Abstract
ReMap (https://remap.univ-amu.fr) aims to provide manually curated, high-quality catalogs of regulatory regions resulting from a large-scale integrative analysis of DNA-binding experiments in Human, Mouse, Fly and Arabidopsis thaliana for hundreds of transcription factors and regulators. In this 2022 update, we have uniformly processed >11 000 DNA-binding sequencing datasets from public sources across four species. The updated Human regulatory atlas includes 8103 datasets covering a total of 1210 transcriptional regulators (TRs) with a catalog of 182 million (M) peaks, while the updated Arabidopsis atlas reaches 4.8M peaks, 423 TRs across 694 datasets. Also, this ReMap release is enriched by two new regulatory catalogs for Mus musculus and Drosophila melanogaster. First, the Mouse regulatory catalog consists of 123M peaks across 648 TRs as a result of the integration and validation of 5503 ChIP-seq datasets. Second, the Drosophila melanogaster catalog contains 16.6M peaks across 550 TRs from the integration of 1205 datasets. The four regulatory catalogs are browsable through track hubs at UCSC, Ensembl and NCBI genome browsers. Finally, ReMap 2022 comes with a new Cis Regulatory Module identification method, improved quality controls, faster search results, and better user experience with an interactive tour and video tutorials on browsing and filtering ReMap catalogs.
INTRODUCTION
Transcriptional regulators (TRs) such as transcription factors (TFs), transcriptional coactivators (TCAs) and chromatin-remodeling factors (CRFs), drive gene transcription and chromatin organization through DNA binding. Since the advent of chromatin immunoprecipitation followed by sequencing (ChIP-seq (1)), it has become possible to study the genome-wide occupancy of DNA-binding proteins. The popularity of ChIP-seq and other ChIP-based sequencing techniques has increased research in genome occupancy maps for various TRs, in various conditions, cells, tissues, and species. This led to an accumulation of functional genomics datasets stored in data repositories such as NCBI GEO (2), ENA EBI (3) or DDBJ (4) providing a unique resource for thousands of DNA-binding sequencing studies. A large-scale integration of these studies would reveal the transcriptional regulatory repertoire as transcription of the Human genome is controlled by about 1600 transcription factors (5,6). The genomic architecture of the regulatory space has started to unfold thanks to large functional genomic consortia (ENCODE (7,8), Roadmap (9)) but more remains to be discovered. Such large-scale integration is challenged by the variety of bioinformatics methods and underlying data formats, the inconsistency in targets, cell types or tissue names, as well as experimental ChIP and sequencing quality. However, such integrative analysis would offer significant insights of the transcriptional regulatory repertoire in different cellular environments.
In 2015, the ReMap project initiated the first large-scale integrative analysis of heterogeneous ChIP-seq revealing the complex architecture of the Human regulatory landscape using dedicated curation and standardized data processing pipeline (10). The manual curation and annotation of DNA-binding sequencing studies are at the foundation of the ReMap project. Each dataset introduced in ReMap is assessed manually to ensure correct target and biotype annotation, as well as experimental metadata curation, making it distinct from other integrative projects (11–14). The 2015 version of ReMap (10) introduced a Human regulatory catalog of 13 million (M) DNA binding regions for 237 TRs across 83 biotypes (cell lines and tissue types) by integrating 395 datasets from GEO and ENCODE. The 2018 ReMap version, followed by the 2020 version, released a Human regulatory catalog of 165M binding regions for 1135 TRs (15,16) by processing ∼5800 datasets. Also, the 2020 ReMap database introduced the first Arabidopsis thaliana regulatory atlas as a result of a large-scale data integration of public data and analysis efforts. Since 2018, the ReMap catalogs are used as one of the input sources for the computation of TF binding profiles for the JASPAR database (17,18).
Here, we present the fourth release of ReMap (‘ReMap2022’), which comes with a major expansion of the Human and Arabidopsis regulatory atlases. Moreover, we introduce two new regulatory atlases for Mus musculus and Drosophila melanogaster. These new catalogs are the results of our continuing efforts in large-scale data integration for these two model species. Faced with large catalogs, this update includes new quality controls to improve the repertoire of binding locations as well as a new method for Cis Regulatory Modules (CRMs) identification. Finally, our database update is backed up by new web functionalities for better community access. The web portal displays an interactive tour and genome track filters are available through track hubs on genome browsers. Taken together the manual metadata curation and large-scale integration engaged in the ReMap project offers a unique and unprecedented collection of DNA-binding regions for four major species.
MATERIALS AND METHODS
Available datasets
New ChIP-seq experiments were retrieved from the NCBI Gene Expression Omnibus (GEO) and ENCODE databases (2,19). For GEO, the query ‘Genome binding/occupancy profiling by high-throughput sequencing’ AND ‘homo sapiens’[organism] AND NOT ‘ENCODE’[project]’ was used to return a list of all potential studies. The same query was used with ‘arabidopsis thaliana’[organism], ‘mus musculus’[organism] and ‘drosophila melanogaster’[organism] to get all the potential datasets for each study. The selected experiments metadata are then manually curated and annotated with official nomenclatures for target names and biotypes. For incomplete metadatas, the materials and methods of associated and published papers are often examined to complete the curation. For Human we used the HUGO Gene Nomenclature Committee (20) (www.genenames.org), the BRENDA Tissue Ontologies for cell lines (21) at the EBI Ontology Lookup Service (22) (www.ebi.ac.uk/ols/ontologies/bto) as well as the Cellosaurus database (23) to homogenize cell and tissue names (e.g. MCF-7 not MCF7, Hep-G2, not HepG2, Hepg2 etc.). For Arabidopsis (A. thaliana) we used gene names from the Ensembl Plant genome annotation (24). Ecotypes and biotypes descriptions were curated and homogenized when the information was available in the metadata or associated publication. For Mouse (M. musculus) we annotated gene names using the official MGI database (25) (http://www.informatics.jax.org/). For Drosophila (D. melanogaster) we used the Flybase database (26) (https://flybase.org/) for gene names. To improve automatic processing of files we removed parentheses and replaced them with hyphens to better handle these names in the pipeline (e.g. E(z) to E-z, See Supplementary Table S12). For Mouse and Drosophila the tissues or cell lines annotation were homogenized using BRENDA Tissue Ontologies as well as the Cellosaurus database. ChIP-seq studies involving RNA polymerases (RNA-Pol2 and RNA-Pol3) were filtered out. When multiple antibodies were pooled (e.g. RUNX1 and RUNX3, GSE17954) targets would be named as RUNX1-3. Also, when a ‘global’ antibody is used to pool a family of targets (eg: RAR, GSE35599) we would name the target as just RAR.
