Abstract
Insecticide-based interventions, notably long-lasting insecticidal nets, against mosquito vectors of malaria are currently threatened by pyrethroid resistance. Here, we contrasted RNAseq-based gene expression profiling of laboratory-resistant (FUMOZ) and susceptible (FANG) strains of the major malaria vector Anopheles funestus. Cytochrome P450 genes were the predominant over-expressed detoxification genes in FUMOZ, with high expression of the duplicated CYP6P9a (fold-change of 82.23 vs FANG) and CYP6P9b (FC 11.15). Other over-expressed P450s belonged to the same cluster of P450s corresponding to the resistance to pyrethroid 1 (rp1) quantitative trait loci (QTL) on chromosome 2R. Several Epsilon class glutathione S-transferases were also over-expressed in FUMOZ, as was the ATP-binding cassette transporter AFUN019220 (ABCA) which also exhibited between-strain alternative splicing events at exon 7. Significant differences in single-nucleotide polymorphism frequencies between strains occurred in resistance QTLs rp1 (CYP6P9a/b and CYP6AA1), rp2 on chromosome 2L (CYP6Z1, CYP6M7, and CYP6Z3), and rp3 on chromosome 3R (CYP9J5, CYP9J4, and CYP9J3). Differences were also detected in CYP4G17 and CYP4G16 genes on the X chromosome, both of which are associated with cuticular resistance in Anopheles gambiae. A close analysis of nonsynonymous diversity at the CYP6P9a/b loci revealed a drastic loss of diversity in FUMOZ with only a single polymorphism and 2 haplotypes vs 18 substitutions and 8 haplotypes in FANG. By contrast, a lowly expressed cytochrome P450 (CYP4C36) did not exhibit diversity differences between strains. We also detected the known pyrethroid resistance conferring amino acid change N384S in CYP6P9b. This study further elucidates the molecular bases of resistance in An. funestus, informing strategies to better manage widespread resistance across Africa.
Keywords: malaria, mosquito, Anopheles funestus, pyrethroid resistance, metabolic resistance, RNAseq, cytochrome P450s
Introduction
Malaria prevention still heavily relies on insecticide-based interventions including long-lasting insecticidal nets (LLINs) and indoor residual spraying. These tools have contributed massively to the reduction of malaria burden observed since 2000 (Bhatt et al. 2015). LLINs have mainly relied on pyrethroid insecticides due to their low toxicity to humans. Unfortunately, major malaria vectors have developed resistance to pyrethroids as a response to the massive scale up of LLINs (Hemingway et al. 2016), and also due to ongoing selection from agricultural use of insecticides (WHO 2012). There is increasing evidence that this resistance is jeopardizing the efficacy of LLINs (Mugenzi et al. 2019; Weedall et al. 2019) and is compromising the success of malaria control programs (Protopopoff et al. 2018). It is vital to implement suitable resistance management strategies to prevent the loss of all the gains recently achieved in reducing malaria burdens (Hemingway et al. 2016). Understanding the molecular mechanisms of insecticide resistance in major malaria vectors such as Anopheles funestus, a major malaria vector in sub-Saharan Africa alongside members of the Anopheles gambiae complex is, therefore, of great importance for tracking and managing this resistance.
There are primarily two major mechanisms by which insecticide resistance occurs in mosquitoes, namely target-site and metabolic resistance (Hemingway and Ranson 2000; Hemingway et al. 2004). Target-site resistance occurs when the mosquito target of an insecticide changes in such a manner as to prevent an insecticide from functioning, for example by reduced binding affinity. Pyrethroid class insecticides target the voltage-gated sodium channel (VGSC) of insects and several mutations of VGSC labeled “knock down” or kdr mutations have been identified in several mosquito species (Hemingway and Ranson 2000; Hemingway et al. 2004). Intriguingly, kdr mutations have yet to be seen in An. funestus (Irving and Wondji 2017) but are common in other Anopheles vectors (Riveron et al. 2018a). Metabolic resistance is the rapid destruction or removal of insecticides by resistant vs susceptible insects and is mediated by more efficient and/or higher levels of detoxifying enzymes (Hemingway et al. 2004). This mechanism is predominant in An. funestus resistance to pyrethroids and occurs through the over-expression of cytochrome P450 class genes (Riveron et al. 2018a). Glutathione S-transferases and carboxylesterases can also act to detoxify various insecticides through elevated expression (Riveron et al. 2018a).
In recent years, increasing resistance to multiple insecticides has been reported in An. funestus species with the first reports emerging from southern Africa (Hargreaves et al. 2000; Casimiro et al. 2006; Cuamba et al. 2010; Riveron et al. 2015). Resistance to pyrethroids has been gradually reported in other regions of Africa from including West (Djouaka et al. 2011, 2016; Samb et al. 2016), Central (Menze et al. 2016, 2018; Riveron et al. 2018b), and East (Morgan et al. 2010; Kawada et al. 2011; Mulamba et al. 2014), which has spurred efforts to generate new insecticides with different modes of action to maintain control of pyrethroid-resistant mosquitoes (Hemingway et al. 2016). However, it is vital to avoid potential cross-resistance between forthcoming novel insecticides and pyrethroids. Hence, it is important to use laboratory pyrethroid resistance strains of malaria vectors which have been well characterized for understanding the molecular basis of resistance, especially fully susceptible laboratory-maintained strains as natural populations are no longer susceptible to pyrethroids.
Two laboratory strains of An. funestus have been successfully established by the Vector Control Reference Laboratory at the National Institute for communicable diseases in South Africa. A pyrethroid-resistant An. funestus strain named FUMOZ was established from mosquitoes collected in southern Mozambique in 2000 (Hunt et al. 2005); this strain is also resistant to carbamates. The pyrethroid-susceptible strain FANG was established from An. funestus mosquitoes collected from southern Angola in 2002 and is fully susceptible to all insecticides. Quantitative trait loci (QTL) approaches demonstrated that pyrethroid resistance in FUMOZ was driven by several cytochrome P450 associated loci, among which one locus named resistance to pyrethroids (rp1) locus was predominantly driving resistance (Wondji et al. 2007, 2009). However, the genome-wide transcription profile associated with resistance specific to the FUMOZ lab strain has not been characterized. Such elucidation will help to identify potential drivers of a cross-resistance between pyrethroids and carbamates resistance. It could also help to establish a correlation between QTLs and transcription profile (Wondji et al. 2007, 2009). Furthermore, a full transcriptomic profiling of the FUMOZ-resistant strain will serve as a useful reference when screening novel insecticide candidates. Such a profile will help explain underlying molecular drivers of any observed cross-resistance, and thus help select novel insecticides and combinations thereof that minimize the risk of quick resistance development.
Here, using an RNAseq-based transcription analysis, we characterized the gene expression profiles of two laboratory-adapted colonies of An. funestus: one selected for resistance to multiple insecticides and the other fully susceptible. This study has revealed a strong association between the over-expression of major resistance genes and the presence of signatures of selective sweep as the duplicated P450 genes CYP6P9a and CYP6P9b are massively over-expressed in FUMOZ while exhibiting a drastically reduced diversity compared to the susceptible FANG strain.
Materials and methods
Mosquito rearing, RNA extraction, and sequencing
Two An. funestus laboratory colonies were used in this study. The FANG colony is a fully insecticide susceptible colony derived from Angola (Hunt et al. 2005). The FUMOZ colony is a multi-insecticide resistant colony derived from southern Mozambique, with additional selection for insecticide resistance (Hunt et al. 2005). Colonies were maintained in insectaries at the Liverpool School of Tropical Medicine at 28°C, 80% humidity, and a 12 h light:dark cycle. Larvae were grown in mineral water in plastic trays and fed daily with ground TetraMin fish food. Adults were kept in 30 × 30 cm cages and provided with 10% sucrose. Adults were arm-fed to allow females to lay eggs.
Total RNA was extracted from pools of 10 individual 3-day-old non-blood fed adult female mosquitoes using the Arcturus PicoPure RNA isolation kit (Life Technologies, Carlsbad, CA, USA). In this study, four pools of FUMOZ RNA were sequenced and one pool of FANG RNA, alongside three FANG RNAseq libraries from a previous study (Weedall et al. 2015). Methods used for RNA extraction, ribosomal RNA-depletion, library preparation, and sequencing were described previously (Weedall et al. 2015, 2019). Library preparation and sequencing were undertaken by the Centre for Genomic Research (CGR), University of Liverpool.
Functional annotation improvement of gene set AfunF3.1
Our analysis used the chromosome-scale An. funestus FUMOZ colony reference genome assembly AfunF3 and annotation gene set AfunF3.1 [downloaded from https://www.vectorbase.org/ (last accessed date: 11/10/2021) June 25, 2019 (Giraldo-Calderon et al. 2015)] (Ghurye et al. 2019, preprint). Gene set AfunF3.1 contains many putative genes with no functional descriptions. To aid interpretation of our results, all AfunF3.1 predicted transcripts were used for similarity-based functional annotation assignment using Blast2Go (Conesa et al. 2005) using the nonredundant (nr) protein database downloaded from NCBI. Blast (BLASTx) searches against the nonredundant (nr) protein database and InterProScan searches of the InterPro protein signature databases were carried out to further annotate the An. funestus protein-coding genes.
