 h after transfection, COS cells were harvested in 1 ml of lysis buffer (1% [vol/vol] NP-40, 10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, and 0.1% protease inhibitors) for 45 min at 4°C, and the amounts of the solubilized proteins were measured (BCA Protein Assay; Pierce, Rockford, Ill). Protein A-Sepharose (Amersham Pharmacia Biotech)-precleared lysates were subjected to immunoprecipitation using a rabbit polyclonal anti-Myc antibody (c-Myc [A-14]; Santa Cruz Biotechnology, Santa Cruz, Calif.). Immune complexes were recovered by binding to protein A-Sepharose beads during the overnight rotation at 4°C, resolved on a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel, and transferred to a nitrocellulose membrane by a semidry technique. The membranes were immunostained with a mouse monoclonal anti-HA antibody (1:2,000; Boehringer Mannheim, Indianapolis, Ind.) followed by a horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G secondary antibody (1:2,000; Gibco-BRL, Rockville, Md.). Blots were developed by chemiluminescence assay (NEN Life Science Products, Boston, Mass.).
In vitro transcription and translation.
The plasmids containing RFXAP (pT7T3-RFXAP), RFX5 (pcDNA3-RFX5), and CIITA (pSV-CIITA) cDNAs were transcribed and translated in vitro using the TnT T3-T7 coupled reticulocyte lysate system (Promega, Madison, Wis.) according to the manufacturer's instructions in the presence or absence of 35S-labeled cysteine (NEN Life Science Products).
In vitro binding assays.
GST fusion proteins were produced in Escherichia coli BL21(DE3)pLysS competent cells (Novagen, Madison, Wis.) during 4 h of induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and purified from total cell lysates with glutathione-Sepharose beads (Amersham-Pharmacia Biotech). For the GST pull-down assay, 10 μg of GST or GST fusion proteins was mixed with 10 μl of in vitro-translated proteins in 300 μl of binding buffer. The composition of the buffer for studying the interaction between RFXANK and CIITA was as follows: 50 mM Tris-HCl (pH 8.0), 5% glycerol, 0.5 mM EDTA, 5 mM MgCl2, 1% bovine serum albumin, 500 mM NaCl, 0.25 Triton X-100, and 0.125% NP-40. When the interaction between RFXAP and RFXANK was studied, the detergent concentrations were increased to 1% Triton X-100 and 0.5% NP-40. After overnight incubation at 4°C, GST-coupled beads were washed five times with 1 ml of binding buffer. Bound proteins were eluted by boiling in SDS sample buffer. Proteins were resolved by SDS–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) and revealed by autoradiography, and the signal was quantified as counts per minute.
EMSA.
Electrophoretic mobility shift assays (EMSAs) were performed as described before (26).
Structure modeling.
The structure of the ankyrin repeat domain of RFXANK (sequence number AAC69883) was modeled with the Swiss-Model approach for automated comparative protein modeling (28). As template files the ankyrin repeat-containing crystal structures of GABPβ (3) (RCBS accession code 1AWC; chain B; 2.15-Å resolution; Swiss-Prot database Q00421) and Swi6 (15) (1SW6; chain A; 2.10-Å resolution; P09959) were used. The sequence homology between the 125-amino-acid fragment of RFXANK (residues 119 to 243) and GABPβ (residues 33 to 157) corresponds to 28.8% identity (62.4% similarity). For RFXANK and Swi6 the sequence identity is about 26.8% (58.2% similarity) for the 67-amino-acid fragment of RFXANK (residues 88 to 154). For structure display and surface evaluation, hydrogen atoms were added to the model coordinates using the program X-PLOR (7). The fragments were assembled by a least-squares fit of the heavy-atom backbone coordinates of the overlapping residues 124 to 146.
RESULTS
RFXANK has four ankyrin repeats.
The 33-kDa RFXANK protein was the last recognized subunit of the RFX complex (22, 24). Besides its N-terminal PEST-like sequence and DNA-binding domain, it contains an ankyrin repeat domain at its C terminus. Three ankyrin repeats were reported to lie in this domain (22, 24), although one report suggested that there might be a fourth one, displaying weak homology to the general ankyrin repeat motif (11).
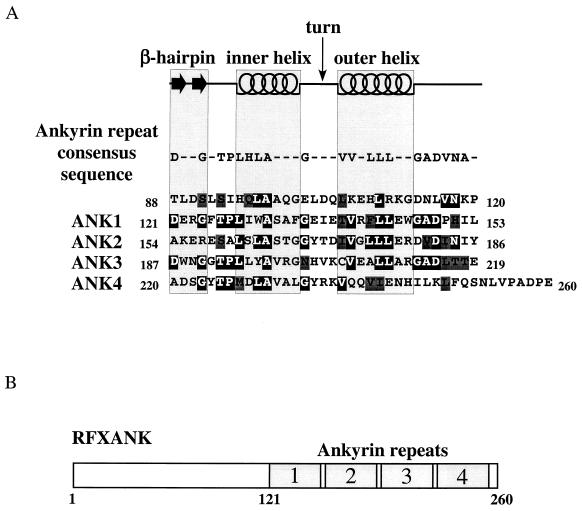
To determine how many ankyrin repeats compose the ankyrin domain of RFXANK, we compared its amino acid sequence to a structure-based ankyrin repeat consensus sequence that has been published recently (30). This sequence keeps the two β-strands of the β-hairpin loop together and therefore better represents the ankyrin repeat as a structural unit. The aspartic acid residue in the β-hairpin stabilizes the loop by hydrogen bonding between its main and side chains. Next, the Thr-Pro-Leu-His (TPLH) peptide forms a turn and initiates the inner helix, while the two conserved glycine residues terminate each of the two helices (Fig. 1A, ankyrin repeat consensus sequence). Conserved hydrophobic residues of both helices are involved in stacking of the repeats, which results in a very stable, nonglobular ankyrin domain structure with a hydrophobic core.
FIG. 1.
RFXANK contains four ankyrin repeats. (A) Sequence analysis of the ankyrin repeat domain of RFXANK. The secondary-structure elements (β-hairpin loops, inner helix, turn, and outer helix) and the ankyrin repeat consensus sequence are displayed above the amino acid sequence of RFXANK residues 88 to 260. Identical and conserved residues relative to the ankyrin repeat consensus sequence are represented by white letters on a black background and by black letters on a dark gray background, respectively. A high degree of sequence similarity to the ankyrin consensus motif sequence can be observed from amino acid V117 to I242, suggesting the formation of four ankyrin repeats in RFXANK. (B) Schematic representation of RFXANK. RFXANK contains 260 amino acid residues and four ankyrin repeats at the C-terminal part of the protein.