We define a dataset as a DNA-binding experiment in a given GEO/AE/ENCODE series (e.g. GSE37345), for a given TR (e.g. FOXA1), and in a particular biotype (e.g. LNCaP, Larva, Leaf, Limb) in a given biological condition (e.g. 45min DMSO, 21d-wt-watered). Datasets are labeled with the concatenation of these informations (e.g. GSE37345.FOXA1.LNCAP_45min-DMSO). For the 2022 update a total of 12 976 new datasets were processed across all four species (Supplementary Table S1). Specifically, we analysed 4121 Human datasets deposited in public repositories from 11 November 2018 to 11 September 2020; 223 Arabidopsis datasets from 3 February 2018 to 9 March 2021; 7317 Mouse datasets from 1 January 2009 to 2 February 2020; and 1308 Drosophila datasets from 1 January 2008 to 17 September 2020 (full list of datasets in Supplementary Tables S3, S5, S7–S9 and S11).
In the 2022 update, as well as in previous updates, the new ENCODE ChIP-seq experiments for TFs, transcriptional and chromatin regulators were re-analysed starting from raw data (FASTQ files) following the same processing pipeline. For Human, we processed data between 6 February 2019 to 18 March 12021, for Mouse, all data until 16 November 2020 and for Drosophila all data until 28 April 2021. We retrieved the list of ENCODE ChIP-seq experiments as FASTQ files from the ENCODE portal (8,19) (https://www.encodeproject.org/) using the following filters: Assay: ‘ChIP-seq’, Organism: ‘Homo sapiens’, Target of assay: ‘TF’, Available data: ‘fastq’ on 6 February 2019. The same filters were used for the Organism: ‘M. musculus’ and ‘D. melanogaster’. Metadata information in JSON format and FASTQ files were retrieved using the Python requests module. We processed 508 Human, 167 Mouse and 525 Drosophila ENCODE datasets, of whom 267 Human, 158 Mouse and 514 Drosophila passed our quality filters. We renamed TR ENCODE aliases using official HGNC and MGI identifiers (e.g. p65 into RELA, see Supplementary Tables S4 and S10) for Human and Mouse respectively, and renamed cell lines to official BRENDA and Cellosaurus conventions (e.g. K562 into K-562, for curated names see Supplementary Tables S4, S6, S10 and S12).
ChIP-seq processing
For each species, the GEO and ENCODE datasets were curated, processed and analysed in the same way. Bowtie 2 (version 2.2.9 (27)) with options -end-to-end -sensitive was used to align all reads on the human genome GRCh38/hg38 assembly, the A. thaliana TAIR10 assembly, the D. melanogaster BDGP6.32/dm6 assembly and the M. musculus GRCm38/mm10 assembly. The GRCm39 assembly was released during the Mouse production. Trim Galore (https://github.com/FelixKrueger/TrimGalore) was used to remove adapters, trimming reads up to 30 bp. Trim Galore is a wrapper around Cutadapt and FastQC to consistently apply adapter and quality trimming to FastQ files. With samtools rmdup polymerase chain reaction duplicates were removed from the alignments. Following the ENCODE ChIP-seq guidelines (28) we used the MACS2 peak-calling tool (29) (version 2.1.1.2) with default thresholds (MACS2 default thresholds, Q-value: 5e–2, -g: with corresponding genome sizes) to identify the TR binding regions. For all the datasets, the corresponding bed file is available for download.
Quality assessment and filters
Because the analysed data comes from a variety of sources and is produced under varied experimental circumstances and platforms, the data quality differs from study to study. Since the first release of ReMap 2015, and unlike similar databases (Supplementary Tables S3, S5, S9 and S11), our pipeline has assessed the quality of each processed dataset. The same quality pipeline and cutoffs were used for ReMap 2022 as they were for ReMap 2020, we named these quality assessments as QC1 in Figure 1. Briefly, we derived a score based on the ENCODE consortium's cross-correlation and FRiP (percent of reads in peaks) metrics (Supplementary Figures S1–S4, ENCODE quality coefficients http://genome.ucsc.edu/ENCODE/qualityMetrics.html) for all species and ChIP-seq datasets for this release. The normalized strand cross-correlation coefficient (NSC), which is a ratio of the maximal fragment-length cross-correlation value to the background cross-correlation value, and the relative strand cross-correlation coefficient (RSC), which is a ratio of the fragment-length cross-correlation to the read-length cross-correlation, are then computed by our pipeline. Datasets that failed QC1 were not included in the catalogs or the BED files available for download. Rejected datasets are listed in (Supplementary Tables S3, S5, S9 and S11).
Figure 1.
