Summary
Background
B cell follicles are immune-privileged sites where intensive HIV-1 replication and latency occur, preventing a permanent cure. Recent study showed that CXCR5+ NK cells in B cell follicles can inhibit SIV replication in African green monkeys, but this has not been reported in HIV-1 infected patients.
Methods
Lymphocytes and tissue sections of lymph node were collected from 11 HIV-1 positive antiretroviral therapy (ART)-naive and 19 HIV-1 negative donors. We performed immunofluorescence and RNA-scope to detect the location of CXCR5+ NK cells and its relationship with HIV-1 RNA, and performed flow cytometry and RNA-seq to analyze the frequency, phenotypic and functional characteristics of CXCR5+ NK cells. The CXCL13 expression were detected by immunohistochemistry.
Findings
CXCR5+ NK cells, which accumulated in LNs from HIV-1 infected individuals, expressed high levels of activating receptors such as NKG2D and NKp44. CXCR5+ NK cells had upregulated expression of CD107a and β-chemokines, which were partially impaired in HIV-1 infection. Importantly, the frequency of CXCR5+NK cells was inversely related to the HIV-1 viral burden in LNs. In addition, CXCL13—the ligand of CXCR5—was upregulated in HIV-1 infected individuals and positively correlated with the frequency of CXCR5+ NK cells.
Interpretation
During chronic HIV-1 infection, CXCR5+ NK cells accumulated in lymph node, exhibit altered immune characteristics and underlying anti-HIV-1 effect, which may be an effective target for a functional cure of HIV-1.
Keywords: HIV-1, CXCR5, NK cells, CXCL13, Lymph node, B cell follicle
Research in context.
Evidence before this study
It is well known that B cell follicles within lymph node (LN) serve as the primary site for HIV-1 replication, partially attributed to the exclusion of immune effector cells in B cell follicles, however, most of the studies investigating the characteristics of immune subsets are performed with blood. The functional cure for HIV-1 may result from a discovery of a new immune subset with potent anti-HIV activities, which can target the immune privilege sites. CXCR5+ NK cells were reported to migrate into and control simian immunodeficiency virus (SIV) replication in lymph node follicles in African green monkeys. However, the characteristics and functions of CXCR5+ NK have not been reported in human studies. The limited access to lymph node tissues from HIV-1 infected patients, especially viremic individuals, poses great challenges to understanding the immune response during HIV-1 infection.
Added value of this study
In this study, viremic HIV-1 infected and HIV-1 negative LN samples were applied to clarify the immune characteristics of follicle-located CXCR5+ NK cells. We showed that CXCR5+ NK cells accumulated during HIV-1 infection and negatively correlated with HIV-1 viral burden in LNs. CXCR5+ NK cells have higher expression of activating receptors such as NKG2D, NKp30, NKp44, and NKp46, and upregulated secretion of CD107a, IFN-γ and β-chemokines (CCL3, CCL4, CCL5). In addition, the upregulated CXCL13 may promote CXCR5+ NK cells accumulation in B cell follicles during HIV-1 infection.
Implications of all the available evidence
Our research, for the first time, demonstrated the accumulation, altered immune characteristics, and underlying anti-HIV-1 effect of CXCR5+ NK cells in LNs during chronic HIV-1 infection, suggesting that CXCR5+ NK cells may be a promising target for functional cure of HIV-1.
Alt-text: Unlabelled box
Introduction
HIV-1 remains a major global public health issue, and, worldwide, has claimed almost 33 million lives to date.1 Although anti-retroviral therapy (ART) effectively suppresses HIV-1 replication, HIV-1 cannot be eliminated by ART due to the persistence of latent reservoirs which are sequestered from treatment.2 B cell follicles, in particular, constitute the major sanctuary for HIV-1 and simian immunodeficiency virus, given the exclusion of immune effector cells and limited penetration of ART.3, 4, 5, 6, 7 A functional, and permanent, cure for HIV-1 may result from the discovery of a new immune subset with potent anti-HIV activities, which can target these sequestered reservoirs of viral hosts.8
NK cells play an important role in innate immune responses to HIV-1 infection, and can inhibit HIV-1 replication via cytotoxic elimination of HIV-1 infected cells, and secrete cytokines and chemokines.9, 10, 11 The role of NK cells in inhibiting HIV-1 replication and delaying disease progression is well documented. Kazer et al. found that two HIV-1 patients who maintained a low plasma viral load without ART for 33 months had a greater proliferation of NK cells within the first week of infection compared with typical progressors.12 Studies have also shown that uninfected HIV-1-exposed individuals show increased NK cell activity.13,14 Additionally, expression of HLA-B and KIR3DS1, genes involved in NK activity which are responsible for leukocyte antigens and killer cell immunoglobulin-like receptors, has been shown to delay the progression of HIV-1 infection to AIDS.15 In a humanized mouse model of HIV-1 infection, an IL-15 (a cytokine which induces NK cell proliferation) superagonist was used to effectively block HIV-1 infection in an NK cell-dependent manner.16 However, the studies mentioned above were mainly performed in PBMCs or animal models.
CXCL13/CXCR5-mediated chemotaxis is crucial for the homeostatic migration of lymphocytes into lymphoid follicles.17, 18, 19 Recently, a subtype of CD8+ T cells expressing CXCR5, termed follicular cytotoxic T cells (Tfcs), were found to accumulate in lymph node B cell follicles during chronic SIV/HIV-1 infection, and appeared to curtail viral infection.20, 21, 22, 23, 24 However, CXCR5 expression was lower in the HIV-1-specific CD8 subset, and most studies have shown that Tfcs have weak cytolytic activity, which indicates that Tfcs may instead have a role in noncytolytic mechanisms and are non-HIV-1 specific.25, 26, 27 Furthermore, compared with rhesus macaques, NK cells in SIV natural host AGMs were more frequently observed in B cell follicles than CD8T cells.28 Huot et al. identified a subset of CXCR5+ NK cells with activity in the B cell follicles of SIV-infected nonpathogenic AGMs, which could explain their lack of disease progression.29 This research extends our knowledge of the control of SIV by follicular NK cells. However, the characteristics and anti-HIV-1 activity of human follicular NK cells have not yet been reported.
Here, we comprehensively characterize the location, phenotype, and function of LN CXCR5+ NK cells in HIV-1 infected individuals. In this study, for the first time, we characterize a subset of follicle-located CXCR5+ NK cells accumulated during HIV-1 infection, which is inversely related to HIV-1 viral burden in LNs. Our data revealed that CXCR5+ NK cells may be a promising target for immune control of HIV-1 infection.
Methods
Ethics statement
This study was approved by the Institutional Review Board and Research Ethics Committees of the Fifth Medical Center of the Chinese PLA General Hospital (2016164D). Written informed consent was obtained from all participants in accordance with the Declaration of Helsinki.
Study participants
In this study, lymph node biopsies were collected from 11 untreated HIV-1 infected individuals and 19 HIV-1 negative individuals. The detailed information of these individuals is shown in Table 1. LNs from HIV-1 infected individuals were mainly from cervical, inguinal, or submandibular tissues. LNs from HIV-1 negative individuals with breast cancer, thyroid cancer, or pleomorphic adenoma were used as controls, and histopathological analysis was performed on cryosections to establish the absence of tumor infiltration in these LNs. Peripheral blood samples were obtained from 53 HIV-1+ untreated individuals and 20 HIV-1− healthy donors from the Fifth Medical Center of the Chinese PLA General Hospital (Supplementary Table 1). Exclusion criteria included pregnancy, coinfection with Hepatitis B (HBV) or Hepatitis C (HCV), and patients in a terminal condition.
Table 1.