We compared the amino acid sequence of RFXANK to the consensus motif containing a single ankyrin repeat (30) with the exposed β-hairpin loop and two antiparallel α-helices (inner and outer helices), connected by the turn region (Fig. 1A, top). This analysis revealed that RFXANK contains four β-hairpin loops with two preceding and two succeeding helices that stabilize the structure (Fig. 1A, bottom). As the alignment shows, the consensus residues located mainly in the inner and outer helices that stabilize the ankyrin repeat fold are well conserved. These residues are hidden inside the structure and are less suitable for mutagenesis since any change will affect the formation and stability of the ankyrin domain but will not directly affect the surface recognition of its binding partners. The degree of conservation of β-hairpin loops shows that the least conserved ankyrin repeat is the second one. This observation might suggest a reduced functional importance of this repeat as well as increased specificity for making contacts with other proteins. We conclude that RFXANK contains four ankyrin repeats that represent a stable ankyrin domain module spanning the C-terminal part of the protein (Fig. 1B).
Prediction of the three-dimensional structure of the ankyrin repeat domain of RFXANK.
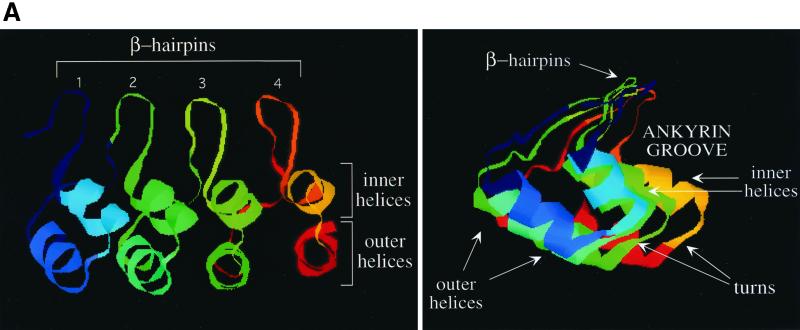
The first ankyrin repeat-containing protein with a determined three-dimensional structure was 53BP2, which interacts with the L-2 loop of the p53 tumor suppressor protein (17). As presented in a general model containing four ankyrin repeats (Fig. 2A, left panel), the ankyrin repeat domain consists of pairs of antiparallel (inner and outer) α-helices that are stacked side by side and connected by a series of intervening β-hairpin loops. The extended β-sheet projects away from the helical pairs almost at right angles, resulting in a characteristic L-shaped cross-section (Fig. 2A, right panel). This assembled structure has been compared to a cupped hand: whereas the β-hairpin loops form the fingers, the concave part, also termed the ankyrin groove, with solvent-exposed residues from the α-helical bundles, forms the palm (30, 32). The structure is further stabilized by extensive intra- and interrepeat hydrogen bonds between the side chains.
FIG. 2.
Secondary-structure prediction of the ankyrin repeat domain of RFXANK. (A) Schematic representation of the secondary-structure elements of the ankyrin repeats in a three-dimensional view. Four ankyrin repeats represent only a general ankyrin domain structure. In the left scheme, β-hairpin loops form the loop structures above the two planes of helices (inner and outer helices). The right scheme represents the same three-dimensional structure from a different perspective (as viewed from the first ankyrin repeat towards the last). The L-shaped structure appears, forming the ankyrin groove. β-Hairpin loops, turns, and inner and outer helices form four different surfaces of the ankyrin repeat domain and are depicted with arrows. (B) Model structure of RFXANK residues 88 to 243. The ankyrin repeat domain of RFXANK is depicted as a ribbon structure, with two exposed variable residues at the very tip of each of the four β-hairpin loops highlighted. This figure was generated with Molscript (19). N, amino terminus; C, carboxy terminus.
To study the most suitable interaction sites on the surface of RFXANK, we took advantage of the three-dimensional structure of the known ankyrin repeats. We modeled the ankyrin repeat domain of RFXANK using the Swiss-Model approach for automated comparative protein modeling (28). A search through the structure database shows that the predictable region in RFXANK ranges from threonine at position 88 to glutamic acid at position 243. However, none of the files displays this region homogeneously as an entity. The structure of the ankyrin repeat-containing β-subunit of the transcriptional regulator GABP protein complex (3) fits best to RFXANK from residue 119 to 243, while transcription factor Swi6 (15) shows the best similarities to RFXANK from residue 88 to 154, which contains the first ankyrin repeat. We assembled both fragments by an overlay and subsequent minimization of the root mean square deviation of the two overlapping sections (residues 124 to 146) to gain a model structure of the ankyrin repeat domain of RFXANK (Fig. 2B).
The β-hairpin loops of the ankyrin repeats of RFXANK are required for binding to RFXAP.
RFXANK and RFXAP, two subunits of the tripartite RFX complex, bind to each other strongly and specifically. We have shown previously that this interaction is the first step in the assembly of RFX (26). The C-terminal region of RFXAP, which contains a glutamine-rich domain, binds to RFXANK (11, 26). However, no mapping has been done on RFXANK.
Deletion mapping of RFXANK was not informative for the interaction between RFXANK and RFXAP (data not shown). However, with our model structure of the ankyrin repeat domain of RFXANK, we were able to select and mutate the surface-exposed residues of the molecule and maintain the ankyrin repeat domain intact structurally. We mutated variable residues on the surface of the ankyrin repeat domain that were the best candidates for making specific contacts with other proteins.