Analysis of RNAseq data
De-multiplexed fastq files were trimmed to remove Illumina adapter sequences (minimum 3 bp match at the 3′ end) using Cutadapt version 1.2.1 (Martin 2011), and low-quality bases trimmed, using Sickle version 1.200 (Joshi and Fass 2011), with a minimum window quality score of 20. After trimming, reads shorter than 10 bp were removed. If both reads from a pair passed this filter, each was included in a forward or reverse reads file. If only one of a read pair passed this filter, it was place in an unpaired reads file.
Strand NGS software, version 3.4 (Strand Life Sciences, Bangalore, India) was used for data analysis. Trimmed R1/R2 read pairs were aligned to the reference sequence (Afun 3.1) using the option ‘transcriptome and genome together’ (with novel splices) which aligns reads to both transcriptome and genome to find the best matches. Reads that remain unaligned after the first-round are then aligned by considering novel splices across known exons and/or novel candidate exons. A custom ‘build’ was generated for An. funestus genome in Strand NGS following the program’s instructions. The best matches for a read were reported if they satisfied the following criteria: a minimum percent identity of 90, a maximum percent gaps between matches of 5, a maximum number of novel splices of 2, a minimum match length of 25, a maximum number of matches to be reported for each read equal to 1, and ignoring reads with matches more than 5 matches. Further quality trimming was performed by removing 3′ ends with average base quality <10.
Strand NGS was used to perform RNA quantification, which reports raw counts per gene by counting the total number of reads that map to each gene and exon in the An. funestus genome. Raw counts were normalized using DESeq’s inbuilt method (Anders and Huber 2010) which accounts for differences in the total number of reads between samples.
Differential gene expression analysis was carried out using DESeq (Anders and Huber 2010) as implemented in Strand NGS. Two conditions (FUMOZ and FANG, each with four biological replicates) were contrasted. The estimated log2 fold-change (FC) for each gene was tested using a moderated t-test which is a modification of the unpaired t-test (Smyth 2004). P-values were adjusted for multiple testing using the false discovery rate (FDR) approach of Storey with bootstrapping (q-value). Differentially expressed genes were defined as those with an FDR-adjusted P-value <5% and FC ≥2.
Gene ontology (GO) enrichment analysis was carried out on differentially expressed gene sets using Strand NGS. A more detailed analysis was performed for detoxification genes of interest over-expressed in FUMOZ to detect sets of transcripts which are similarly expressed and thus potentially acting together in conferring resistance to FUMOZ. This was performed by selecting the list of transcripts significantly over-expressed in FUMOZ followed by the selection of the transcript of interest and then selecting the similarity metric with a minimum set at 0.5 using Strand NGS. Altogether, this option allows to find transcripts, in a specified list of transcripts (such as those upregulated), whose expression profile matches that of the transcript of interest.
Analysis of alternative splicing
Strand NGS identifies differentially spliced genes based on change in transcript proportions across conditions which here is between FANG (susceptible) and FUMOZ (resistant). Here, a threshold of 0.25 was set to detect genes for which the transcript proportion varies between FUMOZ and FANG in which case the gene is termed differentially spliced. The process of determining transcript proportions takes reads which fall within the boundaries of a gene but outside the exon boundaries of any of the associated transcripts and ascribes these reads to an “unknown” transcript. Based on the fraction of reads that get ascribed to this “unknown” transcript, this “unknown” transcript gets a proportion much like any known transcript does. This proportion is checked for variation across conditions exactly as described above. If of all transcripts for the gene, it is the “unknown” transcript that shows the maximum change across conditions Strand NGS labels this gene as “novel”.
Quantitative reverse transcriptase PCR
RNAseq expression patterns of some of the most over-expressed detoxification genes were validated by quantitative reverse transcriptase PCR (qRT-PCR), including CYP6P9a (AFUN015792), CYP6P9b (AFUN015889), GSTe2 (AFUN015809), CYP6P5 (AFUN015888), CYP325A (AFUN015966), and CYP6AA1 (AFUN015786). Total RNA was extracted from 3 batches of 10 mosquitoes for each strain using the picopure RNA extraction kit, as previously described (Riveron et al. 2013). The primers are listed in Supplementary Table S1. Briefly, 1 µg of total RNA was used for cDNA synthesis using Superscript III (Invitrogen) with oligo-dT20 and RNase H according to the manufacturer’s instructions. The qRT-PCR amplification was performed following standard protocol (Kwiatkowska et al. 2013; Riveron et al. 2013) after establishing the standard curves for each gene to assess PCR efficiency and quantitative differences between samples using serial dilution. The relative expression of each gene was calculated according to the 2−ΔΔCT method (Schmittgen and Livak 2008) and compared between the two strains after normalization with the housekeeping genes ribosomal protein S7 (RSP7; AFUN007153) and actin 5C (AFUN006819).
Detection of single-nucleotide polymorphism
The four FUMOZ and four FANG replicates were combined into a single sample to detect the relevant polymorphisms. All variant types [single-nucleotide polymorphisms (SNPs), multiple nucleotide polymorphisms (MNPs), and indels or MNPs] were detected by comparing against the FUMOZ genome using the MAQ independent model implemented in Strand NGS. Variant calling parameters thresholds were a phred-scaled confidence score cutoff of 50 or above. This score represents the confidence in the variant call. Other criteria include a coverage greater than 10, ignoring homopolymer of 10, or higher as well as the locations adjacent to them. A filtering step was performed as implemented in Strand NGS through the “HomPolyfilter” flag which removes indels calls with <25% supporting reads in homopolymer stretches and “IndelSR20” which removes indels with <20% of supporting reads. An SNP multi-sample report was generated which included the allele frequency in the population based on the number of supporting reads for each variant, the zygosity, genotype, score, and other attributes such as % of supporting reads and variants. For each variant, its SNP effect (nonsynonymous, synonymous, 5′ UTR, 3′ UTR etc.) and the affected transcripts were predicted using the gff3 transcript annotation (Afun3.1).
Detection of significant SNPs
To detect SNPs significantly associated with pyrethroid resistance, we took two approaches. Firstly, we used a stringent differential allele frequency-based approach where a variant was considered significant in relation to pyrethroid resistance if the supporting read % range was 0–10% in all four FUMOZ samples, but 90–100% in all the four FANG samples. This is because the AFUN3.1 genome was generated from FUMOZ strain mosquitoes meaning resistance-conferring alleles are present in the reference sequence.
The second approach assessed the significant association between each variant with permethrin resistance by estimating the unpaired t-test of each variant between both and the –Log10 of the resulting P-value used to generate a Manhattan plot for each chromosome. A cutoff of minimum of two samples for plotted SNPs was applied. Furthermore, analysis of the 2R chromosome region spanning the rp1 QTL was comparatively performed between both strains to detect signature of selection in the resistant FUMOZ strain. Briefly, the percentage of supporting reads at each variant was averaged between the four samples of each strain and plotted across the 120 kb region.
Polymorphism of candidate resistance genes CYP6P9a and CYP6P9b
Particular attention was given to the genes CYP6P9a and CYP6P9b which result from a fixed duplication of CYP6P9 in An. funestus compared to other Anopheles malaria vector species (Wondji et al. 2009). Polymorphisms in these genes were comparatively assessed between the two strains by focusing primarily on nonsynonymous SNPs. Each replicate was arbitrarily considered as an individual to facilitate the analysis. Data from the SNP multi-sample report were used to retrieve the polymorphisms observed in each sample and this information was transferred to the full length of each gene retrieved from Vectorbase. Bioedit was used to input various polymorphisms using an ambiguous letter to indicate heterozygote positions. Haplotype reconstruction and polymorphism analysis were performed in DnaSPv5.10 (Librado and Rozas 2009), and MEGA X (Kumar et al. 2018) was used to construct the maximum likelihood phylogenetic tree for each gene.
Results
Differentially expressed genes
A range of 43–65 million reads were obtained for each of the eight samples sequenced and successfully aligned to the An. funestus genome Afun 3.1 as illustrated by the quality metrics (Supplementary Table S2).
Genes over-expressed in FUMOZ
Seven hundred seventeen transcripts were over-expressed in FUMOZ and 1008 downregulated compared to FANG (Figure 1A). Analysis of the list of transcripts upregulated in FUMOZ revealed that among the most over-expressed with FC greater than 10 or more, the transcript with the highest FC at 83 is the AFUN019794 which is an Integrase catalytic domain-containing protein. This list of highly over-expressed genes in FUMOZ also contains the duplicated cytochrome P450s CYP6P9a (FC 82.23) and CYP6P9b (FC 11.15). Other genes belong to serine proteases and others are transposon-related genes (Supplementary Table S3).
