Abstract
Drug response to many diseases varies dramatically due to the complex genomics and functional features and contexts. Cellular diversity of human tissues, especially tumors, is one of the major contributing factors to the different drug response in different samples. With the accumulation of single-cell RNA sequencing (scRNA-seq) data, it is now possible to study the drug response to different treatments at the single cell resolution. Here, we present CeDR Atlas (available at https://ngdc.cncb.ac.cn/cedr), a knowledgebase reporting computational inference of cellular drug response for hundreds of cell types from various tissues. We took advantage of the high-throughput profiling of drug-induced gene expression available through the Connectivity Map resource (CMap) as well as hundreds of scRNA-seq data covering cells from a wide variety of organs/tissues, diseases, and conditions. Currently, CeDR maintains the results for more than 582 single cell data objects for human, mouse and cell lines, including about 140 phenotypes and 1250 tissue-cell combination types. All the results can be explored and searched by keywords for drugs, cell types, tissues, diseases, and signature genes. Overall, CeDR fine maps drug response at cellular resolution and sheds lights on the design of combinatorial treatments, drug resistance and even drug side effects.
INTRODUCTION
Drug response to many diseases varies dramatically due to the complex genomics and functional features and contexts (1–3). As the majority of cellular systems are heterogeneous (4), the bulk RNA sequencing (RNA-seq) technologies may only measure the average expression level that includes a diverse collection of cells concealing cell type specific signatures with different levels of response to treatment (5). Much of the diversity in individual drug response can be attributed to cellular heterogeneity including cell types, proportions, cell-cell communications, and temporal-spatial distributions and compositions of cells in tissues that are critical to diseases (6,7). The inability to efficiently measure transcriptional sensitivities across diverse cell contexts has limited our understanding of how drug response differs across genomic and molecular cell states, which could be critical for predicting the therapeutic outcome of patient tumors (8). The recent advance in single-cell RNA sequencing (scRNA-seq) technologies has made it possible to measure the transcriptomes at the single cell resolution (9,10). Over the past few years, nearly a thousand studies have been conducted using scRNA-seq technologies to study the cell types and related features in bulk tissues. However, a gap has been existing to both experimentally and computationally infer cellular drug response, which would provide references for drug repurposing, drug combination design, and new therapeutic development (11–16).
So far, many efforts have been made to develop computational approaches to predict drug response. The pioneer work led by the Connectivity Map (CMap) project has examined 1309 drugs with different doses in five cell lines to quantitatively measure drug induced gene expression profiles (17). With such a rich resource, many studies implemented the concept of ‘anti-correlation between drug and disease signatures’ for various drug related researches such as drug repurposing and combination (18–20). The underlying assumption is that, for a query transcriptome, candidate drugs can be prioritized if such drugs have drug-induced perturbations (as measured by gene expression) in the opposite direction as the query transcriptome. With the rapid accumulation of transcriptomics data, this method has been adapted and applied in various cases. For example, in a study searching for candidate drugs that can be repurposed for schizophrenia, So et al. searched for drugs that induced ‘anti-correlation’ gene signatures compared to the GWAS-imputed gene expression and interestingly, the set of candidate drugs they identified was significantly enriched with antipsychotics (21). Such an approach has also been implemented to identify candidate drugs in cancer. However, due to the substantial intra- and inter-tumor heterogeneity, advanced procedures are in pressing need to incorporate changes in cell types, cell states, cell–cell communications and proportions and accurately infer drug responses in the precision medicine practice.
To this end, we present CeDR Atlas, a knowledgebase reporting computational inference of cellular drug response for hundreds of cell types from the major organs and tissues in human and mouse. We took advantage of the high-throughput profiling of drug-induced gene expression available through the CMap project as well as hundreds of scRNA-seq data representing cells from a wide variety of organs/tissues, diseases, and conditions. For each cell type, we conducted anti-correlation screening and identified drugs that induced gene expression with the opposite direction compared to the cell transcriptome. CeDR is augmented with user-friendly interfaces and functions, allowing users to easily explore the cell types with associated drugs as well as the corresponding gene signatures. CeDR provides direct references for cellular drug response profiles including not only disease cell types but also normal cell types. The comprehensive cell-type specific drug sets can be used for design of combinatory treatments and identification of drug resistance and even drug side effects.
