Abstract
Insects are the largest group of animals on the planet and have a huge impact on human life by providing resources, transmitting diseases, and damaging agricultural crop production. Recently, a large amount of insect genome and gene data has been generated. A comprehensive database is highly desirable for managing, sharing, and mining these resources. Here, we present an updated database, InsectBase 2.0 (http://v2.insect-genome.com/), covering 815 insect genomes, 25 805 transcriptomes and >16 million genes, including 15 045 111 coding sequences, 3 436 022 3′UTRs, 4 345 664 5′UTRs, 112 162 miRNAs and 1 293 430 lncRNAs. In addition, we used an in-house standard pipeline to annotate 1 434 653 genes belonging to 164 gene families; 215 986 potential horizontally transferred genes; and 419 KEGG pathways. Web services such as BLAST, JBrowse2 and Synteny Viewer are provided for searching and visualization. InsectBase 2.0 serves as a valuable platform for entomologists and researchers in the related communities of animal evolution and invertebrate comparative genomics.
INTRODUCTION
Insects represent one of the largest and most diverse group of animals on earth and play important roles in ecological stability (1), agriculture (2), the economy (3) and human health (4). With rapid technological developments, a sea of insect gene data has been generated, including genomes, transcriptomes, proteomes, metabolomes and chromatin interaction information detected by the Hi-C method (5,6). Although most of these data are available in public databases such as National Center for Biotechnology Information (NCBI) (7), many are not well organized, and some are available only as raw data without annotation information. This hampers the full use of these insect gene resources. Several databases have been constructed to provide well-curated annotations and well-designed data organization in entomological field, including i5k Workspace@NAL (8), Bioinformatics Platform for Agroecosystem Arthropods (BIPAA) (https://bipaa.genouest.org/is/), VectorBase (9), FlyBase (10), LepBase (http://lepbase.org/), Hymenoptera Genome Database (11), Butterfly Genome Database (12), FireflyBase (13), SilkDB (14), KAIKObase (15), KONAGAbase (16), MonarchBase (17), LocustBase (18), BeetleBase (19), etc. Most of these databases focus on only one species or a group of closely related species, and few provide a well-designed and user-friendly platform for curating, visualizing, and sharing insect gene data. To fill this gap, we built InsectBase in 2016, which collected almost all insect genome data available at that time (20).
Due to the emergence of third-generation sequencing technology, the quantity and quality of insect gene and genome data have greatly increased in recent years. Therefore, to provide a revised and more convenient platform, we have updated InsectBase to version 2.0 with three significant improvements: (i) The quantity and quality of insect gene data are significantly increased. In total, InsectBase 2.0 contains >16 million sequences from 815 species with 207 chromosome-level genomes and 134 full-length transcriptomes. (ii) Multi-level gene and genome data are now provided, including RNA–RNA interactions, gene families, KEGG pathways and HGT genes (21). (iii) The user interface features have been enhanced to improve the web server.
MATERIALS AND METHODS
Data source
We collected insect gene and genome data from several databases (as described below) and developed standardized pipelines for annotation and identification of UTRs, miRNAs, lncRNAs, RNA–RNA interactions, gene families, KEGG pathways and genes likely derived from horizontal gene transfer (referred to as ‘potential HGT genes’).
