Abstract
Mitochondria as a signaling platform play crucial roles in deciding cell fate. Many classic anticancer agents are known to trigger cell death through induction of mitochondrial damage. Mitophagy, one selective autophagy, is the key mitochondrial quality control that effectively removes damaged mitochondria. However, the precise roles of mitophagy in tumorigenesis and anticancer agent treatment remain largely unclear. Here, we examined the functional implication of mitophagy in the anticancer properties of magnolol, a natural product isolated from herbal Magnolia officinalis. First, we found that magnolol induces mitochondrial depolarization, causes excessive mitochondrial fragmentation, and increases mitochondrial reactive oxygen species (mtROS). Second, magnolol induces PTEN-induced putative kinase protein 1 (PINK1)‒Parkin-mediated mitophagy through regulating two positive feedforward amplification loops. Third, magnolol triggers cancer cell death and inhibits neuroblastoma tumor growth via the intrinsic apoptosis pathway. Moreover, magnolol prolongs the survival time of tumor-bearing mice. Finally, inhibition of mitophagy by PINK1/Parkin knockdown or using inhibitors targeting different autophagy/mitophagy stages significantly promotes magnolol-induced cell death and enhances magnolol's anticancer efficacy, both in vitro and in vivo. Altogether, our study demonstrates that magnolol can induce autophagy/mitophagy and apoptosis, whereas blockage of autophagy/mitophagy remarkably enhances the anticancer efficacy of magnolol, suggesting that targeting mitophagy may be a promising strategy to overcome chemoresistance and improve anticancer therapy.
KEY WORDS: PINK1‒Parkin-mediated mitophagy, Magnolol, Combination therapy, Apoptosis, Tumor suppression
Graphical abstract
Exposure of magnolol causes mitochondrial dysfunction which induces mitophagy and intrinsic apoptosis. Inhibition of autophagy/mitophagy by distinct inhibitors at different stages promotes cell death and tumor suppression evidenced by further reduction of tumor volume and weight.
1. Introduction
Mitochondria are the most important organelles to provide energy for cells and organisms by producing adenosine triphosphate (ATP). In addition to their role as the “powerhouse of the cells”, mitochondria also participate in a variety of other cellular functions, including regulation of apoptosis and the autophagy signaling pathway, generation of reactive oxygen species (ROS), maintenance of calcium balance, and synthesis of fatty acids1. It is therefore not surprising that mitochondrial dysfunctions are implicated in the pathogenesis of many diseases, including cancer, neurodegenerative diseases, aging, heart failure, and metabolic disorders2,3. Thus, it is crucial to remove dysfunctional mitochondria to keep a healthy mitochondrial network.
Mitophagy, one best-studied type of selective autophagy that specifically eliminates damaged or superfluous mitochondria through lysosomes, is a key mitochondrial quality control system4, 5, 6, 7. In the past decade, the mechanisms and the pathophysiological roles of mitophagy have been extensively explored8,9. Among them, PTEN-induced putative kinase protein 1 (PINK1)‒Parkin-mediated mitophagy is the best characterized mitophagic pathway. When mitochondria are damaged or depolarized, PINK1 is stabilized and activated on the mitochondrial outer membrane10, 11, 12. PINK1, a serine/threonine kinase, phosphorylates ubiquitin at serine 65 (pSer65-Ub)13, 14, 15. Then, pSer65-Ub chains serve as Parkin receptors to recruit Parkin from the cytosol to mitochondria13,16, which leads to the phosphorylation of Parkin by PINK1 also at serine 65 (pSer65-Parkin)17, 18, 19. This in turn results in Parkin's conformational changes that fully activate Parkin20,21. Once activated, Parkin, an E3 ligase, recruits more ubiquitin for PINK1 phosphorylation, and more pSer65-Ub recruits further rounds of Parkin on mitochondria, thus forming the first positive feedforward amplification loop to enable rapid coating of mitochondria with pSer65-Ub and to initiate mitophagy22,23. In addition, pSer65-Ub can recruit primary mitophagy receptors, such as optineurin (OPTN) and CALCOCO2 (NDP52), to mitochondria24,25; OPTN and NDP52 then recruit unc51-like activating kinase 1/2 (ULK1/2) complex on mitochondria to initiate autophagosome formation through the recruitment of autophagy-related 8 proteins (ATG8s) which recruit additional mitophagy receptors to speed autophagosome growth, thus forming the second positive feedforward amplification loop to amplify mitophagy24,26,27.
Accumulating studies are suggesting that mitophagy may serve as a suppressor of cell death and promote cancer progression under cytotoxic stresses through effectively clearing damaged/detrimental mitochondria and thus helping the cancer cells to adjust the microenvironment of limited oxygen or nutrient and develop drug resistance5. For instance, in oncogenic K-Ras-driven transformation, mitophagy promotes cancer development via autophagy-mediated organelle degradation to provide ATP and nutrients for cancer cells when there is insufficient glucose up-take28. Yan et al.29 found that mitophagy was induced in cancer stem cells (CSCs) after doxorubicin treatment, leading to drug resistance. It has also been reported that PINK1 could maintain the human brain tumor stem cell function through the interaction with Notch and promotion of mitochondrial function30. More interesting, Liu et al.31 reported that mitophagy could maintain hepatic CSCs population to promote hepatocarcinogenesis via inhibiting the activity of p53, the most important tumor suppressor, in a PINK1-dependent manner. However, the precise function and regulation of PINK1‒Parkin-mediated mitophagy in anticancer agent treatment remain largely unknown.
Natural compounds from traditional medicine have received increasing attention as potential sources of novel anticancer agents due to their novel biochemical mechanism and few side effects32. Magnolol, a gentle herb with a long history of use in traditional medicine, is isolated from the root and stem bark of the tree Magnolia officinalis. It has been well-established that magnolol exhibits anti-inflammatory, anti-diabetic, anti-microbial, anti-neurodegenerative, and anti-depressant properties33. Moreover, numerous preclinical studies have reported that magnolol displays anticancer effects in various types of cancer, including human glioblastomas34, non-small cell lung cancer35,36, breast cancer37, skin cancer38, and prostate cancer39. The anticancer activities of magnolol have been attributed to induction of apoptosis, inhibition of cell proliferation and suppression of cell metastasis. Due to few side effects on normal human cells and the property of passing through the blood‒brain barrier35,36,39,40, magnolol is probably an ideal anticancer agent, especially for neuroblastoma and glioblastoma. Magnolol has also been reported to play important roles in the regulation of autophagy in cancer cells35,41, 42, 43. Interestingly, it is determined by proteomic approaches that magnolol can target multiple mitochondrial and/or mitophagy related proteins44. However, at present, magnolol's effects on mitophagy have not been investigated.
In this study, we demonstrate that on the one hand, magnolol causes mitochondrial dysfunction and induces intrinsic apoptosis. On the other hand, magnolol can induce mitophagy in a PINK1- and Parkin-dependent manner, which antagonizes its cytotoxicity. More importantly, genetic and pharmacological inhibition of mitophagy dramatically enhances magnolol's anticancer efficacy, both in vitro and in vivo. Understanding the function of magnolol in the regulation of mitophagy and cell death will not only expand the knowledge of mitophagy during anticancer agents treatment and improve the anticancer function of magnolol, but may also provide a foundation of a new strategy in the treatment of cancer-namely targeting mitophagy.
2. Materials and methods
2.1. Reagents and antibodies
Magnolol (M3445), bafilomycin A1 (Baf-A1, B1793), chloroquine (CQ) diphosphate salt (C6628), wortmannin (W1628), Z-VAD-FMK (V116), and bovine serum albumin (BSA, A2153) were purchased from Sigma–Aldrich (USA). Vacuolin-1 (sc-216045) was purchased from Santa Cruz Biotechnology (USA). Ac220 (HY-13001), hydroxychloroquine (HCQ, HY-B1370), ferrostatin-1 (HY-100579), and necrostatin-1 (HY-15760) were from MedChemExpress (USA). Lipofectamine 3000 (L3000075), Lipofectamine RNAiMAX (13778150), propidium iodide (PI, P1304MP), ProLong® Diamond Antifade Mountant (P36970), and ProLong® Diamond Antifade Mountant with 4′,6-diamidino-2-phenylindole (DAPI, 36971) were from Thermo Fisher Scientific (USA). HyperSignal ECL substrate (4AW012-50) was purchased from 4A Biotech Co., Ltd. (China).
The following antibodies were from Cell Signaling Technologies (USA): Parkin (4211), mitofusin-2 (MFN2, 11925), mitofusin-1 (MFN1, 14739), PINK1 (6946), cytochrome c oxidase 4 (COX4, 4850), heat shock protein 60 (HSP60, 12165), glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 2118), caspase-9 (9502), caspase-3 (9662), cleaved caspase-3 (9661), poly ADP ribose polymerase (PARP, 9532) and pTyr591-FLT3 (3461). Tim23 (611223) was from BD Biosciences (USA). Tom20 (FL-145) and ubiquitin (P4D1, 8017) were from Santa Cruz Biotechnology. Microtubule-associated proteins 1A/1B light chain 3B (LC3B, L7543), actin (A5441) and tubulin (T6199) were from Sigma–Aldrich. COX II (ab110258) and NDP52 (ab68588) were from Abcam (USA). pSer65-Parkin (MJF-17-42-4) was from the Michael J. Fox Foundation (Abcam) for Parkinson's Research. pSer65-ubiquitin (ABS1513-I) was from Merck Millipore (USA). OPTN (10827-1-AP) was ordered from Proteintech (China). p62 (SQSTM1, H00008878-M01) was from Abnova (China). Anti-DNA (61014) was from Progen Biotechnik (Germany). Alexa592 goat anti-mouse (A-11032), and Alexa592 goat anti-rabbit (R37117) were from Thermo Fisher Scientific.
2.2. Cell lines and cell culture
Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; HyClone, SH30022.01, USA) with 10% fetal bovine serum (FBS; HyClone, SV30160.03), 100 U/mL penicillin-streptomycin (Pan-Biotech, P06-07100, Germany). All cell lines were maintained in a humidified 5% (v/v) CO2 incubator at 37 °C. HeLa cells stably expressing YFP-Parkin were kind gifts from Dr. Richard Youle (National Institute of Neurological Disorders and Stroke, Bethesda, MD, USA). HeLa cells stably expressing GFP-LC3B were kind gifts from Dr. Noboru Mizushima (University of Tokyo, Tokyo, Japan). HeLa cells and SH-SY5Y were purchased from American Type Culture Collection (USA). All cell lines were tested with MycoAlert PLUS Kits from Lonza (USA) and confirmed to be negative for mycoplasma contamination.
2.3. Cell transfections
PINK1 siRNA (HSS127945, HSS127946, and HSS185707, Thermo Fisher Scientific) and Parkin siRNA (hs.Ri.PARK2.13.1, hs.Ri.PARK2.13.2, and hs.Ri.PARK2.13.3, Integrated DNA Technologies, Singapore) were transfected with Lipofectamine RNAiMAX reagent according to the manufacturer's instructions.
