Abstract
The oncogenic MDM4, initially named MDMX, has been identified as a p53-interacting protein and a key upstream negative regulator of the tumor suppressor p53. Accumulating evidence indicates that MDM4 plays critical roles in the initiation and progression of multiple human cancers. MDM4 is frequently amplified and upregulated in human cancers, contributing to overgrowth and apoptosis inhibition by blocking the expression of downstream target genes of p53 pathway. Disruptors for MDM4-p53 interaction have been shown to restore the anti-tumor activity of p53 in cancer cells. MDM4 possesses multiple splicing isoforms whose expressions are driven by the presence of oncogenes in cancer cells. Some of the MDM4 splicing isoforms lack p53 binding domain and may exhibit p53-independent oncogenic functions. These features render MDM4 to be an attractive therapeutic target for cancer therapy. In the present review, we primarily focus on the detailed molecular structure of MDM4 splicing isoforms, candidate regulators for initiating MDM4 splicing, deregulation of MDM4 isoforms in cancer and potential therapy strategies by targeting splicing isoforms of MDM4.
Keywords: MDM4, p53, splicing, cancer therapy
Introduction
The murine MDM4 protein, also known as MDMX, was first discovered through a screen for p53 binding proteins in 1996 [1]. MDM4 shows high similarity with the MDM2 protein, a well-known E3 ubiquitin-protein ligase [2] that promotes proteasomal degradation of p53 [1]. Studies now established that MDM4 and MDM2 both function as important negative regulators of p53 and both are attractive therapeutic targets to reactivate the tumor suppression function of p53 [3,4]. However, the importance of MDM4 in p53 regulation was not clear for its predicted functional redundancy with MDM2 until revelation in MDM4 knockout mouse studies. MDM4-/- homozygous knockout mice died in utero, while concomitant deletion of the Trp53 gene could avoid the death [5-7], indicating a non-redundant role of MDM4 in regulation of p53. Furthermore, MDM4 was identified to be an MDM2-interacting protein in an unbiased screen [8], which made MDM4 as a potential regulator for MDM2 function in cell. MDM4 gene amplification or overexpression of MDM4 proteins were observed in many cancer types [4] and targeting MDM4 has been shown to be a valid strategy for p53-based cancer therapy [9,10]. Enthusiasm for developing MDM4-targeted cancer therapies is on the rise as a complimentary strategy for MDM2-p53 interaction inhibitors since MDM4 overexpression and p53 mutation poses a challenge to MDM2 inhibitor resistance [11,12]. Importantly, both MDM2 and MDM4 undergo alternative splicing and their splicing isoroms are cancer relevant. While MDM2 splicing has been studied actively and reviewed extensively, MDM4 splicing has drawn attention in recent years. The current review will focus on discussions on the detailed molecular structure of MDM4 splicing isoforms, candidate regulators for initiating MDM4 splicing, deregulation of MDM4 isoforms in cancer, update MDM4-trageting small molecules and implication in MDM4-targeted cancer therapies.
Structure and functions of MDM4
The human MDM4 gene has been mapped to chromosomal locus 1q32 [13]. Currently, about eight isoforms of MDM4 were identified (Figure 1). The longest full-length isoform (MDM4 isoform 1, MDM4-FL, NM_002393) contains 11 exons, and encodes a 490-amino acid protein with a predicted molecular weight at 54 kD. The structure for the mRNA and protein of MDM4 are depicted in Figure 1. In the following part, we mainly focused on four major conserved domains based on the protein structure: the amino-terminal p53-binding domain, the carboxy-terminal RING domain, the zinc-finger domain and the acidic domain (AD).
Figure 1.
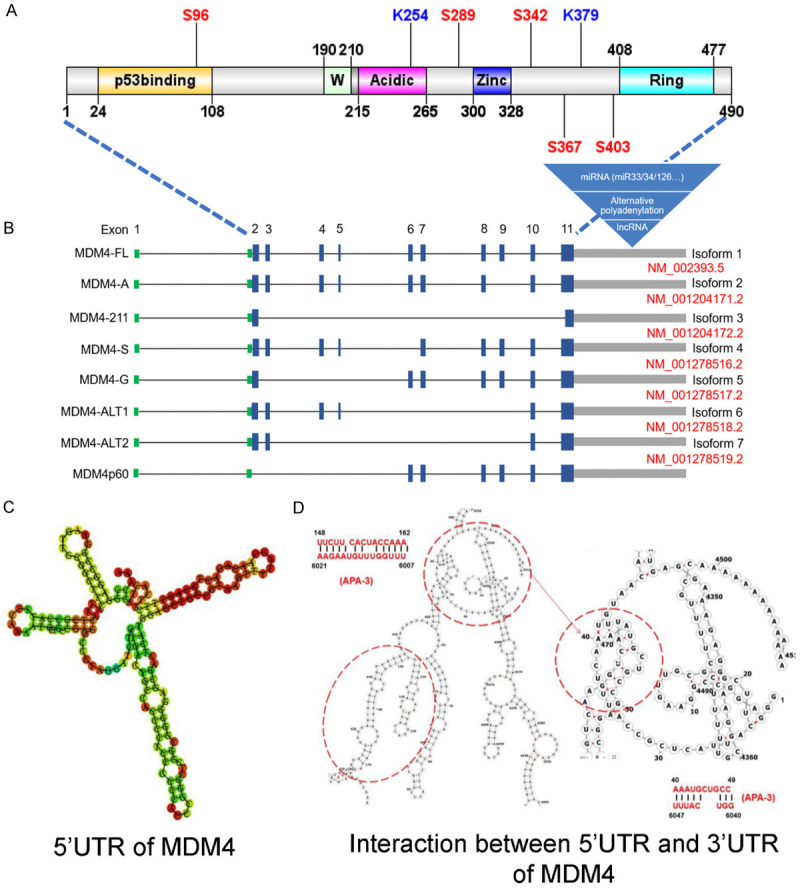
Molecular structures of MDM4 gene and protein. A. A diagram of MDM4 includes known domains and the variants present in this manuscript. B. The 5’UTRs are shown as green bars and the 3’UTRs are shown as grey bars. Separated by introns shown by black lines, exons are indicated by blue solid rectangles. Seven transcripts were listed in NCBI database with indicated accession numbers and one currently identified variant has no accession number. Multiple miRNAs, APA, UPF1 and STAU1 binding motifs were found in 3’UTR region of MDM4 mRNA. C. RNA secondary structure prediction for 5’UTR of MDM4 using online RNA fold program. D. Detailed information about 5’ and 3’UTR interacting regions predicted in MDM4 mRNA.
The amino-terminal p53-binding domain of MDM4
MDM4 protein includes a p53 binding domain located at N-terminus [14], of which the residues associated with p53 binding are strongly conserved in both MDM4 and MDM2 with 62% similarity [15]. Mutation of either residue of Phe19, Trp23 and Leu26 of p53 will abolish the binding between MDM4 and p53 [15], indicating high similarity of binding pockets of MDM4 and MDM2 for the association with p53 [16]. Nevertheless, a crystal structure analysis revealed the differential binding to p53 between MDM4 and MDM2 [17]. MDM4 has a smaller hydrophobic cleft for binding to the transactivation domain of p53 compared to MDM2, which will impact the binding affinity to the p53-TAD [18]. In addition to the binding pocket difference, full-length MDM4, but not MDM2, comprised a regulatory “WWW element”, which could bind to its N-terminal domain and prevent MDM4 from binding to p53 [19]. Some proteins competitively binding to this region may sequester the WWW sequence (aa 190-210, FEEWDVAGLPWWFLGNLRSNY) and unleash the p53-binding power of MDM4. This regulatory mechanism dictates that MDM4 splicing variants lacking the WWW sequence have deregulated high affinity to p53, which explains why they are found to be oncogenic in certain cancer cells [19-21].
The MDM4 RING domain
The second highly conserved motif of MDM4 and MDM2 is RING domain which is located at the carboxy-terminal region of both proteins. The MDM2 RING domain mainly contributes to its ubiquitin ligase activity [22,23]. However, MDM4 RING domain appears to lack this activity [24]. MDM2 can form homo oligomers via its RING domain in a manner dependent on its extreme C-terminal residues [25,26] and form heterodimers with MDM4 via RING-RING interaction [8]. In in vitro studies, MDM4 RING domain not only stimulates the E3 ligase of MDM2 [27], but in fact, is essential for MDM2-mediated p53 polyubiquitination and degradation [28,29]. Mouse genetic studies with MDM4 RING domain deletion or structural point mutation established that MDM4 RING domain is essential for p53 regulation in vivo during development [30,31]. Since MDM2 RING domain mutant mice also suffer from p53-dependent lethality, these studies established that RING-RING interface is key for MDM2 and MDM4 to partner to regulate p53 [32]. Interestingly, MDM4 RING domain can compensate the E3 ligase defects of MDM2 CT mutants for ubiquitination [33] and neddylation [34]. MDM4 RING domain contains a potential nuclear localization signal (NLS) (aa 465-469, RRLKK) which is usually hidden by the intramolecular interaction between RING and AD. DNA damage signaling can uncouple this RING-AD interaction by 14-3-3 with phosphor-MDM4 leading to nuclear localization and degradation of MDM4 in MDM3-dependent manner [35-37].
