Abstract
Sequence compositions of nucleic acids and proteins have significant impact on gene expression, RNA stability, translation efficiency, RNA/protein structure and molecular function, and are associated with genome evolution and adaptation across all kingdoms of life. Therefore, a devoted resource of sequence compositions and associated features is fundamentally crucial for a wide range of biological research. Here, we present CompoDynamics (https://ngdc.cncb.ac.cn/compodynamics/), a comprehensive database of sequence compositions of coding sequences (CDSs) and genomes for all kinds of species. Taking advantage of the exponential growth of RefSeq data, CompoDynamics presents a wealth of sequence compositions (nucleotide content, codon usage, amino acid usage) and derived features (coding potential, physicochemical property and phase separation) for 118 689 747 high-quality CDSs and 34 562 genomes across 24 995 species. Additionally, interactive analytical tools are provided to enable comparative analyses of sequence compositions and molecular features across different species and gene groups. Collectively, CompoDynamics bears the great potential to better understand the underlying roles of sequence composition dynamics across genes and genomes, providing a fundamental resource in support of a broad spectrum of biological studies.
INTRODUCTION
Sequence compositions are intricately implicated with genome evolution and adaptation across all kingdoms of life (1–3). For example, GC content is closely associated with mutational bias (4), DNA recombination (3) and repair (5), mRNA level (6,7) and gene age (8). And codon usage bias (CUB) has significant implications in gene expression (9), translational selection (10), protein structure (11), metabolic ecology (10) and environmental adaptation (12). In addition, sequence composition-derived features, such as coding potential, protein physicochemical property as well as liquid-liquid phase separation (LLPS), impact more directly on molecular functions and biological roles of biomolecules (13–15). Taken together, sequence compositions as well as their derived features are critically essential for better understanding evolutionary processes and molecular mechanisms across all kingdoms of life.
Over the past two decades, several valuable resources have been developed to characterize biomolecular sequence composition (16–21). Among them, representative resources are CUTG (16), CBCB (17), HIVE-CUTs (18) and CUBAP (21). Specifically, CUTG (16), established in 1998, is a widely used database compiling codon usage for 3 027 973 CDSs for 35 799 genomes (including 8,233 chloroplast genomes, 12 271 mitochondrion genomes and 439 plastid genomes). CBCB (17) is a specialized database housing CUB estimates for highly expressed genes in 300+ bacterial genomes. HIVE-CUTs (18) contains 855 412 codon usage tables for 689 420 species and their mitochondrial/plastid genomes derived from GenBank and RefSeq. The webserver CUBAP (21) computes GC content and codon usage for 17 634 human genes and facilitates analyses of CUB across human populations. In addition, there are still several other resources that specialize in integration of protein physicochemical properties (22,23) and phase separation properties (24–26). Albeit great efforts were made by existing resources, they have two major limitations. First, none of them covers the full range of sequence compositions as well as derived features. Second, they do not offer available tools for comparing molecular compositions between gene groupings in terms of protein families and GO terms, with friendly online functionalities for customized data analysis and interactive visualization. Particularly, with the rapid advancement of high-throughput sequencing technologies, an ever-increasing number of high-quality genomes covering a broad diversity of species have been sequenced and well annotated. Therefore, it is in urgent need to build a database incorporating sequence compositions and features based on high-quality genomes and genes.
To fill this gap, we present CompoDynamics (https://ngdc.cncb.ac.cn/compodynamics/), a comprehensive database for characterizing sequence compositions and molecular features across a wide range of species. Based on a large number of high-quality CDSs and genomes derived from RefSeq, CompoDynamics generates a full range of sequence compositions including nucleotide content, codon usage and amino acid usage, as well as derived features of coding potential, protein physicochemical property and phase separation. In addition, CompoDynamics is equipped with a set of online tools for composition analysis, comparison and visualization. Collectively, CompoDynamics has the great potential to become a fundamental resource for better understanding biological significance of sequence composition dynamics.
