Abstract
The United States has a complex regulatory scheme for marketing drugs. Understanding drug regulatory status is a daunting task that requires integrating data from many sources from the United States Food and Drug Administration (FDA), US government publications, and other processes related to drug development. At NCATS, we created Inxight Drugs (https://drugs.ncats.io), a web resource that attempts to address this challenge in a systematic manner. NCATS Inxight Drugs incorporates and unifies a wealth of data, including those supplied by the FDA and from independent public sources. The database offers a substantial amount of manually curated literature data unavailable from other sources. Currently, the database contains 125 036 product ingredients, including 2566 US approved drugs, 6242 marketed drugs, and 9684 investigational drugs. All substances are rigorously defined according to the ISO 11238 standard to comply with existing regulatory standards for unique drug substance identification. A special emphasis was placed on capturing manually curated and referenced data on treatment modalities and semantic relationships between substances. A supplementary resource ‘Novel FDA Drug Approvals’ features regulatory details of newly approved FDA drugs. The database is regularly updated using NCATS Stitcher data integration tool that automates data aggregation and supports full data access through a RESTful API.
INTRODUCTION
The development of new drugs is slow and very expensive. The cost of bringing a single new drug to the market approached a staggering $2.87 billion in 2016, and it keeps climbing (1). However, we do not always need entirely new drugs to address current unmet needs in medicine. The mobilization of tocilizumab (2), dexamethasone (3) and remdesivir (4) for the treatment of COVID-19 reinforced the importance of drug repurposing in translational research. Understanding the regulatory status of these different medicines and how they could be used in new ways was a critical aspect of our pandemic response (5). In fact, the current available pharmacopeia represents an invaluable resource for inspiring the next generation of drug products. In the words of Nobel prize winning chemist Sir James Black, ‘the most fruitful basis for the discovery of a new drug is to start with an old drug’ (6).
A slow upwards trend in new drug approval rates has been observed since 2012, largely due to increased rates of biologics approvals, while the field of synthetic small molecules has begun to show modest signs of improvement (7). Yet, this seemingly low rate for the conversion of small molecules into drugs may be deceiving. First, the highest attrition rate is observed in Phase II (8,9). The development of nearly a quarter of all US-approved drugs on the market was facilitated by serendipity, and for anti-cancer drugs this figure reaches a staggering 35% (10). However, serendipitous discoveries are predicted to become much less likely in the future, in part due to substantial changes in medical practice (11), which may explain the decreasing approval rates and rising costs. Moreover, such a prominent contribution of chance to the drug discovery process is highly suggestive of a substantial unmet need for systematization and overhaul of the existing foundation of the field. As a result of the large number of regulatory agencies worldwide (12), authoritative information related to drug approval and marketing are scattered across numerous different kinds of reports and resources. Therefore, the only way to obtain a single comprehensive list of all marketed substances would be to unite all aforementioned resources. However, such unification is challenging due to the lack of a commonly used universal identifier.
Public access to reliable information has been a long-standing issue in the field of drug development. Manually curated commercial resources can provide high-quality data on the intricate relationships between drugs, targets, and diseases, but these resources are expensive and impose an artificial barrier to translational research by the academic community. Much of the raw data on drug development and regulation is available from the public record, but one has to know where to look and how to interpret the data.
We envisioned a public informatics platform, built on a reliable interconnected standardized set of substances, incorporating data from multiple public resources, including regulatory agencies, and offering manually sourced annotations, providing the most comprehensive and authoritative dataset on drug development. It was clear that we would have to be semantically precise and systematic in our approach. For instance, a simple question such as ‘how many drugs are there?’ is in fact quite challenging and to date, there is a dissonant chorus of competing answers. Researchers still must resort to referencing an authority to indicate a specific number (10). Yet, a standard does exist. The International Organization for Standardization (ISO) has established a scheme for the identification of medicinal products and the ISO 11238 data standard, which precisely defines a ‘substance’, fundamentally a drug product ingredient. Working closely with international regulators, we have implemented the ISO 11238 standard to produce a definitive, regulatory-grade resource for drug substances, the Global Substance Registration System (G-SRS) (13).
Inxight Drugs builds atop this G-SRS dataset to layer on other information about regulatory status, disease biology, and practical use for drug substances. This information is collected and organized using a graph-based data integration tool, NCATS Stitcher (https://github.com/ncats/stitcher), that deterministically incorporates source updates with manual data curations. To date, Inxight Drugs incorporates the most comprehensive subset of substances and related biological mechanisms, targets, and pathways pertaining to translational research, and connects them to the appropriate disease indications and clinical trials. The database offers rigorous structural definition of ingredients in medicinal products, definitive information on the US marketing status, and manually curated data for over 25,000 substances, including succinct summaries, in vitro and in vivo activity, targets, related conditions, pharmacokinetic properties, adverse events, and sourcing.
Inxight Drugs thus enables researchers to access and understand drugs and their roles within the vast, interconnected graph of biomedical entities.
