Abstract
IMGT®, the international ImMunoGeneTics information system®, http://www.imgt.org/, is at the forefront of the immunogenetics and immunoinformatics fields with more than 30 years of experience. IMGT® makes available databases and tools to the scientific community pertaining to the adaptive immune response, based on the IMGT-ONTOLOGY. We focus on the recent features of the IMGT® databases, tools, reference directories and web resources, within the three main axes of IMGT® research and development. Axis I consists in understanding the adaptive immune response, by deciphering the identification and characterization of the immunoglobulin (IG) and T cell receptor (TR) genes in jawed vertebrates. It is the starting point of the two other axes, namely the analysis and exploration of the expressed IG and TR repertoires based on comparison with IMGT reference directories in normal and pathological situations (Axis II) and the analysis of amino acid changes and functions of 2D and 3D structures of antibody and TR engineering (Axis III).
INTRODUCTION
The adaptive immune response appeared with the jawed vertebrates (or Gnathostomata), 450 million years ago. It is characterized by a remarkable immune specificity and memory which are the properties of the B and T cells owing to an extreme diversity of their antigen receptors, immunoglobulins (IG) or antibodies and T cell receptors (TR) (1). In human and other mammals, an IG consists of two identical light chains (Kappa (IGK) or Lambda (IGL)) and two identical heavy chains (IGH) (2), while a TR consists of two chains, either Alpha (TRA) and Beta (TRB), or Gamma (TRG) and Delta (TRD) (3). Each IG and TR chain comprises a variable domain (V-DOMAIN) which determines the specificity for the antigen, and a constant region (C-REGION). The V-DOMAIN results from the genomic DNA rearrangement of variable (V), diversity (D) and joining (J) genes for IGH, TRB and TRD chains (V-D-J-REGION) and from V and J genes for IGK, IGL, TRA and TRG chains (V-J-REGION) (Supplementary Figure S1). Additional mechanisms occurring during the rearrangements (N diversity, somatic hypermutations for the IG) contribute to the extreme diversity of the IG and TR (theoretically 1012 different IG and TR per individual, which is only limited by the number of the B and T cells that an organism is genetically programmed to produce).
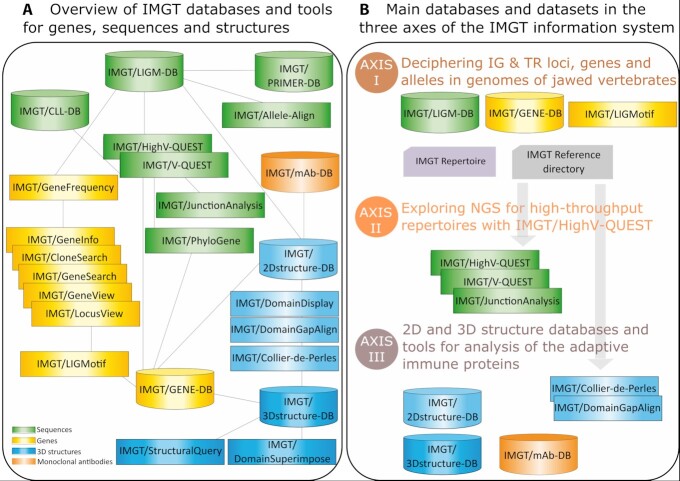
IMGT®, the international ImMunoGeneTics information system® (http://www.imgt.org) (4), was created in 1989 in order to characterize the genes and alleles involved in the IG and TR synthesis of vertebrates. IMGT® is an integrated knowledge system for sequences, genes and structures of the IG or antibodies, TR and major histocompatibility proteins (MH) of the adaptive immune responses, as well as of other proteins of the IG superfamily (IgSF) and MH superfamily (MhSF) of vertebrates and invertebrates. IMGT® comprises 7 databases, 17 online tools (Figure 1A) and >20 000 pages of Web resources.
Figure 1.
IMGT resources. (A) Overview of IMGT databases and tools for genes, sequences and structures. (B) Main databases and datasets in the three axes of IMGT information system.
The accuracy and the consistency of the IMGT® data are based on IMGT-ONTOLOGY (5,6), the first ontology for immunogenetics and immunoinformatics and IMGT Scientific chart rules. IMGT-ONTOLOGY includes the IMGT structured terminology and the annotation rules and is composed of seven axioms. The IDENTIFICATION axiom provides the standardized keywords for the identification of nucleotide and protein sequences and the 3D structures. The DESCRIPTION axiom comprises the IMGT standardized labels for the description and the delimitation of constitutive motifs within sequences and structures. The CLASSIFICATION axiom defines the criteria for IG and TR genes and alleles classification for the setting of the standardized nomenclature. The NUMEROTATION axiom includes the IMGT unique numbering and its graphical 2D representation, the IMGT Collier de Perles. The LOCALIZATION axiom allows to characterize the localization of IG and TR genes. The ORIENTATION axiom defines the orientation of genomic instances (chromosome, locus and gene) of DNA strands. The OBTENTION axiom precises the biological and methodological origins of the IMGT data (5,6).
IMGT® comprises in particular databases which are specialized in nucleotide sequences (IMGT/LIGM-DB) (7), genes and alleles (IMGT/GENE-DB) (8), amino acid sequences and 2D (IMGT/2Dstructure-DB) and 3D structures (IMGT/3Dstructure-DB) (9) and therapeutic monoclonal antibodies (IG, mAb) and other proteins for clinical applications (IMGT/mAb-DB) (4). The four IMGT databases, the related tools and Web resources are described in this manuscript through the three main axes of IMGT research and development: the identification and characterization of IG and TR genes and knowledge of their genomic organization (Axis I), the analysis and exploration of the expressed IG and TR repertoires in normal and pathological situations (Axis II) and the analysis of adaptive immune proteins from antigen receptor to amino acid changes (Axis III) (Figure 1B).
AXIS I Understanding the adaptive immune response: gene characterization and knowledge of their genomic organization
IG and TR chains are encoded by polymorphic multigene families located on different chromosomes. In humans and other mammals, there are seven main loci for IG and TR: three for IG (IGH, IGK and IGL) (2,10) and four for TR (TRA, TRB, TRD and TRG) (3). The V, D, J and constant (C) IMGT gene names were assigned according to the concepts of the CLASSIFICATION axiom (5,6) and were approved by the Human Genome Organization (HUGO) Nomenclature Committee (HGNC) for human (11) in 1999 and were endorsed by the WHO IUIS Nomenclature Subcommittee for IG and TR (12).