MATERIALS AND METHODS
Data collection
Over 127 million coding sequences (*_cds_from_genomic.fna.gz) covering viruses, archaea, bacteria, fungi, protozoa, plants, invertebrates and vertebrates were retrieved from NCBI RefSeq (27) (https://ftp.ncbi.nlm.nih.gov/genomes/refseq/; last access on 2020/5/27). For a species, complete assembly sequence(s) were selected; otherwise, only reference/representative genomes were kept. To enable gene comparisons between different families or functions, protein family/domain annotations were integrated from Pfam (version 34.0) (28), and GO (gene ontology) annotations (29,30) were incorporated using a series of R packages (org.Hs.eg.db, org.Mm.eg.db, org.Dr.eg.db, org.Ss.eg.db, org.Gg.eg.db, org.Mmu.eg.db, org.At.tair.db, org.Dm.eg.db, org.Ag.eg.db, org.Sc.sgd.db, org.EcK12.eg.db). To enable more convenient search, species lineage and alias are integrated from NCBI Taxonomy (31) (https://ftp.ncbi.nih.gov/pub/taxonomy/taxdump_archive/taxdmp_2020_05_01.zip).
Data pre-processing
CDS quality was evaluated by using in-house scripts with consideration of different genetic codes (https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi/). Sequences whose lengths are not multiple of three were removed. Most of CDSs were annotated as canonical, whereas the remaining ones were labelled accordingly if they (i) lack start codon, (ii) lack stop codon or (iii) contain in-frame stop codon. The information including gene locus tag, protein ID, gene name, protein description and genomic location was parsed from CDS fasta files.
Calculation of sequence compositions and features
A full range of sequence compositions and their derived features were computed for each CDS. Among them, gene length, nucleotide content, codon usage, amino acid usage, CDC (Codon Deviation Coefficient) as well as RSCU (Relative Synonymous Codon Usage) were calculated using CAT (32); ENC (Effective Number of Codons) values were computed by CodonW (http://codonw.sourceforge.net/); coding potential was assessed with LGC (33) and CPC2 (34); phase separation features were calculated using ESpritz (35), PLAAC (36) and Pi-Pi (37); and protein physicochemical properties were computed by in-house scripts. Species-specific genetic codes were considered during the above calculations.
For each genome, sequence compositions and features were estimated based on the results of all related CDSs, by adopting the following three strategies: (i) weighted averaging based on CDS length, such as GC content, (ii) direct averaging across all CDSs, such as the averaged CDC and ENC, (iii) calculation based on genome-scale values, such as RSCU. For instance, GC and GC1/2/3 (positional GC content at three codon positions) contents were averaged across all CDSs by weights of CDS lengths (this is equal to calculation by concatenating all the CDSs in a genome) and their distributions were represented as boxplots. Mean values of CUB, viz., averaged CDC and ENC, were calculated and their distributions were represented as boxplots. RSCU values were calculated based on total frequencies of each codon within genome. Detailed descriptions for all sequence compositions and molecular features are available at https://ngdc.cncb.ac.cn/compodynamics/help/.
Implementation
CompoDynamics was built with Spring boot (http://spring.io/), a mature and convention-over-configuration Model-View-Controller (MVC) framework, deployed in a Centos Linux 7.9 environment. In the back-end part, CompoDynamics data was stored in MySQL (https://www.mysql.com/), a free and popular relational database management system. The database was run by Apache ShardingSphere (https://shardingsphere.apache.org/), an open-source ecosystem consisted of a set of distributed database solutions. Web pages were constructed using HTML5 and rendered using Thymeleaf (https://www.thymeleaf.org/). Front-end interfaces were developed by using Bootstrap (https://getbootstrap.com/) with JQuery (https://jquery.com/) to provide responsive and user-friendly web pages. Furthermore, HighCharts (https://www.highcharts.com.cn/), ECharts (https://echarts.apache.org/) and DataTables (https://datatables.net/) were used to perform interactive charting and data visualization.
