Abstract
Cellular de-differentiation is a key mechanism driving cancer progression. Acquisition of mesenchymal features has been associated with drug resistance, poor prognosis, and disease relapse in many tumor types. Therefore, successful targeting of tumors harboring these characteristics is a priority in oncology practice. The SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complex has also emerged as a critical player in tumor progression, leading to the identification of several SWI/SNF complex genes as potential disease biomarkers and targets of anti-cancer therapies. AT-rich interaction domain-containing protein 1A (ARID1A) is a component of SWI/SNF, and mutations in ARID1A represent one of the most frequent molecular alterations in human cancers. ARID1A mutations occur in ~10% of pancreatic ductal adenocarcinomas (PDAC), but whether these mutations confer a therapeutic opportunity remains unclear. Here we demonstrate that loss of ARID1A promotes an epithelial-mesenchymal transition (EMT) phenotype and sensitizes PDAC cells to a clinical inhibitor of HSP90, NVP-AUY922, both in vitro and in vivo. While loss of ARID1A alone did not significantly affect proliferative potential or rate of apoptosis, ARID1A-deficient cells were sensitized to HSP90 inhibition, potentially by promoting the degradation of intermediate filaments driving EMT, resulting in cell death. Our results describe a mechanistic link between ARID1A defects and a quasi-mesenchymal phenotype, suggesting that deleterious mutations in ARID1A associated with protein loss exhibits potential as a biomarker for PDAC patients who may benefit by HSP90-targeting drugs treatment.
Keywords: pancreatic cancer, SWI/SNF, ARID1A, NVP-AUY922, epithelial to mesenchymal transition, HSP90
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer-related death, with a median survival of less than 6 months, and 5-year survival in approximately 9% of patients (1,2). Recently, a consensus molecular subtyping of PDAC based on gene expression identified two to four distinct tumor subtypes of this disease (3–5). There is general agreement in the field to indicate that these subtypes can be broadly grouped into a classical-pancreatic group, which includes the progenitor subtype, and a squamous-basal group, which includes the quasi-mesenchymal (QM) and basal-like subtype (6). Typically, tumors with sustained dependency on oncogenic KRAS signaling appear more epithelial in nature and have significantly improved prognoses, possibly due to their higher sensitivity to currently available treatments (7). In contrast, tumors with the QM/basal-like subtype often exhibit dramatic resistance to chemotherapy and radiation, and are associated with poorer overall survival (3).
We previously reported that the QM phenotype is characterized by depletion of the SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily B, Member 1 (SMARCB1) in QM murine PDAC models, and this phenotype has been similarly identified in human PDAC (8,9). Notably, these mesenchymal tumors are sensitive to proteostasis-targeting drugs, such as HSP90 inhibitors, because of Myc network activation upon SMARCB1 ablation, thus increasing protein metabolism and activating the endoplasmic reticulum stress response pathway (8). Given the many genes involved in the SWI/SNF complex, we speculated that loss of other components of the SWI/SNF complex beyond SMARCB1 may similarly sensitize human cancer cells to HSP90 inhibitors in PDAC.
The human SWI/SNF chromatin remodeling complex members are proteins encoded by genes with known tumor suppressor function, including ARID1A, ARID1B, SMARCA4 and SMARCB1 (10,11). ARID1A, a key non-catalytic component of SWI/SNF complex, has been recently identified as tumor suppressor (12,13), and is one of the most frequently mutated members of the SWI/SNF complex (11,14). It has been reported that ARID1A silencing facilitates migration and invasion in some solid tumors, and improved mobility of tumor cells is a hallmark of the PDAC QM subtype (15,16). However, the mechanism that underlies the relationship between ARID1A alterations and cancer cell migration and invasion remains unclear.
In this study, we identified a predictive biomarker that could be used to expand the subset of PDAC tumors that may respond to clinically actionable small molecule HSP90 inhibitors, targeting the proteostatic machinery. Mutational analysis of the most common genetic alterations of SWI/SNF complex members in PDAC cell lines revealed that loss of ARID1A induces epithelial to mesenchymal transition (EMT) and sensitizes cells to HSP90 inhibition, similar to what has been observed in the context of SMARCB1 deficiency. Our findings suggest that a subset of PDAC patients, whose tumors harbor defective ARID1A, may benefit from HSP90 inhibitor therapy, and encourage further efforts to test these drugs for their clinical effectiveness.
Materials and Methods
Human cell lines.
PANC1, MiaPaCa2, BxPC3, PSN1, AsPC1, CAPAN-1, HPAF-II, HS766T, PANC02.03, CFPAC-1, PANC03.27, PANC10.05, SW1990 were purchased from ATCC. HuPT4, PaTu8988T, DANG were purchased from DSMZ. SUIT2 was purchased from JCRB and SNU324 was purchased from KCLB. Cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (ThermoFiscer Scientific) supplemented with 1% penicillin–streptomycin (Gibco) and 10% fetal bovine serum (FBS) (Gibco). Cell lines were cultured for 3 passages between thawing and use in the described experiments. Mutational profiling via whole-exome sequencing and expression profiling via RNA-Sequencing for these cell lines were obtained from the 20Q1 release of the Cancer Cell Line Encyclopedia via the DepMap Portal. Patient-derived cell lines, PATC53, PATC69, PATC124, PATC148, and PATC153, were isolated and established from the samples of the patients who underwent surgery at the University of Texas MD Anderson Cancer Center (UTMDACC) and cultured in DMEM supplemented with 1% penicillin–streptomycin, 10% FBS as described in “PDAC-PDX and PDX-derived primary cell cohorts” section below. Cells were cultured at 37°C in a humidified atmosphere with 5% CO2. All cell lines were routinely fingerprinted and tested for Mycoplasma contamination (MycoAlert).
PDAC-PDX and PDX-derived primary cell cohorts.
PDAC-PDXs were generated accordingly to Kim MP et al (17). Early passage PDXs (F1) from primary human pancreatic ductal adenocarcinoma were harvested in cold HBSS (Gibco) and tumor cells isolated through mechanical and enzymatic dissociation. Briefly, tumors were minced in very small pieces with scissors under sterile conditions before being processed with the Human Tumor Dissociation Kit (Miltenyi Biotec). After digestion, single isolated cells were seeded at high confluency on collagen-IV-coated plates (Corning) in DMEM/F12 (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco), 1% BSA (Fisher Scientific), 0.5 μM hydrocortisone (Sigma-Aldrich), 10 mM HEPES (Invitrogen), 100 ng/ml cholera toxin (Sigma-Aldrich), 5 ml/l insulin-transferrin-selenium (BD), 100 IU/ml penicillin (Gibco), and 100 μg/ml streptomycin (Gibco). To remove fibroblasts in the culture, we periodically performed brief trypsinization (0.25% Trypsin-EDTA, Gibco). The purity of the human culture derived from PDX was confirmed over time by flow cytometry through the evaluation of HLA-ABC and mouse H-2Kd histocompatibility complex antigens (Becton Dickinson). Isolated human cells were maintained in culture for a maximum of couple of passages before being switch to DMEM 10% FBS and enrolled in phenotypic studies and molecular profiling.
Lentiviral vectors and other plasmids.
