Abstract
Chemosensory systems control the most complex, specialized mode of signal transduction in bacteria and archaea. They are composed of several core and auxiliary protein components that are highly organized in order to deliver a fast response to changing environmental conditions. Chemosensory pathways have been studied in-depth in a handful of model organisms and experimentally characterized at least to some degree in approximately thirty species. However, genome-wide analyses have revealed their presence in thousands of sequenced microbial genomes. Both experimental and computational studies uncovered substantial diversity in system design, functional regulation, cellular localization and phyletic distribution of chemosensory pathways. Here, we summarize advances and expose gaps in our current understanding of the diversity of chemosensory systems.
Introduction
Signal transduction pathways control cellular processes in all living organisms. Based on their component design, these pathways in bacteria and archaea can be classified into three major modes: one-component, two-component and chemosensory systems [1]. The chemosensory system (also called chemotaxis system) is a special case of two-component signal transduction. Its designation as a distinct mode is dictated by a complex system design and unique elements that are not seen in classical two-component pathways (Fig. 1). A pathway controlling chemotaxis in Escherichia coli is the best understood chemosensory system [2,3]. Similar to classical two-component signal transduction systems, the key components of this pathway are a histidine kinase and its cognate response regulator. The CheA histidine kinase lacks an input domain and receives signals from dedicated chemoreceptors (also called methyl-accepting chemotaxis proteins, MCPs) that share a repertoire of input domains with sensor histidine kinases of classical two-component systems. Chemoreceptors, CheA and the CheW scaffolding protein form signaling complexes that assemble into chemosensory arrays [4]. Upon changes in concentration of attractants and repellents, chemoreceptors modulate CheA activity, which ultimately controls flagellar rotation via phosphorylation of the CheY response regulator. The CheR methyltransferase and the CheB methylesterase that covalently modify MCPs by adding and removing methyl groups, correspondingly, comprise an adaptation pathway. The system also involves the CheZ phosphatase, which de-phosphorylates CheY leading to signal termination. Homologous systems were identified and experimentally studied in other bacterial and archaeal species, where they control not only flagellar motility, but also regulate type IV pili (Tfp) based motility, development, biosynthesis, biofilm formation, cell-cell interactions and other cellular functions.
Figure 1. Chemosensory systems as complex type of two-component signal transduction.
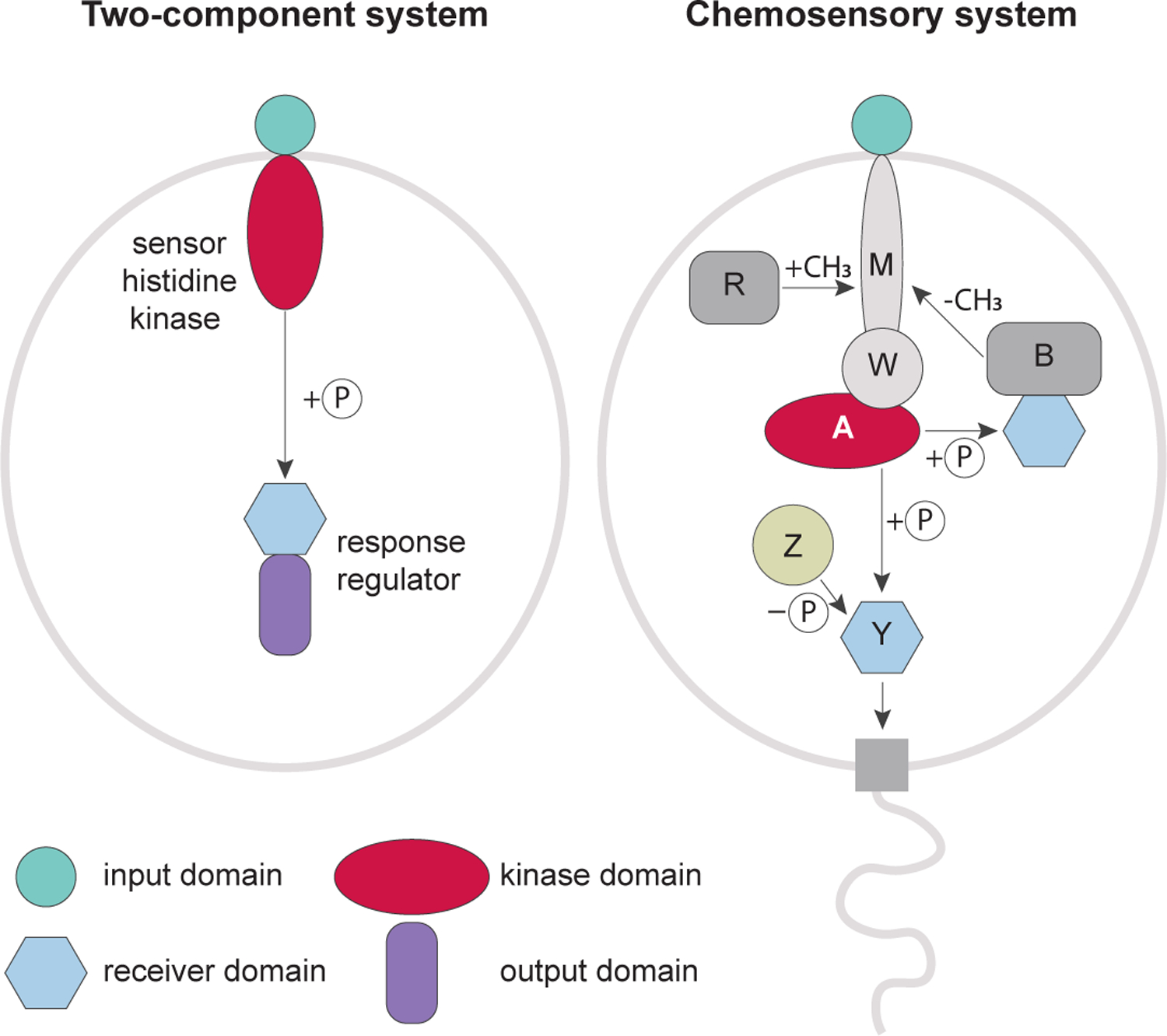
Prototypical two-component systems consists of two proteins: a transmembrane sensor histidine kinase, which contains an input (e.g. ligand-binding) domain and a cognate response regulator, which contains an output (e.g. DNA-binding) domain. A chemosensory system of E. coli has the sensor-less (no input domain) histidine kinase CheA and its response regulator CheY, which lacks an output domain and directly interacts with the flagellar motor. Abbreviations: M, MCP; W, CheW; A, CheA; B, CheB; R, CheR; Y, CheY; Z, CheZ; P, a phosphate group.