Zinc-finger domain and acidic domain
The function of the zinc-finger domain of MDM4 is largely unknown. Based on limited research, both MDM2 and MDM4 could interact with retinoblastoma protein and regulate its expression, however, the important roles of the zinc-finger domain for mediating these effects were only elucidated for MDM2 [38-40]. The cleavage of MDM4 at a canonical caspase cleavage site, close to the Zinc finger motif, has been proved to affect protein stability [41]. The AD domains mediated differential functions of MDM2 and MDM4. In MDM4, this domain is involved in the intramolecular interactions with the p53-binding domain and the RING domain, while the AD domain of MDM2, whose function couldn’t be fulfilled by the analogous domain of MDM4, is essential for p53 ubiquitination [42,43]. The AD in MDM2 is also involved in the interaction with ribosomal proteins and with p14ARF, but none of which were reported to interact with MDM4 so far. However, these proteins can affect the stability of MDM4 through their interaction with MDM2 [44-46].
Alternative splicing of MDM4
Alternative splicing is one of the fundamental mechanisms to regulate gene expression and plays important roles in cell biology including cancer cell biology. Through exon retention or skipping, alternative splicing contributes to protein diversity which directly dictates cellular states [47]. Alternative splicing functions as a powerful mechanism for dynamic regulation of protein stoichiometry involved in many pathways by generating spliced transcripts coupled with NMD degradation in the nucleus [48], unstable protein products [49], or stable truncated proteins lacking certain protein-interaction domains. In humans, approximately 95% of multi-exon genes have alternative splicing, and therefore huge effort is needed to garner a full picture of alternative splicing products as well as understanding the functions of different splicing isoforms of a single gene under normal physiological or pathological conditions [50].
Here, based on literature search and sequence analysis for MDM4 transcripts in UCSC Genome Browser and NCBI Nucleotide database, 8 different splicing isoforms of MDM4 were described, compared with the predominant MDM4 isoform 1 (NM_002393) (Figure 1).
MDM4 isoform 1 (MDM4-full length, MDM4-FL)
As mentioned above, this transcript variant represents the predominant transcript, and encodes the longest isoform (1) containing all the domains described for MDM4 in the literature and contributing to the most important function of MDM4 in p53 regulation together with MDM4-S.
MDM4 isoform 2 (MDM4-A)
MDM4-A was detected in the cervical cancer cell line C33A cells [51]. MDM4-A was generated by the removal of the exon 9 and annotated as MDM4 isoform 2 (NM_001204171) by National Center for Biotechnology Information (NCBI). This variant lacks an in-frame coding exon 9, resulting in a shorter protein missing most of the acidic region. Also, this alternative deletion of exon9 correlates with the MDM2 degradative activity and potentially controls the stability of MDM4-FL. In the meanwhile, the deletion of acidic domain of MDM4 could release the acidic domain of MDM2 and further promote its degradative function especially toward p53 [51]. From this aspect, MDM4-A possibly performs oncogenic roles through regulating the activity of MDM2 to accelerate the degradation of P53. However, no protein evidence of MDM4-A was provided so far.
MDM4 isoform 3 (MDM4-211)
This isoform was previously identified in the thyroid tumor cell line, ARO [52]. It derives from an aberrant splicing event between the canonical donor site in exon 2 and a cryptic acceptor site in exon 11 of the MDM4-FL mRNA (therefore named as MDM4-211), thus lacking eight consecutive exons from 3 to 10 and part of the last exon 11 (Figure 1). The accession number of this isoform in NCBI GenBank was NM_001204172. The open reading frame of this isoform encodes a protein lacking the p53-binding domain, while including the RING-finger domain at the C-terminus. MDM4-211 could only bind and stabilize MDM2 by increasing its half-life [52]. Moreover, the expression of MDM4-211 was limited to cancer types, such as non-small-cell lung cancer [52] and papillary thyroid carcinomas [53], but seldomly observed in normal tissues, indicating that this transcript was generated from an aberrant splicing event occurring only in tumor cells. Even though sharing the similar domains as MDM4-S, the MDM4-211 expression has no relevant to the levels of MDM4-FL transcript suggesting the independency of this splicing event.
MDM4 isoform 4 (MDM4-S)
This isoform is generated by the removal of the internal exon 6 through alternative splicing, which was first reported in rapid growing and transformed cell lines. It was known as MDM4-S, HDMX-S, or HDMX-E previously and annotated as MDM4 isoform 4 (NM_001278516). Since the length of exon 6 is 68 base pairs, this removal produces a shift in the open reading frame (ORF) after codon 114, resulting in the addition of a premature stop codon at amino acid residue 127 [54]. MDM4-S only comprises the p53 binding domain and was proved to be a stronger p53 inhibitor than MDM4-FL [54,55]. Compared with MDM4-FL, MDM4-S misses an auto-inhibitory sequence and shows more efficient nuclear localization [19]. However, the MDM4-S protein is barely detectable, much lower that MDM4-FL protein, even in a context that the mRNA levels for both isoforms are compatible, indicating potential post-transcriptional mechanisms that negatively regulate MDM4-S translation and/or stability. Besides, accumulating evidences support that the effect of exon 6 skipping is to negatively regulate the expression of MDM4-FL [53,56,57].
MDM4 isoform 5 (MDM4-G)
This isoform was detected at the same time as isoform 2 (MDM4-A), and previously known as MDM4-G. It lacks three consecutive exons (3-5) and part of the exon 6. This isoform was annotated as Mdm4 isoform 5 (NM_001278517). MDM4-G contains the same N- and C-termini domains as those of the MDM4-FL, while lacking the integrate p53-binding domain due to the in-frame deletion of aa 27-124 (Figure 1). Interestingly, although this variant can not bind to p53, it can still possess a weak inhibitory activity toward p53. This inhibition has been attributed to the stabilization of MDM2 levels and consequently of its oncogenic properties.
MDM4 isoform 6 (MDM4-XALT1)
In a study on MDM4 alternative splicing in responsive to genotoxic stress, two novel alternative transcripts of MDM4, called XALT1 and XALT2 were identified in several human cancer cell lines [58]. XALT1 derives from a splicing event lacking four consecutive exons (6-9) of MDM4-FL mRNA, resulting in a premature stop codon at the exon 10. Currently, XALT1 has been renamed as isoform 6 of MDM4 (NM_001278518). The predicted protein of XALT1 only comprises the p53 binding domain with 24 amino acids longer than the protein encoded by MDM4-S. XALT1 was speculated to suppress p53 transcriptional activity and inhibit the anti-apoptotic function of p53 [59]. The expression of this variant in non-tumor cells might perform different functions [58]. Overall, this isoform is potentially generated through a general splicing mechanism in response to genotoxic stress.
MDM4 isoform 7 (MDM4-ALT2)
XALT2 was identified in the same project as XALT1 [58] and annotated as MDM4 isoform 7 (NM_001278519). This isoform derives from a splicing between exon 3 and exon 10 and lacks six consecutive exons (4-9) as compared with the MDM4-FL (Figure 1). The predicted protein of XALT2 transcript lacks the p53 binding domain and retains the COOH-terminal RING finger domain. Thus, this variant may bind MDM4-FL and/or MDM2 splicing isoforms and regulate their function. This isoform has been detected in different tumor cell lines. Its presence was supposed to be another layer of regulation of the p53-MDM2-MDM4 network in response to DNA damage.
MDM4-p60
The MDM4p60 isoform was first reported in 2017, which is generated by alternative splicing between exon 2 and exon 6 to remove most part of the exon 2 and three consecutive exons (3-5). The transcript uses an alternative translation initiation codon to generate a protein lacking the N-terminal p53-binding domain [60]. Previous reports demonstrated that MDM4 could both stabilize and destabilize MDM2 [27,61,62]. Besides, with a similarity to the structure of MDM2p76, the MDM4p60 transcript also lacks the N-terminal p53 binding domain and displays a similar function as MDM2p76. MDM4p60 does not have a direct negative effect on p53. On the contrary, it can increase p53 protein levels through blocking the ability of MDM2p90 in mediating p53 degradation. Further investigation will tell whether hMDM4p60 can also play a positive role in the activation of p53 or not.
Candidate regulators for initiating MDM4 splicing
Although eight alternative-spliced isoforms of MDM4 were identified, current studies are mainly focused on two major forms: MDM4-FL (Isoform 1) and MDM4-S (Isoform 4). However, the splicing factors involved in the initiation of MDM4 splicing are still unknown. From a survey of published papers, we summarized candidate regulators involving in the splicing switch from MDM4-FL to MDM4-S isoforms as mechanisms for regulation of MDM4 function (Table 1).
Table 1.