The characterization of genes and alleles for the seven loci of human (Homo sapiens) and mouse (Mus musculus) were published in 2001 and 2005. The organization of the genes within these loci was deduced and built from the complete annotation of the genomic nucleotide sequences and contigs integrated in the IMGT nucleotide sequence database IMGT/LIGM-DB (7) from European Nucleotide Archive (ENA) (13) and GenBank (14). IMGT genes and alleles are managed in the IMGT gene database IMGT/GENE-DB (8) and displayed in IMGT Repertoire (IMGT Web resources) and IMGT tools (http://www.imgt.org/IMGTposters/Poster-10th-Biocuration-Conference2017.pdf).
With the introduction of genome assemblies, which have become available in NCBI assembly (15) and Ensembl (16), IMGT® developed a new approach and new concepts in order to decipher complete IG and TR loci. First of all, IMGT® defines conserved genes that flank the IG and TR loci, designated as ‘IMGT bornes’. IMGT bornes are genes coding for proteins other than IG or TR, which are conserved among species. They are located either upstream of the first IG or TR gene (IMGT_locus_5prime_borne) or downstream of the last IG or TR gene (IMGT_locus_3prime_borne) of the IMGT locus. If the IMGT bornes are identified and are at most 10 kb away from the closest IG or TR genes, they will be included in the locus genomic nucleotide sequences available through IMGT/LIGM-DB.
These IMGT bornes have allowed to set a standardized delimitation of the locus whatever the species and they are helpful for comparative genomics. However, such conserved non IG or TR genes could not be systematically defined (n.d.) up to now, as for example for the IGH locus. In absence of the IMGT borne, the limit of the locus is artificially defined by 10 kb in 5′ upstream of the first IG or TR gene and in 3′ downstream from the last IG or TR gene. TRB is an example of locus with delimited IMGT bornes and can be accessed on the page http://www.imgt.org/IMGTrepertoire/LocusGenes/bornes/bornesTRB.html.
IMGT/LIGM-DB
IMGT/LIGM-DB provides standardized and detailed immunogenetics annotations for IG, TR and MH nucleotide sequences from human and other vertebrate species (7). IMGT/LIGM-DB includes sequences from different steps of IG and TR synthesis and therefore, it integrates: (i) large germline (non-rearranged) genomic DNA (gDNA) sequences, which may involve a complete locus from several hundred kilobases to one (or more) megabase(s); (ii) rearranged gDNA sequences resulting from the recombination of V, J genes or V, D and J genes; and (iii) rearranged V-J-C and V-D-J-C complementary DNA (cDNA) sequences.
Most of the IMGT/LIGM-DB nucleotide sequences come from ENA and from GenBank, using the same accession numbers to facilitate interoperability with the generalist nucleotide databases. More recently, with the extraction of IG and TR loci nucleotide sequences from NCBI genome assemblies, IMGT® created new IMGT/LIGM-DB accession numbers starting with ‘IMGT’ followed by 6 digits. IMGT/LIGM-DB sequences are annotated according to IMGT-ONTOLOGY concepts of the DESCRIPTION axiom (5,6), with IMGT labels (http://www.imgt.org/ligmdb/label) and IMGT qualifiers (http://www.imgt.org/ligmdb/qualifier.action). In order to delimit and annotate a complete IG or TR locus extracted from genome assemblies, a specific IMGT label and a set of IMGT qualifiers has been created for its description (Table 1).
Table 1.
New IMGT concepts and their definitions
| New IMGT concepts | Definition | |
|---|---|---|
| IMGT label | IMGT-LOCUS-UNIT | gDNA of an immunoglobulin (IG) or T cell receptor (TR) IMGT locus unit from chromosome genomic assembly, that starts at the 5 prime (5′) end of the most 5′ IG or TR GENE-UNIT in the locus and ends at the 3 prime (3′) end of the most 3′ IG or TR GENE-UNIT in the locus |
| IMGT qualifiers | IMGT_locus_3prime_borne | Name of the gene identified as the 3′ borne of an IMGT-LOCUS-UNIT |
| IMGT_locus_3prime_gene | IMGT gene name of the most 3′ IG or TR GENE-UNIT of an IMGT-LOCUS-UNIT | |
| IMGT_locus_5prime_borne | Name of the gene identified as the 5′ borne of an IMGT-LOCUS-UNIT | |
| IMGT_locus_5prime_gene | IMGT gene name of the most 5′ IG or TR GENE-UNIT of an IMGT-LOCUS-UNIT | |
| IMGT_locus_length | Length of an IMGT-LOCUS-UNIT in kb or in bp | |
| IMGT_locus_name | Name of an IMGT-LOCUS-UNIT, that includes the Latin genus and species name and the IMGT locus type | |
| IMGT_locus_orientation | Orientation of an IMGT-LOCUS-UNIT on a chromosome, is either forward (FWD) or reverse (REV) | |
| IMGT_locus_positions | Positions of an IMGT-LOCUS-UNIT on a chromosome | |
| IMGT_locus_type | IMGT locus type (in higher vertebrates: IGH, IGK, IGL, TRA, TRB, TRG, TRD) of an IMGT-LOCUS-UNIT | |
IMGT/LIGM-DB interface
The IMGT/LIGM-DB data are accessible via a user-friendly interface described previously in (7). IMGT/LIGM-DB can be queried by: Accession number, IMGT-ONTOLOGY concepts (IDENTIFICATION or Keywords, CLASSIFICATION, DESCRIPTION or labels, OBTENTION), or bibliographical references.
For each nucleotide sequence, IMGT/LIGM-DB provides ‘View details’ displaying an IMGT/LIGM-DB entry according to nine topics: annotations, IMGT flat file, coding regions with protein translation, catalogue and external references, sequence in IMGT/LIGM-DB dump format, sequence in FASTA format, sequence with three reading frames, EMBL flat file, and a direct link to IMGT/V-QUEST (17). As of September 2021, IMGT/LIGM-DB contains 196,516 entries from 358 species and 48,682 IG and TR nucleotide sequences are fully annotated. Weekly release of IMGT/LIGM-DB flat files can be downloaded directly from the IMGT web site (http://www.imgt.org/download/LIGM-DB/) and from ENA (http://ftp.ebi.ac.uk/pub/databases/imgt/LIGM-DB/).
IMGT/GENE-DB
The curated IG and TR genes are entered and managed in IMGT/GENE-DB (8) with all IMGT identified alleles, which highlight the potential high polymorphism of these genes. Each allele is characterized by its IMGT reference allele sequence defined for the coding label V-REGION (with gaps according to the IMGT numbering (18)), D-REGION, J-REGION and C-REGION (or C exons) (with gaps for C-DOMAIN according to the IMGT numbering (19)) of the V, D, J and C genes respectively. An IMGT allele reference sequence is identified by IMGT/LIGM-DB accession number, IMGT gene and allele names, species, allele functionality and IMGT label. IMGT allele reference sequences compose the IMGT reference directories that are used by IMGT sequence analysis tools and by IMGT databases and IMGT Web resources for sequence comparison.