Diversity of system design
A large-scale phylogenomic analysis of chemosensory systems, which was carried out a decade ago, uncovered their remarkable diversity [5]. Merging large volumes of published experimental data with phylogenomic clustering revealed the existence of three principal functional groups of chemosensory systems: those that evolve to control flagellar motility (Fla), Tfp-based motility (Tfp), and alternative (non-motility) cellular functions (ACF). While the Fla group appeared to be very diverse, with seventeen distinct classes (F1-F17), each of the other two groups contained a single class. The core of the chemosensory system includes four essential components (≥ 95% occurrence in genomes with at least one chemosensory system gene [5]) comprising the excitation pathway, as it is seen in E. coli: MCPs, CheA, CheW, and CheY. Two other core components, CheR and CheB, are dispensable (≥ 85% occurrence). For example, they are present in the Tfp system in Pseudomonas aeruginosa but missing from the Tfp system in cyanobacteria.
Various classes of the chemosensory system differ substantially in their repertoire of auxiliary (≤ 60% occurrence) components (Fig. 2). Many chemosensory pathways contain an additional scaffolding protein, CheV, which is a fusion of CheW and receiver (CheY-like) domains [6]. It was proposed that CheV might couple CheA with certain types of chemoreceptors that cannot be effectively accommodated by CheW, and its receiver domain might function as a phosphate sink preventing over-stimulation of CheA [7]. In Vibrio cholerae, a CheW homolog called ParP incorporates into the chemosensory baseplate to promote array formation and localization near the flagellar pole [8]. The diversity of scaffolding CheW homologs is still not fully understood and current automated annotations cannot distinguish between their biological functions Another common auxiliary component is the CheD deamidase that acts on MCPs to augment CheR-mediated methylation, as shown in such distantly related systems as F1 from Bacillus subtilis [9] and F7 (Che2) from P. aeruginosa [10]. The E. coli chemosensory system utilizes the CheZ phosphatase, however, homologous systems in many other prokaryotes do not have CheZ, but instead contain other phosphatases for signal termination [11]. For example, the B. subtilis system has two phosphatases: FliY, which is an integral part of flagellar motor C-ring, and CheC, which localizes with chemoreceptors [9]. Many chemotactic bacteria, including spirochetes Treponema denticola and Borrelia burgdorferi, encode yet another phosphatase, CheX, which is homologous to CheC, but unlike CheC has only one active site and does not require CheD binding [11]. Apart from modification proteins, additional components are found in some classes of the chemosensory system. A small STAS domain-containing protein, CheS, identified in Sinorhizobium meliloti, provides an efficient drainage of the phosphate from CheY1 increasing its binding to CheA more than 100 times [12]. A novel chemotaxis regulator, ChePep, localizes to the flagellar pole and controls swimming direction and switching of flagellar rotation in the Helicobacter pylori [13]. A chemosensory pathway Che7/F13 in Myxococcus xanthus involves a HEAT-repeat protein, which couples aggregation and sporulation [14].
Figure 2. Major classes of the chemosensory system.
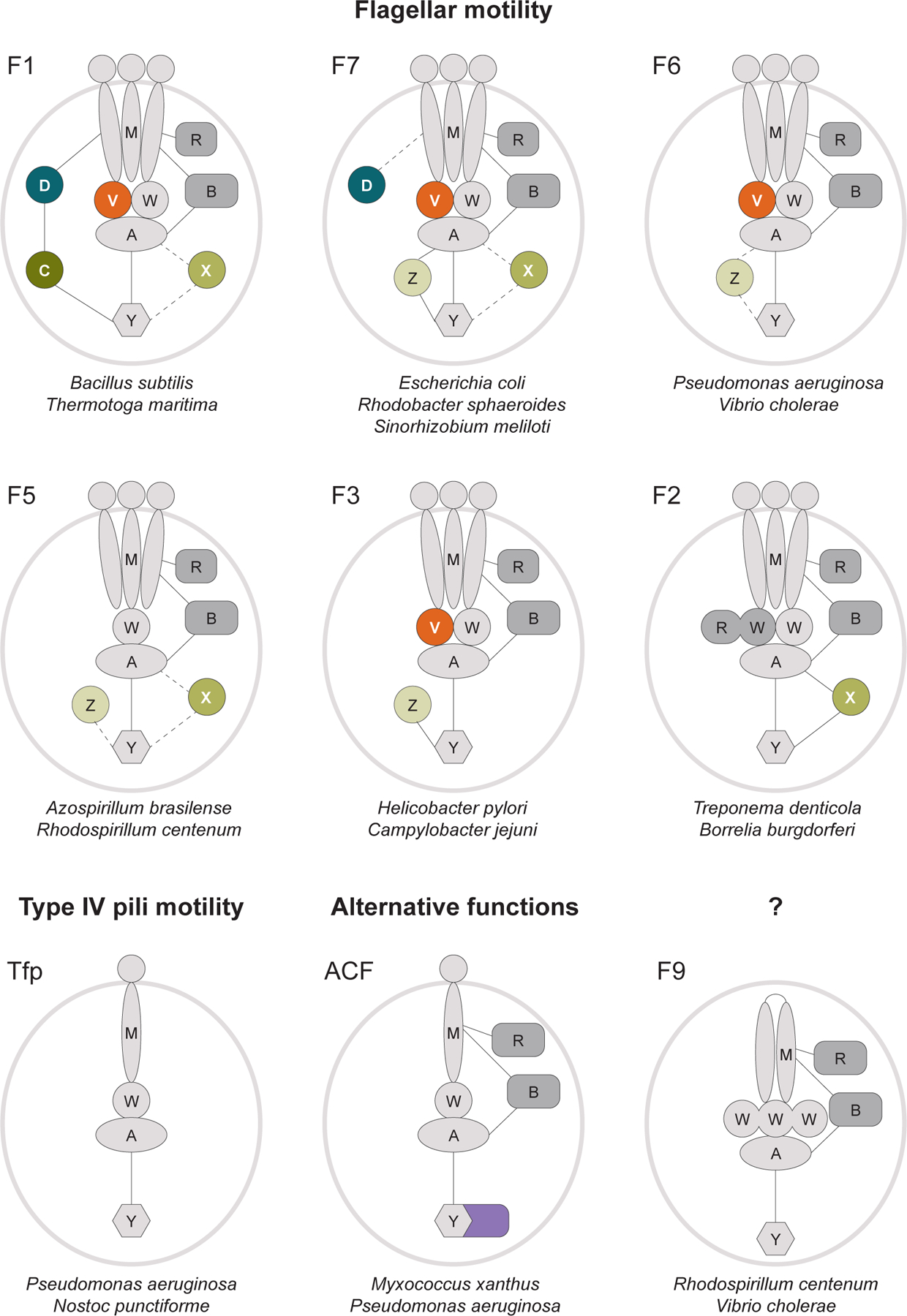
Model organisms in which corresponding chemosensory pathways were studied experimentally are shown for each class. Typical architectures are displayed, but protein repertoire within each class may vary. Tfp, ACF and F9 pathways typically contain a single MCP, whereas multiple MCPs are associated with all other classes. Core essential components (M, MCP; A, histidine kinase CheA; W, scaffolding protein CheW; Y, response regulator CheY) are in light grey; core dispensable components (R, methyltransferase CheR; B, methylesterase CheB) are in dark grey; the auxiliary proteins (D, deamidase CheD; V, scaffolding protein CheV; C, Z and X, phosphatases CheC, CheZ and CheX, respectively) and output domain of response regulator protein from ACF system are shown in color. Experimentally confirmed protein-protein interactions are represented by solid lines; dashed lines indicate predicted interactions.