MATERIALS AND METHODS
Data sources
DailyMed
The DailyMed database (National Library of Medicine, National Institutes of Health, Department of Health & Human Services) is an excellent source for labeling information submitted to the FDA for the FDA-approved drug products that include both small molecules and biologics. As of 16 July 2021, a total of 139 782 drug labels were submitted to the FDA by different companies. The database provides rich prescription information for drug products that includes indications, ingredients, dosage and administration, contraindications, boxed warnings and precautions, adverse reactions, drug interactions and more. We extracted 161 684 entries comprising 110 484 drug products and 5441 unique ingredients.
Drugs@FDA and orange book
Drugs@FDA (https://www.fda.gov/drugsatfda) contains information about products approved by the FDA for human use in the United States, including prescription products and the over-the-counter drugs. The following information is typically available for a product: drug name(s), active ingredient(s), dosage form(s), route(s) of administration, strength(s), the latest FDA-approved labeling and previously approved labeling, and regulatory information. Drugs@FDA is updated every day with new information about approved products. The related Orange Book (https://www.fda.gov/orangebook) identifies which drug products are therapeutically equivalent. Although the Orange Book shares a lot of content with Drugs@FDA, the former resource provides information on patents and exclusivity that are not available from the latter. A total of 3874 drug substances were extracted from these two sources. While Drugs@FDA contains data about modern drug approvals, it can sometimes lack historical data about older drugs and products regulated by the Center for Biologics Evaluation and Research. Further, it references drug substances only by name and does not provide structural data or any other unique identifiers.
DrugBank
DrugBank (14) is a richly annotated online drug database from OMx Personal Health Analytics Inc. that combines detailed drug data with comprehensive drug target and drug action information. Since its first release in 2006 (15), DrugBank was updated several times, each time adding useful information to facilitate in silico drug target discovery, drug design, drug docking or screening, drug metabolism prediction, drug interaction prediction and more. DrugBank aggregates data about drugs from Orange Book as well as drug listings from Health Canada and EMA in addition to data on research tools and products in development. However, it does not rely on a strict definition of a substance and thus does not provide a complete coverage of all marketed substances. The current version of DrugBank comprises a total of 11 898 small molecule and 2651 biotech drug product ingredients. In total, we extracted 13 580 entries from DrugBank.
NCATS pharmaceutical collection
The National Center for Advancing Translational Sciences (NCATS) Pharmaceutical Collection (NPC) is a comprehensive collection of clinically approved drugs that was originally made public in 2011 as the NCGC Pharmaceutical Collection (16). The resource provides a definitive, complete, and nonredundant list of all approved molecular entities both as a freely available electronic resource and a physical collection of small molecules amenable to high-throughput screening. Over the past decade, the NPC has been systematically profiled for activity across an array of pathways and disease models, generating an unparalleled amount of data (17). The NPC contains 14 814 approved and investigational drugs.
Global substance registration system
The FDA and NCATS have collaborated to publish rigorous scientific descriptions of substances relevant to regulated products and populated a database to organize the agency's regulatory submissions and marketed products data. The NCATS has worked with FDA to develop the Global Substance Registration System (G-SRS) (13) and produce a non-proprietary version of the database for public benefit. G-SRS provides a manually curated dataset of ingredients in medicinal products and their scientific definitions for regulatory and translational research. It is also the first database to provide ingredient definitions using the global ISO 11238 data standard adopted by the FDA for the identification of substances in medicinal products. G-SRS efficiently handles substances other than small molecules and its curation process. G-SRS is a major contributor to Inxight Drugs with a core collection of 125 036 substances.
Additional data sources
Apart from the above data, several other data sources were accessed to obtain additional substances and associated data. Data on Over-The-Counter (OTC) products were extracted from the OTC Monographs in the Code of Federal Regulations (CFR) and Federal Register notices of Drug Efficacy Study Implementation (21 CFR parts 310, 356, etc.). Veterinary drugs approved in US (NADA and ANADAs) were extracted from Animal Drugs@FDA and 21 CFR (part516:E, part520, part522, part524, part526, part528, part529, part558:B, part573, part582:B,C,D,E,F,G,H, part584). Any component of a drug product other than the active ingredient is referred to as inactive ingredient as per 21 CFR 210.3(b)(7). The FDA Inactive Ingredient Guide was accessed to locate inactive ingredients present in drug products. Clinical trial data were extracted from PubMed and ClinicalTrials.gov. Additional details on US drug product marketing and FDA regulatory decisions, especially product approvals prior to 1981 were abstracted from multiple independent data sources (see Supplementary Information, S1).
Manual data curation
As of the publication date, 25 445 of approved and marketed drugs and drug candidates have been manually annotated. These annotations span a wide range of properties, including summary descriptions (14 770 substances), known related conditions (9498), biological targets (13 854), underlying mechanisms by which the drugs may act, pharmacokinetics (1509), DDI (1154), toxicity (1314), and sourcing (1963). On average, there are over 140 different properties per fully annotated substance. The maximum number of properties is 4765.