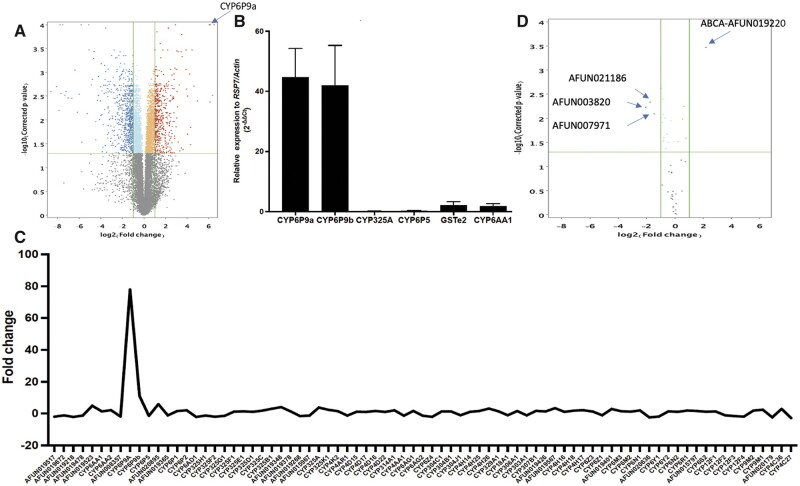
Figure 1.
Comparative transcription profiles of FUMOZ and FANG strains: (A) Volcano plot of differential gene expression between pyrethroid-resistant FUMOZ and susceptible FANG. (B) Gene expression pattern of all cytochrome P450 genes on chromosome 2 highlighting the massive expression of CYP6P9a/b in FUMOZ. (C) qRT-PCR comparison of the expression profile of major detoxification genes associated with pyrethroid resistance between FUMOZ and FANG. The data are shown as mean + SEM (n = 3). (D) Volcano plot showing the differential expression of alternative spliced genes highlighting the ABCA AFUN019220 as the single over-expressed and spliced gene in FUMOZ.
Overall, the list of upregulated genes is dominated by genes belonging to several gene families associated with insecticide resistance including detoxification genes such as cytochrome P450s, glutathione-S-transferases (GSTs), and carboxylesterases (Table 1; Supplementary Figure S1A). Among P450s, the duplicated CYP6P9a and CYP6P9b are by far the most over-expressed. The predominance of CYP6P9a/b is further evident by the fact that no other P450 gene has FC >6 and only three other P450s present expression of >3 notably CYP6P4a (FC 5.9), CYP325J1 (FC 4.9), and CYP4C36. The other P450s have low expression with FC <3 including CYP6M4, CYP6AA2, and CYP6P2 also on the 2R chromosome on the same cluster as CYP6P9a/b.
Table 1.
List of most upregulated detoxification genes in FUMOZ strain
| Gene ID | P | FC | Log FC | Gene symbol | FUMOZ reads | FANG reads | Description |
|---|---|---|---|---|---|---|---|
| Cytochrome P450 | |||||||
| AFUN015792 | 0.0005 | 78.0 | 6.3 | CYP6P9a | 84,176.5 | 822.6 | Cytochrome P450 |
| AFUN015889 | 0.0015 | 11.1 | 3.5 | CYP6P9b | 11,906.6 | 744.2 | Cytochrome P450 |
| AFUN020895 | 0.0065 | 5.9 | 2.6 | CYP6P4a | 1,895.4 | 255.5 | Cytochrome P450 |
| AFUN019523 | 0.0466 | 4.9 | 2.3 | CYP325J1 | 19.6 | 3.4 | Cytochrome P450 |
| AFUN006135 | 0.0049 | 3.0 | 1.6 | CYP4C36 | 987.1 | 261.9 | Cytochrome P450 |
| AFUN019401 | 0.0407 | 2.9 | 1.5 | CYP6M4 | 2,115.5 | 731.1 | Cytochrome P450 |
| AFUN021097 | 0.0491 | 2.5 | 1.3 | CYP4H24 | 2,613.8 | 1,057.2 | Cytochrome P450 |
| AFUN015938 | 0.0042 | 2.5 | 1.3 | CYP9M1 | 1,962.9 | 585.2 | Cytochrome P450 |
| AFUN007549 | 0.0018 | 2.4 | 1.3 | CYP9K1 | 3,991.8 | 1,049.2 | Cytochrome P450 |
| AFUN015785 | 0.0076 | 2.1 | 1.1 | CYP6AA2 | 776.2 | 254.4 | Cytochrome P450 |
| AFUN015801 | 0.0015 | 2.1 | 1.1 | CYP6P2 | 2,106.3 | 718.8 | Cytochrome P450 |
| AFUN002978 | 0.0073 | 2.1 | 1.0 | CYP314A1 | 642.4 | 219.3 | Cytochrome P450 |
| Glutathione S-transferases (GSTs) | |||||||
| AFUN022201 | 0.0081 | 11.8 | 3.6 | AFUN022201 | 54.3 | 5.0 | Glutathione peroxidase |
| AFUN016008 | 0.0072 | 4.0 | 2.0 | GSTE6 | 605.7 | 125.5 | Glutathione S-transferase |
| AFUN015808 | 0.0038 | 3.6 | 1.8 | GSTE3 | 3,351.1 | 772.5 | Glutathione S-transferase |
| AFUN015809 | 0.0013 | 3.3 | 1.7 | GSTE2 | 2,415.9 | 491.8 | Glutathione S-transferase |
| AFUN015839 | 0.0034 | 3.1 | 1.6 | GSTD3 | 1,298.3 | 332.6 | Glutathione S-transferase |
| AFUN015811 | 0.0032 | 2.5 | 1.3 | GSTE5 | 1,185.9 | 332.4 | Glutathione S-transferase |
| AFUN016010 | 0.0031 | 2.4 | 1.3 | GSTD1 | 13,199.4 | 4,246.1 | Glutathione S-transferase |
| AFUN007291 | 0.0217 | 2.4 | 1.2 | GSTT2 | 660.4 | 238.0 | Glutathione S-transferase |
| Carboxylesterases | |||||||
| AFUN016265 | 0.0177 | 7.2 | 2.9 | AFUN016265 | 387.8 | 51.8 | Carboxylesterase |
| AFUN016264 | 0.0119 | 2.4 | 1.3 | AFUN016264 | 240.6 | 73.6 | Carboxylesterase |
| AFUN000374 | 0.0037 | 2.2 | 1.2 | AFUN000374 | 2,557.4 | 846.9 | Carboxylesterase |
| Other detox genes | |||||||
| AFUN004354 | 0.0103 | 2.1 | 1.1 | AFUN004354 | 1,005.3 | 317.5 | UDP-glucuronosyltransferase |
| AFUN020670 | 0.0041 | 2.4 | 1.3 | AFUN020670 | 995.7 | 247.5 | Xanthine dehydrogenase |
| AFUN008239 | 0.0149 | 2.7 | 1.5 | AFUN008239 | 830.9 | 225.8 | Sulfotransferase |
| AFUN016207 | 0.0047 | 4.6 | 2.2 | AFUN016207 | 699.0 | 162.8 | Sulfotransferase |
| AFUN016205 | 0.0246 | 2.8 | 1.5 | AFUN016205 | 1,290.1 | 398.8 | Sulfotransferase |
| AFUN016210 | 0.0054 | 2.4 | 1.3 | AFUN016210 | 297.2 | 91.2 | Sulfotransferase |
| Transporters | |||||||
| AFUN019220 | 0.0020 | 4.7 | 2.2 | AFUN019220 | 4,442.5 | 676.7 | ATP-binding cassette transporter |
| AFUN016412 | 0.0105 | 2.5 | 1.3 | AFUN016412 | 5,425.0 | 1,795.5 | Solute carrier family 15 member |
| AFUN000622 | 0.0099 | 3.8 | 1.9 | AFUN000622 | 1,350.9 | 235.3 | Solute carrier family 23 ( |
| AFUN015826 | 0.0013 | 2.1 | 1.0 | AFUN015826 | 1,479.5 | 496.5 | Solute carrier family 7 |
| AFUN021547 | 0.0087 | 2.8 | 1.5 | AFUN021547 | 513.9 | 144.3 | Sugar transporter SWEET |
| AFUN021546 | 0.0200 | 3.1 | 1.6 | AFUN021546 | 1,490.9 | 362.5 | Sugar transporter SWEET1 |
| Mitochondrial/redox | |||||||
| AFUN010502 | 0.0007 | 8.2 | 3.0 | AFUN010502 | 4,959.9 | 534.9 | ATP synthase subunit d, mitochondrial |
| AFUN011500 | 0.0013 | 2.7 | 1.4 | AFUN011500 | 2,297.9 | 686.9 | Sulfite oxidase |
| AFUN000619 | 0.0025 | 2.4 | 1.3 | GRX2 | 768.9 | 303.6 | Glutaredoxin |
| AFUN011515 | 0.0063 | 2.0 | 1.0 | TPX4 | 3,567.4 | 1,179.5 | Thioredoxin peroxidase 4 |
| AFUN019604 | 0.0042 | 2.7 | 1.4 | AFUN019604 | 30.6 | 10.7 | Glutaredoxin-3 |
| AFUN001952 | 0.0049 | 4.0 | 2.0 | AFUN001952 | 1,159.8 | 267.4 | Uncoupling protein 2, mitochondrial |
| Proteases and others | |||||||
| AFUN016100 | 0.0059 | 2.1 | 1.1 | AFUN016100 | 2,012.7 | 648.1 | Alpha-amylase |
| AFUN019614 | 0.0039 | 3.0 | 1.6 | AFUN019614 | 1,413.7 | 323.6 | Ankyrin repeat |
| AFUN004002 | 0.0021 | 2.7 | 1.4 | AFUN004002 | 17,374.3 | 4,616.8 | Argininosuccinate lyase |
| AFUN019821 | 0.0126 | 2.0 | 1.0 | AFUN019821 | 905.3 | 334.1 | Carboxypeptidase A |
| AFUN021240 | 0.0013 | 17.8 | 4.2 | AFUN021240 | 1,050.2 | 46.2 | Carboxypeptidase Q |
| AFUN021239 | 0.0079 | 11.5 | 3.5 | AFUN021239 | 807.5 | 75.4 | Carboxypeptidase Q |
| AFUN022011 | 0.0200 | 3.2 | 1.7 | AFUN022011 | 50,117.5 | 15,637.6 | Chymotrypsin |
| AFUN007012 | 0.0442 | 5.0 | 2.3 | AFUN007012 | 12.3 | 4.8 | Chymotrypsin-like protease |
| AFUN016591 | 0.0043 | 7.1 | 2.8 | AFUN016591 | 24,258.5 | 2,746.8 | Trypsin |
| AFUN016449 | 0.0222 | 2.3 | 1.2 | AFUN016449 | 3,458.1 | 1,127.3 | Trypsin |
| AFUN020614 | 0.0052 | 3.7 | 1.9 | AFUN020614 | 3,655.9 | 848.5 | Trypsin-6 |
| AFUN022010 | 0.0131 | 4.3 | 2.1 | AFUN022010 | 101,129.8 | 20,343.8 | Serine protease; Trypsin-1 |