The number of individual components per system may also vary. Whereas E. coli has one CheY protein, the chemosensory system of the same F7 class in S. meliloti has two: CheY1 and CheY2. In this pathway, the level of phosphorylated CheY2 responsible for flagellar control is reset by phospho-transfer back to CheA, and then to CheY1, which plays a role of a phosphate sink [15]. The Frz/ACF pathway in M. xanthus contains two CheW proteins with distinct roles in signaling and chemosensory cluster formation [16]. While Campylobacter jejuni has one CheV protein, its close relative H. pylori has three CheV proteins in its single F3 chemosensory pathway [17]. Another dimension of variability in chemosensory systems is brought about by uncommon domain fusions. Although in the vast majority of chemosensory systems CheW is a single domain protein, its homolog from the F9 system in V. cholerae has three CheW domains [18]. The T. denticola F2 chemosensory pathway contains a CheW-CheR fusion protein that plays a critical role in maintaining an extremely curved chemoreceptor array [19]. The CheA protein from Che1/F5 system of Azospirillum brasilense Sp7 contains the N-terminal TMX domain, which anchors the kinase to the membrane [20].
Chemoreceptors are by far the most diverse components of the chemosensory system. Even their highly conserved cytoplasmic domain displays substantial diversity in the number of its helical heptads comprising several distinct classes [21] that are preferentially associated with certain classes of the chemosensory system [5,22]. Truly remarkable diversity of chemoreceptors is seen in the number of different input domains, with nearly one hundred identified to date [23]. Furthermore, the number of chemoreceptors varies dramatically between organisms and, according to our survey using the MiST database [24], ranges from one chemoreceptor per genome, e.g. in a single ACF system of Acinetobacter baumannii, to ninety (!) in Caryophanon latum, which has two chemosensory pathways. Finally, the number of chemosensory pathways per genome also varies substantially [5]. While the model organism for chemotaxis, E. coli, has a single chemosensory pathway, the genome of Cystobacter fuscus encodes twelve such pathways, according to the MiST database survey, which is the largest number of chemosensory pathways identified in a single genome to date.
Diversity of functional regulation
Chemotaxis, which was studied in different organisms, appears to be the main function controlled by chemosensory systems (Table 1). The classical F7 system in E.coli and F1 pathway in B. subtilis regulate flagellar motility through opposing mechanisms of CheA activation. Attractant binding inhibits CheA activity in E. coli [2], but activates it in B. subtilis [9], while leading to the same final result: moving towards attractants and away from repellents. Che4/F7 and Che1/F5 control two aspects of flagellar motility in A. brasilense: flagellar motor reversal frequency and transient increases in swimming speed, respectively [25]. Remarkably, in Rhodobacter sphaeroides two systems, CheOp2/F8 and CheOp3/F7, together control stop frequency and duration of a single subpolar flagellum rotation [26], while another system, CheOp1/F7, controls chemotaxis mediated by several polar flagella [27]. In P. aeruginosa Che1/F6 controls flagella-driven chemotaxis [22], while Che2/F7 regulates the oxygen stress response unrelated to chemotaxis [10].