Genome
We collected and downloaded 815 genomes from NCBI (7), BIPAA (https://bipaa.genouest.org/is/), GigaDB (22), i5k Workspace@NAL (8), InsectBase (20), LepBase (http://lepbase.org/), VectorBase (9), National Genomics Data Center (NGDC) (23), FireflyBase (13), DNA Data Bank of Japan (DDBJ) (24), SilkDB 3.0 (14), Assembled Searchable Giant Arthropod Read Database (ASGARD) (25), DNA Zoo (26), LocustBase (18), DRYAD (https://datadryad.org/stash) and Zenodo (https://zenodo.org/) (Supplementary Tables S1 and S2). Among these, 231 insect genomes were obtained with known annotated official gene sets. A further 482 genomes were annotated using our in-house genome annotation pipeline. First, we identified and masked the repeat sequences by RepeatModeler2 (v.2.0.1) (27) and RepeatMasker (http://www.repeatmasker.org) (v.4.0.7) with both de novo and homology-based methods. Next, three evidences of gene annotation were generated. BRAKER2 (v.2.1.5) (28–34) was used to generate the de novo gene models. HISAT2 (v.2.1.0) (35) and StringTie2 (v.2.1.5) (36) were used for transcripts assembling. And homology-based evidence was generated by GenomeThreader (v.1.7.1) (37). Finally, we integrated three types of evidences by EVidenceModeler (v.1.1.1) (38) to obtain the official gene sets (OGS).
Transcriptome
25 805 transcriptomes of 439 species were downloaded from the NCBI SRA database (Supplementary Table S3) (7). The raw reads were pre-processed using fastp (v.0.21) (39) and mapped to reference genomes with HISAT2 (v.2.1.0) (35). StringTie2 (v.2.1.4) (36) was used for transcript assembly.
ncRNA
1674 small RNA libraries of 60 species were download from the NCBI SRA database (Supplementary Table S4) (7). miRNAs were predicted by miRDeep2 (v.0.1.3) (40) and MapMi (v.1.5.0) (41). TargetScan 70 (42), RNAhybrid (v.2.1.2) (43) and miRanda (v.3.3a) (44) were used for miRNA target prediction. LncRNAs and partner genes were predicted with FEELnc (v.0.2) (45) using the default parameters.
Gene family, KEGG pathway and potential HGT gene
One hundred and sixty-four gene families were annotated by BLASTP against the Swiss-Prot protein database using DIAMOND (v.2.0.0.138) (31,46). For KEGG pathway, the reference KOs of each gene were identified by BLASTP against the KEGG database, and the KEGG pathway genes were obtained by extracting the KO information of each gene (21). Potential HGT genes were filtered by using insect genes to blast against the NCBI non-redundant protein (nr)/nucleotide (nt) database, if at least 15 of the best 20 BLAST hits are from non-insect species, we treated these genes as potential HGT genes (7). It should be noted that this pre-filtering method might have high false positive and further analysis of these genes should consider this.
Insect virus
Genome information of 1524 insect viruses was obtained and organized from the NCBI genome database (7).
Implementation of database
InsectBase 2.0 runs on a nginx (v.1.16.1) web server (http://nginx.org/) based on the CentOS 7.4.1708 platform with a MySQL (v.5.7.17) database (https://dev.mysql.com/). Django (v.3.1.3) framework (https://www.djangoproject.com/) and Vue (v.3.0) JavaScript framework (https://v3.vuejs.org/) were used for the web construction. JBrowse2 (47), the platform for visualizing and integrating biological data, was used for genome visualization. DIAMOND (v.2.0.0.138) (31), NCBI BLAST (v.2.11.0+) and BLAT (v.36) (48) were installed for sequence alignment of genes, proteins, miRNAs and lncRNAs. SynVisio (49) was hosted for visualization of genome synteny files constructed by MCScanX (50).
UPDATES IN INSECTBASE 2.0
More insect gene data with high assembly quality and standard annotations
Recent advances in third-generation sequencing techniques and chromosome conformation capture (3C) methods have provided a valuable platform for generation of high-quality genomes and full-length transcriptomes (5,6). In InsectBase 2.0, we collected 815 genomes from 457 genera and 25 805 well-assembled transcriptomes from 439 species. Among these, 207 genomes were assembled at the chromosome level and 134 full-length transcriptomes from 31 species were generated by nanopore sequencing.
Using an in-house pipeline, we annotated 482 insect genomes, yielding standard official gene sets for these species. In total, we generated 15 045 111 coding sequences of 713 insects, 112 162 miRNAs from 807 insects, 1 293 430 lncRNAs representing 376 insects, 419 KEGG pathways, 7 781 686 UTRs in 374 insects and 164 gene families in 713 insects. Overall, this represents a substantial increase in insect gene and genome data from InsectBase 1.0 (Table 1).
