Abstract
Olfaction is a multi-stage process that initiates with the odorants entering the nose and terminates with the brain recognizing the odor associated with the odorant. In a very intricate way, the process incorporates various components functioning together and in synchronization. OlfactionBase is a free, open-access web server that aims to bring together knowledge about many aspects of the olfaction mechanism in one place. OlfactionBase contains detailed information of components like odors, odorants, and odorless compounds with physicochemical and ADMET properties, olfactory receptors (ORs), odorant- and pheromone binding proteins, OR-odorant interactions in Human and Mus musculus. The dynamic, user-friendly interface of the resource facilitates exploration of different entities: finding chemical compounds having desired odor, finding odorants associated with OR, associating chemical features with odor and OR, finding sequence information of ORs and related proteins. Finally, the data in OlfactionBase on odors, odorants, olfactory receptors, human and mouse OR-odorant pairs, and other associated proteins could aid in the inference and improved understanding of odor perception, which might provide new insights into the mechanism underlying olfaction. The OlfactionBase is available at https://bioserver.iiita.ac.in/olfactionbase/.
Graphical Abstract
Graphical Abstract.
OlfactionBase contains information about odors, odorant, olfactory receptors and odorant-receptor interaction.
INTRODUCTION
Olfaction is a complex, most primeval, volatile, and in comparison, to other senses, less explored multi-sensory mechanism which influences our innate response (1–7). It is highly dynamic, deceptive and complex, capable of detecting and distinguishing between a wide range of small volatile, lighter, hydrophobic compounds (odorants) with diverse chemical and structural properties, even at a very low concentration (parts per trillion) (8–13). The perception of olfaction arises from the interaction of odorants with the highly specific biological machinery, i.e., ORs present in the nasal/ olfactory epithelium (OE) (14–20). A unique pattern of neuronal signals is produced for each odorant, consisting of signal intensity, time, and quality of odorant stimuli which further stimulates a specific population of olfactory sensory neurons (OSNs) present in OE, which process and transduce the signals at the neurological level (21–34). The perception of neural signals produces a representation known as ‘smell/ odor,’ which is semantically represented by various perceptual descriptors such as fruity, rose, woody, etc.
The challenge associated with the olfaction process is daunting because odors are insubstantial, have a complex molecular basis, and are perceived individually (35–39). Odors associated with any substance (flower, plant, etc.) are made up of various odorants, some of which are significant contributors, and others are minor contributors. For example, the main constituent of rose smell is (−)-cis rose oxide, while the minor constituent is beta-damascenone, farnesol, geraniol, etc. The relationship between odorant chemistry and odor perception is the core of olfaction research. Since an odorant can have several odors (eugenol methyl ether associated with 27 odor perceptions), two structurally different chemicals can have virtually the same odor profile (cis-3-hexenol, nonadienal, ligustral exhibits green odor). A slight structural difference can result in distinct odors (carvone enantiomers, (R)-(−)-carvone (spearmint odor) and (S)-(+)-carvone (caraway odor)); this complex relationship is largely unknown until today. Odors are further encoded using a combinatorial approach, in which structurally identical odorants bind to completely different but overlapping olfactory receptors (ORs), hence significantly increasing the problem's complexity (40,41). There is no adequate scientific explanation for how smell is interpreted, particularly in humans. Although some pieces of the puzzle have been discovered in certain species, we still lack a comprehensive understanding of the phenomenon (42,43).
Several databases for odors, chemical compounds, and ORs have been published, but they are still limited in scope and concentrate on specific aspects (Table 1). Some resources focus solely on odorant compounds, others on odorant−OR pairs, and others on odors alone. As a result, these databases are fine but only useful for specific purposes, necessitating creating a robust website with all knowledge available at a single click. Also, there is no database available for odorless compounds. In this study, we created OlfactionBase with the aim of integrating multidimensional aspects of significant components involved in the olfaction mechanism, i.e., odors, chemicals (odorants and odorless), ORs, odorant−ORs interaction, and other associated proteins (odorant-binding proteins (OBPs), pheromone binding proteins (PBPs), and chemosensory proteins). OlfactionBase is a manually curated comprehensive database collating information from various resources, comprised of 106 primary odors, 572 subodor types, 3985 odorant molecules, 1124 odorless compounds, 2067 ORs (human and Mus musculus) compiled from various sources. OlfactionBase stores 874 (408 Human and 466 mouse) odorant−OR interaction information, thereby provides a platform for comparative analysis and quantitative structure odor relationship (QSOR) studies. All the data was manually compiled and extracted from the literature and database searches.
Table 1.
Summary of different databases related to olfaction.