Characteristics of study participants for LN samples.
| Patients No. | Gender | Age (year) | HIV-1 status | Specimen sites | CD4 counts (cells/ul) | pVL (copies/ml) |
|---|---|---|---|---|---|---|
| 1 | M | 24 | Positive | Inguinal | 538 | 11,400 |
| 2 | M | 22 | Positive | Cervical | 325 | 66,818 |
| 3 | M | 37 | Positive | Cervical | 460 | NA |
| 4 | M | 26 | Positive | Cervical | 197 | 18,005 |
| 5 | M | 52 | Positive | Cervical | 408 | 45,556 |
| 6 | M | 20 | Positive | Inguinal | 373 | 3115 |
| 7 | F | 54 | Positive | Submandibular | 308 | 46,220 |
| 8 | M | 65 | Positive | Cervical | 552 | 498,649 |
| 9 | M | 23 | Positive | Cervical | 136 | 1106 |
| 10 | M | 32 | Positive | Cervical | 376 | 3466 |
| 11 | M | 43 | Positive | Cervical | 419 | 24,468 |
| 12 | F | 80 | Negative | Axillary | NA | NA |
| 13 | F | 40 | Negative | Cervical | NA | NA |
| 14 | F | 49 | Negative | Axillary | NA | NA |
| 15 | F | 47 | Negative | Axillary | NA | NA |
| 16 | F | 61 | Negative | Submandibular | NA | NA |
| 17 | F | 63 | Negative | Axillary | NA | NA |
| 18 | F | 36 | Negative | Axillary | NA | NA |
| 19 | F | 46 | Negative | Cervical | NA | NA |
| 20 | F | 68 | Negative | Axillary | NA | NA |
| 21 | F | 37 | Negative | Axillary | NA | NA |
| 22 | M | 52 | Negative | Cervical | NA | NA |
| 23 | F | 28 | Negative | Cervical | NA | NA |
| 24 | F | 60 | Negative | Axillary | NA | NA |
| 25 | F | 73 | Negative | Axillary | NA | NA |
| 26 | F | 48 | Negative | Axillary | NA | NA |
| 27 | M | 26 | Negative | Cervical | NA | NA |
| 28 | F | 33 | Negative | Axillary | NA | NA |
| 29 | F | 31 | Negative | Cervical | NA | NA |
| 30 | F | 37 | Negative | Axillary | NA | NA |
NA: not available; M, male; F, female.
Sample processing and storage
PBMCs were isolated using Ficoll-Paque PLUS (GE Healthcare, Piscataway, NJ, USA) density gradient centrifugation and were used directly. Lymph node mononuclear cells (LNMCs) were collected by mechanical homogenization, all LN samples were processed uniformly. Plasma was stored at −80 °C and LNMCs were cryopreserved in liquid N2.
Flow cytometry
All flow cytometry experiments were performed in the Fifth Medical Center of the Chinese PLA General Hospital. For phenotypic staining, PBMCs or LNMCs were stained with the following antibodies: CD3-APC/Fire750, CD14-APC/Fire750, CD20-APC/Fire750, CD16-PerCP, CD56-PerCP, PD1-BV510, CD69-PE-CY7, TIGIT-PE, CD32-PE, CD57-BV605 and NKp46-BV510. These antibodies were obtained from BioLegend (San Diego, CA, USA); while CD32-FITC, CXCR5-BV421, NKG2D-PE-CY7, NKp30-AF647, and NKp44-PE antibodies were purchased from BD Biosciences (San Jose, CA, USA); NKG2A-FITC, NKG2C-PE, KIR3DL1-PE, and KIR2D-PE from Miltenyi Biotec (Bergisch Gladbach, Germany) and CD56-APC from Tongsheng Shidai (Beijing, China). For transcription factor staining, cells were fixed and permeabilized using the Foxp3/Transcription Factor Fixation/Permeabilization Concentrate and Diluent (eBioscience, San Diego, CA, USA) and stained with the following antibodies: T-BET-FITC (BioLegend), EOMES-APC (eBioscience), BCL-6-PE-CY7 (BioLegend), and TCF-1-PE (BioLegend). For intracellular cytokine staining, PBMCs were co-stimulated with 10 ng/ml IL-12 (Peprotech, Rocky Hill, NJ, USA), 20 ng/ml IL-15 (BioLegend), and 100 ng/ml IL-18 (R&D, Minneapolis, MN, USA) for 24 h. Brefeldin A and CD107a-FITC (BioLegend) were added in the last 6 h. Cells were then permeabilized and stained with the following antibodies: IFN-γ-BV510 (BD Biosciences), Perforin-PE-CY7 (BioLegend), Granzyme B-PE-CY7 (BioLegend), CCL3-FITC (eBioscience), CCL4-APC (eBioscience), and CCL5-BV421 (BD Biosciences). After, cells were stained at room temperature for 20 min and washed with FACS buffer. Flow cytometry was performed using FACS-Canto (BD Biosciences). Data were analyzed using FlowJo version X (FlowJo, Ashland, OR, USA). Detailed information on all antibodies used is provided in Supplementary Table 2.
Image flow cytometry
PBMCs were stained with the following antibodies: CD3-APC/Fire750 (BioLegend), CD14-APC/Fire750 (BioLegend), CD20-APC/Fire750 (BioLegend), CD56-APC (Tongsheng Shidai) and CXCR5-BV421 (BD Biosciences). Data were acquired using an Amnis ImageStreamX Mark II Imaging Flow Cytometer (Amnis, Seattle, WA, USA) and analyzed using the Amnis IDEAS software v6.2 (Amnis).
Enzyme-linked immunosorbent assay (ELISA)
The plasma concentrations of CXCL13 were assessed using CXCL13 ELISA kits (R&D) according to the manufacturer's instructions.
Immunohistochemistry and immunofluorescence
Paraffin-embedded sections of acetone-fixed LN biopsies were stained using the following primary antibodies: CD56 (Abcam, Cambridge, UK), CXCL13 (ZSGB-BIO, Beijing, China), CXCR5 (R&D), and CD20 (ZSGB-BIO). Multiplex immunofluorescence staining was performed using the PANO 7-plex immunochemistry kit (Panovue, Beijing, China) according to the manufacturer's instructions. In brief, sections were stained with a horseradish peroxidase-conjugated antibody, followed by tyramide signal amplification. After labeling all antigens, nuclei were stained with 4–6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, St. Louis, MO, USA). For immunohistochemistry, alkaline phosphatase staining was performed with the Mouse/Rabbit SAP Kit (ZSGB-BIO) and alkaline phosphatase substrate kit (SK-5300, Vector, Burlingame, CA, USA), and horseradish peroxidase staining was performed with HRP-conjugated antibodies and 3,3′-diaminobenzidine (ZSGB-BIO). Both kits were used according to the manufacturer's instructions. The sections were counterstained with hematoxylin for 3 min. Images were acquired with an Aperio VERSA 8 scanning system (Leica Microsystems, Wetzlar, Germany) and CXCL13 average staining intensity was determined using Aperio ImageScope software (Leica). Detailed information for all antibodies used is provided in Supplementary Table 2.
RNAscope in situ hybridization assay
The HIV-1 clade B anti-sense probe (catalog number 317,691) was designed by Advanced Cell Diagnostics (Hayward, CA, USA). The RNAscope analysis was performed as described in a previous publication.30 Images were acquired with an Aperio VERSA 8 scanning system (Leica Microsystems, Wetzlar, Germany) and RNA average immunofluorescence intensity was determined using Aperio ImageScope software (Leica).
Isolation of NK cells
Negative selection was adopted for obtaining NK cells from LNMCs/PBMCs using the NK Cell Isolation Kit (Miltenyi Biotech) according to the manufacturer's instructions. NK cells were then stained with CD3-APC/Fire750, CD14-APC/Fire750, CD20-APC/Fire750, CXCR5-BV421, and CD56-APC antibodies. Stained CXCR5+ NK and CXCR5−NK cells were then sorted using the MA900 Cell Sorter (Sony Biotechnology, Tokyo, Japan).
Transwell migration assays
For the transwell migration assays, 600 μl serum-free medium containing recombinant CXCL13 (Peprotech) at 50 ng/ml was placed in the lower chamber of a 24-well transwell plate with a pore size of 8 μm (Corning, New York, NY, USA). 5 × 105 NK cells isolated from PBMCs were added to 100 μl serum-free medium in the upper chamber. The cells were then incubated for 2 h at 37 °C. The proportion of CXCR5+NK cells in the lower chamber was determined using flow cytometry. Relative migration was calculated as the frequency of CXCR5+ NK cells in the lower chambers of the CXCL13 group divided by the control group.
HIV-1 DNA detection
Total cellular DNA was extracted from LNMCs using the Qiagen QIA Symphony DNA Mini kit (QIAGEN, Valencia, CA, USA). Then, HIV-1 DNA was determined using the SUPBIO HIV Quantitative Detection Kit (SUPBIO, Guangzhou, China). Both kits were used according to the manufacturer's instructions.