Table 1.
Data summary of InsectBase 1.0 and 2.0
| Feature | Units | v1.0 | v2.0 | Fold Increase |
|---|---|---|---|---|
| Genomes | Species | 138 | 815 | 5.9 |
| Transcriptomes | Runs | 116 | 25 805 | 222.4 |
| Coding sequences | Transcripts | 160 905 | 15 045 111 | 93.5 |
| UTRs | - | 678 881 | 7 781 686 | 11.4 |
| miRNAs | - | 7544 | 112 162 | 14.9 |
| lncRNAs | - | 2439 | 1 293 430 | 530.3 |
| Pathways | - | 78 | 419 | 5.4 |
| Gene families | - | 54 | 164 | 3.0 |
| HGT genes | - | - | 215 986 | New |
| Insect viruses | - | - | 1524 | New |
| miRNA–mRNA interactions | - | - | 197 533 | New |
| lncRNA–mRNA interactions | - | - | 5 147 543 | New |
ncRNAs, HGT genes and insect viruses
ncRNAs participate in many important biological processes by interacting with RNAs either directly or indirectly through protein intermediates (51). Here, we predicted 197 533 miRNA–mRNA interactions and identified 1 293 737 lncRNA partner genes. HGT is a key evolutionary force which has constantly reshaped genomes throughout evolution (52). We identified 215 986 potential HGT genes from five kingdoms (Bacteria, Fungi, Metazoa [excluding insecta], Viridiplantae and Virus; Table 1). We also collected 1524 insect viruses which are important pathogens of many arthropod species and are potential microbial control agents. These data will benefit researches in the fields of gene networks, evolution and comparative analysis.
Enhanced user interface features
InsectBase 2.0 contains 12 modules, namely ‘organism’, ‘chromosome’, ‘genome’, ‘transcriptome’, ‘gene’, ‘gene family’, ‘HGT gene’, ‘KEGG pathway’, ‘insect virus’, ‘tools’, ‘links’ and ‘service’ (for searching, browsing, and downloading) (Figure 1).
Figure 1.
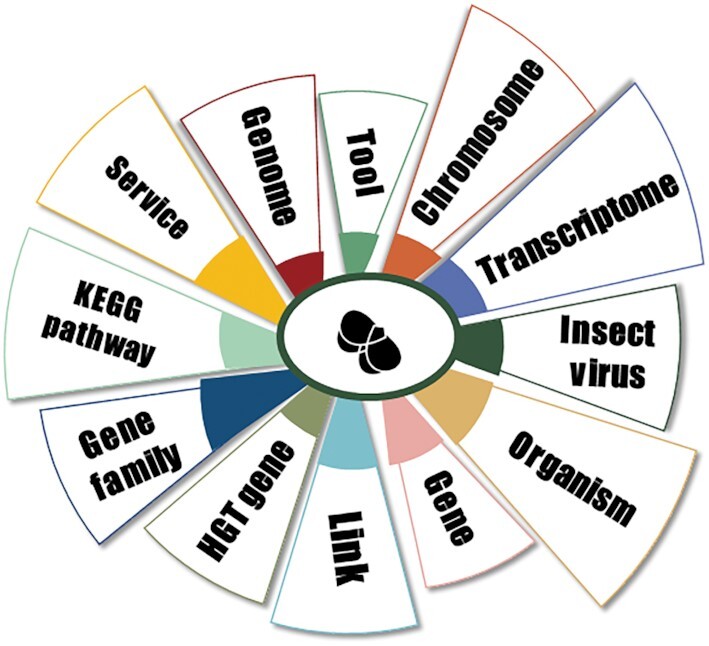
Main modules of InsectBase 2.0. It provides information about an organism, genome, transcriptome, chromosome, gene information about protein coding genes, miRNA and lncRNA, gene family, HGT genes, insect pathways, insect viruses, online tools, links and additional services.