| AFUN020946 | 0.0192 | 23.3 | 4.5 | AFUN020946 | 153.9 | 10.8 | Serine-type endopeptidase -Chymotrypsin-2 |
| Cuticular proteins | |||||||
| AFUN011620 | 0.0154 | 7.8 | 3.0 | AFUN011620 | 92.4 | 11.7 | Cuticle protein |
| AFUN009937 | 0.0101 | 3.9 | 2.0 | AFUN009937 | 5.0 | 1.4 | Cuticular protein RR-1 family |
| AFUN006418 | 0.0235 | 2.8 | 1.5 | AFUN006418 | 282.1 | 72.0 | Cuticular protein RR-2 family |
| AFUN020923 | 0.0259 | 15.7 | 4.0 | AFUN020923 | 57.6 | 3.1 | Cuticular protein RR-2 family |
| AFUN021592 | 0.0155 | 2.4 | 1.3 | AFUN021592 | 194.0 | 54.1 | Larval cuticle protein LCP-30 |
| Gustatory/odorant receptors | |||||||
| AFUN015936 | 0.0107 | 5.5 | 2.5 | Gr20 | 731.4 | 124.2 | Gustatory receptor |
| AFUN015933 | 0.0092 | 2.7 | 1.4 | Gr45 | 24.4 | 7.7 | Gustatory receptor |
| AFUN008090 | 0.0089 | 2.7 | 1.4 | Or36 | 19.9 | 8.1 | Odorant receptor |
| AFUN008522 | 0.0276 | 2.2 | 1.2 | Or39 | 63.3 | 22.0 | Odorant receptor |
| AFUN018655 | 0.0028 | 8.2 | 3.0 | AFUN018655 | 13.0 | 0.4 | Odorant receptor |
| MicroRNA | |||||||
| AFUN017296 | 0.0002 | 4.3 | 2.1 | mir-279 | 4.9 | 0.8 | MicroRNA mir-279 |
| AFUN017073 | 0.0403 | 2.9 | 1.5 | mir-33 | 4.5 | 1.1 | MicroRNA mir-33 |
| Transposon | |||||||
| AFUN019794 | 0.0003 | 83.2 | 6.4 | AFUN019794 | 67,882.0 | 520.4 | BEL12_AG transposon polyprotein |
| AFUN019813 | 0.0053 | 22.9 | 4.5 | AFUN019813 | 4,612.4 | 121.4 | Retrovirus-related Pol polyprotein from transposon TNT 1-94 |
| Transcription factors | |||||||
| AFUN002438 | 0.0039 | 4.2 | 2.1 | AFUN002438 | 446.7 | 102.3 | Transcription elongation factor 1 homolog |
| AFUN019110 | 0.0018 | 2.9 | 1.5 | AFUN019110 | 631.4 | 183.8 | Transcription factor Adf-1 |
| AFUN021815 | 0.0042 | 2.2 | 1.1 | AFUN021815 | 422.6 | 152.5 | Transcription factor grauzone |
| AFUN021263 | 0.0038 | 3.6 | 1.9 | AFUN021263 | 425.8 | 93.9 | Transcription initiation factor TFIID subunit |
| AFUN011301 | 0.0040 | 2.6 | 1.4 | AFUN011301 | 1,434.7 | 388.1 | Transcription initiation factor TFIID subunit 12 |
| AFUN011301 | 0.0040 | 2.6 | 1.4 | AFUN011301 | 1,434.7 | 388.1 | Transcription initiation factor TFIID subunit 12 |
| AFUN021364 | 0.0370 | 2.1 | 1.1 | AFUN021364 | 408.3 | 155.2 | Transcription initiation factor TFIIH subunit |
Description, gene annotation; FC, fold-change; FUMOZ/FANG reads, total read pairs across replicates; Log FC, log2 fold-change; P, FDR-adjusted P-value.
Overexpressed GSTs include four genes from the epsilon class, all with moderate FCs notably GSTe2 (FC 3.3), GSTe3 (FC 3.6), GSTe5 (FC 2.5), and GSTe6 (FC 4.0). Other GSTs include GSTD3 (FC 3.1), GSTD1 (FC 2.4), and GSTT2 (FC 2.4). We also noted that a glutathione peroxidase also exhibits a high FC (FC 11.8) in FUMOZ although the overall number of reads was low (Table 1; Supplementary Figure S1B).
Three carboxylesterases are also over-expressed with AFUN016265 showing an FC of 7.2, an ortholog of AGAP001722 in An. gambiae, whereas the other two have FC <3 (AFUN16264 and AFUN000374, ortholog of AGAP006729 in An. gambiae).
Other detoxification genes over-expressed also have low FCs including an UDP-glucuronosyltransferase (AFUN004354; FC 2.1), a xanthine dehydrogenase (AFUN020670; FC 2.4), and four sulfotransferases (Table 1; Supplementary Figure S1C).
A group of transport-related genes are also over-expressed in FUMOZ among which the ABC transporter AFUN019220 (FC 4.7) and three solute carrier proteins and two sugar transporters with moderate FC (Table 1). Another set of genes over-expressed belong to mitochondrial redox group along with the ATP synthase subunit d (AFUN010502) is the most highly expressed (FC 8.2) as well as two glutaredoxins and thioredoxin peroxidase 4. A group of genes belonging to proteases are also over-expressed including trypsin, chymotrypsin, two carboxypeptidases Q [AFUN021240 (FC 17.8) and AFUN021239 (FC 11.5)], and arginosuccinate lyase (FC 2.7). Other genes families over-expressed include cuticular proteins, gustatory/odorant receptors genes, two microRNAs although with low number of reads (mir-279 and mir-33), and several transcription factors.
Genes downregulated in FUMOZ
Of the 1008 genes significantly downregulated in FUMOZ compared to FANG (Figure 1; Supplementary Table S4), the most downregulated gene is the Group XIIB secretory phospholipase A2-like protein with FC of −602.4. However, this list is dominated by several transcripts belonging to heat shock proteins including heat shock protein 70 A1 (AFUN019775), 70B2 (AFUN019570, AFUN019671, AFUN019513, AFUN019467, and AFUN019289). It also includes Thioredoxin peroxidase 4 (AFUN018693; FC 202.4) which is known to be required for the transcriptional responses to oxidative stress (Ross et al. 2000). Some detoxification genes were also downregulated in FUMOZ including cytochrome P450, GSTs, and carboxylesterases although none of these have been previously associated with resistance to insecticides (Supplementary Table S4).
qRT-PCR
The qRT-PCR performed with some candidate genes was concordant with the results observed with RNAseq with high expression of both CYP6P9a (FC 44.7) and CYP6P9b (FC 41.9), whereas the other genes had far lower expression level. These includes GSTe2, CYP6P5, CYP325A, and CYP6AA1 (Figure 1B).
Gene ontology enrichment
The GO enrichment analysis of the list of 717 transcripts over-expressed in FUMOZ detected 28 enriched GO terms (Supplementary Figure S2A). There was a predominance of terms associated with peptidase activity including GO:0008233 for peptidase activity known to catalyze the hydrolysis of a peptide bond, GO:0004175 for endopeptidase activity known to catalyze the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain, or GO:0008237 for metallopeptidase activity. Another group of GO terms was related to transport including GO:0005215 for transporter activity, GO:0055085 for transmembrane activity, or GO:0030001 for metal ion transport which is known to direct the movement of metal ions into or out of a cell or even between cells (Supplementary Figure S2A). Other GO terms are associated with mitochondrial activity including GO:0005743 for mitochondrial inner membrane and GO:0019866 for organelle inner membrane. The other category of GO relates to hydrolase activity notably GO:0016787 known to catalyze the hydrolysis of various bonds and GO:0017171 for serine hydrolase activity.