2.4. Western blot
After the designed treatments, cells were washed with ice-cold phosphate buffer saline (PBS) and lysed in lysis buffer (62.5 mmol/L Tris, pH 6.8, 25% glycerol, 2% SDS, protease and phosphatase inhibitors, 1 mmol/L dithiothreitol). Equal amounts of proteins were subjected to SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane with wet tank transfer. Immunoblot analysis was performed accordingly. Band intensity was measured with ImageJ software and normalized with control groups.
For the Parkin's E3 ligase activity assay, cells were lysed in lysis buffer [20 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, 0.5% NP-40, 1 mmol/L EDTA (pH 8.0), and 10 mmol/L N-ethylmaleimide].
2.5. Immunofluorescence and time-lapse microscopy
After the designed treatments, cells were fixed with 4% paraformaldehyde (Santa Cruz Biotechnology, sc-281692) for 15 min at room temperature, next permeabilized with 0.25% Triton X-100 for 15 min, and then blocked with 10% BSA for 30 min at 37 °C and incubated with specific primary antibodies overnight at 4 °C, and relative secondary antibodies for 1 h at 37 °C in the next day, finally, mounted the cells with antifade mounting medium and observed by using a confocal microscope (Olympus FV3000 Confocal Laser Scanning Microscope, Olympus, Japan) or a fluorescence microscope (DMi8, Leica, Germany).
For the mitochondrial DNA (mtDNA) staining and quantification, we first used MycoAlert PLUS Kits to confirm that there were no mycoplasma contamination, and then used anti-DNA antibody (Ac-30-10) to perform the staining, which can easily label mitochondria DNA and nuclear DNA (total cellular DNA, TcDNA)45, while DAPI only labels nuclear DNA (nDNA) in an appropriately short incubation time. We performed 3D imaging following the procedures described previously24,46. Briefly, ten image slices through the Z plane were collected by Olympus FV3000 Confocal Laser Scanning Microscope. More than 100 cells were quantified using Imaris 9.1 software (USA). The percentage of mtDNA can be calculated using Eq. (1):
| (1) |
For the quantitative analysis of mitochondrial morphology, the Mitochondrial Network Analysis (MiNA) toolset in Fiji (USA) was used as described previously47. Briefly, after the designed treatments, mitochondria were stained with anti-Tom20 antibody and imaged by confocal microscope. Thirty individual cells were randomly selected for quantification by using the unsharp mask function of MiNA. Mean branch length refers to the average length of all mitochondrial rod/branch length; mean branches per network refer to the average number of mitochondrial branches per network; mitochondrial footprint refers to the total area of mitochondria stained with Tom20.
For the video of Parkin mitochondrial translocation, HeLa-YFP-Parkin cells were plated in cell culture dishes with glass bottom (μ-dish 35 mm, high Glass Bottom, Ibidi, Germany). In the next day, cells were treated with magnolol (100 μmol/L), and the observation and image capture were begun immediately.
2.6. Measurement of mitochondrial membrane potential
The MitoProbe™ DiIC1(5) Assay Kit (M34151, Thermo Fisher Scientific) and TMRE-Mitochondrial Membrane Potential Assay Kit (ab113852, Abcam) were used to determine the mitochondrial membrane potential (MMP). We first treated the cells with magnolol, then measured the MMP by using fluorescence microscopy and/or flow cytometry according to the manufacturer's protocol. In each independent experiment, the fluorescence intensity of the DMSO control groups was normalized to 100%, and the fluorescence intensity of magnolol-treated groups was subsequently compared with control groups to determine the remaining fluorescence intensity. Three independent experiments were performed to get the statistical results.
2.7. Measurement of mitochondrial ROS production
After the designed treatments, the MitoSOX™ Red Mitochondrial Superoxide Indicator Kit (M36008, Thermo Fisher Scientific) was used to evaluate the level of mitochondrial reactive oxygen species (mtROS) with fluorescence microscopy according to the manufacturer's protocol.
For the colocalization of mtROS and mitochondria, live cells were stained with MitoTracker™ Green FM (M7514, Thermo Fisher Scientific) to label mitochondria prior to magnolol treatment, and then mtROS was labelled with MitoSOX™ Red reagent and observed by fluorescence microscopy. MitoSOX fluorescence intensity was measured with ImageJ software and normalized to control groups.
2.8. Transmission electron microscopy
Cells were treated with DMSO or magnolol 100 μmol/L for 4 h. The other related procedures were performed according to our previously described protocols46.
2.9. Detection of cell death
Different methods were used to detect cell death and cell survival, including: (i) morphological changes observed by phase contrast microscopy; (ii) cell death measured by flow cytometry using propidium iodide (PI) exclusion assay. First, medium with dead cells is collected. Second, the remaining cells are detached and harvested with trypsin. Third, combine all cells from each well. Forth, cell pellets are resuspended with PBS which contains 5 μg/mL PI. Finally, each sample is analyzed with FACSCalibur flow cytometry (BD Biosciences) using CellQuest software by counting 10,000 cells.
2.10. Mouse xenograft model
We assessed the efficacy of magnolol or the combination of magnolol and different inhibitors on tumor suppression in BALB/c male nude mice (GemPharmatech Co., Ltd., China). All animal protocols were approved by Sun Yat-sen University (Shenzhen, China), and were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Briefly, 5 × 106 SH-SY5Y cells in 100 μL PBS were suspended in Matrigel (BD Biosciences) at a ratio of 1:1 (v/v), and then subcutaneously injected into the back of nude mice. Tumor volumes were measured every three or four days after injection and calculated from Eq. (2):
| (2) |
After tumor volume reached 80–120 mm3, mice were randomly assigned into several groups (n = 8 for magnolol treatment assay and n = 6 for the combination of magnolol with wortmannin, HCQ, and Ac220 assays) that received vehicle control, magnolol, wortmannin, HCQ, Ac220, and combined agents via intraperitoneal injection, respectively. Mice were euthanized after the treatments, and tumors were harvested and weighed. For survival rate assay, when tumor volume reached 2000 mm3, mice were deemed non-survivable and euthanized.
2.11. Immunohistochemistry
Freshly isolated tumors were fixed in 10% formalin and embedded in paraffin. For immunohistochemistry, tumor sections were deparaffinized, rehydrated, microwaved in 10 mmol/L citrate buffer for 30 min, and incubated in 0.3% Triton X-100 in PBS for 30 min. Sections were blocked using an Avidin-biotin blocking Kit (Abcam), and subsequently incubated with primary antibodies at the dilutions suggested by the manufacturer for 1 h, followed by secondary antibody for 30 min at room temperature. The diaminobenzidine (DAB) detection Kit (Abcam) was used to detect signals. Images were captured by light microscope (Leica DM4B).
2.12. Statistical analysis
All the Western blot data and image data are performed and analyzed from 3 independent experiments. The numeric data are presented as means ± standard deviation (SD) from at least 3 experiments and analyzed by using the Student's t-test, one-way ANOVA, or two-way ANOVA (GraphPad Prism 7, USA). P < 0.05 was considered to be significant.
3. Results
3.1. Magnolol induces mitochondrial dysfunction and ROS production
As previously reported, magnolol is able to induce autophagy in different cancer cell lines including NSCLC cells35, PC-3 cells41, HL-60 cells42, and H460 cells43. Consistent with these findings, Western blot showed that magnolol could significantly increase the level of microtubule associated protein 1 light chain 3 B (MAP1LC3B)-II (LC3-II) in human cervical cancer HeLa cells (Supporting Information Fig. S1A) and human neuroblastoma SH-SY5Y cells (Fig. S1B). To further confirm our results, we checked autophagic flux by using HeLa cells stably expressing GFP-LC3, a well-established cell tool to study autophagy, through co-treatment with magnolol and Baf-A1 (one classic autophagy and lysosome inhibitor). As expected, magnolol markedly increased autophagic flux (Fig. S1A–S1D). More interestingly, we observed that magnolol increased the co-localization of GFP-LC3 with mitochondria, which was further increased after co-treatment with Baf-A1 (Fig. S1E and S1F). As shown in Fig. S1E, We also found that magnolol altered mitochondrial network evidenced by perinuclear clusters of mitochondria which was very similar to the mitochondrial morphology alterations after treatment with carbonyl cyanide 3-chlorophenylhydrazone (CCCP, one well-known mitophagy inducer)48.
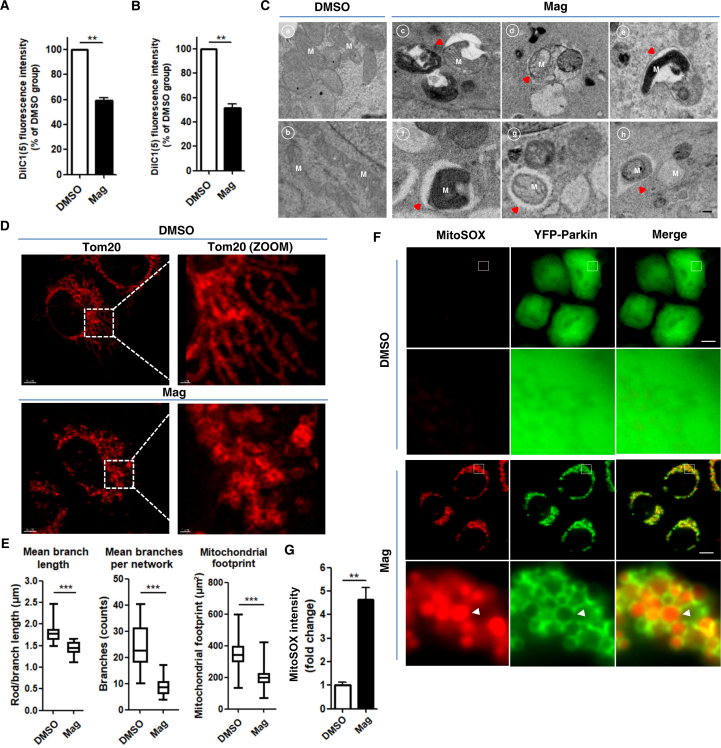
It has been reported that magnolol can target multiple mitochondrial proteins44 and decrease MMP in various cancer cells36,40. We estimated the MMP by using the potential-sensitive dye tetramethylrhodamine ethyl ester (TMRE) to stain mitochondria, which accumulated in active healthy mitochondria but lost the ability to stain depolarized mitochondria. Indeed, we found that the ability of TMRE to label mitochondria was much weaker after magnolol treatment (Supporting Information Fig. S2A–S2C), suggesting that magnolol depolarized mitochondria. Next, we utilized the MitoProbe™ DiIC1(5) Assay Kit to measure the MMP. Consistently, MMP of different cell lines was markedly reduced after magnolol treatment (Fig. 1A and B). To further ascertain the effects of magnolol on mitochondria, we employed transmission electron microscopy to directly examine the morphology of mitochondria. Magnolol treatment resulted in collapse of mitochondrial network, revealed by rupture of mitochondrial membrane, loss of cristae structure and engulfment of mitochondria by different stages of autophagic vesicles (Fig. 1C). Then, we used a high-resolution microscope to confirm our findings that magnolol disrupted the mitochondrial network and caused mitochondrial fragmentation evidenced by reductions of mitochondrial branch length, branch number, and mitochondrial footprint (mitochondrial area) (Fig. 1D and E).