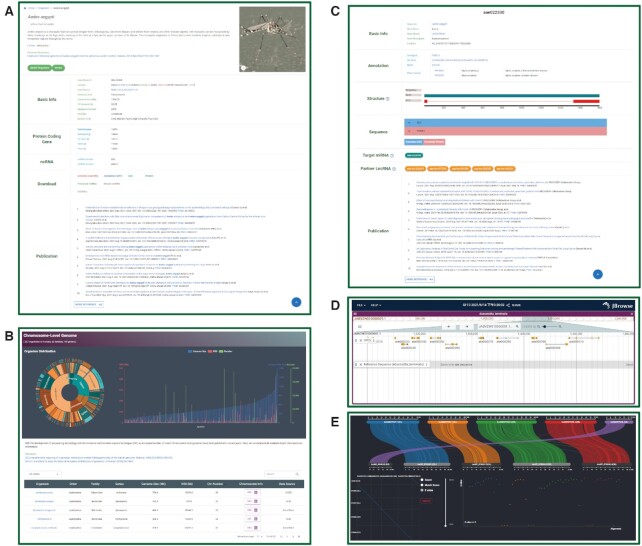
The ‘organism’ module shows a species tree modified from the NCBI Taxonomy common tree (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi) (7). Each order, family, genus, and species are introduced with pictures from public sources such as Wikipedia (https://www.wikipedia.org/) and iNaturalist (https://www.inaturalist.org/). Users can click on the species name to access the species page, which shows information about multiple aspects of the selected species. This includes a basic introduction, genome statistics, gene information, and related publications in PubMed (https://pubmed.ncbi.nlm.nih.gov/) (Figure 2A).
Figure 2.
Enhanced user interface features of InsectBase 2.0. (A) Species: basic information, file downloading and publications related to each species. (B) Chromosome: information for each chromosome. (C) Protein coding gene: detailed information about each protein coding gene. (D) JBrowse2: genome browser of each annotated genome. (E) Genome synteny: visualization of synteny of 155 chromosome-level genomes.
The advent of high-quality genome has greatly advanced the study of entomology. To aid in investigate of chromosome evolution, the ‘chromosome’ module displays 207 genomes with information at the chromosome level (Figure 2B). Chromosomes in 155 genomes are displayed for browsing and downloading.
Transcriptomes are an essential data resource for understanding biological processes under different conditions. The ‘transcriptome’ module contains 25 805 assembled transcriptomes with sample information, including species, gender, tissue, stage, and condition to help researchers conduct genetic investigations with different conditions or treatments.
The ‘gene information’ module allows the user to conduct an advanced search for protein coding genes, miRNAs, and lncRNAs by species, gene name, and gene description. Beyond the basic information of selected gene, gene structure, gene sequence and gene interactions such as mRNA–miRNA and mRNA–lncRNA interactions are displayed. By clicking on the interacting genes, users can access the related gene page (Figure 2C).
Gene families often exhibit apparent expansion or contraction in terms of gene numbers or structures. Gene family analysis is not only essential for uncovering gene functions, but also frequently used in revealing the evolutional mechanism of gene gain and loss. Hence, InsectBase 2.0 analysed 164 gene families by annotating them with an in-house pipeline. The ‘gene family’ module allows users to easily search and download gene families of interest in a given species. In addition to conventional tools such as DIAMOND (31), BLAT (48) and BLAST, we constructed a comprehensive genome browser with all annotated genomes by JBrowse2 (47) (Figure 2D). Moreover, InsectBase 2.0 provides a genome synteny visualization tool. Genome synteny between 155 chromosome-level genomes is visualized for chromosome evolution analysis (Figure 2E).