MATERIALS AND METHODS
Data collection
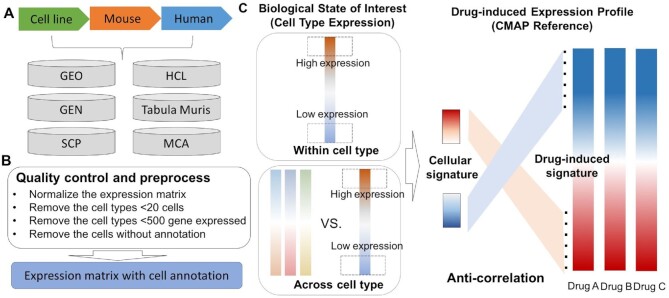
We conducted a comprehensive literature-mining and collected single-cell transcriptome data from hundreds of individual studies (Figure 1A). All the datasets we used in this study were derived from public resources and the detail information for each dataset can be found in Supplementary Data. In the current version, CeDR has included the following data.
Figure 1.
Schematic overview of CeDR. (A) Data curation and collection from different sources. (B) Data quality control and preprocessing. (C) Illustration of the algorithm to infer cellular drug response.
Human cell landscape
The raw expression matrix was downloaded from http://bis.zju.edu.cn/HCL/ (22) consisting of more than 720 000 single cells with cell type annotations from about 50 human tissues. This dataset presents a comprehensive annotation for major human normal organs/tissues.
Gene expression nebulas (GEN)
We collected the human scRNA-seq datasets from the GEN database (https://ngdc.cncb.ac.cn/gen/) (23) generated using the 10× Genomics platform. As a result, a total of 42 projects consisting of 294 high-quality datasets were derived and curated. All of these datasets were processed by using a one-stop analysis pipeline implemented by the CellRanger (24) (v3.1.0) software for quality control, sample de-multiplexing, barcode processing, and generation of feature-barcode matrices. Cell type annotation at the single cell level was performed by using the R package SingleR (25) based on five transcriptomic datasets as built-in references.
Manually curated human scRNA-seq data
We collected publicly available human scRNA-seq data as well as their metadata and cell type annotations by searching PubMed (26) and public databases, such as Gene Expression Omnibus (GEO) (27), Single Cell Portal of Broad Institute (https://singlecell.broadinstitute.org/single_cell) and ArrayExpress Archive of Functional Genomics Data (https://www.ebi.ac.uk/arrayexpress/) (28). As a result, 51 projects with 2 421 320 cells were collected containing data for 55 tissues and 1313 tissue-cell types.
Mouse cell atlas
The datasets in Mouse Cell Atlas (29) were downloaded from GEO with accession ID GSE108097. It contains more than 1 200 000 single cells from about 28 tissues and 170 tissue-cell type clusters.
Tabula muris data
The Tabula Muris (30) mouse atlas expression data and cell annotation files were downloaded from (https://tabula-muris.ds.czbiohub.org/). This dataset contains nearly 100 000 cells from 20 organs and tissues generated by either the FACS-based or droplet-based scRNA-seq technologies.
Manually curated mouse scRNA-seq data
We also manually curated 16 mouse projects with 655 950 cells from GEO, which finally resulted in 25 tissues and 745 tissue-cell types.
Cancer cell line single-cell dataset
The cancer cell line scRNA-seq datasets were downloaded from Single Cell Portal of Broad Institute with the accession ID SCP542 (31). It contains 19 cancer cell lines, including 195 cancer-cell types and 53 298 cells.
Quality control and preprocessing
In the current version of our database, we mainly collected the expression data that have well-annotated cell types. Datasets with unavailable annotations were discarded from further analyses. All datasets were preprocessed using the Python package Scanpy (32) and recorded as an AnnData object in Python. Genes detected in less than five cells were filtered out. Cells with a mitochondrial gene proportion (mtDNA%, the fraction of mitochondrial transcript counts of the total transcript counts) greater than 5% were further filtered out due to the low-quality. Raw count expression matrices were subsequently normalized using the normalize_per_total function and log transformed. For each project, we split it into sub-objects based on the phenotype or disease status. For each single-cell expression sub-object, we kept only the cell types that contained at least 20 cells and further removed the cell types that had less than 500 expressed genes (Figure 1B). In summary, we processed about 120 projects, resulting in 582 data objects, 140 phenotypes and 1250 tissue-specific cell types for human, mouse and cell lines. More detailed information for all studies can be found in Supplementary Table SI.