Figure 1.
Magnolol causes mitochondrial dysfunction and increases ROS production. (A) HeLa cells stably expressing YFP-Parkin (HeLa-YFP-Parkin) were treated with magnolol 100 μmol/L for 6 h. Cells were stained with DiIC1(5) (50 nmol/L) for 20 min and total of 10,000 cells were collected and quantified with a flow cytometer. (B) SH-SY5Y cells were treated as in (A). Data are presented as mean ± SD (n = 3). ∗∗P < 0.01. (C) HeLa-YFP-Parkin cells were treated with DMSO (panels a and b) or magnolol 100 μmol/L (panels c–h) for 4 h. Mitochondrial ultrastructures were observed by transmission electron microscopy. Red arrows denote for the engulfment of mitochondria by early autophagic vesicles (panels c–e) and late autophagic vesicles (panels f–h). M = mitochondria. Scale bar: 0.2 μm. (D) HeLa-YFP-Parkin cells treated with DMSO or magnolol 100 μmol/L for 2 h were stained with anti-Tom20 antibody and observed by confocal fluorescence microscopy. Scale bar: 5 μm and 1 μm (ZOOM). (E) Analysis of mitochondrial network morphology changes in (D) by using MiNA toolset in Fiji (Image J). Mitochondrial morphology parameters are presented as mean ± SD from 30 individual cells (n = 3). ∗∗∗P < 0.001. (F) HeLa-YFP-Parkin cells treated with DMSO or magnolol 100 μmol/L for 3 h were stained by MitoSOX and observed by fluorescent microscopy. White arrows designate the MitoSOX fluorescent signals surrounded by Parkin-ring-like structures. MitoSOX (red), YFP-Parkin (green). Scale bar: 10 μm. (G) Fold changes of relative fluorescence intensity of mitochondrial ROS (mtROS) in (F). Data are presented as fold changes of mean ± SD fluorescence intensity compared with average intensity of DMSO control groups from 150 cells (n = 3). ∗∗P < 0.01. Data are analyzed by using the Student's t-test. Mag, magnolol.
Damaged mitochondria are the major source of intracellular ROS49, which impelled us to test the effects of magnolol on mtROS production. Indeed, we found that magnolol increased the production of mtROS estimated by MitoSOX staining which displayed good co-localization with mitochondria (Supporting Information Fig. S3). Moreover, we observed that the MitoSOX fluorescence signal was engulfed by Parkin-ring-like structures (Fig. 1F and G). Parkin-ring-like structures are suggested to surround fragmented mitochondria50, 51, 52, which further support our mitochondrial fragmentation observation (Fig. 1D and E). Collectively, our data suggest that magnolol treatment causes unambiguous dysfunction of mitochondria.
3.2. Magnolol induces mitophagy
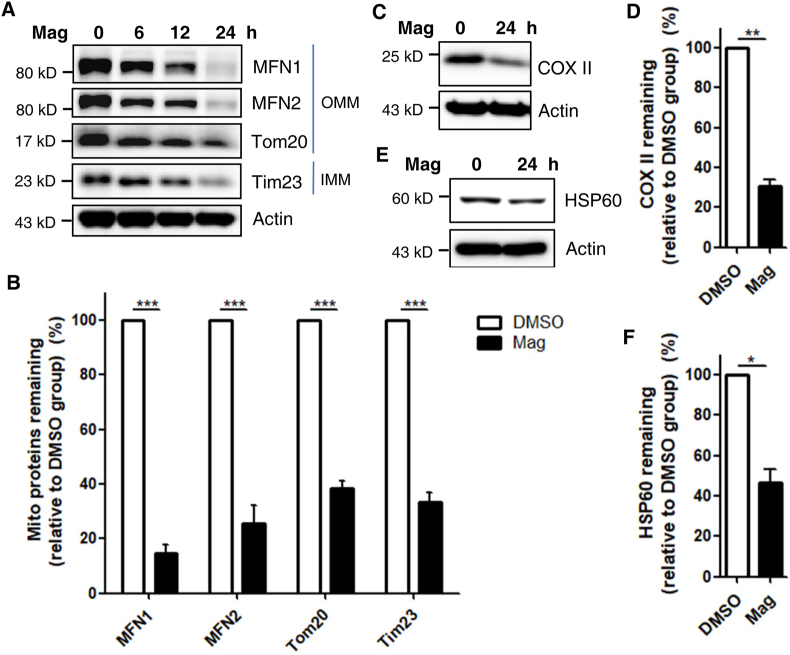
To study the possible regulatory effects of magnolol on mitophagy, we first simultaneously examined the steady-state levels of mitochondrial proteins, including outer mitochondrial membrane (OMM) proteins (MFN1, MFN2, and Tom20) and inner mitochondrial membrane (IMM) proteins (Tim23 and COX4). We observed that magnolol induced significant degradation of multiple mitochondrial proteins in a dose- and time-dependent manner checked by immunoblotting (Fig. 2A and B, and Supporting Information Fig. S4A–S4C). Next, we performed immunostaining for Tom20 (Fig. S4D and S4E) and Tim23 (Fig. S4F and S4G) to detect the effects of magnolol on mitochondrial clearance and found that magnolol could markedly induce mitochondrial elimination.
Figure 2.
Magnolol promotes the degradation of multiple mitochondrial proteins via mitophagy. (A) SH-SY5Y cells were treated with magnolol 100 μmol/L for indicated hours. Whole-cell lysates were analyzed for outer mitochondrial membrane (OMM) proteins [mitofusion1 (MFN1), mitofusion2 (MFN2), and Tom20] and inner mitochondrial membrane (IMM) protein (Tim23) by immunoblotting, and actin was used as control. (B) Quantification of mitochondrial proteins degradation after magnolol 100 μmol/L treatment for 24 h in SH-SY5Y cells. Data are presented as mean ± SD (n = 3). ∗∗∗P < 0.001 (two-way ANOVA). (C) SH-SY5Y cells were treated with magnolol 100 μmol/L for 24 h. Whole-cell lysates were analyzed for mtDNA encoded protein COX II and actin by immunoblotting. (D) Quantification of COX II from (C). Data are presented as mean ± SD (n = 3). ∗∗P < 0.01 (Student's t-test). (E) SH-SY5Y cells were treated with magnolol 100 μmol/L for 24 h. Whole-cell lysates were analyzed for mitochondrial matrix protein HSP60 and actin by immunoblotting. (F) Quantification of HSP60 from (E). Data are presented as mean ± SD (n = 3). ∗P < 0.05 (Student's t-test).
It has been reported that OMM proteins, including MFN1, MFN2, and Tom70, are exclusively degraded via the ubiquitin proteasome system (UPS)24,53. Therefore, to further confirm our conclusion that magnolol is a novel mitophagy inducer, we validated our results by detecting the changes of a mitochondrial DNA (mtDNA)-encoded IMM protein cytochrome C oxidase subunit II (COX II) and mtDNA, which are two well-established markers of mitophagy24. As with other mitochondrial proteins, magnolol could significantly induce the degradation of COX II (Fig. 2C and D, and Supporting Information Fig. S5A and S5B). Next, we performed 3D high-resolution imaging/analyse of mtDNA and found that mtDNA was effectively eliminated by magnolol treatment (Fig. S5C and S5D). In addition, we also observed that mitochondrial matrix protein HSP60 was decreased after 24 h magnolol treatment (Fig. 2E and F, and Fig. S5E–S5H). Taken together, our data demonstrate that magnolol is a novel mitophagy inducer that promotes mitochondrial turnover.
3.3. Magnolol-induced mitophagy is PINK1- and Parkin-dependent
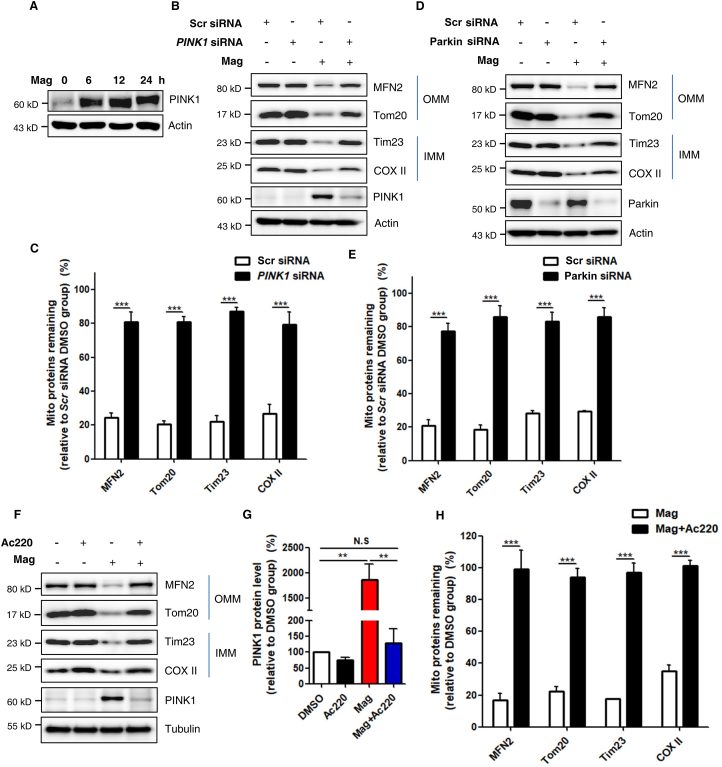
PINK and Parkin are the two most important molecules in the regulation of mitophagy4,23. When mitochondria are healthy, PINK1 is imported into mitochondria and proteolytically cleaved by mitochondrial peptidase and protease, and degraded by N-end rule pathway to keep its low expression level at normal conditions11,54, 55, 56. However, when mitochondria are damaged, PINK1 acting as a mitochondrial stress sensor is stabilized and activated on the OMM12,57. In agreement with this, we found that levels of endogenous PINK1 were quite low in DMSO-treated control groups (Fig. 3A and Supporting Information Fig. S6A). In contrast, PINK1 levels were remarkably increased in a time-dependent manner after magnolol treatment (Fig. 3A and Fig. S6A). To investigate whether magnolol-induced mitophagy is PINK1- and/or Parkin-dependent, we knocked down PINK1 or Parkin by using PINK1 siRNA or Parkin siRNA, respectively. Indeed, we observed that knockdown of either PINK1 (Fig. S6B) or Parkin (Fig. S6C) significantly blocked the degradation of multiple mitochondrial proteins induced by magnolol. To further confirm these findings, we knocked down PINK1 or Parkin in SH-SY5Y cells which express both endogenous PINK1 and Parkin. Consistently, knockdown of endogenous PINK1 or Parkin effectively blocked the degradation of multiple mitochondrial proteins in magnolol-treated groups (Fig. 3B–E).