DISCUSSION AND FUTURE DEVELOPMENT
At present, insect genome and gene data are stored in multiple databases once they are generated (53). InsectBase 2.0 uses standard pipelines to predict protein coding genes, miRNAs, lncRNAs and UTRs, promoting standardisation of comparative genomics. In addition, gene families, KEGG pathways and genes potentially involved in many crucial biological processes (such as pesticide detoxification metabolism and host-seeking) are annotated. In summary, InsectBase 2.0 is a substantially improved database for insect gene resources and serves as a valuable resource to meet the needs of entomologists and the related research communities of animal evolution and invertebrate comparative genomics.
We will continue to add newly-available data and new features. For example, the three-dimensional (3D) organization of genomes plays an essential role in gene regulation. With the development of the 3C technique, such as Hi-C, ChIA-PET, Capture-C and Capture Hi-C, chromosome interaction information has provided an unprecedented opportunity to study spatial organization in a genome-wide fashion (54). We plan to analyse these data and add associated features in the next update. The recently-developed AlphaFold2 (55) predicts protein structure with high accuracy, which would be greatly valuable in investigating protein-protein binding, enzyme active sites, and the functional implications of genetic mutations. We thus plan to integrate this tool in the next update.
DATA AVAILABILITY
All data in InsectBase 2.0 are available for downloading. The database can be accessed at http://v2.insect-genome.com/. The genome annotation pipeline is available at https://github.com/meiyang12/Genome-annotation-pipeline.
Supplementary Material
ACKNOWLEDGEMENTS
We appreciate all researchers sharing public resources in the insect community, and constructing and organizing the databases. We kindly thank Wikipedia, iNaturalist (https://www.inaturalist.org/), the British Natural History Museum (https://www.nhm.ac.uk/), BOLDSYSTEMS (http://www.boldsystems.org/) and EHIME-Fly (https://kyotofly.kit.jp/cgi-bin/ehime/index.cgi) for publicly-available information and images.
Contributor Information
Yang Mei, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Dong Jing, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Shenyang Tang, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Xi Chen, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Hao Chen, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Haonan Duanmu, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Yuyang Cong, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Mengyao Chen, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Xinhai Ye, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Hang Zhou, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Kang He, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
Fei Li, State Key Laboratory of Rice Biology & Ministry of Agricultural and Rural Affairs Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, Zhejiang University, Hangzhou 310058, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National High Technology Research and Development Program of China [2019YFD1002100]; National Science Foundation of China [31972354]; National Science & Technology Fundamental Resources Investigation Program of China [2019FY100400]; Zhejiang National Science Foundation of China [LZ18C060001]; Fundamental Research Funds for the Central Universities [2020QNA6024]. Funding for open access charge: National High Technology Research and Development Program of China [2019YFD1002100]; National Science Foundation of China [31972354]; National Science & Technology Fundamental Resources Investigation Program of China [2019FY100400]; Zhejiang National Science Foundation of China [LZ18C060001]; The Fundamental Research Funds for the Central Universities [2020QNA6024].
Conflict of interest statement. None declared.