Manual data curation interface
To account for one-to-many and many-to-many relationships within the data, achieve annotation consistency, facilitate quality control, and minimize human error, a web-based curation application with a user-friendly interface and an underlying PostgreSQL database was developed and deployed. In the curation application, the sections are laid out as tabs. Each tab has groups of related data fields and, where applicable, tables. Each data field or a group of fields (e.g., PK metrics and associated metadata from the same experiment) has an associated reference (URI) field.
Data fields have interdependencies and safeguards to assist with annotations, namely:
Control for a specific data type where appropriate
Simplify and control input via limiting options (dropdowns and checkboxes) or providing a lookup in a relevant ontology
Allow to clone the data where multiple data elements are expected to have multiple marginally different entries (e.g., DDI, adverse events)
Unambiguously alert a user about the consistency of the overall annotation and missing fields (e.g., values without units, data without references, etc.).
All pertinent fields for which curators were not able to find data within reasonable amount of time, are explicitly marked as ‘Unknown.’ The data are exported from the curation interface in a JSON format and are then consumed by Stitcher (see Data Integration section).
Curated data model
Broadly, the current data model provides annotations in seven largely independent sections that contain the following information:
General information (43 fields): Preferred name, synonyms, structure, originator, notable PubMed articles and patents, a succinct comprehensive description, typical in vitro and in vivo application guides, route of administration, CNS activity, and up to 14 different identifiers from different large-scale databases.
Conditions (30 fields): Condition name, closest matches in Disease Ontology (18) and Medical Subject Headings ontologies, treatment modality (e.g., primary, secondary, palliative, etc.), highest development phase and clinical trials, approved drug product and its current marketing status, approval date, FDA-approved use, off-label use.
Targets (19 fields): The most appropriate identifier (protein, gene, pathway, or biological process), organism, potency and pharmacology.
Pharmacokinetics (54 fields): Information is provided on the per-experiment basis aiming to cover all four key PK metrics (Cmax, T1/2, AUC and Fraction Unbound) and supply all pertinent metadata, including demographics and health status of the study participants, dosage, route of administration, analyte and tissue.
Toxicity (46 fields): Similar to PK, information is provided on the per-experiment basis. For each adverse event, the annotations include liability (condition), frequency, severity, consequences for the study (e.g., discontinuation), and whether it reflects the dose limiting toxicity. All pertinent metadata are provided as well, including demographics and health status of the study participants, dosage, route of administration, treatment duration.
Drug-Drug Interactions (16 fields): Target (specific enzyme), type of interaction (e.g., substrate, inducer, etc.), metabolite, DDI magnitude (e.g., weak, strong, etc.), and availability of clinical support.
Sourcing (6 fields): Source type (vendor or database) and name, source substance identifier and the URL.
Targets and Conditions were explicitly matched if a causal relationship is known. Furthermore, all manually curated data were substantiated with individual references.
Data integration
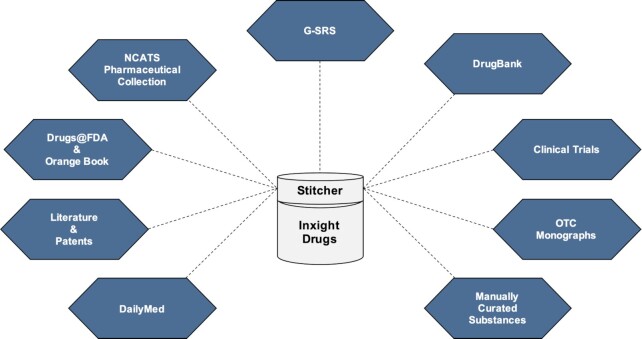
Inxight incorporates data from multiple independent data sources (vide supra) (Figure 1). To unify these data and to deduce how to merge and collapse information when multiple sources are referring to the same substance, NCATS developed a specialized tool known as Stitcher (https://github.com/ncats/stitcher). Stitcher employs entity resolution algorithms to partition entities within a given dataset into disjoint sets such that those within the same set are considered equivalent. Thus, Stitcher is used to untangle a web of connections between entities from multiple sources, form clusters representing unique substances, and thereby locate the unified set of properties for each substance. At the last step, derived variables are computed by traversing the unified property set. For example, the current approval status can be calculated by examining all the different approval status changes and the dates those changes occurred.
Figure 1.
Inxight Drugs: major data sources.
Technical details
The Inxight Drugs resource is built on top of the original G-SRS codebase using the Play framework (https://www.playframework.com). The backend has been extended to support dynamic fetching and indexing of additional data available in Stitcher's Neo4j graph database. This extension provides a robust mechanism by which new data can be attached to the core G-SRS substance records without requiring modifications to the underlying data models. The Inxight Drugs web application is hosted on a virtual server (Debian GNU/Linux 9) accessible at https://drugs.ncats.io. The application also provides a REST API at https://drugs.ncats.io/api/v1. The application is compatible with all modern web browsers, such as Google Chrome, Firefox, Safari, or Microsoft Edge that support JavaScript and HTML5. Inxight Drugs has been available to the community since 2018 and is frequently visited by a large number of users. 661 000 unique visitors accessed the website from January to July 2021.