Figure 3.
Magnolol-induced mitophagy requires PINK1 and Parkin expression. (A) SH-SY5Y cells were treated with magnolol 100 μmol/L for indicated hours. Whole-cell lysates were analyzed for PINK1 by immunoblotting. (B) SH-SY5Y cells were transfected with control (Scr) siRNA and PINK1 siRNA for 48 h and then treated with magnolol 100 μmol/L for 24 h. Immunoblotting for mitochondrial proteins was performed as indicated. (C) Quantification of mitochondrial proteins degradation in (B). Data are presented as mean ± SD (n = 3). ∗∗∗P < 0.001 (two-way ANOVA). (D) SH-SY5Y cells were transfected with Scr siRNA and Parkin siRNA for 48 h and then treated with magnolol 100 μmol/L for 24 h. Immunoblotting for mitochondrial proteins was performed as indicated. (E) Quantification of mitochondrial proteins degradation in (D). Data are presented as mean ± SD (n = 3). ∗∗∗P < 0.001 (two-way ANOVA). (F) SH-SY5Y cells were pretreated with Ac220 10 μmol/L for 2 h and then treated with magnolol 100 μmol/L and magnolol plus Ac220 for 24 h. Immunoblotting for mitochondrial proteins and PINK1 was performed as indicated. (G) Quantification of PINK1 from (F). Data are presented as mean ± SD (n = 3). ∗∗P < 0.01 (one-way ANOVA). N.S, no significant difference. (H) Quantification of mitochondrial proteins degradation in (F). Data are presented as mean ± SD (n = 3). ∗∗∗P < 0.001 (two-way ANOVA).
At present, increasing lines of evidence suggest that PINK1 functions as a central regulatory kinase in PINK1‒Parkin-mediated mitophagy pathway to initiate the first amplification loop and lay the foundations for the following steps6,24. Thus, it is conceivable that inhibition of PINK1 activation can effectively block mitophagy. Intriguingly, one very recent high-through screening identified Ac220 as a powerful inhibitor of PINK1‒Parkin-mediated mitophagy. They found that Ac220 can completely prevent PINK1 accumulation, Parkin mitochondrial translocation, and mitochondrial proteins degradation after CCCP treatment58. Consistent with their study, we also found that Ac220 significantly inhibited magnolol-induced mitophagy evidenced by the following observations: (i) Ac220 blocked PINK1 accumulation (Fig. 3F and G, and Fig. S6D); (ii) Ac220 blocked Parkin mitochondrial translocation and activation (which is detailed in Supporting Information Fig. S7A and S7B); (iii) more importantly, Ac220 dramatically blocked the degradation of multiple mitochondrial proteins (Fig. 3F and H, and Fig. S6E and S6F). Collectively, these data suggest that magnolol-induced mitophagy requires the expression and activation of PINK1 and Parkin.
3.4. Magnolol induces Parkin mitochondrial translocation and activates Parkin
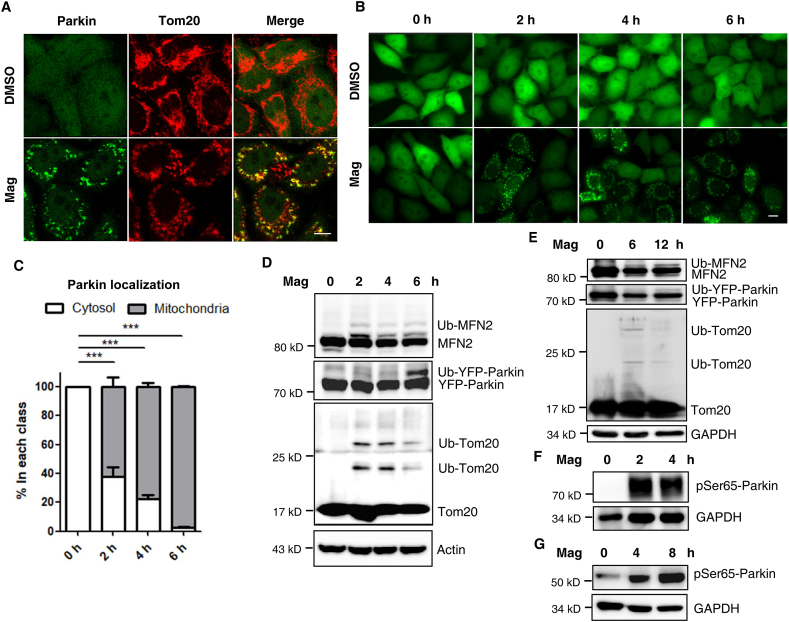
It is well established that, after mitochondrial damage, Parkin is recruited to mitochondria to induce robust mitophagy48,59,60. Earlier observations from our study suggested that magnolol cleared mitochondria in a PINK1- and Parkin-dependent manner. We wondered whether magnolol could regulate Parkin distribution and activity. Indeed, after magnolol treatment, Parkin was frequently observed on the mitochondria, whereas in the control DMSO group, Parkin was distributed in the whole cells (Fig. 4A). We also performed time course analyses to observe the dynamic process of Parkin mitochondrial translocation after magnolol treatment (Fig. 4B and C, and Supporting Information Video 1). Magnolol caused obvious Parkin mitochondrial translocation within 1 h (Supporting Information Video 1). In sharp contrast, Ac220 completely blocked Parkin mitochondrial translocation caused by magnolol at different time points (Fig. S7A and S7B).
Figure 4.
Magnolol induces Parkin mitochondrial translocation and activates Parkin. (A) HeLa-YFP-Parkin cells were treated with magnolol 100 μmol/L for 2 h. Parkin Mitochondrial translocation and co-localization with Tom20 were observed by confocal fluorescence microscopy. YFP-Parkin (green), Tom20 (red). Scale bar: 10 μm. (B) HeLa-YFP-Parkin cells were treated with magnolol 100 μmol/L for indicated time. Parkin mitochondrial translocation was observed by fluorescent microscopy. Scale bar: 10 μm. (C) The percentage of mitochondrial Parkin localization from (B) was quantified by counting at least 300 cells. Data are presented as mean ± SD (n = 3). ∗∗∗P < 0.001 (one-way ANOVA). (D) HeLa-YFP-Parkin cells were treated with magnolol 100 μmol/L for indicated time. Whole-cell lysates were analyzed for MFN2, Parkin, Tom20, and actin by immunoblotting. (E) SH-SY5Y cells transfected with YFP-Parkin were treated with magnolol 100 μmol/L for indicated time. Whole-cell lysates were analyzed by immunoblotting as indicated. (F) HeLa-YFP-Parkin cells were treated with magnolol 100 μmol/L for indicated time. Whole-cell lysates were analyzed for pSer65-Parkin by immunoblotting. (G) SH-SY5Y cells were treated with magnolol 100 μmol/L for indicated time. Whole-cell lysates were analyzed for pSer65-Parkin by immunoblotting.
The following is the supplementary data related to this article:
After mitochondrial translocation, Parkin, as a ubiquitin E3 ligase, ubiquitinates itself60 and multiple mitochondrial substrates, such as MFN2 and Tom2061. Autoubiquitination of Parkin and the polyubiquitination of Parkin's substrates can be used as an indicator for Parkin's E3 ligase activity. We were very interested to know whether magnolol could regulate Parkin's E3 ligase activity. Indeed, magnolol markedly increased the autoubiquitination level of Parkin and polyubiquitination levels of MFN2 and Tom20 (Fig. 4D and E). It has been well-established that phosphorylation of Parkin by PINK1 at Serine 65 (pSer65-Parkin) is a vital step in the activation of Parkin17, 18, 19. Thus, we checked the phosphorylation level of Parkin with a specific anti-pSer65-Parkin monoclonal antibody developed by the Michael J. Fox Foundation and Abcam. We found that pSer65-Parkin was low or undetectable in the control group but significantly increased after magnolol treatment (Fig. 4F and G). Taken together, our data clearly demonstrate that magnolol can activate Parkin via promotion of Parkin's mitochondrial translocation and phosphorylation.
3.5. Magnolol increases pSer65-Ub and recruits mitophagy receptors to mitochondria
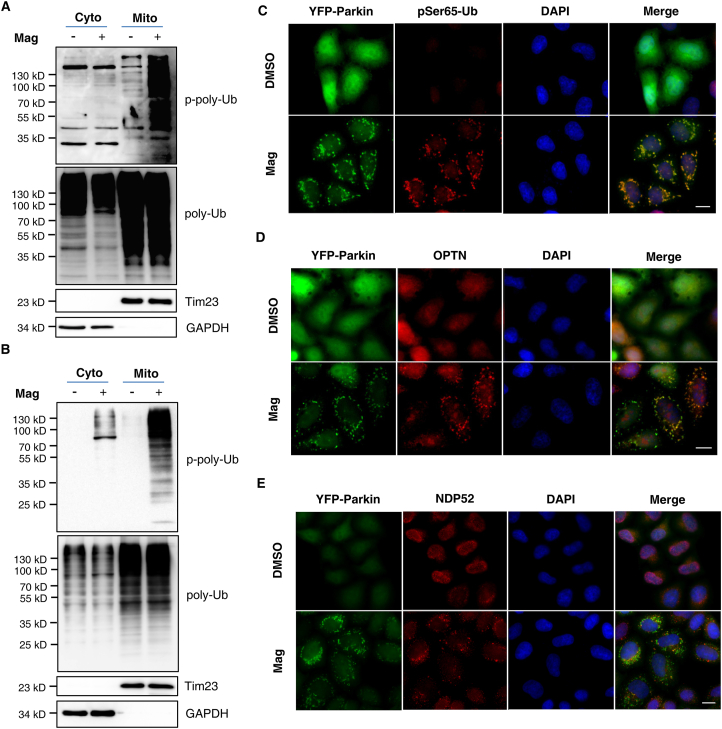
One of the recent major advances in mitophagy study is the understanding that ubiquitin phosphorylation (pSer65-Ub) is mediated by the protein kinase PINK1 when mitochondria are damaged; more importantly, pSer65-Ub chains, PINK1 and Parkin form the first positive feedforward amplification loop to rapidly initiate mitophagy6,7,23. Since we have found that magnolol caused PINK1 stabilization and Parkin mitochondrial translocation, we postulated that magnolol could increase the level of pSer65-Ub. Indeed, immunoblotting showed that the number of pSer65-Ub chains was dramatically increased after magnolol treatment (Supporting Information Fig. S8A and S8B). To determine the subcellular distribution of pSer65-Ub, we performed mitochondrial fractionation assay and found that pSer65-Ub was mainly in the mitochondrial fractions (Fig. 5A and B). We further validated the mitochondrial distribution of pSer65-Ub through the co-localization of pSer65-Ub and mitochondrial Parkin following treatment with magnolol (Fig. 5C).
Figure 5.