| Content type | Database | Description | Website address |
|---|---|---|---|
| Odors and Odorants | AroChemBase | A database of compounds for aroma and chemical analysis using gas chromatography and Kovats index calculations. | http://www.alpha-mos.com/ |
| AromaDB (44) | A database of medicinal and aromatic plant's aroma. | http://bioinfo.cimap.res.in/aromadb/ | |
| EssOilDB (45) | A database of plants essential oils reflecting terpene composition and GC/MS data. | http://nipgr.res.in/Essoildb/ | |
| FlavorBase | A database of flavoring materials and food additives. | http://www.leffingwell.com/flavbase.htm | |
| Flavordb (46) | A database of flavor molecules. | https://cosylab.iiitd.edu.in/flavordb/ | |
| Flavornet (47) | A database of volatile compounds related to human olfaction. | http://www.flavornet.org/flavornet.html | |
| mVOC (48) | A database of microbial volatiles. | http://bioinformatics.charite.de/mvoc | |
| Pubchem (49) | An open chemistry database for small and large molecules with information on chemical structures, chemical, and physical properties, and many others. | https://pubchem.ncbi.nlm.nih.gov/ | |
| ScentBase (50) | A database of floral scent components. | http://www2.dpes.gu.se/SCENTbase.html/ | |
| Sigma Aldrich (51) | A database for flavors, fragrances, and associated chemicals. | https://www.sigmaaldrich.com/industries/flavors-and-fragrances.html | |
| SuperScent (52) | A database of volatille compounds associated with scents, including synthetic compounds. | http://bioinf-applied.charite.de/superscent/ | |
| TGSC | A database of flavor, fragrance, food, and cosmetic industries. | http://www.thegoodscentscompany.com/ | |
| Olfactory receptors | HORDE (53) | A database of human olfactory receptors. | https://genome.weizmann.ac.il/horde/ |
| ORDB (54) | A database of vertebrate OR genes and proteins. | https://senselab.med.yale.edu/ordb/ | |
| UniProt (55) | A comprehensive resource of protein sequence and functional information. | https://www.uniprot.org/ | |
| Odorant-OR interaction | OdoRactor (56) | A web server on odorant identification and olfactory repertoire browse. | http://mdl.shsmu.edu.cn/ODORactor/ |
| OlfactionDB (57) | A database of olfactory receptors and their Ligands | http://molsim.sci.univr.it/OlfactionDB. |
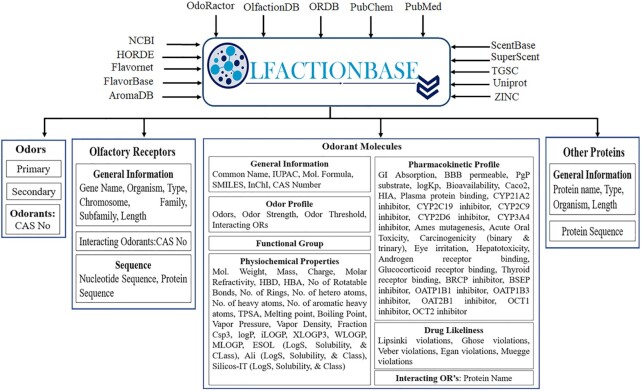
One feature that sets OlfactionBase apart from similar resources is that it presents information in a hierarchy of primary odors, sub-odors, odorants with their odor profile, interacting ORs, and chemical profile, including physicochemical, functional groups, ADMET (absorption, distribution, metabolism, excretion, toxicity) properties. The hierarchy is represented dynamically in the form of Olfaction-Wheel, which offers an integrated platform to explore odor-space, starting from the aroma-wheels suggested by different researchers and terminating at chemicals associated with an odor along with their interacting ORs and chemical information. Hence, OlfactionBase combines different dimensions of odorants constituting the ‘odor space’, ‘interaction space’ and ‘properties space’. Similarly, the general profile of ORs includes sequence, family, organism, length, and interacting odorants. OlfactionBase also houses 2871 entries related to odorant/pheromone binding and chemosensory proteins. OlfactionBase offers a robust dataset backed by an innovative visualization, user-friendly interface, and inter-linked search options for exploring components related to olfaction research at a single platform (Figure 1). OlfactionBase thus paves the way for a better understanding of odor perception due to the dynamic interplay between odors, odorant molecules and their properties with olfactory receptors interaction information, leading to future research directions for scientists.
Figure 1.
OlfactionBase database architecture. OlfactionBase offers a robust dataset of odors, odorants, olfactory receptors information, user-friendly interface and interlinked search options.
DATABASE OVERVIEW
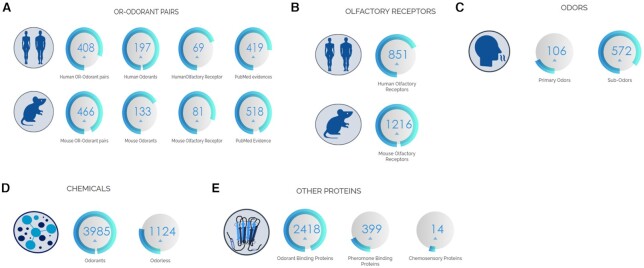
OlfactionBase is a repository with extensive coverage of 5109 chemicals, 2067 ORs, 874 OR-odorant pairs, 106 primary odors and 572 subodors. The chemicals data comprises 3985 odorant and 1124 odorless compounds. Further, the chemical molecules are classified into 30 functional groups and mapped to 572 subodors. OlfactionBase houses the information of ORs from two organisms, i.e., human (851) and Mus musculus (1215). It also lists 2871 OBP/ PBP protein information from 190 species.
OlfactionBase has a simple, user-friendly, and intuitive interface for querying and browsing odors, chemicals, and ORs. Interactive data visualizations such as the Olfaction wheel and interlinked textual and drawing-based (for chemical structure) search options are provided to retrieve relevant information. The Olfaction Wheel allows the user to interactively browse, backward and forward, through the odor classifications to access corresponding odorant molecules and subsequently obtain details of the odorant's chemical profile and interacting ORs. Thus, OlfactionBase provides a broad spectrum of information facilitating insights into the olfaction research through dynamic interface and visualizations.
DATA COMPILATION
The aim of developing OlfactionBase was to organize all information related to olfaction machinery under one umbrella. A list of odors and sub-odors was created from nine classification systems. The overlapping information between classification systems was manually examined and classified into 106 primary odors and 572 subodors.