IMGT/GENE-DB interface
From the IMGT/GENE-DB Query page, search can be performed by IMGT-ONTOLOGY concepts (IDENTIFICATION or keywords, LOCALIZATION, and CLASSIFICATION), LOCALIZATION IN GENOME ASSEMBLIES or IMGT/GENE-DB direct links. IMGT/GENE-DB provides a full access to characterized genes and alleles displaying an IMGT/GENE-DB entry according to six topics: IMGT gene name and definition, Chromosomal localization, IMGT reference alleles, Annotated IMGT/LIGM-DB cDNA and rearranged genomic DNA sequences, Annotated IMGT/3Dstructure-DB structures, and External links.
The section ‘LOCALIZATION IN GENOME ASSEMBLIES’ created in 2015, provides the localizations of the genes and alleles, and IMGT labels in the reference genome assemblies available at NCBI. For each gene, its orientation in the locus is mentioned, and the allele identified in the sequence of the assembly is indicated with its characteristics. The ‘IMGT/GENE-DB direct links’ allows to query dynamically the database, on IMGT gene name, IMGT Group, and to extract labels from the reference sequences of a given gene or gene group. The format for IMGT/GENE-DB direct links is described in http://www.imgt.org/genedb/directlinks.
As of September 2021, IMGT/GENE-DB contains 8,498 genes, 11,349 alleles from human, mouse and other vertebrates. The reference sequences of the IG and TR genes in FASTA format are accessible by group and species from http://www.imgt.org/vquest/refseqh.html#refdir2. IMGT/GENE-DB has a specific section in the ‘IMGT downloads’ section, updated weekly, of the IMGT® portal (http://www.imgt.org/download/GENE-DB/) in different formats.
With the development of new high throughput sequencing technologies for the analysis of IG and TR repertoires, new potential alleles are highlighted by inference from expressed repertoires, particularly in human. Inferred alleles are not systematically integrated within the IMGT databases, because the sequences are not mapped. However, IMGT® can accept inferred alleles if and only if validated by the Working Group (WG) Inferred Allele Review Committee (IARC), within the Adaptive Immune Receptor Repertoire (AIRR) community. IARC ensures that IMGT data quality requirements are met. Nevertheless, reference sequences of inferred alleles are replaced by the corresponding germline DNA sequence once they are characterized (20).
IMGT Repertoire
An overview of IMGT® annotated data is compiled and knowledge pages are made available in IMGT Web Resources ‘IMGT Repertoire’ (http://imgt.org/IMGTrepertoire/), the global ImMunoGeneTics Web Resource for IG, TR, MH of human and other vertebrate species. IMGT Repertoire includes seven organized sections: Locus and genes, Proteins and alleles, 2D and 3D structures, Probes and RFLP, Taxonomy, Gene regulation and expression, Genes and clinical entities. Novel IMGT Repertoire (IG and TR) pages in Locus and genes section were created, focusing on the ‘Locus descriptions’, including Locus bornes, Locus in genome assembly and Locus gene order.
As of September 2021, the number of species present in the IMGT Repertoire reaches 80 species. For each gene analyzed, there are >200 different information fields available in IMGT databases and web pages. Therefore, IMGT Repertoire bridges the gap between curated data resulting from Axis I and IMGT databases and tools (Table 2).
Table 2.
54 fully annotated IG and TR loci are available in IMGT databases and tools, among these 54 loci, 50 have an IMGT locus accession number and 4 (with * in this table) have accession numbers from ENA, NCBI and Ensembl contigs built before the creation of IMGT Locus accession numbers. Note that the IMGT® biocuration of the first two fully annotated species, human (Homo sapiens) and mouse (Mus musculus) are not shown in this table. More information is available in http://www.imgt.org/IMGTrepertoire/LocusGenes/
| Taxon | Species | NCBI Assembly | Locus | Chromosomal localization | NCBI Chromosome Accession numbers | IMGT locus Accession numbers |
|---|---|---|---|---|---|---|
| MAMMALIA EUTHERIA (placentals) | Bos taurus (bovine) Breed: Hereford | ARS-UCD1.2 | IGK | 11 | CM008178.2 | IMGT000047 |
| IGL | 17 | CM008184.2 | IMGT000046 | |||
| TRA | 10 | CM008177.2 | IMGT000049 | |||
| TRD | 10 | CM008177.2 | IMGT000049 | |||
| Bos taurus (bovine) Breed: Holstein | Unknown | IGH | 21q24 | Unknown | * | |
| Bos taurus (bovine) | Unknown | TRG | 4 | Unknown | * | |
| Camelus dromedarius (Arabian camel) | CamDro3 | IGK | 28 | CM016654.2 | IMGT000061 | |
| Canis lupus familiaris (dog) Breed: Boxer | CanFam3.1 | IGH | 8 | CM000008.3 | IMGT000001 | |
| IGK | 17 | CM000017.3 | IMGT000002 | |||
| IGL | 26 | CM000026.3 | IMGT000003 | |||
| TRA | 8 | CM000008.3 | IMGT000004 | |||
| TRB | 16 | CM000016.3 | IMGT000005 | |||
| TRD | 8 | CM000008.3 | IMGT000004 | |||
| TRG | 18 | CM000018.3 | IMGT000006 | |||
| Canis lupus familiaris (dog) Breed: Basenji | Basenji_breed-1.1 | IGK | 17 | CM016447.1 | IMGT000067 | |
| Capra hircus (goat) Breed: San Clemente | ARS1 | IGK | 11 | CM004572.1 | IMGT000009 | |
| IGL | 17 | CM004578.1 | IMGT000033 | |||
| Equus caballus (horse) Breed: Thoroughbred | EquCab3.0 | IGH | 24 | CM009171.1 | IMGT000040 | |
| IGK | 15 | CM009162.1 | IMGT000053 | |||
| Equus caballus (horse) Breed: Thoroughbred | EquCab2.0 | IGK | 15 | CM000391.2 | IMGT000060 | |
| Felis catus (domestic cat) Breed: Abyssinian | Felis_catus_9.0 | IGK | A3 | CM001380.3 | IMGT000050 | |
| IGL | D3 | CM001389.3 | IMGT000038 | |||
| TRA | B3 | CM001383.3 | IMGT000045 | |||
| TRB | A2 | CM001379.3 | IMGT000037 | |||
| TRD | B3 | CM001383.3 | IMGT000045 | |||
| TRG | A2 | CM001379.3 | IMGT000036 | |||
| Macaca fascicularis (crab-eating macaque) | Macaca_fascicularis_5.0 | TRB | 3 | CM001921.1 | IMGT000075 | |