Candidate regulators for initiating MDM4 splicing
| Gene | Function | Cell type | Reference |
|---|---|---|---|
| SRSF3 | necessary, but not sufficient, to promote inclusion of exon 6 in MDM4 mRNA | Melanoma; neural stem/progenitors; Colon Cancer; cutaneous squamous cell carcinoma | [63-65] |
| PRMT5 | Decreases alternative splicing of MDM4 and activates p53 by inhibiting PRMT5 | Melanoma; neural stem/progenitors | [56,66-68] |
| PRMT1 | Decreases alternative splicing of MDM4 and activates p53 by deletion of PRMT1 | Epicardial cells | [69] |
| RBM11 | exogenous RBM11 switches splicing of MDM4 and Cyclin D1 toward the expression of more oncogenic isoforms | Glioblastoma | [70] |
| Zmat3 | Increasing alternative splicing efficiency of MDM4 and inactivate p53 by deletion of Zmat3 | lung adenocarcinoma and Liver cancer | [71] |
| BCAS2 | Decreases alternative splicing of MDM4 and activates p53 by deletion of BCAS2 | Hematopoietic stem and progenitor cells | [72] |
It has been reported that the splicing enhancer serine and arginine rich splicing factor 3 (SRSF3) is a necessary, but not sufficient factor for mediating the inclusion of exon 6 in MDM4 mRNA, whereas SRSF7, SRSF9 and SRSF11 facilitate exclusion of exon 6 using MDM4-FL/MDM4-S ratio change as readout [63]. Of note, one study reported that knockdown of the splicing factor 3b subunit 1 (SF3B1) induced a dramatically changes for MDM2 splicing patterns, but not for MDM4 [64], whereas another study showed that SF3B1 inhibition, either by RNAi or the SF3B1 inhibitor pladienolide B, promotes the altered mRNA splicing and reduced protein expression of both MDM4 and MDM2 leading to p53 increase in cutaneous squamous cell carcinoma [65]. This discrepancy is probably due to low level of normally spliced MDM4 mRNA in the cells used in this study [64].
In addition to SRSF3, another candidate splicing regulator and transcription cofactor protein arginine methyltransferase 5 (PRMT5) triggers alternative splicing of MDM4 in neural stem/progenitor cells (NPCs) [56] and multiple other cancer types [66]. Depletion of PRMT5 by siRNA or PRMT5 inhibitor GSK3203591 promotes a splicing switch from long isoform to short MDM4 leading to shutdown of the expression of MDM4-FL protein and p53 activation. Besides, as a protein arginine methyltransferase and indirect target of CDK4, PRMT5 also plays important roles in response to the treatment of CDK4/6 inhibitors. Combined treatment of CDK4/6 inhibitor palbociclib and the PRMT5 inhibitor GSK3326595 enhances the efficacy of palbociclib and delays the resistance incidence in melanoma [67,68]. These studies uncovered a link between CDK4/6 activity and MDM4 expression via PRMT5 and further extended the mechanism of action of CDK4/6 inhibitors far beyond regulation of the cell cycle. Further investigations need to be performed to check the combined treatment of both CDK4/6 and the PRMT5 inhibitors in other cancer cell models such as breast, pancreatic, and esophageal carcinoma.
Arginine methyltransferase member PRMT1 is another regulator of MDM4 splicing for p53 activation during epicardial EMT and invasion [69]. Currently, there are no reports on PRMT1-dependent splicing regulation of MDM4 in cancer cell models, one can speculate that PRMT1 might play similar roles as PRMT5 in regulation of MDM4 splicing in certain types of cancer. RNA-binding proteins can be important regulators of MDM4 splicing machineries. One study has shown that RNA binding motif protein 11 (RBM11) can mediate switches of alternative splicing of MDM4 and Cyclin D1 to express more oncogenic isoforms [70]. Interestingly, another RNA-binding protein (ZMAT3, zinc finger matrin-type 3) was identified to be a key splicing regulator in the p53 tumor suppression program to maintain p53 tumor suppression functions. ZMAT3 is a p53 downstream gene product and can directly modulate exon inclusion of transcripts of diverse pathways including both p53 inhibitors MDM4 and MDM2. SgRNA-guided knockout of ZMAT3 leads to production of full-length MDM2 and MDM4 leading to p53 degradation and suppression. Importantly, ZMAT3 is a key mediator of p53 tumor suppressor function in mouse Kras (G12D)-driven lung, liver cancers and human carcinoma [71]. Finally, breast carcinoma amplified sequence 2 (BCAS2) was first identified as negative regulator in human cancer cells. Knockout of BCAS2 in zebrafish induces exon-6 skipping with increased production of MDM4-S leading to actvation and p53-mediated apoptosis in hematopoietic stem and progenitor cells [72]. These studies suggest that different MDM4 splicing regulators regulate MDM4 splicing in a tissue-specific manner.
Alternative splicing is a complex process and can be regulated by trans-acting proteins (repressors and activators) and corresponding cis-acting regulatory sites (silencers and enhancers) on the pre-mRNA [73]. Moreover, the secondary structure of the pre-mRNA transcript also plays a role in regulating splicing, such as by bringing together splicing elements or by masking a sequence that would otherwise serve as a binding element for a splicing factor [74,75]. The super-length 3’UTR (8.4 kb) of MDM4 provides plenty of potential binding sites for splicing factors or splicing enhancers, in addition, it can also form stable secondary structure to facilitate the process of splicing. Although several regulatory molecules in MDM4 splicing have been identified (Table 1), more factors are to be uncovered for a better understanding of the underlying splicing mechanisms in the context of disease and cancer types.
Deregulation of MDM4 isoforms in cancer
As a well-known upstream regulator of p53, the functions of MDM4-FL have been intensively investigated. The first report about the aberrant MDM4 expression in malignant gliomas can be tracked back to 1999 that MDM4 gene amplification correlated with high MDM4 expression in 4% of cancer tissues with wild-type p53 [76]. Later, the gene amplification phenomenon of MDM4 was also discovered in other cancer types including breast cancer [77], soft-tissue sarcoma [57], retinoblastomas [78], and cutaneous melanoma [10]. However, no correlation between mRNA expression and protein levels of MDM4 was observed in these cancer samples [79], suggesting that post-transcriptional or post-translational mechanisms might also contribute to the regulation of MDM4 protein levels in tumors.
Although some reports claimed that MDM4-S can be expressed in both normal and tumor cells with higher affinity of p53-binding than full-length MDM4 and the engineered MDM4-S protein primarily showed the nucleus localization [54,55], there is no conclusive evidence supporting the detection of endogenous MDM4-S protein levels in any normal or cancer cell lines. It was suggested that endogenous MDM4-S might be a very unstable and undetectable protein by regular western blotting, and that the switch from MDM4-FL to MDM4-S is one mechanism that cells undergoing stress use to shut down the expression of MDM4-FL protein, and thereby activate p53 [80]. Alternatively, exon 6 might function as an “nonsense-mediated decay (NMD) switch” exon [56,81] and could be recognized by the NMD machinery for degradation of MDM4-S transcripts [82]. To sort out these two possibilities warrants further experimentation.
High expression of MDM4-S has been associated with poor prognosis in multiple cancer types including osteosarcoma, soft tissue sarcoma, breast cancer, glioblastoma, melanoma, and chronic lymphocytic leukemia [63,80,83-85]. The question is whether MDM4-S is tumorigenic on its own, given its potential in p53 inhibition [19]. To answer this question, Lozano group using mouse models and concluded that MDM4-S overexpression is a consequence of splicing defects in tumor cells rather than a cause of tumor evolution since splicing of MDM4 does not promote tumor development or cooperate with other oncogenic insults to alter tumor latency or aggressiveness, yet it can be used as a possible biomarker [86]. Practically, the MDM4-FL/MDM4-S ratio correlates well with overall levels of full-length MDM4 protein and can be used as a reliable predictor of MDM4 protein abundance in panels of several tumor cell lines, including breast cancer, osteosarcomas, and uveal melanoma [4,63].
In addition to splicing regulators for regulation of MDM4 expression in trans, whether the untranslated regions of MDM4 mRNA also contributes to its alternative splicing remains unexplored. It is established that the untranslated regions (5’-UTR and a 3’-UTR) in mRNA can play fundamental roles in regulating the stability, function, and localization of mRNA and shaping the multiple diversity of cellular proteome [87]. This layer of regulation also applies to MDM4 since the MDM4’s 5’-UTR can inhibit ovalbumin synthesis when it is connected to the coding sequence of ovalbumin gene [88]. In addition, 5’-UTR of MDM4 has the potential to form a stable secondary structure (Figure 1C). Furthermore, miR-885-3p was reported to bind the 5’-UTR of MDM4 mRNA to elevate the MDM4 protein level, instead of inhibiting MDM4 expression by other miRNAs binding to 3’-UTR [89]. Besides, the MDM4 mRNA has an exceptionally long 3’-UTR of about 8.5 kb [90], which provides an opportunity for MDM4 mRNA to harbor many candidate miRNA targets within its 3’-UTR. Around 1,276 potential miRNA binding targets of MDM4 have been reported. However, only a small portion of these candidates can exert functional roles in the regulation of MDM4 translation based on cellular experiments. Up to date, around 24 miRNAs have been validated to regulate the expression of MDM4 in multiple cancer types and most of these miRNAs bind to the 3’-UTR of MDM4 and down-regulate the expression of MDM4 (Table 2).
Table 2.