DATABASE FEATURES
Browse drugs
A simple and convenient way to navigate through the substances in the database is to use the ‘Browse Drugs’ feature. The users can choose from a range of filters to narrow down the list of substances of interest. The filters are based on a diverse set of properties such as the development status, approval year, highest phase of clinical development, primary target, treatment modality, CNS activity, originator, substance class, and INN stem. Importantly, due to the nature of the data integration process not all substances are guaranteed to have all of these properties.
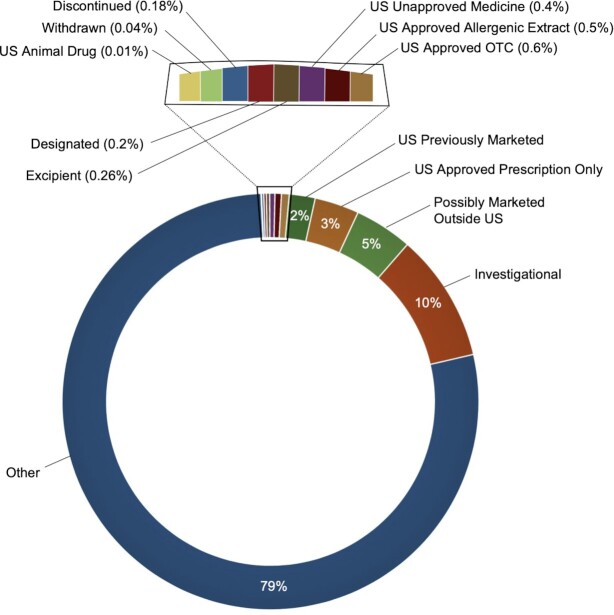
Depending on regulatory status, Inxight Drugs allows users to browse the substances categorized into four major groups: US Approved Drugs, Marketed drugs, Investigational Drugs, All Substances. The application contains other mutually exclusive subsets of substances, such as Previously Marketed Drugs, Drugs Marketed Outside US, Discontinued Drugs, and Withdrawn Drugs, accessible under the ‘Development Status’ filter. References supporting each of these regulatory status classifications is provided as an information tooltip in the substance record. A detailed breakdown of the substances available in Inxight Drugs is presented in Figure 2. The substance counts associated with these categories are provided in the supplementary information (S2).
Figure 2.
Breakdown of the substances in the database.
From the collection of substances that share a common active moiety, the most essential and minimalistic substance form is chosen as ‘Principal Form’. Related substances such as salts, hydrates and esters of the active moiety are not considered as ‘Principal Form’. When browsing substances, users can choose to only view the ‘Principal Form’ of substances using the 'Substance Form’ filter. Thus, the total number of substances varies depending on the representation chosen. Of the 125 036 total substances, 101 996 substances were annotated as ‘Principal Form’ substances, 14 960 substances were found to be salts, hydrates, esters, etc. The remaining entries include so-called ‘alternative definitions’ (representations of a single substance in alternative formats such as a peptide as a small molecule to better support search), and substances of the ‘concept’ class (which capture regulatory terms that are ambiguous in which substance they refer to).
Search drugs
The ‘Search’ feature allows users to search for substances by chemical structure (for small molecules) and sequence (for proteins and nucleic acids). Users can search for small molecules by drawing an exact chemical structure or substructure of interest using the molecule editor provided in the ‘Structure Search’ page. Having specified a query structure, users can choose one of the three different search types: substructure search, similarity search, and exact search. Similarity search option further allows users to define a Tanimoto cutoff (default is 0.8) to find similar small molecules. Furthermore, a name-to-structure feature is implemented on the same page that translates a substance name into chemical structure that could be used for structure search. Inxight Drugs also relies on NCI’s chemical identifier resolver (https://cactus.nci.nih.gov/chemical/structure) and PubChem's REST API (19) to resolve substance names to chemical structures in addition to an internal index based on G-SRS. Systematic chemical names are also resolved using OPSIN (20), open-source software for name-to-structure conversion. The ‘Sequence Search’ feature requires users to provide a query sequence, the desired sequence identity score (default = 0.5), the sequence type (protein or nucleic acid), and the cutoff type for alignment match. The website offers the following options for alignment match: global, local, and contains. Global alignment match attempts to perform an end-to-end alignment of the query sequence with the target sequences. Local alignment match employs a different strategy, where a substring of the query sequence is aligned with a substring of the target sequence to find local regions with highest level of similarity. Contains alignment match checks whether the query sequence is a part of the target sequence. The search results can be modified using the same set of filters described in the ‘Browse Drugs’ section.
Substance record
Each substance is identified by a Unique Ingredient Identifier (UNII). A UNII is generated by the FDA when a new substance satisfying the ISO 11238 standard is registered in the G-SRS. UNIIs are used in regulatory activities at different stages of the product lifecycle that include clinical trials, marketing, and post-marketing surveillance. Another property calculated within Stitcher along with the ‘Development Status’ is the ‘Year of Approval’ in the US and the ‘First Approval Year’. These properties are available for all drug substances that went through corresponding regulatory events at some point in their life cycle.