Two sgRNA constructs provided by Sigma-Aldrich were amplified and cloned into the BmsBI sites of the lentiCRISPR v2 (Addgene, #52961). The oligonucleotide sequences of the sgRNAs specifically targeting ARID1A were the following: sg1, 5’-CAGCAGAACTCTCACGACCA-3’, sg2, 5’- CAGACACATAGAGGCGATAG-3’. The constructs were verified by Sanger sequencing. The two short hairpin RNA (shRNA) targeting VIMENTIN were purchased from Sigma-Aldrich (SHCLNG-NIM_003380) and the negative control vector pLKO.1 puro was purchased from Addgene (Catalog#8453). Infectious viral particles were produced using packaging plasmids psPAX2 and pMD2G from Addgene as previously described (8). Briefly, HEK293T cells were cultured in DMEM containing 10% FBS (Gibco), (100IU/mL) 1% penicillin–streptomycin (Gibco) and transfected using the Polyethylenimine (PEI) method. Virus-containing supernatant was collected 48–72 hours after transfection, spun at 3000 rpm for 10’ and filtered through 0.45um low protein binding filters (Corning). High-titer preps were obtained by ultracentrifugating at 23.000 rpm for 2 hours. Then, the collected preps were added to the culture medium supplemented with 5ug/ml polybrene to improve the infection efficiency. After 48 hours, 10μg/ml puromycin was given to SUIT2 cells, 2μg/ml puromycin to the HuPT4 cells, and 2μg/ml puromycin to the PATC153 cells. The puromycin-supplemented medium was replaced at every passage throughout culturing. After 24 hours of selection, genomic DNA of the infected cells was extracted for PCR and T7 endonuclease I (T7E1) assay. The following primers were used to perform polymerase chain reaction (PCR): F, 5′-AGCCTACGGCTTCGGGCAACCCTA -3′; R, 5′-AGCTGGGCGACGTGAGCAGTTGGTT -3′. The following bands` sizes were observed in ARID1A knocked-out samples: 496 bp, 299 bp and 191 bp.
Cell growth inhibition assay.
Two thousand cells were seeded onto 96-well plates. After 24 hours, cells were exposed to the indicated serial concentrations of drugs, and cell viability was measured 72 hours later using CellTiter-Glo Luminescent Cell Viability Assay (Promega), according to the manufacturer’s instructions. The optical luminescence of each well was detected with the POLARstar omegaTM plate reader. Dose response curves and IC50 values were generated using GraphPad Prism Software (GraphPad Software).
Proliferation assay.
2×103 cells were seeded onto 96-well flat-bottomed plates. Cell proliferation was evaluated using the IncuCyte® Live-Cell Analysis System (Roche Applied Sciences, Indianapolis, IN, USA). The real-time label-free proliferation of those cells was automatically analyzed as the occupied area (% confluence) of cell images over time.
Clonogenic Assay and Acetic acid assay.
Cell viability assays were performed as previously described (18). In brief, cells were seeded in 6-well plate (1.0×103 cells/well) and incubated at 37°C for 24 hours followed or not by 14 days of NVP-AUY-922 treatment. Then, the colonies were washed twice with PBS and stained with 1 mL 0.5% crystal violet in each well for 20 minutes with gentle shaking. After washing with PBS for four times, the cells were air-dry for 2 hours at room temperature. 1 mL of 10% of acetic acid in methanol was added to each well and incubated at room temperature for 20 minutes with gentle shaking. The optical density of each well was detected at 570 nm (OD570) with the POLAR star omegaTM plate reader.
Flow cytometry for apoptosis assay using Annexin V.
Apoptotic rate was determined by flow cytometry with the Annexin V-fluorescein isothiocyanate (V-FITC) apoptosis detection kit (Biovision). Briefly, cells were collected by Accutase and suspended with 500 uL of 1× binding buffer and then treated with 5 μL of Annexin V–FITC and 2.5 μL PI. After 10 minutes incubation on ice, each sample was analyzed immediately using the Gallios Flow Cytometer instrument (BECKMAN COULTER). We defined apoptotic cells as following: unstained cells were classified as “alive”; cells stained by Annexin V only were “early apoptosis”; cells stained by both Annexin V and PI were “late apoptosis”; and cells stained by PI only were “dead” cells
Western Blotting.
Whole cell lysate was obtained by radio immunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) with protease inhibitors, phosphatase inhibitors, and EDTA (Thermo Fisher Scientific). Then, proteins were resolved on 5–15% gradient polyacrylamide SDS/PAGE Tris-HCl gels (Bio-Rad Laboratories) and transferred onto nitrocellulose or PVDF membranes according to manufacturer’s instructions. To reduce non-specific signals, membranes were blocked in 1% milk and 3% BSA, and then incubated with the indicated primary antibodies overnight at 4 degrees Celsius. Membranes were washed in TBST buffer, probed with HRP-linked anti-rabbit or anti-mouse IgG (Cell Signaling Technologies) as secondary antibodies at room temperature for 1 hour and washed again. The antigen-antibody complex was detected with an enhanced chemiluminescent (ECL) substrate (Thermo Fisher Scientific). The bands were obtained using ChemiDoc™ XRS+ with Image lab software version 4.0 (Bio-Rad Laboratories).
Antibodies and chemical reagents.
Primary antibodies used for immunoblotting, co-immunofluorescence (co-IF), and immunohistochemistry: ARID1A/BAF250A (Cell Signaling Technologies, D2A8U), VIMENTIN (Cell Signaling Technologies, D21H3) for co-IF with E-CADHERIN and immunohistochemistry, E-CADHERIN (Cell Signaling Technologies, 4A2), Cytokeratin 19 (Santa Cruz Biotechnologies, A-3), KI-67 (MIB-1) (Dako), Cleaved CASPASE 3 (A175) (Cell Signaling Technologies, 5A1E), HSP90 alpha (Invitrogen, PA3–013), VIMENTIN (Cell Signaling Technologies, 5G3F10) for immunoblotting and co-IF with HSP90. ARID1B/BAF250B (Cell Signaling Technologies, E9J4T), PBRM1/BAF180 (Cell Signaling Technologies, D3F7O), BRM (Cell Signaling Technologies, D9E8B), BRG1 (Cell Signaling Technologies, A52), SMARCC1/BAF155 (Cell Signaling Technologies, D7F8S), SMARCC2/BAF170 (Cell Signaling Technologies, D8O9V), BAF60a (Santa Cruz Biotechnologies, 23), BAF53 (Santa Cruz Biotechnologies, C-7), SMARCB1/BAF47 (Cell Signaling Technologies, D8M1X). Chemical reagents: NVP-AUY-922 (LC Laboratories).
Transcriptional profiling.