| Macaca mulatta (Rhesus monkey) Isolate: AG07107 | Mmul_10 | IGH | 7 | CM014342.1 | IMGT000064 | |
| IGK | 13 | CM014348.1 | IMGT000063 | |||
| IGL | 10 | CM014345.1 | IMGT000062 | |||
| TRB | 3 | CM014338.1 | IMGT000073 | |||
| TRG | 3 | CM014338.1 | IMGT000059 | |||
| Macaca mulatta (Rhesus monkey) Isolate: 17573 | Mmul_8.0.1 | TRA | 7 | CM002991.3 | IMGT000013 | |
| TRB | 3 | CM002984.2 | IMGT000012 | |||
| TRD | 7 | CM002991.3 | IMGT000013 | |||
| Mustela putorius furo (Domestic ferret) Breed: Sable | MusPutFur1.0 | TRB | Unknown | Unplaced genomic scaffold | IMGT000023 | |
| Oryctolagus cuniculus (rabbit) Breed: Thorbecke inbred | OryCun2.0 | TRA | 17 | CM000806.1 | IMGT000031 | |
| TRB | Unknown | Unplaced genomic scaffold | IMGT000032 | |||
| TRD | 17 | CM000806.1 | IMGT000031 | |||
| TRG | 10 | CM000799.1 | IMGT000030 | |||
| Ovis aries (sheep) Breed: Texel | Oar_v4.0 | IGK | 3 | CM001584.2 | IMGT000010 | |
| IGL | 17 | CM001598.2 | IMGT000034 | |||
| Ovis aries (sheep) Breed: Rambouillet | Oar_rambouillet_v1.0 | IGL | 17 | CM008488.1 | IMGT000041 | |
| TRA | 7 | CM008478.1 | IMGT000048 | |||
| TRB | 4 | CM008475.1 | IMGT000042 | |||
| TRD | 7 | CM008478.1 | IMGT000048 | |||
| Rattus norvegicus (Norway rat) Strain: BN; Sprague-Dawley | Rn_Celera Alternate Assembly AC_000074.1 | IGH | 6q32,33 | CM000236.2 | * | |
| Sus scrofa (pig) Breed: Duroc | Sscrofa11.1 | TRB | 18 | CM000829.5 | IMGT000039 | |
| Tursiops truncatus (bottlenose dolphin) | turTru1 (Ensembl assembly) | TRA | Unknown | Ensembl genomic scaffold | IMGT000016 | |
| IMGT000017 | ||||||
| IMGT000018 | ||||||
| IMGT000020 | ||||||
| TRD | Unknown | IMGT000016 | ||||
| IMGT000017 | ||||||
| IMGT000018 | ||||||
| Tursiops truncatus (bottlenose dolphin) Isolate: MMESES2002162SC | NIST Tur_tru v1 | TRG | Unknown | Unplaced genomic scaffold | IMGT000015 | |
| Aves | Gallus gallus (chicken) Breed: Red Jungle fowl | GRCg6 | IGH | 31 | CM003638.2 | IMGT000014 |
| Gallus_gallus-5.0 | Unknown | Unplaced genomic scaffold | IMGT000007 | |||
| Teleostei | Danio rerio (zebrafish) Isolate: Tuebingen | GRCz11 | IGH | 3 | CM002887.2 | * |
| Oncorhynchus mykiss (Rainbow trout) Isolate: Swanson | Omyk_1.0 | IGH | 13 | CM007947.1 | IMGT000043 | |
| 12 | CM007946.1 | IMGT000044 | ||||
| Salmo salar (Atlantic salmon) Breed: double haploid | ICSASG_v2 | IGH | 6 | CM003284.1 | IMGT000028 | |
| 3 | CM003281.1 | IMGT000029 |
IMGT® has recently performed the biocuration of the IG and TR loci of several veterinary species which are useful for biotechnological applications that can also be applied to human medicine (21–27). IMGT Biocuration makes possible the understanding of the gene characterization and the genomic organization of IG and TR, which provide a better understanding of the adaptive immune response.
AXIS II Exploring the expressed IG and TR repertoires
The analysis of the expressed IG and TR repertoires has become an essential step for the study and the understanding of the adaptive response in normal (infectious diseases, vaccination) and pathological situations (autoimmune diseases, cancers) especially since the advent of high throughput sequencing (HTS) over a decade ago. Basically, this analysis relies on the comparison of the expressed V-DOMAIN with the reference sequences of IG and TR genes and alleles. The dedicated and widely used IMGT tools for the IG and TR V-DOMAIN nucleotide sequence analysis are IMGT/V-QUEST (17) and its high throughput version IMGT/HighV-QUEST (28,29).
The IMGT/V-QUEST reference directories used by both tools for sequence comparison are defined of IG and TR gene and allele data from species managed in IMGT/GENE-DB and in the IMGT Web resources. They comprise one sequence per V-REGION, D-REGION, J-REGION of functional, ORF and in-frame pseudogenes V, D and J genes and alleles respectively. V-REGION are gapped according to the IMGT unique numbering (18). Table 3 summarizes the IMGT/V-QUEST reference directories per species and locus available for V-DOMAIN analysis.
Table 3.
IMGT/V-QUEST reference directories for the analysis of rearranged V-DOMAIN (release 202135–4 on 2 September 2021).
| IMGT/V-QUEST reference directories | |||
|---|---|---|---|
| Taxon | Species | IG | TR |
| MAMMALIA EUTHERIA (placentals) | Homo sapiens (human) | IGH, IGK, IGL | TRA, TRB, TRG, TRD |
| Mus musculus (mouse) | IGH, IGK, IGL | TRA, TRB, TRG, TRD | |
| Aotus nancymaae (Ma's night monkey) | TRA, TRG | ||
| Bos taurus (bovine) | IGH, IGK, IGL | TRA, TRG, TRD | |
| Camelus dromedarius (Arabian camel) | IGK | TRB, TRG | |
| Canis lupus familiaris (dog) | IGH, IGK, IGL | TRA, TRB, TRG, TRD | |
| Capra hircus (goat) | IGK, IGL | ||
| Equus caballus (horse) | IGH, IGK | ||
| Felis catus (domestic cat) | IGK, IGL | TRA, TRB, TRG, TRD | |
| Macaca fascicularis (crab-eating macaque) | IGH | TRB | |
| Macaca mulatta (Rhesus monkey) | IGH, IGK, IGL | TRA, TRB, TRG, TRD | |
| Mustela putorius furo (ferret) | TRB | ||
| Oryctolagus cuniculus (rabbit) | IGH, IGK, IGL | TRA, TRB, TRG, TRD | |
| Ovis aries (sheep) | IGH, IGK, IGL | TRA, TRB, TRD | |
| Rattus norvegicus (Norway rat) | IGH, IGK, IGL | ||
| Sus scrofa (pig) | IGH, IGK, IGL | TRB | |
| Tursiops truncatus (bottlenose dolphin) | TRA, TRG, TRD | ||
| Vicugna pacos (alpaca) | IGH | ||
| MAMMALIA PROTHERIA (monotremes) | Ornithorhynchus anatinus (platypus) | IGH | |
| Aves | Gallus gallus (chicken) | IGH, IGL | |
| Teleostei | Danio rerio (zebrafish) | IGH, IGI | TRA, TRD |
| Oncorhynchus mykiss (Rainbow trout) | IGH | TRB | |
| Salmo salar (Atlantic salmon) | IGH | ||
The classical functionalities of IMGT/V-QUEST and IMGT/HighV-QUEST tools have been described previously (17,28–30) and the main results deduced from alignments with the IMGT reference directories by the tools are listed in Table 4.