DATABASE CONTENTS AND FEATURES
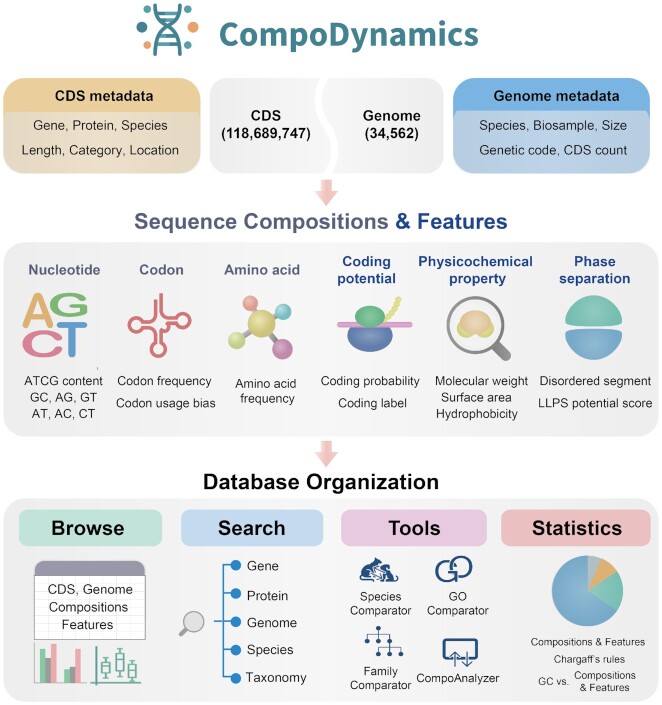
CompoDynamics is a comprehensive database of sequence compositions and features (nucleotide content, codon usage, amino acid usage, coding potential, protein physicochemical property and phase separation) for 118 689 747 high-quality CDSs and 34 562 genomes, covering 1 692 647 genes and 24 995 species. Moreover, CompoDynamics provides interactive and user-friendly tools to perform comparative analysis of composition features across different species and gene groupings in terms of protein families and GO terms, enabling users to investigate composition dynamics across genes and genomes. In CompoDynamics, all these contents are organized in terms of sequences (gene and genome), compositions, features and tools (Figure 1).
Figure 1.
Database contents and organization. The present version of CompoDynamics provides six groups of sequence compositions (nucleotide content, codon usage and amino acid usage) and features (coding potential, protein physicochemical property and phase separation) for 118 689 747 CDSs and 34 562 genomes derived from RefSeq. These contents could be easily browsed, visualized, retrieved and analyzed at both genome and gene levels.
Nucleotide contents in genomes
Taking advantage of the large quantity of genomic data, CompoDynamics presents a whole picture of nucleotide contents for a wide range of species and CDSs, enabling systematic investigations on nucleotide composition dynamics in a cross-species manner. Specifically, detailed compositions of four individual nucleotides (A, T, G, C), nucleotide contents (GC, AG, GT, AT, AC, CT) and their positional contents in the 1st/2nd/3rd codon positions are presented for each genome and CDS in CompoDynamics. Based on 34 562 high-quality genome sequences, we observe that the Chargaff's second parity rule (A = T and C = G) applies in most of the species categories (R2 > 0.99) (Supplementary Figure S1A, B), and positional GC contents (GC1, GC2, GC3) are positively correlated with overall GC content, which is consistent with previous findings (38,39) (Supplementary Figure S1C). In addition, it is observed that GC1 is always higher than GC2, and GC3 has the most variability consistent with our previous observations (40). Taken together, CompoDynamics characterizes nucleotide compositions across a variety of species, facilitating the whole-genome comparative analysis of nucleotide variation.