Candidate miRNAs binding to MDM4 mRNA
| Name | Cell Type | Binding region of MDM4 | Function | Reference |
|---|---|---|---|---|
| miR-34a-5p | Non-Small Cell Lung Cancer Cells | 4238-4259 of MDM4 3’UTR (CATGCCAGCCTCCACACTGCC) | induces apoptosis of NSCLC cells | [119] |
| miR-887-3p | Non-small cell lung cancer patients | rs4245739, 32 of MDM4 3’UTR | inhibition of rs4245739 CC genotype on MDM4 expression | [120] |
| miR-191-5p | Small cell lung cancer patients | rs4245739, 32 of MDM4 3’UTR | MDM4 rs4245739 SNP contributes to SCLC risk | [121] |
| miR-887-3p | ||||
| miR-191-5p | Prostate cancer cells | rs4245739, 32 of MDM4 3’UTR | MDM4 rs4245739 SNP A-allele may be associated with an increased risk for prostate cancer | [122] |
| miR-887 | ||||
| miR-1307 | Breast cancer cell lines MCF-7 and MDA-MB-468 | 6038-6044 of MDM4 3’UTR (CCGGTCG) | modulating apoptosis by targeting MDM4 | [123] |
| miR-766 | Multiple cancer cell lines (MCF7, SBC3 and U2OS) | 1405-1412, 2684-2692, 3237-3245, 5519-5524 of MDM4 3’UTR (GGCTGGAG) | induces p53 accumulation and G2/M arrest | [124] |
| miR-661 | Multiple cancer cell lines (MCF7, A549 and H460) | 9 target regions in MDM4 3’UTR (CCCAGGC) | downregulates both MDM2 and MDM4 to activate p53 | [125] |
| miR-766 | Colon cancer | 918-932 of the MDM4 3’UTR (CATCCAAGCTGGAGT) | induces apoptosis of human colon cancer cells | [126] |
| miR-34a | Colorectal cancer cells | NA | miR-34a negatively regulates MDM4 gene expression to inhibit LOVO cell proliferation | [127] |
| miR-34a | Multiple cancer cell lines (H1299, MCF7 and U2OS) | 6038-1045 of Mdm4 exon 11 (GTCTGATATCACTGCCA) | modulates MDM4 expression via a Target Site in exon 11 | [128] |
| miR-205 | Colorectal cancer cells | 570-577 of the MDM4 3’UTR (ATGAAGGA) | suppresses cell migration, invasion and EMT of colon cancer | [129] |
| miR-128 | Pancreatic carcinoma cell line MIA PaCa-2 | 4617-4623 of MDM4 3’UTR (ACUGUG) | induces pancreas cancer cell apoptosis | [130] |
| miR-191 | Ovarian cancer cells | rs4245739, 32 of MDM4 3’UTR | delaying ovarian carcinoma progression and tumor-related death | [131] |
| miR-191 | Oropharyngeal cancer cells | rs4245739, 32 of MDM4 3’UTR | associated with HPV16-positive tumors and survival of oropharyngeal cancer | [132] |
| miR-10a | Acute myeloid leukemia | 511-531 of MDM4 3’UTR (AGGTGTGGGGCGACAGGGT) | associated with MDM4 downregulation in intermediate-risk acute myeloid leukemia | [133] |
| Let-7 | Glioma cells | 464-483 of MDM4 exon 7 (CCCCACACTGCCTACCTCA) | let-7 binding to MDM4 is implicated in the DNA damage response | [134] |
| Let-7 | Extravillous trophoblast cell* (htr-8/svneo) | 1543-1549 of the MDM4 3’UTR (TTGTACA) | inhibits the migration and invasion of extravillous trophoblast cell | [135] |
| miR-33a | Renal cell cancer | 4436-4442 of the MDM4 3’UTR (CAATGCAA) | inhibits cell growth in renal cancer | [136] |
| miR-301a-5p | Kidney cell* (HK2) | 89-96 of MDM4 3’UTR (GTCAGAGA) | induces kidney cell apoptosis | [137] |
| miR-885-3p | Squamous cell carcinomas cells | 30-54 of MDM4 5’UTR (ACTCGCCATTTCAAAATGCTGCCG) | miR-885-3p might contribute to the regulation of cell viability, apoptosis and/or autophagy in squamous cell carcinoma cells upon cisplatin exposure | [89] |
| miR-126 | Cervical cancer | 6389-6396 of MDM4 3’UTR (GTAATAAT) | the upregulation of miR-126 resulted in suppressed proliferation, accompanied by the induced apoptosis of CC cells | [138] |
| miR-150-5p | Cervical cancer | 8180-8186 of MDM4 3’UTR (TTGGGAG) | hsa_circ_0000263 can regulate the expression of MDM4 by affecting miR-150-5p | [139] |
represent non-cancer cell type;
NA: Not Available.
In addition to providing binding sites for miRNAs, the long 3’UTR of MDM4 also contains several alternative polyadenylation (APA) sites with conserved motif sequences (AATAAA). These APA sites do not alter the protein coding frame, while they might affect mRNA stability and translation efficiency [91-94]. Interestingly, the binding motifs of UPF1 and STAU1 can be found in 3’UTR of MDM4 [95]. UPF1 plays essential roles in both Staufen 1 (STAU1)-mediated mRNA decay (SMD) and nonsense-mediated mRNA decay (NMD) [96]. Stau1 functions as a splicing regulator and can regulate the alternative splicing of exon 11 of the human insulin receptor gene (INSR) through binding to Alu elements located in intron 10 [97]. Therefore, it is possible that STAU1 can be a regulator of MDM4/MDM4-S ratio by specifically regulating stability of either isoforms. Another possible mechanism involves the effect of the secondary structure of UTR sequences on mRNA translation. One such example is from p53 studies. The base-pairing interactions between 5- and 3-UTR of p53 positively regulate the translational efficiency of p53 mRNA in a RPL26-dependent manner [98]. Whether MDM4 mRNA uses the similar mechanism is an open question. We tried an online tool named RNA fold (http://www.tbi.univie.ac.at/RNA) to predict the secondary structure of human MDM4 mRNA based on the minimum free energy [99] and got a potential dsRNA structure comprising a complementary sequence located at both 5’ and 3’-UTR (Figure 1D). Taken together, MDM4 regulation through its long untranslated regions add another layer of complexity of MDM4 splicing and expression.
MDM4, the hub to restore p53 activity
Despite existence of multiple MDM4 isoforms, MDM4-FL is the only established key negative regulator of p53. Mouse models studies further established that MDM4 is a drug target for p53 reactivation therapy [9]. Up to date, several MDM4-targeting strategies have been attempted, including (1) directly blocking p53-MDM4 interaction, (2) degrading MDM4 protein and (3) inhibiting MDM4 expression. Currently, more than 10 inhibitors targeting directly to the MDM4-p53 binding interface have been reported, which could be divided into two main categories, short peptide inhibitors, such as SAH-p53-8 [100], mSF-SAH [101], ATSP-7041 [102] and ALRN-6924 [103]; and small molecular inhibitors, such as Compound B1 [104], CTX1 [105], K-178 [106], Protoporphyrin IX [107], SJ-172550 [108], RO-5963 [109], Pyrrolopyrimidine 3a [110] and WK298 [111] (Table 3). Some of these inhibitors show they hit the target leading to p53 activation in vitro and some inhibitors displayed promising anticancer efficacy and safety profiles in preclinical models in vivo [112]. Currently, only one short peptide inhibitor, ALRN-6924 was advanced to Phase 2 clinical trial study for lymphoma treatment (NCT02264613), Phase 1 for lung cancer (NCT04022876), Phase 1 for advanced or metastatic malignant solid neoplasm (NCT03725436), and Phase 1 for pediatric Cancer (NCT03654716). These MDM4-p53 disruptor drugs can also be used to combat resistance to MDM2 inhibitors. However, one limitation of targeting the MDM4-p53 interaction strategy is that it will not inhibit the p53-independent oncogenic activity of MDM4.
Table 3.
Inhibitors target for p53-MDMX binding
| Inhibitors | Mechanisms of action | References |
|---|---|---|
| Short peptide | ||
| SAH-p53-8 | Binds to p53-binding pocket of MDMX and blocks the p53-MDMX binding | [100] |
| mSF-SAH | Covalently binds to p53-binding pocket of MDMX and blocks the p53-MDMX binding | [101] |
| ATSP-7041 | Binds to MDM2 and MDMX and blocks the p53-MDM2/MDMX bindings | [102] |
| ALRN-6924 | Binds to MDM2 and MDMX and blocks the p53-MDM2/MDMX bindings | [103] |
| Molecular inhibitors | ||
| Compound B1 | Binds to p53-binding pocket of MDMX and blocks the p53-MDMX binding | [104] |
| CTX1 | Binds to MDMX, blocks the p53-MDMX binding, and activates p53 | [105] |
| K-178 | Inhibits the p53-MDMX binding and activates p53 | [106] |
| Protoporphyrin IX | Blocks the p53-MDM2/MDMX bindings and stabilizes p53 and TAp73 | [107] |
| SJ-172550 | Covalently but reversibly binds to p53-binding pocket of MDMX | [108] |
| RO-5963 | dual p53-MDM2 and p53-MDMX inhibitor | [109] |
| Pyrrolopyrimidine 3a | dual p53-MDMX/p53-MDM2 inhibitor | [110] |
| WK298 | Inhibition the binding of p53-MDMX | [111] |
As an alternative approach, targeting MDM4 protein abundance can be an effective way to overcome such limitation. In this approach, one strategy is to target MDM4 protein for degradation or inhibit MDM4 expression. Since MDM2-MDM4 heterodimers not only plays critical role for p53 degradation but also for MDM4 degradation [29]. Some compounds such as camptothecin analog FL118 and HSP90 inhibitor 17AAG can induce MDM2-dependent degradation of MDM4 [113,114]. Therefore, they can be used to overcome MDM4-mediated oncogenic activities. Another strategy to decrease MDM4 abundance is to manipulate MDM4 alternative splicing: i.e. to induce exon 6-skipping of MDM4 splicing which will introduce a premature stop codon in the mRNA leading to switch from MDM4-FL transcripts to MDM4-S transcripts that is not used for protein translation. This strategy is very effective in shutting down MDM4 expression at splicing level leading to p53 stabilization and reactivation. As mentioned above, several splicing related factors, such as SRSF3 and PRMT5 were reported to regulate MDM4 splicing (Table 1). Targeting SRSF3 and PRMT5 will induce MDM4 exon 6-skipping to shut down MDM4 expression. Elegantly, Gerhart et al. have shown that PRMT5 inhibition by a specific inhibitor GSK3203591 induces the alternative splicing of MDM4 and activates the p53 pathway as one of the mechanisms underlying the antitumor activity of PRMT5 inhibitors [66]. Similarly, SF3B1 inhibitor pladienolide B also causes wild-type p53 up-regulation through inefficient mRNA splicing that brought down protein expression of both MDM4 and MDM2, contributing to the antitumor effect of pladienolide B in cutaneous squamous cell carcinoma [65]. Another strategy for splicing manipulation is to use specific antisense oligonucleotides (ASOs). ASO against exon-intron boundaries of MDM4 exon 6 can mimic the in vivo splicing event and promotes the exon 6 skipping to decrease the MDM4 abundance (Figure 2). Up to date, around 100 different antisense drugs against various targets have been enrolled into various phase II and III clinical trials, including three oral ASOs (fomivirsen, mipomersen and inotersen) [115-117]. Specific ASOs that target MDM4 the exon-intron boundaries of MDM4 exon 6 also demonstrated effectiveness in silencing MDM4 expression in preclinical models [63]. Given the rapid development of ASO-related therapeutics, one can expect a fast advancement of this type of cancer therapeutics to clinic than other chemical entities.