Table 4.
IMGT/V-QUEST reference directory based alignment results for nucleotide V-DOMAIN analysis
| IMGT/V-QUEST reference directory sets | IMGT tools | Results for IG and TR V-DOMAIN |
|---|---|---|
| V, D, J reference sequences per species and per locus | IMGT/V-QUEST IMGT/HighV-QUEST | 1. Introduction of IMGT gaps according to the IMGT unique numbering (18) |
| 2. Identification of the closest germline V, D and J genes and alleles | ||
| 3. Delimitation of the FR-IMGT and CDR-IMGT | ||
| Closest germline V gene and allele | 5. Identification of indels and their corrections (optional) (17) | |
| 6. Evaluation of the percentage of identity for the V-REGION | ||
| 7. Description of mutations and amino acid (AA) changes (transitions, transversions, codon change, qualification of AA change according to the eleven IMGT AA classes (31), localisation of mutation hotspot motifs) | ||
| Closest V, D, J genes and alleles | Performed by the integrated IMGT/JunctionAnalysis (32). | 8. Analysis of the Junction |
| IMGT/V-QUEST IMGT/HighV-QUEST | 9. Evaluation of the V-DOMAIN functionality | |
| Performed by the integrated IMGT/Automat (33) | 10. Complete V-DOMAIN annotation (33) | |
| IMGT/V-QUEST | 11. Advanced functionality for Clinical application: search for CLL subsets #2 and #8 (optional) (34,35) |
It should be noticed that the V-DOMAIN analysis based on the IMGT/V-QUEST directories has been extended to two new advanced functionalities, one related to the antibody engineering for analysis and annotation of scFv (sequences comprising 2 IG or TR V-DOMAIN covalently linked by a linker) (30) and the second one related to clinical applications with identification of sequences that could be assigned to stereotyped subsets 2 and 8 of Chronic Lymphocytic Leukemia (CLL), related to a non-favourable prognostic outcome (34,35). Interestingly, the characterization of the IMGT clonotypes (AA) and the evaluation of profiles for clonal diversity and expression (36) performed by statistic module of IMGT/HighV-QUEST and the subsequent statistical analysis (37) also rely on the results deduced from the alignment of the IMGT/V-QUEST reference directory sets.
IMGT reference directory sets are used by other external tools dedicated to IG and TR analysis based on sequence comparison such as IgBLAST (38) and MiXCR (39). The IMGT/V-QUEST reference directory sets are regularly enriched with the results of Axis I, whether it is the integration of a new species or the upgrade of existing repertoires. Each update gives rise to a new IMGT/V-QUEST reference directory release (see http://www.imgt.org/IMGT_vquest/data_releases). Links to the IMGT/V-QUEST reference directory sets per species, locus and gene type are available in IMGT reference directory in FASTA format (IG and TR) from http://www.imgt.org/vquest/refseqh.html#VQUEST and from the IMGT/V-QUEST Welcome page.
AXIS III IMGT 2D and 3D structure databases and tools for analysis of the adaptive immune proteins
Considering the great complexity of the immune proteins, their interactions with the antigens and their high number of published sequences, the classification and the detailed annotation are very difficult tasks, especially at the structural level. Therefore, a specialized 3D immune protein database was established to identify the genes and alleles encoding these proteins through alignment against the amino acid IMGT reference directory, provided by Axis I.
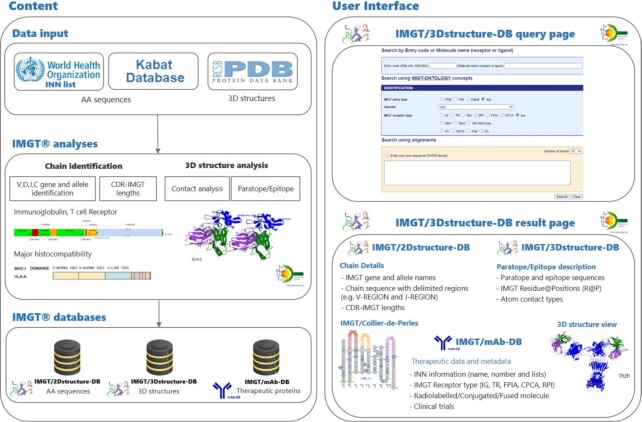
Since 2001, IMGT/3Dstructure-DB (9) has provided IMGT annotations and contact analysis for immune proteins structural data. From 2008 onwards, AA sequences of mAb and fusion proteins for immune applications from World Health Organization (WHO) - International Nonproprietary Names (INN) programme (40,41) are being incorporated in IMGT/2Dstructure-DB, a section of IMGT/3Dstructure-DB. To bring together information about therapeutic proteins and to facilitate their access, IMGT/mAb-DB was made available online in 2010. IMGT/mAb-DB extends 2D and 3D annotations with a unique resource on mAbs and relevant therapeutic metadata. Figure 2 provides a schematic representation of the whole procedure.
Figure 2.
Axis III workflow overview. The left panel displays all analysis processes: the data input panel shows the sources of the data present in the Axis III databases. The data analysis procedure is done by IMGT experts and it includes the analysis of the amino acid sequences and the 3D structures. Data are stored in IMGT/2Dstructure-DB and IMGT/3Dstructure-DB and linked to IMGT/mAb-DB. The right panel shows the user interface that provides flexible ways to query the data sets. The result page panel shows the online tools, such as IMGT/DomainGapAlign (Chain Details) and IMGT/Collier-de-Perles included in IMGT/2Dstructure-DB and Paratope/Epitope description and 3D structure incorporated in IMGT/3Dstructure-DB. IMGT/mAb-DB centralizes the information present in the other databases and adds several metadata for therapeutic proteins.
IMGT/3Dstructure-DB functionalities
The IMGT/3Dstructure-DB structural data are extracted from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) (42) and annotated according to the IMGT Scientific chart rules based on the IMGT-ONTOLOGY concepts (5,6,43). IMGT/3Dstructure-DB integrates the IMGT/DomainGapAlign tool (44), which aligns the AA sequences per domain, creates gaps according to the IMGT unique numbering and highlights differences with the closest reference genes and alleles found in the IMGT reference directory. 3D structure analysis includes chain annotation, paratope/epitope description of IG/antigen and TR/pMH complexes and contact analysis.