Each chemical molecule was mapped to 30 functional groups. We provided the standard CAS (Chemical Abstract Service) for each chemical mapped to their corresponding Pubchem and ZINC IDs. Since CAS numbers are degenerate and often point to multiple molecules, PubChem IDs were used as the unique primary key for every chemical. Using PubChem Id, compounds identifiers (such as IUPAC name, common name, physicochemical properties, 2D images, and structure (.mol) files) were obtained from PubChem FTP service. The odor profile of odorants was created by compiling information and mapped using data collected from AromaDB (44), FlavorBase (http://www.leffingwell.com/flavbase.htm), Flavornet (47), PubChem (49), Sigma Aldrich (51), SuperScent (52), TGSC (http://www.thegoodscentscompany.com/), Odoractor (56), and OlfactionDB 57), followed by redundancy removal. Further ADMET, few physicochemical properties and Drug likeness for all 5109 chemicals were obtained from admetsar2 web server (58).
The information of human and mouse ORs was collected from UniProt, HORDE, ORDB database. We provide its UniProt ID, GenBank Accession number, ORDB, and HORDE identifier, and links to respective databases for each receptor. Using UniProt IDs, receptor information such as length, family, subfamily, chromosome number, protein, and nucleotide sequences were obtained. The other proteins related to olfaction are collected using keyword search (‘odorant-binding protein’, chemosensory protein, and pheromone binding protein) in the UniProt database. The OR-odorant interaction information was collected from an extensive literature survey (59–90) and database search. The supporting literature evidence from PubMed for odorant-OR pairs was provided for each entry. Figure 2 refers to the statistics of entities of OlfactionBase.
Figure 2.
Statistics of OlfactionBase. (A) Numbers of OR-odorant pairs in humans and mouse, (B) numbers of olfactory receptors, (C) numbers of odors, (D) numbers of chemicals, (E) number of other proteins related to olfaction.
DATABASE ARCHITECTURE AND WEB INTERFACE
OlfactionBase is a free, open-access web server powered by MySQL relational database management system. For better performance, data is kept in interrelated tables logically (Figure 3). All the tables are optimized to serve their purpose. The webserver has been built using the PHP Laravel framework, to deploy dynamic pages onto the server. Two JavaScript libraries (D3.js and Vue.js) have been used to develop the Olfaction Wheel and inner graphs. Other applications like composer, git, CSS3 and HTML were also used to create the entire web application of OlfactionBase.
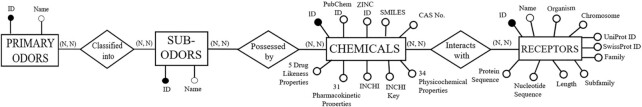
Figure 3.
Relational scheme. Each type of entry, i.e., odor, chemical, or receptor, is represented by a different field, depending on the nature of the molecule. Odors are linked to chemicals, while chemicals are linked to receptors using the interaction information from published literature.
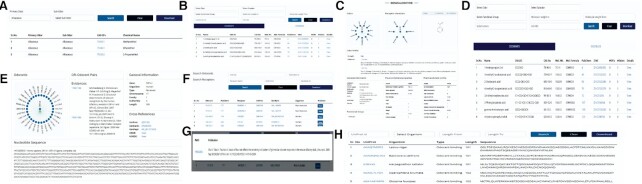
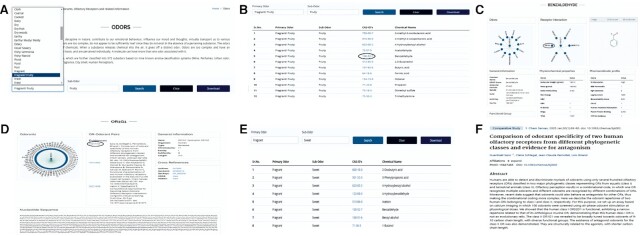
OlfactionBase provides a user-friendly web interface to browse odorants, odors, olfactory receptors, and odorant-OR pairs. On the ‘Home’ page, users can find a summary of OlfactionBase, including the total number of OR-odorant pairs with evidence for human and M. musculus, ORs, chemicals and other proteins. ‘Odors’ page lists out all primary and subodors along with their CAS No. of odorants in a tabular format (Figure 4A). Users can select primary odor and subodor information from the dropdown menus to obtain a list of related compounds. ‘Chemical page list out odorants and odorless compounds in separate tables. General information related to chemicals like Common name, SMILES, CAS No., molecular weight, molecular formula, PubChem ID, ZINC ID, number of interacting ORs and number of odors is given in a tabular format (Figure 4B). By clicking on ‘View’, users can view detailed information about chemicals (odorant/odorless) (Figure 4C). The various search options allow the user to search for a chemical compound within the database based on: (a) CAS No, (b) odors, (c) substructure (SMILES based textual search), (d) functional groups, (e) molecular weight (using lower and upper limit), (f) JSME applet to draw a chemical structure. ‘Receptors’ page contains general information about human and M. musculus olfactory receptors like name, organism, length, family, UniProt ID, GenBank ID, number of odorants, etc., in a tabular format (Figure 4D). Users can view nucleotide sequence, protein sequence, and other information related to ORs by clicking on ‘View’ on the ‘Receptors’ page (Figure 4E). The receptor can be searched using various search options given page like: (a) organism, (b) GenBank accession no., (c) chromosome, (d) family and (e) length of the sequence. On ‘OR−odorant pairs’ page, users can find information ORs interacting with odorants (Figure 4F). Related evidences for each OR-odorant pair can be viewed by clicking on the ‘View’ button (Figure 4G). Users can explore proteins other than ORs related to olfaction on the ‘OBP/PBP’ page. It summarises the general information about OBPs, PBPs and chemosensory proteins from 190 species (Figure 4H). User can search for proteins using UniProt ID, organism, and length. All pages are interlinked, enabling users to easily navigate the pages to gain more information about the desired entity. Figure 5 shows how to look for the ‘fruity’ sub-odor and its related odorants, as well as their interacting ORs.
Figure 4.
