Abstract
SATB1 is expressed primarily in thymocytes and orchestrates temporal and spatial expression of a large number of genes in the T-cell lineage. SATB1 binds to the bases of chromatin loop domains in vivo, recognizing a special DNA context with strong base-unpairing propensity. The majority of thymocytes are eliminated by apoptosis due to selection processes in the thymus. We investigated the fate of SATB1 during thymocyte and T-cell apoptosis. Here we show that SATB1 is specifically cleaved by a caspase 6-like protease at amino acid position 254 to produce a 65-kDa major fragment containing both a base-unpairing region (BUR)-binding domain and a homeodomain. We found that this cleavage separates the DNA-binding domains from amino acids 90 to 204, a region which we show to be a dimerization domain. The resulting SATB1 monomer loses its BUR-binding activity, despite containing both its DNA-binding domains, and rapidly dissociates from chromatin in vivo. We found this dimerization region to have sequence similarity to PDZ domains, which have been previously shown to be involved in signaling by conferring protein-protein interactions. SATB1 cleavage during Jurkat T-cell apoptosis induced by an anti-Fas antibody occurs concomitantly with the high-molecular-weight fragmentation of chromatin of ∼50-kb fragments. Our results suggest that mechanisms of nuclear degradation early in apoptotic T cells involve efficient removal of SATB1 by disrupting its dimerization and cleavage of genomic DNA into loop domains to ensure rapid and efficient disassembly of higher-order chromatin structure.
SATB1 is a cell type-restricted protein expressed predominantly in thymocytes and is essential for T-cell development (2, 12). SATB1 binds in a specialized DNA context wherein one strand consists of mixed A's, T's, and C's, but not G's (ATC sequences). Clustered ATC sequences have a high propensity to unwind by extensive base unpairing when placed under a negative superhelical strain. Such base-unpairing regions (BURs), which are not more than 150 to 200 bp in length, are typically identified in genomic segments known as matrix or scaffold attachment regions (MARs or SARs; the term MARs is used here). Within BURs, the core unwinding element can often be identified, and mutation within such an element abolishes the base-unpairing potential of the BUR within a MAR (36). SATB1 was originally cloned by employing a specific sequence containing the core unwinding element derived from the BUR (12, 36) located within the MAR 3′ of the immunoglobulin heavy chain (IgH) gene enhancer (8). BURs are most likely the critical sequences for MARs. This is because the high unwinding capability of BURs has been shown to be important for MAR activity, e.g., by conferring high-affinity binding to the nuclear matrix in vitro and augmenting the activity of a reporter gene in a stably transformed cell line. When a BUR is mutated to abrogate its unwinding capability, these activities are either lost or reduced for the MAR containing the mutated BUR (4).
MARs, originally identified as DNA fragments with high affinity to salt-extracted and DNase I-digested nuclei (called nuclear matrix), have been postulated to contain sequences that form the bases of chromosomal loops in both interphase nuclei and metaphase chromosomes and thus play an important role in the organization of higher-order chromatin structure (7, 28, 47; reviewed in reference 22). To address whether SATB1 binds to genomic DNA anchored to the underlying structure of nuclei, a series of genomic sequences that bind to SATB1 in vivo in human Jurkat lymphoblastic cells were cloned and used as probes for fluorescence in situ hybridization. It was found that SATB1's target sequences are tightly associated with the nuclear matrix and located at the bases of chromatin loop domains and that SATB1 itself is bound to these sites inside cells (11). Thus, SATB1 was characterized as a thymocyte and a T-cell-specific in vivo MAR/BUR-binding protein (we describe SATB1 as a BUR-binding protein in this paper).
Recent transgenic-mouse studies have demonstrated the biological significance of certain MARs in tissue-specific gene expression and chromatin structure. In particular, studies on MARs flanking the IgH enhancer showed that these sequences are essential for the B-lymphocyte-specific transcription of a rearranged μ gene (20). These MARs have also been shown to collaborate with the μ enhancer to generate long-range chromatin accessibility to transcription factors. This phenomenon correlates with extended demethylation of the gene locus in a transcription-independent manner (29). In addition, by using a transfected cell line, it was found that B-cell-specific demethylation at the Ig(κ) gene locus requires both the intronic kappa enhancer and the nearby MAR (35, 43). Furthermore, recent evidence suggests that the function of MARs in mediating long-range chromatin accessibility involves generation of an extended domain of histone acetylation (18). The potential role of MARs in DNA recombination was studied for certain MARs (reviewed in reference 59).
The biological roles of MAR/BUR-binding proteins in specific cell lineages were unknown. Recently, the role of SATB1 in the T-cell lineage was studied using SATB1 knockout mice (2). SATB1 was found to be essential for proper T-cell development and T-cell activation. At the molecular level, multiple genes (at least 2% of the total genes), including a proto-oncogene, cytokine receptor genes, and apoptosis-related genes, were derepressed at inappropriate stages of T-cell development in SATB1 null mice. This is consistent with earlier findings that SATB1 can act as a transcriptional repressor mediated by BUR sequences (38, 44). SATB1 is crucial in coordinating the temporal and spatial expression of genes during T-cell development, thereby ensuring the proper development of this lineage. These data from SATB1 knockout mice suggest that BUR-binding proteins can act as global regulators of cell function in specific cell lineages (2).
Within the thymus, an estimated 99% of all thymocytes undergo apoptosis, mainly due to lack of positive selection based on the failure to make a productive T-cell receptor (TCR) gene rearrangement and, to a lesser degree, due to negative selection for producing autoreactive TCRs (60, 63). Since SATB1 is bound to the bases of chromatin loop domains presumably contributing to the higher-order chromatin structure in thymocytes, SATB1 may be an early target of degradation to efficiently disassemble chromatin. In this report, we describe the fate of SATB1 in Jurkat T cells and mouse thymocytes in response to apoptotic stimuli. A MAR-binding domain and homeodomain were hitherto identified as being essential for recognition of the core unwinding element within BURs (13, 50, 67); we show that SATB1 exists as a homodimer and that dimerization is essential for its DNA-binding activity. The dimerization of SATB1 is disrupted due to cleavage by a caspase 6-like protease during apoptosis. Once SATB1 becomes a monomer, even if the MAR-binding domain and the homeodomain remain intact, it readily dissociates from chromatin in vivo concomitant with the cleavage of its target sequences by apoptotic endonuclease(s).
MATERIALS AND METHODS
Reagents.
Tosyl-l-lysine chloromethyl ketone (TLCK) and tosyl-l-phenylalanine chloromethyl ketone (TPCK) were purchased from Sigma Chemical Co. (St. Louis, Mo.). Leupeptin was purchased from Boehringer Mannheim (Indianapolis, Ind.). Acetyl-Val-Ala-Asp-fluoromethyl ketone (Z-VAD-fmk) and acetyl-Val-Glu-Ile-Asp-fluoromethyl ketone (Z-VEID-fmk) were obtained from Enzyme Systems Products (Livermore, Calif.). Acetyl-Asp-Glu-Val-Asp-aldehyde (Ac-DEVD-CHO) was procured from Pharmingen (La Jolla, Calif.). Stock solutions of these inhibitors were prepared according to the manufacturer's instructions. All other molecular-biology grade reagents were purchased from Sigma. Double-stranded poly(dI-dC) was purchased from Amersham Pharmacia Biotech (Piscataway, N.J.).
Cell culture and induction of apoptosis.
Jurkat cells (American Type Culture Collection, Manassas, Va.) were maintained in RPMI 1640 medium containing 2 mM pyruvate (GIBCO-BRL Life Technologies, Burlington, Ontario, Canada) and 10% fetal calf serum (Tissue Culture Biologicals, Tulare, Calif.) at 37°C with 5% CO2 in a humidified incubator. For induction of apoptosis, Jurkat cells were grown to 106 cells/ml and incubated with 100 ng of anti-Fas antibody (monoclonal human anti-Fas clone CH-11; MBL International Corp., Watertown, Mass.)/ml for various times prior to harvesting. The initial seeding density and time of culture varied. The conditions that we typically employed involved either seeding at a low density of 5 × 104 cells/ml and incubation for 8 days essentially as described by Washo-Stultz et al. (68) or seeding at a higher density of 2 × 105 cells/ml and incubation for 2 days. Mouse thymocytes were maintained in the same medium as Jurkat cells but were induced by adding 2 μM dexamethasone (Sigma Chemical Co.). For protease inhibitor assays, Jurkat cells were preincubated with the respective inhibitors for 30 min. Apoptosis was then induced by the addition of anti-Fas. Cells were harvested 4 h after induction of apoptosis.
Nuclear extracts, total cellular lysates, and Western blotting.
Approximately 5 × 106 to 10 × 106 cells were used to prepare nuclear extracts for each time point. Briefly, cells were collected, washed twice in ice-cold phosphate-buffered saline (PBS), and stored overnight at −80°C. Cell pellets were thawed on ice the next day and resuspended at 5 × 106 cells per 100 μl of buffer C (0.42 M NaCl, 10% glycerol, 20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) (14), followed by centrifugation for 15 min at 10,000 × g. Total cellular lysates were prepared from approximately 1 × 106 to 2 × 106 cells by lysing the cells directly in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer without dye. Protein concentrations were determined by a Bio-Rad protein assay using bovine serum albumin (BSA) as a standard. Thirty micrograms of nuclear extract or 50 μg of total cellular lysate was separated by SDS–10% PAGE and analyzed by Western blotting as described previously (37). Antibodies used were polyclonal anti-PARP-1 (H-250) (Santa Cruz Biotechology, Santa Cruz, Calif.) and anti-SATB1 (12). Antibodies were detected by enhanced chemiluminescence using a SuperSignal West Pico detection kit (Pierce Chemical Co., Rockford, Ill.).
Southwestern blotting.
Southwestern blotting was performed essentially as described previously (37). Briefly, 40 μg of nuclear extract was incubated with SDS-PAGE sample buffer at 37°C for 10 min. The proteins were then separated by SDS–10% PAGE such that the dye front ran out. Under this condition, histone H1 and high-mobility-group (HMG) protein I/Y were removed from the gel. The gel was then transferred onto an Immobilon-P membrane (Millipore Corporation, Bedford, Mass.). The membrane was incubated in renaturation and binding buffer containing 20 mM Tris, pH 7.4, 50 mM NaCl, 1 mM DTT, 0.1% Tween 20, and 5% BSA for 1 h at room temperature to allow refolding of proteins in situ. The blot was then incubated with competitor DNA followed by 32P-labeled wild-type (25)7 [WT (25)7] probe in binding buffer. After being washed the membrane was exposed to X-ray film for visualization of BUR-binding activity.
Immunofluorescence.