Table 1.
Experimentally studied chemosensory systems in Bacteria
| Organism | Taxonomya | System class/CheA accession number |
Function controlled |
|---|---|---|---|
| Escherichia coli | Gammaproteobacteria | F7 NP_416402.1 |
Flagellar motility (chemotaxis) |
|
Salmonella enterica
serovar Typhimurium |
Gammaproteobacteria | F7 NP_460878.1 |
Flagellar motility (chemotaxis) |
| Bacillus subtilis | Firmicutes | F1 NP_389525.2 |
Flagellar motility (chemotaxis) |
| Thermotoga maritima | Thermotogota | F1 NP_228511.1 |
Unknown |
| Helicobacter pylori | Campylobacterota | F3 WP_108169127.1 |
Flagellar motility (chemotaxis) |
| Campylobacter jejuni | Campylobacterota | F3 YP_002343725.1 |
Flagellar motility (chemotaxis) |
| Myxococcus xanthus | Myxococcota | F1 (Dif) WP_011556617.1 |
Tfp-mediated social (S) motility, EPS production |
| ACF (Frz, che1) WP_011554143.1 |
Adventurous (A) and social (S) motility |
||
| ACF (che3) WP_011555118.1 |
Gene expression, S motility |
||
| ACF (che4) WP_011552754.1 |
S motility | ||
| ACF (che5) WP_011555978.1 |
S motility, development | ||
| ACF (che6) WP_011556872.1 |
S motility, development | ||
| F10 (che8) WP_011554745.1 |
unknown | ||
| F13 (che7) WP_020480737.1 |
S motility | ||
| Pseudomonas aeruginosa | Gammaproteobacteria | F6 (Che1) NP_250149.1 |
Flagellar motility (chemotaxis) |
| F7 (Che2) NP_248868.1 |
Stress response, unrelated to chemotaxis |
||
| ACF (Wsp) NP_252393.1 |
c-di-GMP levels, biofilm formation |
||
| Tfp (Chp) NP_249104.1 |
Tfp motility, cAMP levels |
||
| Vibrio cholerae | Gammaproteobacteria | F6 (cluster II) | Flagellar motility (chemotaxis) |
| F7 (cluster III) | Unknown | ||
| F9 (cluster I) | Unknown | ||
|
Rhodobacter
sphaeroides |
Alphaproteobacteria | F7 (cheOp1) WP_002719591.1 |
Flagellar motility (chemotaxis) |
| F7 (cheOp3) WP_002720190.1 WP_002720197.1 |
Flagellar motility (chemotaxis) | ||
| F8 (cheOp2) WP_002722331.1 |
Flagellar motility (chemotaxis) |
||
| Sinorhizobium meliloti | Alphaproteobacteria | F7 (Che1) WP_014528988.1 |
Flagellar motility (chemotaxis) |
| ACF (Che2) WP_014531470.1 |
Unknown | ||
| Azospirillum brasilense | Alphaproteobacteria | F5 (Che1) WP_035678528.1 | Flagellar motility (chemokinesis), cell length |
| F7 (Che4) WP_059399028.1 |
Flagellar motility (chemotaxis) |
||
| F9 (Che2) WP_059399521.1 | Unknown; not expressed under laboratory conditions |
||
| ACF (Che3) WP_059399396.1 |
Flocculation | ||
| Rhodospirillum centenum | Alphaproteobacteria | F5 (Che1) WP_012566940.1 |
Flagellar motility (chemotaxis) |
| F9 (Che2) WP_012565576.1 |
Lateral flagellum biosynthesis |
||
| ACF (Che3) WP_012567300.1 |
Cyst development | ||
| Caulobacter crescentus | Alphaproteobacteria | F7 NP_419252.1 | Flagellar motility (chemotaxis), biofilm formation |
| F5 NP_419412.1 |
Biofilm formation | ||
| Azorhizobium caulinodans | Alphaproteobacteria | F5 WP_012169192.1 | Flagellar motility (chemotaxis), biofilm formation |
| Borrelia burgdorferi | Spirochaetota | F2 (CheA2) NP_212803.1 |
Flagellar motility (chemotaxis) |
| F8 (CheA1) NP_212701.1 |
Unknown | ||
| Treponema denticola | Spirochaetota | F2 NP_972097.1 |
Flagellar motility (chemotaxis) |
| Synechocystis sp. PCC 6803 | Cyanobacteria | Tfp (tax1/pix) WP_010874009.1 |
Tfp motility (phototaxis) |
| Tfp (tax2) WP_010871827.1 |
Unknown | ||
| Tfp (tax3/Pil) WP_010873252.1 |
Pilus biogenesis, Tfp motility |
||
| Nostoc punctiforme | Cyanobacteria | Tfp (hmp/cl 1) WP_012412198.1 |
Tfp motility, EPS secretion |
| Tfp (pix) WP_012412241.1 |
Unknown | ||
| Tfp/ptx WP_012408770.1 |
Tfp motility (phototaxis) | ||
| Tfp (cl 3) WP_012411886.1 |
Morphogenesis | ||
| ACF WP_012407058.1 |
Unknown | ||
| Comamonas testosteroni | Gammaproteobacteria | F7 (che) WP_012836878.1 |
Flagellar motility, biofilm formation |
| Tfp (Flm) WP_012839726.1 |
Biofilm formation | ||
| Magnetospirillum gryphiswaldense | Alphaproteobacteria | F5 (cheOp1) WP_106003281.1 |
Flagellar motility (chemotaxis) |
| ACF (cheOp2) WP_106001668.1 |
Unknown | ||
| ACF (cheOp3) WP_106002789.1 |
Unknown | ||
| F7 (cheOp4) WP_106001879.1 |
Unknown |
According to the Genome Taxonomy Database [50].