RNA-seq
Total RNA was extracted from sorted CXCR5+ and CXCR5− NK cells using a PicoPure RNA Isolation Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. Whole-transcriptome amplification and tagmentation-based library preparation were performed using SMART-seq2, followed by sequencing on a NextSeq 500 system (Illumina, San Diego, CA, USA). Transcript abundance was quantified using RSEM software (v1.2.22) supported by STAR aligner software (STAR 2.5.1b) and aligned to the GRCh38 human genome. The transcripts per million values were normalized among all samples using the upper-quantile normalization method.
Statistics
Statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software, CA, USA). The Mann-Whitney U test (unpaired) and Wilcoxon signed-rank test (paired) were used to compare median values when comparing two groups. Nonparametric Spearman correlation coefficient was used to assess the correlation between the two variables. The numbers of samples per group (n) are specified in the figure legends. In all analyses, P values < 0.05 were considered statistically significant.
Role of funding source
The funders had no role in study design, data collection, data analyses, interpretation, or writing of the report.
Results
The frequency of CXCR5+ NK cells in HIV-1 negative LNs
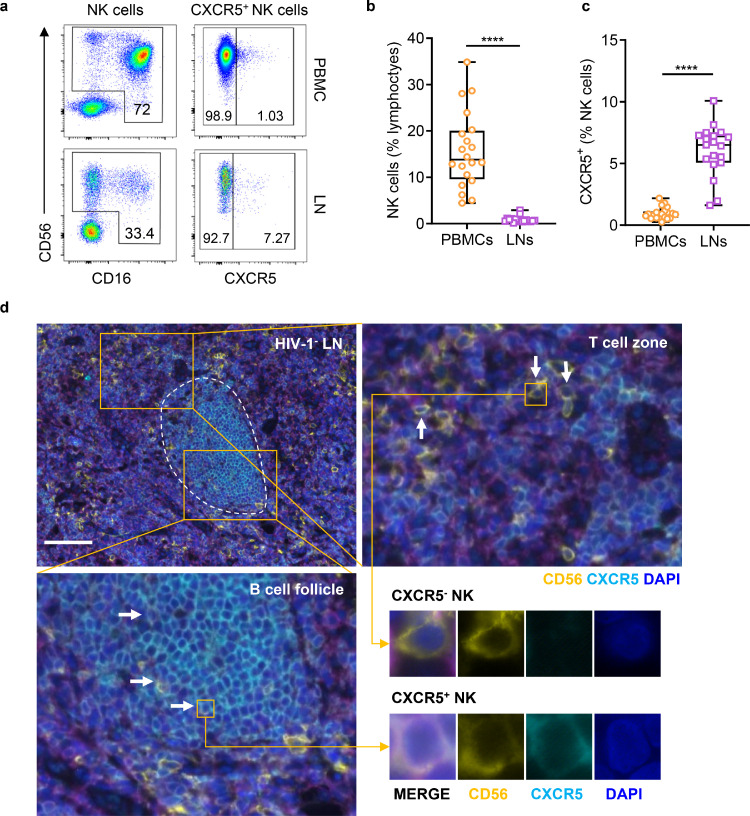
The frequency and location of CXCR5+ NK cells in human LNs are still not well understood. In this study, 16 HIV-1 negative (HIV-1−) LNs were enrolled and biopsied. Flow cytometry was used to analyze the frequency of CXCR5+NK cells. The detailed gating strategies for CXCR5+ NK cells are shown in Supplementary Fig. 1a. We found a significantly lower frequency of NK cells (in lymphocytes, P < 0.0001) and a higher frequency of CXCR5+NK cells (in total NK cells, P < 0.0001) in LNs than in PBMCs (Figure. 1a–c). Representative images from image flow cytometry of CXCR5+ NK cells from an HIV-1− individual are shown in Supplementary Fig. 1b. Multiplex immunofluorescence staining demonstrated that CXCR5+ NK cells were mainly located in B cell follicles, while their CXCR5− counterparts were instead mainly located in T cell zones (Figure. 1d).
Figure 1.
Frequency and location of CXCR5+ NK cells in HIV-1 negative LNs. (a) Representative flow cytometry plots showing the frequencies of NK cell and CXCR5+ NK cells in PBMC (upper) and LN (lower) from HIV-1− individuals. Numbers indicate the percentage of gated cells. (b) The frequencies of NK cells (in lymphocytes) from PBMCs (n = 20) and LNs (n = 17) of HIV-1− individuals. (c) The frequencies of CXCR5+ (in NK cells) from PBMCs (n = 20) and LNs (n = 19) of HIV-1− individuals. (d) Representative immunofluorescence image of HIV-1− LN (patient No. 20). Yellow boxes indicate areas with higher magnifications. Circled areas represent B cell follicle limits. White arrows indicate individual CXCR5+ NK cells. Scale bar = 100 μm. Significant differences in b, c were calculated using Mann-Whitney U test. ****P < 0.0001.
Phenotypic and functional characteristics of CXCR5+ NK cells in HIV-1 negative LNs
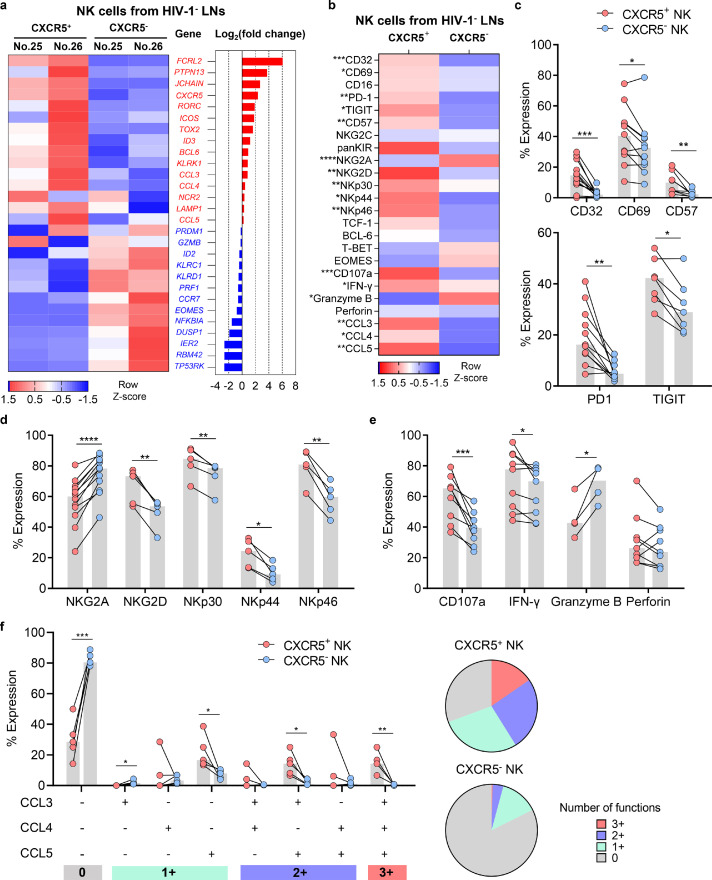
Transcriptional profiling using RNA-seq revealed that CXCR5+ NK and CXCR5− NK cells in HIV-1− LNs had distinct gene signatures (Figure. 2a). Notably, CXCR5+ NK cells expressed higher levels of genes for NK activating receptors (KLRK1), chemokines (CCL3), and lower levels of inhibitory receptors (KLRD1) and cytotoxic molecules (PRF1) than CXCR5− NK cells (Figure. 2a). It is well established that CXCR5 expression is regulated by a diverse array of transcriptional factors. The transcription factors Blimp1 and E2A directly promote the expression of CXCR5, while BCL6, TCF1, and E2A inhibitory proteins ID2 and ID3 jointly regulate the differentiation and development of Tfcs.20,21,31 Here, we also found that CXCR5+ NK cells partially shared a transcriptional profile with follicle-located cells such as Tfc and T follicular helper cell (Tfh), manifested by the higher expression of BCL6 and ID3, and the lower expression of ID2 and CCR7 (Figure. 2a).
Figure 2.