IMGT/2Dstructure-DB functionalities
The IMGT/2Dstructure-DB data include AA sequences of immune proteins, which are retrieved from WHO-INN programme (41) and from Kabat database (45). The AA sequences are analysed with the IMGT® criteria of the standardized IDENTIFICATION axiom, DESCRIPTION axiom, CLASSIFICATION axiom and NUMEROTATION axiom (5,6), and the V, C and G domain sequences are numbered according to the IMGT unique numbering (18,19,44).
Amino acid sequences from the WHO-INN programme have been provided since 2008 (IMGT entry type INN). This programme provides names for pharmaceutical substances recognized worldwide in biannual lists. The IMGT INN data include mAb, fusion proteins for immune application (FPIA), composite proteins for clinical applications (CPCA) and related proteins of the immune system (RPI). The INN name, INN number, common name, commercial name, Proposed and Recommended lists are available for each entry, along with the IMGT receptor description, the target and the molecule species. Recently, AA sequences of CAR-T (chimeric antigen receptor T cell) and TR were made available in IMGT/2Dstructure-DB, also from WHO-INN, after translating the nucleotide sequences and analysing them according to standardized IMGT information on chains and domains by IMGT experts.
IMGT/2Dstructure-DB and IMGT/3Dstructure-DB use the same interface via which amino acid sequences and 3D structures for immunological proteins can be queried and analysed. Currently, their algorithms have been revisited and they are more robust and efficient. Around 100 new structures are automatically retrieved from PDB per month. As of September 2021, the IMGT/3Dstructure-DB and IMGT/2Dstructure-DB have 7,657 entries, 6,533 PDB, 788 INN and 336 KAB.
IMGT/mAb-DB for therapeutic proteins
IMGT/mAb-DB provides a unique resource on mAbs and other therapeutic proteins. This database facilitates access to the therapeutic proteins present in IMGT/2Dstructure-DB and IMGT/3Dstructure-DB. The database is updated twice per year, in line with WHO-INN lists. In addition, metadata are constantly enriched from regulatory agencies as FDA and EMA. As of September 2021, the IMGT/mAb-DB contains 1,189 entries (1,033 IG, 53 RPI, 62 CPCA, 36 FPIA and 5 TR).
The IMGT/mAb-DB provides information about many therapeutic metadata. The ‘Specificity target name’ allows to select mAbs that bind to a particular antigen, for instance SARS-CoV-2. Results are returned in a table format, i.e. nine entries (eight mAbs and one CPCA) are shown for ‘Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)’ specificity target query. The common name, the INN name and number, as well as the Proprietary name (when available) are listed in the first columns. Following AA sequence analysis by IMGT® experts, the molecule information as receptor type, IG species, IG class and subclass are shown within the table. A standardized graphical format of the molecule, based on antibody INN definition, that facilitates the visualization of the molecule, is available in the database. Links to AA sequences (IMGT/2Dstructure-DB) and 3D structures (IMGT/3Dstructure-DB) are shown. The gene name of the target is linked to HGNC or VGNC pages that assign standardized names and unique symbols to genes for human or vertebrate loci, respectively (11). Other therapeutic metadata such as ‘Company’, ‘Clinical trials’ and ‘Authority decisions’ are also accessible in the result table.
The therapeutic monoclonal antibody engineering field represents a real promising potential in medicine (46–48). The rich, precise and standardized information available via IMGT/mAb-DB provides a unique and useful resource to the scientific community.
CONCLUSION
IMGT® provides to the scientific community a huge amount of knowledge and curated data in the field of immunogenetics, from genome to proteome through IMGT databases, IMGT tools and IMGT Web resources, which represent >20 000 html pages. To our knowledge, the richness of the website is still unmatched in 2021. IMGT metadata in the IMGT databases, tools and Web resources are based on IMGT-ONTOLOGY, the first ontology in immunogenetics and immunoinformatics. IMGT research and development rely on three main axes which correspond to the deciphering of the IG and TR loci, genes and alleles in the genomes of jawed vertebrates (Axis I), the exploration of the expressed IG and TR repertoires (Axis II), and the analysis of the 2D and 3D structures of the adaptive immune proteins (Axis III).
We focussed on the most recent data integrated in IMGT/LIGM-DB and IMGT/GENE-DB, the extraction of the complete IG and TR loci from genome assemblies and on the creation of terminology and new concepts for their annotation. A new section in IMGT/GENE-DB was created to provide links between genes and alleles of the IG and TR loci and their localization in genome assemblies (for interoperability with genome sites). IMGT tools and IMGT reference directories for the analysis of expressed IG and TR repertoire are regularly updated. Regarding the importance of the chemical interactions in the antibody specificity, affinity and half-life, the IMGT/2Dstructure-DB, IMGT/3Dstructure-DB and IMGT/mAb-DB provide an integrated and standardized approach for the description of new engineered antibody formats. This approach can be used for the construction and expression of engineered antibodies towards targeted and customized therapy in the context of personalized medicine.
The three IMGT axes are heavily interconnected and there is a constant flow of information among them. IMGT® is continuing the standardization efforts and the improvement of application of the FAIR principles (49) in order to enhance the quality, findability, accessibility, interoperability and reusability of IMGT data and metadata. To be Findable, IMGT databases use unique and persistent identifiers (IMGT/LIGM-DB, IMGT/2Dstructure-DB, IMGT/3Dstructure-DB and IMGT/mAb-DB) and are described with rich metadata based on IMGT-ONTOLOGY and IMGT Scientific chart rules. To be Accessible, IMGT data and metadata are freely available for academics. In addition, IMGT/GENE-DB can be dynamically queried through HTML direct links. To be Interoperable and Reusable, IMGT data and metadata have links to their sources and related databases, all IMGT sequence data are available in FASTA format, widely accepted by many bioinformatics programs and are described with their relevant attributes. Furthermore, the IMGT download sections for the IMGT reference directories ensure the follow up of new releases and facilitate the extraction and the reusability of the data by external tools.
DATA AVAILABILITY
IMGT® is freely available online for academics and non-profit use at http://www.imgt.org/. All the databases and tools referred to in this article are accessible from IMGT® webpage.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to Marie-Paule Lefranc, IMGT® founder in 1989, for her great expertise, daily assistance and continuous contribution to the research and development axes at IMGT®. We thank Gérard Lefranc and all members of the IMGT® team for their expertise and constant motivation. IMGT® is a registered trademark of CNRS. IMGT® is a member of the Confederation of Laboratories for Artificial Intelligence Research in Europe (CLAIRE), https://claire-ai.org/network/. IMGT® is a member of the International Medical Informatics Association (IMIA), https://imia-medinfo.org/wp/ and a member of the Global Alliance for Genomics and Health (GA4GH), https://www.ga4gh.org/. IMGT® is currently supported by the Centre National de la Recherche Scientifique (CNRS), the Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation (MESRI), the University of Montpellier, and the French Infrastructure Institut Français de Bioinformatique (IFB) ANR-11-INBS-0013. IMGT® is a member of BioCampus https://www.biocampus.cnrs.fr/index.php/fr/, MabImprove https://mabimprove.univ-tours.fr/en/ and IBiSA https://www.ibisa.net/.