RNA from triplicate samples were collected for all conditions. RNA libraries were prepared with the Lexogen QuantSeq 3’ mRNA-Seq FWD Kit, multiplexed and sequenced on the NextSeq 500 (single-end, 75 bp reads). Transcript compatibility counts were obtained with kallisto (v0.44.0) running the pseudo mode with Gencode 23 transcript annotations (19). Gene counts were obtained by summing all reads that uniquely mapped, and differential expression analysis was carried out using DESeq2’s default settings. Genes were considered differentially expressed if there was more than 1.5x fold-change and FDR < 0.05 between experimental conditions. Functional annotation of differentially expressed genes was performed using the weighted pre-ranked mode of the Gene Set Enrichment Analysis (GSEA)(20) software over the MSigDB Hallmark Gene Set with 10,000 permutations(21). Genes are only included in the enrichment step if they are expressed (DESeq2 FDR is defined), protein-coding, and has unique gene names. Genes were then ranked by the DESeq2 T-statistic of the gene. GSEA was specifically deployed on differential expression profiles from the SUIT2, HUPT4 and PATC153 models 24 hours after starting ARID1A-knock-out clones’ selection, against the parental cell lines.
Immunohistochemistry.
Tumor samples were fixed in 4% formaldehyde for 24 hours at room temperature, moved in 70% ethanol for 48 h, and then embedded in paraffin (Leica ASP300S). After cutting (Leica RM2235), baking and deparaffinization, slides were treated with Citra-Plus Solution (BioGenex) according to manufacturer’s instructions. For immunohistochemistry staining, endogenous peroxidases were inactivated by 3% hydrogen peroxide for 18 minutes. Non-specific signals were blocked using 10% FBS and 5% BSA for 1 hour. Tumor samples were stained with primary antibodies for 12 hours at 4°C according to manufacturer’s instructions. For immuno-staining, ImmPress (Vector Lab) was used as secondary antibodies and Nova RED (Vector Lab) for detection. Images were captured with a Nikon DS-Fi1 digital camera using a wide-field Nikon EclipseCi microscope.
Image quantification.
To quantify immunohistochemistry staining, representative fields with 20X-magnification from each biological replicates in each experimental group were analyzed with Immunoratio software of ImageJ (22). Images used for quantification were captured with a Cool-SNAP ES2 digital camera using a wide-field Nikon Eclipse-Ti microscope.
Co-immunofluorescence.
Cells (5000 cells/well) were plated on glass coverslips in 24-wells plate for indicated time. Then, cells were washed three times with 1X PBS and fixed using 4.0% formaldehyde for 20 minutes at room temperature. Fixed cells were washed with PBS 1X and permeabilized by using 0.2% Triton X-100 in PBS 1X for 10 minutes. After permeabilization, cells were washed 3 times with PBS 1X and then blocked using 5% goat serum in PBS 1X for 60 minutes at room temperature. Cells were washed three times with PBS 1X and then were incubated with primary antibodies overnight at 4 degrees Celsius. Then, cells were washed three times with 1X PBS and incubated with both an anti-mouse and an-anti rabbit secondary antibody (Alexa Flour 488, Alexa Flour 555-Thermo Fisher Scientific) diluted 1:400 in antibody dilution buffer for 120’ at room temperature in the dark. After three washes with PBS 1X, cells were stained with DAPI (Invitrogen) for 10 minutes. Then cells were washed with PBS 1X and coverslips where mounted on the microscopy slides with antifade mounting medium (VECTASHIELD). After 24 hours fixed images were captured with a Hamamatsu C11440 digital camera, using a wide-field Nikon Eclipse Ni microscope.
In vivo studies and treatment schedules.
For pre-clinical studies, CD-1 nude female mice were subcutaneously injected with 3×106 cells resuspended in 200ul of 1:1 Matrigel (BD Biosciences, 356231) and media mixture. Once tumors reached approximately 150mm3, NVP-AUY922 or Gemcitabine were administered. NVP-AUY922 dissolved in 10% DMSO/ 25% water/ 65% PEG400 solution and administered via intraperitoneal injection at 75mg/kg every day for 15 days. Gemcitabine was dissolved in saline solution with 0.9 percent sodium chloride and administered via intraperitoneal injection at 100mg/kg every four day for 16 days. Control mice received DMSO vehicle alone. Tumor volume was measured every three days by caliper, and the body weight was also monitored. Tumor volume (mm3) was calculated using the following formula: V (mm3) = L (mm) × W (mm) 2 /2, where W is tumor width and L is tumor length. Mice were sacrificed when tumors were ulcerating according to approved guidelines of the Institutions Animal Ethics Committee. Tumors were harvested and fixed in formalin for histological analysis. All animal studies and procedures were approved by the UTMDACC Institutional Animal Care and Use Committee.
Statistical analysis.
In vitro and in vivo data are presented as the mean ± s.d. of biological replicates. Statistical analyses were performed using a two-tailed Student’s t-test. For in vivo studies, mice were assigned randomly to experimental arms. P-values < 0.05 was considered statistically significant.
Results
ARID1A deficiency predicts sensitivity to NVP-AUY922 in PDAC cell lines
We previously reported that SMARCB1 deletion confers sensitivity to HSP90 inhibitors and other drugs targeting the proteostatic machinery in PDAC (8, 9). However, genetic mutations affecting SMARCB1 are not a common event in PDAC. Thus, we focused our research on novel biomarkers to stratify PDAC tumors that may respond to HSP90 inhibition, on the hypothesis that defects in other SWI/SNF complex genes or proteins might result in similar responses observed in SMARCB1-deficient PDAC models.