Phenotypic and functional profiles of CXCR5+ and CXCR5− NK cells in HIV-1 negative LNs. (a) RNA-seq were performed to investigate the relative expression of indicated genes in CXCR5+ and CXCR5− NK cells from two HIV-1− LNs. (b) Flow cytometry were performed to investigate the protein expression phenotypes of CXCR5+ and CXCR5− NK cells from HIV-1− LNs. (c) The expression of CD69 (n = 11), CD32 (n = 12), CD57 (n = 8), PD-1 (n = 11) and TIGIT (n = 7) on CXCR5+ and CXCR5− NK cells from HIV-1− LNs. (d) The expression of NK surface receptors (NKG2A, n = 12; NKG2D, n = 5; NKp30, n = 5; NKp44, n = 5; NKp46, n = 5) on CXCR5+ and CXCR5− NK cells from HIV-1− LNs. (e-f) The expression of (e) CD107a (n = 9), IFN-γ (n = 9), granzyme B (n = 4), perforin (n = 9) and (f) β-chemokines (CCL3, CCL4 and CCL5, n = 5) in CXCR5+ and CXCR5− NK cells which were stimulated with IL-12 (10 ng/ml), IL-15 (20 ng/ml) and IL-18 (100 ng/ml) for 16 h. Significant differences in b, c, d, e, and f were calculated using Wilcoxon signed-rank test. *P < 0.05, ** P < 0.01, ***P < 0.001, ****P < 0.0001.
Using flow cytometry (Figure. 2b), we observed increased expression in HIV-1− LNs of CD69 (P = 0.042), CD32 (P = 0.001), CD57 (P = 0.0078), PD-1 (P = 0.02), and TIGIT (P = 0.0313) in CXCR5+NK cells compared to their CXCR5− counterparts (Figure. 2c). As NK cell function is modulated by a variety of activating/inhibitory receptors such as natural cytotoxic receptor (NCR) and the NKG2/CD94 heterodimer family (C-type lectin-like receptors),32,33 we surprisingly observed higher expression of NK cell-activating receptors in CXCR5+ NK cells, including NKG2D (P = 0.0047), NKp30 (P = 0.0033), NKp44 (P = 0.0103), and NKp46 (P = 0.0013), while expression of the inhibitory receptor NKG2A was lower (P < 0.0001) (Figure. 2d). In vitro stimulation found that, compared with their CXCR5− counterparts, CXCR5+ NK cells produced higher levels of CD107a (P = 0.0007) and IFN-γ (P = 0.0385), as well as lower levels of granzyme B (P = 0.0196) (Figure. 2e). Interestingly, we also observed significantly higher expression of CCL3, CCL4, and CCL5 in CXCR5+ NK cells (Figure. 2b, f). These β-chemokines are natural ligands of CCR5, and can impede HIV-1 infection by competitively binding to CCR5.34, 35, 36 Thus, our results demonstrate that CXCR5+ NK cells displayed robust secretory function and increased expression of activating receptors compared to their CXCR5− counterparts.
In addition, we characterized CXCR5+ NK cells in HIV-1− PBMCs. Compared with CXCR5− NK cells, the phenotypic and functional properties of CXCR5+ NK cells were almost consistent with NK cells in HIV-1− LNs, other than a significantly lower expression of perforin and CD16, while NKG2A and CD57 was not significant in PBMCs (Supplementary Fig. 2a).
As NK cells are highly heterogeneous, with diverse immune functions which largely depend on their tissue residency,37,38 the characteristics of CXCR5+ NK cells from PBMCs and LNs were compared. We found increased expression of CD69, CD32, NKG2A, NKp44, and CD107a in CXCR5+ NK cells from LNs than from PBMCs, while the expression of CD16, CD57, NKG2C, granzyme B and perforin was lower in the former (Supplementary Fig. 2b).
Collectively, these results demonstrate that CXCR5+ NK cells from HIV-1 individuals have distinct phenotypic and functional characteristics compared to CXCR5− NK cells, and might exert potent anti-HIV-1 activity in LNs.
Altered frequency, phenotypic and functional characteristics of CXCR5+ NK cells in HIV-1 infection
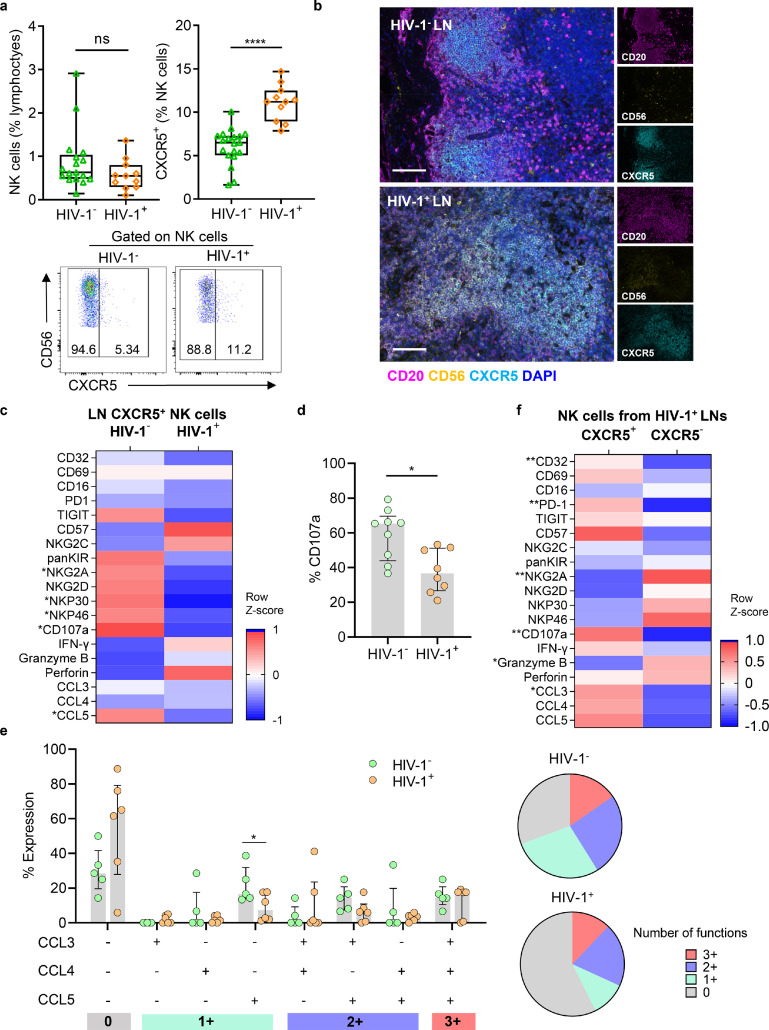
To clarify the effects of HIV-1 infection on CXCR5+ NK cells, we further compared the frequencies of LN CXCR5+ NK cells from untreated HIV-1 infected (HIV-1+) and HIV-1− individuals. Although the overall frequencies of LN NK cells were similar between the two groups, there was a significantly higher frequency of LN CXCR5+ NK cells in HIV-1+ individuals than in HIV-1− individuals (Fig. 3a). Notably, CXCR5+ NK cells had accumulated in the B cell follicles of HIV-1+ individuals and not of HIV-1− individuals (Figure. 3b). In addition, we found that NK number per area of GC was higher in HIV-1+ individuals than HIV-1− individuals (Supplementary Fig. 3a). Consistently, we also found that an increase in the frequency of peripheral CXCR5+ NK cells correlated with HIV-1 infection (Supplementary Fig. 3b).
Figure 3.
Effect of HIV-1 infection on frequency, phenotypic and functional features of CXCR5+ NK cells in LNs. (a) The frequencies of NK cells (in lymphocytes, left) and CXCR5+ (in NK cells, right) from HIV-1− (n = 19) and HIV-1+ LNs (n = 11). Representative flow cytometry plots showing the frequencies of CXCR5+ NK cells from HIV-1− LN (left) and HIV-1+LN (right). Numbers indicate the percentage of gated cells. (b) Representative immunofluorescence images showing the distribution of CXCR5+ NK cells in HIV-1− (left, patient No. 20) and HIV-1+ (right, patient No. 4) LNs. (c) Flow cytometry were performed to investigate the protein expression phenotypes of CXCR5+ NK cells from HIV-1− and HIV-1+ LNs. (d) The expression of CD107a in CXCR5+ NK cells from HIV-1− and HIV-1+ LNs, data represented as median with interquartile range. (e) Co-expression of β-chemokines (CCL3, CCL4 and CCL5) in CXCR5+ NK cells between HIV-1− and HIV-1+ LNs, data represented as median with interquartile range. (f) Flow cytometry were performed to investigate the phenotypic and functional features of CXCR5+ and CXCR5− NK cells from HIV-1+ LNs. Scale bar = 100 μm. Significant differences in a, d, and e were calculated using Mann-Whitney U test, and g, f were calculated using Wilcoxon signed-rank test. *P < 0.05, ** P < 0.01, ****P < 0.0001.