Contributor Information
Taciana Manso, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Géraldine Folch, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Véronique Giudicelli, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Joumana Jabado-Michaloud, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Anjana Kushwaha, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Viviane Nguefack Ngoune, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Maria Georga, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Ariadni Papadaki, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Chahrazed Debbagh, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Perrine Pégorier, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Morgane Bertignac, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Saida Hadi-Saljoqi, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Imène Chentli, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Karima Cherouali, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Safa Aouinti, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Amar El Hamwi, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Alexandre Albani, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Merouane Elazami Elhassani, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Benjamin Viart, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Agathe Goret, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Anna Tran, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Gaoussou Sanou, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Maël Rollin, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Patrice Duroux, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
Sofia Kossida, IMGT®, the international ImMunoGeneTics Information System®, Scientific Research National Center (CNRS), Institute of Human Genetics (IGH), University of Montpellier (UM), Montpellier, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
IMGT® was funded in part by the BIOMED1 [BIOCT930038]; Biotechnology BIOTECH2 [BIO4CT960037]; 5th PCRDT Quality of Life and Management of Living Resources [QLG2-2000-01287]; 6th PCRDT Information Science and Technology [ImmunoGrid, FP6 IST-028069] programmes of the European Union (EU); IMGT® received financial support from the GIS IBiSA, the Agence Nationale de la Recherche (ANR) Labex MabImprove [ANR-10-LABX-53-01]; Région Occitanie Languedoc-Roussillon (Grand Plateau Technique pour la Recherche (GPTR), BioCampus Montpellier; IMGT® is granted access to the High Performance Computing (HPC) resources of Meso@LR and of the Centre Informatique National de l’Enseignement Supérieur (CINES), to Très Grand Centre de Calcul (TGCC) of the Commissariat à l’Energie Atomique et aux Énergies Alternatives (CEA) and Institut du développement et des ressources en informatique scientifique (IDRIS) [036029 (2010-2022)] made by GENCI (Grand Equipement National de Calcul Intensif).
Conflict of interest statement. The IMGT® software and data are provided to the academic users and NPO’s (Not for Profit Organization(s)) under the CC BY-NC-ND 4.0 license. Any other use of IMGT® material, from the private sector, needs a financial arrangement with CNRS. The authors declare that they do not have any conflict of interest for the work carried out within IMGT.
REFERENCES
- 1. Lefranc M.-P. Immunoglobulin and T cell receptor genes: IMGT® and the birth and rise of immunoinformatics. Front. Immunol. 2014; 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lefranc M.-P., Lefranc G.. The Immunoglobulin FactsBook. 2001; London, UK: Academic Press. [Google Scholar]
- 3. Lefranc M.-P., Lefranc G.. The T Cell Receptor FactsBook. 2001; London, UK: Academic Press. [Google Scholar]
- 4. Lefranc M.-P., Giudicelli V., Duroux P., Jabado-Michaloud J., Folch G., Aouinti S., Carillon E., Duvergey H., Houles A., Paysan-Lafosse T.et al.. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015; 43:D413–D422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duroux P., Kaas Q., Brochet X., Lane J., Ginestoux C., Lefranc M.-P., Giudicelli V.. IMGT-Kaleidoscope, the formal IMGT-ONTOLOGY paradigm. Biochimie. 2008; 90:570–583. [DOI] [PubMed] [Google Scholar]
- 6. Giudicelli V., Lefranc M.-P.. IMGT-ONTOLOGY 2012. Front. Genet. 2012; 3:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giudicelli V., Duroux P., Ginestoux C., Folch G., Jabado-Michaloud J., Chaume D., Lefranc M.-P.. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006; 34:D781–D784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giudicelli V., Chaume D., Lefranc M.-P.. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005; 33:D256–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ehrenmann F., Kaas Q., Lefranc M.-P.. IMGT/3Dstructure-DB and IMGT/DomainGapAlign: a database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res. 2010; 38:D301–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lefranc M.-P., Lefranc G.. Immunoglobulins or Antibodies: IMGT® Bridging Genes, Structures and Functions. Biomedicines. 2020; 8:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tweedie S., Braschi B., Gray K., Jones T.E.M., Seal R.L., Yates B., Bruford E.A.. Genenames.org: the HGNC and VGNC resources in 2021. Nucleic Acids Res. 2021; 49:D939–D946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lefranc M.-P. WHO-IUIS Nomenclature Subcommittee for immunoglobulins and T cell receptors report. Immunogenetics. 2007; 59:899–902. [DOI] [PubMed] [Google Scholar]
- 13. Amid C., Alako B.T.F., Balavenkataraman Kadhirvelu V., Burdett T., Burgin J., Fan J., Harrison P.W., Holt S., Hussein A., Ivanov E.et al.. The European Nucleotide Archive in 2019. Nucleic Acids Res. 2020; 48:D70–D76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2020; 48:D84–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kitts P.A., Church D.M., Thibaud-Nissen F., Choi J., Hem V., Sapojnikov V., Smith R.G., Tatusova T., Xiang C., Zherikov A.et al.. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 2016; 44:D73–D80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J.et al.. Ensembl 2021. Nucleic Acids Res. 2021; 49:D884–D891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brochet X., Lefranc M.-P., Giudicelli V.. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008; 36:W503–W508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lefranc M.-P., Pommié C., Ruiz M., Giudicelli V., Foulquier E., Truong L., Thouvenin-Contet V., Lefranc G.. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003; 27:55–77. [DOI] [PubMed] [Google Scholar]
- 19. Lefranc M.-P., Pommié C., Kaas Q., Duprat E., Bosc N., Guiraudou D., Jean C., Ruiz M., Da Piédade I., Rouard M.et al.. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev. Comp. Immunol. 2005; 29:185–203. [DOI] [PubMed] [Google Scholar]