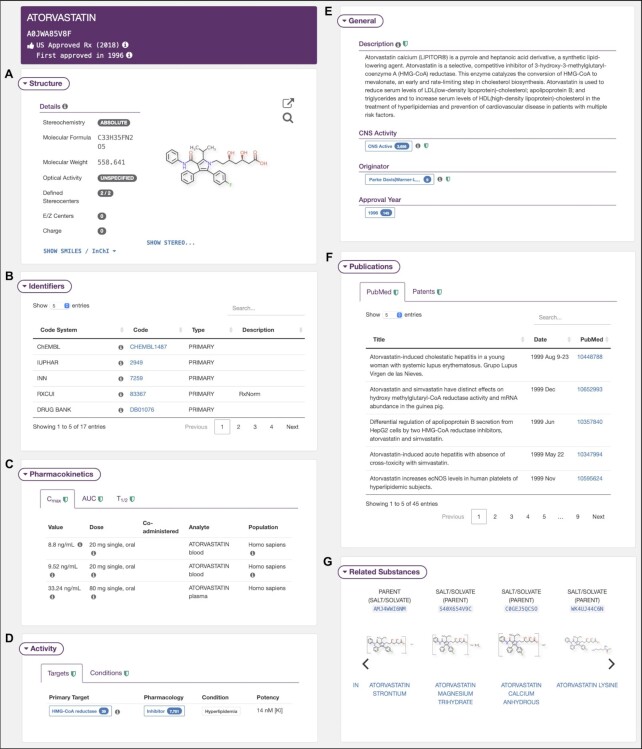
For a typical drug substance record (Figure 3), a wide range of details are provided that include chemical structure information, general regulatory information, activity data, indications, pharmacokinetic data, vendor information, literature and patents, synonyms, classification, links to external resources, and related substances. An information tooltip is provided when references and additional relevant information are available for a particular annotation. The two-dimensional representation of small molecules does not include explicit absolute stereochemistry labels by default, but users can choose to display the stereocenters when corresponding data are available. It was previously shown that >53% of the total substances in G-SRS core collection are achiral, 30% are enantiomers, 12% are racemic mixtures and 5% are more complex cases (13). Structural information is followed by manually curated description of the substance, CNS Activity annotation, and originator information. Activity information includes Primary Target and Condition. The former contains data on Pharmacology and Potency, while the latter specifies Treatment Modality (Primary, Secondary, Palliative, etc.), Highest Phase of clinical development, and if any Products are available. A Target may be explicitly related to a Condition, in which case a link is provided in the corresponding row of the Targets table and vice-versa in the Conditions table.
Figure 3.
Different types of data available for atorvastatin: (A) chemical structure and properties; (B) identifiers and links to external databases; (C) pharmacokinetic data; (D) targets, activity data and treatment modalities; (E) general description, CNS activity, originator, and approval year; (F) scientific literature and patents; (G) semantically related substances.
Manually curated pharmacokinetic data includes several key metrics—the peak plasma concentration (Cmax), elimination half-life (T1/2), area under curve (AUC), and fraction unbound—along with comprehensive metadata annotations. Relevant literature from PubMed and patent information for multiple sources such as USPTO and JPO are available in the Publications section. For most drugs, manually curated in vitro and in vivo usage guides are provided. Those illustrate typical application scenarios along with dosages and administration routes. Drug classification information is available from different code systems such as WHO’s Anatomical Therapeutic Chemical (ATC) Classification System, National Drug File - Reference Terminology (NDF-RT) and NCI Thesaurus along with hyperlinks to the respective resources. Inxight Drugs provides external identifiers that connect drugs with a range of drug and compound databases and regulatory resources via the code systems rigorously implemented in G-SRS. Finally, for each substance, all related substances that are linked via semantic relationships (e.g., active moiety, prodrug, metabolite, etc.) are listed.
Semantic relationships
Substances in the Inxight Drugs database are linked by a wide range of semantic relationships. Presence of a salt or a solvate is a critical aspect of drug product ingredients. Thus, accurate representation of chemical structure is essential for certain analyses in drug discovery. Inxight Drugs distinguishes substances based on this criterion and provides a total of 9,413 ‘PARENT → SALT/SOLVATE’ relationships. Active metabolites are sometimes more potent than their parent drugs. In such cases, it is the active metabolite that is responsible for much of the therapeutic action. In this context, the database provides 2,625 ‘PARENT → METABOLITE’ relationships. The complete list of different relationships and the corresponding number of substances is provided in the supplementary information (S3). For each relationship, the list of associated substances can be browsed on the website using the URL: https://drugs.ncats.io/substances?facet=Relationships/{REL}; where REL is any of the relationships listed in the supplementary information. For example, https://drugs.ncats.io/substances?facet=Relationships/PARENT→METABOLITE lists all 2625 substances that have an associated parent or metabolite.