Drug induced gene expression data
To systematically analyze the cellular drug sensitivities across different tissues and perturbations, we assembled drug induced gene signatures from the CMap database (version: build 02) (17) for 1309 drugs with different doses originally generated using the Affymetrix Gene Chips. We downloaded the rank matrix based on the differentially expressed gene analyses of drug treatments (drug-treated versus no drug-treated). The Affymetrix (33) probe set identifiers were mapped with gene symbols for the following analyses.
Cell type-drug response analysis (CeDR)
To predict cellular drug response, we implemented the anti-correlation screening procedure. Specifically, for each query transcriptome (which can be from bulk tissues or single cells), candidate drugs can be prioritized if they have drug-induced perturbations (as measured by gene expression) in the opposite direction as the query transcriptome. To this end, for each cell type, we first rank the expressed genes (defined as those with non-zero average expression values) according to their average expression across all cells for the same type. The k most highly expressed genes and the k most lowly expressed genes were then selected as the signature gene set for the cell type (Figure 1C, ‘within cell type’). Next, we conducted differentially expressed gene (DEG) analysis for each cell type as compared to other cell types, which is a standard procedure in scRNA-seq analysis. DEGs identified in this way are deemed as cell-type specific genes. We similarly selected the k most highly specifically expressed genes and the k most lowly specifically expressed genes as the signature gene set for the ‘across cell type’ information (Figure 1C). The ‘within cell type’ gene set and the ‘across cell type’ gene set were combined to define the cellular signature genes. The parameter k can be defined manually. In this work, we used k = 300 and the cellular signature genes contain a total of 1200 genes.
For each compound, we also defined a signature gene set based on their rank as provided by the CMap data, i.e. the top k and bottom k genes. In this case, we selected k = 600 to match the size of cellular signature genes. Following the concept of ‘anti-correlation’, we next constructed contingency tables using the down-regulated signature genes of each drug and the cellular signature genes that were highly (or highly specifically) expressed, followed by the chi-square test for drug and cell type association test (denoted as P-value 1). Similarly, we constructed contingency tables using the up-regulated signature genes of each drug and the cellular signature genes that were lowly expressed or lowly specifically expressed, followed by the chi-square test (denoted as P-value 2). The two contingency tables should be constructed separately to ensure that the chi-square test was conducted purposely to identify the anti-correlation relationship. Moreover, we required that the expression of the overlapping genes from the two signature sets of the drug and the cell type should also be significantly anti-correlated, which is examined by Spearman correlation coefficient.
For each data object, to provide a relatively comprehensive resource, we provide in CeDR the drug and cell type associations with nominal significance (P-value 1 < 0.05 and P-value 2 < 0.05). In addition, we present the enrichment and anti-correlation P-values for each associated pair, as well as the signature genes. The functional enrichment analysis for the corresponding signature genes were performed by GSEA (34).
Description of the website and tools
CeDR aims to provide references for cell type and drug associations across different tissues and species. Table 1 summarized the data sets and results deposited in CeDR. Currently, CeDR organized all data into 109 projects, including 93 from human, 15 from mouse, and one from cell lines. These projects contained a total of 582 datasets (460 for human, 102 for mouse, and 20 for cell lines), providing information for 140 phenotypes (human: 93, mouse: 27, cell line: 20) and 1250 cell types (human: 684, mouse: 370, cell line: 196). As aforementioned, we selected associations with nominal significance based on both the chi-square test and the Spearman correlation analysis. This resulted in a total of 241 116 associations in CeDR.
Table 1.
Data summary of CeDR
| Source | Projects | Subjects | Phenotype | Tissue-cell type | Association |
|---|---|---|---|---|---|
| Human | 93 | 460 | 93 | 684 | 188 157 |
| Mouse | 15 | 102 | 27 | 370 | 42 660 |
| CCLE | 1 | 20 | 20 | 196 | 10 299 |
| Total | 109 | 582 | 140 | 1250 | 241 116 |
Web interface
As shown in Figure 2, the CeDR interface allows users to intuitively browse and search for any data in the database. The home page provides an overview of phenotypes across different tissues and a quick search function for users to query the database for species, tissues, cell types, or phenotypes conveniently (Figure 2A and B). Datasets have been classified by tissues where users can select relevant dataset in a pop-up window to retrieve the corresponding phenotypes.