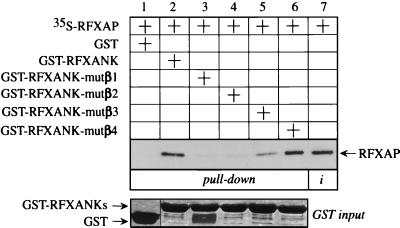
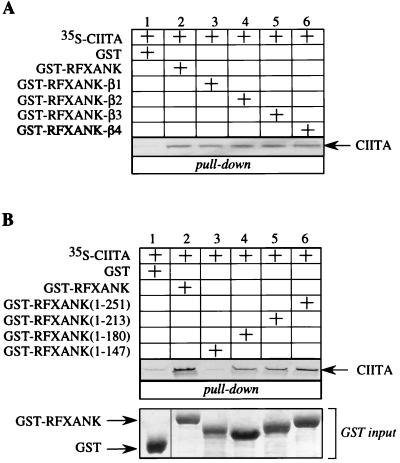
The first group of residues that fulfilled these criteria were the four exposed residues at the tips of each of the four β-hairpin loops. By using alanine mutagenesis with the GST-RFXANK fusion protein as a template, we created four mutant chimeras. In the mutant hybrid GST-RFXANK-mutβ1 to -4 proteins, four amino acids of each β-hairpin loop were changed to alanines (see Materials and Methods). Wild-type and mutant GST chimeras were expressed in E. coli, and the wild-type, 35S-labeled RFXAP protein was transcribed and translated in vitro by using rabbit reticulocyte lysate. Next, RFXAP was combined with the GST fusion proteins in a GST pull-down assay (Fig. 3). As established before, RFXAP interacted with the wild-type GST-RFXANK fusion protein but did not interact with GST alone, showing the specificity of this interaction (Fig. 3, compare lanes 1 and 2). In comparison to the input (Fig. 3, lane 7), approximately 25% of RFXAP was retained by the hybrid GST-RFXANK protein. However, when the mutant GST-RFXANK fusion proteins with mutations in the first two β-hairpin loops were used, no binding was observed with RFXAP (Fig. 3, lanes 3 and 4). In contrast, the mutant GST-RFXANK fusion protein with mutations in the β-hairpin loop of the third ankyrin repeat retained some of its binding to RFXAP (Fig. 3, lane 5). Interestingly, the mutant GST-RFXANK fusion protein with mutations in the β-hairpin loop of the last ankyrin repeat was able to bind to RFXAP at the same level as the wild-type fusion protein (Fig. 3, compare lanes 2 and 6). The input amounts of all bacterially produced proteins were equivalent (Fig. 3, GST input). We conclude that RFXANK binds to RFXAP via its β-hairpin loops and that the first three loops are important for this binding.
FIG. 3.
β-Hairpin loops of the ankyrin repeats of RFXANK are required for binding to RFXAP in vitro. The GST-RFXANK protein links 260 amino acids of RFXANK to GST and was used as a template for alanine mutagenesis. Four amino acid residues at the tips of the four β-hairpin loops of RFXANK were mutated into alanines, and the resulting proteins were named GST-RFXANK-mutβ1 to−4 (see Materials and Methods). In a GST pull-down assay, 35S-labeled RFXAP was incubated with GST alone or the wild-type and mutant GST-RFXANK fusion proteins and selected on glutathione-Sepharose beads. Bound proteins were separated by SDS-PAGE and revealed by autoradiography. RFXAP that was retained on the beads is depicted with an arrow. Lanes 1 to 6, results of the binding assay; lane 7, 25% of the input 35S-labeled RFXAP. Pluses above the autoradiographs indicate the presence of different proteins in the assay. GST alone (lane 1) and the wild-type (lane 2) and mutant (lanes 3 to 6) GST-RFXANK fusion proteins were equivalent and are presented in a Coomassie blue-stained SDS-polyacrylamide gel (GST input).
RFXANK binds simultaneously to RFXAP and CIITA in cells.
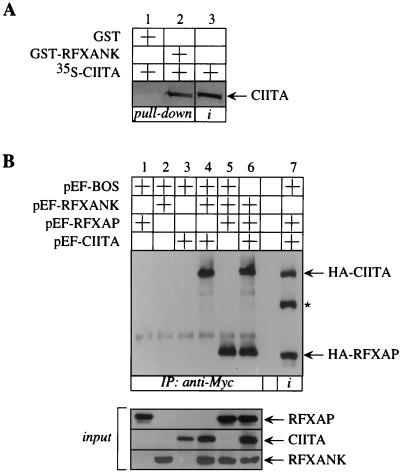
In our preliminary studies we asked whether RFXANK interacts with proteins other than RFXAP. Under stringent conditions in vitro, we could not detect its binding to RFX5 (26). However, in vitro-transcribed and -translated CIITA was able to bind to bacterially produced hybrid GST-RFXANK protein in a GST pull-down assay (Fig. 4A, lane 2). The specificity of this binding was established because CIITA did not bind to GST alone (Fig. 4A, lane 1). In vitro studies for the binding of RFXAP and CIITA to RFXANK were done under more and less stringent binding conditions, respectively. In addition, only about 10% of input CIITA was retained by the hybrid GST-RFXANK protein (Fig. 4A, compare lanes 2 and 3). We conclude that although both can bind to RFXANK in vitro, RFXAP does so with higher affinity than CIITA.
FIG. 4.
RFXANK binds to CIITA both in vitro and in vivo. (A) RFXANK binds to CIITA in vitro. 35S-labeled CIITA was incubated with GST alone or the GST-RFXANK fusion protein and selected on glutathione-Sepharose beads. Retained CIITA is depicted with an arrow. Lanes 1 and 2, results of the binding assay; lane 3, 10% of the input 35S-labeled CIITA. Amounts of GST alone and GST-RFXANK fusion protein were the same as in Fig. 3 (lanes 1 and 2, GST input) and are therefore not presented. (B) RFXANK coprecipitates RFXAP and CIITA from the cells. The N terminus of RFXANK was linked to a Myc epitope tag, and the N termini of RFXAP and CIITA were linked to an HA epitope tag. Epitope-tagged proteins were expressed alone (lanes 1 to 3) or in different combinations (lanes 4 to 6) in COS cells. Precleared total cell lysates were immunoprecipitated (IP) with the anti-Myc antibody and protein A-Sepharose beads and examined for the presence of RFXAP and CIITA by Western blotting with the anti-HA antibody; 10% of precleared total cell lysates was analyzed for the presence of RFXANK, RFXAP, and CIITA (input). The same amount of total cell lysate from lane 6 was applied to lane 7 for easier identification of HA-tagged proteins (depicted with arrows). The asterisk depicts the unspecific band (lane 7).