Thymocytes were plated onto poly-l-lysine-coated coverslips (Sigma Chemical Co.), fixed for 5 min in 3% paraformaldehyde (J. B. EM Services Inc., Pointe Claire, Dorval, Quebec, Canada), and permeabilized for 20 min in 0.2% Triton X-100 (Chromatographic Specialties Inc., Nepean, Ontario, Canada). The cells were incubated for 60 min with primary antibodies and 45 min with secondary antibodies and were counterstained for 1 min with 1 μg of Hoechst 33258 dye (Sigma Chemical Co.)/ml. All incubations were performed at room temperature. For double staining, the antibodies were applied sequentially and the blocking step with 0.15% (wt/vol) gelatin (Bio-Rad, Mississauga, Ontario, Canada) was added before application of the second antibody (69). The following antibodies were used for immunofluorescence staining: a mouse monoclonal IgG1 anti-lamin B (dilution 1:50; clone 119D5-F1; provided by Y. Raymond, Institut du Cancer de Montreal, Montreal, Quebec, Canada), fluorescein isothiocyanate-conjugated goat anti-mouse IgG heavy-chain- and light-chain-specific antibodies (dilution, 1:300; Sigma), rabbit polyclonal anti-SATB1 (dilution, 1:300, batch 1583), and CY3-conjugated goat anti-mouse IgG (Jackson Labs). The cells were examined using an Olympus Bmax fluorescence microscope and Photosystems.
In vivo cross-linking of chromatin.
In vivo cross-linking of DNA and protein was carried out as described by Wedrychowski et al. (70). Briefly, mouse thymocytes were isolated, washed in PBS, and treated with 2 μM dexamethasone. Thirty million cells were used per sample and were treated with 3 mM cis-diaminedichloro-platinum II (cis-DDP; Aldrich Chemical Co., Milwaukee, Wis.) for 2 h at 37°C. Cells were washed in cold PBS and solubilized in 10 ml of 4% SDS–50 mM Tris-HCl, pH 7.5–1 mM PMSF by rotation for 3 h at room temperature. Samples were homogenized in a Dounce with a loosely fitting pestle and centrifuged for 16 h at 100,000 × g at 20°C in an SW 41 rotor. The pellets were resuspended in 5 M urea–4% SDS–50 mM Tris-HCl, pH 7.5–1 mM PMSF, and centrifugation as described above was repeated. The DNA pellets were briefly air dried and resuspended in 0.5 ml of 2 mM Tris-HCl, pH 7.5–1 mM PMSF using a wide-bore pipette tip. The solution was sonicated, and DNA was quantitated by spectrophotometric absorption. Samples were precipitated with acetone, resuspended in 40 μl of 2 mM Tris-HCl, pH 7.5–1 mM MgCl2–1 mM PMSF, and digested with 15 mg of bovine pancreatic DNase I (Boehringer Mannheim Corp.)/ml for 1 h at 37°C. The reaction was stopped by adding SDS sample buffer containing 10% β-mercaptoethanol and boiling for 5 min. For Western blots, one-fourth of the reaction mixture was loaded on an SDS–7.5% polyacrylamide gel, which corresponded to approximately 60 μg of DNA as determined before nuclease digestion or to chromatin from approximately 7.5 × 106 cells.
In vitro cleavage of SATB1.
SATB1 was purified from mouse thymus by passing the 0.42 M extract successively through the mutated and wild-type BUR affinity columns as described previously (37). One hundred nanograms of purified native SATB1 or 40 μg of Jurkat cell extracts was incubated with 1 μM recombinant caspase 3, 6, or 7 (kind gift from Guy Salvesen, The Burnham Institute, La Jolla, Calif.) for 1 h at 37°C in caspase activity buffer in a 20-μl reaction volume (62). Reactions were terminated by adding an equal volume of 2× Laemmli SDS-PAGE sample buffer and heating the samples at 95°C for 5 min, or the samples were directly transferred to the band shift assay mixture. Preparative scale digestion of SATB1 for N-terminal sequencing of the 65-kDa fragment was performed by incubating 5 μg of purified native SATB1 with 10 μM purified recombinant caspase 6 for 1 h in caspase activity buffer in a 100-μl reaction mixture.
Pulsed-field gel electrophoresis.
Jurkat cells were induced for apoptosis using anti-Fas antibody (clone CH-11) as described above. At defined time points (0.5 to 9 h) postinduction aliquots of cells were removed, washed in chilled PBS, resuspended in PBS, and embedded in 1.2% low-melting-point agarose (Bio-Rad, Hercules, Calif.). The agarose plugs were lysed and deproteinized by incubation in 0.5 M EDTA–0.5-mg/ml proteinase K–1% Sarkosyl at 56°C for 16 h. The plugs were then washed extensively with 10 mM Tris–1 mM EDTA, pH 7.5. The digested plugs were then loaded into a 1% pulsed-field-certified agarose (Bio-Rad) gel. High-molecular-weight DNA was resolved by pulsed-field gel electrophoresis in a CHEF DR-III apparatus (Bio-Rad) at 120° included angle and 14°C for 22 h. The switch time was set to 50 to 100 s, and the voltage was kept constant at 6 V/cm. A phage lambda concatemeric DNA ladder and Saccharomyces cerevisiae chromosomal DNA were used as molecular size markers (Bio-Rad). The gels were stained with Sybr gold dye (Molecular Probes, Eugene, Oreg,) and photographed under UV illumination.
Isolation of low-molecular-weight DNA.
Low-molecular-weight (nucleosomal) DNA fragments were prepared from anti-Fas antibody-treated Jurkat cells as described by Park and Patek (55). Briefly, 106 cells were washed with PBS and lysed by resuspension in Tris-EDTA lysis buffer containing 0.1% NP-40. Lysed cells were treated sequentially with RNase A and proteinase K to remove RNA and proteins, respectively. The resulting DNA solution was loaded directly on a 1.6% agarose gel and electrophoresed for 3 h at 4 V/cm. The gels were stained with Sybr gold dye and photographed under UV illumination.
Generation of caspase-resistant mutant and in vitro translation.
Replacement of aspartate at position 254 in the human SATB1 primary sequence with alanine was carried out by overlapping PCR. Briefly, a 1.1-kb fragment containing the site to be mutagenized was subcloned into pBluescript KS(+) (Stratagene, La Jolla, Calif.). Two oligonucleotides spanning the site were synthesized (MUT I, 5′GGTTGAAATGGCTAGCCTTTCTGAGC3′; MUT II, 5′GCTCAGAAAGGCTAGCCATTTCAACC3′). The oligonucleotides were engineered in such a way that the amino acid sequence would be conserved but a new NheI site would be introduced to facilitate the screening of mutagenized clones. PCR was used to amplify an 800-bp fragment using the M13 forward and MUT II primers and a 300-bp fragment using the M13 reverse and MUT I primers. The fragments were mixed at equal molar concentrations, and a 1.1-kb fragment was amplified using the M13 forward and reverse primers. The PCR product was digested with EcoRI and XbaI and subcloned in pBluescript KS(+). The mutagenesis was confirmed by automated sequencing using the T3 primer. The mutagenized 1.1-kb EcoRI-XbaI fragment was then cloned into the parental plasmid (pAT1146) to obtain full-length D254A-SATB1 cDNA.
In vitro translation was performed using the coupled TNT-T3 reticulocyte lysate system (Promega Corporation, Madison, Wis.) and [35S]methionine (Redivue; Amersham Pharmacia Biotech). The products of translation reactions were heated in the presence of SDS-PAGE sample buffer and loaded directly on SDS–10% polyacrylamide gels unless mentioned otherwise. The 35S-labeled proteins were visualized by autoradiography of dried gels.
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed basically as described previously (37, 50). Binding reactions were performed in a 10-μl total volume containing 10 mM HEPES (pH 7.9), 1 mM DTT, 50 mM KCl, 2.5 mM MgCl2, 10% glycerol, 0.5 μg of double-stranded poly(dI-dC), 10 μg of BSA, and 1 μl of the 35S-labeled in vitro translation reaction mixture. Samples were preincubated at room temperature for 5 min prior to addition of 32P-labeled WT (25)7 synthetic BUR DNA substrate (37). After 15 min of incubation at room temperature, the products of such binding reactions were then resolved by 6% native polyacrylamide gel electrophoresis. The gels were dried under vacuum and exposed to two layers of X-ray film. The top film was developed to detect the 32P-specific signal.
Yeast two-hybrid analysis.
We subcloned the DraI fragment of SATB1 cDNA that codes for most of the protein except the N-terminal 55 amino acids in yeast two-hybrid expression vectors that allowed low-level expression of the fusion proteins. We used residues 56 to 763 of SATB1 fused to the GAL4 DNA-binding domain (DBD) in the pGBT9 vector (Clontech Laboratories Inc., Palo Alto, Calif.) as a bait for delineating the dimerization domain of SATB1. The DraI fragment and various constructs with truncations from the N-terminal region of SATB1 were fused with the GAL4 activation domain (AD) in pGAD424 vector (Clontech). The AD and DBD fusion constructs were cotransformed in a pairwise fashion in yeast strain CG-1945 (Clontech) as described previously (R. Agatep, R. D. Kirkpatrick, D. L. Parchliuk, R. A . Woods, and R. D. Gietz, Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ssDNA/PEG) protocol, Technical Tips Online, http://tto.trends.com, 1998) and assayed for protein-protein interaction using standard protocols, with HIS3 as the reporter gene.
RESULTS
SATB1 dissociates from chromatin early during thymocyte apoptosis.
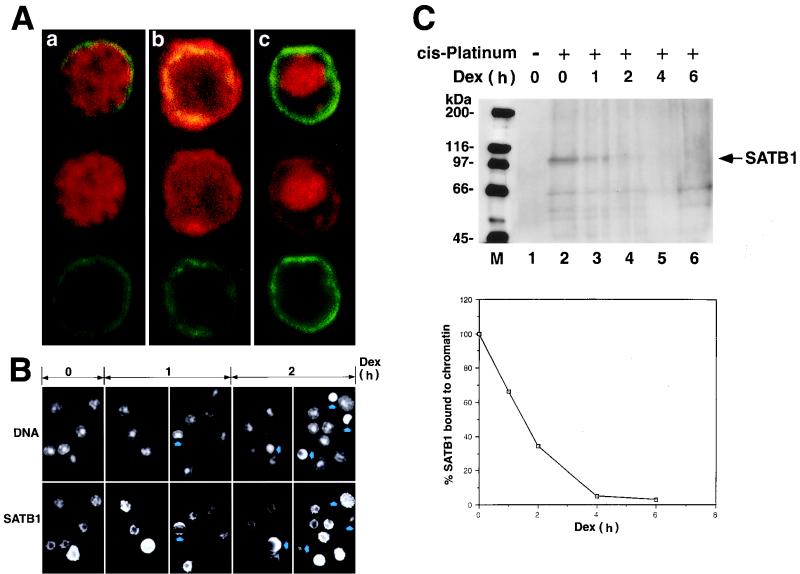
Thymocytes as well as peripheral lymphocytes are known to undergo spontaneous apoptosis in response to various physiological stimuli (33). BUR-binding protein SATB1 is predominantly and abundantly expressed in thymocytes and activated T cells and is an important regulatory protein for proper T-cell development (2). We studied the fate of SATB1 in T cells and thymocytes, first focusing on its intracellular localization. The subcellular localization of SATB1 in early stages of dexamethasone-induced apoptotic thymocytes was studied by immunostaining and visualization by confocal microscopy (Fig. 1A). Thymocytes exhibited a continuous rim of peripheral nuclear staining when labeled with anti-lamin B as previously reported (69). Untreated thymocytes contain SATB1 exclusively in their nuclei as evidenced by double staining with anti-lamin B and SATB1 antibodies (Fig. 1A, a). As apoptosis progressed, SATB1 relocalization became apparent due to the fact that some SATB1 migrated out from the nucleus to the cytoplasm (Fig. 1A, b). At later stages of apoptosis, some SATB1 remained in the nuclei for most thymocytes (Fig. 1A, c). To examine the relative locations of SATB1 and DNA, double staining of a thymocyte population with Hoechst 33258 dye (for DNA) and an anti-SATB1 antibody was performed at 0, 1, and 2 h after dexamethasone treatment. The results show that SATB1 immediately circumscribes the DNA stained regions in nonapoptotic thymocytes, and the two stained regions remain for the most part mutually exclusive (Fig. 1B). This remained true for apoptotic cells, but there was an apparent decrease in the levels of SATB1 for most of these cells with chromatin condensation.