Figure 2.
Screenshots of web pages for CeDR. (A) The web images in the home page allow users to switch sources and tissues with detailed data summary information and phenotypes. (B) Quick search functions in the home page. (C) The browse page allows users to browse and search all the project, tissues, phenotypes and other detailed information. (D) Summary of the selected dataset and cell type-drug association network. (E) Detailed results for each cell type with associated drugs and corresponding signature genes.
Users can also navigate the whole database through ‘Browse’ functions. The ‘Browse’ page displays a general table of all sub-objects and provides data advanced search functions (Figure 2C). The visible part of the table row contains the major meta information of the data, including Source, Project ID, Tissue, Phenotype, Cell type, Drug and other information specific to this sub-object. Users can select a sub dataset of interest and clicking the corresponding ‘Dataset ID’ button will enable the browse of detailed results for tissue specific cell type-drug associations. In particular, cell types-drug associations with significant P-values will be returned. The result page contains an intuitive table with more detailed information about the sub-object. Moreover, the visualization of cell types, fraction and predicted association network will be displayed (Figure 2D). Moving the mouse to a point in the UMap diagram, and the coordinate of the cell-type information will be displayed. Clicking the ‘Detail’ button for each association, the gene signature with GSEA prerank result in drug and single cell expression data will be further displayed (Figure 2E).
We also provide multiple search function pages for users to identify cell type, drug, disease or tissue of interest. For a user-input query string, we will search both the short names and the full names of interest. In addition, users can download all data via the ‘Download’ page. We also built a detailed tutorial for the usage in the documentation page to briefly describe the data collection, preprocessing, and analysis (https://ngdc.cncb.ac.cn/cedr/documentation).
Application of CeDR: pancreas as an example
CeDR provides direct references for cellular drug response profiles including not only disease cell types but also normal cell types which can be further used for design of combinatory treatments, drug resistance and even drug side effects. As shown in Figure 3A, treatment with a single drug on normal cells may lead to side effects; on the other hand, treatment on disease cells may not be always as efficient as expected due to the sample heterogeneity. Investigating drug response at single-cell level can provide insight into combination treatment which can maximize the efficiency by killing disease cells and minimize the side effect to the normal cells.
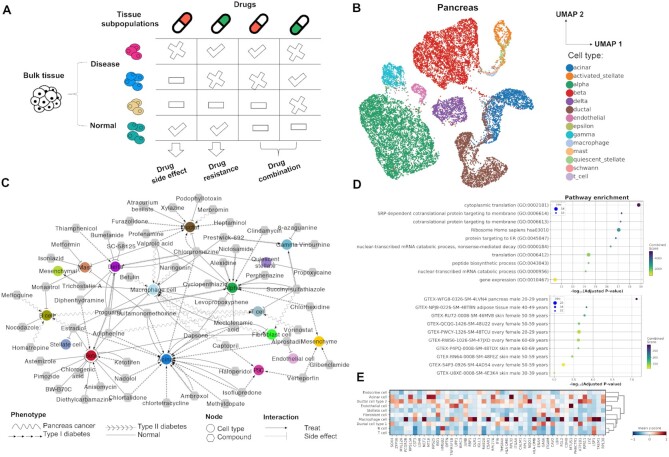
Figure 3.
Drug response at the single cell Level: Pancreas as an example. (A) Tissue subpopulations maintain diverse response to different drugs. (B) UMap visualization for major cell types across different datasets. (C) Cell type-drug associated network. The circular node represents cell types and the hexagon denotes associated drugs. Different interactions refer to treatments or side effects and the shape of the edge denotes the corresponding phenotypes. Here, only the top 10 drugs with corresponding cell types were shown in (C). (D) Functional enrichment analysis for signature genes referring to acinar-dapsone association. (E) Matrix plot of signature genes in pancreas cancer scRNA-seq dataset referring to buetulin-macrophage association.