Magnolol increases the level of pSer65-Ub and recruits mitophagy/autophagy receptors to mitochondria. (A) SH-SY5Y cells were treated with magnolol 100 μmol/L for 6 h. Cell fractions were performed to isolate mitochondria. Tim23 and GAPDH were used as mitochondrial (Mito) and cytosolic (Cyto) fraction markers, respectively. pSer65-Ub were analyzed by immunoblotting. (B) HeLa-YFP-Parkin cells were treated with magnolol 100 μmol/L for 4 h. Cell fractions were performed to isolate mitochondria. Tim23 and GAPDH were used as mitochondrial (Mito) and cytosolic (Cyto) fraction markers, respectively. pSer65-Ub were analyzed by immunoblotting. (C) HeLa-YFP-Parkin cells were treated with magnolol 100 μmol/L for 2 h. pSer65-Ub was stained by anti-pSer65-Ub antibody and observed by fluorescent microscopy. pSer65-Ub (red), YFP-Parkin (green), nucleus (DAPI, blue). Scale bar: 10 μm. (D, E) HeLa-YFP-Parkin cells were treated with magnolol 100 μmol/L for 2 h. OPTN (D) and NDP52 (E) were stained by anti-OPTN and anti-NDP52 antibody, respectively. Cells were observed by fluorescent microscopy. OPTN or NDP52 (red), YFP-Parkin (green), nucleus (DAPI, blue). Scale bar: 10 μm.
After mitophagy initiation, mitophagy/autophagy receptors, such as OPTN, NDP52, SQSTM1/p62, and NBR1, are recruited to mitochondria and serve as a bridge to connect the ubiquitinated mitochondrial cargos with LC3-coated autophagosomes via their LC3 interacting region (LIR) and ubiquitin binding domain5. More recently, the second positive feedforward amplification loop of mitophagy between OPTN, NDP52, and ATG8s has been clarified26. We then systematically examined, by immunostaining, the distribution of these receptors after magnolol treatment. We frequently observed all three receptors (OPTN, NDP52, and p62) on the damaged mitochondria of the treatment groups, whereas all three receptors were diffusely distributed in the cytoplasm of the control DMSO groups (Fig. 5D and E, and Fig. S8C). Therefore, data from this part of our study suggest that magnolol can increase the number of pSer65-Ub chains and recruit mitophagy receptors to damaged mitochondria to promote the feedforward mechanisms, thus finally inducing robust mitophagy.
3.6. Magnolol elicits cytotoxicity in cancer cells and inhibits tumor growth in vivo
In this part of our study, we attempted to explore the functional importance of magnolol in the regulation of cancer cell viability and tumor growth. After 24 h incubation, magnolol caused significant cell death in human neuroblastoma SH-SY5Y cells (Fig. 6A and B). We also observed that magnolol induced apoptosis through the intrinsic apoptosis pathway by checking the apoptotic hallmarks, including cleaved-caspase9, cleaved-caspase3, and cleaved-PARP (Fig. 6C and Supporting Information Fig. S9A). In addition, Z-VAD-FMK-pan-caspase inhibitor (ZVAD) was able to block cell death induced by magnolol (Fig. S9B–S9E) while ferroptosis inhibitor ferrostatin-1 or necroptosis inhibitor necrostatin-1 could not prevent cell death (Fig. S9F).
Figure 6.
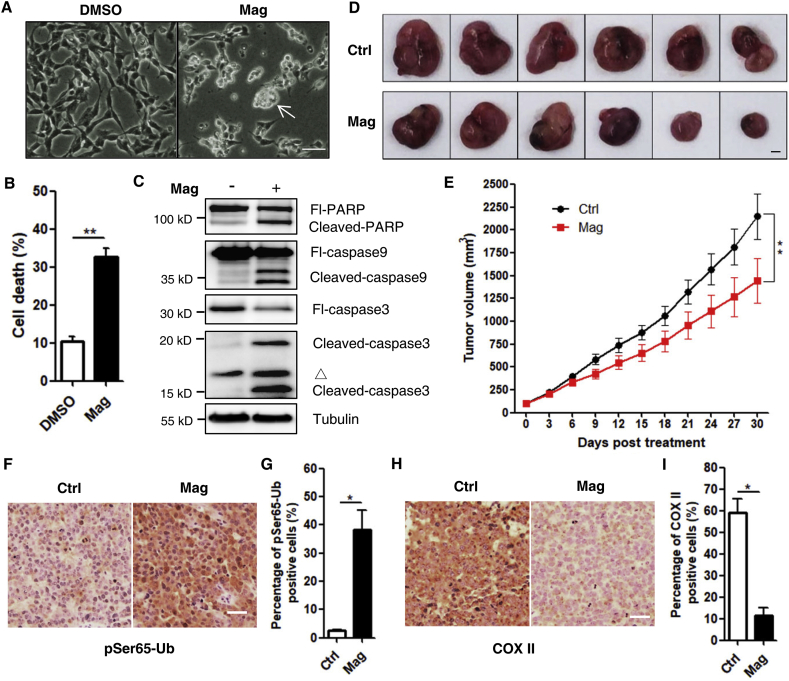
Magnolol induces cell death in SH-SY5Y cells and inhibits tumor growth in vivo. (A) The bright field images of SH-SY5Y cells treated with DMSO and magnolol 100 μmol/L for 24 h. Morphological changes resembling cell death (white arrow) were observed in magnolol-treated group. Scale bar: 200 μm. (B) Cell pellets of (A) were subsequently collected and cell death was quantified using PI live exclusion staining. Data are presented as mean ± SD (n = 3). ∗∗P < 0.01 (Student's t-test). (C) SH-SY5Y cells were treated as in (A) for 24 h. Whole-cell lysates were analyzed for full-length PARP (Fl-PARP), cleaved-PARP, full-length caspase 9 (Fl-caspase 9), cleaved-caspase 9, full-length caspase 3 (Fl-caspase 3), cleaved-caspase 3, and tubulin by immunoblotting. ▵, non-specific bands. (D) Representative photos of SH-SY5Y xenografted tumors. Mice were intraperitoneally treated with saline (Ctrl) or magnolol (35 mg/kg) for 30 days. Scale bar: 5 mm. (E) Tumor volumes were measured every three days following the treatment in (D). Values are represented as mean ± SD from at least 6 tumors in each group. ∗∗P < 0.01 (two-way ANOVA). (F) pSer65-Ub was assessed with immunohistochemistry by using anti-pSer65-Ub antibody in tumor sections from each group. Scale bar: 20 μm. (G) Quantification of pSer65-Ub in (F). Data are presented as mean ± SD from three random fields per sample. ∗P < 0.05 (Student's t-test). (H) COX II was assessed with immunohistochemistry by using anti-COX II antibody in tumor sections from each group. Scale bar: 20 μm. (I) Quantification of COX II in (H). Data are presented as mean ± SD from three random fields per sample. ∗P < 0.05 (Student's t-test).
Next, we evaluated the potential therapeutic effects of magnolol in vivo by using a xenograft mouse model. SH-SY5Y cells were implanted into 6-week-old male nude mice. One week after cell inoculation, mice bearing visible tumors (80–120 mm3) were treated once-daily with saline (control) or magnolol. Tumor volume and mice body weight were monitored every three days. We found that the growth of tumors was significantly reduced by 32.81% (control vs. magnolol-treatment) (Fig. 6D and E). Meanwhile, magnolol treatment did not affect the body weight of the mice (Fig. S9G), suggesting that there were no side effects. Notably, in a separate study, we found that magnolol prolonged the life of mice bearing tumors (Fig. S9H). These data indicate that magnolol significantly inhibits tumor growth in vivo and improves life span outcomes. Additionally, we were very interested to know whether magnolol could induce mitophagy in vivo. Indeed, we found that magnolol treatment markedly increased the level of pSer65-Ub in tumor sections from xenografts (Fig. 6F and G), while the level of COX II was significantly reduced (Fig. 6H and I), indicating that magnolol increased mitophagy in vivo.
3.7. Suppression of autophagy/mitophagy enhances the anticancer efficacy of magnolol
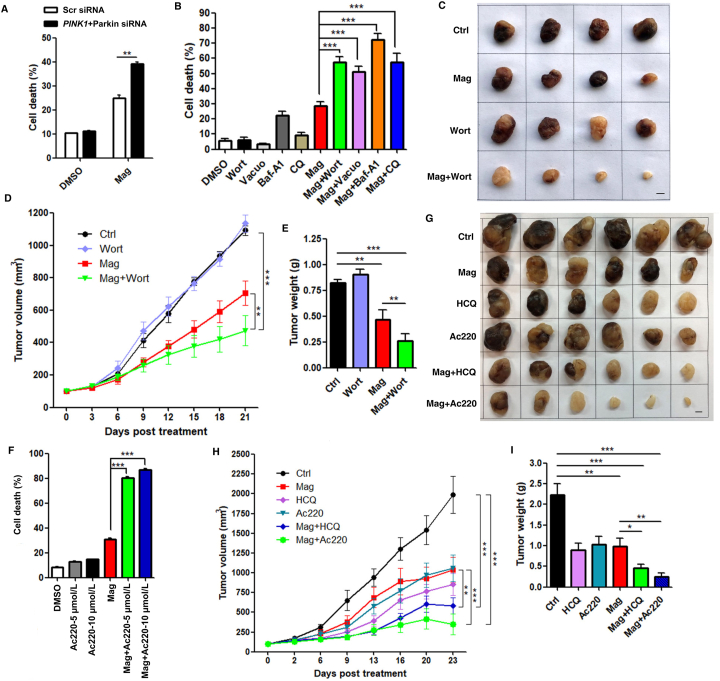
Although Parkin has been suggested as a tumor suppressor at steady state, it is still unclear or controversial how Parkin determines cell fate after mitochondrial damage or during mitophagy. It has been reported that during PINK1‒Parkin-mediated mitophagy, Parkin can inhibit intrinsic apoptosis through targeting the key mitochondrial apoptotic effectors of BAK and BAX62. We then proceeded to explore the role of magnolol-induced mitophagy in the regulation of cell viability and tumor growth. We first knocked down PINK1 and Parkin by using siRNA, and found that knockdown of PINK1 and Parkin significantly increased cell death compared with control cells with non-targeting siRNA under magnolol treatment (Fig. 7A and Supporting Information Fig. S10A). Furthermore, to better determine the translatability of targeting mitophagy during cancer progression, we systematically inhibited autophagy/mitophagy at different stages by using multiple inhibitors, including wortmannin, Baf-A1, CQ, and vacuolin-1. All these inhibitors effectively blocked mitophagy in different cancer cell lines (Fig. S10B and S10C), which is consistent with the reports of other studies26,53. Noticeably, the cell death caused by magnolol was remarkably increased after autophagy/mitophagy inhibition (Fig. 7B and Fig. S10D–S10F), implying that magnolol-induced autophagy/mitophagy plays a cell protective role in this process.
Figure 7.