FIG. 1.
SATB1 dissociates from chromatin early during apoptosis. (A) Confocal analysis of immunofluorescence staining of lamin B and SATB1. Rat thymocytes treated with 2 μM dexamethasone for 2 h were double immunostained with anti-SATB1 antibody (red) and anti-lamin B antibody (green). Individual cells in very early (a), middle (b), and late stages of apoptosis (c) are shown. (B) Double staining of genomic DNA and SATB1 with Hoechst 33258 dye (top) and anti-SATB1 antibody (bottom) from thymocytes treated with dexamethasone (dex) for 0, 1, and 2 h. Arrows indicate apoptotic cells with apparent morphological alteration. Differences in SATB1 staining pattern and the sizes of cells reflect the different developmental stages of the thymocyte (our unpublished result). (C) (Top) Dissociation of SATB1 from chromatin early in apoptosis. Dexamethasone-treated thymocytes at different time points were incubated with 50 mM cis-DDP and solubilized in 4% SDS, and DNA-protein complexes (cross-linked genomic DNA fraction) were pelleted by ultracentrifugation. The pellets were resuspended in 5 M urea–2% SDS, and untracentrifugation was repeated to isolate DNA-cross-linked proteins. The pellets were sonicated and treated with DNase I. The solubilized protein fractions were subjected to SDS–7.5% PAGE and Western blot analysis using anti-SATB1 polyclonal serum. Positions of the molecular mass markers (in kilodaltons) are indicated on the left. Arrow, band corresponding to full-length SATB1. Other faint bands, representing nonspecific cross-reactivity of the anti-SATB1 antibody, became visible only after longer exposure of a Western blot. (Bottom) Densitometric analysis of the SATB1 signals shown at the top. Intensity of the band corresponding to full-length SATB1 was quantitated using a laser densitometer and plotted as a function of time after dexamethasone treatment. All values were normalized to the intensity of the SATB1 signal in the absence of dexamethasone treatment (lane 2) and were expressed as percentages of the zero time value.
Change in the intracellular localization of SATB1 early during apoptosis suggests dissociation of SATB1 from genomic DNA in vivo. To address this question, mouse thymocytes were treated with dexamethasone to induce apoptosis and, at different time points, cells were removed and incubated in the presence of cis-DDP to cross-link SATB1 to genomic DNA in vivo. Under these conditions, only the proteins that are bound to DNA were expected to be cross-linked. The genomic DNA cross-linked to DNA-binding proteins (cross-linked DNA-protein fraction) was isolated, resuspended in buffer, and fragmented by sonication. DNA was quantitated and digested with DNase I. To analyze SATB1 that was cross-linked to DNA on a Western blot, proteins isolated from a defined amount of cross-linked DNA (60 μg) were loaded in each lane. As a control, genomic DNA was also prepared similarly from cells that were not treated with cis-DDP. Figure 1C shows results of Western blot analysis of SATB1 in the cross-linked genomic DNA fraction using an anti-SATB1 antibody. Although the amount of SATB1 is small, probably due to nonquantitative cross-linking and loss during repeated suspension and centrifugation, SATB1 is clearly detected in the cross-linked DNA-protein preparations. The predicted molecular mass of SATB1 is 85.9 kDa, but it migrates anomalously at ∼103 kDa on SDS-polyacrylamide gels (50). In the control sample (Fig. 1C, top, lane 1), there was no detectable SATB1 signal, indicating that non-cross-linked SATB1 was separated from DNA during the preparation. On the contrary, the cross-linked DNA-protein sample shows a prominent band at 103 kDa, corresponding to full-length SATB1 (Fig. 1C, top, lane 2). The intensity of the signal corresponding to the full-length SATB1 protein rapidly decreases upon onset of apoptosis, and, by 2 h following dexamethasone treatment, approximately 65% of SATB1 was already dissociated from chromatin (Fig. 1C, top, lane 4). Virtually all SATB1 (over 90%) was dissociated by 4 h after treatment (Fig. 1C, top, lane 5). The amount of SATB1 bound to genomic DNA was measured by densitometric analysis of the 103-kDa signal on the immunoblot (Fig. 1C, bottom).
SATB1 is cleaved early during T-cell apoptosis.
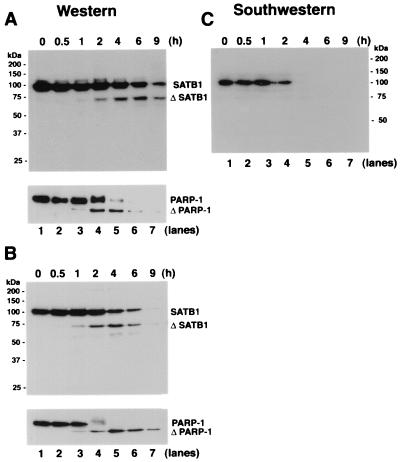
The rapid dissociation of SATB1 from the genomic DNA early during apoptosis in vivo might be due to specific cleavage of SATB1 to abrogate its DNA-binding activity. The integrity of SATB1 was monitored by immunoblot analysis of extracts from dexamethasone-treated mouse thymocytes prepared at different time intervals using anti-SATB1 polyclonal serum 1583. As shown in Fig. 2A, the antibody detects a prominent band of 103 kDa corresponding to full-length SATB1 (Fig. 2A; lane 1). Within 0.5 h posttreatment, no appreciable change in the amount of SATB1 was detected (Fig. 2A, lane 2). However, at 1 h, a new SATB1 cleavage product started to appear as a faint band at a position corresponding to an estimated molecular mass of 65 kDa, and this band became more intense after 2 h posttreatment (Fig. 2A, lanes 3 to 7). We refer to the 65-kDa band as the signature apoptotic fragment of SATB1. The complementary cleavage product, which is estimated to be an approximately 20-kDa polypeptide, was not detectable with polyclonal serum 1583. By raising a polyclonal antibody against a C-terminal peptide of SATB1, we found that the 65-kDa band contains the C terminus of SATB1 (data not shown), and polyclonal serum 1583 fails to detect the N-terminal 254 amino acids (as described below). A virtually identical pattern and rate of SATB1 cleavage occurred for human Jurkat cells treated with anti-CD95 (Fas) antibody, at least until 6 h posttreatment (Fig. 2B). Antibody-mediated ligation of CD95 molecules on the surfaces of Jurkat human lymphoblastic T cells follows a rapid cell death pathway leading to the serial activation of caspase 8 and then caspases 3 and 6 (reviewed in references 16 and 61). Similar to SATB1 in either apoptotic mouse thymocytes or human Jurkat cells, poly(ADP-ribose) polymerase (PARP-1), which is known to be cleaved by caspase 3 (40) and caspase 7 (reviewed in reference 16), was cleaved, giving rise to an 89-kDa signature fragment starting at 1 h posttreatment (Fig. 2A and B, bottom, lanes 3). At later time points of apoptosis, the overall signal of SATB1 declines, indicating total destruction of the protein. This is also true for PARP-1 (Fig. 2A and B, bottom), indicating protein degradation due to nonspecific proteolysis, which occurs after the specific cleavage.
FIG. 2.
Proteolytic cleavage of SATB1 early during T-cell apoptosis. (A) Cleavage of SATB1 in mouse thymocytes. Thymocytes were collected from a 3-week-old mouse, treated with 2 μM dexamethasone to induce apoptosis, and harvested. Nuclear extracts were prepared as described in Materials and Methods, and 20 μg of protein was resolved by SDS–10% PAGE, transferred to a polyvinylidene difluoride membrane, and probed with anti-SATB1. Left, positions of the molecular weight markers; right, positions of the intact and cleaved (Δ) SATB1. Bottom, immunoblot analysis of the same extracts using anti-PARP-1. Positions of intact and cleaved (Δ) PARP-1 are indicated. (B) Cleavage of SATB1 in a human lymphoblastic T-cell line. Jurkat cells were grown continuously for 8 days in culture as described in Materials and Methods and treated with 100 ng of anti-Fas antibody (clone CH-11)/ml to induce apoptosis. Cells were harvested at the indicated times (0.5 to 9 h) thereafter. Nuclear extracts were prepared, and 20 μg of protein was resolved by SDS–10% PAGE and analyzed for SATB1 by Western blotting. Bottom, status of PARP-1 in identical extracts. (C) Loss of BUR-binding activity during apoptosis. The same series of proteins used in panel A were separated on a 10% polyacrylamide gel except that the gel was run longer than for panel A and subjected to Southwestern analysis as described in Materials and Methods using a radiolabeled WT (25)7-mer probe. The autoradiogram shows a signal corresponding to the BUR-binding activity of intact SATB1. The various time points after dexamethasone treatment (in hours) are indicated above each lane.
In contrast to what was found for PARP-1 (Fig. 2A, bottom), the full-length SATB1 protein remained at significant levels even after 6 h post-dexamethasone treatment of thymocytes (Fig. 2A, lane 6). A similar result was obtained for anti-Fas antibody-induced Jurkat cells (Fig. 2A and B). This was unexpected in light of our results from the in vivo cross-linking analysis, which indicated that SATB1 bound to chromatin was lost in thymocytes by 4 h after dexamethasone treatment (Fig. 1C). These results suggest that full-length SATB1 that remained at 4 and 6 h after dexamethasone treatment may not have BUR-binding activity. To examine this point further, we monitored the BUR-binding activity of SATB1 during apoptosis by Southwestern blot analysis with the same series of thymocyte proteins as that used for the Western blot analysis shown in Fig. 2A. When the blot was probed with a radiolabeled synthetic BUR probe [WT (25)7] (12), we detected BUR-binding activity only up to 2 h (Fig. 2C, lanes 1 to 4); beyond 4 h, the BUR-binding activity due to SATB1 was undetectable (Fig. 2C, lanes 5 to 7). This is consistent with the results obtained from the in vivo cross-linking study (Fig. 1C). Apparently, the 65-kDa major apoptotic breakdown product of SATB1 does not bind to the labeled BUR probe (Fig. 2C). When this Southwestern data were directly compared with the Western blot data shown in Fig. 2A, especially for 2, 4, and 6 h posttreatment, it was clear that at least two groups of SATB1s exist, one with BUR-binding activity and the other without. The results of the Southwestern analysis and the in vivo cross-linking study and the timing of the appearance of the 65-kDa fragment indicated by Western analysis suggest that during apoptosis SATB1 with BUR-binding activity is cleaved and that, as a result, SATB1 loses its binding activity. This hypothesis was tested and shown to be true as described below. SATB1 is a phosphorylated protein (our unpublished observation). However, the exact biochemical difference(s) between the two groups of SATB1 and their relative amounts remain to be determined.