Overview of the OlfactionBase web interface. Snapshot of (A) odors page, (B) chemical's page, (C) detailed information regarding chemical, (D) olfactory receptors page, (E) detailed information of OR, (F) OR−odorant pairs page, (G) evidence for each OR−odorant pair, (H) information related to OBP, PBP and chemosensory proteins.
Figure 5.
Explore OlfactionBase to look for a ‘fruity’ odor. (A) Using the dropdown menu on the odors page, enter the primary odor and sub-odor. (B) Search results listing odorants with a ‘fruity’ sub-odor. (C) Click on CAS no. gives detailed information about the odorant ‘Benzaldehyde’, including its odors graph, Interacting ORs graph, odor profile, physicochemical parameters, etc. (D) Click on OR1G1 node in Interaction graph gives detailed information about the OR1G1 receptor. (E) Users can explore different odors linked to ‘Benzaldehdye’ and associated odorants by clicking on any node (‘Sweet’) in the odors graph. (F) Experimental evidence of OR−odorant interaction (OR1G1-benzaldehyde).
OLFACTION WHEEL
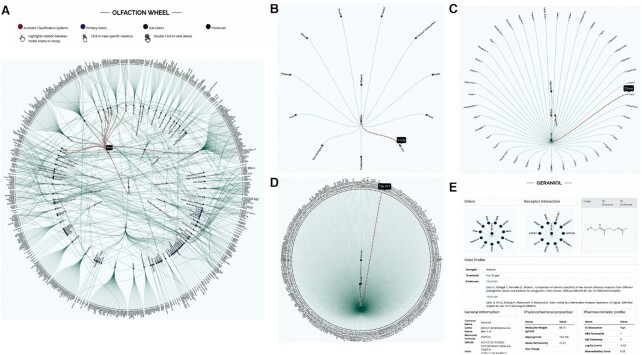
Various industry-dependent odor classification systems are developed for conveniently learning and remembering the odors in the form of circular diagrams/wheels. These visual odor wheels are constructed to make it easy to understand the relationship between complex and simple odors. The available aroma wheels and work done by Dr Kate Mclean (91) inspired the development of the Olfaction Wheel. Olfaction Wheel, a dynamic circular diagram, shows the inferred relationships among known aroma classification systems, odorants, and how smells are categorized. Odor classification from known classification systems is combined to design an Olfaction wheel, revealing the connection between all classification systems and chemicals possessing the odors. The nine seminal classification systems were chosen from research in established fields: City Smell (92), Compost (93), Drinking Water (94), Fragrance (95), Human Perception (96), Perfumery (97), Urban (98), Waste-water (99) and Wine (100). Odors verbatim not covered in the nine classification systems were added as ‘Other.’ The 106 primary odors were aggregated in alphabetic order, connected to 572 sub-odors, 30 functional groups and 3985 odorants.
The olfaction wheel comprises four concentric circles (Figure 6), the innermost circle is derived from compiling information from 10 aroma classification systems. The second and third circles list the primary and subodors and the relation between them. The fourth circle comprises odorants and is linked to sub-odors associated with them. The first three circles (classification system, primary odors, subodors) appear on the Olfaction Wheel homepage, while the fourth circle is visible in the subgraph, which appears on navigating deeper into the graph. All nodes are interlinked, making the graph's exploration (forward and backward) easy to understand visually and conceptually. A single click on a node explores the relationship/ connectivity between the node in question, its parents, and child nodes and helps navigate the wheel (forward and backward). Double click on any node lands on the page containing detailed information related to the node.
Figure 6.
Olfaction Wheel is a dynamic interlinked graph to visualize the relation between primary odors, subodors, and odorants. A single click on the node is for backward or forward navigation, while a double click lands on a page with information about the node in question. (A) Olfaction Wheel showing the relation between classification systems (red), primary odors (blue), subodors (green). Mouse hover on Wine highlights the associated primary odors, (B) Click on Wine (classification system, red) node generates the subgraph showing its relation to primary odors (blue). (C) Click on Fruity (primary odor, green) generates a subgraph showing its linkage to the classification system (red) and sub-odors (blue). (D) Click on Citrus (subodor, green) generates a subgraph containing CAS No. (black) of all compounds associated with it. (E) Double click on CAS No. (odorant, black) opens up a page containing detailed information about odorants chemical, physiochemical, odor profile, etc.
COMPARISON WITH ODORACTOR
OdoRactor (56) and OlfactionBase appear to have overlapping information. OdoRactor is a web server that predicts the ORs for small-molecule compounds by combining two separate techniques; characterising an odorant and then predicting its candidate ORs. However, OlfactionBase is an open-access database that brings together experimental knowledge on different aspects related to olfaction mechanism at one place from a variety of sources, as mentioned above in data compilation section. The data statistics in both databases are shown in Table 2.
Table 2.
Summary and comparison of OlfactionBase with OdoRactor (56).
| Data | Odoractor | OlfactionBase |
|---|---|---|
| OR | 1608 | 2067 |
| Odorants | 3038 | 3985 |
| Odorless compounds | 0 | 1124 |
| OR-odorant pair | 627 | 874 |
| Odors | No | Yes (primary odors: 106 sub-odors: 572) |
| Dynamic representation of odors-odorants relation | No | Yes |
| Download option | No | Yes |
CONCLUSIONS
The current version of OlfactionBase contains comprehensive information for (a) 3985 odorant compounds, (b) 1124 odorless compounds, (c) 2871 OBP/PBP proteins. It also includes 408 and 466 Odorant-OR interaction pairs for humans and M. musculus, respectively. The odorant−OR pairs in OlfactionBase include 197 odorants, 67 receptors, and 419 associations among Humans and 133 ligands, 81 receptors and 518 associations among M. musculus (Figure 2). All the data were extracted and annotated from published articles; the majority of data was indeed extracted from the seminal work of Saito et al. (69, 83). A typical entry of a chemical compound offers a chemical characterization of the compound, including H-bond donors and acceptors, molecular weight and mass, log P and log K values among 34 physicochemical properties, 31 pharmacokinetic based properties and five drug-likeness violations. A typical entry for a receptor includes UniProt, GenPept, GenBank accession code, name, FASTA sequence (nucleotide and protein), location, family, and organism; an entry for odors includes primary odor classification and a list of compounds possessing a particular subodor. Moreover, direct links to the corresponding databases (UniProt, PubChem, ZINC, HORDE, ORDB, PubMed) entries are given in each of the entry pages. The database's relational scheme, as shown in Figure 3, on which the current database is based, is specifically designed to capture OR−odorant interaction pairs and odorant−odor preferences as stated in scientific literature and databases.