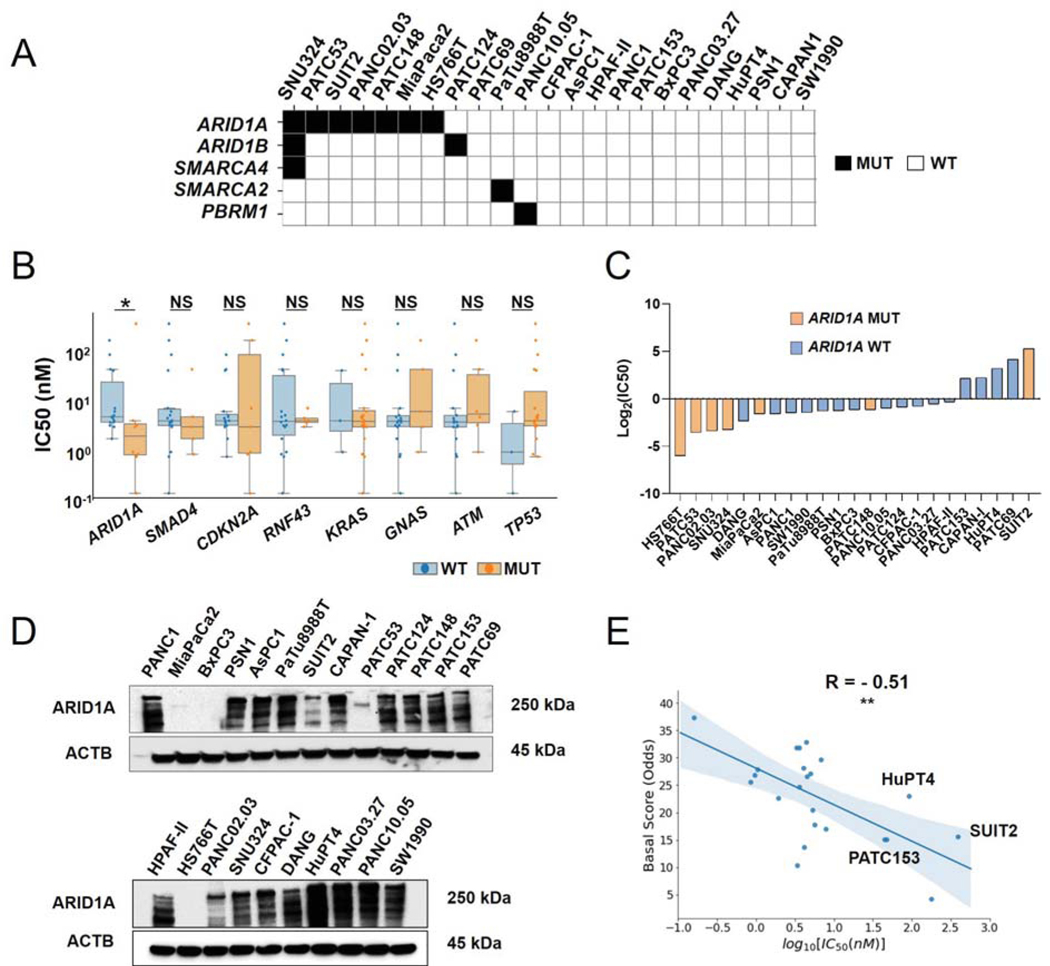
We analyzed the mutational status of key gene members of the SWI/SNF complex for 5 patient-derived cell models and 18 human cell lines available from ATCC and found that 11 of these 23 models harbored at least one non-synonymous mutations in ARID1A, ARID1B, SMARCA4, SMARCA2, or PBMR1 (Fig. 1A) (23). Notably, in line with patient tumor data, the most mutated gene among members of the SWI/SNF complex was ARID1A (Supp. Fig. 1A). We then sought to identify whether any of the most commonly mutated genes in PDAC tumors (24), including ARID1A, and identified in at least 5 of our 23 cell lines, could stratify cell line response to HSP90 inhibition. After treating all 23 cell lines with the HSP90 inhibitor NVP-AUY922, we found that among all common PDAC mutations we examined, only ARID1A mutation status was significantly associated with NVP-AUY922 sensitivity. Conversely, a trend for TP53 mutation conferring resistance to the NVP-AUY922 was also observed (Fig. 1B, Suppl. Table S1). Specifically, we identified 5 cell lines resistant to the drug (IC50>10nM): CAPAN-1, HuPT4, SUIT2, PATC69, and PATC153 (Fig. 1C, Table 1). Interestingly, all of these resistant models harbored a wild-type ARID1A gene, except SUIT2, whose ARID1A protein expression is still detectable, although at low level, potentially due to the heterozygous nature of its mutation (Fig. 1D, Supp. Fig. 1A, B). In line with previous reports, we also observed a significant correlation between a basal-like signature and sensitivity to NVP-AUY922 (Suppl. Fig. 1C and Fig. 1E) (4). These evidence suggest that mesenchymal cell features, along with ARID1A loss, may be involved in the mechanism of action of HSP90 inhibitor NVP-AUY922.
Figure 1. Mutation status of ARID1A and its level of protein expression are associated with sensitivity to NVP-AUY922 in PDAC cell lines.
A. Non-synonymous, exonic mutations of BAF complex members` genes, across a panel of 18 commercially available PDAC cell lines and 5PDX-derived cell lines. B. Box plot comparing IC50 for NVP-AUY922 among all cell lines in study, based on the mutation status of each recurrently mutated gene typically found in PDAC tumors (WT: wild type; MUT: mutant). Genes were included if N ≥ 3 models were found harboring a non-synonymous, exonic mutations for it. C. Bar chart showing IC50 for NVP-AUY922 values and ARID1A mutational status across all models analyzed in the study. D. Immunoblotting analysis of ARID1A protein in 23 PDAC cell lines. E. Sperman correlation of Moffit Basal Score vs log10(NVP-AUY922 IC50) in PDAC cell lines. Data represent mean ± SD. *, P <0.05; **, P <0.01; NS: not significant.
Table 1.
IC50 for NVP-AUY922 among 18 commercially available PDAC cell lines and 5 PDX derived cell lines.
| Cell Line | IC50 (nM) |
|---|---|
| PANC1 | 3.56 |
| MiaPaCa2 | 3.33 |
| BxPC3 | 4.45 |
| PSN1 | 4.18 |
| AsPC1 | 3.35 |
| PaTu8988T | 4.1 |
| SUIT2 | 385.4 |
| CAPAN-I | 47.1 |
| HPAF-II | 7.88 |
| HS766T | 0.158 |
| PANC02.03 | 0.95 |
| SNU324 | 1.05 |
| CFPAC-1 | 5.69 |
| DANG | 1.94 |
| HuPT4 | 92.5 |
| PANC03.27 | 6.79 |
| PANC10.05 | 4.98 |
| SW1990 | 3.62 |
| PATC53 | 0.85 |
| PATC69 | 178.9 |
| PATC124 | 5.3 |
| PATC148 | 4.46 |
| PATC153 | 44.8 |
ARID1A deletion sensitizes pancreatic cancer cells to NVP-AUY922
To evaluate whether ARID1A loss may sensitize PDAC resistant cells to NVP-AUY922, we knocked out ARID1A, using the CRISPR-Cas9 gene editing tool, in 3 of the 5 cell lines most resistant to NVP-AUY922. In particular, we examined 2 ARID1A wild-type models, HuPT4 and PATC153, and one heterozygous ARID1A mutant model, SUIT2, in order to investigate whether further decrease of ARID1A protein level might confer sensitivity to NVP-AUY922. Successful deletion of ARID1A was confirmed by T7 endonuclease I mismatch assay (Suppl. Fig. 2A) and immunoblot analysis, where a reduction of ARID1A levels was readily appreciated across all cell lines tested (Suppl. Fig. 2B). No significant differences in cellular proliferation rate were observed comparing parental cells with isogenic ARID1A-null cells, in either short-term proliferation or long-term clonogenic assays (Suppl. Fig. 2C–D). Moreover, only minor differences in growth kinetics and apoptosis were observed between all parental and ARID1A-KO cells (Suppl. Fig. 2E–F). In contrast, when treated with NVP-AUY922, a robust increase in sensitivity to NVP-AUY922 was observed in all three ARID1A-null isogenic cell lines (IC50 values between 6.1 nM and 11.3 nM) compared with parental cells (IC50 values between 44.8 nM and 385.4 nM). A consistent and significant reduction in clonogenicity and proliferative potential was also readily detected in the ARID1A-KO cells after treatment (Fig. 2A–C). Furthermore, 48-hour exposure to NVP-AUY922 at 25nM concentration markedly increased apoptosis in ARID1A-KO cells as compared to parental cell lines (Fig. 2D). Thus, our results strongly suggest that ARID1A loss can increase PDAC cell lines sensitivity to NVP-AUY922 by affecting cell growth and survival.
Figure 2. ARID1A deletion sensitizes PDAC cells to NVP-AUY922.