Analysis of the GO enrichment from the list of downregulated transcripts provided a profile like that of upregulated genes (Supplementary Figure S2B) with similar groups of GO terms observed although with different % of enrichment.
General expression patterns of major detoxification families using all entities
The entire list of detoxification genes was analyzed in detail to identify clusters of genes co-located on chromosomal regions of the genome. Analysis of the expression patterns of the entire P450 list revealed a peak of expression around CYP6P9a/b on 2R chromosome which is very similar in profile to the QTL peak previously detected in FUMOZ for the rp1 (resistance to permethrin-1) locus (Wondji et al. 2007, 2009) (Figure 1C). No other major peak was detected for P450s either on chromosome 3 or X (Supplementary Figure S3, A and B). No major peak was also observed for GSTs (Supplementary Figure S3C), ABC transporters (Supplementary Figure S3D), and carboxylesterases (Supplementary Figure S3E) with all transcripts exhibiting FC <5.
Alternative splicing
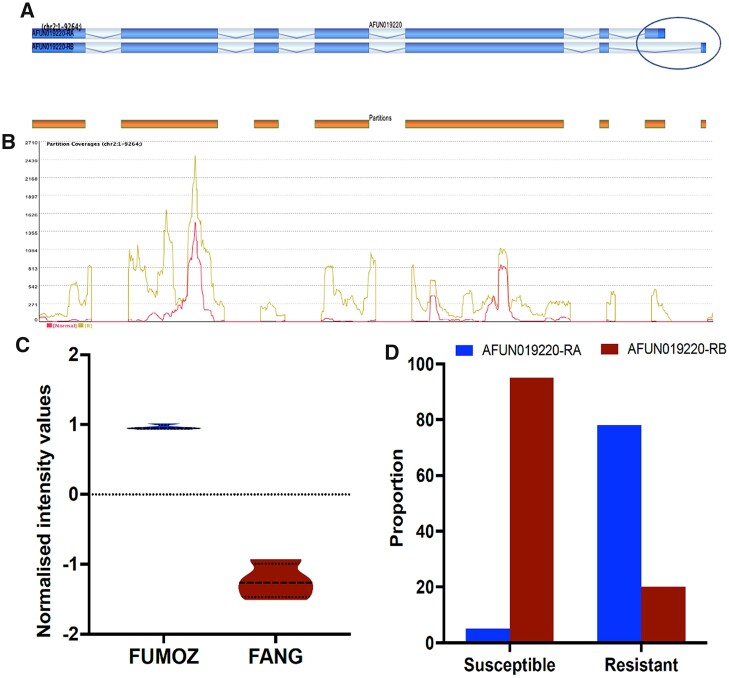
Strand NGS identified 27 genes exhibiting differentially alternative spliced events between FUMOZ and FANG (Figure 1D;Supplementary Table S5). The gene with highest index of alternative splicing was AFUN021186 encoding for a DEAD-box ATP-dependent RNA helicase 30 on chromosome 3 which is over-expressed in the susceptible FANG strain [splicing index (SI) = 0.77]. Among the genes upregulated in FUMOZ, the ABC transporter AFUN019220 had the highest splicing index (0.75). AFUN019220 has 7 exons and two isoforms (see https://vectorbase.org/vectorbase/app/record/gene/AFUN019220; last accessed date: 11/10/2021) with exon 7 exhibiting alternative splicing in this experiment (Figure 2A). Analysis of the partition coverage across the 7 exons supports this difference at exon 7 between the two strains (Figure 2B). Gene-wide expression of this ABC gene is greater in the resistant FUMOZ strain (Violin plot, Figure 2C). A greater proportion of the isoform AFUN019220-RA (78.1%) is observed in the FUMOZ-resistant strain whereas a greater proportion of the other transcript AFUN019220-RB is seen in the susceptible strain (94.6%) (Figure 2D). The other detoxification gene of this list was the carboxylesterase AFUN000373 (SI = 0.33), whereas other genes do not have clear association with resistance mechanisms.
Figure 2.
Alternative splicing event at the over-expressed ABC transporter ABCA (AFUN019220). (A) Schematic representation of the transcript organization of this gene with 7 exons for each transcript with last exon 7 having an alternative splice version in AFUN019220-RA and AFUN019220-RB. (B) Comparative read coverage for each exon in each strain. (C) Violin plot showing the greater expression of AFUN019220 in FUMOZ (R) than FANG (Normal). (D) Differential expression of spliced transcripts AFUN019220-RA and -RB in FUMOZ and FANG.
Patterns of co-expression for major resistance genes
Analysis of the list of transcripts with similar expression to CYP6P9a detected 17 transcripts (Supplementary Table S6) with an expression similarity score >0.6 among which the only other detoxification gene was CYP6P9b P450 gene (score = 0.63). This could be explained by the very high expression of CYP6P9a which thus stands as an outlier compared to other detoxification genes.
CYP6P9b in contrast to CYP6P9a had similar expression patterns to more genes (Supplementary Table S6). Among detoxification genes, the most similar gene being ABCA (AFUN019220) (SI = 0.77) followed by CYP6P4a P450. Other detox genes included the GSTs GSTe2 and GSTe6 and a carboxylesterase AFUN016265 (Supplementary Table S6). The list of entities with expression similar to GSTe2 was longer with the closest entity being a xanthine dehydrogenase (SI = 0.91) including six GSTs (with three GST epsilon and three GST Delta class), five P450s (CYP4C36, CYP9K1, CYP6N1, CYP6P2, and CYP9M1), three sulfotransferases, and a carboxylesterase (AFUN016264). The list of entity with expression similar to ABCA (AFUN019220) had GSTe2 as the closest (SI = 0.89) with six other GSTs found. It also has five P450 with CYP4C36 been the closest (Supplementary Table S6).
SNPs detection: differential significant SNPs in general and for key detoxification gene families
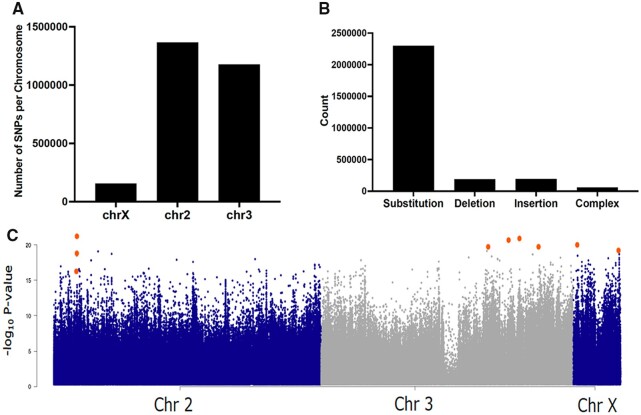
A total of 2,793,727 SNPs were detected between FUMOZ and FANG after comparison to the reference AFUN3.1 genome (from FUMOZ strain) and distributed across the three chromosomes from various variant types (Figure 3, A and B). The frequency-based filtering approach, considering only variant with supporting reads percentage between 0–10% for FUMOZ (close to reference genome) and 90–100% for FANG, detected 17,976 variants significantly different between the two strains across 3,599 genes. Of these, 1,596 were nonsynonymous variants and 6,354 synonymous and the remainder occur in 5′ UTR, 3′ UTR, and intron and intergenic regions. When a more stringent selection criteria of consistency in all four samples of each strain was applied, only 2,217 variants were detected including 254 nonsynonymous and 821 synonymous substitutions. A summary of differential SNPs predicted in detoxification genes is presented in Supplementary Table S7. Overall, analysis of SNPs located in detoxification genes highlighted a cluster of SNPs around the genomic region spanning the rp1 QTL on 2R chromosome belonging to highly over-expressed CYP450s, notably CYP6P9a and CYP6P9b (Supplementary Table S7). Other detoxification genes with significant SNPs belonging to cytochrome P450s located in rp1 included CYP6AA1, CYP6AA2, CYP6P4a, CYP6P1, and CYP6P2 (Supplementary Table S8). A second cluster of SNPs is associated with rp2 QTL on chromosome 2L including CYP6Z1, CYP6M7, CYP6Z3 all previously shown to be conferring pyrethroid resistance (Irving et al. 2012; Ibrahim et al. 2016). Another cluster of SNPs belong to P450s located on 3R chromosome spanning the previously described rp3 QTL including CYP9J5 (previously CYP9J11) shown to metabolize pyrethroids (Riveron et al. 2017) and CYP9J4 and CYP9J3. Notably, significant SNPs were also detected on chromosome X in CYP4G17 and CYP4G16. Interestingly, significant SNPs also belong to several ABC transporters including the alternatively spliced ABCA (AFUN019220). When looking more specifically at those significant nonsynonymous SNPs in the P450 genes, it was observed that they include a known asparagine to serine change at amino acid 384 (N384S) from CYP6P9b (Supplementary Table S8).
Figure 3.
Detection of pyrethroid associated SNPs. (A) Distribution of all detected variants across the 3 chromosomes. (B) Distribution of the variant types detected between strains. (C) Manhattan plot of significant variants associated between FUMOZ and FANG and possibly associated with pyrethroid resistance using an unpaired t-test between RNAseq samples of both strains. Large orange circles represent SNPs of interest as discussed in the text and listed in Supplementary Table S9. Figure was created in qqman R package using T-test P-values as input. Chr 2 = Chromsome 2 (blue), Chr 3 = Chromsome 3 (grey), Chr X = Chromosome X (blue).