- 20. Ohlin M., Scheepers C., Corcoran M., Lees W.D., Busse C.E., Bagnara D., Thörnqvist L., Bürckert J.-P., Jackson K.J.L., Ralph D.et al.. Inferred Allelic Variants of Immunoglobulin Receptor Genes: A System for their Evaluation, Documentation, and Naming. Front. Immunol. 2019; 10:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pégorier P., Bertignac M., Nguefack Ngoune V., Folch G., Jabado-Michaloud J., Giudicelli V., Duroux P., Lefranc M.-P., Kossida S.. IMGT® Biocuration and Comparative Analysis of Bos taurus and Ovis aries TRA/TRD Loci. Genes. 2020; 12:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pégorier P., Bertignac M., Chentli I., Nguefack Ngoune V., Folch G., Jabado-Michaloud J., Hadi-Saljoqi S., Giudicelli V., Duroux P., Lefranc M.-P.et al.. IMGT® Biocuration and Comparative Study of the T cell Receptor Beta Locus of Veterinary Species Based on Homo Sapiens. Front. Immunol. 2020; 11:821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Linguiti G., Kossida S., Pierri C.L., Jabado-Michaloud J., Folch G., Massari S., Lefranc M.-P., Ciccarese S., Antonacci R.. The T Cell Receptor (TRB) Locus in Tursiops truncatus: From sequence to structure of the Alpha/Beta Heterodimer in the Human/Dolphin Comparison. Genes. 2021; 12:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magadan S., Mondot S., Palti Y., Gao G., Lefranc M.P., Boudinot P.. Genomic analysis of a second rainbow trout line (Arlee) leads to an extended description of the IGH VDJ gene repertoire. Dev. Comp. Immunol. 2021; 118:103998. [DOI] [PubMed] [Google Scholar]
- 25. Radtanakatikanon A., Keller S.M., Darzentas N., Moore P.F., Folch G., Nguefack Ngoune V., Lefranc M.-P., Vernau W.. Topology and expressed repertoire of the Felis catus T cell receptor loci. BMC Genomics. 2020; 21:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Magadan S., Krasnov A., Hadi-Saljoqi S., Afanasyev S., Mondot S., Lallias D., Castro R., Salinas I., Sunyer O., Hansen J.et al.. Standardized IMGT® Nomenclature of Salmonidae IGH Genes, the Paradigm of Atlantic Salmon and Rainbow Trout: from Genomics to Repertoires. Front. Immunol. 2019; 10:2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mondot S., Lantz O., Lefranc M.-P., Boudinot P.. The T cell receptor (TRA) locus in the rabbit (Oryctolagus cuniculus): Genomic features and consequences for invariant T cells. Eur. J. Immunol. 2019; 49:2146–2158. [DOI] [PubMed] [Google Scholar]
- 28. Alamyar E., Giudicelli V., Li S., Duroux P., Lefranc M.-P.. IMGT/HighV-Quest: the IMGT® web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 2012; 8:26. [Google Scholar]
- 29. Alamyar E., Duroux P., Lefranc M.-P., Giudicelli V.. IMGT® tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 2012; 882:569–604. [DOI] [PubMed] [Google Scholar]
- 30. Giudicelli V., Duroux P., Kossida S., Lefranc M.-P.. IG and TR single chain fragment variable (scFv) sequence analysis: a new advanced functionality of IMGT/V-QUEST and IMGT/HighV-QUEST. BMC Immunol. 2017; 18:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pommié C., Levadoux S., Sabatier R., Lefranc G., Lefranc M.-P.. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J. Mol. Recognit. 2004; 17:17–32. [DOI] [PubMed] [Google Scholar]
- 32. Yousfi Monod M., Giudicelli V., Chaume D., Lefranc M.-P.. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004; 20:i379–85. [DOI] [PubMed] [Google Scholar]
- 33. Giudicelli V., Chaume D., Jabado-Michaloud J., Lefranc M.-P.. Immunogenetics Sequence Annotation: the Strategy of IMGT based on IMGT-ONTOLOGY. Stud. Health Technol. Inform. 2005; 116:3–8. [PubMed] [Google Scholar]
- 34. Agathangelidis A., Darzentas N., Hadzidimitriou A., Brochet X., Murray F., Yan X.-J., Davis Z., van Gastel-Mol E.J., Tresoldi C., Chu C.C.et al.. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood. 2012; 119:4467–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Agathangelidis A., Chatzidimitriou A., Gemenetzi K., Giudicelli V., Karypidou M., Plevova K., Davis Z., Yan X.-J., Jeromin S., Schneider C.et al.. Higher-order connections between stereotyped subsets: implications for improved patient classification in CLL. Blood. 2021; 137:1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li S., Lefranc M.-P., Miles J.J., Alamyar E., Giudicelli V., Duroux P., Freeman J.D., Corbin V.D.A., Scheerlinck J.-P., Frohman M.A.et al.. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat. Commun. 2013; 4:2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aouinti S., Giudicelli V., Duroux P., Malouche D., Kossida S., Lefranc M.-P.. IMGT/StatClonotype for Pairwise Evaluation and Visualization of NGS IG and TR IMGT Clonotype (AA) Diversity or Expression from IMGT/HighV-QUEST. Front. Immunol. 2016; 7:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ye J., Ma N., Madden T.L., Ostell J.M.. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013; 41:W34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M.. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015; 12:380–381. [DOI] [PubMed] [Google Scholar]
- 40. Lefranc M.-P. Antibody nomenclature: from IMGT-ONTOLOGY to INN definition. MAbs. 2011; 3:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. World Health Organization (WHO) International Nonproprietary Names (INN) for biological and biotechnological substances (a review). 2016; World Health Organization; INN Working Document 05.179. [Google Scholar]
- 42. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The protein data bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaas Q., Lefranc M.-P.. T cell receptor/peptide/MHC molecular characterization and standardized pMHC contact sites in IMGT/3Dstructure-DB. In Silico Biol. 2005; 5:505–528. [PubMed] [Google Scholar]
- 44. Ehrenmann F., Lefranc M.-P.. IMGT/DomainGapAlign: the IMGT® tool for the analysis of IG, TR, MH, IgSF, and MhSF domain amino acid polymorphism. Methods Mol. Biol. 2012; 882:605–633. [DOI] [PubMed] [Google Scholar]
- 45. Johnson G., Wu T.T.. Kabat database and its applications: 30 years after the first variability plot. Nucleic Acids Res. 2000; 28:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ciardiello F., Normanno N.. HER2 signaling and resistance to the anti-EGFR monoclonal antibody cetuximab: a further step toward personalized medicine for patients with colorectal cancer. Cancer Discov. 2011; 1:472–474. [DOI] [PubMed] [Google Scholar]
- 47. Schmid A.S., Neri D.. Advances in antibody engineering for rheumatic diseases. Nat. Rev. Rheumatol. 2019; 15:197–207. [DOI] [PubMed] [Google Scholar]
- 48. Shepard H.M., Phillips G.L., Thanos D.C., Feldmann M.. Developments in therapy with monoclonal antibodies and related proteins. Clin. Med. 2017; 17:220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E.et al.. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
IMGT® is freely available online for academics and non-profit use at http://www.imgt.org/. All the databases and tools referred to in this article are accessible from IMGT® webpage.