The dysfunction of circulating NK cells in HIV-1 infection has been widely reported, but the effects of HIV-1 infection on NK cells in LNs remain unclear. We compared the phenotypic and functional profiles of CXCR5+ NK cells from HIV-1+ and HIV-1− LNs (Figure. 3c). Significantly lower expression of CD107a (P = 0.0111) and CCL5 (P = 0.0482) were found in HIV-1+ LN CXCR5+ NK cells than in HIV-1− LN NK cells (Figure. 3d, e). We also observed a lower expression of CCL5 in HIV-1+ peripheral CXCR5+ NK cells than in their HIV-1− counterparts (Supplementary Fig. 3c, d). Besides, CXCR5+ NK cells in HIV-1+ PBMCs had lower expression of IFN-γ but higher expression of TIGIT, perforin, and CCL4 than in HIV-1− PBMCs (Supplementary Fig. 3c, d). Notably, the expression of CD107a in peripheral CXCR5+ NK cells was negatively correlated with plasma HIV-1 viral load, while the correlation was not significant in CXCR5− NK cells (Supplementary Fig. 3e).
We have previously compared the features of CXCR5+ and CXCR5− NK cells from HIV-1− LNs (Figure. 2b), but given the alterations in CXCR5+ NK cell functionality during HIV-1 infection, further characterization of CXCR5+ and CXCR5− NK cells from HIV-1+ LNs was necessary. Consistent with HIV-1− LNs, we also found higher expression levels of CD32, PD-1, CD107a, and CCL3; and lower expression of NKG2A and granzyme B on CXCR5+ NK cells from HIV-1+ LNs compared with their CXCR5− counterparts, while the expression of CD69, TIGIT, CD57, IFN-γ, CCL4, and CCL5 were not significantly different (Figure. 3f). In addition, we compared the frequency and characteristics of CXCR5+cells between HIV-1+ PBMCs and LNs. Consistent with HIV-1− individuals, we also observed a higher CXCR5+ NK cell frequency in HIV-1+ LNs than paired PBMCs (Supplementary Fig. 3f), and found increased expression of CD69, CD32, NKG2A, and CD107a; and decreased expression of granzyme B and perforin (Supplementary Fig. 3g). In summary, our results indicate that, although HIV-1 provokes a dysfunction of cytokine production in CXCR5+ NK cells, production is still significantly higher than in CXCR5− NK cells.
CXCR5+ NK cells negatively correlated with HIV-1 viral burden in LNs
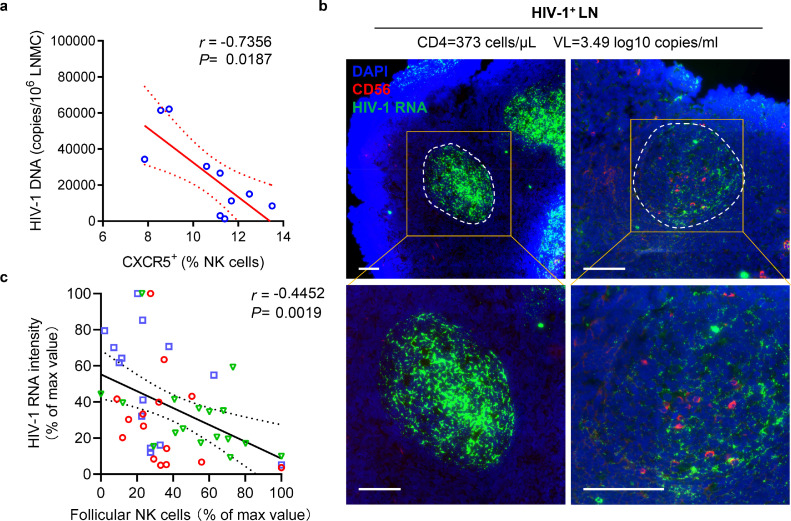
Since HIV-1 is mainly replicated in B cell follicles, we examined the association of CXCR5+ NK cells with HIV-1 viral load in LNs. We found that there was a negative correlation between CXCR5+ NK frequencies and HIV-1 DNA levels (P = 0.0187, r = −0.7356; Figure. 4a). Furthermore, by performing HIV-1 RNAscope and immunofluorescence staining, we confirmed that HIV-1 RNA-positive cells were enriched in B cell follicles (Supplementary Fig. 4), which was consistent with our previous work.39 Importantly, we observed a negative correlation between NK cell count and HIV-1 RNA signal intensity inside the follicles (Figure. 4b, c). NK cells in B cell follicles exclusively express CXCR5, so are regarded as CXCR5+NK cells. These results suggest a relation between the accumulation of CXCR5+ NK cells and the suppression of HIV-1 in B cell follicles.
Figure 4.
Frequency of CXCR5+ NK cells in LNs is inversely related to virus burden in chronic HIV-1 infection. (a) Inverse correlation between CXCR5+ NK frequencies and HIV-1 DNA levels (n = 10). (b) Representative immunofluorescence images showing the distribution of follicular NK cells (red) and viral RNA (green) in HIV-1+ LNs (patient No. 7). (c) Correlation between average NK count and HIV-1 RNA MFI in follicles (n = 46) from three HIV-1+ LNs. HIV-1 RNA intensity and NK cell count per follicle area were expressed as the percentage of max value, each color representing a sample. Correlation in a, c were calculated using Spearman correlation coefficient. P and r values are presented. Scale bar = 100 μm.
Upregulated CXCL13 was associated with CXCR5+ NK cells during HIV-1 infection
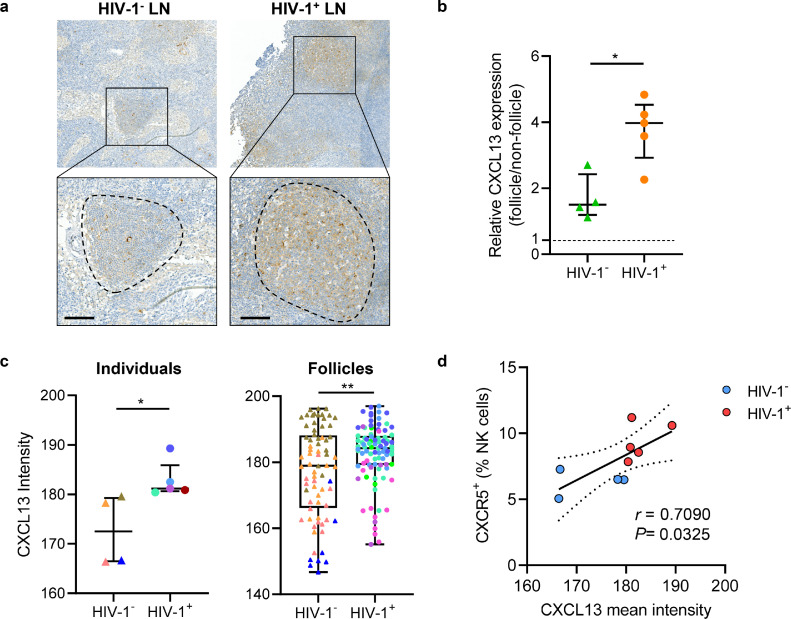
CXCL13, a ligand of CXCR5, plays an important role in the migration of follicular cells into B cell follicles.19 An in vitro transwell assay confirmed that CXCR5+ NK cells can migrate towards CXCL13 (Supplementary Fig. 5a). Immunohistochemical staining (Figure. 5a) showed CXCL13 expression inside B cell follicles is about 1.5 folds higher than outside, and the difference was more pronounced in HIV-1 infected LNs (Figure. 5b). Furthermore, there was greater expression of CXCL13 in the total LN or follicle area in HIV-1+ sections than in HIV-1− sections (Figure. 5c). Since CXCR5+ NK cells accumulated during HIV-1 infection, we further explored the relationship between CXCL13 and CXCR5+ NK cells, and found that the intensity of CXCL13 expression positively correlated with the frequency of LN CXCR5+ NK cells (P = 0.0325, r = 0.7090; Figure. 5d). In addition, we also found increased plasma CXCL13 levels in HIV-1+ individuals than in HIV-1− individuals (P < 0.001) (Supplementary Fig. 5b). There was also a positive correlation between plasma CXCL13 concentration and the peripheral CXCR5+ NK cell frequency (P = 0.0012, r = 0.6717; Supplementary Fig. 5c). Our results suggest that expression of CXCL13 was induced in LNs upon HIV-1 infection, and the increased concentration gradient of CXCL13 in B cell follicles may promote CXCR5+ NK cell enrichment through chemotaxis.