High-molecular-weight chromatin fragmentation occurs concomitant with SATB1 cleavage.
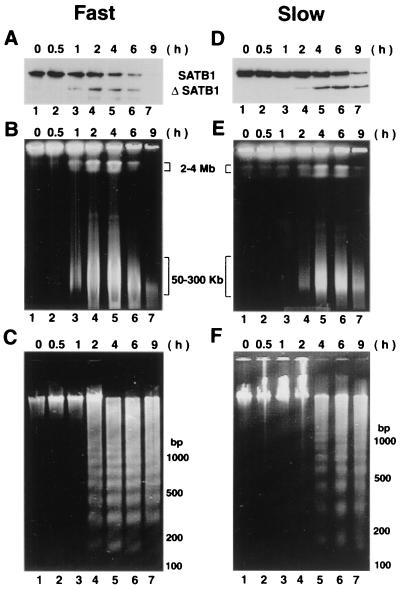
We examined the genomic DNA fragmentation for anti-Fas-treated Jurkat cells by pulsed-field gel electrophoresis to determine if there was any correlation regarding the timing of SATB1 cleavage and DNA fragmentation. The cells treated with anti-Fas antibody in the same manner as that shown in Fig. 2B exhibited the major appearance of large DNA fragmentation in the range of 2 to 4 Mb as well as 50 to 300 kb at around 1 h posttreatment (Fig. 3B, lane 3). At this early time point, the nucleosomal ladder was hardly detected (Fig. 3C, lane 3). Only after 2 to 4 h posttreatment did we detect the nucleosomal ladder (Fig. 3C, lanes 4 to 7) together with larger-size DNA fragmentation (Fig. 3B, lanes 4 to 7). This result shows that the timing of the cleavage of SATB1 correlates well with the initial prominent signals for the large-size DNA fragmentation.
FIG. 3.
High-molecular-weight DNA fragmentation coincides with SATB1 cleavage. (A and D) Immunoblot analysis of SATB1 in apoptotic Jurkat cells. Jurkat cells were treated with anti-Fas, and the fate of SATB1 was monitored by immunoblot analysis of cell extracts as described in Materials and Methods. (B and E) Cleavage of genomic DNA into 50- to 300-kb chromatin loops. Pulsed-field gel electrophoretic separation of apoptotic Jurkat cell DNA was performed as described in Materials and Methods. Jurkat cells were induced for apoptosis using anti-Fas antibody (clone CH-11). At defined time points (0.5 to 9 h) postinduction aliquots of cells were removed and embedded in LMP agarose. The agarose plugs were lysed and deproteinized as described in Materials and Methods. The digested plugs were then loaded onto a 1% agarose gel, and the high-molecular-weight DNA was resolved by pulsed-field gel electrophoresis as described in Materials and Methods. Positions of the 50- to 300-kb chromatin loops and 2- to 4-Mb giant DNA fragments are indicated. (C and F) Low-molecular-weight DNA fragmentation. Low-molecular weight DNA was prepared from apoptotic Jurkat cells as described previously (55). For all panels, time (in hours) after the anti-Fas treatment of cells in culture is indicated on top. Two different seeding densities were used to culture Jurkat cells as described in Materials and Methods. Fast (A to C), apoptotic cleavage profiles from an 8-day continuous culture seeded at 5 × 104 cells/ml; slow (D to F), apoptotic cleavage profiles from a 2-day culture seeded at 2 × 105 cells/ml.
To verify this observation, we examined the Jurkat cells under different cell culture conditions so that these cells would undergo apoptosis at a slower rate. It is known that time in culture plays a critical role in determining the rate of apoptosis (68). In fact, when Jurkat cell cultures that were seeded at high density (2 × 105 cells per ml) and grown for 2 days until a density of 106 cells per ml was reached, apoptosis proceeded more slowly (Fig. 3D to F) than for cells seeded at 5 × 104 per ml and continuously cultured for 8 days (Fig. 3A to C). For Jurkat cells cultured for 2 days, SATB1 cleavage started only after approximately 2 h posttreatment (Fig. 3D, lane 5). Moreover, SATB1 cleavage was also not complete until 9 h posttreatment (Fig. 3D, lane 7). For these cells, the first visible large-size DNA fragmentation signals (2 to 4 Mb and 50 to 300 kb) occurred only after 2 h (Fig. 3E, lane 4), and this timing agrees with the timing of the onset of SATB1 cleavage. The nucleosomal ladder was not detected at 2 h, but it becomes prominent at 4 h posttreatment (Fig. 3F, lanes 4 and 5). The onset and kinetics of PARP-1 cleavage were similar to those of SATB1 under the respective culture conditions (data not shown). Even though we cannot determine if SATB1 cleavage precedes large-scale DNA fragmentation or vice versa from these experiments, it is of interest that these two events occur at a very similar timing during apoptosis.
Identification of the protease that cleaves SATB1.
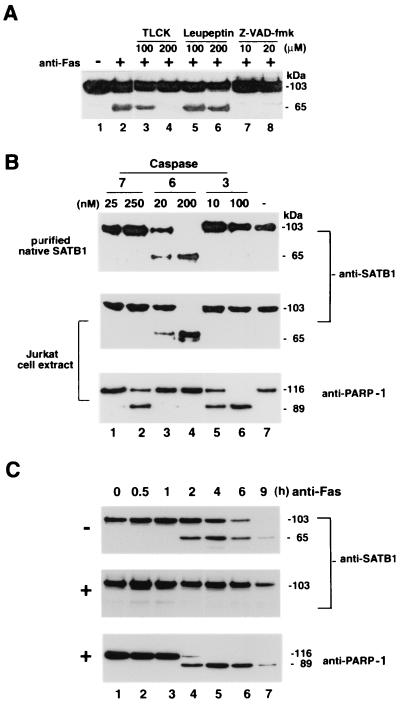
Serine proteases are the most common proteases in cells, and cysteine-specific aspartate proteases or caspases constitute the central component of the death machinery (reviewed in reference 16). We employed a series of inhibitors to identify the class of protease(s) involved in the cleavage of SATB1 (Fig. 4A). Jurkat T cells were treated with a spectrum of protease inhibitors. Cells pretreated in such manner were then induced for apoptosis using a Fas monoclonal antibody (CH-11). As a control, we used cells that were not exposed to any of the inhibitors. Western blot analysis of cell extracts revealed that the proteolytic degradation of SATB1 was affected neither by 100 or 200 μM leupeptin (Fig. 4A, lanes 5 and 6, respectively) nor by 100 μM TLCK (Fig. 4A, lane 3), both of which are inhibitors of serine proteases. However, at a concentration of 200 μM, TLCK completely abolished cleavage of SATB1 (Fig. 4A, lane 4). TLCK at concentrations greater than 200 μM induces necrosis in Jurkat cells and abolishes the features of apoptosis (9). Next, we tested broad-range caspase inhibitor Z-VAD-fmk at 10 and 20 μM (Fig. 4A, lanes 7 and 8, respectively). At both these concentrations Z-VAD-fmk effectively abolished the proteolytic cleavage of SATB1. These results indicated that cleavage of SATB1 during apoptosis is dependent on the proteolytic activity of a caspase.
FIG. 4.
Identification of protease that cleaves SATB1 during apoptosis. (A) Effect of protease inhibitors on in vivo SATB1 cleavage in Jurkat cells treated with anti-Fas antibody. Jurkat cells were preincubated with control solvent (−) or with specific protease inhibitors for 30 min as indicated. An anti-CD95 monoclonal antibody (clone CH-11) was then added (+) to a final concentration of 100 ng/ml, the cells were incubated further for 3 h, and SATB1 proteolysis was analyzed by immunoblotting. The 65-kDa band represents the major proteolytic degradation product of SATB1. (B) In vitro cleavage by purified caspases 7, 6, and 3. Purified native SATB1 from mouse thymus (top) and Jurkat whole-cell extract (middle and bottom) were incubated with indicated amounts of caspase 7 (lanes 1 and 2), 6 (lanes 3 and 4), or caspase 3 (lanes 5 and 6) or without caspase (lane 7) for 1 h and examined for SATB1 (top and middle) and PARP-1 (bottom) cleavage by Western blotting using the appropriate antibody. (C) In vivo inhibition of SATB1 cleavage by a caspase 6 inhibitor. Jurkat cells were preincubated with solvent dimethyl sulfoxide alone (−; top) or with 10 μM Z-VEID-fmk (+; middle and bottom) for 30 min. Anti-Fas antibody was then added to a final concentration of 100 ng/ml. Aliquots of cells were removed at indicated times, and protein extracts were prepared as described in Materials and Methods. SATB1 proteolysis was analyzed by immunoblotting as described in Materials and Methods. As a marker for apoptosis, the Western blot (middle) was stripped and reprobed (bottom) with anti-PARP-1 (H-250).
To further elaborate the caspase(s) involved in the proteolysis of SATB1 during apoptosis, we asked whether SATB1 either in the form of purified native protein or as a component of whole-cell extract could be cleaved by activated recombinant executioner caspase 3, 6, or 7 in vitro and, if so, whether the pattern of cleavage resembled that observed in vivo. We found that caspase 7 and 3 (Fig. 4B, top, lanes 1, 2, 5, and 6) failed to cleave purified SATB1 from mouse thymus. However, caspase 6 effectively cleaved SATB1 to its signature 65-kDa major fragment at 20 and 200 nM (Fig. 4B, lanes 3 and 4). Treatment of Jurkat whole-cell extracts with purified caspases also yielded a similar pattern of specificity of cleavage (Fig. 4B, middle). As a control, we probed this Western blot with an anti-PARP-1 antibody to demonstrate that caspases 3 and 7 are indeed active. Incubation of Jurkat cell extract with caspase 7 (Fig. 4B, bottom, lanes 1 and 2) and caspase 3 (Fig. 4B, bottom, lanes 5 and 6) led to the characteristic cleavage pattern of PARP-1. However, PARP-1 is not cleaved by caspase 6 (Fig. 4B, bottom, lanes 3 and 4), confirming the specificity of target selection by different executioner caspases.
The involvement of a caspase 6-like protease in apoptotic cleavage of SATB1 in vivo was demonstrated by treating the cells with Z-VEID-fmk, a cell-permeable peptide inhibitor for caspase 6-like proteases (45). Immunoblot analysis of extracts from cells that were not treated with the peptide inhibitor prior to antibody-mediated CD95 ligation (Fig. 4C, top) showed a time-dependent decrease in the signal corresponding to full-length SATB1. For up to 1 h post-anti-Fas treatment there was no apparent cleavage of SATB1 (Fig. 4C, top, lane 3). However, by 2 h approximately 30% of SATB1 was cleaved to generate its signature 65-kDa major apoptotic fragment, referred to as ΔSATB1 (Fig. 4C, top, lane 4). At 4 and 6 h after induction of apoptosis, approximately equal amounts of full-size SATB1 and the 65-kDa fragment were detected (Fig. 4C, top, lane 7). In contrast, pretreatment of Jurkat cells with 10 μM Z-VEID-fmk in culture prior to the addition of anti-Fas resulted in a complete inhibition of SATB1 cleavage for up to 6 h postinduction (Fig. 4C, compare top and bottom lanes 7). In contrast, PARP-1, which is known to be cleaved by caspase 3 (40) and caspase 7 (reviewed in reference 16), was cleaved in the presence of Z-VEID-fmk. These data show the involvement of caspase 6-like protease in anti-Fas-mediated apoptotic cleavage of SATB1 in T cells.