Figure 2.
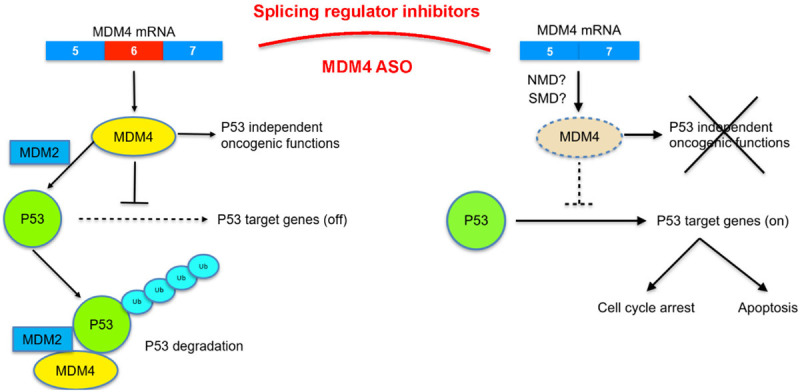
Targeting MDM4 splicing in cancer therapy. In normal adult tissues, exon6 of MDM4 tends to be skipped due to unproductive splicing. This will produce unstable transcript which is prone to be degraded through NMD or SMD pathway and decrease the abundance of MDM4-FL protein, which further transactivates and represses a number of target genes that function in apoptosis, cell cycle, and DNA repair (right part). While MDMX protein is highly expressed in embryonic tissues and in cancers due to the enhanced exon 6 inclusion. MDM4 can work together with MDM2 to mediate p53 degradation and perform both p53-dependent and independent oncogenic functions. Current strategy is that splicing regulator inhibitors (such as pladienoide b or GSK591) or MDM4 ASO can be used to induce MDM4 skipping to decrease the MDM4 abundance, and further inhibit p53-dependent or independent MDM4 oncogenic functions. Either splicing regulator inhibitors or antisense oligonucleotides (ASOs) are very specific, efficient, and clinically compatible approach.
Concluding remarks
Studies in the past 25 years established that up-regulation of MDM4 contributes to p53 inactivation and tumor progression and drug resistance to MDM2 inhibitors. Recent advances highlight the importance of alternative splicing in MDM4 regulation in the context of tumor development via p53 downstream gene ZMAT3 and manipulation of MDM4 splicing for targeted therapies. Although the biological effect of different alternatively spliced MDM4 isoforms is not well understood, MDM4 alternative splicing emerges as an effective regulatory mechanism to shutdown the expression of MDM4-FL mRNA and eliminate MDM4 protein expression. Consequently, MDM4-specific manipulation of splicing with MDM4 exon 6-targted ASO will be an attractive therapeutic strategy for MDM4-targted p53 reactivation therapy. This strategy also provides advantage over other inhibitors that alter splicing of diverse pathways. Finally, although strategies targeting MDM4 splicing is promising, two issues should be considered. First, the increased p53 activity caused by MDM4-FL inhibition will cause toxicity to normal cells and tissues [50,118]. Second, cancer cells with reduced MDM4-FL expression due to alternative MDM4 splicing often express a mutant p53 which will not respond to such strategies. Therefore, stratification of cancer patients into MDM4-high and MDM4-low expression groups in clinical trials will help identify patients who will likely benefit from this MDM4-targted therapies.
Acknowledgements
This work was supported in part by grants from R01CA208352 (XW), Roswell Park Alliance Foundation (XW) and by funding in China from the Key Research and Development Project of Deyang science and Technology Bureau (21ZDYF0011); Special Fund for Incubation Projects of People’s Hospital of Deyang City (FHG202004) and Projects of Department of Science and Technology of Sichuan Province (2018JY0389).
Disclosure of conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, Jochemsen AG. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 2.Haupt Y, Maya R, Kazaz A, Oren M. MDM2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 3.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 4.Marine JC, Jochemsen AG. MDMX (MDM4), a promising target for p53 reactivation therapy and beyond. Cold Spring Harb Perspect Med. 2016;6:a026237. doi: 10.1101/cshperspect.a026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in MDM4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 6.Migliorini D, Denchi EL, Danovi D, Jochemsen A, Capillo M, Gobbi A, Helin K, Pelicci PG, Marine JC. MDM4 (MDMX) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22:5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finch RA, Donoviel DB, Potter D, Shi M, Fan A, Freed DD, Wang CY, Zambrowicz BP, Ramirez-Solis R, Sands AT, Zhang N. MDMX is a negative regulator of p53 activity in vivo. Cancer Res. 2002;62:3221–3225. [PubMed] [Google Scholar]
- 8.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 9.Garcia D, Warr MR, Martins CP, Swigart LB, Passegué E, Evan GI. Validation of MDMX as a therapeutic target for reactivating p53 in tumors. Genes Dev. 2011;25:1746–1757. doi: 10.1101/gad.16722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, Zwolinska A, Haupt S, De Lange J, Yip D. MDM4 is a key therapeutic target in cutaneous melanoma. Nature Med. 2012;18:1239–1247. doi: 10.1038/nm.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman-Luca CG, Yang CY, Lu J, Ziazadeh D, McEachern D, Debussche L, Wang S. Significant differences in the development of acquired resistance to the MDM2 inhibitor SAR405838 between in vitro and in vivo drug treatment. PLoS One. 2015;10:e0128807. doi: 10.1371/journal.pone.0128807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem. 2006;281:33030–33035. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 13.Shvarts A, Bazuine M, Dekker P, Ramos YF, Steegenga WT, Merckx G, van Ham RC, van der Houven van Oordt W, van der Eb AJ, Jochemsen AG. Isolation and identification of the human homolog of a new p53-binding protein, MDMx. Genomics. 1997;43:34–42. doi: 10.1006/geno.1997.4775. [DOI] [PubMed] [Google Scholar]
- 14.Böttger V, Böttger A, Garcia-Echeverria C, Ramos YF, van der Eb AJ, Jochemsen AG, Lane DP. Comparative study of the p53-MDM2 and p53-MDMX interfaces. Oncogene. 1999;18:189–199. doi: 10.1038/sj.onc.1202281. [DOI] [PubMed] [Google Scholar]
- 15.Popowicz G, Czarna A, Holak T. Structure of the human MDMX protein bound to the p53 tumor suppressor transactivation domain. Cell cycle. 2008;7:2441–2443. doi: 10.4161/cc.6365. [DOI] [PubMed] [Google Scholar]
- 16.Popowicz GM, Czarna A, Rothweiler U, Szwagierczak A, Krajewski M, Weber L, Holak TA. Molecular basis for the inhibition of p53 by MDMX. Cell Cycle. 2007;6:2386–2392. doi: 10.4161/cc.6.19.4740. [DOI] [PubMed] [Google Scholar]
- 17.Worrall EG, Wawrzynow B, Worrall L, Walkinshaw M, Ball KL, Hupp TR. Regulation of the E3 ubiquitin ligase activity of MDM2 by an N-terminal pseudo-substrate motif. J Chem Biol. 2009;2:113–129. doi: 10.1007/s12154-009-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy MA, Gesell JJ, Senior MM, Wyss DF. Flexible lid to the p53-binding domain of human MDM2: implications for p53 regulation. Proc Natl Acad Sci U S A. 2003;100:1645–1648. doi: 10.1073/pnas.0334477100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bista M, Petrovich M, Fersht AR. MDMX contains an autoinhibitory sequence element. Proc Natl Acad Sci U S A. 2013;110:17814–17819. doi: 10.1073/pnas.1317398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Borcherds W, Wu S, Becker A, Schonbrunn E, Daughdrill GW, Chen J. Autoinhibition of MDMX by intramolecular p53 mimicry. Proc Natl Acad Sci U S A. 2015;112:4624–4629. doi: 10.1073/pnas.1420833112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S, Chen L, Becker A, Schonbrunn E, Chen J. Casein kinase 1α regulates an MDMX intramolecular interaction to stimulate p53 binding. Mol Cell Biol. 2012;32:4821–4832. doi: 10.1128/MCB.00851-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. MDM2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 23.Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 24.Marine JC, Jochemsen AG. MDMX as an essential regulator of p53 activity. Biochem Biophys Res Commun. 2005;331:750–760. doi: 10.1016/j.bbrc.2005.03.151. [DOI] [PubMed] [Google Scholar]