Figure 5.
Increased CXCL13 expression level in LNs during HIV-1 chronic infection. (a) Representative immunohistochemistry staining of CXCL13 in HIV-1− (left, patient No. 20) and HIV-1+ (right, patient No. 6) LNs, circled areas denoting a B cell follicle. (b) Relative CXCL13 expression were calculated as the ratio of the average intensity per unit area between follicle area and non-follicle area. (c) CXCL13 expression of HIV-1− and HIV-1+ individuals (left) and follicles (right). The follicles from the same individual are displayed in the same color, data represented as median with interquartile range (left). (d) Correlation between mean CXCL13 intensity and CXCR5+ NK frequencies from HIV-1+ (red) and HIV-1− (blue) LNs. Scale bar = 100 μm. Significant differences in b, c were calculated using Mann-Whitney U test. Correlation in d were calculated using Spearman correlation coefficient. *P < 0.05, ** P < 0.01.
Discussion
Although NK cells play an important role during HIV-1 infection, there is a relative scarcity of research focusing on the anti-HIV-1 effect of NK cells in LNs due to the lack of access to HIV-1 infected samples. Previous studies have identified a population of CXCR5+ NK cells in B cell follicles that could inhibit SIV replication and potentially contribute to the lack of disease progression in nonpathogenic AGMs.29 This has promise for the development of novel HIV-1 treatments, and requires further characterization of CXCR5+ NK cells in human LNs. In this study, we demonstrated the location and functional profiles of CXCR5+ NK cells in untreated HIV-1 infected and HIV-1 negative individuals, revealing that CXCR5+ NK cells may inhibit HIV-1 virus proliferation in B cell follicles in a noncytolytic manner. Our findings showed that CXCR5+ NK cells may be a promising target for the future development of a functional cure for HIV-1.
The CXCR5/CXCL13 axis is required for the migration of immune cells into B cell follicles. In our study, we observed a concomitant increase in the frequency of CXCR5+ NK cells and CXCL13 expression levels in LNs from HIV-1+ individuals compared to HIV-1− individuals. Although CXCL13 expression is induced in many cell types under inflammatory conditions,19 Tfh cells were reported to uniquely produce CXCL13 in HIV-1-infected secondary lymphoid tissues.40 Consistently, our results indicate that CXCL13 expression is mainly confined to B cell follicles. Furthermore, we observed a positive correlation between the frequency of CXCR5+ NK cells and the intensity of CXCL13 expression. As CXCL13 could induce the expression of CXCR5 mRNA in PBMCs,41 the accumulation of CXCR5+ NK cells may be attributed, at least in part, to the increased CXCL13 in HIV-1 infected LNs. In addition, other CXCR5-expressing immune cells such as Tfh and Tfc cells have also been reported to accumulate in LNs upon HIV-1 infection.24,42 Chronic HIV-1 infection is characterized by persistent immune activation and inflammation, and the inflammatory milieu may also promote the expansion of CXCR5 expressing immune cells during chronic HIV-1 infection. TGF-β has been shown to co-opt STAT3-STAT4 signaling to promote Tfh cell differentiation.43 TGF-β can also significantly increase the expression of CXCR5 in CD8+ T cells.23,44 In addition, chronic inflammation may also induce the generation and expansion of adaptive NK cells expressing TCF7, a transcription factor involved in the regulation of CXCR5.45 Taken together, the inflammation which accompanies HIV-1 replication or microbial translocation may lead to the accumulation of CXCR5+ NK cells in LNs, to a certain degree.
Abundant research has delineated the effect of HIV-1 infection on peripheral NK cell functions and phenotypes, but there are comparatively few studies concerning the role of LN NK cells in HIV-1 infected patients. In addition, there are many NK cell subtypes, as their phenotypes vary according to their tissue origin and NK cells are widely distributed throughout the human body.37,46 In this study, we found that CXCR5+NK cells have a tissue-resident phenotype, characterized by greater expression of CD69 and PD-1, and stronger functional activity with higher expression of CD107a, which is consistent with the phenotype of CXCR5+ NK cells in SIV-infected AGMs.29 CD69 has been confirmed to play an important role in maintaining immune cells in tissues.47,48 As high levels of CD69 are expressed by NK cells in several tissues, it can be considered a relevant marker for tissue-resident NK cells.49 PD-1 and TIGIT are reported to be highly co-expressed in lymphatic tissue, and we observed greater expression in CXCR5+ NK cells.50 Notably, we also observed greater expression of the activating receptors NKG2D, NKp30, NKp44, and NKp46. Collectively, these results demonstrate that CXCR5+ NK cells display strong anti-HIV-1 and tissue-resident phenotypes. However, HIV-1 infection has been reported to induce the dysfunction of circulating NK cells, characterized by the expansion of a dysfunctional CD56negative subset, downregulation of surface activating receptors, impaired cytotoxicity, and inhibited secretion of chemokines and cytokines.51, 52, 53 In our study, we also found that the expression of CD107a and β-chemokines in LN CXCR5+NK cells decreased during HIV-1 infection. Despite the functional abnormalities, in HIV-1+LNs, we still found higher expression of CD32, CD107a, and CCL3, and lower expression of NKG2A in CXCR5+ NK cells than in their CXCR5− counterparts (Figure. 3f).
There was an inverse relationship between CXCR5+ NK cell count and HIV-1 DNA/RNA levels within B cell follicles, suggesting that this subset may exert an inhibitory effect on the HIV-1 virus. In addition to the natural cytotoxicity regulated by surface receptors, NK cells also exert anti-HIV-1 activity by producing cytokines such as IFN-γ and β-chemokines. Here, we observed a greater secretion of IFN-γ and β-chemokines in CXCR5+ NK cells than in their CXCR5− counterparts, while granzyme B expression were observed to be lower in CXCR5+ NK cells. Thus, we speculate that CXCR5+ NK cells may inhibit the HIV-1 virus via a noncytolytic mechanism. Noncytolytic mechanisms of immune control have been previously described in both HIV-1 and SIV infections.27,54 Higher IFN-γ production by NK cells was associated with delayed HIV-1 disease progression in long-term non-progressors (LTNPs),55 and secretion of βchemokines by NK cells hampers the HIV-1 infectivity of target cells. In addition, studies have shown that β-chemokines and IFN-γ promote the accumulation of NK cells in stimulated lymphatic tissues.56, 57, 58 Even though our results indicated that CXCR5+ NK cells exhibit robust secretion functions, the mechanism by which CXCR5+ NK cells inhibit HIV-1 remains to be further explored. Although we observed the negative correlation between follicular NK cells with HIV-DNA/RNA levels within lymph nodes, the presence of CXCR5+ NK cells is not sufficient to stall disease progression. As some patients have quite advanced disease progression and HIV viral RNA was observed in all assessed lymph nodes. This is in stark contrast to the virtual absence of viral RNA in AGM lymph node, and the presence of CXCR5+ NK cells has been linked to viral control within the lymph node and a lack of disease progression in SIV-infected AGM.29 Thus, additional immune factors may be present in AGM, but absent in HIV-1-infected humans, are required to prevent disease progression which warrants further investigation.
Our study has several limitations. First, there are gender and regional differences in the source of lymph node samples. Moreover, although we confirmed the absence of tumor cells in HIV-1 negative LNs, we cannot rule out the possibilities that cancer would impact the result. In addition, limited access to enough LN CXCR5+ NK cells impedes the in vitro co-culture assay to validate anti-HIV-1 effect of CXCR5+ NK cells. Furthermore, we lacked longitudinal LN samples that could provide definitive data concerning the effect of HIV-1 infection on CXCR5+ NK cells. Besides, the effect of CXCR5+ NK cells in different phases of infection, especially in the early stage, and in other HIV-1-enriched tissues, requires further exploration.
In this study, using untreated HIV-1 infected LN samples, we clarified for the first time the locational, phenotypic, and functional characteristics of human CXCR5+ NK cells, and revealed their dynamics during chronic HIV-1 infection. Our results indicate that B cell follicle-located CXCR5+ NK cells accumulate during HIV-1 infection and their cell count negatively correlates with HIV-1 viral load in LNs. This research extends our understanding of HIV-1 inhibition by follicular NK cells in secondary lymphoid tissues, providing a novel target for a functional cure of HIV-1.