The second approach used to detect significant variants using an unpaired t-test using percentage of supporting reads, identified sets of SNPs statistically with contrasted between both strains. Several hits were detected spanning the three chromosomes (Figure 3C). The genomic region with the highest P-value was in rp1 region on 2R chromosome. A detailed analysis showed that within this region the most significant SNPs corresponded to CYP6P9a gene (P = 4.53 × 10−22) with two other significant SNPs located in the same gene (Supplementary Table S9; Figure 3). Other highly significant variants were detected on chromosome 3 all in 3′ UTR regions of genes such as cytochrome b5 reductase (AFUN007146), CLIP-domain serine protease (AFUN022249), and an Ecdysone-induced protein 75B (AFUN020438) (Figure 3C). However, none of these genes were differentially expressed between the two strains. Two other SNPs were detected in chromosome X located in 5′ UTR of a cytochrome c oxidase (AFUN019973) and the other with no annotation (Supplementary Table S9; Figure 3C).
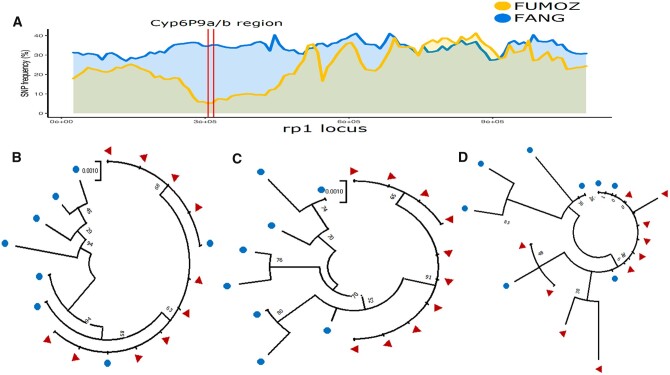
The reduced diversity observed around rp1 and the consistency of significant SNPs detected in CYP6P9a and CYP6P9b led us to further analyze the diversity of both genes (Figure 4A). Another P450 gene CYP4C36, upregulated in FUMOZ but at lower FC (3.0), was also analyzed in comparison to the CYP6P9a/b. Analysis of nonsynonymous substitutions of CYP6P9a revealed a drastic loss of diversity in FUMOZ with only a single polymorphism and two haplotypes vs 18 substitutions and eight haplotypes in FANG which was also reflected with lower genetic diversity π in FUMOZ (0.53) than in FANG (7) (Figure 4B; Supplementary Table S10). A similar pattern was also observed for CYP6P9b (Figure 4C; Supplementary Table S10). By contrast, CYP4C36 did not show any difference between strains with similar estimates of genetic diversity for both and we therefore inferred that no selection at this locus (Figure 4D; Supplementary Table S10). Phylogenetic analysis of the three genes further supported this observation with FUMOZ samples clustering together alone for both CYP6P9a/b on just two haplotypes, whereas FANG is more diverse (Figure 4). Whereas for CYP4C36, samples from both strains are intermingled.
Figure 4.
Polymorphism patterns on the genomic regions spanning the rp1 QTL regions including CYP6P9a and b. (A) Comparative SNP frequency along the 2R chromosome region spanning rp1 showing a marked reduced diversity in FUMOZ notably at the vicinity of the highly over-expressed CYP6P9a and CYP6P9b genes. (B) Maximum likelihood phylogenetic tree of CYP6P9a sequences generated from RNAseq data between FUMOZ and FANG. (C) Is for CYP6P9b and (D) is for CYP4C36.
Discussion
Elucidating mechanisms of resistance to insecticides in malaria vectors is a prerequisite for a successful implementation of resistance management strategies. Here, we took advantage of two phenotypically contrasted laboratory colonies of the major malaria vector An. funestus to further decipher the underlying molecular basis of resistance to pyrethroids using RNAseq-based transcription analysis.
Transcription pattern matches QTL mapping
The transcription profile of both lab strains correlates strongly to previous results obtained through QTL mapping using advanced inter-crossed lines from reciprocal crosses between FUMOZ and FANG (Wondji et al. 2007, 2009; Irving et al. 2012). Indeed, the most highly overexpressed detoxification gene from this analysis are the duplicated CYP6P9a and CYP6P9b cytochrome P450s with FCs far greater than other overexpressed genes, previously shown to drive resistance in this strain (Amenya et al. 2008; Wondji et al. 2009). This is in line with QTL mapping which detected three QTLs associated with pyrethroid resistance with the major one rp1, harboring CYP6P9a and b, explaining 86% of genetic variance of pyrethroid resistance (Wondji et al. 2009). This observation suggests that in a species such as An. funestus in which resistance to insecticides is mainly driven by metabolic resistance, combining RNAseq-based transcription analysis to QTL mapping provides a robust methodology for elucidating the molecular basis of resistance. Furthermore, the over-expression of other P450 genes on chromosome 2L and 3R correspond to two other QTL, rp2 and rp3 which were shown to be minor contributors well in line with the lower expression of these genes. The predominant role of CYP6P9a/b in FUMOZ also correlates with the high expression of these genes in wild populations across southern Africa (Riveron et al. 2013; Barnes et al. 2017a; Weedall et al. 2019). This suggests that the colonization of this strain in the laboratory has not significantly altered its genetic background in relation to resistance.
Interestingly, in all three QTL regions, over-expression extends to multiple genes in the gene cluster including in rp1 where CYP6P4a, CYP6AA2, and CYP6P2 were also overexpressed. Such a pattern suggests a similar mechanism of transcriptional regulation applies to the cluster, rather than to an individual gene. While cis-regulation of transcription of individual genes likely plays a role (e.g., CYP6P9a and b in FUMOZ are much more over-expressed than are other genes in the cluster), other mechanisms such as chromatin remodeling (Clapier et al. 2017) around the gene cluster could allow transcriptional co-regulation of all of the genes. As could differential micro-RNA control of gene expression (Feng et al. 2018), which we observed for mir-33 and mir-279. Among upregulated P450s, those belonging to the CYP6 family were predominant as reported previously for insecticide-resistant mosquitoes (Djouaka et al. 2008; Riveron et al. 2018a), although not for Aedes aegypti where CYP9 genes are more abundantly over-expressed (Bariami et al. 2012; Ishak et al. 2017).
GST genes were also over-expressed, including GSTe2 and other epsilon class GSTs previously shown to metabolize permethrin, although the 119F resistant allele detected in West/central African populations is absent in FUMOZ (Riveron et al. 2013; 2017). Other members of this epsilon class such as GSTe4 and GSTe5 have been shown to confer pyrethroid resistance either in An. funestus (Kouamo et al. 2021) and An. gambiae (Wilding et al. 2015). Altogether, this evidence provides further support a key role for GST enzyme family in pyrethroid resistance.
Besides cytochrome P450 monooxygenases and glutathione S-transferases, a number of genes from other detoxification-associated gene families were also overexpressed in FUMOZ as is often the cases for metabolic resistance (Puinean et al. 2010; Bariami et al. 2012; Riveron et al. 2013). These genes belonging to carboxylestases, sulfotransferases, UDP-glycosyltransferases, and ATP-binding cassette transporter warrant further study to assess their roles in metabolic insecticide resistance which has lagged behind cytochrome P450s. Both GO enrichment and detection of genes showing similar expression patterns indicate that these other genes probably act together with the primary genes as part of the same metabolic detoxification pathway.
The upregulation of transposon-related genes in FUMOZ including the AFUN019794, (BEL12_AG transposon polyprotein; FC 83.2), is another feature of the differential expression between the two strains. We have previously reported that insertion of transposable elements is associated with the upregulation of detoxification genes in An. funestus (Weedall et al. 2020). Similar observations have been made in Drosophila melanogaster with the insertion the Accord transposable element into the 5′ end of the CYP6G1 gene which drives its over-expression (Daborn et al. 2002). Transposable element insertions are commonly associated with disruption of gene function (Bourque et al. 2018), but in the case of An. funestus, they probably facilitate resistance by introducing additional cis-regulatory elements. Differential suppression of transposable elements could control the frequency at which insertions occur (Bourque et al. 2018). In most contexts, weaker suppression would be deleterious, and we speculate here for future exploration, that such an extreme strategy could reflect the strength of selection imposed by insecticides on mosquitoes.
Interestingly, a case of alternative splicing in the ABCA gene (AFUN019220) was strongly contrasted between strains suggesting a possible association with resistance as this gene has been shown to be over-expressed in pyrethroid-resistant populations Africa-wide (Weedall et al. 2019). Although this difference in isoform preference may also reflect the different geographic origins of the two strains, hence, further evidence is required to link this observation to insecticide resistance. A similar case of alternative splicing driving resistance to insecticide was reported in the tomato leaf miner, Tuta absoluta and shown to confer resistance to Spinosad insecticide through an exon skipping mechanism in the nicotinic acetylcholine receptor (nAChR) α6 subunit (Berger et al. 2016). The effect of differing alternative last exons (ALEs) in insects is poorly understood (Lee et al. 2021), however, in mice ALE choice directs subcellular localization of mRNAs (Taliaferro et al. 2016). Further work is needed to assess the association of the alternative splicing event at ABCA (AFUN019220) and pyrethroid resistance.