Identification of the caspase cleavage site.
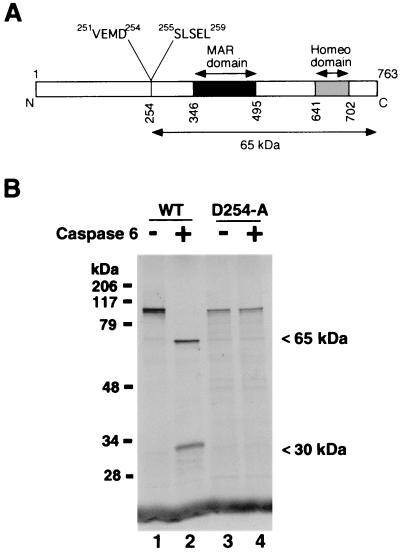
Analysis of the primary structure of SATB1 for a potential caspase 6 cleavage site revealed only one stretch of four amino acids, VEMD, that is highly similar to optimal consensus recognition site VEID for caspase 6 (65). While this work was in progress, it was reported that SATB1 is cleaved in a caspase-dependent manner and VEMD was predicted as a candidate cleavage recognition sequence. However, this study did not identify the protease or confirm the cleavage site (24). The potential cleavage site aspartate is located at amino acid position 254, and cleavage at this site would yield two proteolytic fragments with calculated molecular masses of 20 and 65 kDa. The predicted size of the larger fragment corresponds exactly with the estimated molecular weight of the SATB1 cleavage product that we observed in vivo and in vitro. The polyclonal antibody that we raised against full-length SATB1 does not recognize the smaller cleavage product.
To ascertain that the cleavage indeed occurred after the aspartate at position 254, we performed preparative-scale digestion of purified native SATB1 using purified recombinant caspase 6. The digestion products were then resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane, and the N-terminal sequence of the 65-kDa band was determined by an N-terminal microsequencing method. The sequence was 255Ser-Leu-Ser-Glu-Leu259, which confirmed the predicted position of the cleavage site. The position of this sequence with respect to the caspase 6 cleavage site and DBDs in SATB1 is depicted schematically in Fig. 5A.
FIG. 5.
Caspase 6 cleaves after aspartate 254 in SATB1. (A) Schematic representation of various known functional domains in SATB1. Black and gray boxes, MAR-binding domain and homeodomain (13), respectively. The caspase 6 cleavage site is located at amino acid (aa) position 254. The caspase 6 recognition sequence (aa 251 to 254) and the N-terminal sequence of the 65-kDa cleavage product (aa 255 to 259) are indicated. (B) VEMD-to-VEMA mutation abolishes the cleavage by caspase 6. Wild-type SATB1 (lanes 1 and 2) and the caspase-resistant mutant SATB1-D254A (lanes 3 and 4) were transcribed and translated in vitro and digested with recombinant activated caspase 6 (lanes 2 and 4), as described in Materials and Methods. The intact proteins and fragments were separated by SDS-4 to 15% gradient PAGE (Bio-Rad) and visualized by autoradiography. Positions of the cleavage products are indicated on the right.
We then mutated the aspartate residue in the P4 position (254) to alanine using an in vitro overlapping-PCR mutagenesis strategy. The mutagenized SATB1 is referred to as D254A-SATB1. When we digested the in vitro-translated wild-type (Fig. 5B, lane 1) and D254A-35S-SATB1 (Fig. 5B, lane 3) proteins with caspase 6, we observed no degradation of D254A-SATB1 (Fig. 5B, lane 4), whereas wild-type SATB1 was completely cleaved to generate the 65-kDa major fragment and ∼30-kDa minor fragment (Fig. 5B, lane 2). The smaller cleavage product apparently migrates anomalously since it is expected be a 20-kDa peptide. In conclusion, the results of in vivo as well as in vitro proteolysis studies strongly argue in favor of caspase 6 or a caspase 6-like proteinase, and not caspase 3 and/or caspase 7, in mediating the cleavage of SATB1 during apoptosis to generate the 65-kDa fragment.
Cleavage by caspase 6 abolishes the DNA-binding activity of SATB1 in vitro.
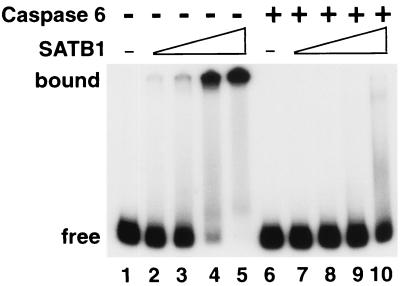
All the evidence gathered so far strongly suggests that cleavage by caspase 6 alters the DNA-binding ability of SATB1. To test this directly, we employed an in vitro system wherein a 32P-labeled BUR DNA substrate was incubated with increasing amounts of purified SATB1 with or without prior incubation with purified recombinant caspase 6. The products of such binding reactions were then resolved by native polyacrylamide gel electrophoresis. As depicted in Fig. 6, untreated SATB1 bound the WT (25)7-mer DNA substrate in a dose-dependent manner (Fig. 6, lanes 2 to 5). However, when preincubated with caspase 6, SATB1 completely lost its DNA-binding activity (Fig. 6, lanes 7 to 10) as judged by the total lack of a slow-migrating protein-DNA complex at the highest concentration of SATB1 used (Fig. 6, lane 10). A parallel Western blot analysis of caspase 6-treated SATB1 indicated that virtually all of SATB1 was cleaved under the conditions employed (data not shown). Results of this in vitro binding study strongly indicate that the cleavage mediated by caspase 6 causes loss of the DNA-binding activity of SATB1.
FIG. 6.
Cleavage by caspase 6 abolishes the DNA-binding activity of SATB1. Wild-type SATB1 was transcribed and translated in vitro and digested with recombinant activated caspase 6 as described in Materials and Methods. 32P-labeled BUR probe WT (25)7 was prepared as described previously (37). The binding reactions were performed as described in Materials and Methods and then resolved by 6% native PAGE. Free, position of the labeled DNA substrate alone (lanes 1 and 6). Mock-treated (lanes 2 to 5) and caspase 6-treated (lanes 7 to 10) in vitro translation mixtures were serially diluted in EMSA buffer and incubated with the labeled WT (25)7-mer. One microliter of either 20-fold-diluted (lanes 2 and 7), 10-fold-diluted (lanes 3 and 8), or 5-fold-diluted (lanes 4 and 9) or undiluted (lanes 5 and 10) translation mixture was used for each binding reaction. The positions of SATB1-DNA complexes (bound) and the free DNA probe (free) are indicated on the left.
Delineation of an N-terminal domain necessary for SATB1 binding to BURs.
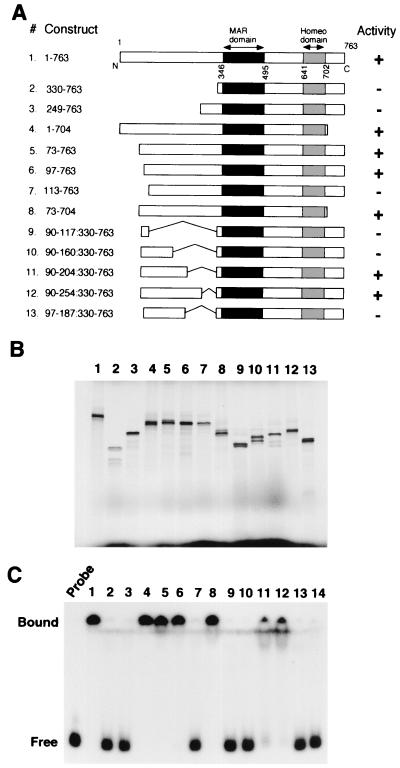
The 65-kDa major apoptotic fragment of SATB1, which results from the cleavage at position 254 by caspase 6, still contains both the MAR-binding domain (residues 346 to 495) and homeodomain (residues 641 to 702) previously characterized (13, 50). These two domains together are essential for specific recognition of the core unwinding element of BUR and confer full DNA-binding activity when fused with either glutathione S-transferase (GST) protein (13) or protein A (50). Therefore, it was unexpected that the apoptotic fragment containing both domains completely lost BUR-binding activity. It is clear from our data, however, that these two domains are not sufficient for BUR binding and that an additional domain necessary for BUR binding must exist in the N-terminal region. In vitro-translated SATB1 (amino acids 1 to 495) containing the N-terminal region and the MAR-binding domain and the MAR-binding domain fused with GST protein or protein A confers BUR-binding activity (13, 50). Therefore, there must be a domain with a common activity among fused peptides and the N-terminal region of SATB1. First, a series of truncations were constructed to delineate this additional domain in the N-terminal region, as shown in Fig. 7A.
FIG. 7.
Identification of an N-terminal domain that is required for the DNA-binding activity of SATB1. (A) Schematic representation of SATB1 N- and C-terminal and internal deletions. Various truncated versions of SATB1 cDNA that were used as templates for coupled in vitro transcription and translation are shown. Construct 1 depicts all the known functional domains in SATB1. All the constructs are named according to the amino acids encoded by the full-length cDNA that they represent. Black boxes, MAR-binding domain; gray boxes, homeodomain. The result of DNA-binding studies using these constructs is summarized in the “Activity” column. +, DNA-binding activity comparable to that of the full-length protein; −, total lack of DNA binding. (B) SDS-PAGE analysis of in vitro translation products. Coupled in vitro transcription and translation of SATB1 with various terminal and internal deletions as depicted in panel A were performed as described in Materials and Methods. The 35S-labeled translation products were resolved by SDS-10% PAGE and visualized by autoradiography. The numbers above each lane correspond to the constructs depicted in panel A. The dark patch at the bottom of the gel indicates position of the dye front. (C) EMSA analysis. The DNA-binding activity of each of the above constructs was monitored by EMSA analysis using a 32P-labeled WT (25)7 probe as described in Materials and Methods. Numbers on top of lanes correspond to those of the constructs in panel A. Lane 14, control binding reaction using vector (pBlueScript)-translated lysate. Free and bound, positions of the unbound and protein-bound DNA probes, respectively.