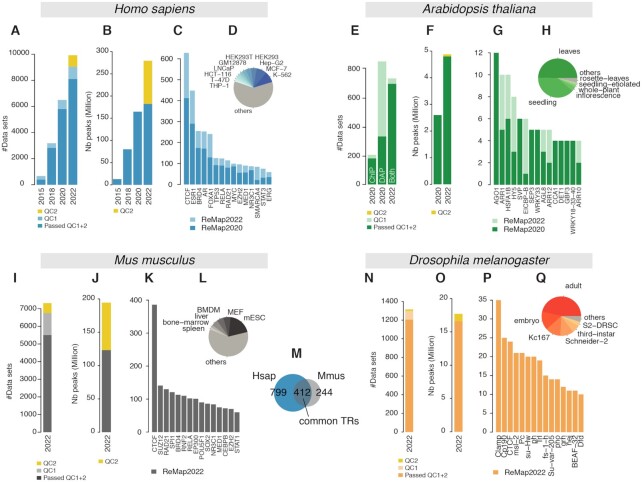
Overview of the ReMap 2022 database growth. (A) Analysed Human datasets growth in ReMap 2022 compared to 2020, 2018 and 2015, removed datasets from the new quality control (QC2) in yellow. (B) Human ChIP-seq peaks growth in ReMap 2022 compared to previous releases, yellow corresponds to removed peaks. (C, D) Evolution of the number of datasets across the top 15 transcriptional regulators (TRs) and biotypes between ReMap 2022 and 2020. (E) Analysed Arabidopsis thaliana datasets in 2022 compared to 2020. (F) Arabidopsis regulatory peaks growth in ReMap 2022 compared to 2020, yellow corresponds to removed peaks (G, H) Evolution of the number of datasets across the top 15 transcriptional regulators and biotypes between the two Arabidopsis ReMap catalogs. (I) Analysed ChIP-seq datasets for the ReMap 2022 Mus musculus catalog, removed datasets from QC2 in yellow. (J) Size of the Mus musculus regulatory catalog, before and after QC2 (yellow). (K, L) Number of datasets for the top 15 TRs and top 6 biotypes. (M) Transcriptional regulators shared between the Human and Mouse regulatory catalogs. (N) Analysed ChIP-seq datasets for the ReMap 2022 Drosophila melanogaster catalog, removed datasets from QC2 in yellow. (O) Size of the Drosophila melanogaster regulatory catalog, before and after QC2 (yellow). (P,Q) Number of datasets for the top 15 TRs and top 6 biotypes.
In 2022, two new sets of filters were added in our assessment steps, named QC2 in Figure 1. These filters were applied to the new catalogs, and retroactively to previous data for Human and Arabidopsis. We discarded datasets having less than 100 peaks or more than two times the number of annotated genes according to the Ensembl gene annotation statistics. As of late 2021, about 20 000 coding and non-coding genes are identified for Drosophila giving a cutoff of a maximum of 40 000 peaks, about 30 000 annotated genes for Arabidopsis giving a cutoff of 60 000 peaks, and about 40 000 annotated genes for Human or Mouse giving a cutoff of 80 000 peaks (Supplementary Figure S5). For the second filter, within each dataset we discarded peaks that fall outside a base pair length range. The range is defined as 50bp minimum and an upper cutoff in which we have 99% of catalogue (Supplementary Figure S6A). These upper cutoffs are rounded to 1.5 kb for Human and Mouse, 2kb for Drosophila and Arabidopsis (see Supplementary Figure S6B). Full data with rejected peaks are made available in the download table as ‘Permissive peaks’ in the Supplementary tab.
Open ReMap pipeline
The code of the ReMap pipeline is available on Github in the ReMap Github organisation (https://github.com/remap-cisreg). The pipeline uses Snakemake (30) in a Conda (https://conda.io) or Singularity (https://sylabs.io/) environment, depending on the High-Performance Computing (HPC) resources available, and both Torque and Slurm managers are supported. You can also find multiple python and shell scripts used to format the datasets. The repository contains information in the Github wiki on the utilisation of the pipeline.
Non-redundant peak sets
ReMap provides non-redundant binding regions (NR peaks) for each target, a unique feature not seen in other databases (Supplementary Table S2). This gives an accurate genomic location of peaks regardless of biotypes, in a multicell manner. All peak lengths for a TR were trimmed to the median size of all peaks in that TR. Then, we used BedTools to intersect overlapping truncated peaks across multiple datasets to discover clusters of duplicate peaks (with at least 25% overlap, both ways). After the clusters of overlapping peaks have been identified, non-redundant peaks are computed by averaging start, end and summits coordinates of all peaks in a cluster using original ReMap peaks lengths. The non-redundant peak set across all experiments for a particular factor consists of calculated non-redundant peaks plus singletons and is available for download from the ReMap website.
Cis regulatory modules
To accurately delineate CRMs in the 2022 release, we have developed a new methodology that relies on detecting peak density along the genome. Briefly, we delineate CRMs at each local minimum of NR peak density function (See Supplementary Figure S7B). A NR peak is assigned to a CRM if its peak summit falls in between the identified flanking local minimas. Finally, the CRM boundaries are reduced using the 5′ and 3′ coordinates of the flanking NR peaks. The ‘peakMerge’ code is available at https://github.com/remap-cisreg/peakMerge.