CRediT authorship contribution statement
An-Liang Guo: Conceptualization, Writing – original draft, Visualization, Methodology, Formal analysis, Supervision. Yan-Mei Jiao: Resources, Data curation, Conceptualization. Qi-Wen Zhao: Resources, Data curation, Conceptualization. Hui-Huang Huang: Resources, Data curation, Conceptualization. Jian-Ning Deng: Resources, Data curation, Conceptualization. Chao Zhang: Project administration, Conceptualization. Xing Fan: Methodology, Project administration, Conceptualization. Ruo-Nan Xu: Project administration, Conceptualization. Ji-Yuan Zhang: Project administration, Conceptualization. Cheng Zhen: Project administration, Conceptualization. Zhi-Man Xie: Resources, Data curation, Conceptualization. Ying-Mei Qin: Resources, Data curation, Conceptualization. Jian-Qing Xu: Resources, Data curation, Conceptualization. Yu Yang: Resources, Data curation, Conceptualization. Ming Shi: Project administration, Conceptualization. Lei Huang: Conceptualization, Writing – original draft, Resources, Data curation. Jin-Wen Song: Conceptualization, Writing – original draft, Visualization, Methodology, Formal analysis, Supervision. Fu-Sheng Wang: Conceptualization, Writing – original draft.
Declaration of Competing Interest
The authors have declared that no conflict of interest exists.
Acknowledgments
Data sharing
Data from this study is available from the corresponding author upon reasonable request.
Acknowledgments
We thank Song-Shan Wang, Chun-Bao Zhou, Jin-Hong Yuan, and Xin-Xin Yang (The Fifth Medical Center of the PLA General Hospital) for their technical support with flow cytometry and pathological analysis. This study was supported by National Natural Science Foundation of China (grant no. 82101837), Innovation Groups of the National Natural Science Foundation of China (grant no. 81721002), and Peking University Clinical Scientist Program Special (grant no. BMU2019LCKXJ013).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103794.
Contributor Information
Lei Huang, Email: huangleiwa@sina.com.
Jin-Wen Song, Email: songjinwenchina@yeah.net.
Fu-Sheng Wang, Email: fswang302@163.com.
Appendix. Supplementary materials
References
- 1.UNAIDS . UNAIDS; 2021. Global HIV & AIDS Statistics-Fact Sheet Global HIV & AIDS Statistics-Fact Sheet.https://www.unaids.org/en/resources/fact-sheet [Available from. [Google Scholar]
- 2.Siliciano J.D., Kajdas J., Finzi D., Quinn T.C., Chadwick K., Margolick J.B., et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9(6):727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 3.Perreau M., Savoye A.L., De Crignis E., Corpataux J.M., Cubas R., Haddad E.K., et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210(1):143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banga R., Procopio F.A., Noto A., Pollakis G., Cavassini M., Ohmiti K., et al. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med. 2016;22(7):754–761. doi: 10.1038/nm.4113. [DOI] [PubMed] [Google Scholar]
- 5.Boritz E.A., Darko S., Swaszek L., Wolf G., Wells D., Wu X., et al. Multiple origins of virus persistence during natural control of HIV infection. Cell. 2016;166(4):1004–1015. doi: 10.1016/j.cell.2016.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connick E., Mattila T., Folkvord J.M., Schlichtemeier R., Meditz A.L., Ray M.G., et al. CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue. J Immunol. 2007;178(11):6975–6983. doi: 10.4049/jimmunol.178.11.6975. [DOI] [PubMed] [Google Scholar]
- 7.Fukazawa Y., Lum R., Okoye A.A., Park H., Matsuda K., Bae J.Y., et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21(2):132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitman M.C., Lau J.S.Y., McMahon J.H. Lewin SR. Barriers and strategies to achieve a cure for HIV. Lancet HIV. 2018;5(6):e317–ee28. doi: 10.1016/S2352-3018(18)30039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altfeld M., Gale M. Innate immunity against HIV-1 infection. Nat Immunol. 2015;16(6):554–562. doi: 10.1038/ni.3157. [DOI] [PubMed] [Google Scholar]
- 10.Florez-Alvarez L., Hernandez J.C., Zapata W. NK cells in hiv-1 infection: from basic science to vaccine strategies. Front Immunol. 2018;9:2290. doi: 10.3389/fimmu.2018.02290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjorkstrom N.K., Strunz B., Ljunggren H.G. Natural killer cells in antiviral immunity. Nat Rev Immunol. 2021 doi: 10.1038/s41577-021-00558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazer S.W., Aicher T.P., Muema D.M., Carroll S.L., Ordovas-Montanes J., Miao V.N., et al. Integrated single-cell analysis of multicellular immune dynamics during hyperacute HIV-1 infection. Nat Med. 2020;26(4):511–518. doi: 10.1038/s41591-020-0799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott-Algara D., Truong L.X., Versmisse P., David A., Luong T.T., Nguyen N.V., et al. Cutting edge: increased NK cell activity in HIV-1-exposed but uninfected Vietnamese intravascular drug users. J Immunol. 2003;171(11):5663–5667. doi: 10.4049/jimmunol.171.11.5663. [DOI] [PubMed] [Google Scholar]
- 14.Tiemessen C.T., Shalekoff S., Meddows-Taylor S., Schramm D.B., Papathanasopoulos M.A., Gray G.E., et al. Cutting edge: unusual NK cell responses to HIV-1 peptides are associated with protection against maternal-infant transmission of HIV-1. J Immunol. 2009;182(10):5914–5918. doi: 10.4049/jimmunol.0900419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M.P., Gao X., Lee J.H., Nelson G.W., Detels R., Goedert J.J., et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31(4):429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 16.Seay K., Church C., Zheng J.H., Deneroff K., Ochsenbauer C., Kappes J.C., et al. In vivo activation of human NK cells by treatment with an interleukin-15 superagonist potently inhibits acute in vivo HIV-1 infection in humanized mice. J Virol. 2015;89(12):6264–6274. doi: 10.1128/JVI.00563-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cyster J.G. Chemokines and cell migration in secondary lymphoid organs. Science. 1999;286(5447):2098–2102. doi: 10.1126/science.286.5447.2098. [DOI] [PubMed] [Google Scholar]
- 18.Reif K., Ekland E.H., Ohl L., Nakano H., Lipp M., Forster R., et al. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature. 2002;416(6876):94–99. doi: 10.1038/416094a. [DOI] [PubMed] [Google Scholar]
- 19.Allen C.D., Okada T., Cyster J.G. Germinal-center organization and cellular dynamics. Immunity. 2007;27(2):190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leong Y.A., Chen Y., Ong H.S., Wu D., Man K., Deleage C., et al. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol. 2016;17(10):1187–1196. doi: 10.1038/ni.3543. [DOI] [PubMed] [Google Scholar]
- 21.He R., Hou S., Liu C., Zhang A., Bai Q., Han M., et al. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature. 2016;537(7620):412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 22.Im S.J., Hashimoto M., Gerner M.Y., Lee J., Kissick H.T., Burger M.C., et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537(7620):417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mylvaganam G.H., Rios D., Abdelaal H.M., Iyer S., Tharp G., Mavigner M., et al. Dynamics of SIV-specific CXCR5+ CD8 T cells during chronic SIV infection. Proc Natl Acad Sci USA. 2017;114(8):1976–1981. doi: 10.1073/pnas.1621418114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrovas C., Ferrando-Martinez S., Gerner M.Y., Casazza J.P., Pegu A., Deleage C., et al. Follicular CD8 T cells accumulate in HIV infection and can kill infected cells in vitro via bispecific antibodies. Sci Transl Med. 2017;9(373) doi: 10.1126/scitranslmed.aag2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuter M.A., Del Rio Estrada P.M., Buggert M., Petrovas C., Ferrando-Martinez S., Nguyen S., et al. HIV-specific CD8+ T cells exhibit reduced and differentially regulated cytolytic activity in lymphoid tissue. Cell Rep. 2017;21(12):3458–3470. doi: 10.1016/j.celrep.2017.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miles B., Miller S.M., Folkvord J.M., Levy D.N., Rakasz E.G., Skinner P.J., et al. Follicular Regulatory CD8 T cells impair the germinal center response in SIV and ex vivo HIV infection. PLoS Pathog. 