A series of constructs with N-terminal deletion mutations (constructs 2 to 7) as well as internal deletions (constructs 9 to 13) that spanned an ∼160-amino-acid region immediately upstream of the caspase 6 cleavage site were prepared. In addition, truncated constructs lacking the C-terminal amino acids downstream of the homeodomain (constructs 4 and 8) were tested. All of these truncated versions were cloned into the pBluescript vector (Stratagene) under the control of the T3 promoter, and the in vitro-translated proteins were tested for their BUR-binding activity. The SDS-PAGE analysis of the various translation products revealed that all of the truncated proteins were translated efficiently, yielding comparable amounts of the proteins and very few incomplete translation products (Fig. 7B). The BUR-binding activity of each of these translation products was determined by gel shift assay using a labeled WT (25)7 probe (a synthetic BUR DNA) in the presence of excess competitor DNA as described previously (50). Full-length SATB1 binds very strongly to the WT (25)7 probe, and virtually all of the probe is shifted in the form of a high-molecular-weight complex, indicating DNA binding (Fig. 7C, lane 1). On the other hand, the in vitro-translated MAR-binding domain alone (amino acids 345 to 495) (data not shown) or together with the homeodomain and the C-terminal region (amino acids 330 to 763) (Fig. 7A, construct 2) failed to bind the DNA (Fig. 7C, lane 2). A similar result was obtained with the translation product that contained all of the residues downstream of the caspase 6 cleavage site, including the cleavage recognition sequence itself (Fig. 7C, lane 3). The extreme C-terminal 60 amino acid residues were found to be unnecessary for the DNA-binding activity of SATB1 (Fig. 7C, lane 4 and 8). Similarly, removal of 73 (Fig. 7C, lane 5) and 97 (Fig. 7C, lane 6) amino acids from the N terminus of SATB1 did not have any measurable effect on its DNA-binding activity. Interestingly, removal of 113 amino acid residues from the N terminus resulted in complete loss of DNA-binding activity (Fig. 7C, lane 7), suggesting that the region encompassing residues 97 to 113 is required for the DNA-binding activity of SATB1. Although this region is necessary, it is not sufficient for restoring the DNA-binding activity of the MAR-binding domain of SATB1 since the fusion product is not active (Fig. 7C, lane 9).
The C-terminal boundary of the additional domain in the N-terminal region must therefore lie upstream of the caspase 6 cleavage site. To delineate the C-terminal boundary, we created a series of MAR domain fusion proteins by serially adding approximately 50 residues to the region spanning residues 97 to 113 (Fig. 7A, constructs 9 to 12). Our data show that the region spanning residues 90 to 204 is the minimal region that can restore full binding activity of SATB1 (Fig. 7C, lane 11), while a shorter protein with residues 90 to 160 fused with the MAR-binding domain is not active (Fig. 7C, lane 10). To further examine the C-terminal boundary of this newly discovered domain, we created another fusion in which residues 97 to 187 were fused with the MAR-binding domain (Fig. 7A, construct 13). This protein is inactive (Fig. 7C, lane 13), suggesting that the C-terminal boundary of this domain lies between a short stretch spanning residues 187 and 204. As a control, a vector (pBluescript)-translated lysate did not bind to the probe (Fig. 7C, lane 14). The results of these BUR-binding studies are summarized as activities in Fig. 7A.
The N-terminal region of SATB1 is responsible for protein dimerization.
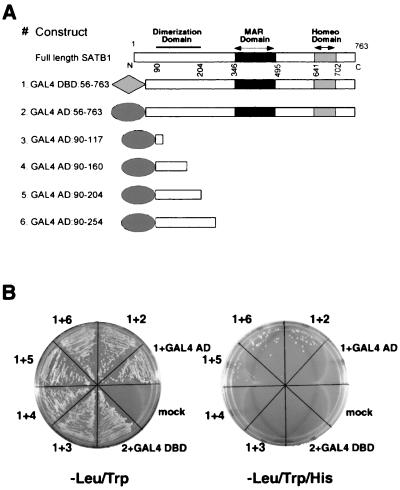
Next, we employed the yeast two-hybrid system (19) to analyze whether the domain identified by the in vitro DNA-binding studies is involved in mediating protein-protein interaction. The various constructs that were used for transformation are depicted in Fig. 8A. We found that expression of full-length SATB1 is toxic to yeast (our unpublished observation). However, expression of the DraI fragment of SATB1 cDNA that codes for most of the protein except the N-terminal 55 amino acids was tolerated, as indicated by using vectors pGBT9 and pGAD624 (Clontech) that allowed low-level expression of the cDNAs. We used residues 56 to 763 of SATB1 fused to the GAL4 DBD as a bait for delineating the dimerization domain of SATB1. Both the AD and DBD constructs were transformed in a pairwise fashion in yeast strain CG-1945 (Clontech) and assayed for protein-protein interaction. Figure 8B depicts results of the yeast two-hybrid analysis. Cells carrying both the transformed plasmids were those that grew on a plate lacking Leu and Trp (Fig. 8B, left). Any type of protein-protein interaction between the fusion proteins would bring the GAL4 DBD and AD in close proximity, thus activating transcription of four reporter genes. We chose HIS3 as a reporter gene and scored for colony formation on media lacking three amino acids, viz., Trp, Leu, and His. Coexpression of a SATB1 DraI restriction fragment fused to the GAL4 DBD (GAL4 DBD:56–763) and to the GAL4 AD (GAL4 AD:56–763) resulted in transcriptional activation of the HIS3 reporter gene (Fig. 8B, right), indicating that SATB1 polypeptides interact with each other, most probably by forming a homodimer. Expression of any of the fusion protein along with either the GAL4 AD or GAL4 DBD was not sufficient to achieve transcriptional activation of the reporter gene (Fig. 8B). To further delineate the dimerization domain, we tested a series of truncated versions of SATB1 for interaction with GAL4 DBD-SATB1. In agreement with the results of our in vitro DNA-binding studies, we found that GAL4 AD fusions with residues 90 to 117 and 90 to 160 were unable to interact with the bait (GAL4 DBD-SATB1); however, fusions with residues 90 to 204 and 90 to 254 yielded transcriptional activation of HIS3, resulting in growth of colonies (Fig. 8B). The results of these interaction studies demonstrated that the N-terminal region comprising amino acids 97 to 204 alone is both essential and sufficient for dimerization.
FIG. 8.
SATB1 is a homodimer. (A) Schematic representation of SATB1 with N- and C-terminal deletions used in the yeast two-hybrid assay. Various truncated versions of SATB1 that were used as baits for the yeast two-hybrid assay are shown. Two nearly full-length fusion constructs of SATB1 were used to monitor the dimerization potential of SATB1 (constructs 1 and 2). Constructs 3 to 6 represent various truncations that were used to map the dimerization domain of SATB1. (B) Yeast two-hybrid assay. Nearly full-length SATB1 cDNA (encoding amino acids 56 to 763) fused with the GAL4 DBD was cotransformed in yeast strain CG1945 with one of the GAL4 AD-SATB1 fusion constructs (constructs 2 to 6) as described in Materials and Methods. The transformation mixtures were streaked in defined sectors on minimal-medium plates lacking either Leu and Trp (left) or Leu, Trp, and His (right). The numbers of constructs used for cotransformation are indicated outside of each sector. Mock, transformation mixture in which water was added instead of DNA.
The dimerization domain of SATB1 is similar to the PDZ domain.
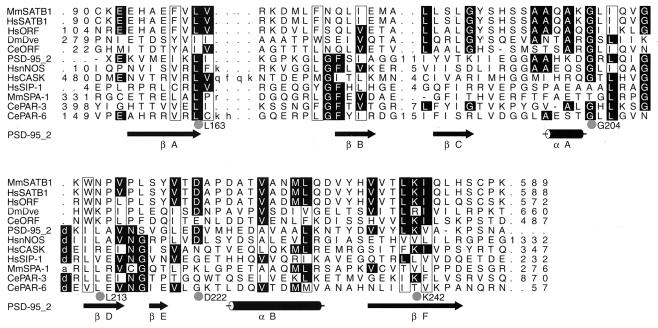
The dimerization domain of mouse and human SATB1 was found by BLAST search (1) to be highly homologous to the two hypothetical proteins encoded by human and Caenorhabditis elegans cDNAs and the Drosophila melanogaster Dve protein encoded by a defective proventriculus (dve) gene. Dve is a homeodomain protein that is required for midgut specification under the control of different extracellular signals (51). The significance of the region in Dve that is homologous to the SATB1 dimerization domain has not been assigned. To further investigate whether the dimerization region of SATB1 is similar to any domains of a known function, the 90- to 204-amino-acid-peptide was used as the query sequence for a search against the Conserved Domain Database (version 1.01). The program compared the SATB1 dimerization sequence to 3,019 position-specific score matrices prepared from domains derived from the Smart and Pfam collections. Two significant alignments were found to be the PcrB family (code pfam01884; E value = 0.16) and the PDZ domain (pfam00595; E value = 1.8). The function of the homologous sequences in the PcrB family is unknown (30). The PDZ domain, on the other hand, is a well-studied protein-protein interaction domain found in proteins that mediate targeting and clustering of channels, receptors, cell adhesion proteins, and other signaling enzymes at the specific sites of cell-cell contact, including synapses (reviewed in reference 17). Canonical PDZ domains contain ∼80 to 100 amino acid residues. The PDZ domains form a compact, globular structure consisting of a six-stranded antiparallel β-barrel flanked by two α-helices (15, 48). We performed hidden-Markov model (HMM)-based analysis of the sequence homologs of SATB1 with several known PDZ domain-containing proteins including the second PDZ domain of postsynaptic density-95 (PSD-95 2; RCSB code 1QLC). PSD-95 is a neuronal-membrane-associated guanylate kinase that associates with receptors and cytoskeletal elements at synapses (reviewed in reference 42). The HMM-generated alignment of these proteins is shown in Fig. 9. PDZ domains are known to dimerize with other PDZ domains forming heterodimers or to bind to the carboxyl termini of interacting proteins (reviewed in reference 17). However, the requirement of a PDZ domain-mediated homodimerization for protein function has not been reported to date. For SATB1, the putative PDZ domain is responsible for forming homodimers, and, as a result of this dimerization, SATB1 binds genomic DNA specifically recognizing BUR sequences.
FIG. 9.
SATB1 dimerization domain is homologous to PDZ domains. Shown is an HMM-generated alignment of the dimerization domain of SATB1 and related sequences (MMSatb1 to CeORF) and selected PDZ domains (PSD-95 2 to CePAR-6). Columns containing residues that are conserved in 6 or more of the 12 sequences are highlighted. Dots, columns that are most conserved (10 of 12 sequences) and residues that might be functionally important owing to their spatial proximity on the surface of the PDZ domain based on the alignment in the context of the three-dimensional structure. Hydrophobic columns are boxed. Numbers denote the numbers of amino acids that are not shown. Arrows and cylinders, β-strands and α-helices taken from the nuclear magnetic resonance structure of the second PDZ domain of PSD-95 (PSD-95–2; PDB-RCSB code 1QLC). Amino acids in lowercase within the alignment correspond to residues aligned to the insert state of the HMM. The sequences shown are MmSATB1 (NP 033148), HsSATB1 (NP 002962.1), HsORF (Homo sapiens hypothetical protein KIAA1034; BAA82986.1), DmDve (CAA09729.1), CeORF (hypothetical protein ZK1193.5; T27710), PD-95–2 (second PDZ domain of PSD-95), HsnNOS (P29475), HsCASK (AAB88125), HsSIP-1 (AAB53042), MmSPA-1 (BAA01973), CePAR-3 (T34302), and CePAR-6 (T43216). Species abbreviations are as follows: Mm, Mus musculus; Hs, H. sapiens; Ce, C. elegans; Dm, D. melanogaster.