Novel FDA drug approvals
Inxight Drugs features ‘Novel FDA Drug Approvals’ as a supplementary resource that provides regulatory aspects of new molecules approved by FDA annually starting from 2010. Comprehensive manually curated data are provided for each drug with the most important information being the regulatory details such as the date of approval and the clinical development time (in years) at https://drugs.ncats.io/newdrugs. Additional details include active ingredients, biological target (if available), and disease/indication. Specific annotations are provided to indicate the type of FDA approval (priority review, fast track approval, first-in-class, accelerated approval, etc.), designations (FDA breakthrough therapy, orphan product, etc.) and whether the product is a diagnostic imaging agent or labelled with a black box warning. The data can also be browsed using the interactive distributions of approved drugs by substance class and therapeutic class rendered as pie charts. This resource also presents the trend of FDA’s new drug approvals from 1941 to 2020. Additionally, the distribution of new drug approvals by substance class and year is presented in Figure 4.
Figure 4.
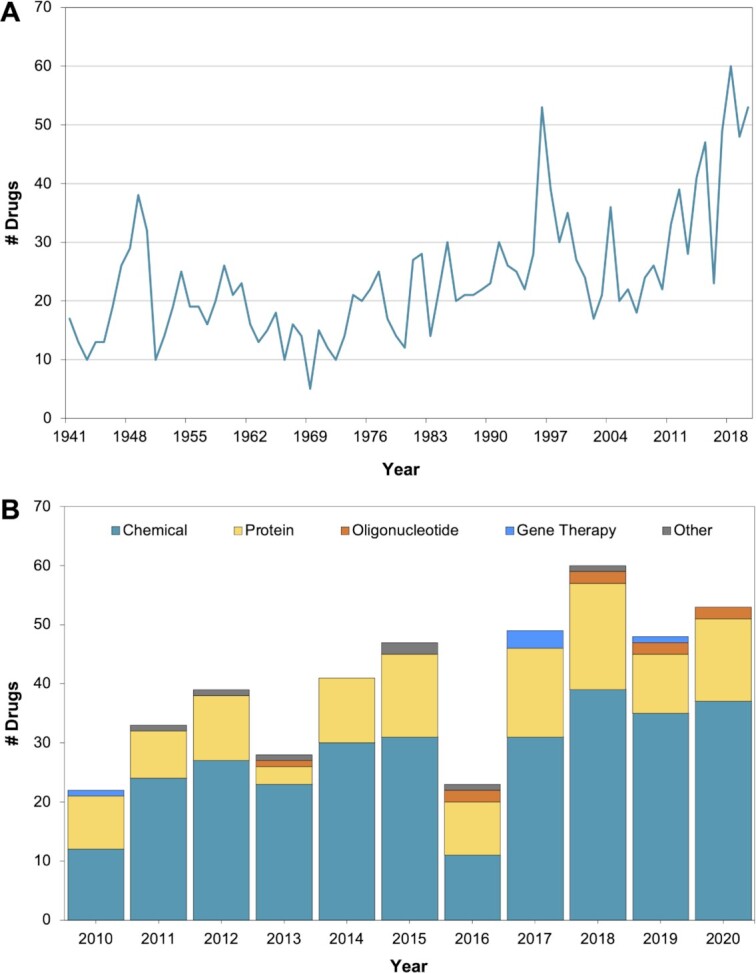
The trends of FDA new drug approvals: (A) Number of drugs approved in each year starting from 1941 to 2020. (B) Annual drug approvals by substance class starting from 2010 to 2020.
DISCUSSION
Research in the fields of bio- and cheminformatics led to a rapid rise in the amount of data generated in the last two decades. These data are commonly stored in publicly accessible databases or otherwise made commercially available. A tremendous amount of work has been invested in the past several years to bridge the gaps that emerged due to the decentralized distribution of these data. Several databases were created with emphasis on different molecular entities (small molecules (15,21–25), proteins (26,27), nucleic acids (28,29), etc.) and research areas (genomics (30,31), proteomics (32), metabolomics (33), etc.). Information gaps among the public databases is a widely acknowledged phenomenon, which highlights the importance of manual curation in filling these gaps. In addition, none of the previously published drug databases have adopted international standards for substance classification. On the other hand, the process of approval and regulation of pharmaceuticals has rapidly evolved and increased in complexity in the last four decades (34). Only a handful of databases focused on drugs tried to systematically address these key issues and provide comprehensive and most up-to-date information rigorously extracted from scientific literature and regulatory resources that can boost translational drug discovery.
In this context, NCATS Inxight Drugs was launched as a comprehensive portal for drugs and other substances with a focus on rigorous definitions and manual curation. The database offers three key benefits. First, it is using a complete set of substances, as per the ISO 11238 standard definition, as its core parent collection. Second, Inxight Drugs implements novel algorithms to integrate information across various public sources, including data from the FDA. Third, a large fraction of the most important substances, such as drugs and drug candidates, has been provided with thorough manual annotations. This approach ensures the research community has access to the most comprehensive and accurate information. Technical content includes, among others, marketing and regulatory status, rigorous drug ingredient definitions, clinical use information with examples and links to trials, biological targets, and underlying mechanisms by which the drugs may act.