Codon/amino acid usage in genomes
Considering that CUB is a complex interplay between mutation and selection, CompoDynamics houses several estimates of codon usage and CUB across a broad spectrum of organisms. CUB is represented by two popular measures viz., ENC (41) and CDC (32). The former evaluates the randomness of observed codon usage relative to uniformed codon usage (ranging from 20 for maximum bias to 61 for no bias) and the latter considers background compositions and thus reflects the strength of selection on synonymous codon usage (ranging from 0 for no bias to 1 for maximum bias). At the genome level, CUB estimates for each genome are averaged over all CDSs (see details in Materials and Methods). CompoDynamics presents codon usage tables and CUB estimates for all collected genomes and genes and offers visualization functionalities to investigate their distributions in bar plots and box plots.
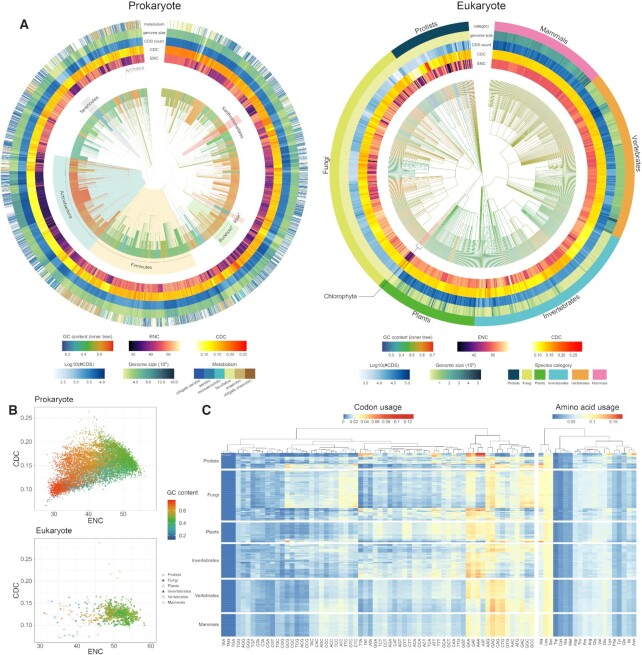
Based on all collected genomes in CompoDynamics, we investigate the dynamics of codon usage, CUB, GC content as well as genome size across clades (Figure 2). Consistent with extensive reports (42,43), prokaryotes differ greatly in GC content and CUB (Figure 2A and B). Notably, prokaryotes with extreme high (such as the majority of Actinobacteria) or low (such as Tenericutes and Rickettsiales) GC contents, tend to show lower ENC and CDC estimates (Figure 2A), indicating stronger CUB on their genomes presumably caused by mutational pressure. Likewise, Burkholderiales and Xanthomonadales seem to present lower ENC estimates and relatively higher CDC estimates (Figure 2B), suggesting stronger CUB on their genomes more likely due to selection. Different from prokaryotes, eukaryotes always exhibit moderate ENC/CDC with a narrow range of GC content especially for more complex organisms (Figure 2B). Intriguingly, it is consistently found that extreme GC contents and biased codon usages are mostly observed in less complex organisms, such as protists, fungi and green algae (Figure 2B). More complex organisms, such as Mammalia (mammals), Aves and Actinopteri, show high uniformity in codon and amino acid usage (Figure 2C), which coincides with their narrow GC/ENC/CDC estimates. And not surprisingly, amino acid usage shows better uniformity than codon usage (Figure 2C).
Figure 2.
Codon usage dynamics across prokaryote and eukaryote genomes. (A) CUB distributions in prokaryote and eukaryote genomes. CUB (represented by ENC and CDC), GC content of genomic coding region, genome size, CDS number and metabolism type are visualized by different color palettes. For prokaryote, organisms with CDS count ≥100 are displayed in the cladogram. Several clades are highlighted to exemplify different kinds of strong CUBs. Burkhold*: Burkholderiales; Rick*: Rickettsiales. (B) Relationship between ENC and CDC for different GC values in prokaryote and eukaryote. (C) Codon usage and amino acid usage across six species categories in eukaryote.