OlfactionBase offers several navigation and data retrieval options. For compounds, one can perform (a) CAS ID, (b) molecular weight, c) substructure based, (d) odor and (e) functional group-based searches. Similarly, for ORs, (a) GenBank accession number, (b) organism, (c) chromosome, (d) family and (e) length-based search options are available. One can explore compounds possessing specific odors based on primary and sub-group odors. OlfactionBase is a useful and informative resource for investigating odors-odorants and odorant−OR interactions in one place, and it may be a helpful tool for deciphering olfaction mechanisms. It could be valuable for both academic olfaction research and odorant discovery in the industry.
DATA AVAILABILITY
The website of OlfactionBase is available at https://bioserver.iiita.ac.in/olfactionbase/.
ACKNOWLEDGEMENTS
The authors acknowledge the logistic support of Systems Biology Lab − Department of Applied Science, Indian Institute of Information Technology – Allahabad (IIITA), Parayagraj, Uttar Pradesh, India. The authors are also thankful to the Ministry of Human Resource Development (MHRD), Government of India, for providing financial support as a monthly Research Scholarship.
Contributor Information
Anju Sharma, Department of Applied Science, Indian Institute of Information Technology, Allahabad, Uttar Pradesh 211015, India.
Bishal Kumar Saha, Gentryx Technologies, West Bengal, Kolkata, India.
Rajnish Kumar, Amity Institute of Biotechnology, Amity University Uttar Pradesh, Lucknow Campus, Uttar Pradesh 226028, India.
Pritish Kumar Varadwaj, Department of Applied Science, Indian Institute of Information Technology, Allahabad, Uttar Pradesh 211015, India.
FUNDING
No external funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Sela L., Sobel N.. Human olfaction: a constant state of change-blindness. Exp. Brain Res. 2010; 205:13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoskison E.E. Olfaction, pheromones and life. J. Laryngol. Otol. 2013; 127:1156–1159. [DOI] [PubMed] [Google Scholar]
- 3. Hayden S., Teeling E.C.. The molecular biology of vertebrate olfaction. Anat. Rec. (Hoboken). 2014; 297:2216–2226. [DOI] [PubMed] [Google Scholar]
- 4. Amoore J.E. Molecular Basis of Odor. 1970; Springfield, IL: C.C. Thomas Pub. [Google Scholar]
- 5. Mombaerts P. How smell develops. Nat. Neurosci. 2004; 4:1192–1198. [DOI] [PubMed] [Google Scholar]
- 6. Wright R.H. The Sense of Smell. 1982; Boca Raton, FL: CRC Press. [Google Scholar]
- 7. Lancet D., Ben-Arie N.. Olfactory receptors. Curr. Biol. 1993; 3:668–674. [DOI] [PubMed] [Google Scholar]
- 8. Heinz B. Olfactory receptors: molecular basis for recognition and discrimination of odors. Anal. Bioanal. Chem. 2003; 377:427–433. [DOI] [PubMed] [Google Scholar]
- 9. Sephr M., Munger S.D.. Olfactory receptors: G protein-coupled receptors and beyond. J. Neurochem. 2009; 109:1570–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Touhara K., Vosshall L.B.. Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 2009; 71:307–332. [DOI] [PubMed] [Google Scholar]
- 11. Walliczek-Dworschak U., Hummel T.. The human sense of olfaction. Facial Plast. Surg. 2017; 33:396–404. [DOI] [PubMed] [Google Scholar]
- 12. Majid A. Human olfaction at the intersection of language, culture, and biology. Trends Cogn. Sci. 2021; 25:111–123. [DOI] [PubMed] [Google Scholar]
- 13. Pinto J.M. Olfaction. Proc. Am. Thorac. Soc. 2011; 8:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Breer H., Fleischer J., Strotmann J.. The sense of smell: multiple olfactory subsytems. Cell Mol. Life Sci. 2006; 63:1465–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strotman J., Levai O., Fleischer J., Schwarzenbacher K., Breer H.. Olfactory receptor proteins in axonal processes of chemosensory neurons. J. Neurosci. 2004; 24:7754–7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Menco B.M., Bruch R.C., Dau B., Danho W.. Ultrastructural localization of olfactory transduction components: the G protein subunit Gold alpha and type III adenylyl cyclase. Neuron. 1992; 8:441–453. [DOI] [PubMed] [Google Scholar]
- 17. Mombaerts P. How smell develops. Nat. Neurosci. 2004; 4:1192–1198. [DOI] [PubMed] [Google Scholar]
- 18. Lodovich C., Bellusico L.. Odorant Receptors in the formation of the olfactory bulb circuitry. Physiology. 2012; 2:200–212. [DOI] [PubMed] [Google Scholar]
- 19. Kosaka T., Kosaka K.. Olfactory Anatomy. Reference Module in Biomedical Research. 2014; Elsevier. [Google Scholar]
- 20. Mombaerts P. Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 2014; 286:707–711. [DOI] [PubMed] [Google Scholar]
- 21. Sharma A., Kumar R., Aier I., Semwal R., Tyagi P., Varadwaj P.. Sense of smell: structural, functional, mechanistic advancements and challenges in human olfactory research. Curr. Neuropharmacol. 2019; 17:891–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kato A., Touhara K.. Mammmalian olfactory receptors: pharmacology, G protein coupling and desensitization. Cell. Mol. Life. Sci. 2009; 66:3743–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bradley J., Reisert J., Frings S.. Regulation of cyclic nucleotide-gated channels. Curr. Opin. Neurobiol. 2005; 15:343–349. [DOI] [PubMed] [Google Scholar]