Inhibition of mitophagy enhances the anticancer efficacy of magnolol. (A) SH-SY5Y cells were transiently transfected with Scr siRNA and mixed PINK1 plus Parkin siRNA for 48 h, and then treated with DMSO and magnolol 100 μmol/L for 24 h. Cell pellets were subsequently collected, and cell death was quantified using PI live exclusion staining. Data are presented as mean ± SD (n = 3). ∗∗P < 0.01 (two-way ANOVA). (B) SH-SY5Y cells treated with indicated treatments [DMSO, magnolol 100 μmol/L, wortmannin (wort) 5 μmol/L, vacuolin-1 1 μmol/L, Baf-A1 100 nmol/L, CQ 20 μmol/L, and combined agents] for 24 h were collected and cell death was quantified using PI live exclusion staining. Data are presented as mean ± SD (n = 3). ∗∗∗P < 0.001 (one-way ANOVA). (C) Representative photos of SH-SY5Y xenografted tumors. Mice were intraperitoneally treated with saline (Ctrl), magnolol (35 mg/kg), wortmannin (0.35 mg/kg), magnolol + wortmannin for 21 days. Scale bar: 5 mm. (D) Tumor volumes were measured every three days following the treatment in (B). Values are represented as mean ± SD from at least 5 tumors in each group. ∗∗P < 0.01, ∗∗∗P < 0.001 (two-way ANOVA). (E) Tumor weights were analyzed after 21 days' treatment in (B). Values are represented as mean ± SD from at least 5 tumors in each group. ∗∗P < 0.01, ∗∗∗P < 0.001 (Student's t-test). (F) SH-SY5Y cells were pre-treated with 5 or 10 μmol/L Ac220 for 2 h and then treated with DMSO, Ac220 (5 or 10 μmol/L), magnolol 100 μmol/L and combined agents for 24 h. Cells were collected, and cell death was quantified using PI live exclusion staining. Data are presented as mean ± SD (n = 3). ∗∗∗P < 0.001 (one-way ANOVA). (G) Representative photos of SH-SY5Y xenografted tumors. Mice were intraperitoneally treated with saline (Ctrl), magnolol (35 mg/kg), HCQ (50 mg/kg), Ac220 (1.5 mg/kg), magnolol + HCQ, magnolol + Ac220 for 23 days. Scale bar: 5 mm. (H) Tumor volumes were measured every three or four days following the treatment in (F). Values are represented as mean ± SD from 6 tumors in each group. ∗∗P < 0.01, ∗∗∗P < 0.001 (two-way ANOVA). (I) Tumor weights were analyzed after 23 days' treatment in (F). Values are represented as mean ± SD from 6 tumors in each group. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (Student's t-test).
To further validate our aforementioned findings, we used SH-SY5Y cells to generate xenograft mouse model, and employed three distinct inhibitors to target the different steps of mitophagic processes. First, we chose wortmannin, a well-established autophagy inhibitor that blocks the formation of autophagosomes via inhibition of the class III phosphatidylinositol 3-kinase PIK3C3/Vps34 pathway63,64. Another, more important reason is that, according to a recent study, wortmannin is able to efficiently block mitophagy through inhibition of LIR-mediated recruitment of primary mitophagy receptors, including OPTN and NDP52, thus to disrupt the second positive feedforward amplification loop of mitophagy and thus block mitophagy at the early stage26. In our studies, co-treatment with wortmannin significantly enhanced the antitumor efficacy of magnolol, as shown by comparison of tumor volume (Fig. 7C and D) and tumor weight (Fig. 7E). Tumor volume reduction increased from 32.25% (magnolol-treated) to 56.60% (magnolol + wortmannin-treated) of control; while tumor weight reduction increased from 43.05% (magnolol-treated) to 68.50% (magnolol + wortmannin-treated) of control. And there were no differences of the mice body weight between the co-treatment of magnolol with wortmannin and the single treatment (Supporting Information Fig. S11A).
Second, we utilized HCQ, a derivative of CQ, to block mitophagy at the late stage. HCQ can effectively inhibit lysosomal acidification and fusion of mitophagosomes with lysosomes to block the degradation of mitochondria65, which has been widely used in phase I and phase II clinical trials in combination with other anticancer agents66. Consistently, combination of HCQ and magnolol significantly increased the reduction the tumor volume (47.88%, magnolol-treated vs. 70.66%, magnolol + HCQ-treated; Fig. 7G and H) and tumor weight (55.65%, magnolol-treated vs. 79.28%, magnolol + HCQ-treated; Fig. 7I), compared with control groups.
Third and more importantly, to further confirm the importance of mitophagy in the above processes, we used Ac220 to block PINK1 accumulation and Parkin mitochondrial translocation, thus inhibiting mitophagy as previous study done58. We first observed that the combination of Ac220 and magnolol increased significant cell death (Fig. 7F and Fig. S11B–S11D). Second, the combination of Ac220 and magnolol exerted potent antitumor activity. Tumor volume reduction increased from 47.88% (magnolol-treated) to 82.28% (magnolol + Ac220-treated) of control (Fig. 7G and H); while tumor weight reduction increased from 55.65% (magnolol-treated) to 88.78% (magnolol + Ac220-treated) of control (Fig. 7I). Third, we also confirmed that different inhibitors clearly blocked mitophagy in vivo evidenced by the changes of mitophagy marker COX II (Supporting Information Fig. S12A and S12B), whilst there were no differences of the mice body weight among different treated groups (Fig. S12C). Taken together, our data demonstrate that suppression of autophagy/mitophagy can greatly promote magnolol's anticancer efficacy.
4. Discussion
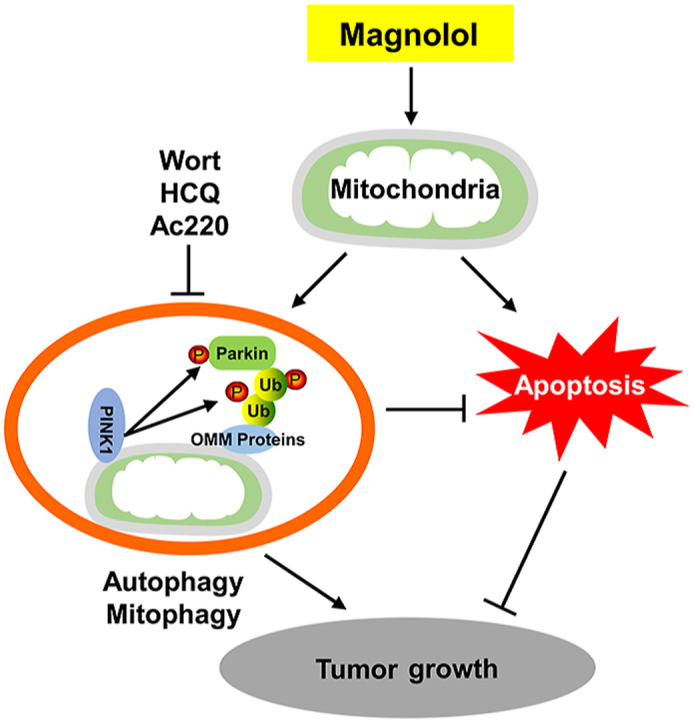
Multiple studies have reported that mitophagy plays essential roles in the pathogenesis of neurodegeneration diseases to effectively and specifically remove dysfunctional mitochondria8,9. However, the function of mitophagy in anticancer therapy and/or chemoresistance is largely unclear. In this study, we found that magnolol causes mitochondrial dysfunction, increases mtROS production, induces autophagy/mitophagy and mitochondrial apoptosis; blockage of autophagy/mitophagy through genetic or pharmacological approaches promotes cell death rather than attenuates cell death; furthermore, in vivo, inhibition of autophagy/mitophagy at different stages significantly enhances magnolol's anticancer efficacy. Therefore, our study provides a rational basis for manipulating mitophagy to improve the potency and efficacy of anticancer agents.
At present, chemotherapy is still the most widely used treatment of cancer. However, the major obstacle with chemotherapy is that cancer cells develop drug resistance which ultimately results in therapeutic failure and patient death. Since most anticancer agents elicit cytotoxicity through induction of mitochondrial damage or dysfunction67, to compensate, the cancer cells possibly initiate mitophagy to rapidly clear the dysfunctional mitochondria and reduce detrimental ROS, thereby evading cell death and develop chemoresistance. Thus, at this stage, exploration of the novel function of mitophagy in cancer and the development of novel therapeutic intervention through administration of combined agents represent unmet urgent clinical need for cancer drug resistance, which have been highlighted in recent excellent reviews68,69.
To explore the function of magnolol in the regulation of mitophagy, we first provided compelling evidence to demonstrate that magnolol could induce autophagy in different types of cancer cells; this is consistent with previous reports35,41, 42, 43. Intriguingly, we also observed that magnolol could increase the mitochondrial distribution of LC3-II and resulted in perinuclear clusters of mitochondria; importantly, co-treatment with Baf-A1 further increased LC3-II accumulation on mitochondria, suggesting that magnolol possibly increased mitophagic flux. To provide more direct evidence, we used different methods, including transmission electron microscopy and immunostaining, to examine the structure and morphology of mitochondria. We found that magnolol resulted in collapse of the mitochondrial network and caused mitochondrial fragmentation, which was accompanied by the engulfment of mitochondria within different stages of autophagic vesicles, indicating the possibility that mitophagy was induced after magnolol treatment.
It is well-known that the normal morphology and distribution of mitochondria are essential for their function, and dysfunctional mitochondria are the major source of intracellular ROS49,70. Indeed, we found that magnolol treatment reduced MMP and increased mtROS production, which is in accordance with many other studies40,71,72. Interestingly, another school of thoughts suggest magnolol displays antioxidant properties73, 74, 75, mainly focusing on the neuron or cardiovascular protective potential of magnolol, suggesting the complicated mechanism and function of natural products. Future work in this area is still needed.
Next, we utilized various established methods to check the effects of magnolol on mitophagy, including immunoblotting and immunostaining to detect the degradation of multiple mitochondrial proteins with different subcellular distribution, and staining of mtDNA. All these data consistently demonstrate that magnolol induces PINK1- and Parkin-dependent mitophagy. Mechanistically, magnolol can positively regulate two rounds of feedforward amplification loops of mitophagy. The first one is PINK1‒pSer65-Ub‒Parkin to mediate the initiation stage of mitophagy; the second one is LC3‒OPTN/NDP52 to mediate the recognition and sequestration stages of mitophagy.
After establishing the positive regulatory effects of magnolol on mitophagy, we were very interested to investigate the biological functions of mitophagy induced by magnolol. Since Parkin is undetectable in most cancer cell lines, HeLa cell line with stable expression of YFP-Parkin has become the most commonly used cell system to study PINK1‒Parkin-mediated mitophagy. However, this causes limitations for physiological functional studies. Thus, in this part of our study, we mainly used a physiologically relevant human neuroblastoma SH-SY5Y cell line which expresses endogenous Parkin, especially in the in vivo xenograft mouse model. And we put HeLa-YFP-Parkin cell line in Supporting Information figures as evidence for the mechanistic study as previous studies done58,76. We first observed that magnolol treatment caused significant cell death through the intrinsic apoptosis pathway, which could be attenuated by caspase inhibitor ZVAD but not ferroptosis inhibitor ferrostatin-1 or necroptosis inhibitor necrostatin-1. Next and more importantly, our in vivo study revealed that magnolol remarkedly inhibited tumor growth and prolonged the survival time of the mice bearing tumors with no side effects. It is worthy to note that we also found that magnolol increased the level of mitophagy in the tumor tissues, which was indeed consistent with our in vitro data and also raised an important question to us: what is the function of mitophagy in the magnolol's anticancer effects?