Sequence compositions and features in genes
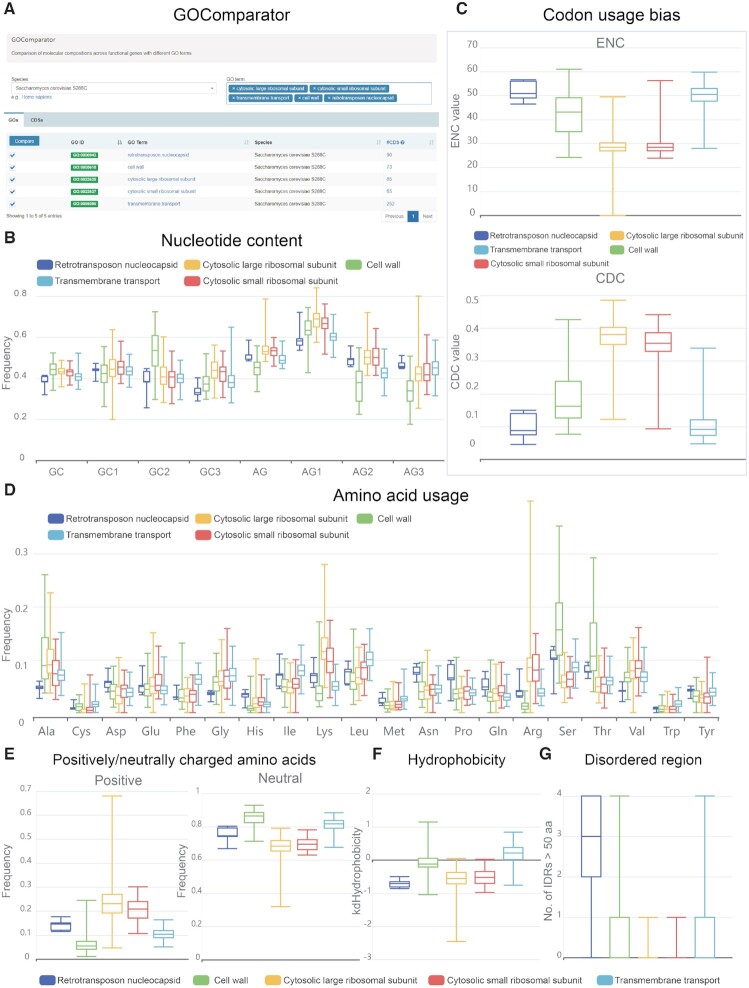
Different genes, in respect of their families or functions, are under different selective pressures and may exhibit different compositional patterns. Regarding this, CompoDynamics provides a wealth of CDS-level compositions and molecular features for 118 689 747 CDSs across 24 995 species and supports online analysis for composition comparison between gene groupings with different families or GO terms (Figure 3). Taking Saccharomyces cerevisiae S288C as an example, five groups of yeast genes, according to GO terms, namely, ‘cytosolic large ribosomal subunit’, ‘cytosolic small ribosomal subunit’, ‘transmembrane transport’, ‘cell wall’ and ‘retrotransposon nucleocapsid’, are selected for composition comparison (Figure 3A). Online tools in CompoDynamics enable the investigation of nucleotide content variation in these five groups (Figure 3B). Among them, genes with the GO terms of ‘cytosolic large ribosomal subunit’ and ‘cytosolic small ribosomal subunit’, exhibit more biased codon usage (represented by low ENC and high CDC) (Figure 3C) as well as amino acid usage (Figure 3D) and are positively charged (Figure 3E), conforming with a previous report (44). In addition, genes with terms of ‘cell wall’ and ‘transmembrane transport’ are observed to be mostly neutrally charged and highly hydrophobic, which coincides well with their specialized functions (Figure 3E and F). Furthermore, by estimating a series of indices for liquid-liquid phase separation potential, we observe that ‘retrotransposon nucleocapsid’ associated genes harbour detectable disordered regions and show relatively strong signals for phase separation potential (Figure 3G). In short, CompoDynamics features online functionalities for composition comparison and visualization, paving the way for in-depth investigation of different genes.