Recently, CIITA has been shown to interact with multiple proteins of the MHC II transcriptosome in vivo, namely, RFX5 and RFXANK from the RFX complex and NFYB and NFYC from the NFY complex (33). These data confirmed our notion that CIITA is the second binding partner for RFXANK. However, we wanted to show that RFXAP and CIITA could bind to RFXANK simultaneously. To this end, COS cells were transfected with plasmids which directed the expression of the N-terminally HA epitope-tagged RFXAP and CIITA as well as the Myc epitope-tagged RFXANK proteins alone or in different combinations. Precleared total cell lysates were incubated with anti-Myc antibody, and immunoprecipitates were examined for the presence of HA-tagged proteins by Western blotting with anti-HA antibody. When either of the three plasmids alone was transfected into COS cells, no RFXAP or CIITA was detected in the immunoprecipitates (Fig. 4B, lanes 1 to 3). However, when RFXANK was coexpressed with either CIITA or RFXAP alone, both proteins were detected separately in our immunoprecipitates (Fig. 4B, lanes 4 and 5, respectively). Most importantly, immunoprecipitates from cell lysates containing all three proteins revealed the coprecipitation of RFXAP and CIITA (Fig. 4B, lane 6). Ten percent of precleared total cell lysate from a triple cotransfection was revealed separately (Fig. 4B, lane 7). All three proteins were expressed at comparable levels (Fig. 4B, input). We conclude that RFXANK can bind simultaneously to RFXAP and CIITA in vivo.
Ankyrin repeats as structural modules are required for the binding of RFXANK to CIITA.
To determine which part of RFXANK interacts with CIITA, we used the same approach as previously for studying its interaction with RFXAP. Ankyrin repeats were again the most likely candidate for the binding to CIITA. First, the mutant GST-RFXANK fusion proteins with substituted β-hairpin residues were combined with in vitro- transcribed and -translated CIITA in a GST pull-down assay. As already shown in Fig. 4A, CIITA bound to the wild-type GST-RFXANK fusion protein but did not bind to GST alone (Fig. 5A, lanes 1 and 2). The binding persisted when all four mutant GST-RFXANK fusion proteins were used instead of the wild-type GST-RFXANK fusion protein. We conclude that CIITA does not interfere with the binding of RFXAP to the β-hairpin loops of RFXANK.
FIG. 5.
The ankyrin repeat domain of RFXANK also binds to CIITA. (A) β-Hairpin loops of RFXANK ankyrin repeats are not involved in binding to CIITA. The mutant GST-RFXANK-β1 to −4 fusion proteins were used in a GST pull-down assay. 35S-labeled CIITA was incubated with GST alone or with the wild-type and mutant GST-RFXANK fusion proteins and selected on glutathione-Sepharose beads. Retained CIITA is depicted with an arrow. Lanes 1 to 6, results of the binding assay. Pluses above the autoradiographs indicate the presence of different proteins in the assay. Amounts of GST alone together with the wild-type and mutant GST-RFXANK fusion proteins were the same as in Fig. 3 (GST input) and are therefore not presented; 10% input 35S-labeled CIITA was the same as in Fig. 4A, lane 3. (B) Ankyrin repeats as structural units are required for CIITA binding. The first 251, 213, 180, and 147 amino acid residues of RFXANK represent the mutant RFXANK proteins that retain four, three, two, and one (the first) ankyrin repeat(s), respectively, and are fused to GST. All four C-terminal deletion mutants of GST-RFXANK were used in a GST pull-down assay similar to that described for panel A. Lanes 1 to 6, results of the binding assay. The amounts of all of the GST fusion proteins were the same and are shown in the Coomassie blue-stained gel at the bottom of the panel (GST input).
CIITA could bind to another surface of ankyrin repeats or, alternatively, could bind to a region outside the ankyrin repeat domain of RFXANK. To distinguish between these two possibilities, we first created C-terminal deletion mutants of GST-RFXANK fusion protein and expressed them in E. coli. The mutant GST-RFXANK fusion proteins contain the first 251, 213, 180, and 147 residues of RFXANK fused to GST. Therefore, they contain all four, the first three, the first two, and only the first ankyrin repeat(s), respectively. Four deletion mutants were used in a GST pull-down assay, where they were combined with in vitro-transcribed and -translated CIITA protein. As before, CIITA bound specifically to the wild-type GST-RFXANK fusion protein but not to GST alone (Fig. 5B, lanes 1 and 2). When the longest deletion mutant, the hybrid GST-RFXANK(1–251) protein, was used, the binding to CIITA was preserved (Fig. 5B, lane 6). Interestingly, when the ankyrin repeats of RFXANK were sequentially removed, the binding of CIITA decreased gradually (Fig. 5B, lanes 4 and 5) until it was completely abolished with the mutant GST-RFXANK(1–147) fusion protein. From these data, we conclude that the last three ankyrin repeats of RFXANK are important for its binding to CIITA, although the most critical repeat seems to be the second one.
The inner helix of the third ankyrin repeat of RFXANK contacts RFXAP.
So far we had determined the residues of the ankyrin repeat domain of RFXANK that bind to RFXAP and shown that the ankyrin repeats also bind to CIITA. To map precisely the residues of RFXANK that bind to CIITA, we performed another series of alanine mutageneses with the GST-RFXANK fusion protein as a template. Recently, a study was performed that included some mutant RFXANK proteins with point mutations in the ankyrin repeat domain (11). However, in that study the conserved structural residues of ankyrin repeats were mutated to alanines, causing a destruction of the ankyrin repeat domain. In contrast, we wanted to mutate variable residues of ankyrin repeats that should elucidate additional specific interactions between RFXANK and its binding partners in the context of the intact ankyrin repeat domain.