To answer the above-mentioned question, we effectively blocked mitophagy by first silencing PINK1 and Parkin by using siRNA. We found that PINK1 and Parkin knockdown sensitized magnolol-induced cell death. Then, we co-treated the cells with magnolol and different autophagy/mitophagy inhibitors, and confirmed that combined treatment inhibited mitophagy. Interestingly, combined treatment markedly increased cell death, suggesting a protective role of mitophagy. Furthermore, to determine the translatability of targeting autophagy/mitophagy for future cancer treatment, we used three different inhibitors to block autophagy/mitophagy at different stages, including (1) wortmannin to block formation of autophagosomes/mitophagosomes and the second feedforward amplification loop of mitophagy (Inhibition of autophagy/mitophagy at early stage); (2) HCQ to block the degradation of mitochondria and formation of mitolysosomes (inhibition of autophagy/mitophagy at late stage); and (3) Ac220 to block PINK1 accumulation and Parkin mitochondrial translocation (inhibition of the initiation of PINK1‒Parkin-mediated mitophagy). In line with the previous cell death results, in our in vivo studies, both the combined magnolol‒wortmannin treatment and the combined magnolol‒HCQ were much more effective in reducing tumor growth than magnolol alone. However, although wortmannin, CQ, and/or HCQ have been widely used to inhibit mitophagy in many studies26,65,77, these inhibitors can also affect general autophagy or other molecular pathways. Thus, specific inhibitors, especially targeting PINK1‒Parkin-mediated mitophagy are in high demand. We and others58 found that Ac220 can effectively inhibit mitophagy. First, Ac220 dramatically blocked PINK1 accumulation. Second, Ac220 completely blocked Parkin mitochondrial translocation and activation. Third, Ac220 significantly inhibited magnolol-induced mitophagy in vitro and in vivo. Fourth, compared with magnolol alone, the combination of Ac220 and magnolol did not affect the phosphorylation level of FLT3 (a known target of Ac220). More importantly, Ac220 sensitized magnolol-induced cell death. In line with these results, the combination of Ac220 and magnolol further reduced tumor volume and tumor weight compared with single treatment. Overall, these results indicate that mitophagy protects cancer cells from magnolol-induced cytotoxicity.
Although our study and previous study provided convincing evidence that Ac220 can effectively inhibit PINK1‒Parkin-mediated mitophagy during mitochondrial damage, it will be interesting and necessary to further study whether Ac220 is a specific inhibitor of PINK1 or Parkin. For example, future studies are needed to explore whether Ac220 can directly bind PINK1 or Parkin. In addition, since autophagy/mitophagy plays crucial roles in maintaining cellular homeostasis, healthy longevity, cancer, and neuroprotection78, 79, 80, 81, it is important to note the possible side effects after autophagy/mitophagy inhibition. Thus, we can consider the following aspects to reduce possible severe side effects when we use autophagy/mitophagy inhibition strategy for cancer treatment: (1) treat the cancer patients with high autophagy/mitophagy level; (2) develop tumor targeting agents to reduce the toxicity to other tissues; (3) increase the therapeutic index, shorten the chemotherapy time, and space the treatments at intervals to give the body some to recover; and (4) combine with other beneficial drugs to reduce the side effects. For example, we can additionally administer NAD+ precursors to protect the neurons82. Further studies are thus needed.
5. Conclusions
Our study demonstrates that inhibition of PINK1‒Parkin-mediated mitophagy significantly enhances magnolol's anticancer efficacy. Our study has three noteworthy findings. First, it helps elucidate the protective role of PINK1‒Parkin-mediated mitophagy during anticancer agents' treatment and apoptosis. Second, it predicts that the combination of magnolol with autophagy/mitophagy inhibitors is a promising therapeutic strategy for cancer treatment. Third, manipulating autophagy/mitophagy will probably provide a solution to overcome chemoresistance in cancer. Therefore, our proposed therapeutic strategy whereby inhibitors block autophagy/mitophagy to enhance anticancer efficacy of magnolol warrants further clinical studies. In addition, further studies are needed to identify more specific mitophagy inhibitors and explore how to best combine these inhibitors with anticancer agents in different animal models and clinical trials which will provide new therapeutic approaches.
Acknowledgments
This work was supported by research grants from Innovation and Technology Fund (PRP/036/20FX, China) and Health and Medical Research Fund (MHRF-16170251, China) of Hong Kong to Hu-Biao Chen, Singapore Ministry of Education (MOE) Tier 2 (MOE2018-T2-1-060, Singapore) to Han-Ming Shen, National Natural Science Foundation of China (82074123 to Hu-Biao Chen; 31501116 to Yingying Lu; 82071441 to Liming Wang). We thank members of Chen's laboratory and Shen's laboratory for valuable discussion. We gratefully thank the support from Dr. Richard Youle for providing the YFP-Parkin-HeLa cells; Dr. Noboru Mizushima for providing the GFP-LC3B-HeLa cells. We thank Dr. Martha Dahlen for polishing this manuscript.
Author contributions
Yancheng Tang, Liming Wang, Yingying Lu, Han-Ming Shen, and Hu-Biao Chen conceived the study and designed the experiments. Yancheng Tang, Liming Wang, Yingying Lu, Tao Yi, Jun Xu, Jiang-Jiang Qin, Qilei Chen, Ka-Man Yip, Yihang Pan, and Peng Hong performed the experiments. Yancheng Tang, Liming Wang, Yingying Lu, Han-Ming Shen, and Hu-Biao Chen analysed the data and wrote the paper.
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.06.007.
Contributor Information
Yingying Lu, Email: Luyy39@sysu.edu.cn.
Han-Ming Shen, Email: phsshm@nus.edu.sg.
Hu-Biao Chen, Email: hbchen@hkbu.edu.hk.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Pfanner N., Warscheid B., Wiedemann N. Mitochondrial proteins: from biogenesis to functional networks. Nat Rev Mol Cell Biol. 2019;20:267–284. doi: 10.1038/s41580-018-0092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy M.P., Hartley R.C. Mitochondria as a therapeutic target for common pathologies. Nat Rev Drug Discov. 2018;17:865–886. doi: 10.1038/nrd.2018.174. [DOI] [PubMed] [Google Scholar]
- 4.Pickles S., Vigie P., Youle R.J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol. 2018;28:R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palikaras K., Lionaki E., Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20:1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 6.Wang L., Qi H., Tang Y., Shen H.M. Post-translational modifications of key machinery in the control of mitophagy. Trends Biochem Sci. 2020;45:58–75. doi: 10.1016/j.tibs.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Harper J.W., Ordureau A., Heo J.M. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol. 2018;19:93–108. doi: 10.1038/nrm.2017.129. [DOI] [PubMed] [Google Scholar]
- 8.Williams J.A., Ding W.X. Mechanisms, pathophysiological roles and methods for analyzing mitophagy — recent insights. Biol Chem. 2018;399:147–178. doi: 10.1515/hsz-2017-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou G., Palikaras K., Lautrup S., Scheibye-Knudsen M., Tavernarakis N., Fang E.F. Mitophagy and neuroprotection. Trends Mol Med. 2019;26:8–20. doi: 10.1016/j.molmed.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazarou M., Jin S.M., Kane L.A., Youle R.J. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okatsu K., Oka T., Iguchi M., Imamura K., Kosako H., Tani N., et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat Commun. 2012;3:1016. doi: 10.1038/ncomms2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 15.Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K., et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okatsu K., Koyano F., Kimura M., Kosako H., Saeki Y., Tanaka K., et al. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J Cell Biol. 2015;209:111–128. doi: 10.1083/jcb.201410050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiba-Fukushima K., Imai Y., Yoshida S., Ishihama Y., Kanao T., Sato S., et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iguchi M., Kujuro Y., Okatsu K., Koyano F., Kosako H., Kimura M., et al. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J Biol Chem. 2013;288:22019–22032. doi: 10.1074/jbc.M113.467530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H.I., Campbell D.G., Gourlay R., et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gladkova C., Maslen S.L., Skehel J.M., Komander D. Mechanism of parkin activation by PINK1. Nature. 2018;559:410–414. doi: 10.1038/s41586-018-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sauve V., Sung G., Soya N., Kozlov G., Blaimschein N., Miotto L.S., et al. Mechanism of parkin activation by phosphorylation. Nat Struct Mol Biol. 2018;25:623–630. doi: 10.1038/s41594-018-0088-7. [DOI] [PubMed] [Google Scholar]
- 22.Ordureau A., Sarraf S.A., Duda D.M., Heo J.M., Jedrychowski M.P., Sviderskiy V.O., et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen T.N., Padman B.S., Lazarou M. Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol. 2016;26:733–744. doi: 10.1016/j.tcb.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heo J.M., Ordureau A., Paulo J.A., Rinehart J., Harper J.W. The PINK1–PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padman B.S., Nguyen T.N., Uoselis L., Skulsuppaisarn M., Nguyen L.K., Lazarou M. LC3/GABARAPs drive ubiquitin-independent recruitment of Optineurin and NDP52 to amplify mitophagy. Nat Commun. 2019;10:408. doi: 10.1038/s41467-019-08335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vargas J.N.S., Wang C., Bunker E., Hao L., Maric D., Schiavo G., et al. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol Cell. 2019;74:347–362. doi: 10.1016/j.molcel.2019.02.010. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J.H., Kim H.Y., Lee Y.K., Yoon Y.S., Xu W.G., Yoon J.K., et al. Involvement of mitophagy in oncogenic K-Ras-induced transformation: overcoming a cellular energy deficit from glucose deficiency. Autophagy. 2011;7:1187–1198. doi: 10.4161/auto.7.10.16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan C., Luo L., Guo C.Y., Goto S., Urata Y., Shao J.H., et al. Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Cancer Lett. 2017;388:34–42. doi: 10.1016/j.canlet.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Lee K.S., Wu Z., Song Y., Mitra S.S., Feroze A.H., Cheshier S.H., et al. Roles of PINK1, mTORC2, and mitochondria in preserving brain tumor-forming stem cells in a noncanonical Notch signaling pathway. Genes Dev. 2013;27:2642–2647. doi: 10.1101/gad.225169.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu K., Lee J., Kim J.Y., Wang L., Tian Y., Chan S.T., et al. Mitophagy controls the activities of tumor suppressor p53 to regulate hepatic cancer stem cells. Mol Cell. 2017;68:281–292. doi: 10.1016/j.molcel.2017.09.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobili S., Lippi D., Witort E., Donnini M., Bausi L., Mini E., et al. Natural compounds for cancer treatment and prevention. Pharmacol Res. 2009;59:365–378. doi: 10.1016/j.phrs.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 33.Ranaware A.M., Banik K., Deshpande V., Padmavathi G., Roy N.K., Sethi G., et al. Magnolol: a neolignan from the magnolia family for the prevention and treatment of cancer. Int J Mol Sci. 2018;19:2362. doi: 10.3390/ijms19082362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Y.C., Tsao M.J., Chiu C.Y., Kan P.C., Chen Y. Magnolol inhibits human glioblastoma cell migration by regulating N-cadherin. J Neuropathol Exp Neurol. 2018;77:426–436. doi: 10.1093/jnen/nly021. [DOI] [PubMed] [Google Scholar]