- 25.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. The MDM2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karni-Schmidt O, Lokshin M, Prives C. The roles of MDM2 and MDMX in cancer. Annu Rev Pathol. 2016;11:617–44. doi: 10.1146/annurev-pathol-012414-040349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linares LK, Hengstermann A, Ciechanover A, Müller S, Scheffner M. HDMX stimulates HDM2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci U S A. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain-mediatedinteraction is a requirement for MDM2’s E3 ligase activity. Cancer Res. 2007;67:6026–6030. doi: 10.1158/0008-5472.CAN-07-1313. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Wang J, Jiang X. MDMX protein is essential for MDM2 protein-mediated p53 polyubiquitination. J Biol Chem. 2011;286:23725–23734. doi: 10.1074/jbc.M110.213868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L, Yan Z, Liao X, Li Y, Yang J, Wang ZG, Zuo Y, Kawai H, Shadfan M, Ganapathy S. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci U S A. 2011;108:12001–12006. doi: 10.1073/pnas.1102309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pant V, Xiong S, Iwakuma T, Quintás-Cardama A, Lozano G. Heterodimerization of MDM2 and MDM4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and MDM2 stability. Proc Natl Acad Sci U S A. 2011;108:11995–12000. doi: 10.1073/pnas.1102241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X. p53 regulation: teamwork between RING domains of MDM2 and MDMX. Cell Cycle. 2011;10:4225–4229. doi: 10.4161/cc.10.24.18662. [DOI] [PubMed] [Google Scholar]
- 33.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh RK, Iyappan S, Scheffner M. Hetero-oligomerization with MDMX rescues the ubiquitin/Nedd8 ligase activity of RING finger mutants of MDM2. J Biol Chem. 2007;282:10901–10907. doi: 10.1074/jbc.M610879200. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto K, Kashima K, Pereg Y, Ishida M, Yamazaki S, Nota A, Teunisse A, Migliorini D, Kitabayashi I, Marine JC. DNA damage-induced phosphorylation of MDMX at serine 367 activates p53 by targeting MDMX for MDM2-dependent degradation. Mol Cell Biol. 2005;25:9608–9620. doi: 10.1128/MCB.25.21.9608-9620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, Biton S, Teunisse AF, Lehmann WD, Jochemsen AG. Phosphorylation of HDMx mediates its HDM2-and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci U S A. 2005;102:5056–5061. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LeBron C, Chen L, Gilkes DM, Chen J. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J. 2006;25:1196–1206. doi: 10.1038/sj.emboj.7601032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, He Y, Jin A, Tikunov AP, Zhou L, Tollini LA, Leslie P, Kim TH, Li LO, Coleman RA. Ribosomal protein-MDM2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc Natl Acad Sci U S A. 2014;111:E2414–E2422. doi: 10.1073/pnas.1315605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchida C, Miwa S, Isobe T, Kitagawa K, Hattori T, Oda T, Yasuda H, Kitagawa M. Effects of MDMX on MDM2-mediated downregulation of pRB. FEBS Lett. 2006;580:1753–1758. doi: 10.1016/j.febslet.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Hu L, Qiu W, Deng T, Zhang Y, Bergholz J, Xiao Z. MDMX exerts its oncogenic activity via suppression of retinoblastoma protein. Oncogene. 2015;34:5560–5569. doi: 10.1038/onc.2015.11. [DOI] [PubMed] [Google Scholar]
- 41.Gentiletti F, Mancini F, D’Angelo M, Sacchi A, Pontecorvi A, Jochemsen AG, Moretti F. MDMX stability is regulated by p53-induced caspase cleavage in NIH3T3 mouse fibroblasts. Oncogene. 2002;21:867–877. doi: 10.1038/sj.onc.1205137. [DOI] [PubMed] [Google Scholar]
- 42.Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol. 2003;23:4939–4947. doi: 10.1128/MCB.23.14.4939-4947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meulmeester E, Frenk R, Stad R, De Graaf P, Marine JC, Vousden KH, Jochemsen AG. Critical role for a central part of MDM2 in the ubiquitylation of p53. Mol Cell Biol. 2003;23:4929–4938. doi: 10.1128/MCB.23.14.4929-4938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Gilkes D, Li B, Cheng Q, Pernazza D, Lawrence H, Lawrence N, Chen J. Abnormal MDMX degradation in tumor cells due to ARF deficiency. Oncogene. 2012;31:3721–3732. doi: 10.1038/onc.2011.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daftuar L, Zhu Y, Jacq X, Prives C. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the MDM2-p53-MDMX network. PLoS One. 2013;8:e68667. doi: 10.1371/journal.pone.0068667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 48.Ge Y, Porse BT. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. Bioessays. 2014;36:236–243. doi: 10.1002/bies.201300156. [DOI] [PubMed] [Google Scholar]
- 49.Hao Y, Colak R, Teyra J, Corbi-Verge C, Ignatchenko A, Hahne H, Wilhelm M, Kuster B, Braun P, Kaida D. Semi-supervised learning predicts approximately one third of the alternative splicing isoforms as functional proteins. Cell Rep. 2015;12:183–189. doi: 10.1016/j.celrep.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 50.Bardot B, Toledo F. Targeting MDM4 splicing in cancers. Genes. 2017;8:82. [Google Scholar]
- 51.De Graaf P, Little NA, Ramos YF, Meulmeester E, Letteboer SJ, Jochemsen AG. HDMX protein stability is regulated by the ubiquitin ligase activity of MDM2. J Biol Chem. 2003;278:38315–38324. doi: 10.1074/jbc.M213034200. [DOI] [PubMed] [Google Scholar]
- 52.Giglio S, Mancini F, Gentiletti F, Sparaco G, Felicioni L, Barassi F, Martella C, Prodosmo A, Iacovelli S, Buttitta F. Identification of an aberrantly spliced form of HDMX in human tumors: a new mechanism for HDM2 stabilization. Cancer Res. 2005;65:9687–9694. doi: 10.1158/0008-5472.CAN-05-0450. [DOI] [PubMed] [Google Scholar]
- 53.Prodosmo A, Giglio S, Moretti S, Mancini F, Barbi F, Avenia N, Di Conza G, Schünemann HJ, Pistola L, Ludovini V. Analysis of human MDM4 variants in papillary thyroid carcinomas reveals new potential markers of cancer properties. J Mol Med. 2008;86:585–596. doi: 10.1007/s00109-008-0322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rallapalli R, Strachan G, Cho B, Mercer WE, Hall DJ. A novel MDMX transcript expressed in a variety of transformed cell lines encodes a truncated protein with potent p53 repressive activity. J Biol Chem. 1999;274:8299–8308. doi: 10.1074/jbc.274.12.8299. [DOI] [PubMed] [Google Scholar]
- 55.Rallapalli R, Strachan G, Tuan RS, Hall DJ. Identification of a domain within MDMX-S that is responsible for its high affinity interaction with p53 and high-level expression in mammalian cells. J Cell Biochem. 2003;89:563–575. doi: 10.1002/jcb.10535. [DOI] [PubMed] [Google Scholar]
- 56.Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, Guccione E. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for MDM4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013;27:1903–1916. doi: 10.1101/gad.219899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartel F, Schulz J, Böhnke A, Blümke K, Kappler M, Bache M, Schmidt H, Würl P, Taubert H, Hauptmann S. Significance of HDMX-S (or MDM4) mRNA splice variant overexpression and HDMX gene amplification on primary soft tissue sarcoma prognosis. Int J Cancer. 2005;117:469–475. doi: 10.1002/ijc.21206. [DOI] [PubMed] [Google Scholar]
- 58.Chandler DS, Singh RK, Caldwell LC, Bitler JL, Lozano G. Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4. Cancer Res. 2006;66:9502–9508. doi: 10.1158/0008-5472.CAN-05-4271. [DOI] [PubMed] [Google Scholar]
- 59.Shmueli A, Oren M. MDM2: p53’s lifesaver? Mol Cell. 2007;25:794–796. doi: 10.1016/j.molcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 60.Tournillon AS, López I, Malbert-Colas L, Naski N, Olivares-Illana V, Fåhraeus R. The alternative translated MDMXp60 isoform regulates MDM2 activity. Cell Cycle. 2015;14:449–458. doi: 10.4161/15384101.2014.977081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- 62.Stad R, Ramos YF, Little N, Grivell S, Attema J, van der Eb AJ, Jochemsen AG. HDMX stabilizes MDM2 and p53. J Biol Chem. 2000;275:28039–28044. doi: 10.1074/jbc.M003496200. [DOI] [PubMed] [Google Scholar]
- 63.Dewaele M, Tabaglio T, Willekens K, Bezzi M, Teo SX, Low DH, Koh CM, Rambow F, Fiers M, Rogiers A. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest. 2016;126:68–84. doi: 10.1172/JCI82534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allende-Vega N, Dayal S, Agarwala U, Sparks A, Bourdon J, Saville M. p53 is activated in response to disruption of the pre-mRNA splicing machinery. Oncogene. 2013;32:1–14. doi: 10.1038/onc.2012.38. [DOI] [PubMed] [Google Scholar]