DISCUSSION
Disruption of dimerization is the cause of SATB1 dissociation from chromatin.
SATB1 is tightly bound to genomic DNA at the base of chromatin loop domains in Jurkat cells (11). Therefore, to ensure rapid disassembly of higher-order chromatin structure, SATB1 is expected to be an early target for degradation during apoptosis. We analyzed SATB1 during anti-Fas antibody-induced T-cell- and dexamethasone-induced thymocyte apoptosis and found that it dissociates from chromatin in vivo early during apoptosis. Dissociation of SATB1 from chromatin is associated with its cleavage at amino acid position 254 to generate a 65-kDa fragment containing both the MAR-binding domain and the homeodomain. Although these two domains, when fused with GST protein, were previously shown to be sufficient to confer both high specificity and affinity toward the core unwinding elements of BURs, the 65-kDa fragment itself totally lacks DNA-binding activity. The present study showed that an additional region (amino acids 90 to 204) is essential for the two domains to confer BUR-binding activity, and this region was identified as a dimerization domain. This was demonstrated by a yeast two-hybrid assay system, which has been previously employed for demonstrating the in vivo dimerization of proteins including GCN4 (26) and TRF1 (3). Our data show that SATB1 dissociates from chromatin due to its cleavage, which separates the dimerization domain from the DBDs, thus becoming a nonfunctional monomeric protein.
SATB1 is a target of caspase 6-like protease during T-cell apoptosis.
Caspases are the central component of the death machinery (reviewed in references 16 and 61). We have shown that SATB1 is site-specifically cleaved at an aspartate at position 254 by caspase 6 or a caspase 6-like protease. Although many cytoplasmic and nuclear proteins have been reported as targets for the effector caspases 3 and 7 (reviewed in reference 16), only a few target proteins for caspase 6 have been identified so far. These include lamin A (54, 64), lamin B1 (10), keratin 18 (6), the amyloid precursor protein-binding protein (APP-BP1) (41, 56), and Huntingtin proteins (71). Our data add SATB1 to this short list as the first transcription factor that is cleaved by a caspase 6-like protease.
Two proteins with binding specificity similar to that of SATB1 are also cleaved at an early stage during apoptosis. The first is PARP-1, a DNA damage-sensing protein known to preferentially bind nicked DNA. Using closed-circle double-stranded DNA templates, PARP-1 was found to specifically recognize BUR elements (21) and is cleaved simultaneously with SATB1 early during apoptosis, but by caspase 3 and caspase 7 (reviewed in reference 16) instead of caspase 6. Scaffold attachment region-binding protein SAF-A (hnRNPU), a ubiquitous MAR-binding protein, is also cleaved by caspase 3 early during apoptosis and dissociates from chromatin (23). Among sequences within MARs, SAF-A also confers specificity to BUR elements (S. Galande, C. C. Lee, and T. Kohwi-Shigematsu, unpublished results). Therefore, two chromatin-binding proteins of similar binding specificities, PARP-1 and SAF-A, are targets of caspase 3, whereas SATB1 is uniquely cleaved by caspase 6. However, not all BUR-binding proteins are cleaved during apoptosis. The Ku70/86 heterodimer, which has been shown to exhibit strong binding affinity and specificity toward BURs using closed-circle DNA templates (21), remains intact at least up to 9 h after the start of apoptosis (data not shown). PARP-1 and Ku70/86 were shown to form a protein complex in the absence of DNA, and the BUR-binding affinity of this protein complex is synergistically augmented (21). Therefore, once PARP-1 is cleaved, Ku70/86 may also disassemble from chromatin.
Our attempts at studying the effects of expressing caspase 6-resistant SATB1 in a cell culture system were hampered because expression of either wild-type or mutated SATB1 in transfected T cells at levels any higher than those of endogenous SATB1 induced cell death. We are currently taking an alternative approach, which is to establish transgenic mice in a SATB1 null background and examine the effects of caspase 6-resistant SATB1 on T-cell development. Whether SATB1 cleavage by caspase 6-like protease plays an important role in selection of thymocytes must await further experimentation.
Degradation of genomic DNA into the loop domain size fragments occurs concomitantly with SATB1 cleavage during apoptosis.
The DNA degradation into a specific pattern of fragments is a characteristic feature of apoptosis (reviewed in reference 49). These include 2- to 4-Mb giant-size fragments, 50- to 300-kb fragments, and a DNA ladder consisting of multimers of approximately 200 bp (nucleosomal ladder). These distinct sizes of DNA fragments most likely reflect the structural organization of chromatin in the nucleus. It has been demonstrated that the first appearance of chromatin degradation to 50- to 300-kb fragments occurs just prior to internucleosomal fragmentation in various apoptotic cells (53; reviewed in reference 66), and therefore genomic DNA degradation starts with disassembly of higher-order chromatin structure. We compared the timing of large-scale chromatin fragmentation with that of SATB1 cleavage during apoptosis in two different growth phases of Jurkat T-cell culture. It is known that the susceptibility of cultured cells to apoptosis heavily depends on their growth phase. In late log phase, cell cultures exhibit an increased susceptibility to apoptosis compared with cultures in early log growth phase, and this difference is independent of cell density. In the experimental system employed, where early and late log phases differ greatly with respect to timing of DNA fragmentation, we observed that the timing for the start of SATB1 cleavage matched with that of the ∼50-kb DNA fragmentation just preceding nucleosomal ladder generation. These data suggest that, early during anti-Fas antibody-induced Jurkat T-cell apoptosis, a higher-order chromatin structure is first disassembled by removing SATB1 from the bases of chromatin loop domains, and these domains are cleaved and digested further. Whether SATB1, which is an abundant protein in thymocytes (2.5 × 104 to 5 × 104 molecules/cell, data not shown), binds chromatin at virtually all of their loop attachment sites or only a subset of them remains to be investigated. Furthermore, whether there is any effect of SATB1 ablation on disassembly of higher-order chromatin structure during apoptosis in SATB1-deficient thymocytes will be studied in the future.
Identification of PDZ-like domain of SATB1 and its potential biological significance.
The newly identified dimerization domain of SATB1 (90 to 204 amino acids), which is essential for its BUR-binding activity, was found to be homologous to PDZ domains. PDZ domains are modular protein-binding domains that have at least two distinct mechanisms for binding. PDZ domains can bind to specific recognition sequences at the carboxyl termini of proteins (31, 32, 39, 46, 52), or they can bind with other PDZ domains forming heterodimers (5). Owing to these capabilities, PDZ domain-containing proteins can form mutimeric protein complexes. Many PDZ domain-containing proteins identified to date are associated with the plasma membrane, and accumulated evidence suggests that PDZ domains are involved in recruiting signaling proteins to protein complexes at the membrane. For example, the second PDZ domain of PSD-95 has been shown to bind directly to PDZ domains within neuronal nitric oxide synthase at synapses, thus coupling Ca2+ entry through N-methyl-d-aspartate channels to NO synthesis (5, 58). Although most PDZ domain proteins are found at the plasma membrane, two nuclear proteins that possess a domain similar to PDZ are known to date. One is SIP-1, of unknown function, which interacts with human Y-linked testis-determining gene SRY-encoded protein (57), and the other is Spa1, with a Ran GTPase-activating domain (25). Recent evidence shows that CASK, a PDZ domain-containing protein, is concentrated at neuronal synapses, enters the nucleus, and interacts with a defined transcription factor to regulate transcription (27). Although this interaction is mediated by its guanylate kinase domain and not by the PDZ domain, it provides the evidence for the translocation of membrane-associated PDZ domain-containing proteins to the nucleus. The fact that chromatin-associated protein SATB1 contains a putative PDZ domain has important biological implications. For SATB1, the putative PDZ domain is responsible for homodimerization and is necessary to manifest BUR-binding activity to the protein. It can be speculated that some PDZ-interacting proteins can relay cell surface information to the nucleus and derepress multiple genes by disrupting dimerization of SATB1. Recently, a protein domain called the SAF box, which is structurally related to the homeodomain, has been identified in various MAR-binding proteins (34). Whether a PDZ domain that has an activity similar to that found in SATB1 is present in other BUR-binding proteins awaits future investigation.
ACKNOWLEDGMENTS
We thank Guy Salvesen for kindly providing purified recombinant caspases and valuable discussion, Christein E. Carson for the immunostaining analysis of thymocytes, and Yves Raymond for kindly providing anti-lamin B antibody.
The initial part of this work was supported by National Institutes of Health RO1(CA39681), and the latter part was supported by RO1(GM59901) (to T.K-S.)