Figure 3.
Sequence composition and feature comparisons between genes with different GO terms. (A) Five groupings of yeast genes, according to GO terms, namely, ‘cytosolic large ribosomal subunit’, ‘cytosolic small ribosomal subunit’, ‘transmembrane transport’, ‘cell wall’ and ‘retrotransposon nucleocapsid’, are selected for comparison with the online tool GOComparator in CompoDynamics. The comparison results are illustrated for (B) nucleotide content, (C) codon usage bias, (D) amino acid usage, (E) positively/neutrally charged amino acids, (F) hydrophobicity and (G) intrinsically disordered regions.
Data organization and presentation
CompoDynamics is organized to enable easy and efficient data browse, search, comparison and statistics of sequence compositions and derived features (Figure 1). In the homepage, CompoDynamics offers a fast and case-insensitive search functionality, allowing users to search simply by specifying any term (including species, taxonomy, assembly, gene and protein) and by setting advanced filtering options. Once a search is submitted, relevant results are displayed in the web page where genome/CDS-level compositions and features are provided. Clicking any individual genome/CDS can direct to the corresponding page that contains detailed results of compositions and features in terms of basic information, nucleotide content, codon usage, amino acid usage, coding potential, physicochemical property and phase separation of each genome/CDS. Additionally, CompoDynamics offers browse functionalities to help users easily retrieve compositions and features for any genome/CDS of interest.
Importantly, CompoDynamics provides several interactive online tools for analyzing molecular compositions and features across different species (SpeciesComparator), different gene families (FamilyComparator) and different GO terms (GOComparator). By selecting genomes or gene groups of interest, these tools allow users to perform comparisons in terms of nucleotide composition, codon usage, amino acid usage, coding potential, physicochemical properties and phase separation. In addition, CompoAnalyzer is designed to accept user-input sequences for custom analyses of compositions and features. Moreover, a series of data statistics is presented to help users obtain an overview of compositions and features, Chargaff's rules and the relationships between GC content and other compositions/features across various species categories.
DISCUSSION AND FUTURE DEVELOPMENTS
Based on the high-quality data, CompoDynamics presents a wealth of sequence compositions and molecular features for 118 689 747 CDSs and 34 562 genomes covering 1 692 647 genes and 24 995 species. Consequently, CompoDynamics yields a whole picture of sequence compositions and features across all kingdoms of life, greatly facilitating users to investigate variation dynamics at the genome level, as well as providing important insights on better understanding molecular sequence evolution. Also, CompoDynamics is equipped with several useful tools for online analysis and visualization, thus enabling users to perform comparative analysis of sequence compositions and features across various species and gene groups. Future developments of CompoDynamics include periodical update (at least once a year) of high-quality genomes and CDSs from NCBI RefSeq, GENCODE, and Genome Warehouse (45) in NGDC (46). Moreover, untranslated genomic regions, such as the untranslated regions (UTRs), mRNA introns and noncoding RNAs, will be added to characterize composition dynamics in a larger scale. Furthermore, CompoDynamics will incorporate more curated metadata and molecular features (e.g. di-nucleotides, optimal codons, etc.) and enhance online tools for better analysis and visualization. Collectively, CompoDynamics bears great utility to help users better understand sequence composition dynamics of genes and genomes and thus to serve as a fundamental resource in support of global biological research.
DATA AVAILABILITY
CompoDynamics is a user-friendly database for characterizing sequence composition dynamics and can be accessed directly at https://ngdc.cncb.ac.cn/compodynamics/.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dong Zou, Qianpeng Li and Zhao Li for reporting bugs and providing suggestions as well as Zhuojing Fan for her assistance in web design of our database.