- 65.Hepburn LA, McHugh A, Fernandes K, Boag G, Proby CM, Leigh IM, Saville MK. Targeting the spliceosome for cutaneous squamous cell carcinoma therapy: a role for c-MYC and wild-type p53 in determining the degree of tumour selectivity. Oncotarget. 2018;9:23029. doi: 10.18632/oncotarget.25196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gerhart SV, Kellner WA, Thompson C, Pappalardi MB, Zhang XP, Montes de Oca R, Penebre E, Duncan K, Boriack-Sjodin A, Le B, Majer C, McCabe MT, Carpenter C, Johnson N, Kruger RG, Barbash O. Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci Rep. 2018;8:1–15. doi: 10.1038/s41598-018-28002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.AbuHammad S, Cullinane C, Martin C, Bacolas Z, Ward T, Chen H, Slater A, Ardley K, Kirby L, Chan KT. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc Natl Acad Sci U S A. 2019;116:17990–18000. doi: 10.1073/pnas.1901323116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sheppard KE, AbuHammad S. CDK4/6 inhibition in cancer: the cell cycle splicing connection. Mol Cell Oncol. 2019;6:e1673643. doi: 10.1080/23723556.2019.1673643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jackson-Weaver O, Ungvijanpunya N, Yuan Y, Qian J, Gou Y, Wu J, Shen H, Chen Y, Li M, Richard S. PRMT1-p53 pathway controls epicardial EMT and invasion. Cell Rep. 2020;31:107739. doi: 10.1016/j.celrep.2020.107739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pavlyukov MS, Yu H, Bastola S, Minata M, Shender VO, Lee Y, Zhang S, Wang J, Komarova S, Wang J. Apoptotic cell-derived extracellular vesicles promote malignancy of glioblastoma via intercellular transfer of splicing factors. Cancer Cell. 2018;34:119–135. e110. doi: 10.1016/j.ccell.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bieging-Rolett KT, Kaiser AM, Morgens DW, Boutelle AM, Seoane JA, Van Nostrand EL, Zhu C, Houlihan SL, Mello SS, Yee BA. Zmat3 is a key splicing regulator in the p53 tumor suppression program. Mol Cell. 2020;80:452–469. e459. doi: 10.1016/j.molcel.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu S, Jiang T, Jia D, Han Y, Liu F, Huang Y, Qu Z, Zhao Y, Tu J, Lv Y. BCAS2 is essential for hematopoietic stem and progenitor cell maintenance during zebrafish embryogenesis. Blood. 2019;133:805–815. doi: 10.1182/blood-2018-09-876599. [DOI] [PubMed] [Google Scholar]
- 73.Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14:153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 74.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci. 2010;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reid DC, Chang BL, Gunderson SI, Alpert L, Thompson WA, Fairbrother WG. Next-generation SELEX identifies sequence and structural determinants of splicing factor binding in human pre-mRNA sequence. RNA. 2009;15:2385–2397. doi: 10.1261/rna.1821809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Riemenschneider MJ, Büschges R, Wolter M, Reifenberger J, Boström J, Kraus JA, Schlegel U, Reifenberger G. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 1999;59:6091–6096. [PubMed] [Google Scholar]
- 77.Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, De Graaf P, Francoz S, Gasparini P, Gobbi A. Amplification of MDMX (or MDM4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 79.McEvoy J, Ulyanov A, Brennan R, Wu G, Pounds S, Zhang J, Dyer MA. Analysis of MDM2 and MDM4 single nucleotide polymorphisms, mRNA splicing and protein expression in retinoblastoma. PLoS One. 2012;7:e42739. doi: 10.1371/journal.pone.0042739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lenos K, Jochemsen AG. Functions of MDMX in the modulation of the p53-response. J Biomed Biotechnol. 2011;2011:876173. doi: 10.1155/2011/876173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29:63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lenos K, Grawenda AM, Lodder K, Kuijjer ML, Teunisse AF, Repapi E, Grochola LF, Bartel F, Hogendoorn PC, Wuerl P. Alternate splicing of the p53 inhibitor HDMX offers a superior prognostic biomarker than p53 mutation in human cancer. Cancer Res. 2012;72:4074–4084. doi: 10.1158/0008-5472.CAN-12-0215. [DOI] [PubMed] [Google Scholar]
- 84.Liu L, Fan L, Fang C, Zou ZJ, Yang S, Zhang LN, Li JY, Xu W. S-MDM4 mRNA overexpression indicates a poor prognosis and marks a potential therapeutic target in chronic lymphocytic leukemia. Cancer Sci. 2012;103:2056–2063. doi: 10.1111/cas.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grawenda AM, Møller EK, Lam S, Repapi E, Teunisse AF, Alnæs GI, Børresen-Dale AL, Kristensen VN, Goding CR, Jochemsen AG. Interaction between p53 mutation and a somatic HDMX biomarker better defines metastatic potential in breast cancer. Cancer Res. 2015;75:698–708. doi: 10.1158/0008-5472.CAN-14-2637. [DOI] [PubMed] [Google Scholar]
- 86.Pant V, Larsson CA, Aryal N, Xiong S, You MJ, Quintas-Cardama A, Lozano G. Tumorigenesis promotes MDM4-S overexpression. Oncotarget. 2017;8:25837. doi: 10.18632/oncotarget.15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tournillon A, Lopez I, Malbert-Colas L, Findakly S, Naski N, Olivares-Illana V, Karakostis K, Vojtesek B, Nylander K, Fåhraeus R. p53 binds the MDMX mRNA and controls its translation. Oncogene. 2017;36:723–730. doi: 10.1038/onc.2016.236. [DOI] [PubMed] [Google Scholar]
- 89.Huang Y, Chuang AY, Ratovitski EA. Phospho-ΔNp63α/miR-885-3p axis in tumor cell life and cell death upon cisplatin exposure. Cell Cycle. 2011;10:3938–3947. doi: 10.4161/cc.10.22.18107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 92.Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18:18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Berkovits BD, Mayr C. Alternative 3’UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen W, Jia Q, Song Y, Fu H, Wei G, Ni T. Alternative polyadenylation: methods, findings, and impacts. Genomics Proteomics Bioinformatics. 2017;15:287–300. doi: 10.1016/j.gpb.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Lucas S, Oliveros JC, Chagoyen M, Ortín J. Functional signature for the recognition of specific target mRNAs by human Staufen1 protein. Nucleic Acids Res. 2014;42:4516–4526. doi: 10.1093/nar/gku073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bondy-Chorney E, Crawford Parks TE, Ravel-Chapuis A, Jasmin BJ, Côté J. Staufen1s role as a splicing factor and a disease modifier in Myotonic Dystrophy Type I. Rare Dis. 2016;4:e1005827. doi: 10.1080/21675511.2016.1225644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen J, Kastan MB. 5’-3’-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev. 2010;24:2146–2156. doi: 10.1101/gad.1968910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hofacker IL. RNA secondary structure analysis using the vienna RNA package. Curr Protoc Bioinformatics. 2009 doi: 10.1002/0471250953.bi1202s26. Chapter 12: Unit12.2. [DOI] [PubMed] [Google Scholar]
- 100.Bernal F, Wade M, Godes M, Davis TN, Whitehead DG, Kung AL, Wahl GM, Walensky LD. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoppmann C, Wang L. Proximity-enabled bioreactivity to generate covalent peptide inhibitors of p53-MDM4. Chem Commun. 2016;52:5140–5143. doi: 10.1039/c6cc01226d. [DOI] [PubMed] [Google Scholar]
- 102.Chang YS, Graves B, Guerlavais V, Tovar C, Packman K, To KH, Olson KA, Kesavan K, Gangurde P, Mukherjee A. Stapled α-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc Natl Acad Sci U S A. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carvajal LA, Neriah DB, Senecal A, Benard L, Thiruthuvanathan V, Yatsenko T, Narayanagari S-R, Wheat JC, Todorova TI, Mitchell K. Dual inhibition of MDMX and MDM2 as a therapeutic strategy in leukemia. Sci Transl Med. 2018;10:eaao3003. doi: 10.1126/scitranslmed.aao3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boltjes A, Huang Y, van de Velde R, Rijkee L, Wolf S, Gaugler J, Lesniak K, Guzik K, Holak TA, Dömling A. Fragment-based library generation for the discovery of a peptidomimetic p53-Mdm4 inhibitor. ACS Comb Sci. 2014;16:393–396. doi: 10.1021/co500026b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Karan G, Wang H, Chakrabarti A, Karan S, Liu Z, Xia Z, Gundluru M, Moreton S, Saunthararajah Y, Jackson MW. Identification of a small molecule that overcomes HDMX-mediated suppression of p53. Mol Cancer Ther. 2016;15:574–582. doi: 10.1158/1535-7163.MCT-15-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uesato S, Matsuura Y, Matsue S, Sumiyoshi T, Hirata Y, Takemoto S, Kawaratani Y, Yamai Y, Ishida K, Sasaki T. Discovery of new low-molecular-weight p53-MDMX disruptors and their anti-cancer activities. Bioorg Med Chem. 2016;24:1919–1926. doi: 10.1016/j.bmc.