UPDATE OF THE HUMAN AND ARABIDOPSIS CATALOGS
Human transcriptional regulatory expansion
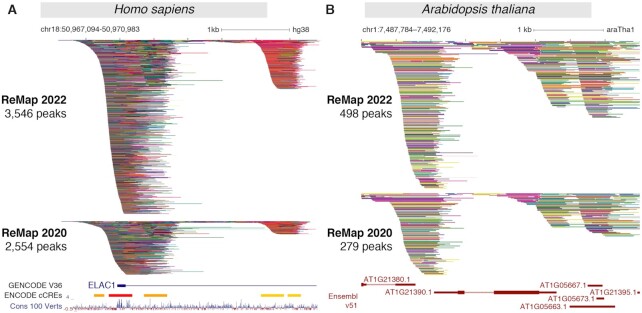
This fourth release of ReMap comes with a significant update of the Human regulatory catalog by adding a large number of new datasets, new transcriptional regulators and biotypes. We curated, processed and analysed 4121 ChIP-seq datasets targeted against TRs collected from GEO and ENCODE databases since the last release. Since 2015 we provide consistency and comparability between datasets by processing raw data through our standardized ReMap pipeline, which includes read filtering, read mapping, peak calling, and quality controls (See ‘Materials and methods’). In contrast to other databases, the manual curation and annotation of studies we process are at the foundation of the ReMap project. It involves analysing the warehouse study design descriptions and reading submitted materials and methods from GEO or in the manuscripts to curate heterogeneous experimental information. Furthermore, to correct the diverse quality of DNA binding experiments the pipeline contains quality controls and filtering steps (See ‘Materials and methods’). After applying our quality filters, we retained 2828 datasets (65%) from the 4121 new deposited ChIP-seq datasets (Supplementary Figures S1, S5–S6). As a result, the updated Human regulatory atlas contains 182 416 820 peaks, derived from 8,103 datasets (Figure 1A), which includes 1210 TRs (Figure 1B). More precisely 181 426 344 peaks spread over 1002 TRs come from ChIP-seq studies, while 990 476 peaks spread over 208 TRs come from ChIP-exo studies. A large ChIP-exo study was processed (GSE151287), but the resulting peaks were inconsistent with published regions, and not added in this release. When compared to ReMap 2020, the large data gain is dispersed over practically all TRs (Figure 1C, light blue bars). We observe that the most studied transcription factors (e.g. ESR1, AR, FOXA1, TP53), transcriptional repressors (e.g. CTCF), and CRFs (e.g. BRD4) display, as expected, more datasets than other DNA-binding proteins. Nonetheless, all of the top 15 TRs show additional datasets integrated in ReMap 2022 (Figure 1C, light blue bars). The top 10 biotypes present in the human catalog correspond to the most common cell lines used in genomics (e.g. MCF-7, K-562; Figure 1D). Our uniform data processing contributes to an updated ReMap 2022 human regulatory atlas of 182M binding regions revealing an unprecedented view of the regulatory landscape and complexity. To illustrate this complexity with dense co-localizations of peaks creating tight clusters (CRMs), we have been tracking the Human ELAC1 promoter since 2015. This genomic region (chr18:50 967 094–50 970 983) highlights the 2022 catalog expansion compared with 2020 (Figure 2A), and across all four catalogs with 229, 1037, 2554 and 3546 binding regions respectively (Supplementary Figure S8). Three clusters of peaks can be observed, one large at the promoter embedding a second cluster after the transcription start site (TSS) at about +500 bp and +2 kb from the Gencode TSS (31). Interestingly these clusters concords with locations of candidate Cis-Regulatory Elements (cCREs) derived from ENCODE data (8). The third cluster located further up the first intron has been described in depth in precedent ReMap papers (10,15,16) to illustrate how combining data from various sources enhances genome annotations. Indeed, this third cluster contains 179 FOXA1 peaks in the 2022 catalog (93 in 2020, 60 in 2018, 15 in 2015) including one peak from ENCODE (7) (Supplementary Figure S9). The ReMap database provides three majors atlases, the main catalog containing all binding regions, a non-redundant set and a CRMs atlas. Indeed, to show a discrete repertoire of binding regions in the genome, the redundant binding regions are merged for identical TRs resulting in a multi-cell, multi-tissue regulatory map of 68M non-redundant binding regions (see ‘Materials and Methods’ for details). The CRM atlas (3.4M regions) has been improved with a new methodology that relies on detecting peak density along the genome, reinforcing the identification of regulatory hotspots by the integration of ChIP-seq from various cells, antibodies, and laboratories. Overall, this 2022 update of the human catalog expands the genome regulatory space revealing complex regulatory architectures strengthening the identification of DNA bound regions across thousands of experimental evidences.
Figure 2.
Updated ReMap 2022 regulatory atlas for Human and Plant. (A) ReMap 2022 Human DNA-protein binding pattern of 8103 valid datasets. This genome browser example of the DNA-binding peak depth of ReMap 2022 atlas is compared to ReMap 2020 at the vicinity of the ELAC1 promoter (chr18:50 967 094–50 970 983). The tracks displayed are compacted to thin lines so the depth of ReMap 2022 bindings can be compared to 2020. Around this ELAC1 location ReMap 2022 displays 3546 peaks, while the 2020 version contains 2554 peaks. The following genome tracks correspond to the GENCODE v36 annotation, the ENCODE candidate Cis Regulatory Elements (cCREs, red promoters, orange proximal enhancer-like, yellow distal enhancer-like) and the 100 vertebrates base-wise conservation showing regions predicted to be conserved (positive scores in blue). (B) A genome browser view of the ReMap 2022 Arabidopsis TF a atlas compared to the ReMap 2020 version at the vicinity of the AT1G21390.1 gene model (chr1:7 487 784–7 492 176). The annotation genome track corresponds to the latest Ensembl Plants v51 TAIR10 gene annotation. All peaks have been compacted for rendering.