Chemosensory systems also control Tfp- based twitching motility across solid surfaces. The Tfp system (Fig. 2) modulating twitching motility is best studied in P. aeruginosa [28] but it was also described in other gamma-proteobacteria, including human and plant pathogens [29–31] and in cyanobacteria. Paralogous Tfp systems in Synechocystis sp. PCC 6803 and Nostoc punctiforme control not only phototaxis [32,33], but also other cellular functions related to development and biosynthesis [32,34]. Interestingly, the Tfp-based motility in M. xanthus is regulated by Frz and Dif chemosensory pathways [35] that belong not to Tfp, but to ACF and F1 classes, respectively. Many non-motility functions are regulated by chemosensory systems that belong to the ACF class (Fig. 2). The Che3/ACF pathway regulates formation of stress-resistant cysts in R. centenum [36] and the Che3/ACF pathway in the closely-related A. brasilense controls flocculation in liquid cultures [37]. Three ACF pathways in M. xanthus, namely Che4, Che5 and Che6, interact to form a large chemosensory module regulating fruiting body formation [38], while the fourth member of the ACF class in this bacterium, Che3, regulates expression of genes important for fruiting body formation [35]. In P. aeruginosa the Wsp/ACF chemosensory system mediates biofilm formation through modulation of c-di-GMP production [39]. Regulation of biofilm formation has recently emerged as yet another common feature of chemosensory systems [40] and in several instances non-ACF pathways were implicated in this function. For example, in Comamonas testosteroni, the Tfp pathway termed Flm regulates biofilm formation [41], and in Caulobacter crescentus two pathways that belong to F5 and F7 classes appeared to be involved in this process [42]. Intriguingly, a cross talk between systems controlling chemotaxis and biofilm formation was reported in both organisms [41, 42] as well as in a plant pathogen P. syringae [43]. The F5 pathway of Azorhizobium caulinodans that is important for plant colonization was also reported to mediate biofilm formation [44]. The exact function of some other classes of chemosensory systems remain poorly understood. The Che2/F9 pathway in R. centenum was found to regulate flagella biosynthesis [36]; however, the exact function of orthologous pathways in A. brasilense [25] and V. cholerae [18] awaits elucidation.
The phylogenomic classification of chemosensory systems [5] should not be taken as an absolute predictor of their function. As shown in several examples here, in some organisms, pathways that originally controlled flagellar motility might have switched their output targets to mediate other functions. During the course of evolution one chemosensory pathway may also “take over” the original function of another pathway to control the same output target [45].
Diversity of cellular localization
Typical MCPs are transmembrane and in the homodimer form assemble into trimers-of-dimers that together with CheA and CheW form functional signaling units [2,3]. A network of such units is arranged into a hexagonal array, as observed in many distantly related bacteria [46], but their cellular localization varies [4]. Chemoreceptor arrays in E. coli, Magnetospirillum magneticum, R. sphaeroides, T. primitia, T. maritima, and Listeria monocytogenes localize to the poles of the cell. In H. hepaticus and C. jejuni the arrays are also polar, but the chemoreceptors surround the gap at the apex occupied by the flagellar motor. In V. cholerae and C. crescentus polar receptors are localized to the convex side of the crescent shaped cells. In Acetonema longum and B. burgdorferi the arrays were shown to be typically subpolar but inconsistently positioned [4].
In addition to transmembrane MCPs, many bacteria have cytoplasmic chemoreceptors [47]. In the V. cholerae F9 chemosensory pathway, an unusual receptor DosM containing two signaling domains forms two cytoplasmic hexagonal arrays of trimers-of-dimers sandwiched between two CheA and CheW baseplates thus stabilizing the complex [48]. Transmembrane arrays are curved as they follow the curvature of the cell membrane whereas the cytoplasmic arrays can be either flexible as seen in R. sphaeroides and Methanoregula formicicum [49] or rigid as in V. cholerae due to the stabilizing effect of DosM [48]. In M. xanthus, chemoreceptor arrays of the Frz pathway assemble at the nucleoid [16]. Their exact arrangement with CheA and CheW is unknown, but it is speculated that similarly to transmembrane receptors they may form a monolayer running parallel to the nucleoid surface.
Phyletic diversity
Chemosensory systems have been studied experimentally in approximately 30 bacterial species, according to our literature analysis. These organisms represent only eight bacterial phyla (Fig. 3) out of more than one hundred bacterial phyla defined by the latest release of the Genome Taxonomy database [50]. The majority of bacteria with experimentally studied chemosensory systems are Proteobacteria and systems from several major classes, namely F5, F6, F7, and F9, are described only for bacteria in this phylum (Table 1, Fig. 3). Overall, there is a tendency for chemosensory systems to occur most commonly in certain taxonomic groups [5]. For example, F1 system is found in Firmicutes, Thermotogota, and Archaea, but not in Proteobacteria, whereas F6 system is found exclusively in Proteobacteria. F2 and F3 systems are found in Spirochaetota and Campylobacterota, respectively (Fig. 3). The F5 system emerges as the second major chemosensory system in Alphaproteobacteria, in addition to F7 system [25, 36, 42, 44, 51]. Except for Gammaproteobacteria, the twitching motility controlling Tfp system is described only in Cyanobacteria. The F4 system has only been studied in Desulfovibrio vulgaris [52], and F10 and F13 systems have been explored exclusively in M. xanthus and their function remains largely unknown. Flagellar systems F11 (Ruminiclostridium thermocellum), F12 (Geobacter uraniireducens), and F17 (Thermodesulfovibrio yellowstonii) have never been studied experimentally.
Figure 3. Phyletic distribution of experimentally studied bacterial chemosensory systems.