A. Growth inhibition assay of parental cells and ARID1A knock-out cells, measured as cell viability after treatment with NVP-AUY922 for 72 hours. Data represent mean ± SD (n=4). *, P <0.05. B. Clonogenic assay (top panel) and acetic acid assay (bottom panel) in parental cells and ARID1A knock-out cells after treatment with NVP-AUY922 for 14 days. Data represent mean ± SD (n=3). ***, P <0.001. C. Proliferation curves of parental cells and ARID1A knock-out cell lines treated or not with with NVP-AUY922. Data represent mean ± SD (n=6). ***, P <0.001; **, P <0.01; *, P <0.05. D. Annexin V apoptosis assay in parental cells and ARID1A knock-out cell lines treated or not with NVP-AUY922 for 48 hours. Data represent mean ± SD (n=3). ***, P <0.001; **, P <0.01.
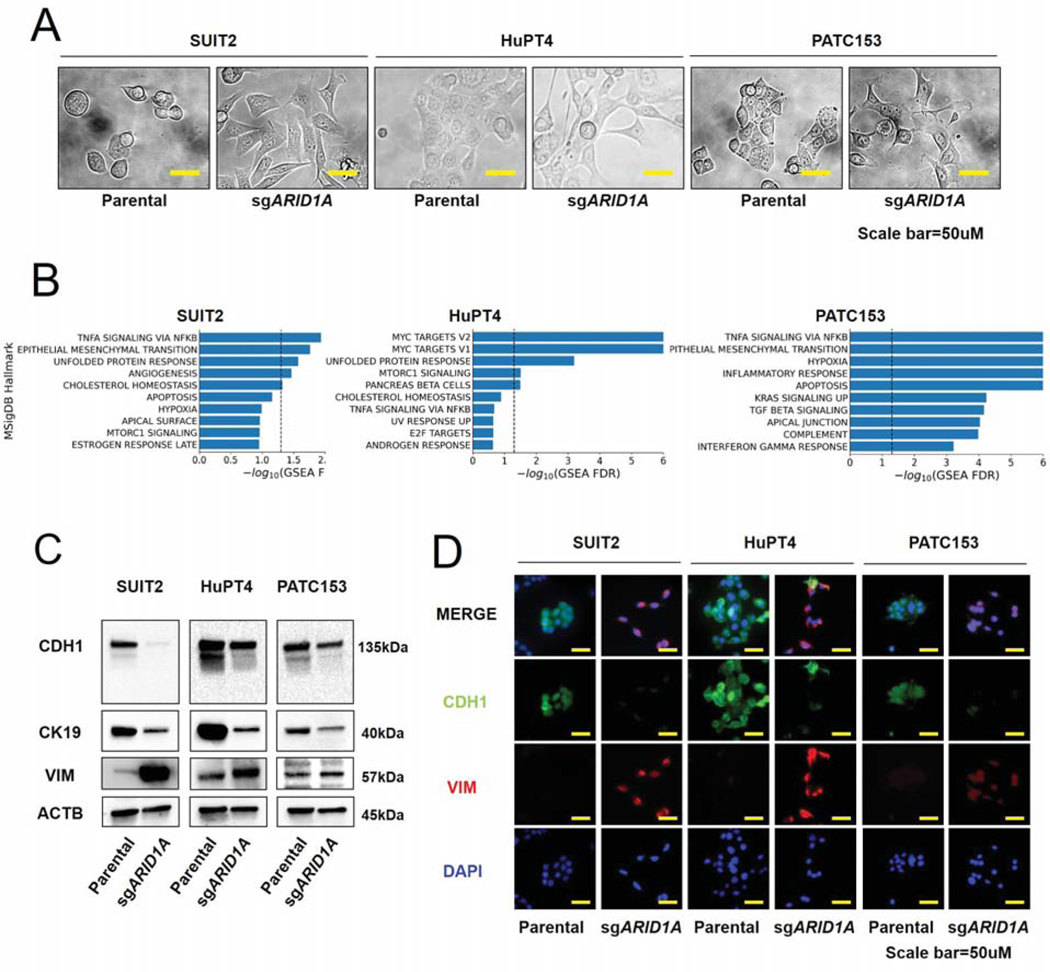
ARID1A-deleted cells activate EMT pathways
Given the significant correlation we observed between a basal-like signature and sensitivity to NVP-AUY922 in PDAC cells in study (Fig. 1E), we also investigated EMT-driven features in both ARID1A wild-type and knock-out models. Interestingly, we noted a prominent spindle-like morphology in all ARID1A-deleted cells compared to the parental cells: a biological effect of ARID1A knock-out consistent with the activation of mesenchymal programs (Fig. 3A) (25). Moreover, we compared the transcriptomic profiles of ARID1A-deleted and isogenic parental cell lines using the MSigDB Hallmark gene set collection. In line with our hypothesis, gene set enrichment analysis (GSEA) revealed a significant overrepresentation of pathways involved in Unfolded protein response, TNFα signaling via NFKβ, and EMT in at least 2 out of 3 models of ARID1A-null cell lines compared with ARID1A-proficient parental cells (Fig. 3B, Suppl. Table S2). Consistently, immunoblotting and immunofluorescence analysis for classical EMT markers demonstrated a decrease in the epithelial markers E-CADHERIN and cytokeratin-19 and an increase in the mesenchymal marker VIMENTIN upon ARID1A ablation (Fig. 3C–D) (25). Overall, these results further suggest a role of ARID1A in preserving an epithelial phenotype in PDAC cells.
Figure 3. Epithelial-mesenchymal transition is upregulated in ARID1A-ablated PDAC cell lines.
A. Morphologies of parental cells and ARID1A knock-out cells. Scale bars, 50 um. B. Top 10 positively enriched MSigDB Hallmark gene sets via GSEA for differential expression profiles of each ARID1A knock-out cell line compared to isogenic parental cells. Dotted line represent -log10(FDR<0.05). C. Immunoblotting analysis of E-CADHERIN (CDH1), cytokeratin 19 (CK19), VIMENTIN (VIM), and β-ACTIN (ACTB) in parental cells and ARID1A knock-out cells. D. Co-immunofluorescence staining for E-CADHERIN, VIMENTIN, and DAPI. Scale bars, 50 um.