Here, we select human pancreas as an example to indicate the application of CeDR in drug treatment. Pancreas is part of the human endocrine system and mainly produces hormones and digestive enzymes. To study the major cell type in pancreas, we integrated nine scRNA-seq data sets from different studies using our recently developed method INSCT which based on batch-aware triplet neural networks (35,36). These data sets included four normal pancreas tissues, three type I diabetes, one type II diabetes and one pancreatic cancer dataset and were expected with strong batch effects. By applying INSCT, we successfully corrected the potential batch effects and reported the primary cell types in pancreas such as acinar, duct, alpha, beta, delta, gamma, and epsilon cells (37,38) (Figure 3B). With CeDR, we screened the compounds in CMap across all the cell types based on the signature genes defined above. Among our results, some of the top ranked drugs have already been reported sensitive to pancreas. To better visualize the results, we construct a cell type-drug association network (Figure 3C).
In normal cells, we particularly examined the reversed activity. Our results showed that the drug dapsone had adverse effects with normal beta cells (P-value = 4.81 × 10–14), macrophage (P-value = 1.13 × 10–10), alpha cells (P-value = 1.15 × 10–12), acinar cells (P-value = 1.26 × 10–9) and fibroblast cells (P-value = 6.55 × 10–11). Dapsone is an antibiotics and anti-inflammatory medication typically used for skin disorders. Previous reports have indicated that dapsone can induce pancreatitis, especially acute pancreatitis (AP) (39), and beta cell dysfunction is involved in the early stages in pancreatitis (40). In addition, it is well known that macrophage cells can secrete proinflammatory cytokines and further expedite the formation of pancreatic fibrosis in chronic pancreatitis (CP) (41). Moreover, premature activation of digestive enzymes in acinar cells may lead to the onset of AP. Thus, the cell types we identified well supported previous studies. Importantly, the signature genes associated with dapsone and cell types are also significantly enriched in GTEx (42) pancreas and adipose tissues as well as cytoplasmic translation pathways (Figure 3D). Although the functions of pancreas associated adipose tissue (PAT) are still unknown, recent studies have indicated that PAT released adipokines may protect the pancreas against the dysfunction of metabolism (43). These results provided insights into understanding the potential mechanism of the dapsone induced side effect in pancreatitis.
On the contrary, by comparing the results in normal and tumor cells, CeDR results can be used to infer the potential drugs that can be repurposed individually for the treatment of pancreas cancer. For example, numerous reports have indicated that B cells can promote pancreatic tumorigenesis and have been investigated as a potential target in cancer treatment (44). In our results, the compound mefloquine, previously used to treat malaria, was suggested to be effective for B cells (P-value = 3.65 × 10–12). This is consistent with previous reports that malaria drugs may improve the effectiveness in cancer therapies. Moreover, tumor-associated macrophages (TAM) as versatile immune cells can lead to a variety of malignant changes in pancreatic cancer (45). Our results suggested that betulin and naringenin are potential drugs associated with TAM for pancreatic cancer treatment. Indeed, betulin has already been suggested as a potent anticancer agent based on a comprehensive review and naringenin was reported to decrease invasiveness and metastasis (46,47). Among the signature genes in pancreatic cancer scRNA-seq, there were well-known genes highly expressed in pancreatic cancer, such as OLR1, LSP1, FGL2 (Figure 3E) (48). Interestingly, our results can also be used to infer drug combinations in the treatment of cancer. For instance, the combined treatment with naringenin and mefloquine may potentially inhibit the growth and metastasis of pancreatic cancer cells without damaging the normal cells. Notably, naringenin has already been identified as a combination therapy with hesperetin for pancreatic cancer (49). Collectively, these results demonstrated that CeDR can be used as a valuable resource for better investigation of tissue heterogeneity and drug response at the single cell level.
DATABASE DESIGN AND UPDATES
CeDR is hosted by a local server with a Centos Linux 7.4 environment. It is constructed using the Java Spring Boot (https://spring.io/projects/spring-boot) as a back-end RESTful web service framework. The cell type and drug association results are hosted by the MySQL Database service (https://www.mysql.com). Front-end user interfaces are designed using the React (https://reactjs.org) and Umi (https://umijs.org) frameworks, which are scalable enterprise-class front-end application frameworks allowing flexible maintenance and extension in future. Ant Design (https://ant.design) is used as the UI library which contains a set of high-quality components and demos. Furthermore, we used the charting library available through HighCharts (https://www.highcharts.com/) to implement interactive charting and data visualization. The CeDR resource will be updated twice per year depending on the number of newly published scRNA-seq, especially those with cell type annotations.