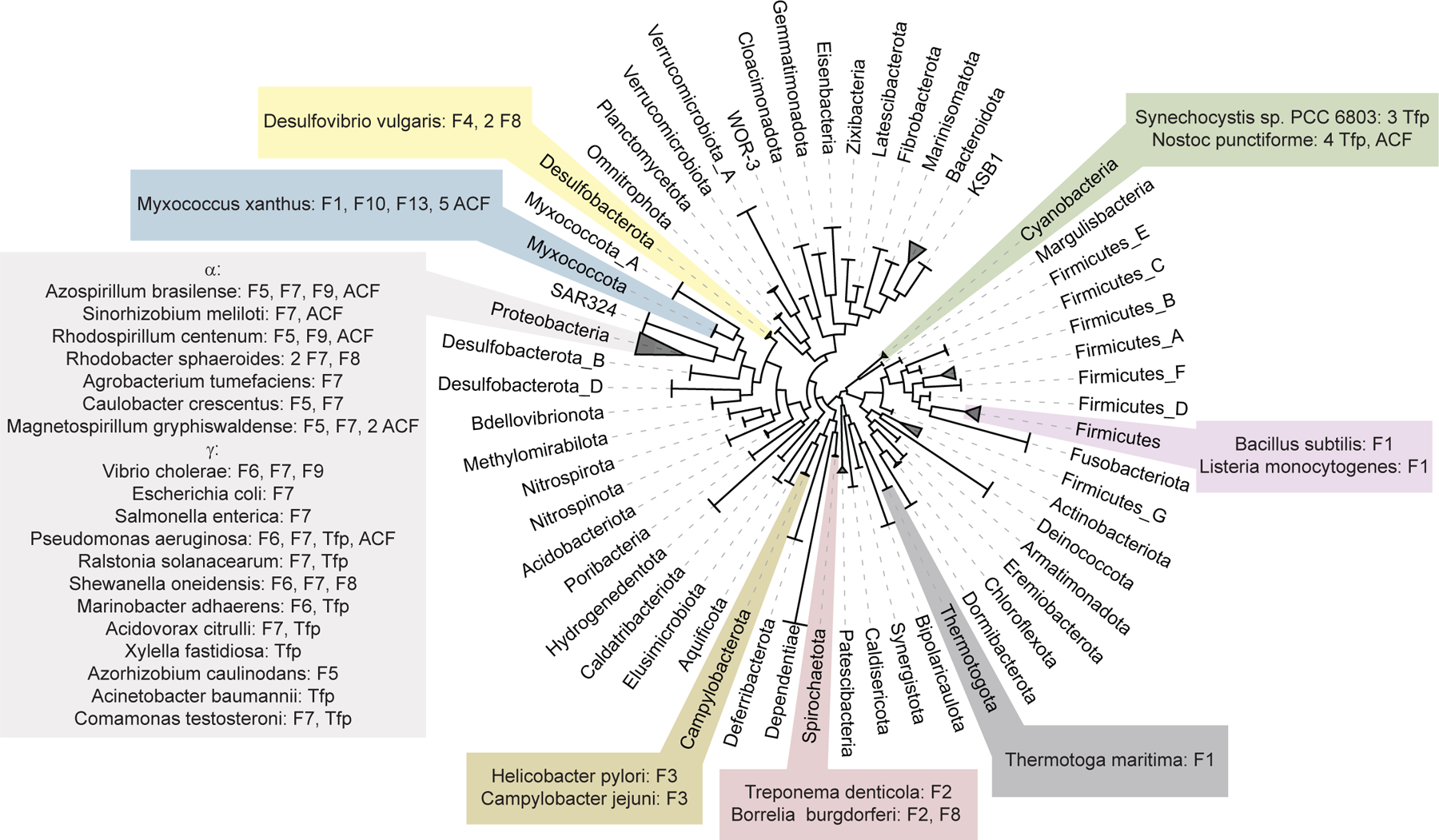
The genome tree showing all bacterial phyla with at least 10 sequenced genomes was adapted from the Genome Taxonomy Database [49]. Organisms with experimentally studied chemosensory systems are mapped to their respective phyla. All known types of chemosensory systems are listed for each organism.
Conclusions
A tremendous amount of information on bacterial chemosensory systems was generated in the past twenty years but most of the in-depth knowledge comes from a handful of model bacteria. The precise evolutionary history of these pathways is poorly understood. What was the original role of the ancestral chemosensory system: regulation of swimming motility, twitching motility or some other cellular function? What is the extent of horizontal gene transfer in chemosensory systems? Can a function of a chemosensory system be predicted from genome information? Can efficient computational approaches be developed to distinguish CheY from other one domain response regulators and CheW from other CheW domain-containing proteins? These and other questions still await to be answered. The latest improvements in bacterial taxonomy together with comparative genomic approaches and newly generated experimental evidence should enable further advances in our understanding of function and evolution of these remarkable regulatory pathways.
Acknowledgements
This work was supported by the National Institutes of Health [R35GM131760].
Footnotes
Conflict of Interest
Authors have no conflicts of interest to declare.
References and recommended reading
- 1.Wuichet K, Cantwell BJ, Zhulin IB: Evolution and phyletic distribution of two-component signal transduction systems. Curr Opin Microbiol 2010, 13:219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkinson JS, Hazelbauer GL, Falke JJ: Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 2015, 23:257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colin R, Sourjik V: Emergent properties of bacterial chemotaxis pathway. Curr Opin Microbiol 2017, 39:24–33. [DOI] [PubMed] [Google Scholar]
- ··4. Yang W, Briegel A: Diversity of Bacterial Chemosensory Arrays. Trends Microbiol 2020, 28:68–80. This recent thorough review summarizes all currently available knowledge on cellular localization of chemosensory pathways.