Contributor Information
Shuai Jiang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Qiang Du, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Changrui Feng, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Lina Ma, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Zhang Zhang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Special Investigation on Science and Technology Basic Resources of the MOST [2019FY100102]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB38030400, XDA19050302]; National Natural Science Foundation of China [32030021, 31871328, 32100520]; Youth Innovation Promotion Association of Chinese Academy of Sciences [2019104]. Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Simon D., Cristina J., Musto H.. Nucleotide composition and codon usage across viruses and their respective hosts. Front Microbiol. 2021; 12:646300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lassalle F., Perian S., Bataillon T., Nesme X., Duret L., Daubin V.. GC-Content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLoS Genet. 2015; 11:e1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pessia E., Popa A., Mousset S., Rezvoy C., Duret L., Marais G.A.. Evidence for widespread GC-biased gene conversion in eukaryotes. Genome Biol Evol. 2012; 4:675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu H., Zhang Z., Hu S., Yu J.. On the molecular mechanism of GC content variation among eubacterial genomes. Biol. Direct. 2012; 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lind P.A., Andersson D.I.. Whole-genome mutational biases in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:17878–17883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kudla G., Lipinski L., Caffin F., Helwak A., Zylicz M.. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006; 4:e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Courel M., Clement Y., Bossevain C., Foretek D., Vidal Cruchez O., Yi Z., Benard M., Benassy M.N., Kress M., Vindry C.et al.. GC content shapes mRNA storage and decay in human cells. Elife. 2019; 8:e49708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yin H., Wang G., Ma L., Yi S.V., Zhang Z.. What signatures dominantly associate with gene age?. Genome Biol. Evol. 2016; 8:3083–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Z., Dang Y., Zhou M., Li L., Yu C.H., Fu J., Chen S., Liu Y.. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6117–E6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. LaBella A.L., Opulente D.A., Steenwyk J.L., Hittinger C.T., Rokas A.. Signatures of optimal codon usage in metabolic genes inform budding yeast ecology. PLoS Biol. 2021; 19:e3001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao F., Yu C.H., Liu Y.. Codon usage regulates protein structure and function by affecting translation elongation speed in Drosophila cells. Nucleic Acids Res. 2017; 45:8484–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Botzman M., Margalit H.. Variation in global codon usage bias among prokaryotic organisms is associated with their lifestyles. Genome Biol. 2011; 12:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Statello L., Guo C.J., Chen L.L., Huarte M.. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021; 22:96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hyman A.A., Weber C.A., Julicher F.. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014; 30:39–58. [DOI] [PubMed] [Google Scholar]
- 15. Uversky V.N. Protein intrinsic disorder and structure-function continuum. Prog. Mol. Biol. Transl. Sci. 2019; 166:1–17. [DOI] [PubMed] [Google Scholar]
- 16. Nakamura Y., Gojobori T., Ikemura T.. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000; 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hilterbrand A., Saelens J., Putonti C.. CBDB: the codon bias database. BMC Bioinformatics. 2012; 13:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Athey J., Alexaki A., Osipova E., Rostovtsev A., Santana-Quintero L.V., Katneni U., Simonyan V., Kimchi-Sarfaty C.. A new and updated resource for codon usage tables. BMC Bioinformatics. 2017; 18:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meiler A., Klinger C., Kaufmann M.. ANCAC: amino acid, nucleotide, and codon analysis of COGs–a tool for sequence bias analysis in microbial orthologs. BMC Bioinformatics. 2012; 13:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zaneveld J., Hamady M., Sueoka N., Knight R.. CodonExplorer: an interactive online database for the analysis of codon usage and sequence composition. Methods Mol. Biol. 2009; 537:207–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hodgman M.W., Miller J.B., Meurs T.E., Kauwe J.S.K.. CUBAP: an interactive web portal for analyzing codon usage biases across populations. Nucleic Acids Res. 2020; 48:11030–11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurotani A., Yamada Y., Shinozaki K., Kuroda Y., Sakurai T.. Plant-PrAS: a database of physicochemical and structural properties and novel functional regions in plant proteomes. Plant Cell Physiol. 