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez J D, Yasui D H, Niida H, Joh T, Loh D Y, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bode J, Kohwi Y, Dickinson L A, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 5.Brenman J E, Chao D S, Gee S H, McGee A W, Craven S E, Santillano D R, Wu Z, Huang F, Xia H, Peters M F, Froehner S C, Bredt D S. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 6.Caulín C, Salvesen G S, Oshima R G. Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis. J Cell Biol. 1997;138:1379–1394. doi: 10.1083/jcb.138.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cockerill P N, Garrard W T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 8.Cockerill P N, Yuen M-H, Garrard W T. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987;262:5394–5397. [PubMed] [Google Scholar]
- 9.Cryns V L, Bergeron L, Zhu H, Li H, Yuan J. Specific cleavage of α-Fodrin during Fas- and tumor necrosis factor-induced apoptosis is mediated by an interleukin 1-β converting enzyme/Ced-3 protease distinct from the poly (ADP-ribose) polymerase protease. J Biol Chem. 1996;271:31277–31282. doi: 10.1074/jbc.271.49.31277. [DOI] [PubMed] [Google Scholar]
- 10.Cuvillier O, Rosenthal D S, Smulson M E, Spiegel S. Sphingosine 1-phosphate inhibits activation of caspases that cleave poly (ADP-ribose) polymerase and lamins during Fas- and ceramide-mediated apoptosis in Jurkat T lymphocytes. J Biol Chem. 1998;273:2910–2916. doi: 10.1074/jbc.273.5.2910. [DOI] [PubMed] [Google Scholar]
- 11.de Belle I, Cai S, Kohwi-Shigematsu T. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivoin Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J Cell Biol. 1998;141:335–348. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickinson L A, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 13.Dickinson L A, Dickinson C D, Kohwi-Shigematsu T. The nuclear matrix attachment region (MAR)-binding protein SATB1 contains a homeodomain that promotes specific recognition of the core unwinding element of a MAR. J Biol Chem. 1997;272:11463–11470. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- 14.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle D A, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 16.Earnshaw W C, Martins L M, Kaufmann S H. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 17.Fanning A S, Anderson J M. Protein-protein interactions: PDZ domain networks. Curr Biol. 1996;6:1385–1388. doi: 10.1016/s0960-9822(96)00737-3. [DOI] [PubMed] [Google Scholar]
- 18.Fernández L A, Winkler M, Grosschedl R. Matrix attachment region-dependent function of the immunoglobulin μ enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol Cell Biol. 2001;21:196–208. doi: 10.1128/MCB.21.1.196-208.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 20.Forrester W C, van Genderen C, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglobulin mu gene on nuclear matrix attachment regions. Science. 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- 21.Galande S, Kohwi-Shigematsu T. Poly (ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J Biol Chem. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- 22.Gasser S M, Laemmli U K. A glimpse at chromosomal order. Trends Genet. 1987;3:16–22. [Google Scholar]
- 23.Göhring F, Schwab B L, Nicotera P, Leist M, Fackelmayer F O. The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J. 1997;16:7361–7371. doi: 10.1093/emboj/16.24.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gotzmann J, Meissner M, Gerner C. The fate of the nuclear matrix-associated-region-binding protein SATB1 during apoptosis. Cell Death Differ. 2000;7:425–438. doi: 10.1038/sj.cdd.4400668. [DOI] [PubMed] [Google Scholar]
- 25.Hattori M, Tsukamoto N, Nur-e-Kamal M S, Rubinfeld B, Iwai K, Kubota H, Maruta H, Minato N. Molecular cloning of a novel mitogen-inducible nuclear protein with a Ran GTPase-activating domain that affects cell cycle progression. Mol Cell Biol. 1995;15:552–560. doi: 10.1128/mcb.15.1.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hope I A, Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsueh Y P, Wang T F, Yang F C, Sheng M. Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature. 2000;404:298–302. doi: 10.1038/35005118. [DOI] [PubMed] [Google Scholar]
- 28.Jackson D A, Dolle A, Robertson G, Cook P R. The attachments of chromatin loops to the nucleoskeleton. Cell Biol Int Rep. 1992;16:687–696. doi: 10.1016/s0309-1651(05)80013-x. [DOI] [PubMed] [Google Scholar]
- 29.Jenuwein T, Forrester W C, Fernandez-Herero L A, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 30.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 31.Kim E, Cho K O, Rothschild A, Sheng M. Heteromultimerization and NMDA receptor-clustering activity of Chapsyn-110, a member of the PSD-95 family of proteins. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 32.Kim E, Niethammer M, Rothschild A, Jan Y N, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 33.King L B, Ashwell J D. –1995. Thymocyte and T cell apoptosis: is all death created equal? Thymus. 1994;23:209–230. [PubMed] [Google Scholar]
- 34.Kipp M, Göhring F, Ostendorp T, van Drunen C M, van Driel R, Przybylski M, Fackelmayer F O. SAF-box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol Cell Biol. 2000;20:7480–7489. doi: 10.1128/mcb.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirillov A, Kistler B, Mostsolasky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-κB-cell-specific demethylation of the Igklocus. Nat Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 36.Kohwi-Shigematsu T, Kohwi Y. Torsional stress stabilizes extended base unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry. 1990;29:9551–9560. doi: 10.1021/bi00493a009. [DOI] [PubMed] [Google Scholar]
- 37.Kohwi-Shigematsu T, deBelle I, Dickinson L A, Galande S, Kohwi Y. Identification of base-unpairing region (BUR)-binding proteins and characterization of their in vivobinding sequences. Methods Cell Biol. 1998;53:323–354. doi: 10.1016/s0091-679x(08)60885-7. [DOI] [PubMed] [Google Scholar]
- 38.Kohwi-Shigematsu T, Maass K, Bode J. A thymocyte factor SATB1 suppresses transcription of stably integrated matrix-attachment region-linked reporter genes. Biochemistry. 1997;36:12005–12010. doi: 10.1021/bi971444j. [DOI] [PubMed] [Google Scholar]
- 39.Kornau H C, Schenker L T, Kennedy M B, Seeburg P H. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 40.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 41.LeBlanc A, Liu H, Goodyer C, Bergeron C, Hammond J. Caspase-6 role in apoptosis of human neurons, amyloidogenesis, and Alzheimer's disease. J Biol Chem. 1999;274:23426–23436. doi: 10.1074/jbc.274.33.23426. [DOI] [PubMed] [Google Scholar]
- 42.Lee S H, Sheng M. Development of neuron-neuron synapses. Curr Opin Neurobiol. 2000;10:125–131. doi: 10.1016/s0959-4388(99)00046-x. [DOI] [PubMed] [Google Scholar]
- 43.Lichtenstein M, Keini G, Cedar H, Bergman Y. B cell-specific demethylation: a novel role for the intronic kappa chain enhancer sequence. Cell. 1994;76:913–923. doi: 10.1016/0092-8674(94)90365-4. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross S R, Dudley J P. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol Cell Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martins L M, Kottke T, Mesner P W, Basi G S, Sinha S, Frigon N, Jr, Tatar E, Tung J S, Bryant K, Takahashi A, Svingen P A, Madden B J, McCormick D J, Earnshaw W C, Kaufmann S H. Activation of multiple interleukin-1 converting enzyme homologues in cytosol and nuclei of HL-60 cells during etoposide-induced apoptosis. J Biol Chem. 1997;272:7421–7430. doi: 10.1074/jbc.272.11.7421. [DOI] [PubMed] [Google Scholar]
- 46.Matsumine A, Ogai A, Senda T, Okumura N, Satoh K, Baeg G H, Kawahara T, Kobayashi S, Okada M, Toyoshima K, Akiyama T. Binding of APC to the human homolog of the Drosophila discs large tumor suppressor protein. Science. 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- 47.Mirkovitch J, Mirault M-E, Laemmli U K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;9:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 48.Morais Cabral J H, Petosa C, Sutcliffe M J, Raza S, Byron O, Poy F, Marfatia S M, Chishti A H, Liddington R C. Crystal structure of a PDZ domain. Nature. 1996;382:649–652. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- 49.Nagata S. Apoptotic DNA fragmentation. Exp Cell Res. 2000;256:12–18. doi: 10.1006/excr.2000.4834. [DOI] [PubMed] [Google Scholar]
- 50.Nakagomi K, Kohwi Y, Dickinson L A, Kohwi-Shigematsu T. A novel DNA-binding motif in the nuclear matrix attachment DNA-binding protein SATB1. Mol Cell Biol. 1994;14:1852–1860. doi: 10.1128/mcb.14.3.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakagoshi H, Hoshi M, Nabeshima Y, Matsuzaki F. A novel homeobox gene mediates the Dpp signal to establish functional specificity within target cells. Genes Dev. 1998;12:2724–2734. doi: 10.1101/gad.12.17.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oberhammer F, Wilson J W, Dive C, Morris I D, Hickman J A, Wakeling A E, Walker P R, Sikorska M. Apoptotic death in epithelial cells; cleavage of DNA to 300 and/or 50kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 1993;12:3679–3684. doi: 10.1002/j.1460-2075.1993.tb06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orth K, Chinnaiya A M, Garg M, Froelich C J, Dixit V M. The CED-3/ICE-like protease Mch2 is activated during apoptosis and cleaves the death substrate lamin A. J Biol Chem. 1996;271:16443–16446. [PubMed] [Google Scholar]
- 55.Park D J, Patek P Q. Detergent and enzyme treatment of apoptotic cells for the observation of DNA fragmentation. BioTechniques. 1998;24:558–560. doi: 10.2144/98244bm07. [DOI] [PubMed] [Google Scholar]
- 56.Pellegrini L, Passer B J, Tabaton M, Ganjei J K, D'Adamio L. Alternative, nonsecretase processing of Alzheimer's-amyloid precursor protein during apoptosis by caspase-6 and -8. J Biol Chem. 1999;274:21011–21016. doi: 10.1074/jbc.274.30.21011. [DOI] [PubMed] [Google Scholar]
- 57.Poulat F, Barbara P S, Desclozeaux M, Soullier S, Moniot B, Bonneaud N, Boizet B, Berta P. The human testis determining factor SRY binds a nuclear factor containing PDZ protein interaction domains. J Biol Chem. 1997;272:7167–7172. doi: 10.1074/jbc.272.11.7167. [DOI] [PubMed] [Google Scholar]
- 58.Sattler R, Xiong Z, Lu W Y, Hafner M, MacDonald J F, Tymianski M. Specific coupling of NMDA receptor activation to nitric oxide neurotoxicity by PSD-95 protein. Science. 1999;284:1845–1848. doi: 10.1126/science.284.5421.1845. [DOI] [PubMed] [Google Scholar]
- 59.Scheuermann R H, Garrard W T. MARs of antigen receptor and co-receptor genes. Crit Rev Eukaryot Gene Expr. 1999;9:295–310. doi: 10.1615/critreveukargeneexpr.v9.i3-4.140. [DOI] [PubMed] [Google Scholar]
- 60.Shortman K, Egerton M, Spangrude G J, Scollay R. The generation and fate of thymocytes. Semin Immunol. 1990;2:3–12. [PubMed] [Google Scholar]
- 61.Stennicke H R, Salvesen G S. Caspases—controlling intracellular signals by protease zymogen activation. Biochim Biophys Acta. 2000;1477:299–306. doi: 10.1016/s0167-4838(99)00281-2. [DOI] [PubMed] [Google Scholar]
- 62.Stennicke H R, Salvesen G S. Caspases: preparation and characterization. Methods. 1999;17:313–319. doi: 10.1006/meth.1999.0745. [DOI] [PubMed] [Google Scholar]
- 63.Surh C D, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- 64.Takahashi A, Alnemri E S, Lazebnik Y A, Fernandes-Alnemri T, Litwack G, Moir R D, Goldman R D, Poirier G G, Kaufmann S H, Earnshaw W C. Cleavage of lamin A by Mch2 alpha but not CPP32: multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Natl Acad Sci USA. 1996;93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, Chapman K T, Nicholson D W. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 66.Walker R P, Sikorska M. Endonuclease activities, chromatin structure, and DNA degradation in apoptosis. Biochem Cell Biol. 1994;72:615–623. doi: 10.1139/o94-081. [DOI] [PubMed] [Google Scholar]
- 67.Wang B, Dickinson L A, Koivunen E, Ruoslahti E, Kohwi-Shigematsu T. A novel matrix attachment region DNA binding motif identified using a random phage peptide library. J Biol Chem. 1995;270:23239–23242. doi: 10.1074/jbc.270.40.23239. [DOI] [PubMed] [Google Scholar]
- 68.Washo-Stultz D, Crowley C, Payne C M, Bernstein C, Marek S, Gerner E W, Bernstein H. Increased susceptibility of cells to inducible apoptosis during growth from early to late log phase: an important caveat for in vitro apoptosis research. Toxicol Lett. 2000;116:199–207. doi: 10.1016/s0378-4274(00)00225-3. [DOI] [PubMed] [Google Scholar]
- 69.Weaver V M, Carson C E, Walker P R, Chaly N, Lach B, Raymond Y, Brown D L, Sikorska M. Degradation of nuclear matrix and DNA cleavage in apoptotic thymocytes. J Cell Sci. 1996;109:45–56. doi: 10.1242/jcs.109.1.45. [DOI] [PubMed] [Google Scholar]
- 70.Wedrychowski A, Schmidt W N, Hnilica L S. The in vivocross-linking of proteins and DNA by heavy metals. J Biol Chem. 1986;261:3370–3376. [PubMed] [Google Scholar]
- 71.Wellington C L, Singaraja R, Ellerby L, Savill J, Roy S, Leavitt B, Cattaneo E, Hackam A, Sharp A, Thornberry N, Nicholson D W, Bredesen D E, Hayden M R. Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J Biol Chem. 2000;275:19831–19838. doi: 10.1074/jbc.M001475200. [DOI] [PubMed] [Google Scholar]