Arabidopsis thaliana regulatory update
The A. thaliana 2022 release focuses on updating the regulatory catalog as illustrated in Figure 1E–H and in Figure 2. In this update, we curated, processed and analysed 223 new ChIP-seq datasets against TRs submitted in GEO since our previous release (Figure 1E). These datasets were processed uniformly using the ReMap pipeline. After applying quality controls and filters, 185 datasets (79%) were retained then integrated with the current catalogue leading to a total of 694 datasets (Figure 1E, Supplementary Figure S2). The 2022 Arabidopsis regulatory catalog contains 4.8M binding regions for 423 TRs in 23 biotypes and 14 ecotypes (Figure 1F–H). This update shows a growth in both the number of peaks and the number of TRs. In fact, the number of peaks is almost 2-fold superior to the 2020 Arabidopsis regulatory catalog (Figure 1F). The top three most represented immunoprecipitated DNA-binding proteins are Argonaute protein AGO1 (mRNA and chromatin binding), the two-component response regulator ARR1 (Transcription activator), the Heat stress transcription factor HSFA1B (Figure 1G), while the two most represented biotypes are leaves and seedlings (Figure 1H). This ReMap Arabidopsis regulatory catalog reveals an unprecedented view of the landscape and complexity of a Plant transcription factors occupancy map. This complex architecture in a plant genome is illustrated in the vicinity of the AT1G21390.1 gene model (emb2170; chr1:7 487 784–7 492 176) revealing the data growth between 2022 and 2020 catalogs (498 and 279 peaks respectively, Figure 2B). The genomic view highlights how the regulatory repertoire can be complemented by uniform processing of public studies. The 2022 Arabidopsis release comes with the usual three catalogs, all peaks, non-redundant peaks and CRMs identifications. To date, this Arabidopsis ReMap catalog is still the first to provide a global view of all detected TRs binding in a wide variety of biological contexts and a variety of experiments.
NEW REGULATORY CATALOGS FOR MOUSE AND DROSOPHILA
Mouse transcriptional regulatory catalog
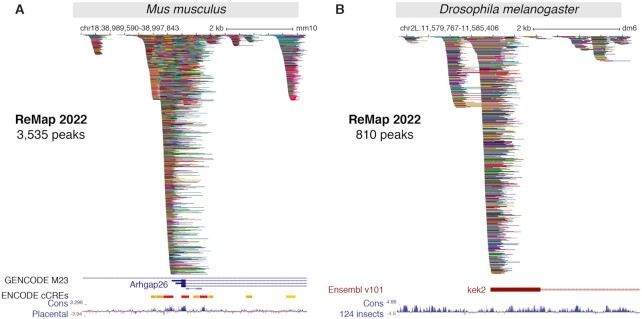
A new regulatory catalog for M. musculus is included in this fourth release of ReMap, as a result of a one-of large-scale integration of public murine DNA-binding assays. While few Mouse regulatory atlases from consortiums (8,32–34) are available for viewing in genome browsers, none offer significant details and depth of transcription factors occupancy map. To create this unique Mouse regulatory atlas we have collected, curated, uniformly processed, and analysed 7317 ChIP-seq datasets against TRs, precisely 7050 from GEO and 167 from Mouse ENCODE (Figure 1I). After applying quality controls and filters 5503 datasets (76%) were retained (Supplementary Figure S3). Our analyses lead to a final Mouse regulatory atlas of 123 207 170 binding regions for 648 TRs in 373 different murine biotypes (Figure 1J–L). The top three most represented TRs are the transcriptional repressor Ctcf, the polycomb protein Suz12 and the double-strand-break repair protein Rad21 homolog Rad21 (also a member of the cohesin complex) (Figure 1K). The most commonly represented biotypes (cells or tissues) are the mouse Embryonic Stem Cells (mESC), Mouse Embryonic Fibroblast (MEF) and Bone Marrow Derived Macrophage (BMDM) (Figure 1L). A recent study (35) has experimentally quantitatively identified over 60% of Mouse TFs (n = 941) out of the approximated 1500 TFs encoded in the mammalian genome (6). The complex regulatory architecture of the Mouse ReMap catalog is illustrated in the vicinity of the Rho GTPase-activating protein 26 (Arhgap26) gene (Figure 3A), combined with the Mouse ENCODE cCREs annotation (8). We observe a good correlation between cCREs regions and ReMap peak clusters (99.6% overlap), with some clusters (e.g. first cluster) yet undescribed by ENCODE. While our catalog contains 648 transcriptional regulators (TRs), which may represent ∼44% of the current TF census (6), it provides a unique collection of manually curated and uniformly processed ChIP-seq datasets from heterogeneous sources. When comparing with the Human atlas, 412 transcriptional regulators are found in common with the Mouse catalogs (Figure 1M), allowing exploration of evolutionary conservation of cis-regulatory modules (36,37). We present a unique collection of regulatory regions in Mouse as a result of a large-scale integrative analysis of ChIP-seq experiments for hundreds of transcriptional regulators.
Figure 3.
New ReMap 2022 regulatory atlas for Mus musculus and Drosophila melanogaster. (A) A genome browser view of the first Mouse ReMap 2020 atlas for transcriptional regulators at the vicinity of the Arhgap26 gene (chr18:38 989 590–38 997 843). Genome tracks correspond to the GENCODE M23 gene annotation, the ENCODE candidate Cis Regulatory Elements for mouse (cCREs, red promoters, orange proximal enhancer-like, yellow distal enhancer-like) and the Placental Mammal Basewise Conservation showing regions predicted to be conserved (positive scores in blue). (B) The Drosophila ReMap 2022 regulatory atlas at the vicinity of the kekkon 2 promoter (kek2, FBgn0265689, chr2L:11 579 767–11 585 406). The genome tracks correspond to the Ensembl v101 Drosophila melanogaster gene annotation and the 124 insects Basewise Conservation showing regions predicted to be conserved (positive scores in blue). All peaks have been compacted for rendering.