2015; 56:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arun P.V., Bakku R.K., Subhashini M., Singh P., Prabhu N.P., Suzuki I., Prakash J.S.. CyanoPhyChe: a database for physico-chemical properties, structure and biochemical pathway information of cyanobacterial proteins. PLoS One. 2012; 7:e49425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meszaros B., Erdos G., Szabo B., Schad E., Tantos A., Abukhairan R., Horvath T., Murvai N., Kovacs O.P., Kovacs M.et al.. PhaSePro: the database of proteins driving liquid-liquid phase separation. Nucleic Acids Res. 2020; 48:D360–D367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Q., Peng X., Li Y., Tang W., Zhu J., Huang J., Qi Y., Zhang Z.. LLPSDB: a database of proteins undergoing liquid-liquid phase separation in vitro. Nucleic Acids Res. 2020; 48:D320–D327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ning W., Guo Y., Lin S., Mei B., Wu Y., Jiang P., Tan X., Zhang W., Chen G., Peng D.et al.. DrLLPS: a data resource of liquid-liquid phase separation in eukaryotes. Nucleic Acids Res. 2020; 48:D288–D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D.et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T.et al.. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gene Ontology, C. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021; 49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., McVeigh R., O’Neill K., Robbertse B.et al.. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020; 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Z., Li J., Cui P., Ding F., Li A., Townsend J.P., Yu J.. Codon Deviation Coefficient: a novel measure for estimating codon usage bias and its statistical significance. BMC Bioinformatics. 2012; 13:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang G., Yin H., Li B., Yu C., Wang F., Xu X., Cao J., Bao Y., Wang L., Abbasi A.A.et al.. Characterization and identification of long non-coding RNAs based on feature relationship. Bioinformatics. 2019; 35:2949–2956. [DOI] [PubMed] [Google Scholar]
- 34. Kang Y.J., Yang D.C., Kong L., Hou M., Meng Y.Q., Wei L., Gao G.. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017; 45:W12–W16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walsh I., Martin A.J., Di Domenico T., Tosatto S.C.. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012; 28:503–509. [DOI] [PubMed] [Google Scholar]
- 36. Lancaster A.K., Nutter-Upham A., Lindquist S., King O.D.. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014; 30:2501–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vernon R.M., Chong P.A., Tsang B., Kim T.H., Bah A., Farber P., Lin H., Forman-Kay J.D.. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife. 2018; 7:e31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Z., Yu J.. Modeling compositional dynamics based on GC and purine contents of protein-coding sequences. Biol. Direct. 2010; 5:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu J. A content-centric organization of the genetic code. Genomics Proteomics Bioinformatics. 2007; 5:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu J., Zhao X., Zhang Z., Yu J.. Compositional dynamics of guanine and cytosine content in prokaryotic genomes. Res. Microbiol. 2007; 158:363–370. [DOI] [PubMed] [Google Scholar]
- 41. Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990; 87:23–29. [DOI] [PubMed] [Google Scholar]
- 42. Reichenberger E.R., Rosen G., Hershberg U., Hershberg R.. Prokaryotic nucleotide composition is shaped by both phylogeny and the environment. Genome Biol Evol. 2015; 7:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Supek F., Skunca N., Repar J., Vlahovicek K., Smuc T.. Translational selection is ubiquitous in prokaryotes. PLos Genet. 2010; 6:e1001004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin K., Kuang Y., Joseph J.S., Kolatkar P.R.. Conserved codon composition of ribosomal protein coding genes in Escherichia coli, Mycobacterium tuberculosis and Saccharomyces cerevisiae: lessons from supervised machine learning in functional genomics. Nucleic Acids Res. 2002; 30:2599–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen M., Ma Y., Wu S., Zheng X., Kang H., Sang J., Xu X., Hao L., Li Z., Gong Z.et al.. Genome warehouse: a public repository housing genome-scale data. Genomics Proteomics Bioinformatics. 2021; 10.1016/j.gpb.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Res. 2021; 49:D18–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CompoDynamics is a user-friendly database for characterizing sequence composition dynamics and can be accessed directly at https://ngdc.cncb.ac.cn/compodynamics/.