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 107.Jiang L, Malik N, Acedo P, Zawacka-Pankau J. Protoporphyrin IX is a dual inhibitor of p53/MDM2 and p53/MDM4 interactions and induces apoptosis in B-cell chronic lymphocytic leukemia cells. Cell Death Discov. 2019;5:1–11. doi: 10.1038/s41420-019-0157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bista M, Smithson D, Pecak A, Salinas G, Pustelny K, Min J, Pirog A, Finch K, Zdzalik M, Waddell B. On the mechanism of action of SJ-172550 in inhibiting the interaction of MDM4 and p53. PLoS One. 2012;7:e37518. doi: 10.1371/journal.pone.0037518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Graves B, Thompson T, Xia M, Janson C, Lukacs C, Deo D, Di Lello P, Fry D, Garvie C, Huang KS. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci U S A. 2012;109:11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee JH, Zhang Q, Jo S, Chai SC, Oh M, Im W, Lu H, Lim HS. Novel pyrrolopyrimidine-based α-helix mimetics: cell-permeable inhibitors of protein-protein interactions. J Am Chem Soc. 2011;133:676–679. doi: 10.1021/ja108230s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lemos A, Leao M, Soares J, Palmeira A, Pinto M, Saraiva L, Sousa ME. Medicinal chemistry strategies to disrupt the p53-MDM2/MDMX interaction. Med Res Rev. 2016;36:789–844. doi: 10.1002/med.21393. [DOI] [PubMed] [Google Scholar]
- 112.Yu DH, Xu ZY, Mo S, Yuan L, Cheng XD, Qin JJ. Targeting MDMX for cancer therapy: rationale, strategies and challenges. Front Oncol. 2020;10:1389. doi: 10.3389/fonc.2020.01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ling X, Xu C, Fan C, Zhong K, Li F, Wang X. FL118 induces p53-dependent senescence in colorectal cancer cells by promoting degradation of MDMX. Cancer Res. 2014;74:7487–7497. doi: 10.1158/0008-5472.CAN-14-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vaseva A, Yallowitz A, Marchenko N, Xu S, Moll U. Blockade of Hsp90 by 17AAG antagonizes MDMX and synergizes with Nutlin to induce p53-mediated apoptosis in solid tumors. Cell Death Dis. 2011;2:e156–e156. doi: 10.1038/cddis.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Raal FJ, Santos RD, Blom DJ, Marais AD, Charng MJ, Cromwell WC, Lachmann RH, Gaudet D, Tan JL, Chasan-Taber S. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 116.Hair P, Cameron F, McKeage K. Mipomersen sodium: first global approval. Drugs. 2013;73:487–493. doi: 10.1007/s40265-013-0042-2. [DOI] [PubMed] [Google Scholar]
- 117.Monteleone G, Neurath MF, Ardizzone S, Di Sabatino A, Fantini MC, Castiglione F, Scribano ML, Armuzzi A, Caprioli F, Sturniolo GC. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn’s disease. N Engl J Med. 2015;372:1104–1113. doi: 10.1056/NEJMoa1407250. [DOI] [PubMed] [Google Scholar]
- 118.Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine JC. MDM4 and MDM2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jiang ZQ, Li MH, Qin YM, Jiang HY, Zhang X, Wu MH. Luteolin inhibits tumorigenesis and induces apoptosis of non-small cell lung cancer cells via regulation of MicroRNA-34a-5p. Int J Mol Sci. 2018;19:447. doi: 10.3390/ijms19020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang Y, Gao W, Ding X, Xu W, Liu D, Su B, Sun Y. Variations within 3’-UTR of MDM4 gene contribute to clinical outcomes of advanced non-small cell lung cancer patients following platinum-based chemotherapy. Oncotarget. 2017;8:16313. doi: 10.18632/oncotarget.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gao F, Xiong X, Pan W, Yang X, Zhou C, Yuan Q, Zhou L, Yang M. A regulatory MDM4 genetic variant locating in the binding sequence of multiple microRNAs contributes to susceptibility of small cell lung cancer. PLoS One. 2015;10:e0135647. doi: 10.1371/journal.pone.0135647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stegeman S, Moya L, Selth LA, Spurdle AB, Clements JA, Batra J. A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer. Endocr Relat Cancer. 2015;22:265–276. doi: 10.1530/ERC-15-0013. [DOI] [PubMed] [Google Scholar]
- 123.Wang X, Zhu J. Mir-1307 regulates cisplatin resistance by targeting MDM4 in breast cancer expressing wild type P53. Thorac Cancer. 2018;9:676–683. doi: 10.1111/1759-7714.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Q, Selth LA, Callen DF. MiR-766 induces p53 accumulation and G2/M arrest by directly targeting MDM4. Oncotarget. 2017;8:29914. doi: 10.18632/oncotarget.15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hoffman Y, Bublik D, Pilpel Y, Oren M. miR-661 downregulates both MDM2 and MDM4 to activate p53. Cell Death Differ. 2014;21:302–309. doi: 10.1038/cdd.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen W, Cai G, Liao Z, Lin K, Li G, Li Y. miRNA-766 induces apoptosis of human colon cancer cells through the p53/Bax signaling pathway by MDM4. Exp Ther Med. 2019;17:4100–4108. doi: 10.3892/etm.2019.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li S, Pei Y, Zhang W, Sun L, Jiang M, Shang G, Niu S, Ma Z. Effects of MDM4 gene regulation by miR-34a on LOVO cell proliferation. Ann Clin Lab Sci. 2020;50:342–347. [PubMed] [Google Scholar]
- 128.Mandke P, Wyatt N, Fraser J, Bates B, Berberich SJ, Markey MP. MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS One. 2012;7:e42034. doi: 10.1371/journal.pone.0042034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fan Y, Wang K. miR-205 suppresses cell migration, invasion and EMT of colon cancer by targeting mouse double minute 4. Mol Med Rep. 2020;22:633–642. doi: 10.3892/mmr.2020.11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Han H, Wang L, Xu J, Wang A. miR-128 induces pancreas cancer cell apoptosis by targeting MDM4. Exp Ther Med. 2018;15:5017–5022. doi: 10.3892/etm.2018.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wynendaele J, Böhnke A, Leucci E, Nielsen SJ, Lambertz I, Hammer S, Sbrzesny N, Kubitza D, Wolf A, Gradhand E. An illegitimate microRNA target site within the 3’UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010;70:9641–9649. doi: 10.1158/0008-5472.CAN-10-0527. [DOI] [PubMed] [Google Scholar]
- 132.Zhang Y, Sturgis EM, Wei P, Liu H, Wang Z, Ma Y, Liu C, Gu KJ, Wei Q, Li G. A genetic variant within MDM4 3’UTR miRNA binding site is associated with HPV16-positive tumors and survival of oropharyngeal cancer. Mol Carcinog. 2019;58:2276–2285. doi: 10.1002/mc.23116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ovcharenko D, Stölzel F, Poitz D, Fierro F, Schaich M, Neubauer A, Kelnar K, Davison T, Müller-Tidow C, Thiede C. miR-10a overexpression is associated with NPM1 mutations and MDM4 downregulation in intermediate-risk acute myeloid leukemia. Exp Hematol. 2011;39:1030–1042. e1037. doi: 10.1016/j.exphem.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 134.Xie C, Chen W, Zhang M, Cai Q, Xu W, Li X, Jiang S. MDM4 regulation by the let-7 miRNA family in the DNA damage response of glioma cells. FEBS Lett. 2015;589:1958–1965. doi: 10.1016/j.febslet.2015.05.030. [DOI] [PubMed] [Google Scholar]
- 135.Zhang L, Wang K, Wu Q, Jin L, Lu H, Shi Y, Liu L, Yang L, Lv L. Let-7 inhibits the migration and invasion of extravillous trophoblast cell via targeting MDM4. Mol Cell Probes. 2019;45:48–56. doi: 10.1016/j.mcp.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 136.Jiang K, Sun F, Zhu J, Luo G, Ban Y, Zhang P. miR-33a inhibits cell growth in renal cancer by downregulation of MDM4 expression. Mol Genet Genomic Med. 2019;7:e833. doi: 10.1002/mgg3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang J, Li H, Qiu S, Dong Z, Xiang X, Zhang D. MBD2 upregulates miR-301a-5p to induce kidney cell apoptosis during vancomycin-induced AKI. Cell Death Dis. 2017;8:e3120–e3120. doi: 10.1038/cddis.2017.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tian Y, Xu Z, Fu J. CircularRNA-9119 promotes the proliferation of cervical cancer cells by sponging miR-126/MDM4. Mol Cell Biochem. 2020;470:53–62. doi: 10.1007/s11010-020-03745-3. [DOI] [PubMed] [Google Scholar]
- 139.Cai H, Zhang P, Xu M, Yan L, Liu N, Wu X. Circular RNA hsa_circ_0000263 participates in cervical cancer development by regulating target gene of miR-150-5p. J Cell Physiol. 2019;234:11391–11400. doi: 10.1002/jcp.27796. [DOI] [PubMed] [Google Scholar]