Drosophila transcriptional regulatory catalog
Finally, this new ReMap release allows the Fruit Fly community to browse and study the regulatory landscape of the D. melanogaster genome as we present a regulatory catalog of Drosophila ChIP-seq studies in diverse experimental conditions and various tissues and cells. A total of 1308 datasets were processed with 790 datasets from GEO and 525 from ENCODE (461 modERN, 69 modENCODE) (38,39). These datasets were manually curated and annotated using the Flybase database (26) for the official gene name convention. After applying our quality controls and filters 1,205 datasets (92%) were retained (692 from GEO and 514 from ENCODE, Supplementary Figure S4). Our analyses lead to a final Drosophila regulatory atlas of 16 634 486 binding regions for 550 TRs in 17 different fly biotypes (Figure 1N-O). These biotypes correspond to either cell lines (e.g. Schneider-2) or developmental stages (e.g. embryo, first-instar, second-instar, third-instar and adult), where names were standardized across studies. The top three most represented biotypes in our atlas are the adult fly, the embryo and the cell line Kc167 (Figure 1Q). Regarding TRs the top three TRs are the chromatin-linked adaptor for MSL proteins Clamp, a component of the gypsy chromatin insulator complex Cp190 and the transcription factor CTCF (Figure 1P). Like all ReMap catalogs from previous or current releases, the Drosophila regulatory atlas can be browsed with major Genome Browsers (40–42). We illustrate the complexity of the Fly regulatory landscape around the kek2 gene with 810 peaks forming various clusters at the kek2 promoter and up/downstream of the TSS (Figure 3B). We present here a unique Fly regulatory occupancy map forming complex architecture revealed by a large-scale integration of public D. melanogaster DNA-binding experiments.
CATALOG IMPROVEMENTS
Less is more
In this release we focused on improving the content of the ReMap catalogs by extending our filtering procedures. With updates adding thousands of published datasets, the ReMap catalogs have expanded dramatically for Human, but also for the new Mouse catalog. With a range of 100–200M peaks, a small fraction of spurious peaks or datasets may influence or bias the characterisation of the regulatory landscape. Indeed, ReMap has reached a point where the identification of regulatory occupancy regions by adding large quantities of datasets may have been achieved. High quality redundant ChIP-seq evidence, illustrated by the FOXA1 peaks in ELAC1 gene (Supplementary Figure S9), allows to improve the identification of ReMap occupancy regions. To remove spurious peaks, two sets of controls were added in our quality control steps, named QC2 in Figure 1. These filters were applied to new catalogs, and retroactively for Human and Arabidopsis. We discarded datasets with less than 100 peaks or with more than two times the number of annotated genes, according to the Ensembl gene annotation statistics (Supplementary Figure S5). A few datasets passing our initial quality controls would generate an unusual amount of peaks, potentially affecting the occupancy repertoire. The second filter removes peaks whose length are outside set cutoffs. Ranges were defined as a minimum of 50bp and a maximum upper cutoff for which we have 99% of catalogue, either 1.5kb or 2kb (See Material and Method, Supplementary Figure S6). This discards large peaks spanning multiple regulatory regions, those peaks not being informative for the transcription factor repertoire identification, or definition of CRMs. In Human, these filters (QC2) remove 97M peaks as they are applied retroactively to the entire catalog (279M without QC2), 71M peaks removed in Mouse, 1.1M in Drosophila and 0.1M in Arabidopsis. However, following the open science principles, the peaks filtered out are available in the download section as ‘filtered out’ peaks. We believe a conservative approach will benefit the ReMap catalogs by removing uninformative peaks and datasets, leading to clearer regulatory catalogs.
Improved CRM identification
With the Human ReMap catalog reaching ∼182M peaks, the identification of clusters of peaks located often in close proximity, or tightly grouped around a TSS, can be problematic (Figure 2A). In this update, we applied a newly developed method to better identify Cis-Regulatory Modules (CRMs). The initial method consisted in merging overlapping non-redundant peaks (NR peaks), but this approach is only applicable when the number of peaks remains small. When dealing with catalogs of >100 millions of peaks, the genome coverage becomes too high, making the initial but simple approach unable to distinguish CRMs properly in high density regions such as in the ELAC1 example (Supplementary Figure S7A). Thus, to accurately identify CRMs, we have developed a new methodology that detects individual regions in tight clusters of peaks by defining CRMs at each local minimum of NR peak density function (Supplementary Figure S7B). An NR peak is assigned to a CRM if its peak summit falls in between the identified flanking local minimas. The CRM boundaries are reduced using the 5′ and 3′ coordinates of the flanking NR peaks. This newly developed method better defines the genomic organization of our atlas by better identifying dense co-localizations of non-redundant peaks forming tight clusters of transcriptional regulators.
AN IMPROVED USER EXPERIENCE
Interactive tour and fast search
The ReMap 2022 release comes with an improved user experience. On the ReMap homepage, as well as on the header of the site, we provide an interactive tour walking users through the main features of the website. The tour is activated by clicking on the ‘Tour’ button right in the middle of the homepage or header bar. The tour dynamically shows the different types of catalogs available on the website (all peaks, non redundant and CRMs), how to browse the ReMap catalogs, download and search the database. This newly introduced interactive tour is a useful feature to better understand the ReMap database, the data content and its functionalities. The ReMap database can be browsed by using the navigation links on the left sidebar, or searched for individual TRs, specific biotypes (cell lines or tissues) using the simple or advanced search box. With an increased number of datasets and species in 2022 the search engine has been rewritten allowing for faster search queries. Search results are returned in a paginated table along with TRs aliases, TF classification, experiment IDs and data source. Also this responsive search results table can be dynamically searched to refine results. Finally, the ReMap database can be queried programmatically using the RESTful API, which contains added functions in 2022.
Genomic tracks and videos
Since ReMap 2015 an annotation track is available on the UCSC Genome browser website (41,43) within public sessions or public hubs (Figures 2 and 3). These genome track hubs are an essential tool for the visualization of the expanding ReMap catalogs. Track hubs are a convenient, efficient, mechanism for importing the large ReMap collections of regulatory features, providing standards for data tracks across genome-browsing platforms. For each ReMap species, and in different assemblies (hg38, hg19, mm39, mm10, dm6, TAIR10), track hubs have been added in the public hubs listing of the UCSC Genome browser as well as deposited to the EMBL-EBI Track Hub Registry (https://trackhubregistry.org/, Supplementary Figure S10). This ReMap release comes along with the new track hub filtering options available from the UCSC Genome browser. ReMap users can now filter which peaks are displayed by selecting specific biotypes and/or by specific TRs (e.g. FOXA1 in MCF-7). This new feature enables better flexibility especially as the ReMap database increases in size. Additionally, the peak names displayed on the browser can be adapted for a better visualisation. To illustrate and explain these browsing options we added multiple videos. These will guide users to make the best of the ReMap catalogs. Furthermore, to represent the depth of regulatory regions, we added a density track on top of the ReMap catalog track. By adding these browsing features, our objective is to make it easier for researchers to explore the variety and the complexity of ReMap catalogs when combined with other biological tracks.