CONCLUDING REMARKS AND FUTURE DEVELOPMENT
A large number of scRNA-seq datasets have been generated to decode the cell type compositions and expression heterogeneity across different tissues. With these available data, CeDR provides direct references for cellular drug response profiles and has implications in various future applications. First, many cells deposited in our database were identified from disease tissues, such as tumor tissues from cancer patients, post-mortem human brain samples from Alzheimer's disease, and pancreatic tissues from patients with type 2 diabetes. We identified candidate drugs that were consistent with previous reports. Second, the comprehensive cell-type specific drug sets can be used for design of combinatory treatments. In cancer samples, cellular heterogeneity plays important roles in shaping the actual drug response and eventually leads to drug resistance. Identification of drugs, or drug combinations, that can kill cancer cells but not normal cells would be of high interest in precision medicine. Third, CeDR includes not only disease cell types but also normal cell types. The cellular drug response to the normal cell types would provide new insights for drug side effects.
Due to the limitation of well-known tumor-related scRNA-seq as well as the cell annotation information, the drug response analysis for tumors did not include all tissues. However, with the rapidly evolving technologies in next-generation sequencing, we expect more data at the single cell level to be generated in the near future, especially tumor related datasets. We will continuously collect and curate the emerging single-cell transcriptome data and conduct cellular drug response analyses for different tissues and phenotypes. In addition to scRNA-seq, other single cell techniques, such as scDNA-seq, scRNA-seq and scATAC-seq, are also valuable resources to investigate drug response. We will incorporate more drug treatment application tools for various types of data and develop CeDR as a comprehensive database for cellular drug response. Future database updates will thus include and integrate the genomic and transcriptome profiles of individual cells. We believe that CeDR will be a useful resource for the single-cell and drug design research community.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank NGDC for providing computational resource support in our data process. We would like to thank all the members of Laboratory for Precision Health (LPH) for valuable discussion.
Contributor Information
Yin-Ying Wang, CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Hongen Kang, CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Tianyi Xu, China National Center for Bioinformation, Beijing 100101, China; National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China.
Lili Hao, China National Center for Bioinformation, Beijing 100101, China; National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China.
Yiming Bao, China National Center for Bioinformation, Beijing 100101, China; National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China.
Peilin Jia, CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Strategic Priority Research Program of Chinese Academy of Sciences [XDB38010400]; Science and Technology Service Network Initiative of Chinese Academy of Sciences [KFJ-STS-QYZD-2021-08-001]; Y.B. was supported by the Genomics Data Center Construction of Chinese Academy of Sciences [WX145XQ07-04]. Funding for open access charge: Strategic Priority Research Program of Chinese Academy of Sciences [XDB38010400 to P.J.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wang R., Jin C., Hu X.. Evidence of drug-response heterogeneity rapidly generated from a single cancer cell. Oncotarget. 2017; 8:41113–41124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu S., Sun C., Li Y., Wang T., Jia L., Lai S., Yang Y., Luo P., Dai D., Yang Y.Q.et al.. GMrepo: a database of curated and consistently annotated human gut metagenomes. Nucleic Acids Res. 2020; 48:D545–D553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lai S., Jia L., Subramanian B., Pan S., Zhang J., Dong Y., Chen W.H., Zhao X.M.. mMGE: a database for human metagenomic extrachromosomal mobile genetic elements. Nucleic Acids Res. 2021; 49:D783–D791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altschuler S.J., Wu L.F.. Cellular heterogeneity: do differences make a difference. Cell. 2010; 141:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dagogo-Jack I., Shaw A.T.. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018; 15:81–94. [DOI] [PubMed] [Google Scholar]