REFERENCES
- 1. Losey J.E., Vaughan M.. The economic value of ecological services provided by insects. Bioscience. 2006; 56:311–323. [Google Scholar]
- 2. Meier R., Lim G.S.. Conflict, convergent evolution, and the relative importance of immature and adult characters in endopterygote phylogenetics. Annu. Rev. Entomol. 2009; 54:85–104. [DOI] [PubMed] [Google Scholar]
- 3. Robinson G.E., Hackett K.J., Purcell-Miramontes M., Brown S.J., Evans J.D., Goldsmith M.R., Lawson D., Okamuro J., Robertson H.M., Schneider D.J.. Creating a buzz about insect genomes. Science. 2011; 331:1386. [DOI] [PubMed] [Google Scholar]
- 4. Lounibos L.P. Invasions by insect vectors of human disease. Annu. Rev. Entomol. 2002; 47:233–266. [DOI] [PubMed] [Google Scholar]
- 5. Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O.et al.. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009; 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Berkum N.L., Lieberman-Aiden E., Williams L., Imakaev M., Gnirke A., Mirny L.A., Dekker J., Lander E.S.. Hi-C: a method to study the three-dimensional architecture of genomes. J. Vis. Exp. 2010; 39:1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sayers E.W., Beck J., Bolton E.E., Bourexis D., Brister J.R., Canese K., Comeau D.C., Funk K., Kim S., Klimke W.et al.. Database resources of the National Center for Biotechnology Information. Nucleic. Acids. Res. 2021; 49:D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poelchau M., Childers C., Moore G., Tsavatapalli V., Evans J., Lee C.Y., Lin H., Lin J.W., Hackett K.. The i5k Workspace@NAL–enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Res. 2015; 43:D714–D719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giraldo-Calderón G.I., Emrich S.J., MacCallum R.M., Maslen G., Dialynas E., Topalis P., Ho N., Gesing S., VectorBase Consortium, Madey G.et al.. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015; 43:D707–D713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Larkin A., Marygold S.J., Antonazzo G., Attrill H., Dos-Santos G., Garapati P.V., Goodman J.L., Gramates L.S., Millburn G., Strelets V.B.et al.. FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021; 49:D899–D907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elsik C.G., Tayal A., Diesh C.M., Unni D.R., Emery M.L., Nguyen H.N., Hagen D.E.. Hymenoptera Genome Database: integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 2016; 44:D793–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davey J.W., Chouteau M., Barker S.L., Maroja L., Baxter S.W., Simpson F., Merrill R.M., Joron M., Mallet J., Dasmahapatra K.K.et al.. Major improvements to the Heliconiusmelpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3 (Bethesda). 2016; 6:695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fallon T.R., Lower S.E., Chang C.H., Bessho-Uehara M., Martin G.J., Bewick A.J., Behringer M., Debat H.J., Wong I., Day J.C.et al.. Firefly genomes illuminate parallel origins of bioluminescence in beetles. eLife. 2018; 7:e36495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu F., Wei Z., Luo Y., Guo H., Zhang G., Xia Q., Wang Y.. SilkDB 3.0: visualizing and exploring multiple levels of data for silkworm. Nucleic Acids Res. 2019; 48:D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang C., Yokoi K., Yamamoto K., Jouraku A.. An update of KAIKObase, the silkworm genome database. Database (Oxford). 2021; 2021:baaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jouraku A., Yamamoto K., Kuwazaki S., Urio M., Suetsugu Y., Narukawa J., Miyamoto K., Kurita K., Kanamori H., Katayose Y.et al.. KONAGAbase: a genomic and transcriptomic database for the diamondback moth, Plutellaxylostella. BMC Genomics. 2013; 14:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhan S., Reppert S.M.. MonarchBase: the monarch butterfly genome database. Nucleic Acids Res. 2013; 41:D758–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X., Fang X., Yang P., Jiang X., Jiang F., Zhao D., Li B., Cui F., Wei J., Ma C.et al.. The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 2014; 5:2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim H.S., Murphy T., Xia J., Caragea D., Park Y., Beeman R.W., Lorenzen M.D., Butcher S., Manak J.R., Brown S.J.. BeetleBase in 2010: revisions to provide comprehensive genomic information for Triboliumcastaneum. Nucleic Acids Res. 2010; 38:D437–D442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yin C., Shen G., Guo D., Wang S., Ma X., Xiao H., Liu J., Zhang Z., Liu Y., Zhang Y.et al.. InsectBase: a resource for insect genomes and transcriptomes. Nucleic Acids Res. 2016; 44:D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiao S.Z., Armit C., Edmunds S., Goodman L., Li P., Tuli M.A., Hunter C.I.. Increased interactivity and improvements to the GigaScience database, GigaDB. Database (Oxford). 2019; 2019:baz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. National Genomics Data Center Members and Partners. Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 2020; 48:D24–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fukuda A., Kodama Y., Mashima J., Fujisawa T., Ogasawara O.. DDBJ update: streamlining submission and access of human data. Nucleic Acids Res. 2021; 49:D71–D75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zeng V., Extavour C.G.. ASGARD: an open-access database of annotated transcriptomes for emerging model arthropod species. Database (Oxford). 2012; 2012:bas048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dudchenko O., Batra S.S., Omer A.D., Nyquist S.K., Hoeger M., Durand N.C., Shamim M.S., Machol I., Lander E.S., Aiden A.P.et al.. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017; 356:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Flynn J.M., Hubley R., Goubert C., Rosen J., Clark A.G., Feschotte C., Smit A.F.. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brůna T., Hoff K.J., Lomsadze A., Stanke M., Borodovsky M.. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinform. 2021; 3:lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoff K.J., Lomsadze A., Borodovsky M., Stanke M.. Whole-genome annotation with BRAKER. Methods Mol. Biol. 2019; 1962:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brůna T., Lomsadze A., Borodovsky M.. GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genom. Bioinform. 2020; 2:lqaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 32. Gotoh O. A space-efficient and accurate method for mapping and aligning cDNA sequences onto genomic sequence. Nucleic Acids Res. 2008; 36:2630–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iwata H., Gotoh O.. Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features. Nucleic Acids Res. 2012; 40:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stanke M., Diekhans M., Baertsch R., Haussler D.. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008; 24:637–644. [DOI] [PubMed] [Google Scholar]
- 35. Zhang Y., Park C., Bennett C., Thornton M., Kim D.. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 2021; 31:1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kovaka S., Zimin A.V., Pertea G.M., Razaghi R., Salzberg S.L., Pertea M.. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019; 20:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gremme G., Brendel V., Sparks M.E., Kurtz S.. Engineering a software tool for gene structure prediction in higher organisms. Inf. Softw. Technol. 2005; 47:965–978. [Google Scholar]
- 38. Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., White O., Buell C.R., Wortman J.R.. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008; 9:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen S., Zhou Y., Chen Y., Gu J.. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Friedländer M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N.. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012; 40:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guerra-Assunção J.A., Enright A.J.. MapMi: automated mapping of microRNA loci. BMC Bioinformatics. 2010; 11:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B.. Prediction of mammalian microRNA targets. Cell. 2003; 115:787–798. [DOI] [PubMed] [Google Scholar]
- 43. Krüger J., Rehmsmeier M.. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006; 34:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S.. MicroRNA targets in Drosophila. Genome Biol. 2003; 5:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wucher V., Legeai F., Hédan B., Rizk G., Lagoutte L., Leeb T., Jagannathan V., Cadieu E., David A., Lohi H.et al.. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017; 45:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L.et al.. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bandi V., Gutwin C.. Interactive exploration of genomic conservation. Proc. Graph. Interface. 2020; 74–83. [Google Scholar]
- 50. Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T., Jin H., Marler B., Guo H.et al.. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012; 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Engreitz J.M., Sirokman K., McDonel P., Shishkin A., Surka C., Russell P., Grossman S.R., Chow A.Y., Guttman M., Lander E.S.. RNA-RNA Interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 2014; 159:188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Husnik F., McCutcheon J.P.. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 2018; 16:67–79. [DOI] [PubMed] [Google Scholar]
- 53. Li F., Zhao X., Li M., He K., Huang C., Zhou Y., Li Z., Walters J.R.. Insect genomes: progress and challenges. Insect Mol. Biol. 2019; 28:739–758. [DOI] [PubMed] [Google Scholar]
- 54. Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N.K., Li Y., Hu M.et al.. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018; 19:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., Qin C., Žídek A., Nelson A.W.R., Bridgland A.et al.. Improved protein structure prediction using potentials from deep learning. Nature. 2020; 577:706–710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in InsectBase 2.0 are available for downloading. The database can be accessed at http://v2.insect-genome.com/. The genome annotation pipeline is available at https://github.com/meiyang12/Genome-annotation-pipeline.