- 24. Liman E.R., Buck L.B.. A second subunit of the olfactory cyclic nucleotide-gated channel confers high sensitivity to cAMP. Neuron. 1994; 13:611–621. [DOI] [PubMed] [Google Scholar]
- 25. Prasad B.C., Reed R.R.. Chemosensation: molecular mechanisms in worms and mammals. Trends Genet. 1999; 15:150–153. [DOI] [PubMed] [Google Scholar]
- 26. Schild D., Restrepo D.. Transduction mechanisms in vertebrate olfactory receptor cells. Physiol. Rev. 1998; 78:429–466. [DOI] [PubMed] [Google Scholar]
- 27. Gabriela A., Sebastiao A.M., de Souza F.M.S.. Mechanisms of regulation of olfactory transduction and adaptation in the olfactory cilium. PLoS One. 2014; 9:e105531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kurahashi T., Menini A.. Mechanism of odorant adaptation in the olfactory receptor cell. Nature. 1997; 385:725–729. [DOI] [PubMed] [Google Scholar]
- 29. Barnea G., O’Donnell S., Mancia F., Sun X., Nemes A., Mendelsohn M., Axel R.. Odorant receptors on axon termini in the brain. Science. 2004; 304:1468. [DOI] [PubMed] [Google Scholar]
- 30. Buck L.B. The search for odorant receptors. Cell. 2004; 116:S117–119. [DOI] [PubMed] [Google Scholar]
- 31. Eggan K., Baldwin K., Tackett M., Osborne J., Gogos J., Chess A., Axel R., Jaenisch R.. Mice cloned from olfactory sensory neurons. Nature. 2004; 428:44–49. [DOI] [PubMed] [Google Scholar]
- 32. Benovic J.L., Kuhn H., Weyand I., Codina J., Caron M.G., Lefkowitz R.J.. Functional desensitization of the isolated beta-adrenergic receptor by the beta-adrenergic receptor kinase: potential role of an analog of the retinal protein arrestin. Proc. Natl. Acad. Sci. U.S.A. 1986; 84:8879–8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lohse M.J., Benovic J.L., Codina J., Caron M.G., Lefkowiz R.J.. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990; 248:1547–1550. [DOI] [PubMed] [Google Scholar]
- 34. Boekhoff I., Inglese J., Schleicher S., Koch W.J., Lefkowitz R.J., Breer H.. Olfactory desensitization requires membrane targeting of receptor kinase mediated by beta gamma-subunits of heterotrimeric G proteins. J. Biol. Chem. 1994; 269:37–40. [PubMed] [Google Scholar]
- 35. Kermen F., Chakirian A., Sezillem C., Joussain P., Goff L.G., Ziessel A., Chastrette M., Mandairon N., Didier A., Rouby C.et al.. Molecular complexity determines the number of olfactory notes and the pleasantness of smells. Sci. Rep. 2011; 1:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keller A., Vosshall L.B.. Olfactory perception of chemically diverse molecules. BMC Neurosci. 2016; 17:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chastrette M. Rouby C., Schaal B., Dubois D., Gervais R., Holley A.. Classification of Odors and Structure–Odor Relationships. Olfaction, Taste, and Cognition. 2002; Cambridge: Cambridge University Press; 100–116. [Google Scholar]
- 38. Brookes J.C., Horsfield A.P., Stoneham A.M.. Odour character differences for enantiomers correlate with molecular flexibility. J R. Soc. Interface. 2009; 6:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rossiter K.J. Structure-odor relationships. Chem. Rev. 1996; 96:3201–3240. [DOI] [PubMed] [Google Scholar]
- 40. Sharma A., Kumar R., Semwal R., Aier I., Tyagi P., Varadwaj P.. DeepOlf: Deep neural network-based architecture for predicting odorants and their interacting Olfactory Receptors. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020; 10.1109/TCBB.2020.3002154. [DOI] [PubMed] [Google Scholar]
- 41. Sharma A., Kumar R., Ranjta S., Varadwaj P.. SMILES to smell: Decoding the structure-odor relationship of chemical compounds using the deep neural network approach. J. Chem. Inf. Model. 2021; 61:676–688. [DOI] [PubMed] [Google Scholar]
- 42. Sell C.S. On the unpredictability of odor. Angew. Chem. Int. Ed. Engl. 2006; 45:6254–6261. [DOI] [PubMed] [Google Scholar]
- 43. Kraft P., Bajgrowicz J.A., Denis C., Frater G.. Odds and trends: recent developments in the chemistry of odorants. Angew. Chem. Int. Ed. Engl. 2000; 39:2980–3010. [DOI] [PubMed] [Google Scholar]
- 44. Kumar Y., Prakash O., Tripathi H., Tandon S., Gupt M.M., Rahman L.U., Lal R.K., Semwal M., Darokar M.P., Khan F.. AromaDb: a database of medicinal and aromatic plant's aroma molecules with phytochemistry and therapeutic potentials. Front. Plant Sci. 2018; 13:1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumari S., Pundhir S., Priya P., Jeena G., Punetha A., Chawla K., Firdos J.Z, Mondal S., Yadav G.. EssOilDB: a database of essential oils reflecting terpene composition and variability in the plant kingdom. Database (Oxford). 2014; 22:bau120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garg N., Sethupathy A., Tuwani R., Nk R., Dokania S., Iyer A., Gupta A., Agrawal S., Singh N., Shukla S.et al.. FlavorDB: a database of flavor molecules. Nucleic Acids Res. 2018; 46:D1210–D1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arn H., Acree T.E.. Flavornet: a database of aroma compounds based on odor potency in natural products. Dev. Food Sci. 1998; 40:27. [Google Scholar]