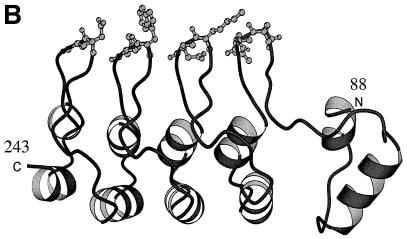
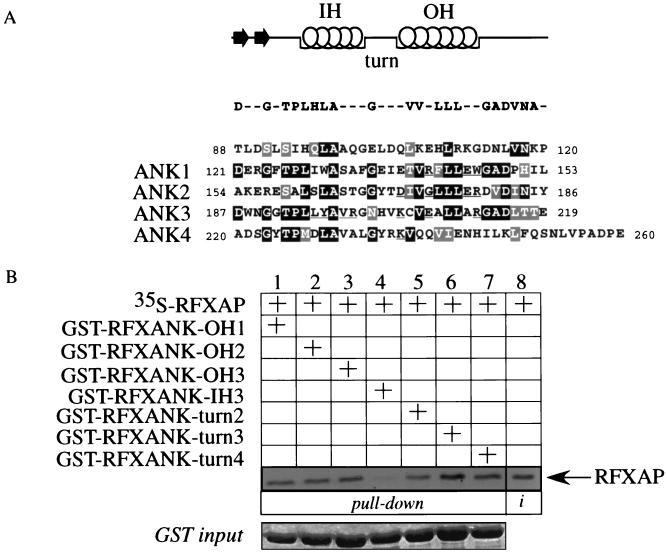
Besides the β-hairpin loops, we determined the exposed residues on three other surfaces of the ankyrin repeat domain of RFXANK by looking at its model structure (Fig. 2B) with the RasMol program. We performed single and clustered point mutations of nonconserved residues in three outer helices of the first three ankyrin repeats, the inner helix of the third ankyrin repeat, and a turn region of the last three ankyrin repeats (see Materials and Methods). The residues of RFXANK in seven mutant GST-RFXANK fusion proteins were successfully replaced with alanines (underlined in Fig. 6A).
FIG. 6.
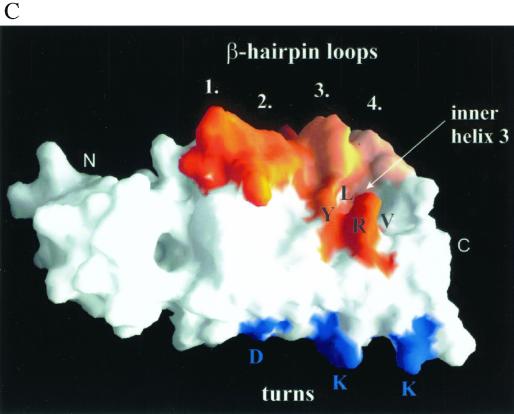
RFXAP binds to RFXANK at an additional contact point. (A) Mutagenesis scheme for exposed variable amino acid residues in the ankyrin repeat domain of RFXANK. The GST-RFXANK fusion protein was a template for another series of alanine mutagenesis reactions. Exposed amino acid residues (other than the ones from the β-hairpin loops) were selected on the basis of the predicted three-dimensional structure of the ankyrin repeat domain of RFXANK. All mutated residues are underlined. The GST-RFXANK fusion proteins with mutations in the outer helices (OH) of the first, second, and third ankyrin repeats were named GST-RFXANK-OH1, -OH2, and -OH3, respectively. The fusion proteins with mutations in the turn regions of the second, third, and fourth repeats were named GST-RFXANK-turn2, -turn3, and -turn4, respectively. The inner helix (IH) of the third ankyrin repeat was mutated to yield mutant GST-RFXANK-IH3 fusion protein. The point mutations were introduced as single mutations (turns 2 to 4) or in clusters (OH1 to−3 and IH3). (B) Cluster mutations in the inner helix of the third ankyrin repeat of RFXANK abolish its binding to RFXAP. Seven mutants depicted in panel A were used in a GST pull-down assay. They were mixed with 35S-labeled RFXAP, selected on glutathione-Sepharose beads, separated by SDS-PAGE, and revealed by autoradiography. Retained RFXAP is depicted with an arrow. Lanes 1 to 7, results of the binding assay; lane 8, 25% of the input 35S-labeled RFXAP. The mutant GST-RFXANK fusion proteins were equivalent and are presented in a Coomassie blue-stained SDS-polyacrylamide gel (GST input). (C) Display of the sites of mutational analysis on the surface of the modeled ankyrin repeat domain of RFXANK. Displayed in orange are mutated residues in four β-hairpin loops (1 to 4) at the top of the model and residues in the inner helix of the third ankyrin repeat (inner helix 3) in the middle part of the model. L, Y, V, and R represent four mutated residues in inner helix 3, which is involved in binding to RFXAP. Displayed in blue are mutated turn residues (D, K, and K [bottom of the model]). This figure was generated with GRASP (27). N, amino terminus; C, carboxy terminus.
Next, we combined the mutant GST-RFXANK fusion proteins with in vitro-transcribed and -translated CIITA in a GST pull-down assay. All of the mutants retained the binding to CIITA (data not shown). To determine whether the mutant GST-RFXANK fusion proteins bind normally to RFXAP, we performed another series of pull-down assays. Interestingly, while none of the mutant proteins containing introduced alanine residues in outer helices or the turn region showed a changed pattern of binding to RFXAP (Fig. 6B, lanes 1 to 3 and 5 to 7), the mutant GST-RFXANK-IH3 fusion protein did not bind to RFXAP (Fig. 6B, lane 4). The input amounts of GST proteins were equivalent (Fig. 6B, GST input). Although it seems that the intensity of the binding differs between different mutants in the turn region (Fig. 6B, lanes 5 to 7), these intensities differed slightly from experiment to experiment. In contrast, the lack of binding to RFXAP for the mutant GST-RFXANK-IH3 fusion protein was highly reproducible. We conclude that besides binding to β-hairpin loops of RFXANK, RFXAP also contacts the inner helix of the third ankyrin repeat of RFXANK. Both surfaces lie on the same face of the ankyrin repeat domain of RFXANK (Fig. 6C; mutated residues displayed in orange). The turn region (Fig. 6C; mutated residues displayed in blue) and the outer helices (not displayed) lie on the opposite face of the ankyrin repeat domain of RFXANK.
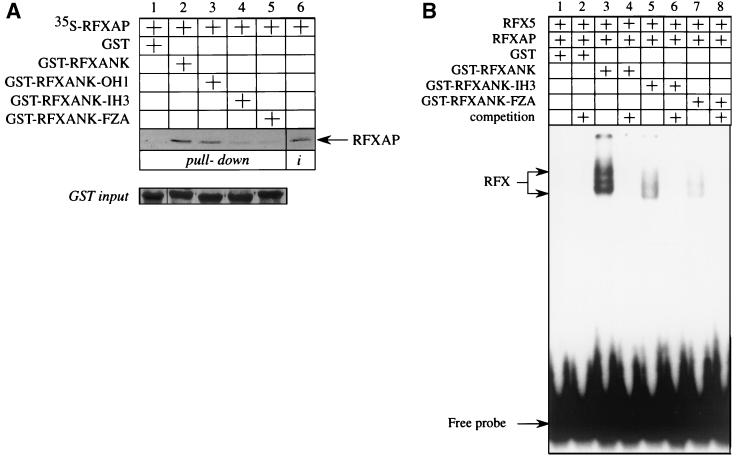
The point mutation in RFXANK from the FZA BLS patient abolishes its binding to RFXAP.