- 5.Wuichet K, Zhulin IB: Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 2010, 3:ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexander RP, Lowenthal AC, Harshey RM, Ottemann KM: CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol 2010, 18:494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortega DR, Zhulin IB: Evolutionary Genomics Suggests That CheV Is an Additional Adaptor for Accommodating Specific Chemoreceptors within the Chemotaxis Signaling Complex. PLoS Comput Biol 2016, 12:e1004723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarado A, Kjaer A, Yang W, Mann P, Briegel A, Waldor MK, Ringgaard S: Coupling chemosensory array formation and localization. Elife 2017, 6:e31058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao CV, Glekas GD, Ordal GW: The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol 2008, 16:480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ··10. Orillard E, Watts KJ: Deciphering the Che2 chemosensory pathway and the roles of individual Che2 proteins from Pseudomonas aeruginosa. Mol Microbiol 2020. This work provides a deep insight into a classical type of chemosensory pathway with unusual function.
- 11.Silversmith RE: Auxiliary phosphatases in two-component signal transduction. Curr Opin Microbiol 2010, 13:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ·12. Dogra G, Puschke FG, Wagner V, Haslbeck M, Kriehuber T, Hughes JG, Van Tassell ML, Gilbert C, Niemeyer M, Ray WK, et al. : Sinorhizobium meliloti CheA complexed with CheS exhibits enhanced binding to CheY1, resulting in accelerated CheY1 dephosphorylation. J Bacteriol 2012, 194:1075–1087. This works identifies a STAS-domain protein as a component of a chemosensory system.
- 13.Johnson KS, Ottemann KM: Colonization, localization, and inflammation: the roles of H. pylori chemotaxis in vivo. Curr Opin Microbiol 2018, 41:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell CL, Wilson JM, Tiwari N, Fuentes EJ, Kirby JR: Chemosensory regulation of a HEAT-repeat protein couples aggregation and sporulation in Myxococcus xanthus. J Bacteriol 2014, 196:3160–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sourjik V, Schmitt R: Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 1998, 37:2327–2335. [DOI] [PubMed] [Google Scholar]
- ·16. Guiseppi A, Vicente JJ, Herrou J, Byrne D, Barneoud A, Moine A, Espinosa L, Basse MJ, Molle V, Mignot T, et al. : A divergent CheW confers plasticity to nucleoid-associated chemosensory arrays. PLoS Genet 2019, 15:e1008533. This study describes an unusual version of the scaffold protein and its role in cellular localization in a bacterium with multiple chemosensory pathways.
- 17.Lertsethtakarn P, Ottemann KM, Hendrixson DR: Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 2011, 65:389–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortega DR, Kjaer A, Briegel A: The chemosensory systems of Vibrio cholerae. Mol Microbiol 2020, 114:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ··19. Muok AR, Ortega DR, Kurniyati K, Yang W, Maschmann ZA, Mabrouk AS, Li C, Crane BR, Briegel A: Atypical chemoreceptor arrays accommodate high membrane curvature. Nat Commun 2020, 11:5763. The first insight into a chemosensory array arrangement in spirochetes.
- 20.Gullett JM, Bible A, Alexandre G: Distinct Domains of CheA Confer Unique Functions in Chemotaxis and Cell Length in Azospirillum brasilense Sp7. J Bacteriol 2017, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander RP, Zhulin IB: Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A 2007, 104:2885–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortega DR, Fleetwood AD, Krell T, Harwood CS, Jensen GJ, Zhulin IB: Assigning chemoreceptors to chemosensory pathways in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 2017, 114:12809–12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ortega A, Zhulin IB, Krell T: Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev 2017, 81:E00033–00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ··24. Gumerov VM, Ortega DR, Adebali O, Ulrich LE, Zhulin IB: MiST 3.0: an updated microbial signal transduction database with an emphasis on chemosensory systems. Nucleic Acids Res 2020:48:D459–D464. This work describes the only comprehensive database on bacterial and archaeal signal transduction specifically focusing on chemosensory systems.
- ·25. O’Neal L, Gullett JM, Aksenova A, Hubler A, Briegel A, Ortega DR, Kjaer A, Jensen GJ, Alexandre G: Distinct Chemotaxis Protein Paralogs Assemble into Chemoreceptor Signaling Arrays To Coordinate Signaling Output. mBio 2019, 10:e01757–01719. This study disentangles properties, function, and localization of pathways controlling chemotaxis and chemokinesis in a bacterium with multiple chemosensory systems.