Patterns of SNP polymorphism correlate with transcription profiles with reduced diversity in rp1
Analysis of the genetic diversity patterns between both strains revealed striking differences with marked reduced diversity in FUMOZ in genomic regions spanning major resistance loci notably rp1 in the 2R chromosome. This result further supports the usefulness of RNAseq data to detect phenotypic signatures of selective sweep as the reduced diversity in FUMOZ using RNAseq correlates well with signatures previously detected with whole-genome sequencing in An. funestus (Barnes et al. 2017b; Weedall et al. 2020). The detection of the N384S mutation from CYP6P9b previously reported to drive an allelic variation conferring pyrethroid resistance to this species (Ibrahim et al. 2015) further shows the robustness of the frequency filtering approach used here in detecting phenotypically meaningful variants from RNAseq data. Analysis of the polymorphism patterns of major resistance genes CYP6P9a and CYP6P9b confirms that the reduced diversity in FUMOZ is likely associated with pyrethroid resistance since other genes such as CYP4C36 did not show the same difference in polymorphisms. Similar correlation between patterns of genetic diversity from RNAseq and genome sequencing has been reported recently in the almond agroecosystem (Calla et al. 2021).
One of the significant observations was the correlation between the reduced diversity around rp1 and the high over-expression of CYP6P9a and CYP6P9b supporting that metabolic resistance in An. funestus operates through both over-expression of detoxification genes but also through the selection of a highly metabolically active allele in line with the role of allelic variation shown previously for the duplicated CYP6P9a/b genes (Ibrahim et al. 2015). Similar correlation between over-expression and strong signatures of selective sweep have been reported before in An. funestus for other detoxification genes including GSTe2 (Riveron et al. 2014; Weedall et al. 2020), for CYP9K1 (Weedall et al. 2020), CYP6 cluster on 2R chromosome in An. gambiae as revealed by the 1000Ag genomes study (Anopheles gambiae 1000 Genomes Consortium 2017), for the CYP6G1 in Drosophila (Schlenke and Begun 2004) or in the almond agroecosystem (Calla et al. 2021).
Not all significant SNPs in detoxification genes correlate with differential expression. For example, the CYP6Z1, CYP6M7, CYP6Z3 genes of the resistance to pyrethroid 2 (rp2) locus all previously shown to confer pyrethroid resistance (Irving et al. 2012; Ibrahim et al. 2016). These genes are particularly intriguing as the rp2 locus has not been predominant in resistant in natural populations across Africa, including samples from southern Africa sampled more recently than the FUMOZ strain (Weedall et al. 2019). Neither were the CYP4G17 and CYP4G16 P450 genes shown to be associated with cuticular resistance in An. gambiae (Balabanidou et al. 2016). Perhaps the most important role for these P450s is in the larval and pupal stage and we may have missed the peak of expression for these genes in our adult samples. Alternatively, as their expression is restricted to cuticular hydrocarbon secreting oenocytes (Balabanidou et al. 2016) any resistance-associated signal was swamped by our whole organism approach. As cuticular resistance is thought likely to confer cross-resistance by creating a barrier to insecticide permeability (Balabanidou et al. 2018), the variants identified here warrant further investigation.
Conclusion
The comparative analysis of the transcription profiles of two phenotypically contrasted laboratory strains of the malaria vectors An. funestus has highlighted specific features associated with pyrethroid resistance. These include the predominance of cytochrome P450 genes in association with previous QTL studies; the strong association between over-expression and reduced diversity of major resistance genes; a potential role of alternative splicing from ABC transporter and the co-expression of several genes belonging to different families to confer resistance to mosquitoes. The detection of SNPs of interest and signatures of selective sweep and using RNAseq have also been achieved supporting the usefulness of this method beyond transcription profiling. This study will further facilitate future studies on the molecular basis of resistance in field populations of mosquitoes.
Data availability
RNAseq data submitted to the European Nucleotide Archive under PRJEB45224.
Supplementary material is available at G3 online.
Supplementary Material
Acknowledgments
The authors would like to thank Drs Jacob Riveron and Sulaiman Ibrahim for their insight into this study.
Funding
This work was supported by a Wellcome Trust Senior Research Fellowships in Biomedical Sciences (101893/Z/13/Z and 217188/Z/19/Z to C.S.W.) and Bill and Melinda Gates foundation Investment ID INV-006003 to C.S.W.
Author contributions
C.S.W. conceived and designed the study. H.I., G.W., and C.S.W. performed sample preparation for all RNAseq sequencing. G.W. and C.S.W. analyzed next-generation sequencing data with assistance from J.H. M.J.W. performed qRT-PCR. C.S.W. wrote the paper with assistance from J.H. All authors read and approved the final draft of the manuscript.
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Amenya DA, Naguran R, Lo TC, Ranson H, Spillings BL, et al. 2008. Over expression of a cytochrome P450 (CYP6P9) in a major African malaria vector, Anopheles funestus, resistant to pyrethroids. Insect Mol Biol. 17:19–25. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anopheles gambiae 1000 Genomes Consortium. 2017. Genetic diversity of the African malaria vector Anopheles gambiae. Nature. 552:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabanidou V, Grigoraki L, Vontas J.. 2018. Insect cuticle: a critical determinant of insecticide resistance. Curr Opin Insect Sci. 27:68–74. [DOI] [PubMed] [Google Scholar]
- Balabanidou V, Kampouraki A, MacLean M, Blomquist GJ, Tittiger C, et al. 2016. Cytochrome P450 associated with insecticide resistance catalyzes cuticular hydrocarbon production in Anopheles gambiae. Proc Natl Acad Sci USA. 113:9268–9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariami V, Jones CM, Poupardin R, Vontas J, Ranson H.. 2012. Gene amplification, ABC transporters and cytochrome P450s: unraveling the molecular basis of pyrethroid resistance in the dengue vector, Aedes aegypti. PLoS Negl Trop Dis. 6:e1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes KG, Irving H, Chiumia M, Mzilahowa T, Coleman M, et al. 2017a. Restriction to gene flow is associated with changes in the molecular basis of pyrethroid resistance in the malaria vector Anopheles funestus. Proc Natl Acad Sci USA. 114:286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes KG, Weedall GD, Ndula M, Irving H, Mzihalowa T, et al. 2017b. Genomic footprints of selective sweeps from metabolic resistance to pyrethroids in African malaria vectors are driven by scale up of insecticide-based vector control. PLoS Genet. 13:e1006539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M, Puinean AM, Randall E, Zimmer CT, Silva WM, et al. 2016. Insecticide resistance mediated by an exon skipping event. Mol Ecol. 25:5692–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, et al. 2015. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 526:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, et al. 2018. Ten things you should know about transposable elements. Genome Biol. 19:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calla B, Demkovich M, Siegel JP, Viana JPG, Walden KKO, et al. 2021. Selective sweeps in a nutshell: the genomic footprint of rapid insecticide resistance evolution in the almond agroecosystem. Genome Biol Evol. 13:evaa234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro S, Coleman M, Mohloai P, Hemingway J, Sharp B.. 2006. Insecticide resistance in Anopheles funestus (Diptera: Culicidae) from Mozambique. J Med Entomol. 43:267–275. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Iwasa J, Cairns BR, Peterson CL.. 2017. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 18:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Cuamba N, Morgan JC, Irving H, Steven A, Wondji CS.. 2010. High level of pyrethroid resistance in an Anopheles funestus population of the Chokwe District in Mozambique. PLoS One. 5:e11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, et al. 2002. A single p450 allele associated with insecticide resistance in Drosophila. Science. 297:2253–2256. [DOI] [PubMed] [Google Scholar]
- Djouaka R, Irving H, Tukur Z, Wondji CS.. 2011. Exploring mechanisms of multiple insecticide resistance in a population of the malaria vector Anopheles funestus in Benin. PLoS One. 6:e27760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouaka RF, Bakare AA, Coulibaly ON, Akogbeto MC, Ranson H, et al. 2008. Expression of the cytochrome P450s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria. BMC Genomics. 9:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouaka RJ, Atoyebi SM, Tchigossou GM, Riveron JM, Irving H, et al. 2016. Evidence of a multiple insecticide resistance in the malaria vector Anopheles funestus in South West Nigeria. Malar J. 15:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Zhou S, Wang J, Hu W.. 2018. microRNA profiles and functions in mosquitoes. PLoS Negl Trop Dis. 12:e0006463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghurye J, Koren S, Small ST, Redmond S, Howell P, et al. 2019. A chromosome-scale assembly of the major African malaria vector Anopheles funestus. BioRxiv, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- Giraldo-Calderon GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, et al. ; VectorBase Consortium. 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43:D707–D713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves K, Koekemoer LL, Brooke BD, Hunt RH, Mthembu J, et al. 2000. Anopheles funestus resistant to pyrethroid insecticides in South Africa. Med Vet Entomol. 14:181–189. [DOI] [PubMed] [Google Scholar]
- Hemingway J, Hawkes NJ, McCarroll L, Ranson H.. 2004. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem Mol Biol. 34:653–665. [DOI] [PubMed] [Google Scholar]
- Hemingway J, Ranson H.. 2000. Insecticide resistance in insect vectors of human disease. Annu Rev Entomol. 45:369–389. [DOI] [PubMed] [Google Scholar]
- Hemingway J, Ranson H, Magill A, Kolaczinski J, Fornadel C, et al. 2016. Averting a malaria disaster: will insecticide resistance derail malaria control? Lancet. 387:1785–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt RH, Brooke BD, Pillay C, Koekemoer LL, Coetzee M.. 2005. Laboratory selection for and characteristics of pyrethroid resistance in the malaria vector Anopheles funestus. Med Vet Entomol. 19:271–275. [DOI] [PubMed] [Google Scholar]
- Ibrahim SS, Ndula M, Riveron JM, Irving H, Wondji CS.. 2016. The P450 CYP6Z1 confers carbamate/pyrethroid cross-resistance in a major African malaria vector beside a novel carbamate-insensitive N485I acetylcholinesterase-1 mutation. Mol Ecol. 25:3436–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim SS, Riveron JM, Bibby J, Irving H, Yunta C, et al. 2015. Allelic variation of cytochrome P450s drives resistance to bednet insecticides in a major malaria vector. PLoS Genet. 11:e1005618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving H, Riveron JM, Ibrahim SS, Lobo NF, Wondji CS.. 2012. Positional cloning of rp2 QTL associates the P450 genes CYP6Z1, CYP6Z3 and CYP6M7 with pyrethroid resistance in the malaria vector Anopheles funestus. Heredity (Edinb). 109:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving H, Wondji CS.. 2017. Investigating knockdown resistance (kdr) mechanism against pyrethroids/DDT in the malaria vector Anopheles funestus across Africa. BMC Genet. 18:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishak IH, Kamgang B, Ibrahim SS, Riveron JM, Irving H, et al. 2017. Pyrethroid resistance in Malaysian populations of dengue vector Aedes aegypti is mediated by CYP9 family of cytochrome P450 genes. PLoS Negl Trop Dis. 11:e0005302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NA, Fass JN.. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files. https://githubcom/najoshi/sickle (Version 1.33) [Software].