The mutant GST-RFXANK-IH3 fusion protein that was not able to bind to RFXAP (Fig. 6B) was created by alanine mutagenesis of a cluster of residues in the inner helix of the third ankyrin repeat of RFXANK. Four residues were replaced with alanines, namely, Leu 195, Tyr 196, Val 198, and Arg 199 (see also Fig. 6C). All of these residues have protruding side chains and could be involved in contacting RFXAP. Interestingly, the recently described FZA patient from the complementation group B of BLS has a point mutation in RFXANK that changes leucine at position 195 into proline, resulting in the loss of expression of MHC II molecules on the surface of the patient's immune cells (25). Since the proline residue is a so-called helix breaker, we had to extend this structural change to the other residues in the inner helix of the third ankyrin repeat. We speculated that a single point mutation in the FZA patient was responsible for the loss of binding to RFXAP.
To test this possibility, we created the mutant RFXANK protein as present in the FZA patient and fused it to GST to get the mutant GST-RFXANK-FZA fusion protein. Next, we combined this mutant protein with in vitro-transcribed and -translated RFXAP protein and tested their interaction in a GST pull-down assay. As shown before, the wild-type RFXAP protein interacted with the wild-type GST-RFXANK and the mutant GST-RFXANK-OH1 fusion proteins (Fig. 7A, lanes 2 and 3) but did not interact with GST alone (Fig. 7A, lanes 1) or the mutant GST-RFXANK-IH3 fusion protein (Fig. 7A, lane 4). Importantly, RFXAP was also unable to bind to the mutant GST-RFXANK-FZA fusion protein (Fig. 7A, lane 5).
FIG. 7.
Intact inner helix 3 of RFXANK is critical for the assembly of the RFX complex. (A) The FZA patient from BLS complementation group B carries a mutation in the inner helix of the third ankyrin repeat of RFXANK. The leucine residue at position 195 in RFXANK was mutated into a proline residue, and the mutant protein was fused to GST to yield the mutant GST-RFXANK-FZA fusion protein. The mutant protein was used together with the wild-type and mutant GST-RFXANK-OH1 and -IH3 fusion proteins in a GST pull-down assay similar to that for Fig. 6B. Retained RFXAP is depicted with an arrow. Lanes 1 to 5, results of the binding assay; lane 6, 25% of the input 35S-labeled RFXAP. Pluses above the autoradiographs indicate the presence of different proteins in the assay. GST alone (lane 1) and the wild-type (lane 2) and mutant (lanes 3 to 5) GST-RFXANK fusion proteins were equivalent and are presented in a Coomassie blue-stained SDS-polyacrylamide gel (GST input). (B) The mutation in RFXANK from the FZA patient blocks the assembly of the RFX complex on DNA. Wild-type RFX5 and RFXAP proteins were transcribed and translated in vitro using the rabbit reticulocyte system. Wild-type and mutant GST-RFXANK fusion proteins were produced in bacteria, mixed in different combinations with the other two subunits of RFX, and incubated with the 32P-labeled oligonucleotide containing the S and X boxes of the DRA promoter. Inputs of GST proteins were equal and the same as presented in Fig. 7 (GST input).
In our previous work we established a direct correlation between in vitro binding of RFXANK to RFXAP and the RFX complex assembly (26). The mutant RFXANK and RFXAP proteins that lacked domains required for their interaction were not able to assemble the RFX complex. However, despite extensive washing, a weak background signal from radiolabeled RFXAP was detected when the mutant GST-FRXANK-IH3 and -FZA fusion proteins were used in a GST pull-down assay (Fig. 7A, lanes 4 and 5), suggesting that a low percentage of input RFXANK and RFXAP proteins still interacted and could possibly assemble a small amount of the RFX complex in vivo. This binding could explain the previously observed residual expression of HLA-DR on the surface of less than 1% of lymphocytes from the FZA patient (25).
Our next experiment confirmed the above speculations. RFX5 and RFXAP were transcribed and translated in vitro and mixed with GST alone or the wild-type and mutant GST-RFXANK fusion proteins. EMSAs were performed by mixing different combinations of proteins with 32P-labeled SX oligonucleotide. As reported before, the DNA-RFX complex formed only when all three subunits were present (Fig. 7B, lane 3). GST alone did not have any effect on the complex formation (Fig. 7B, lane 2). Importantly, the mutant GST-RFXANK-IH3 and GST-RFXANK-FZA fusion proteins supported the assembly of only trace amounts of the complex (Fig. 7B, lanes 5 and 7, respectively). The competition with the unlabeled SX oligonucleotide completely abolished the formation of the complex, showing that the binding of RFX to DNA was specific (Fig. 7B, lanes 4, 6, and 8). These data clearly demonstrate that the mutation in RFXANK from the FZA patient blocks the assembly of the RFX complex.
DISCUSSION
In this study, we defined four ankyrin repeats of RFXANK. Next, we modeled and studied its three-dimensional structure on the basis of other known ankyrin repeat-containing proteins. Exposed variable residues were replaced with alanines. In this way, we were able to determine the surfaces of ankyrin repeats that interact with its two binding partners, RFXAP and CIITA. These surfaces are composed of scattered residues rather than continuous amino acid stretches. RFXAP contacts two surfaces of RFXANK: β-hairpin loops of the first three ankyrin repeats and one helix in the ankyrin groove. Contact points are limited and were clearly pinpointed. In contrast, CIITA binds RFXANK via multiple residues in the outer helices of the last three ankyrin repeats, which are located on the opposite side from the ankyrin groove of RFXANK. Alanine mutagenesis successfully positioned the binding partners of RFXANK into a complex protein network on MHC II promoters. Finally, we connected our binding studies to a disease. The FZA patient with BLS carries a single point mutation within RFXANK (25), resulting in an amino acid change within the inner helix of the third ankyrin repeat that is required for the binding to RFXAP. This mutation blocked the interaction between RFXANK and RFXAP in binding assays in vitro and the assembly of the RFX complex on DNA in EMSA. Thus, our mapping elucidates the background of yet another BLS mutation, which is responsible for the absence of MHC II determinants on the surface of B cells.