- 6. Wu Z., Lawrence P.J., Ma A., Zhu J., Xu D., Ma Q.. Single-Cell techniques and deep learning in predicting drug response. Trends Pharmacol. Sci. 2020; 41:1050–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao W., Dovas A., Spinazzi E.F., Levitin H.M., Banu M.A., Upadhyayula P., Sudhakar T., Marie T., Otten M.L., Sisti M.B.et al.. Deconvolution of cell type-specific drug responses in human tumor tissue with single-cell RNA-seq. Genome Med. 2021; 13:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adam G., Rampasek L., Safikhani Z., Smirnov P., Haibe-Kains B., Goldenberg A.. Machine learning approaches to drug response prediction: challenges and recent progress. NPJ Precis. Oncol. 2020; 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Olsen T.K., Baryawno N.. Introduction to single-cell RNA sequencing. Curr Protoc Mol Biol. 2018; 122:e57. [DOI] [PubMed] [Google Scholar]
- 10. Song L., Pan S., Zhang Z., Jia L., Chen W.H., Zhao X.M.. STAB: a spatio-temporal cell atlas of the human brain. Nucleic Acids Res. 2021; 49:D1029–D1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao X.M., Iskar M., Zeller G., Kuhn M., van Noort V., Bork P.. Prediction of drug combinations by integrating molecular and pharmacological data. PLoS Comput. Biol. 2011; 7:e1002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aissa A.F., Islam A., Ariss M.M., Go C.C., Rader A.E., Conrardy R.D., Gajda A.M., Rubio-Perez C., Valyi-Nagy K., Pasquinelli M.et al.. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat. Commun. 2021; 12:1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y.Y., Chen W.H., Xiao P.P., Xie W.B., Luo Q., Bork P., Zhao X.M.. GEAR: a database of Genomic Elements Associated with drug Resistance. Sci. Rep. 2017; 7:44085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J.H., Zhao X.M., Yoon I., Lee J.Y., Kwon N.H., Wang Y.Y., Lee K.M., Lee M.J., Kim J., Moon H.G.et al.. Integrative analysis of mutational and transcriptional profiles reveals driver mutations of metastatic breast cancers. Cell Discov. 2016; 2:16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Y.G., Wang Y.Y., Zhao X.M.. A survey on computational approaches to predicting adverse drug reactions. Curr. Top. Med. Chem. 2016; 16:3629–3635. [DOI] [PubMed] [Google Scholar]
- 16. Wang Y.Y., Bai H., Zhang R.Z., Yan H., Ning K., Zhao X.M.. Predicting new indications of compounds with a network pharmacology approach: Liuwei Dihuang Wan as a case study. Oncotarget. 2017; 8:93957–93968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lamb J. The connectivity map: a new tool for biomedical research. Nat. Rev. Cancer. 2007; 7:54–60. [DOI] [PubMed] [Google Scholar]
- 18. Qiu K., Lee J., Kim H., Yoon S., Kang K.. Machine learning based anti-cancer drug response prediction and search for predictor genes using cancer cell line gene expression. Genomics Inform. 2021; 19:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi K., Lin W., Zhao X.. Identifying molecular biomarkers for diseases with machine learning based on integrative omics. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020; 10.1109/TCBB.2020.2986387. [DOI] [PubMed] [Google Scholar]
- 20. Wang Y.Y., Cui C., Qi L., Yan H., Zhao X.M.. DrPOCS: drug repositioning based on projection onto convex sets. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019; 16:154–162. [DOI] [PubMed] [Google Scholar]
- 21. So H.C., Chau C.K., Chiu W.T., Ho K.S., Lo C.P., Yim S.H., Sham P.C.. Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry. Nat. Neurosci. 2017; 20:1342–1349. [DOI] [PubMed] [Google Scholar]
- 22. Han X., Zhou Z., Fei L., Sun H., Wang R., Chen Y., Chen H., Wang J., Tang H., Ge W.et al.. Construction of a human cell landscape at single-cell level. Nature. 2020; 581:303–309. [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y., Zou D., Zhu T., Xu T., Chen M., Niu G., Zong W., Pan R., Jing W., Sang J.et al.. Gene Expression Nebulas (GEN): a comprehensive data portal integrating transcriptomic profiles across multiple species at both bulk and single-cell levels. Nucleic Acids Res. 2022; https://doi.org/10.1093/nar/gkab878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J.et al.. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017; 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R.et al.. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019; 20:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts R.J. PubMed Central: The GenBank of the published literature. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:381–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clough E., Barrett T.. The gene expression omnibus database. Methods Mol. Biol. 2016; 1418:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sarkans U., Fullgrabe A., Ali A., Athar A., Behrangi E., Diaz N., Fexova S., George N., Iqbal H., Kurri S.et al.. From arrayexpress to biostudies. Nucleic Acids Res. 2021; 49:D1502–D1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F.et al.. Mapping the mouse cell atlas by Microwell-Seq. Cell. 2018; 173:1307. [DOI] [PubMed] [Google Scholar]