By the virtue of incorporating multiple sources of drug approval authority, Inxight Drugs enumerates and describes approved, marketed, and investigational drugs and therapies, and thus is intended to be the most definitive source of drug development information for the translational research community. Specifically, Inxight Drugs offers:
Authority
Inxight Drugs is the only resource that provides a complete drug development profile, including a rigorous, ISO 11238-compliant definition of the substance, context of use including activity, targets, conditions, and references for its clinical development status.
Precision
A substance, fundamentally a drug product ingredient, is precisely defined by the rigorous ISO 11238 standard, and Inxight Drugs is the only biomedical resource that is using a complete list of all known substances as its core entity.
Quality
Inxight Drugs incorporates large volumes of manually curated data, including regulatory data from the FDA Substance Registration System, with over 120 000 current entries, as well as comprehensive annotations from Rancho BioSciences for over 25 000 substances, with >140 different properties per entry, on average.
Aggregation
Inxight Drugs incorporates data from multiple independent data sources. The use of data unification and de-duplication software enables first-in-class automated data aggregation from a virtually unlimited number of sources. The software identifies all ‘sufficiently similar’ entities as one substance; their properties are unified and key derivative variables, such as approval status, are then calculated.
FUTURE WORK
At its core, Inxight Drugs application was developed as a branch of the G-SRS software, and since that split the two projects have further separated in function and form. Currently, Inxight Drugs relies on the G-SRS substance index and does not present data for substances that are not mapped to a UNII even though many hundreds of such substances are present in other data sources in the underlying Inxight Drugs database. Expansion of Inxight Drugs to include these substances as a separate collection would facilitate their identification for the purposes of registration and allow for a higher cadence of the release schedule.
As a part of this effort, we envision making the application much more modular, easier to update and expand, more responsive, and even possible to repurpose for other projects of similar nature that involve NCATS Stitcher.
Insofar as the data are concerned, NCATS continues the manual curation and the data integration efforts. Substances are routinely prioritized for manual annotation based on multiple criteria (current development phase, ongoing internal projects at NCATS, etc.). The next data release is scheduled for February 2022. We also continue to identify additional data sources, including from ChEMBL, IUPHAR Guide to Pharmacology, Health Canada, European Medicines Agency, etc. that we hope will gradually be integrated into the existing database.
DATA AVAILABILITY
The contents of the database are available at https://drugs.ncats.io. The latest release of data occurred on July 30, 2021, and comprises 125,036 substances. The “About” page on the website (https://drugs.ncats.io/about) provides a detailed list of data sources, definitions of key entities and concepts, as well as direct download links for certain subsets of data. The data can also be accessed through a RESTful API (https://drugs.ncats.io/api/v1).
Supplementary Material
Notes
Present address: Tyler Peryea, Office of the Commissioner, US Food and Drug Administration, Silver Spring, MD, USA.
Present address: Ajit Jadhav, Worldwide Research, Development and Medical, Pfizer Inc., USA.
Contributor Information
Vishal B Siramshetty, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD, USA.
Ivan Grishagin, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD, USA.
Ðắc-Trung Nguyễn, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD, USA.
Tyler Peryea, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD, USA.
Yulia Skovpen, Rancho BioSciences, San Diego, CA, USA.
Oleg Stroganov, Rancho BioSciences, San Diego, CA, USA.
Daniel Katzel, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD, USA.
Timothy Sheils, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD, USA.
Ajit Jadhav, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD, USA.
Ewy A Mathé, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD, USA.