At the beginning of our mapping studies we were unable to detect an interaction between RFX5 and RFXANK in a stringent in vitro system (26), although these two subunits coimmunoprecipitated within the RFX complex from cells (data not shown). Thus, RFX5 requires a combinatorial surface of RFXANK and RFXAP to form a stable RFX complex. In contrast, direct interactions with RFXANK were obvious for RFXAP and CIITA. Therefore, we concentrated on the interaction between RFXANK and RFXAP for its essential role in the assembly of the RFX complex and on CIITA, which bound to a different surface of the ankyrin repeats. Although the ankyrin repeats of RFXANK were required for the binding to CIITA, no single or clustered point mutation abolished it. Moreover, CIITA is held on MHC II promoters by multiple interactions (33), suggesting that each one is relatively weak. Thus, an already weak interaction between CIITA and RFXANK is the sum of multiple contacts with its last three ankyrin repeats, a feature that makes their fine mapping an extremely difficult if not impossible task. Therefore, attempts to combine single and/or clustered point mutations to map this interaction precisely will most probably remain uninformative.
In this study, we combined direct binding assays with principles of structural biology that provided an advantage of looking at the protein as a module that can be changed without affecting its stability and conformation. Therefore, prediction of the three-dimensional structure of RFXANK represented a more reliable system for fine mapping of protein-protein interactions with its binding partners. The mutant protein from the FZA patient with BLS confirmed the importance of the conserved secondary-structure elements within the ankyrin repeats of RFXANK. Indeed, the leucine at position 195 does not play a structure-determining role by itself but is exposed on the surface of the inner helix 3 and is involved in the binding to RFXAP (Fig. 6B and C). However, the point mutation in the FZA patient that changes this residue to a proline destabilized the inner helix of the third ankyrin repeat and severely impaired the binding to RFXAP, which prevented normal nucleation of the RFX complex. Therefore, our mutagenesis distinguished between mutations that abolished the binding to the ankyrin repeat domain directly without affecting its overall secondary structure, as in the case of binding via β-hairpin loops, or indirectly by influencing its secondary structure.
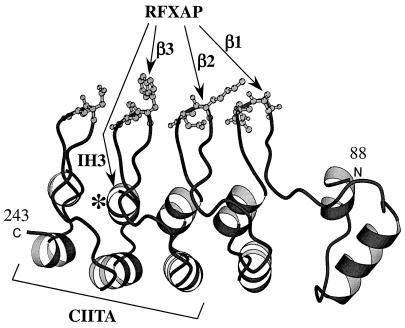
Our data show that RFXAP binds to two different surfaces of RFXANK (Fig. 8). These two surfaces are located on the same face of the ankyrin repeat domain and comprise the ankyrin groove that is shielded from the upper side by the cluster of four β-hairpin loops. It is easy to speculate that RFXAP fits into this groove much like a key fits into a lock and is stabilized in this position by interactions with the β-hairpin loops. In sharp contrast, CIITA does not bind to the β-hairpin loops of RFXANK but requires its last three ankyrin repeats. Therefore, CIITA binds to the opposite surface of RFXANK, which is composed of outer helices and turns.
FIG. 8.
Binding surfaces for RFXAP and CIITA on the model structure of the ankyrin repeat domain of RFXANK (residues 88 to 243). The arrows depict the secondary-structure elements of the ankyrin repeat domain that are required for binding to RFXAP. The predicted surface that contacts CIITA is also shown. β1, β2, and β3, β-hairpin loops of the first, second, and third ankyrin repeats, respectively; IH3, inner helix of the third ankyrin repeat; N, amino terminus; C, carboxy terminus; ∗, position of the L195P mutation in the FZA patient.
Although there are no specific secondary-structure elements within the ankyrin repeats that would be required for the binding of partner proteins, some common features exist. In the literature, there are many examples of other ankyrin repeat-binding proteins with the same binding pattern as that between RFXANK and RFXAP. For example, β-hairpin loops are a very common interaction site of ankyrin repeats. Since the inner two residues of this highly exposed motif (DxxG) are variable (Fig. 1A), they generate the specificity required for the recognition of different binding partners. All β-hairpin loops are involved in the interaction between the ankyrin-containing GABPβ and its DNA-binding partner protein GABPα (3). On the other hand, only the fourth β-hairpin loop is involved in the interaction between the ankyrin-containing 53BP2 and its binding partner p53 (17). Another group of nonconserved residues are those lying on the exposed face of α-helices in the ankyrin groove. Interactions between GABPα and GABPβ as well as Cdk kinase activity inhibitors p16INK4a and p19INK4d that bind to Cdk6 are examples of this type of interaction (6, 29).
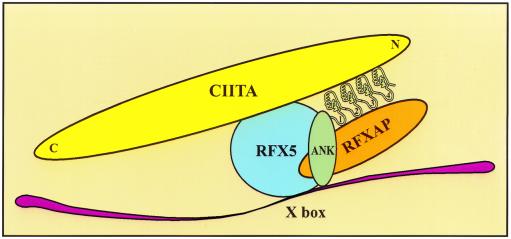
In addition, there are many ankyrin repeat-containing proteins with multiple binding partners. For example, the dimeric transcription factor NF-κB interacts with its inhibitor I-κB, which contains six ankyrin repeats (18). Two different domains of p65 as well as p50 bind to the ankyrin groove and β-hairpin loops of I-κB, respectively. Similarly, outer helices of ankyrin repeats can mediate protein-protein interactions (21). Therefore, all of the surfaces of ankyrin repeats of RFXANK that contact its binding partners have been verified in other systems. The architecture of the DNA-bound complex between RFX and CIITA and the central role of RFXANK in its assembly are summarized in our model in Fig. 9.
FIG. 9.
A model for RFXANK-mediated protein complex assembly on the X box of MHC II promoters. The β-hairpin loops of the first three ankyrin repeats of RFXANK interact with RFXAP, while the outer helices of the last three ankyrin repeats on the opposite face of the ankyrin repeat domain contact CIITA. The interaction between RFXAP and the inner helix of the third ankyrin repeat of RFXANK is not shown for simplicity of the model. Ankyrin repeats of RFXANK are depicted with numbers 1 to 4. N, amino terminus; C, carboxy terminus.