- 30. Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018; 562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kinker G.S., Greenwald A.C., Tal R., Orlova Z., Cuoco M.S., McFarland J.M., Warren A., Rodman C., Roth J.A., Bender S.A.et al.. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 2020; 52:1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wolf F.A., Angerer P., Theis F.J.. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018; 19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu G., Loraine A.E., Shigeta R., Cline M., Cheng J., Valmeekam V., Sun S., Kulp D., Siani-Rose M.A.. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003; 31:82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang J., Zhang X., Wang Y., Zeng F., Zhao X.M.. MAT2: manifold alignment of single-cell transcriptomes with cell triplets. Bioinformatics. 2021; https://doi.org/10.1093/bioinformatics/btab250. [DOI] [PubMed] [Google Scholar]
- 36. Simon M.L., Wang Y.Y., Zhao Z.. Integration of millions of transcriptomes using batch-aware triplet neural networks. Nat. Mach. Intell. 2021; 3:705–715. [Google Scholar]
- 37. Peng J., Sun B.F., Chen C.Y., Zhou J.Y., Chen Y.S., Chen H., Liu L., Huang D., Jiang J., Cui G.S.et al.. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019; 29:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muraro M.J., Dharmadhikari G., Grun D., Groen N., Dielen T., Jansen E., van Gurp L., Engelse M.A., Carlotti F., de Koning E.J.et al.. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016; 3:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jha S.H., Reddy J.A., Dave J.K.. Dapsone-induced acute pancreatitis. Ann. Pharmacother. 2003; 37:1438–1440. [DOI] [PubMed] [Google Scholar]
- 40. Sasikala M., Talukdar R., Pavan kumar P., Radhika G., Rao G.V., Pradeep R., Subramanyam C., Nageshwar Reddy D.. beta-Cell dysfunction in chronic pancreatitis. Dig. Dis. Sci. 2012; 57:1764–1772. [DOI] [PubMed] [Google Scholar]
- 41. Hu F., Lou N., Jiao J., Guo F., Xiang H., Shang D.. Macrophages in pancreatitis: mechanisms and therapeutic potential. Biomed. Pharmacother. 2020; 131:110693. [DOI] [PubMed] [Google Scholar]
- 42. GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quiclet C., Dittberner N., Gassler A., Stadion M., Gerst F., Helms A., Baumeier C., Schulz T.J., Schurmann A.. Pancreatic adipocytes mediate hypersecretion of insulin in diabetes-susceptible mice. Metabolism. 2019; 97:9–17. [DOI] [PubMed] [Google Scholar]
- 44. Roghanian A., Fraser C., Kleyman M., Chen J.. B cells promote pancreatic tumorigenesis. Cancer Discov. 2016; 6:230–232. [DOI] [PubMed] [Google Scholar]
- 45. Yang S., Liu Q., Liao Q.. Tumor-associated macrophages in pancreatic ductal adenocarcinoma: origin, polarization, function, and reprogramming. Front. Cell Dev. Biol. 2020; 8:607209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krol S.K., Kielbus M., Rivero-Muller A., Stepulak A.. Comprehensive review on betulin as a potent anticancer agent. Biomed. Res. Int. 2015; 2015:584189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park H.J., Choi Y.J., Lee J.H., Nam M.J.. Naringenin causes ASK1-induced apoptosis via reactive oxygen species in human pancreatic cancer cells. Food Chem. Toxicol. 2017; 99:1–8. [DOI] [PubMed] [Google Scholar]
- 48. Yang G., Xiong G., Feng M., Zhao F., Qiu J., Liu Y., Cao Z., Wang H., Yang J., You L.et al.. OLR1 promotes pancreatic cancer metastasis via increased c-Myc expression and transcription of HMGA2. Mol. Cancer Res. 2020; 18:685–697. [DOI] [PubMed] [Google Scholar]
- 49. Lee J., Kim D.H., Kim J.H.. Combined administration of naringenin and hesperetin with optimal ratio maximizes the anti-cancer effect in human pancreatic cancer via down regulation of FAK and p38 signaling pathway. Phytomedicine. 2019; 58:152762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.