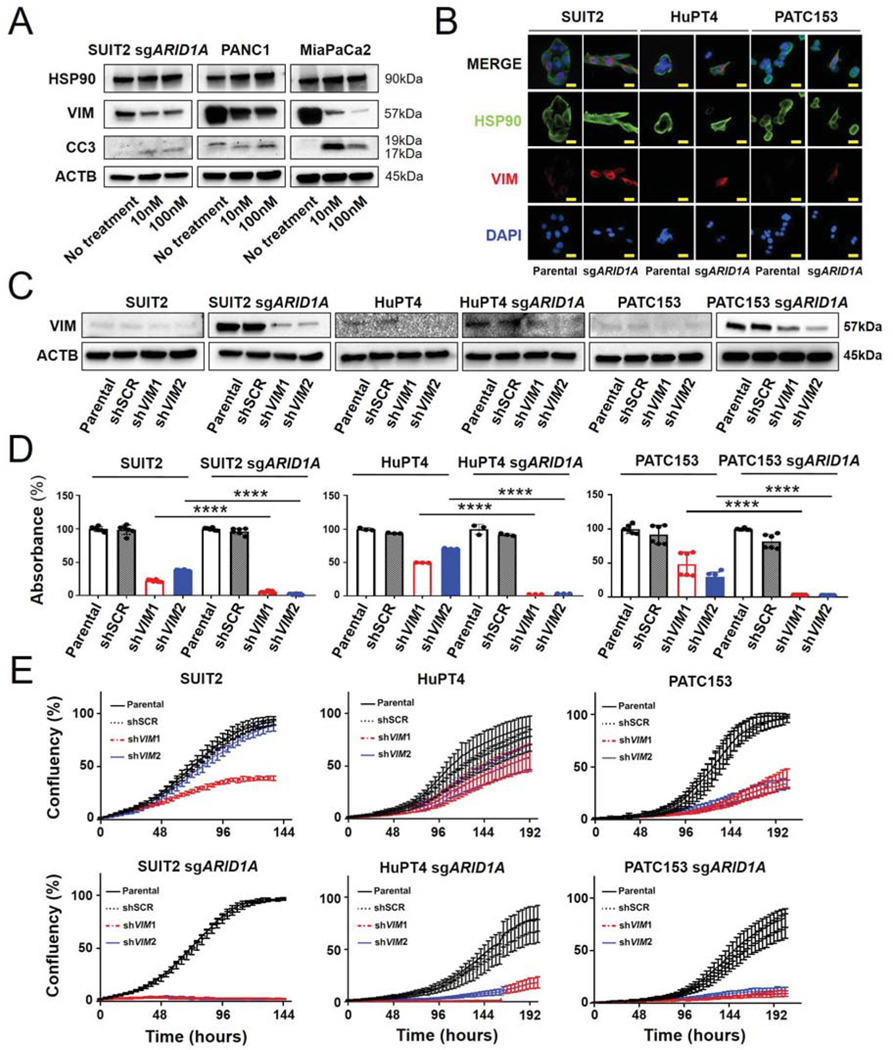
HSP90 inhibition destabilizes VIMENTIN and induces apoptosis in vitro
Given the chaperone role of HSP90 (26), and the correlation observed between QM features and sensitivity to NVP-AUY922, we posited that HSP90 might regulate one or more proteins necessary for mesenchymal cells survival. Specifically, we hypothesized that VIMENTIN may be a “client protein” for HSP90, and its dysregulation could play a role in enhancing sensitivity of ARID1A-null cells to HSP90 inhibition. To test this, we examined two NVP-AUY922-sensitive cell lines, PANC1 and MiaPaCa2 cells (27), as well as SUIT2, sensitized by engineering ARID1A loss. We treated these cells with NVP-AUY922 and evaluated protein levels of HSP90, VIMENTIN, and cleaved CASPASE 3, before and after 48 hours of treatment. Consistent with our hypothesis, in all three lines, NVP-AUY922 caused a significant decrease in VIMENTIN protein expression, along with increased protein levels of cleaved CASPASE 3, as marker of apoptosis (Fig. 4A). Furthermore, immunofluorescence analysis of resistant cells demonstrated a pronounced co-localization of HSP90 and VIMENTIN upon ARID1A loss, confirming protein-protein interaction (Fig, 4B). Next, to investigate whether VIMENTIN is required for survival in the ARID1A-null context, we infected three ARID1A-deleted cell lines and their parental strains with either of two independent lentiviral vectors expressing shRNA constructs targeting VIMENTIN (Fig. 4C). Interestingly, we found that when the expression of VIMENTIN is transiently abrogated, clonogenicity and proliferative cells capacities were negatively impacted more in ARID1A-KO cells than in ARID1A-proficient controls (Fig. 4D–E), suggesting that, in the absence of ARID1A, VIMENTIN is critical for PDAC cell line expansion.
Figure 4. VIMENTIN is required for survival in ARID1A knock-out cells.
A. Immunoblotting analysis for HSP90, VIMENTIN, Cleaved CASPASE 3 (CC3), and β-ACTIN in ARID1A-deleted SUIT2, PANC1, and MiaPaCa2 cells. Cells were treated with NVP-AUY922 for 48 hours. B. Co-immunofluorescence assay of HSP90 and VIMENTIN upon ARID1A loss in SUIT2, HuPT4, and PATC153 and in parental cells. C. Immunoblotting analysis for VIMENTIN and β-ACTIN in ARID1A-deficient SUIT2, HuPT4, and PATC153 cells infected and not with shRNA targeting VIMENTIN (shVIM1, shVIM2) or scrambled construct (shSCR). D. Clonogenicity analyzed by measuring absorbance of colony formation assay with acetic acid assay in ARID1A knock-out cells compared to isogenic parental cells (n=3) and E. Proliferation curves of ARID1A knock-out cells compared to isogenic parental cells (n=6). ****, P <0.0001.
NVP-AUY922 inhibits growth of ARID1A-null tumors in vivo
To evaluate whether ARID1A deletion promotes tumor cells sensitivity to NVP-AUY922 in vivo, we injected SUIT2, HupT4, PATC153 ARID1A-null cells or their parental cell lines subcutaneously in CD-1 nude mice. After tumor establishment, animals were assigned to receive 75 mg/kg NVP-AUY922 or vehicle for 15 days, and tumor size was measured every 3 days, up to 28 days. Tumor kinetics were similar between parental and matched ARID1A-null models. NVP-AUY922 was well tolerated and it did not induce any significant weight loss or other physical/behavioral change to require euthanasia (Suppl. Fig. 3A). At the end of study, animals harboring ARID1A-null tumors that received NVP-AUY922 had significantly smaller tumors compared to vehicle controls. Differences in growth trend were not detectable among parental cells-derived tumors treated or not with HSP90 inhibitor, all increased in size (Fig. 5A–C). IHC staining confirmed very low ARID1A expression in tumors derived from sgRNA-engineered cells, in contrast to the high protein level observed from parental cells (Suppl. Fig. 3B). We also investigated the expression of proliferation marker KI-67 (28), cleaved CASPASE 3, and VIMENTIN. Upon treatment with NVP-AUY922, KI-67 and Cleaved CASPASE 3 signals were significantly reduced and increased, respectively, in ARID1A-deleted tumors, compared to parental cell line-derived tumors (Fig. 5D–E). These results suggest that, in line with our in vitro findings, treatment with NVP-AUY922 attenuates cell proliferation and increases apoptosis in ARID1A-deficient PDAC tumors. Furthermore, we also observed a significant increase in VIMENTIN expression in ARID1A-null versus ARID1A-wild type tumors treated with vehicle, a feature no longer observed after NVP-AUY922 treatment, whose action restored the expression level observed in the ARID1A-wild type tumors (Fig. 5D–E). Finally, we also evaluated whether ARID1A loss enhances sensitivity to gemcitabine treatment. We observed that ARID1A status did not affect either in vitro cell viability (Supp. Fig. 3C) or in vivo tumor growth (Supp. Fig 3D), strongly supporting the role of ARID1A deficiency in sensitizing PDAC cells specifically to agents perturbing the proteostatic machinery.
Figure 5. NVP-AUY922 inhibits tumor growth in ARID1A-deleted subcutaneous xenograft models.