CONCLUSION AND FUTURE DIRECTIONS
This fourth release of the ReMap database pursues its commitment to provide manually curated datasets and high-quality regulatory catalogs for the research community. For this 2022 update, we processed 13 000 ChIP-seq datasets starting from raw data. With four species, this version of ReMap continues the long-term goal of maintaining accessible, browseable, high-quality regulatory catalogs. The ReMap 2022 database comes with many updates, (i) a substantially expanded human regulatory catalog followed by (ii) an update of the Arabidopsis thaliana regulatory catalog; (iii) a new regulatory atlas for Mus musculus with more than 7,000 datasets curated and processed; (iv) the first Drosophila melanogaster regulatory atlas for the Fly scientific community; (v) an improved CRM identification method, (vi) a conservative filtering approach to improve our catalogs; (vii) an improved user experience with an interactive tour and (viii) updated genomic track hubs in different assemblies for better visualization in genome browser. Finally, since 2018 each ReMap update is incorporated into the JASPAR pipeline to infer new and updated TF binding profiles (17,18).
Overall, the ReMap database reaches 327M binding regions across four regulatory atlases. We anticipate that upcoming functional studies will provide additional experimental regulatory evidence giving rise to even larger catalogs. While manual curation and annotation are the foundation of ReMap, we intend to evolve to more conservative approaches in future releases, such as directing our efforts towards redundant peaks to build robust regulatory regions. Our long term goal of providing qualitative high quality catalogs will focus on redundant occupancy evidence for new releases, rather than releasing quantitative catalogs consisting in incremental inventories. Depending on outcomes, future ReMap releases may be crossed with other regulatory catalogs (DNAse, Histone marks) to strengthen and filter the regulatory space.
FEEDBACK
We thank our users for past and future feedback to make ReMap useful for the community. The ReMap team welcomes your feedback on the catalogs, use of the website and use of the downloadable files. Please contact benoit.ballester@inserm.fr for development requests.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Nathalie Arquier and Laurent Perrin for their expertises and scientific discussions regarding the curation and annotation of Drosophila melanogaster ChIP-seq data. We would like to thank the JASPAR Team led by Anthony Mathelier from NCMM Norway for constant scientific feedback on the ReMap catalogues. Finally, we would like to thank Maximilian Haeussler, Gerardo Perez and the UCSC Genome informatics groups for their help with public track hubs and their latest hub development, also the Ensembl and Ensembl Plant group for their help with the Human, Mouse, Fly and Arabidopsis track hubs.
Contributor Information
Fayrouz Hammal, Aix Marseille Univ, INSERM, TAGC, Marseille, France.
Pierre de Langen, Aix Marseille Univ, INSERM, TAGC, Marseille, France.
Aurélie Bergon, Aix Marseille Univ, INSERM, TAGC, Marseille, France.
Fabrice Lopez, Aix Marseille Univ, INSERM, TAGC, Marseille, France.
Benoit Ballester, Aix Marseille Univ, INSERM, TAGC, Marseille, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
PhD Fellowship to F.H. from the Provence-Alpes-Côte d’Azur Regional Council (Région SUD); Institut National de la Santé et de la Recherche Médicale (INSERM); PhD Fellowship to P.D.L. from the French Ministry of Higher Education and Research (MESR); HPC resources of Aix-Marseille Université financed by the project Equip@Meso [ANR-10-EQPX-29-01] of the program ‘Investissements d’Avenir’ supervised by the Agence Nationale de la Recherche. Funding for Open Access charge: INSERM, MarMaRa, this project has received funding from the Excellence Initiative of Aix-Marseille University - A*Midex a French “Investissements d’Avenir programme”- Institute MarMaRa AMX-19-IET- 007.
Conflict of interest statement. None declared.