- Kawada H, Dida GO, Ohashi K, Komagata O, Kasai S, et al. 2011. Multimodal pyrethroid resistance in malaria vectors, Anopheles gambiae s.s., Anopheles arabiensis, and Anopheles funestus s.s. in Western Kenya. Plos One. 6:e22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouamo MFM, Ibrahim SS, Hearn J, Riveron JM, Kusimo M, et al. 2021. Genome-wide transcriptional analysis and functional validation linked a cluster of epsilon glutathione S-transferases with insecticide resistance in the major malaria vector Anopheles funestus across Africa. Genes (Basel). 12:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska RM, Platt N, Poupardin R, Irving H, Dabire RK, et al. 2013. Dissecting the mechanisms responsible for the multiple insecticide resistance phenotype in Anopheles gambiae s.s., M form, from Vallee du Kou, Burkina Faso. Gene. 519:98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wei L, Zhang B, Goering R, Majumdar S, et al. 2021. ELAV/Hu RNA binding proteins determine multiple programs of neural alternative splicing. PLoS Genet. 17:e1009439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J.. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12. [Google Scholar]
- Menze BD, Riveron JM, Ibrahim SS, Irving H, Antonio-Nkondjio C, et al. 2016. Multiple insecticide resistance in the malaria vector Anopheles funestus from Northern Cameroon is mediated by metabolic resistance alongside potential target site insensitivity mutations. PLoS One. 11:e0163261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menze BD, Wondji MJ, Tchapga W, Tchoupo M, Riveron JM, et al. 2018. Bionomics and insecticides resistance profiling of malaria vectors at a selected site for experimental hut trials in central Cameroon. Malar J. 17:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JC, Irving H, Okedi LM, Steven A, Wondji CS.. 2010. Pyrethroid resistance in an Anopheles funestus population from Uganda. PLoS One. 5:e11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugenzi LMJ, Menze BD, Tchouakui M, Wondji MJ, Irving H, et al. 2019. Cis-regulatory CYP6P9b P450 variants associated with loss of insecticide-treated bed net efficacy against Anopheles funestus. Nat Commun. 10:4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulamba C, Riveron JM, Ibrahim SS, Irving H, Barnes KG, et al. 2014. Widespread pyrethroid and DDT resistance in the major malaria vector Anopheles funestus in East Africa is driven by metabolic resistance mechanisms. PLoS One. 9:e110058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopoff N, Mosha JF, Lukole E, Charlwood JD, Wright A, et al. 2018. Effectiveness of a long-lasting piperonyl butoxide-treated insecticidal net and indoor residual spray interventions, separately and together, against malaria transmitted by pyrethroid-resistant mosquitoes: a cluster, randomised controlled, two-by-two factorial design trial. Lancet. 391:1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puinean AM, Foster SP, Oliphant L, Denholm I, Field LM, et al. 2010. Amplification of a cytochrome P450 gene is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. PLoS Genet. 6:e1000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron JM, Chiumia M, Menze BD, Barnes KG, Irving H, et al. 2015. Rise of multiple insecticide resistance in Anopheles funestus in Malawi: a major concern for malaria vector control. Malar J. 14:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron JM, Ibrahim SS, Mulamba C, Djouaka R, Irving H, et al. 2017. Genome-wide transcription and functional analyses reveal heterogeneous molecular mechanisms driving pyrethroids resistance in the major malaria vector Anopheles funestus across Africa. G3 (Bethesda). 7:1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron JM, Irving H, Ndula M, Barnes KG, Ibrahim SS, et al. 2013. Directionally selected cytochrome P450 alleles are driving the spread of pyrethroid resistance in the major malaria vector Anopheles funestus. Proc Natl Acad Sci USA. 110:252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron JM, Tchouakui M, Mugenzi LMJ, Menze BD, Chiang M, Wondji CS.. 2018a. Insecticide resistance in malaria vectors: an update at a global scale. In: Manguin S, Dev V, editors. Towards Malaria Elimination: A Leap Forward. IntechOpen. [Google Scholar]
- Riveron JM, Watsenga F, Irving H, Irish SR, Wondji CS.. 2018b. High plasmodium infection rate and reduced bed net efficacy in multiple insecticide-resistant malaria vectors in Kinshasa, Democratic Republic of Congo. J Infect Dis. 217:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron JM, Yunta C, Ibrahim SS, Djouaka R, Irving H, Menze BD, et al. 2014. A single mutation in the GSTe2 gene allows tracking of metabolically-based insecticide resistance in a major malaria vector. Genome Biol. 15:R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SJ, Findlay VJ, Malakasi P, Morgan BA.. 2000. Thioredoxin peroxidase is required for the transcriptional response to oxidative stress in budding yeast. Mol Biol Cell. 11:2631–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samb B, Konate L, Irving H, Riveron JM, Dia I, et al. 2016. Investigating molecular basis of lambda-cyhalothrin resistance in an Anopheles funestus population from Senegal. Parasit Vectors. 9:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ.. 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc Natl Acad Sci USA. 101:1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ.. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 3:Article3. [DOI] [PubMed] [Google Scholar]
- Taliaferro JM, Vidaki M, Oliveira R, Olson S, Zhan L, et al. 2016. Distal alternative last exons localize mRNAs to neural projections. Mol Cell. 61:821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedall GD, Irving H, Hughes MA, Wondji CS.. 2015. Molecular tools for studying the major malaria vector Anopheles funestus: improving the utility of the genome using a comparative poly(A) and Ribo-Zero RNAseq analysis. BMC Genomics. 16:931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedall GD, Mugenzi LMJ, Menze BD, Tchouakui M, Ibrahim SS, et al. 2019. A cytochrome P450 allele confers pyrethroid resistance on a major African malaria vector, reducing insecticide-treated bednet efficacy. Sci Transl Med. 11:eaat7386. [DOI] [PubMed] [Google Scholar]
- Weedall GD, Riveron JM, Hearn J, Irving H, Kamdem C, et al. 2020. An Africa-wide genomic evolution of insecticide resistance in the malaria vector Anopheles funestus involves selective sweeps, copy number variations, gene conversion and transposons. PLoS Genet. 16:e1008822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2012. Global Plan for Insecticide Resistance Management (GPIRM). Geneva, Switzerland: World Health Organization. [Google Scholar]
- Wilding CS, Weetman D, Rippon EJ, Steen K, Mawejje HD, et al. 2015. Parallel evolution or purifying selection, not introgression, explains similarity in the pyrethroid detoxification linked GSTE4 of Anopheles gambiae and An. arabiensis. Mol Genet Genomics. 290:201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji CS, Irving H, Morgan J, Lobo NF, Collins FH, et al. 2009. Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome Res. 19:452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji CS, Morgan JC, Coetzee M, Hunt R, Steen K, et al. 2007. Mapping a quantitative trait locus conferring pyrethroid resistance in the African malaria vector Anopheles funestus. BMC Genomics. 8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data submitted to the European Nucleotide Archive under PRJEB45224.
Supplementary material is available at G3 online.