- 35.Shen J., Ma H., Zhang T., Liu H., Yu L., Li G., et al. Magnolol inhibits the growth of non-small cell lung cancer via inhibiting microtubule polymerization. Cell Physiol Biochem. 2017;42:1789–1801. doi: 10.1159/000479458. [DOI] [PubMed] [Google Scholar]
- 36.Tsai J.R., Chong I.W., Chen Y.H., Hwang J.J., Yin W.H., Chen H.L., et al. Magnolol induces apoptosis via caspase-independent pathways in non-small cell lung cancer cells. Arch Pharm Res. 2014;37:548–557. doi: 10.1007/s12272-013-0232-1. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y., Cao W., Zhang B., Liu Y.Q., Wang Z.Y., Wu Y.P., et al. The natural compound magnolol inhibits invasion and exhibits potential in human breast cancer therapy. Sci Rep. 2013;3:3098. doi: 10.1038/srep03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chilampalli C., Guillermo R., Zhang X., Kaushik R.S., Young A., Zeman D., et al. Effects of magnolol on UVB-induced skin cancer development in mice and its possible mechanism of action. BMC Cancer. 2011;11:456. doi: 10.1186/1471-2407-11-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee D.H., Szczepanski M.J., Lee Y.J. Magnolol induces apoptosis via inhibiting the EGFR/PI3K/Akt signaling pathway in human prostate cancer cells. J Cell Biochem. 2009;106:1113–1122. doi: 10.1002/jcb.22098. [DOI] [PubMed] [Google Scholar]
- 40.Wen H., Zhou S., Song J. Induction of apoptosis by magnolol via the mitochondrial pathway and cell cycle arrest in renal carcinoma cells. Biochem Biophys Res Commun. 2019;508:1271–1278. doi: 10.1016/j.bbrc.2018.12.087. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S., Guru S.K., Pathania A.S., Kumar A., Bhushan S., Malik F. Autophagy triggered by magnolol derivative negatively regulates angiogenesis. Cell Death Dis. 2013;4:e889. doi: 10.1038/cddis.2013.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S., Kumar A., Pathania A.S., Guru S.K., Jada S., Sharma P.R., et al. Tiron and trolox potentiate the autophagic cell death induced by magnolol analog Ery5 by activation of Bax in HL-60 cells. Apoptosis. 2013;18:605–617. doi: 10.1007/s10495-013-0805-y. [DOI] [PubMed] [Google Scholar]
- 43.Li H.B., Yi X., Gao J.M., Ying X.X., Guan H.Q., Li J.C. Magnolol-induced H460 cells death via autophagy but not apoptosis. Arch Pharm Res. 2007;30:1566–1574. doi: 10.1007/BF02977326. [DOI] [PubMed] [Google Scholar]
- 44.Cassiano C., Esposito R., Tosco A., Casapullo A., Mozzicafreddo M., Tringali C., et al. Chemical proteomics-guided identification of a novel biological target of the bioactive neolignan magnolol. Front Chem. 2019;7:53. doi: 10.3389/fchem.2019.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheer U., Messner K., Hazan R., Raska I., Hansmann P., Falk H., et al. High sensitivity immunolocalization of double and single-stranded DNA by a monoclonal antibody. Eur J Cell Biol. 1987;43:358–371. [PubMed] [Google Scholar]
- 46.Wang L., Cho Y.L., Tang Y., Wang J., Park J.E., Wu Y., et al. PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1–Parkin-mediated mitophagy. Cell Res. 2018;28:787–802. doi: 10.1038/s41422-018-0056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valente A.J., Maddalena L.A., Robb E.L., Moradi F., Stuart J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119:315–326. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polster B.M., Nicholls D.G., Ge S.X., Roelofs B.A. Use of potentiometric fluorophores in the measurement of mitochondrial reactive oxygen species. Methods Enzymol. 2014;547:225–250. doi: 10.1016/B978-0-12-801415-8.00013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai Q., Zakaria H.M., Simone A., Sheng Z.H. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol. 2012;22:545–552. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamano K., Wang C., Sarraf S.A., Münch C., Kikuchi R., Noda N.N., et al. Endosomal Rab cycles regulate Parkin-mediated mitophagy. Elife. 2018;7 doi: 10.7554/eLife.31326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puri R., Cheng X.T., Lin M.Y., Huang N., Sheng Z.H. Mul1 restrains Parkin-mediated mitophagy in mature neurons by maintaining ER-mitochondrial contacts. Nat Commun. 2019;10:3645. doi: 10.1038/s41467-019-11636-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rakovic A., Ziegler J., Martensson C.U., Prasuhn J., Shurkewitsch K., Konig P., et al. PINK1-dependent mitophagy is driven by the UPS and can occur independently of LC3 conversion. Cell Death Differ. 2018;26:1428–1441. doi: 10.1038/s41418-018-0219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deas E., Plun-Favreau H., Gandhi S., Desmond H., Kjaer S., Loh S.H., et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin S.M., Lazarou M., Wang C., Kane L.A., Narendra D.P., Youle R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamano K., Youle R.J. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okatsu K., Uno M., Koyano F., Go E., Kimura M., Oka T., et al. A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J Biol Chem. 2013;288:36372–36384. doi: 10.1074/jbc.M113.509653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moskal N., Riccio V., Bashkurov M., Taddese R., Datti A., Lewis P.N., et al. ROCK inhibitors upregulate the neuroprotective Parkin-mediated mitophagy pathway. Nat Commun. 2020;11:88. doi: 10.1038/s41467-019-13781-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L., et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernardini J.P., Brouwer J.M., Tan I.K., Sandow J.J., Huang S., Stafford C.A., et al. Parkin inhibits BAK and BAX apoptotic function by distinct mechanisms during mitophagy. EMBO J. 2019;38 doi: 10.15252/embj.201899916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blommaart E.F., Krause U., Schellens J.P., Vreeling-Sindelarova H., Meijer A.J. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 64.Wu Y.T., Tan H.L., Shui G., Bauvy C., Huang Q., Wenk M.R., et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qu F., Wang P., Zhang K., Shi Y., Li Y., Li C., et al. Manipulation of mitophagy by “all-in-one” nanosensitizer augments sonodynamic glioma therapy. Autophagy. 2019;16:1413–1435. doi: 10.1080/15548627.2019.1687210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeh H.J., Bahary N., Boone B.A., Singhi A.D., Miller-Ocuin J.L., Normolle D.P., et al. A randomized phase II preoperative study of autophagy inhibition with high-dose hydroxychloroquine and gemcitabine/nab-paclitaxel in pancreatic cancer patients. Clin Cancer Res. 2020;26:3126–3134. doi: 10.1158/1078-0432.CCR-19-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan C., Li T.S. Dual role of mitophagy in cancer drug resistance. Anticancer Res. 2018;38:617–621. doi: 10.21873/anticanres.12266. [DOI] [PubMed] [Google Scholar]
- 68.Vasan N., Baselga J., Hyman D.M. A view on drug resistance in cancer. Nature. 2019;575:299–309. doi: 10.1038/s41586-019-1730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gustafsson Å.B., Dorn G.W., 2nd Evolving and expanding the roles of mitophagy as a homeostatic and pathogenic process. Physiol Rev. 2019;99:853–892. doi: 10.1152/physrev.00005.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim G.D., Oh J., Park H.J., Bae K., Lee S.K. Magnolol inhibits angiogenesis by regulating ROS-mediated apoptosis and the PI3K/AKT/mTOR signaling pathway in mES/EB-derived endothelial-like cells. Int J Oncol. 2013;43:600–610. doi: 10.3892/ijo.2013.1959. [DOI] [PubMed] [Google Scholar]
- 72.Zhou Y., Bi Y., Yang C., Yang J., Jiang Y., Meng F., et al. Magnolol induces apoptosis in MCF-7 human breast cancer cells through G2/M phase arrest and caspase-independent pathway. Pharmazie. 2013;68:755–762. [PubMed] [Google Scholar]
- 73.Xie Z., Zhao J., Wang H., Jiang Y., Yang Q., Fu Y., et al. Magnolol alleviates Alzheimer's disease-like pathology in transgenic C. elegans by promoting microglia phagocytosis and the degradation of beta-amyloid through activation of PPAR-γ. Biomed Pharmacother. 2020;124:109886. doi: 10.1016/j.biopha.2020.109886. [DOI] [PubMed] [Google Scholar]
- 74.Huang S.Y., Tai S.H., Chang C.C., Tu Y.F., Chang C.H., Lee E.J. Magnolol protects against ischemic-reperfusion brain damage following oxygen-glucose deprivation and transient focal cerebral ischemia. Int J Mol Med. 2018;41:2252–2262. doi: 10.3892/ijmm.2018.3387. [DOI] [PubMed] [Google Scholar]
- 75.Ho J.H., Hong C.Y. Cardiovascular protection of magnolol: cell-type specificity and dose-related effects. J Biomed Sci. 2012;19:70. doi: 10.1186/1423-0127-19-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bingol B., Tea J.S., Phu L., Reichelt M., Bakalarski C.E., Song Q., et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 77.Georgakopoulos N.D., Wells G., Campanella M. The pharmacological regulation of cellular mitophagy. Nat Chem Biol. 2017;13:136–146. doi: 10.1038/nchembio.2287. [DOI] [PubMed] [Google Scholar]
- 78.Hansen M., Rubinsztein D.C., Walker D.W. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol. 2018;19:579–593. doi: 10.1038/s41580-018-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang E.F., Scheibye-Knudsen M., Brace L.E., Kassahun H., SenGupta T., Nilsen H., et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD+/SIRT1 reduction. Cell. 2014;157:882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fleming A., Rubinsztein D.C. Autophagy in neuronal development and plasticity. Trends Neurosci. 2020;43:767–779. doi: 10.1016/j.tins.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 81.Chao X., Qian H., Wang S., Fulte S., Ding W.X. Autophagy and liver cancer. Clin Mol Hepatol. 2020;26:606–617. doi: 10.3350/cmh.2020.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fang E.F., Hou Y., Palikaras K., Adriaanse B.A., Kerr J.S., Yang B., et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat Neurosci. 2019;22:401–412. doi: 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.