- 48. Lemfack M.C., Gohlke B.O., Toguem S.M.T., Preissner S., Piechulla B., Preissner R.. mVOC 2.0: a database of microbial volatiles. Nucleic Acids Res. 2018; 46:D1261–D1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A.et al.. PubChem substance and compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Knudsen J.T., Eriksson R., Gershenzon J., Ståhl B.. Diversity and distribution of floral scent. Bot. Rev. 2006; 72:1–120. [Google Scholar]
- 51. Sigma-Aldrich Flavors and Fragrances Products Catalog. 2016; Darmstadt, Germany: Merck KGaA. [Google Scholar]
- 52. Dunkel M., Schmid U., Struck S., Berger L., Gruening B., Hossbach J., Jaeger I.S., Effmert U., Piechulla B., Eriksson R.et al.. SuperScent—a database of flavors and scents. Nucleic Acids Res. 2008; 37:D291–D294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Olender T., Nativ N., Lancet D.. HORDE: comprehensivere source for olfactory receptor genomics. Methods Mol. Biol. 2013; 1003:23–38. [DOI] [PubMed] [Google Scholar]
- 54. Healy M.D., Smith J.E., Singer M.S., Nadkarni P.M., Skoufos E., Miller P.L., Shepherd G.M.. Olfactory receptor database (ORDB): a resource for sharing and analyzing published and unpublished data. Chem. Senses. 1997; 22:321–326. [DOI] [PubMed] [Google Scholar]
- 55. The UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic. Acids. Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu X., Su X., Wang F., Huang Z., Wang Z., Li Z., Zhang R., Wu L., Pan Y., Chen Y.et al.. ODORactor: a web server for deciphering olfactory coding. Bioinformatics. 2011; 27:2302–2303. [DOI] [PubMed] [Google Scholar]
- 57. Modena D., Trentini M., Corsini M., Bombaci A., Giorgetti A.. OlfactionDB: a database of olfactory receptors and their ligands. Adv. Life Sci. 2011; 1:1–5. [Google Scholar]
- 58. Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y.. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019; 35:1067–1069. [DOI] [PubMed] [Google Scholar]
- 59. Malnic B., Hirono J., Sato T., Buck L.B.. Combinatorial receptor codes for odors. Cell. 1999; 96:713–723. [DOI] [PubMed] [Google Scholar]
- 60. Floriano W.B., Vaidehi N., Goddard W.A., Singer M.S., Shepher G.M.. Molecular mechanisms underlying differential odor responses of a mouse olfactory receptor. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:10712–10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kajiya K., Inaki K., Tanaka M., Haga T., Kataoka H., Touhara K.. Molecular bases of odor discrimination: reconstitution of olfactory receptors that recognize overlapping sets of odorants. J. Neurosci. 2001; 21:6018–6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gaillard I., Rouquier S., Pin J.P., Mollar P., Richard S., Barnabe C., Demaille J., Giorgi D.. A single olfactory receptor specifically binds a set of odorant molecules. Eur. J. Neurosci. 2002; 15:409–418. [DOI] [PubMed] [Google Scholar]
- 63. Bozza T., Feinstein P., Zheng C., Mombaerts P.. Odorant receptor expression defines functional units in the mouse olfactory system. J. Neurosci. 2002; 22:3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Spehr M., Gisselmann G., Poplawski A., Riffell J.A., Wetzel C.H., Zimmer R., Hatt H.. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003; 299:2054–2058. [DOI] [PubMed] [Google Scholar]
- 65. Oka Y., Omura M., Kataoka H., Touhara K.. Olfactory receptor antagonism between odorants. EMBO J. 2004; 23:120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Araneda R.C., Peterlin Z., Zhang X., Chesler A., Firestein S.. A pharmacological profile of the aldehyde receptor repertoire in rat olfactory epithelium. J. Physiol. 2004; 555:743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Godfrey P.A., Malnic B., Buck L.B.. The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:2156–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Malnic B., Godfrey P.A., Buck L.B.. The human olfactory receptor gene family. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:2584–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Saito H., Kubota M., Roberts R.W., Chi Q., Matsunami H.. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004; 119:679–691. [DOI] [PubMed] [Google Scholar]
- 70. Shirokova E., Schmiedeberg K., Bedner P., Niessen H., Willecke K., Raguse J.D., Meyerhof W., Krautwurst D.. Identification of specific ligands for orphan olfactory receptors. G protein-dependent agonism and antagonism of odorants. J. Biol. Chem. 2005; 280:11807–11815. [DOI] [PubMed] [Google Scholar]
- 71. Sanz G., Schlegel C., Pernollet J.C., Briand L.. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem. Senses. 2005; 30:69–80. [DOI] [PubMed] [Google Scholar]
- 72. Matarazzo V., Clot-Faybesse O., Marcet B., Guiraudie-Capraz G., Atanasove B., Devauchelle G., Cerutti M., ETievant P., Ronin C.. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem. Senses. 2005; 30:195–207. [DOI] [PubMed] [Google Scholar]
- 73. Jacquier V., Pick H., Vogel H.. Characterization of an extended receptive ligand repertoire of the human olfactory receptor OR17-40 comprising structurally related compounds. J. Neurochem. 2006; 97:537–544. [DOI] [PubMed] [Google Scholar]
- 74. Neuhaus E.M., Mashukova A., Zhang W., Barbour J., Hatt H.. A specific heat shock protein enhances the expression of mammalian olfactory receptor proteins. Chem. Senses. 2006; 31:445–452. [DOI] [PubMed] [Google Scholar]