2016;12(10) doi: 10.1371/journal.ppat.1005924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen S., Deleage C., Darko S., Ransier A., Truong D.P., Agarwal D., et al. Elite control of HIV is associated with distinct functional and transcriptional signatures in lymphoid tissue CD8(+) T cells. Sci Transl Med. 2019;11(523) doi: 10.1126/scitranslmed.aax4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrando-Martinez S., Moysi E., Pegu A., Andrews S., Nganou Makamdop K., Ambrozak D., et al. Accumulation of follicular CD8+ T cells in pathogenic SIV infection. J Clin Invest. 2018;128(5):2089–2103. doi: 10.1172/JCI96207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huot N., Jacquelin B., Garcia-Tellez T., Rascle P., Ploquin M.J., Madec Y., et al. Natural killer cells migrate into and control simian immunodeficiency virus replication in lymph node follicles in African green monkeys. Nat Med. 2017;23(11):1277–1286. doi: 10.1038/nm.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song J.W., Zhang C., Fan X., Meng F.P., Xu Z., Xia P., et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat Commun. 2020;11(1):3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu D., Ye L. A portrait of CXCR5(+) follicular cytotoxic CD8(+) T cells. Trends Immunol. 2018;39(12):965–979. doi: 10.1016/j.it.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Lanier L.L. NK cell receptors. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 33.Bonaparte M.I., Barker E. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood. 2004;104(7):2087–2094. doi: 10.1182/blood-2004-02-0696. [DOI] [PubMed] [Google Scholar]
- 34.Fehniger T.A., Herbein G., Yu H., Para M.I., Bernstein Z.P., O'Brien W.A., et al. Natural killer cells from HIV-1+ patients produce C-C chemokines and inhibit HIV-1 infection. J Immunol. 1998;161(11):6433–6438. [PubMed] [Google Scholar]
- 35.Oliva A., Kinter A.L., Vaccarezza M., Rubbert A., Catanzaro A., Moir S., et al. Natural killer cells from human immunodeficiency virus (HIV)-infected individuals are an important source of CC-chemokines and suppress HIV-1 entry and replication in vitro. J Clin Invest. 1998;102(1):223–231. doi: 10.1172/JCI2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner L., Yang O.O., Garcia-Zepeda E.A., Ge Y., Kalams S.A., Walker B.D., et al. Beta-chemokines are released from HIV-1-specific cytolytic T-cell granules complexed to proteoglycans. Nature. 1998;391(6670):908–911. doi: 10.1038/36129. [DOI] [PubMed] [Google Scholar]
- 37.Hashemi E., Malarkannan S. Tissue-resident NK cells: development, maturation, and clinical relevance. Cancers. 2020;12(6) doi: 10.3390/cancers12061553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huot N., Rascle P., Petitdemange C., Contreras V., Palgen J.L., Stahl-Hennig C., et al. Non-human primate determinants of natural killer cells in tissues at steady-state and during simian immunodeficiency virus infection. Front Immunol. 2020;11:2134. doi: 10.3389/fimmu.2020.02134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C., Song J.W., Huang H.H., Fan X., Huang L., Deng J.N., et al. NLRP3 inflammasome induces CD4+ T cell loss in chronically HIV-1-infected patients. J Clin Invest. 2021;131(6) doi: 10.1172/JCI138861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Havenar-Daughton C., Lindqvist M., Heit A., Wu J.E., Reiss S.M., Kendric K., et al. CXCL13 is a plasma biomarker of germinal center activity. Proc Natl Acad Sci USA. 2016;113(10):2702–2707. doi: 10.1073/pnas.1520112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cagigi A., Mowafi F., Phuong Dang L.V., Tenner-Racz K., Atlas A., Grutzmeier S., et al. Altered expression of the receptor-ligand pair CXCR5/CXCL13 in B cells during chronic HIV-1 infection. Blood. 2008;112(12):4401–4410. doi: 10.1182/blood-2008-02-140426. [DOI] [PubMed] [Google Scholar]
- 42.Lindqvist M., van Lunzen J., Soghoian D.Z., Kuhl B.D., Ranasinghe S., Kranias G., et al. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J Clin Invest. 2012;122(9):3271–3280. doi: 10.1172/JCI64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitt N., Liu Y., Bentebibel S.E., Munagala I., Bourdery L., Venuprasad K., et al. The cytokine TGF-beta co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat Immunol. 2014;15(9):856–865. doi: 10.1038/ni.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H.G., Jiao Y.M., Huang H.H., Zhang C., Zhang J.Y., Xu R.N., et al. Transforming growth factor-beta promotes the function of HIV-specific CXCR5(+) CD8 T cells. Microbiol Immunol. 2020;64(6):458–468. doi: 10.1111/1348-0421.12789. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Lifshitz L., Gellatly K., Vinton C.L., Busman-Sahay K., McCauley S., et al. HIV-1-induced cytokines deplete homeostatic innate lymphoid cells and expand TCF7-dependent memory NK cells. Nat Immunol. 2020;21(3):274–286. doi: 10.1038/s41590-020-0593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J., Tian Z., Peng H. Tissue-resident NK cells and other innate lymphoid cells. Adv Immunol. 2020;145:37–53. doi: 10.1016/bs.ai.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Shiow L.R., Rosen D.B., Brdickova N., Xu Y., An J., Lanier L.L., et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440(7083):540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 48.Mackay L.K., Braun A., Macleod B.L., Collins N., Tebartz C., Bedoui S., et al. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J Immunol. 2015;194(5):2059–2063. doi: 10.4049/jimmunol.1402256. [DOI] [PubMed] [Google Scholar]
- 49.Bjorkstrom N.K., Ljunggren H.G., Michaelsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immunol. 2016;16(5):310–320. doi: 10.1038/nri.2016.34. [DOI] [PubMed] [Google Scholar]
- 50.Blessin N.C., Simon R., Kluth M., Fischer K., Hube-Magg C., Li W., et al. Patterns of TIGIT expression in lymphatic tissue, inflammation, and cancer. Dis Markers. 2019;2019 doi: 10.1155/2019/5160565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mavilio D., Benjamin J., Daucher M., Lombardo G., Kottilil S., Planta M.A., et al. Natural killer cells in HIV-1 infection: dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc Natl Acad Sci U S A. 2003;100(25):15011–15016. doi: 10.1073/pnas.2336091100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fauci A.S., Mavilio D., Kottilil S. NK cells in HIV infection: paradigm for protection or targets for ambush. Nat Rev Immunol. 2005;5(11):835–843. doi: 10.1038/nri1711. [DOI] [PubMed] [Google Scholar]
- 53.Mavilio D., Lombardo G., Benjamin J., Kim D., Follman D., Marcenaro E., et al. Characterization of CD56-/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci U S A. 2005;102(8):2886–2891. doi: 10.1073/pnas.0409872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong J.K., Strain M.C., Porrata R., Reay E., Sankaran-Walters S., Ignacio C.C., et al. In vivo CD8+ T-cell suppression of siv viremia is not mediated by CTL clearance of productively infected cells. PLoS Pathog. 2010;6(1) doi: 10.1371/journal.ppat.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang Y., Zhou F., Tian Y., Zhang Z., Kuang R., Liu J., et al. Higher NK cell IFN-gamma production is associated with delayed HIV disease progression in LTNPs. J Clin Immunol. 2013;33(8):1376–1385. doi: 10.1007/s10875-013-9930-1. [DOI] [PubMed] [Google Scholar]
- 56.Maghazachi A.A., al-Aoukaty A., Schall T.J. C-C chemokines induce the chemotaxis of NK and IL-2-activated NK cells. role for G proteins. J Immunol. 1994;153(11):4969–4977. [PubMed] [Google Scholar]
- 57.Inngjerdingen M., Damaj B., Maghazachi A.A. Expression and regulation of chemokine receptors in human natural killer cells. Blood. 2001;97(2):367–375. doi: 10.1182/blood.v97.2.367. [DOI] [PubMed] [Google Scholar]
- 58.Watt S.V., Andrews D.M., Takeda K., Smyth M.J., Hayakawa Y. IFN-gamma-dependent recruitment of mature CD27(high) NK cells to lymph nodes primed by dendritic cells. J Immunol. 2008;181(8):5323–5330. doi: 10.4049/jimmunol.181.8.5323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.