REFERENCES
- 1. Johnson D.S., Mortazavi A., Myers R.M., Wold B.. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007; 316:1497–1502. [DOI] [PubMed] [Google Scholar]
- 2. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sarkans U., Füllgrabe A., Ali A., Athar A., Behrangi E., Diaz N., Fexova S., George N., Iqbal H., Kurri S.et al.. From ArrayExpress to BioStudies. Nucleic Acids Res. 2021; 49:D1502–D1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fukuda A., Kodama Y., Mashima J., Fujisawa T., Ogasawara O.. DDBJ update: streamlining submission and access of human data. Nucleic Acids Res. 2021; 49:D71–D75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T.. The human transcription factors. Cell. 2018; 172:650–665. [DOI] [PubMed] [Google Scholar]
- 6. Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M.. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009; 10:252–263. [DOI] [PubMed] [Google Scholar]
- 7. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. ENCODE Project Consortium Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A.et al.. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020; 583:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roadmap Epigenomics Consortium Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J.et al.. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Griffon A., Barbier Q., Dalino J., van Helden J., Spicuglia S., Ballester B.. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res. 2015; 43:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kolmykov S., Yevshin I., Kulyashov M., Sharipov R., Kondrakhin Y., Makeev V.J., Kulakovskiy I.V., Kel A., Kolpakov F.. GTRD: an integrated view of transcription regulation. Nucleic Acids Res. 2021; 49:D104–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng R., Wan C., Mei S., Qin Q., Wu Q., Sun H., Chen C.-H., Brown M., Zhang X., Meyer C.A.et al.. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019; 47:D729–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., Kawaji H., Nakaki R., Sese J., Meno C.. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018; 19:e46255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou K.-R., Liu S., Sun W.-J., Zheng L.-L., Zhou H., Yang J.-H., Qu L.-H.. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017; 45:D43–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chèneby J., Gheorghe M., Artufel M., Mathelier A., Ballester B.. ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments. Nucleic Acids Res. 2018; 46:D267–D275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chèneby J., Ménétrier Z., Mestdagh M., Rosnet T., Douida A., Rhalloussi W., Bergon A., Lopez F., Ballester B.. ReMap 2020: a database of regulatory regions from an integrative analysis of human and Arabidopsis DNA-binding sequencing experiments. Nucleic Acids Res. 2020; 48:D180–D188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Castro-Mondragon J.A., Riudavets-Puig R., Rauluseviciute I., Berhanu Lemma R., Turchi L., Blanc-Mathieu R., Lucas J., Boddie P., Khan A., Manosalva Pérez N.. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2021; 10.1093/nar/gkab1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gheorghe M., Sandve G.K., Khan A., Cheneby J., Ballester B., Mathelier A.. A map of direct TF-DNA interactions in the human genome. Nucleic Acids Res. 2019; 47:7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K.et al.. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tweedie S., Braschi B., Gray K., Jones T.E.M., Seal R.L., Yates B., Bruford E.A.. Genenames.org: the HGNC and VGNC resources in 2021. Nucleic Acids Res. 2021; 49:D939–D946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang A., Schomburg I., Placzek S., Jeske L., Ulbrich M., Xiao M., Sensen C.W., Schomburg D.. BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res. 2015; 43:D439–D446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Côté R., Reisinger F., Martens L., Barsnes H., Vizcaino J.A., Hermjakob H.. The Ontology Lookup Service: bigger and better. Nucleic Acids Res. 2010; 38:W155–W160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bairoch A. The Cellosaurus, a Cell-Line Knowledge Resource. J. Biomol. Tech. JBT. 2018; 29:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Howe K.L., Contreras-Moreira B., De Silva N., Maslen G., Akanni W., Allen J., Alvarez-Jarreta J., Barba M., Bolser D.M., Cambell L.et al.. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020; 48:D689–D695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Law M., Shaw D.R.. Mouse Genome Informatics (MGI) is the international resource for information on the laboratory mouse. Methods Mol. Biol. 2018; 1757:141–161. [DOI] [PubMed] [Google Scholar]
- 26. Larkin A., Marygold S.J., Antonazzo G., Attrill H., Dos Santos G., Garapati P.V., Goodman J.L., Gramates L.S., Millburn G., Strelets V.B.et al.. FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021; 49:D899–D907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P.et al.. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012; 22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nussbaum C., Myers R.M., Brown M., Li W.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Köster J., Rahmann S.. Snakemake–a scalable bioinformatics workflow engine. Bioinforma. Oxf. Engl. 2012; 28:2520–2522. [DOI] [PubMed] [Google Scholar]
- 31. Frankish A., Diekhans M., Jungreis I., Lagarde J., Loveland J.E., Mudge J.M., Sisu C., Wright J.C., Armstrong J., Barnes I.et al.. GENCODE 2021. Nucleic Acids Res. 2021; 49:D916–D923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stamatoyannopoulos J.A., Snyder M., Hardison R., Ren B., Gingeras T., Gilbert D.M., Groudine M., Bender M., Kaul R., Canfield T.et al.. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 2012; 13:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lesurf R., Cotto K.C., Wang G., Griffith M., Kasaian K., Jones S.J.M., Montgomery S.B., Griffith O.L.. ORegAnno 3.0: a community-driven resource for curated regulatory annotation. Nucleic Acids Res. 2016; 44:D126–D132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zerbino D.R., Wilder S.P., Johnson N., Juettemann T., Flicek P.R.. The ensembl regulatory build. Genome Biol. 2015; 16:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou Q., Liu M., Xia X., Gong T., Feng J., Liu W., Liu Y., Zhen B., Wang Y., Ding C.et al.. A mouse tissue transcription factor atlas. Nat. Commun. 2017; 8:15089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schmidt D., Wilson M.D., Ballester B., Schwalie P.C., Brown G.D., Marshall A., Kutter C., Watt S., Martinez-Jimenez C.P., Mackay S.et al.. Five-vertebrate ChIP-seq reveals transcription factor binding. Science. 2010; 328:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ballester B., Medina-Rivera A., Schmidt D., Gonzàlez-Porta M., Carlucci M., Chen X., Chessman K., Faure A.J., Funnell A.P., Goncalves A.et al.. Multi-species, multi-transcription factor binding highlights conserved control of tissue-specific biological pathways. Elife. 2014; 3:e02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kudron M.M., Victorsen A., Gevirtzman L., Hillier L.W., Fisher W.W., Vafeados D., Kirkey M., Hammonds A.S., Gersch J., Ammouri H.et al.. The ModERN resource: genome-wide binding profiles for hundreds of Drosophila and Caenorhabditis elegans transcription factors. Genetics. 2018; 208:937–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. modENCODE Consortium Roy S., Ernst J., Kharchenko P.V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L.et al.. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010; 330:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J.et al.. Ensembl 2021. Nucleic Acids Res. 2021; 49:D884–D891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Navarro Gonzalez J., Zweig A.S., Speir M.L., Schmelter D., Rosenbloom K.R., Raney B.J., Powell C.C., Nassar L.R., Maulding N.D., Lee C.M.et al.. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 2021; 49:D1046–D1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rangwala S.H., Kuznetsov A., Ananiev V., Asztalos A., Borodin E., Evgeniev V., Joukov V., Lotov V., Pannu R., Rudnev D.et al.. Accessing NCBI data using the NCBI Sequence Viewer and Genome Data Viewer (GDV). Genome Res. 2021; 31:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T., Nguyen N., Paten B., Zweig A.S., Karolchik D.et al.. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014; 30:1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.