BLS is a unique genetic disease with a highly heterogeneous genetic background resulting in severe combined immunodeficiency. In general, different BLS mutations result in a disease that is more or less severe, depending on the amount of residual MHC II molecules on the surface of patient's B cells. This polymorphism could result from residual binding and activity of mutated proteins. This possibility was confirmed by our in vitro binding assays and EMSA with the mutant RFXANK protein from the FZA patient (Fig. 7). Moreover, the overexpression of CIITA can increase the surface expression of MHC II molecules on gamma interferon-treated FZA fibroblasts (25). On the other hand, large deletions of proteins that are common in most BLS patients cannot be compensated for unless mutated proteins are replaced by their wild-type counterparts. Finally, BLS has taught us a lot about the assembly of regulatory complexes on MHC II promoters and eukaryotic transcription. Although mutations in ankyrin repeats had been connected to a disease, namely, cancer (17, 30), they arose in somatic cells. To our knowledge, BLS is the first congenital disease that targets the ankyrin repeats.
ACKNOWLEDGMENTS
We thank Paula Zupanc-Ecimovic for secretarial assistance and other members of the laboratory for helpful discussions and comments on the manuscript.
Matthias Geyer acknowledges support from the Peter and Traudl Engelhorn Stiftung. This work was supported by a grant from the Nora Eccles Treadwell Foundation.
REFERENCES
- 1.Alberts A S, Treisman R. Activation of RhoA and SAPK/JNK signalling pathways by the RhoA-specific exchange factor mNET1. EMBO J. 1998;17:4075–4085. doi: 10.1093/emboj/17.14.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axton J M, Shamanski F L, Young L M, Henderson D S, Boyd J B, Orr-Weaver T L. The inhibitor of DNA replication encoded by the Drosophila gene plutonium is a small, ankyrin repeat protein. EMBO J. 1994;13:462–470. doi: 10.1002/j.1460-2075.1994.tb06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batchelor A H, Piper D E, de la Brousse F C, McKnight S L, Wolberger C. The structure of GABPalpha/beta: an ETS domain-ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 4.Benoist C, Mathis D. Regulation of major histocompatibility complex class-II genes: X, Y and other letters of the alphabet. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- 5.Boss J M. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 6.Brotherton D H, Dhanaraj V, Wick S, Brizuela L, Domaille P J, Volyanik E, Xu X, Parisini E, Smith B O, Archer S J, Serrano M, Brenner S L, Blundell T L, Laue E D. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature. 1998;395:244–250. doi: 10.1038/26164. [DOI] [PubMed] [Google Scholar]
- 7.Brünger A T. X-PLOR version 3.1: a system for X-ray crystallography and NMR. New Haven, Conn: Yale University Press; 1992. [Google Scholar]
- 8.Caretti G, Cocchiarella F, Sidoli C, Villard J, Peretti M, Reith W, Mantovani R. Dissection of functional NF-Y-RFX cooperative interactions on the MHC class II Ea promoter. J Mol Biol. 2000;302:539–552. doi: 10.1006/jmbi.2000.4028. [DOI] [PubMed] [Google Scholar]
- 9.Cogswell J P, Zeleznik-Le N, Ting J P. Transcriptional regulation of the HLA-DRA gene. Crit Rev Immunol. 1991;11:87–112. [PubMed] [Google Scholar]
- 10.Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu Rev Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 11.DeSandro A M, Nagarajan U M, Boss J M. Associations and interactions between bare lymphocyte syndrome factors. Mol Cell Biol. 2000;20:6587–6599. doi: 10.1128/mcb.20.17.6587-6599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontes J D, Jiang B, Peterlin B M. The class II trans-activator CIITA interacts with the TBP-associated factor TAFII32. Nucleic Acids Res. 1997;25:2522–2528. doi: 10.1093/nar/25.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontes J D, Kanazawa S, Nekrep N, Peterlin B M. The class II transactivator CIITA is a transcriptional integrator. Microbes Infect. 1999;1:863–869. doi: 10.1016/s1286-4579(99)00232-4. [DOI] [PubMed] [Google Scholar]
- 15.Foord R, Taylor I A, Sedgwick S G, Smerdon S J. X-ray structural analysis of the yeast cell cycle regulator Swi6 reveals variations of the ankyrin fold and has implications for Swi6 function. Nat Struct Biol. 1999;6:157–165. doi: 10.1038/5845. [DOI] [PubMed] [Google Scholar]
- 16.Glimcher L H, Kara C J. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 17.Gorina S, Pavletich N P. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs M D, Harrison S C. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 19.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 20.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 21.Mandiyan V, Andreev J, Schlessinger J, Hubbard S R. Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein beta. EMBO J. 1999;18:6890–6898. doi: 10.1093/emboj/18.24.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez J C, Hochstrasser D F, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 23.Michaely P, Bennett V. The membrane-binding domain of ankyrin contains four independently folded subdomains, each comprised of six ankyrin repeats. J Biol Chem. 1993;268:22703–22709. [PubMed] [Google Scholar]
- 24.Nagarajan U M, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss J M. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- 25.Nagarajan U M, Peijnenburg A, Gobin S J, Boss J M, van den Elsen P J. Novel mutations within the RFX-B gene and partial rescue of MHC and related genes through exogenous class II transactivator in RFX-B-deficient cells. J Immunol. 2000;164:3666–3674. doi: 10.4049/jimmunol.164.7.3666. [DOI] [PubMed] [Google Scholar]
- 26.Nekrep N, Jabrane-Ferrat N, Peterlin B M. Mutations in the bare lymphocyte syndrome define critical steps in the assembly of the regulatory factor X complex. Mol Cell Biol. 2000;20:4455–4461. doi: 10.1128/mcb.20.12.4455-4461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholls A, Sharp K A, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 28.Peitsch M C. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 29.Russo A A, Tong L, Lee J O, Jeffrey P D, Pavletich N P. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 30.Sedgwick S G, Smerdon S J. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 31.Steimle V, Durand B, Barras E, Zufferey M, Hadam M R, Mach B, Reith W. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Devarajan P, Dorfman A L, Morrow J S. Structure of the ankyrin-binding domain of alpha-Na,K-ATPase. J Biol Chem. 1998;273:18681–18684. doi: 10.1074/jbc.273.30.18681. [DOI] [PubMed] [Google Scholar]
- 33.Zhu X S, Linhoff M W, Li G, Chin K C, Maity S N, Ting J P. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol Cell Biol. 2000;20:6051–6061. doi: 10.1128/mcb.20.16.6051-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]