- 26.De Beyer JA, Szollossi A, Byles E, Fischer S, Armitage JP: Mechanism of Signalling and Adaptation through the Rhodobacter sphaeroides Cytoplasmic Chemoreceptor Cluster. Int J Mol Sci 2019, 20:5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez-Valle J, Domenzain C, de la Mora J, Poggio S, Dreyfus G, Camarena L: The Master Regulators of the Fla1 and Fla2 Flagella of Rhodobacter sphaeroides Control the Expression of Their Cognate CheY Proteins. J Bacteriol 2017, 199:e00670–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inclan YF, Persat A, Greninger A, Van Dollen J, Johnson J, Krogan N, Gitai Z, Engel JN: A scaffold protein connects type IV pili with the Chp chemosensory system to mediate activation of virulence signaling in Pseudomonas aeruginosa. Mol Microbiol 2016, 101:590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corral J, Perez-Varela M, Barbe J, Aranda J: Direct interaction between RecA and a CheW-like protein is required for surface-associated motility, chemotaxis and the full virulence of Acinetobacter baumannii strain ATCC 17978. Virulence 2020, 11:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cursino L, Galvani C, Athinuwat D, Zaini PA, Li Y, De La Fuente L, Hoch HC, Burr TJ, Mowery P: Identification of an operon, Pil-Chp, that controls twitching motility and virulence in Xylella fastidiosa. Mol Plant Microbe Interact 2011, 24:1198–1206. [DOI] [PubMed] [Google Scholar]
- 31.Corral J, Sebastia P, Coll NS, Barbe J, Aranda J, Valls M: Twitching and Swimming Motility Play a Role in Ralstonia solanacearum Pathogenicity. mSphere 2020, 5:e00740–00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell EL, Hagen KD, Chen R, Risser DD, Ferreira DP, Meeks JC: Genetic analysis reveals the identity of the photoreceptor for phototaxis in hormogonium filaments of Nostoc punctiforme. J Bacteriol 2015, 197:782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhaya D: Light matters: phototaxis and signal transduction in unicellular cyanobacteria. Mol Microbiol 2004, 53:745–754. [DOI] [PubMed] [Google Scholar]
- 34.Yoshihara S, Geng X, Ikeuchi M: pilG Gene cluster and split pilL genes involved in pilus biogenesis, motility and genetic transformation in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 2002, 43:513–521. [DOI] [PubMed] [Google Scholar]
- 35.Mauriello EM, Jones C, Moine A, Armitage JP: Cellular targeting and segregation of bacterial chemosensory systems. FEMS Microbiol Rev 2018, 42:462–476. [DOI] [PubMed] [Google Scholar]
- 36.He K, Bauer CE: Chemosensory signaling systems that control bacterial survival. Trends Microbiol 2014, 22:389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bible AN, Khalsa-Moyers GK, Mukherjee T, Green CS, Mishra P, Purcell A, Aksenova A, Alexandre G: Metabolic adaptations of Azospirillum brasilense to oxygen stress by cell-to-cell clumping and flocculation. Appl Environ Microbiol 2015, 81:8346–8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moine A, Agrebi R, Espinosa L, Kirby JR, Zusman DR, Mignot T, Mauriello EM: Functional organization of a multimodular bacterial chemosensory apparatus. PLoS Genet 2014, 10:e1004164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huangyutitham V, Guvener ZT, Harwood CS: Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. MBio 2013, 4:e00242–00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corral-Lugo A, de la Torre J, Matilla MA, Fernandez M, Morel B, Espinosa-Urgel M, Krell T: Assessment of the contribution of chemoreceptor-based signaling to biofilm formation. Environ Microbiol 2016, 18:3355–3372. [DOI] [PubMed] [Google Scholar]
- ·41. Huang Z, Wang Y-H, Zhu H-Z, Andrianova EP, Jiang CY, Li D, Ma L, Feng J, Liu Z-P, Xiang H, et al. : Cross Talk between Chemosensory Pathways That Modulate Chemotaxis and Biofilm Formation. mBio 2019, 10:e02876–02818. This study suggests sites of potential cross talk between two chemosensory systems to coordinate chemotaxis and biofilm formation depending on changing environmental conditions.
- 42.Berne C, Brun YV: The Two Chemotaxis Clusters in Caulobacter crescentus Play Different Roles in Chemotaxis and Biofilm Regulation. J Bacteriol 2019, 201:e00071–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerna-Vargas JP, Santamaria-Hernando S, Matilla MA, Rodriguez-Herva JJ, Daddaoua A, Rodriguez-Palenzuela P, Krell T, Lopez-Solanilla E: Chemoperception of Specific Amino Acids Controls Phytopathogenicity in Pseudomonas syringae pv. tomato. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W, Sun Y, Shen R, Dang X, Liu X, Sui F, Li Y, Zhang Z, Alexandre G, Elmerich C, et al. : A Chemotaxis-Like Pathway of Azorhizobium caulinodans Controls Flagella-Driven Motility, Which Regulates Biofilm Formation, Exopolysaccharide Biosynthesis, and Competitive Nodulation. Mol Plant Microbe Interact 2018, 31:737–749. [DOI] [PubMed] [Google Scholar]
- ·45. Ortega DR, Yang W, Subramanian P, Mann P, Kjaer A, Chen S, Watts KJ, Pirbadian S, Collins DA, Kooger R, et al. : Repurposing a chemosensory macromolecular machine. Nat Commun 2020, 11:2041. This study suggests how one chemosensory system can displace another during the course of evolution.
- 46.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, et al. : Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A 2009, 106:17181–17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collins KD, Lacal J, Ottemann KM: Internal sense of direction: sensing and signaling from cytoplasmic chemoreceptors. Microbiol Mol Biol Rev 2014, 78:672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Briegel A, Ortega DR, Mann P, Kjaer A, Ringgaard S, Jensen GJ: Chemotaxis cluster 1 proteins form cytoplasmic arrays in Vibrio cholerae and are stabilized by a double signaling domain receptor DosM. Proc Natl Acad Sci U S A 2016, 113:10412–10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Briegel A, Ortega DR, Huang AN, Oikonomou CM, Gunsalus RP, Jensen GJ: Structural conservation of chemotaxis machinery across Archaea and Bacteria. Environ Microbiol Rep 2015, 7: 414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ··50. Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P: A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 2018, 36:996–1004. A fundamental bacterial and archaeal taxonomy based on evolutionary analysis of more than a hundred genes conserved across prokaryotes.
- 51.Popp F, Armitage JP, Schuler D. Polarity of bacterial magnetotaxis is controlled by aerotaxis through a common sensory pathway. Nat Commun 2014, 5:5398. [DOI] [PubMed] [Google Scholar]
- 52.Ray J, Keller KL, Catena M, Juba TR, Zemla M, Rajeev L, Knierim B, Zane GM, Robertson JJ, Auer M, et al. : Exploring the role of CheA3 in Desulfovibrio vulgaris Hildenborough motility. Front Microbiol 2014, 5:77. [DOI] [PMC free article] [PubMed] [Google Scholar]