Noel T Southall, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health, National Center for Advancing Translational Sciences. Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Center for Advancing Translational Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. DiMasi J.A., Grabowski H.G., Hansen R.W.. Innovation in the pharmaceutical industry: New estimates of R&D costs. J. Health Econ. 2016; 47:20–33. [DOI] [PubMed] [Google Scholar]
- 2. Stone J.H., Frigault M.J., Serling-Boyd N.J., Fernandes A.D., Harvey L., Foulkes A.S., Horick N.K., Healy B.C., Shah R., Bensaci A.M.et al.. Efficacy of tocilizumab in patients hospitalized with Covid-19. N. Engl. J. Med. 2020; 383:2333–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. RECOVERY Collaborative Group Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A.et al.. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021; 384:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S.et al.. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020; 383:1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sultana J., Crisafulli S., Gabbay F., Lynn E., Shakir S., Trifirò G.. Challenges for drug repurposing in the COVID-19 pandemic era. Front. Pharmacol. 2020; 11:588654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Raju T.N. The Nobel chronicles. 1988: James Whyte Black, (b 1924), Gertrude Elion (1918-99), and George H Hitchings (1905-98). Lancet. 2000; 355:1022. [DOI] [PubMed] [Google Scholar]
- 7. Smietana K., Siatkowski M., Møller M.. Trends in clinical success rates. Nat. Rev. Drug Discov. 2016; 15:379–380. [DOI] [PubMed] [Google Scholar]
- 8. Hay M., Thomas D.W., Craighead J.L., Economides C., Rosenthal J.. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014; 32:40–51. [DOI] [PubMed] [Google Scholar]
- 9. Ban T.A. The role of serendipity in drug discovery. Dialogues Clin. Neurosci. 2006; 8:335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hargrave-Thomas E., Yu B., Reynisson J.. Serendipity in anticancer drug discovery. World J. Clin. Oncol. 2012; 3:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klein D.F. The loss of serendipity in psychopharmacology. JAMA. 2008; 299:1063–1065. [DOI] [PubMed] [Google Scholar]
- 12. World Health Organization List of Globally identified Websites of Medicines Regulatory Authorities. 2015; [Google Scholar]
- 13. Peryea T., Southall N., Miller M., Katzel D., Anderson N., Neyra J., Stemann S., Nguyễn Ð.-T., Amugoda D., Newatia A.et al.. Global Substance Registration System: consistent scientific descriptions for substances related to health. Nucleic. Acids. Res. 2021; 49:D1179–D1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z.et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J.. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006; 34:D668–D672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang R., Southall N., Wang Y., Yasgar A., Shinn P., Jadhav A., Nguyen D.-T., Austin C.P.. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Transl. Med. 2011; 3:80ps16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang R., Zhu H., Shinn P., Ngan D., Ye L., Thakur A., Grewal G., Zhao T., Southall N., Hall M.D.et al.. The NCATS Pharmaceutical Collection: a 10-year update. Drug Discov. Today. 2019; 24:2341–2349. [DOI] [PubMed] [Google Scholar]
- 18. Schriml L.M., Arze C., Nadendla S., Chang Y.-W.W., Mazaitis M., Felix V., Feng G., Kibbe W.A.. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012; 40:D940–D946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S., Thiessen P.A., Cheng T., Yu B., Bolton E.E.. An update on PUG-REST: RESTful interface for programmatic access to PubChem. Nucleic Acids Res. 2018; 46:W563–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lowe D.M., Corbett P.T., Murray-Rust P., Glen R.C.. Chemical name to structure: OPSIN, an open source solution. J. Chem. Inf. Model. 2011; 51:739–753. [DOI] [PubMed] [Google Scholar]
- 21. Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M.. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012; 40:D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ursu O., Holmes J., Knockel J., Bologa C.G., Yang J.J., Mathias S.L., Nelson S.J., Oprea T.I.. DrugCentral: online drug compendium. Nucleic Acids Res. 2017; 45:D932–D939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thorn C.F., Klein T.E., Altman R.B.. PharmGKB: the Pharmacogenomics Knowledge Base. Methods Mol. Biol. 2013; 1015:311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M.P., Mosquera J.F., Mutowo P., Nowotka M.et al.. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Armstrong J.F., Faccenda E., Harding S.D., Pawson A.J., Southan C., Sharman J.L., Campo B., Cavanagh D.R., Alexander S.P.H., Davenport A.P.et al.. The IUPHAR/BPS Guide to PHARMACOLOGY in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV Guide to MALARIA PHARMACOLOGY. Nucleic Acids Res. 2020; 48:D1006–D1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M.et al.. UniProt: the Universal Protein knowledgebase. Nucleic. Acids. Res. 2004; 32:D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Betel D., Wilson M., Gabow A., Marks D.S., Sander C.. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008; 36:D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mashima J., Kodama Y., Kosuge T., Fujisawa T., Katayama T., Nagasaki H., Okuda Y., Kaminuma E., Ogasawara O., Okubo K.et al.. DNA data bank of Japan (DDBJ) progress report. Nucleic Acids Res. 2016; 44:D51–D57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clough E., Barrett T.. The Gene Expression Omnibus database. Methods Mol. Biol. 2016; 1418:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smigielski E.M., Sirotkin K., Ward M., Sherry S.T.. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000; 28:352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hsu F., Pringle T.H., Kuhn R.M., Karolchik D., Diekhans M., Haussler D., Kent W.J.. The UCSC Proteome Browser. Nucleic Acids Res. 2005; 33:D454–D458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wishart D.S., Tzur D., Knox C., Eisner R., Guo A.C., Young N., Cheng D., Jewell K., Arndt D., Sawhney S.et al.. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007; 35:D521–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Darrow J.J., Avorn J., Kesselheim A.S.. FDA approval and regulation of pharmaceuticals, 1983-2018. JAMA. 2020; 323:164–176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The contents of the database are available at https://drugs.ncats.io. The latest release of data occurred on July 30, 2021, and comprises 125,036 substances. The “About” page on the website (https://drugs.ncats.io/about) provides a detailed list of data sources, definitions of key entities and concepts, as well as direct download links for certain subsets of data. The data can also be accessed through a RESTful API (https://drugs.ncats.io/api/v1).