A-C. Tumor burden of mice engrafted with ARID1A-knock-out or parental cells, treated with NVP-AUY922 (75 mg/kg) or vehicle every day for 15 days after tumor establishment, via intraperitoneal injection. Tumor volume was measured every 3 days after starting treatment until day 24 in SUIT2 models, until day 28 in HuPT4 models, and until day 24 in PATC153 models. Data represent mean ± SD (n=5). *, P <0.05; NS: not significant. D. Immunohistochemistry of KI-67, Cleaved CASPASE 3, and VIMENTIN in each group. Scale bars, 200 um. E. Staining`s quantification by measurement of NOVARED positive area/Nuclear area. Data represent mean ± SD (n=5). **, P <0.01; *, P <0.05.
Discussion
Given our observation that sensitivity to HSP90 inhibition was positively correlated with a QM phenotype and ARID1A mutation in PDAC cell lines, we hypothesized that we could modulate EMT through the deletion of ARID1A in PDAC models. Our results show acquisition of spindle-like morphology, depletion of epithelial markers, and increase of mesenchymal markers upon ARID1A loss. Our group and others have previously demonstrated enrichment of mesenchymal features and an aggressive growth phenotype in tumors with deleted or depleted function of SWI/SNF components (29) (8). More specifically, we reported that SMARCB1 loss drives mesenchymal reprogramming in PDAC and renders cells highly sensitive to proteotoxic stress, suggesting high dependence on a functional proteostatic machinery for survival. In other gastrointestinal cancers, ARID1A loss leads to more mesenchymal phenotypes and is associated with poor prognosis, while in breast and ovarian cancer, loss of SWI/SNF genes promotes dedifferentiation (15,16). In this study, we conducted transcriptomic analysis that uncovered an upregulation of EMT programs in PDAC models upon sgRNA-mediated deletion of ARID1A compared with isogenic parental cells; these data were then confirmed by protein expression analysis. Specifically, we found a critical downregulation of cytokeratin-19 and E-CADHERIN, along with a robust upregulation of VIMENTIN after ablation of ARID1A. These data are consistent with recent reports demonstrating that EMT can be modulated by post-transcriptional mechanisms (30–32).
Similar to our finding of increased sensitivity to proteostatic machinery perturbations in PDAC models affected by SMARCB1 loss (16), ARID1A mutation status also showed a significant association with sensitivity to the HSP90 inhibitor, NVP-AUY922. In particular, we found that ARID1A-mutant cells are significantly more sensitive to NVP-AUY922 compared to ARID1A wild-type cells and among a panel of gene mutations commonly occurring in PDAC, only ARID1A mutation status correlates with sensitivity to NVP-AUY922. Tumors derived from ARID1A-deleted cell lines also showed marked sensitivity to NVP-AUY922 in vivo, whereas ARID1A-proficient tumors did not respond to treatment. While the ARID1A-mutant cell lines harboring mutations resulting in loss of protein expression examined in this study were overall sensitive to NVP-AUY922, we also identified cell line models with wild-type ARID1A that displayed sensitivity to NVP-AUY922. This suggests that other independent mechanisms can sensitize PDAC cells to HSP90 inhibition, including mutation of histone methylation regulators, like KDM6A (33), though other unknown mechanisms may also be at play. Loss of ARID1A expression is typically induced by nonsense mutations, insertions, and deletions in the gene-coding region, which lead to mRNA decay or sequence truncation (14). A recent study suggests that promoter hyper-methylation may also result in reduced expression of ARID1A (34). Additional reports have identified ARID1A-defective cancers as exquisite responders to ATR or PARP inhibitors (35,36). Thus, deleterious mutations in ARID1A resulting in significant changes in protein expression, may serve as a biomarker to predict sensitivity to several targeted therapies. Further research may clarify the mechanism by which ARID1A and other SWI/SNF components might alter cellular functions, such as differentiation, proliferation and apoptosis.
HSP90 is an ATP-dependent molecular chaperone protein involved in the stabilization of many membrane and intracellular proteins, including some proteins regulating EMT reprogramming (37–39). NVP-AUY922 acts by inhibiting HSP90 ATPase activity, thus preventing the formation of a multi-chaperone complex, which may lead to the misfolding of client proteins. Our current study reveals that HSP90 inhibition results in VIMENTIN protein degradation. Moreover, we demonstrated that ARID1A-depleted PDAC cells are dependent on VIMENTIN for their growth. This result suggests that the role of HSP90 stabilizing VIMENTIN may be a mechanism underlying the sensitization of ARID1A knock-out cells to NVP-AUY922. Other reports, describing VIMENTIN expression as restricted to mesenchymal cells, and that disruption of its filamentous structure promotes apoptosis, are also consistent with our findings (40–42). Further work is required to fully describe the interactions between HSP90 and VIMENTIN in PDAC, and to understand how perturbing their association may induce cell death. However, in our study, we demonstrated that loss of ARID1A drives a switch towards a stable mesenchymal state, at the expenses of an increased dependency on chaperone-mediated protein folding mechanisms to sustain it. These results are particularly relevant in the PDAC molecular landscape, in which mesenchymal features are associated with aggressive tumors and poor prognosis.
Taken together, our data strongly suggest the protein folding machinery as an attractive therapeutic target in a subset of pancreatic tumors with mutations affecting ARID1A expression and functionality. This expands the population of patients who may benefit from HSP90 inhibitor therapy, or treatment with other drugs targeting the proteostatic machinery, beyond those patients affecting by SMARCB1 mutations, as we previously described. Our identification of models with functional ARID1A that respond to HSP90 inhibition, indicates that sensitivity to the drug may also be conferred by other mutations, including those affecting other members of the SWI/SNF complex, as well as other genes for which the functional relationship with the proteostatic machinery is not well understood yet. Moreover, interestingly, our results suggest no increase in response to standard chemotherapy in ARID1A-deficient PDAC models, paving the way for a potential therapeutic strategy with HSP90 inhibitor drugs to treat some of the most aggressive QM/basal-like tumors resistant to more traditional therapies. In conclusion, our data support further preclinical and clinical development of HSP90 inhibitor therapy to treat a biomarker-informed patient population affected by PDAC.
Supplementary Material
Statement of Significance.
This study identifies ARID1A loss as a promising biomarker for the identification of PDAC tumors that are potentially responsive to treatment with proteotoxic agents.
Acknowledgements.
We thank Jintan Liu and I-Lin Ho for technical help. We wish to thank members of the Genovese lab and the Draetta lab for discussions and reagents. We wish to thank the MDACC Department of Veterinary Medicine and the UTMDACC Flow Facility. F.C. wassupported by the AIRC fellowship for abroad 2018/2020. L.P. was supported by Ermenegildo Zegna Founder’s Scholarship 2019/2020. J.R. received support from the Paula-Altman Goldstein Discovery Fellowship and from P01 CA117969 12. N.M.T. was supported by the Ransom Horne Jr. Professorship for Cancer Research. G.G. was supported by the Barbara Massie Memorial Fund, the MDACC Moonshot FIT Program, the Bruce Krier Endowment Fund, and the Lyda Hill Foundation. G.F.D. and G.G were supported by the 2014 Pancreatic Cancer Action Network-AACR Research Acceleration Network Grant, in memory of Skip Viragh, Grant Number 14-90-25-DRAE. G.F.D. received support from the Sewell Family Chairmanship in Genomic Medicine.