- 75. Abaffy T., Matsunami H., Luetje C.W.. Functional analysis of a mammalian odorant receptor subfamily. J. Neurochem. 2006; 97:1506–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Oka Y., Katada S., Omura M., Suwa M., Yoshihara Y., Touhara K.. Odorant receptor map in the mouse olfactory bulb: in vivo sensitivity and specificity of receptor-defined glomeruli. Neuron. 2006; 52:857–869. [DOI] [PubMed] [Google Scholar]
- 77. Schmiedeberg K., Shirokova E., Weber H.P., Schillin B., Meyerhof W., Krautwurst D.. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J. Struct. Biol. 2007; 159:400–412. [DOI] [PubMed] [Google Scholar]
- 78. Keller A., Zhuang H., Chi Q., Vosshall L.B., Matsunami H.. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007; 449:468–472. [DOI] [PubMed] [Google Scholar]
- 79. Menashe I., Abaffy T., Hasin Y., Goshen S., Yahalom V., Luetje C.W., Lancet D.. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007; 5:e284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nguyen M.Q., Zhou Z., Marks C.A., Ryba N.J.P., Belluscio L.. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007; 131:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yoshikawa K., Touhara K.. Myr-Ric-8A enhances G(alpha15)-mediated Ca2+ response of vertebrate olfactory receptors. Chem. Senses. 2009; 34:15–23. [DOI] [PubMed] [Google Scholar]
- 82. Repicky S.E., Luetje C.W.. Molecular receptive range variation among mouse odorant receptors for aliphatic carboxylic acids. J. Neurochem. 2009; 109:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Saito H., Chi Q., Zhuang H., Matsunami H., Mainland J.D.. Odor coding by a mammalian receptor repertoire. Sci. Signal. 2009; 2:ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cook B.L., Steuerwald D., Kaiser L., Graveland-Bikker J., Vanberghem M., Berke A.P., Herlihy K., Pick H., Zhang S.. Large-scale production and study of a synthetic G protein-coupled receptor: human olfactory receptor 17-4. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:11925–11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Oka Y., Takai Y., Touhara K.. Nasal airflow rate affects the sensitivity and pattern of glomerular odorant responses in the mouse olfactory bulb. J. Neurosci. 2009; 29:12070–12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Olsson P., Laska M.. Human male superiority in olfactory sensitivity to the sperm attractant odorant bourgeonal. Chem. Senses. 2010; 35:427–432. [DOI] [PubMed] [Google Scholar]
- 87. Kurland M.D., Newcomer M.B., Peterlin Z., Ryan K., Firestein S., Batista V.S.. Discrimination of saturated aldehydes by the rat I7 olfactory receptor. Biochemistry. 2010; 49:6302–6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gonzalez-Kristeller D.C., Nascimento J.B., Galante P.A., Malnic B.. Identification of agonists for a group of human odorant receptors. Front. Pharmacol. 2015; 6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Harini K., Sowdhamini R.. Computational approaches for decoding select odorant-olfactory receptor interactions using mini-virtual screening. PLoS One. 2015; 10:e0131077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mainland J.D., Li Y.R., Zhou T., Liu W.L., Matsunami H.. Human olfactory receptor responses to odorants. Sci. Data. 2015; 2:150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. McLean K. Kobayashi A. Sensory Maps. International Encyclopaedia of Human Geography. 2019; 2nd ednElsevier. [Google Scholar]
- 92. Quercia D., Schifanella R., Aiello L., Mc Lean K.. Smelly Maps: The Digital Lives of Urban Smellscapes. 2015; Oxford: AAAI Publications, Ninth International AAAI Conference on Web and Social Media. [Google Scholar]
- 93. Suffet I.H., Decottignies V., Senante E., Bruchet A.. Sensory assessment and characterization of odor nuisance emissions during the composting of waste water biosolids. Water Environ. Res. 2009; 81:670–679. [DOI] [PubMed] [Google Scholar]
- 94. Suffet I.H., Khiari D., Bruchet A.. The drinking water taste and odor wheel for the millennium: beyond geosmin and 2-methylisoborneol. Water Sci. Technol. 1999; 40:1–13. [Google Scholar]
- 95. Edwards M. Fragrances of the World 2008. 2008; Michael Edwards & Co; ISBN 978-0-9756097-3-6. [Google Scholar]
- 96. Castro J.B., Ramanathan A., Chennubhotla C.S.. Categorical dimensions of human odor descriptor space revealed by non-negative matrix factorization. PLoS One. 2013; 8:e73289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Carl-Von L. Dissertation Medica Odores Medicamentorum Exhibens. 1752; United States: Kessinger Publishing. [Google Scholar]
- 98. Henshaw V. Urban Smellscapes: Understanding and Designing City Smell Environments. 2013; 2013:NY: Routledge. [Google Scholar]
- 99. Suffet M., Burlingame G., Rosenfeld P., Bruchet A.. The value of an odor-quality-wheel classification scheme for wastewater plants. Water Sci. Technol. 2004; 50:25–32. [PubMed] [Google Scholar]
- 100. Herdenstam A., Hammarén M., Ahlström R., Wiktorsson P.A.. The professional language of wine: perception, training and dialogue. J. Wine Res. 2009; 20:53–84. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The website of OlfactionBase is available at https://bioserver.iiita.ac.in/olfactionbase/.