Grant Support: G.F.D. and G.G. received support from the AACR Pancreatic Cancer Action Network grant 14-90-25-DRAE. G.F.D. received support from the Sewell Family Chairmanship in Genomic Medicine.
Footnotes
Disclosure of Potential Conflicts of Interest:
G.F.D. reports personal fees from and stock ownership in Forma Therapeutics, Metabomed, BiovelocITA, Nurix, Frontier Medicines and Orionis Biosciences; and personal fees from Blueprint Medicines, Frontier Medicines, BiovelocITA, Taiho Pharmaceutical Company.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research 2014;74:2913–21 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians 2019;69:7–34 [DOI] [PubMed] [Google Scholar]
- 3.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nature medicine 2011;17:500–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nature genetics 2015;47:1168–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52 [DOI] [PubMed] [Google Scholar]
- 6.Collisson EA, Bailey P, Chang DK, Biankin AV. Molecular subtypes of pancreatic cancer. Nature reviews Gastroenterology & hepatology 2019;16:207–20 [DOI] [PubMed] [Google Scholar]
- 7.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer cell 2009;15:489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genovese G, Carugo A, Tepper J, Robinson FS, Li L, Svelto M, et al. Synthetic vulnerabilities of mesenchymal subpopulations in pancreatic cancer. Nature 2017;542:362–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carugo A, Minelli R, Sapio L, Soeung M, Carbone F, Robinson FS, et al. p53 Is a Master Regulator of Proteostasis in SMARCB1-Deficient Malignant Rhabdoid Tumors. Cancer cell 2019;35:204–20.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America 2012;109:E252–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics 2013;45:592–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oike T, Ogiwara H, Nakano T, Yokota J, Kohno T. Inactivating mutations in SWI/SNF chromatin remodeling genes in human cancer. Japanese journal of clinical oncology 2013;43:849–55 [DOI] [PubMed] [Google Scholar]
- 13.Wu RC, Wang TL, Shih Ie M. The emerging roles of ARID1A in tumor suppression. Cancer biology & therapy 2014;15:655–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer discovery 2013;3:35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan HB, Wang XF, Zhang Q, Tang ZQ, Jiang YH, Fan HZ, et al. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of E-cadherin transcription. Carcinogenesis 2014;35:867–76 [DOI] [PubMed] [Google Scholar]
- 16.Sun X, Wang SC, Wei Y, Luo X, Jia Y, Li L, et al. Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer. Cancer cell 2017;32:574–89.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nature protocols 2009;4:1670–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feoktistova M, Geserick P, Leverkus M. Crystal Violet Assay for Determining Viability of Cultured Cells. Cold Spring Harbor protocols 2016;2016:pdb.prot087379 [DOI] [PubMed] [Google Scholar]
- 19.Yi L, Pimentel H, Bray NL, Pachter L. Gene-level differential analysis at transcript-level resolution. Genome biology 2018;19:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuominen VJ, Ruotoistenmaki S, Viitanen A, Jumppanen M, Isola J. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast cancer research : BCR 2010;12:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019;569:503–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer cell 2017;32:185–203.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871–90 [DOI] [PubMed] [Google Scholar]
- 26.Buchner J. Hsp90 & Co. - a holding for folding. Trends in biochemical sciences 1999;24:136–41 [DOI] [PubMed] [Google Scholar]
- 27.Gradiz R, Silva HC, Carvalho L, Botelho MF, Mota-Pinto A. MIA PaCa-2 and PANC-1 - pancreas ductal adenocarcinoma cell lines with neuroendocrine differentiation and somatostatin receptors. Scientific reports 2016;6:21648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. Journal of cellular physiology 2000;182:311–22 [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Friedland SC, Guo B, O’Dell MR, Alexander WB, Whitney-Miller CL, et al. ARID1A, a SWI/SNF subunit, is critical to acinar cell homeostasis and regeneration and is a barrier to transformation and epithelial-mesenchymal transition in the pancreas. Gut 2019;68:1245–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nature cell biology 2010;12:286–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diepenbruck M, Tiede S, Saxena M, Ivanek R, Kalathur RKR, Luond F, et al. miR-1199–5p and Zeb1 function in a double-negative feedback loop potentially coordinating EMT and tumour metastasis. Nature communications 2017;8:1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shelton PM, Duran A, Nakanishi Y, Reina-Campos M, Kasashima H, Llado V, et al. The Secretion of miR-200s by a PKCzeta/ADAR2 Signaling Axis Promotes Liver Metastasis in Colorectal Cancer. Cell reports 2018;23:1178–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe S, Shimada S, Akiyama Y, Ishikawa Y, Ogura T, Ogawa K, et al. Loss of KDM6A characterizes a poor prognostic subtype of human pancreatic cancer and potentiates HDAC inhibitor lethality. International journal of cancer 2019;145:192–205 [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Sun Q, Shan M, Niu M, Liu T, Xia B, et al. Promoter hypermethylation of ARID1A gene is responsible for its low mRNA expression in many invasive breast cancers. PloS one 2013;8:e53931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williamson CT, Miller R, Pemberton HN, Jones SE, Campbell J, Konde A, et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nature communications 2016;7:13837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park Y, Chui MH, Suryo Rahmanto Y, Yu ZC, Shamanna RA, Bellani MA, et al. Loss of ARID1A in Tumor Cells Renders Selective Vulnerability to Combined Ionizing Radiation and PARP Inhibitor Therapy. Clinical cancer research : an official journal of the American Association for Cancer Research 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang MH, Lee JS, Kim HJ, Jin DI, Kim JI, Lee KJ, et al. HSP90 protects apoptotic cleavage of vimentin in geldanamycin-induced apoptosis. Molecular and cellular biochemistry 2006;281:111–21 [DOI] [PubMed] [Google Scholar]
- 38.Meng J, Chen S, Lei YY, Han JX, Zhong WL, Wang XR, et al. Hsp90beta promotes aggressive vasculogenic mimicry via epithelial-mesenchymal transition in hepatocellular carcinoma. Oncogene 2019;38:228–43 [DOI] [PubMed] [Google Scholar]
- 39.Chong KY, Kang M, Garofalo F, Ueno D, Liang H, Cady S, et al. Inhibition of Heat Shock Protein 90 suppresses TWIST1 Transcription. Molecular pharmacology 2019;96:168–79 [DOI] [PubMed] [Google Scholar]
- 40.van Engeland M, Kuijpers HJ, Ramaekers FC, Reutelingsperger CP, Schutte B. Plasma membrane alterations and cytoskeletal changes in apoptosis. Experimental cell research 1997;235:421–30 [DOI] [PubMed] [Google Scholar]
- 41.Byun Y, Chen F, Chang R, Trivedi M, Green KJ, Cryns VL. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell death and differentiation 2001;8:443–50 [DOI] [PubMed] [Google Scholar]
- 42.Bollong MJ, Pietila M, Pearson AD, Sarkar TR, Ahmad I, Soundararajan R, et al. A vimentin binding small molecule leads to mitotic disruption in mesenchymal cancers. Proceedings of the National Academy of Sciences of the United States of America 2017;114:E9903-e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.